Abstract
The effects of Newcastle disease virus (NDV) fusion (F) glycoprotein cleavage mutants on the cleavage and syncytium-forming activity of the wild-type F protein were examined. F protein cleavage mutants were made by altering amino acids in the furin recognition region (amino acids 112 to 116) in the F protein of a virulent strain of NDV. Four mutants were made: Q114P replaced the glutamine residue with proline; K115G replaced lysine with glycine; double mutant K115G, R113G replaced both a lysine and an arginine with glycine residues; and a triple mutant, R112G, K115G, F117L, replaced three amino acids to mimic the sequence found in avirulent strains of NDV. All mutants except Q114P were cleavage negative and fusion negative. However, addition of exogenous trypsin cleaved all mutant F proteins and activated fusion. As expected for an oligomeric protein, the fusion-negative mutants had a dominant negative phenotype: cotransfection of wild-type and mutant F protein cDNAs resulted in an inhibition of syncytium formation. The presence of the mutant F protein did not inhibit cleavage of the wild-type protein. Furthermore, evidence is presented that suggests that the mutant protein and the wild-type protein formed heterooligomers. By measuring the syncytium-forming activity of the wild-type protein at various ratios of expression of mutant and wild-type protein, results were obtained that are most consistent with the notion that the size of the functionally active NDV F protein in these assays is a single oligomer, likely a trimer. That a larger oligomer, containing a mix of both wild-type and mutant F proteins, has partial activity cannot, however, be ruled out.
Paramyxoviruses such as Newcastle disease virus (NDV) are a class of enveloped viruses which fuse with cell plasma membranes at neutral pH (reviewed in reference 7). Because fusion occurs at neutral pH, cells infected with these viruses can fuse with adjacent cells, resulting in giant, multinucleated cells, or syncytia (7). Most paramyxovirus-mediated fusion is directed by the combined action of the two virus-encoded glycoproteins, the attachment protein (hemagglutinin-neuraminidase protein, or HN) and the fusion (F) protein (reviewed in reference 9). Expression of both of these proteins in transfected cells is necessary and sufficient for syncytium formation. While the HN protein is required for attachment as well as another undefined role in fusion, the fusion protein is thought to be directly involved in fusion and, in one case, can mediate fusion independent of the HN protein (1).
The NDV F protein is synthesized as a precursor, F0, which, to be fusogenic, must be cleaved at a site 116 amino acids from the amino terminus, resulting in disulfide-linked F2 and F1 (21, 22). Cleavage is accomplished by host cell proteases (reviewed in reference 10). Fusion proteins encoded by the virulent strains of NDV have amino acid sequences at the cleavage site which are substrates for the furin family of proteases and are thus cleaved intracellularly in the trans-Golgi membranes. Fusion proteins encoded by avirulent strains do not have this sequence motif but rather encode single basic residues and are cleaved by extracellular enzymes, such as a protease released by Clara cells in the respiratory tract or a factor X-like activity in the allantoic fluid of embryonated eggs (reviewed in reference 10). Cleavage of these fusion proteins in tissue culture depends upon the addition of exogenous trypsin (16).
Paramyxovirus fusion proteins exist as noncovalently linked oligomers. It has been reported that both the Sendai virus F protein (23) and the respiratory syncytial virus F protein are tetramers (4). However, other reports support the notion that the oligomer is a trimer composed of three disulfide-linked F1-F2 molecules (17, 18). All of these studies, however, measured the physical size of the fusion protein on sucrose gradients or in polyacrylamide gels. In no case has the functionally active size of the fusion protein oligomer been investigated. In several systems, it has been suggested that large complexes of several glycoprotein oligomers may be required for fusion. For example, it has been proposed that initial stages in fusion mediated by the influenza virus hemagglutinin require a complex of at least three trimers on the basis of effects on fusion of different concentrations of cell surface hemagglutinin protein (5). Other studies have proposed even larger complexes (2). In human immunodeficiency virus-infected cells, through comparisons of the activity of wild-type protein in the presence of various amounts of a dominant negative mutant, it has been suggested that several oligomers of gp41 are required to mediate membrane fusion (6).
The effects of NDV fusion-negative mutant proteins on the wild-type protein activity were characterized. Mutations were introduced into the cleavage site of the F protein derived from a virulent strain of NDV in order to generate uncleaved and, therefore, inactive forms of the F protein. As expected for an oligomeric protein, the fusion-negative proteins had a dominant negative phenotype. By measuring the syncytium-forming activity of the wild-type protein at various ratios of expression of mutant and wild-type protein, results were obtained that are consistent with the notion that the size of the functionally active NDV F protein in syncytium formation is an oligomer, likely a trimer. A very similar approach for determining the active unit of the influenza virus M2 protein has been recently reported (20).
MATERIALS AND METHODS
Cells, vectors, and viruses.
Cos-7 cells, obtained from the American Type Culture Collection, were maintained in Dulbecco’s modified Eagle medium (DMEM) supplemented with nonessential amino acids, vitamins, penicillin-streptomycin, sodium bicarbonate, and 10% fetal calf serum.
NDV HN and F genes, characterized previously (25), were expressed in Cos cells with pSVL obtained from Pharmacia.
Site-specific mutagenesis.
Mutagenesis reactions were done with the mutagenesis kit from 5 Prime→3 Prime, Inc. The appropriate oligomers (22 to 26 nucleotides in length) were used for each mutation. The mutations isolated are shown in Fig. 1. With one exception, mutants are named, in the single-letter code, with the amino acid in the wild type and with the position of the change and the amino acid in the mutant. Thus Q114P is a mutant which has a proline substituted for a glutamine at amino acid 114 in the F protein sequence. The triple mutant R113G, K115G,F117L was named AF (for avirulent F) for ease of presentation. The mutants AF and K115G were modified by insertion of the Flag sequence (DYKDDDDK) just prior to the chain termination codon at the carboxy terminus of the protein to produce AF-Flag and K115G-Flag.
FIG. 1.
Location of cleavage site mutations. Shown at the top is the sequence at the cleavage site of the F protein of the virulent NDV strain AV (wild type [WT]). The arrow indicates the site of cleavage. The amino acid changes (in boldface) and the name of each mutant are shown below the wild-type sequence.
Transfections.
Transfections with Lipofectin were done essentially as recommended by the manufacturer, BRL GIBCO. Cos cells were plated at 3 × 105 per 35-mm-diameter plate. Twenty to 24 h later, the cells were transfected. For each 35-mm-diameter plate, a mix of DNA (up to 2 μg) in 0.1 ml of OptiMem (BRL GIBCO) and 10 μg of Lipofectin in 0.1 ml of OptiMem was incubated at room temperature for 15 min and then diluted with 0.7 ml of OptiMem and added to a plate previously washed with OptiMem. Cells were incubated with the Lipofectin-DNA for 4 to 5 h, which was then replaced with 2 ml of Cos cell media.
Transfections with Lipofectamine were accomplished as recommended by the manufacturer essentially as described above. Transfections were also accomplished with DOPE (dioleoyl-l-α phosphatidylethanolamine) prepared as described by Campbell (3). This reagent was used in protocols identical to that described above for Lipofectin.
Antibody, radiolabeling, and immunoprecipitation of protein.
The antibody used for immunoprecipitation of the fusion protein was anti-Ftail (27). This antibody is a polyclonal rabbit antibody raised against a synthetic peptide with the sequence of the cytoplasmic tail of the fusion protein as described by Wang et al. (28) and was prepared by the Peptide Core Facility of the University of Massachusetts Medical School. Anti-Flag antibody (M2) was purchased from Kodak.
Transfected cells were radiolabeled for 2 to 4 h at 37°C in Dulbecco modified Eagle medium lacking methionine and cysteine but containing 100 μCi of [35S]methionine and [35S]cysteine (New England Nuclear Express 35S) per ml and chased in nonradioactive media for 4 h. Cells were then washed in phosphate-buffered saline (PBS) and lysed in RSB buffer (0.01 M Tris-HCl [pH 7.4], 0.01 M NaCl) containing 1% Triton X-100, 0.5% sodium deoxycholate, and 2 mg of iodoacetamide per ml as previously described (25–27). Nuclei were removed by centrifugation. Immunoprecipitation of NDV proteins was accomplished as previously described (11, 12). Precipitated proteins were resolved on 10% polyacrylamide gels as previously described (11, 12).
In some experiments, cells were incubated for 10 min at room temperature in 5 μg of acetylated trypsin per ml in OptiMem after plates had been washed with OptiMem. After trypsin digestion, monolayers were washed in OptiMem containing soybean trypsin inhibitor (7.5 μg/ml).
Fusion assays.
Fusion activity was determined by syncytium formation. For syncytium formation, Cos cells were cotransfected with wild-type or mutant fusion protein genes and the wild-type HN protein gene with Lipofectin. At 24 and 48 h posttransfection, the number of nuclei in 40 fusion areas were counted to determine the average size of syncytia at each time point as previously described (25). Values obtained after transfection of the vector alone were subtracted.
Chemical cross-linking.
Cross-linking with 3,3′-dithiobis(sulfosuccinimidyl proprionate) (DTSSP) (Pierce) was accomplished by a modification of the protocol described by Russell et al. (18). For cross-linking of cell surface molecules, radioactivity labeled cells were washed in PBS (pH 8.5) and then incubated with 1 mM DTSSP in PBS at 4°C overnight. The cross-linker was quenched in 50 mM glycine for 5 min, and the cells were washed with PBS and then lysed at 4°C in 1% Triton X-100. For cross-linking of cell extracts, radioactively labeled cells were lysed in 1% Triton X-100 and then incubated in 1 mM DTSSP for 2 h at 4°C. After being quenched with glycine, the radioactively labeled, cross-linked proteins were precipitated as described above.
RESULTS
Expression of mutant proteins.
Figure 1 shows the amino acid sequence of the cleavage site of the AV strain (virulent) of NDV (wild type). Four new mutants were made in this region with the goal of eliminating cleavage of the expressed F protein. First, the glutamine at amino acid 114 was changed to proline to determine if this residue could eliminate intracellular cleavage by altering the conformation of the cleavage site. Two other mutants altered the furin consensus site either by substituting the lysine at amino acid 115 with glycine or substituting glycine residues for both lysine 115 and arginine 113. Glycine was substituted because the avirulent F protein contains glycine at position 115 as well as three glycine residues at positions 110, 111, and 112 (19). Mutant AF contained the sequence found at the cleavage site of avirulent strains of NDV (19). The mutant F117L has been previously described (14) and was included here for comparative purposes.
The mutant proteins were expressed in Cos-7 cells with simian virus 40-based vector as previously described (25). Cells transfected with either wild-type or mutant fusion protein genes were radioactively labeled with [35S]methionine and [35S]cysteine in a pulse-chase labeling protocol, and labeled protein was precipitated with anti-Ftail. In this and other experiments, all mutant proteins were expressed in amounts equivalent to those of the wild type (Fig. 2). F protein precipitated from wild-type transfected cells contained cleaved fusion protein (F1) as well as small amounts of uncleaved F0 (Fig. 2B, −trypsin). F2 is not visible on 10% gels (17). Mutant Q114P protein was also cleaved to the same extent as the wild-type protein (Fig. 2B). Thus, the nonconservative mutation at this position had no effect on furin recognition. Since cleavage likely occurs in the trans-Golgi membranes (15), the cleavage of this protein also indicates that the mutation has no effect on intracellular transport. In contrast to the Q114P mutation, all mutations which altered the furin recognition motif reduced cleavage of F0 to less than 20% (Fig. 2B, −trypsin; Table 1). The F117L protein was also minimally cleaved as previously reported (14).
FIG. 2.
Expression of mutant proteins in the absence and presence of added trypsin. In duplicate, Cos cells, transfected with vector, pSVL (SVL), wild-type F DNA (WT), or mutant DNAs, were radioactively labeled with [35S]methionine and [35S]cysteine for 2 h at approximately 48 h posttransfection and then subjected to a nonradioactive chase for 2 h. One set of cells was incubated with trypsin (indicated at the top of the figure), as described in Materials and Methods, prior to lysis. Proteins present in equivalent amounts of cell extract were precipitated with excess anti-Ftail antibody, and the resulting precipitated proteins were resolved on 10% polyacrylamide gels in the absence (−β-ME) (A) or presence (+β-ME) (B) of reducing agent β-mercaptoethanol. The mutant DNA used for each transfection is indicated at the top of the figure. Fnr, nonreduced fusion protein containing both F0 and F1+F2; F0, uncleaved fusion protein; F1, the cleaved fusion protein. F2 is not resolved on this gel. Marker, infected cell extract.
TABLE 1.
Cleavage of mutant proteins in the presence and absence of trypsin
| DNA | % Cleavagea
|
|
|---|---|---|
| Without trypsin | With trypsin | |
| Wild type | 0.73 | 0.79 |
| Q114P | 0.73 | 0.82 |
| K115G | 0.16 | 0.71 |
| F117L | 0.36 | 0.77 |
| R113G,K115G | 0.10 | 0.74 |
| AF | 0.06 | 0.75 |
Results are the average of two experiments, both of which yielded very similar values.
All mutants, however, were readily cleaved if trypsin was added to intact cells prior to cell lysis (Fig. 2B, +trypsin; Table 1). The amount of cleaved F1 was approximately 70 to 80%, that of wild-type protein. Because trypsin was added only to intact cells, these results clearly demonstrated that all mutant proteins were transported to the cell surface as efficiently as wild-type proteins. Addition of trypsin to cells expressing the wild-type protein did not significantly increase the amount of cleaved F protein, a result that suggests that the F0 detected was likely due to residual intracellular protein.
Fusion activity of mutant proteins in the absence and presence of trypsin.
To measure the ability of these mutant proteins to direct membrane fusion, Cos cells were transfected with the mutant fusion protein genes as well as the wild-type HN gene. The fusion activities of these mutant proteins were determined as previously described (17, 24, 25, 27) and are shown as a percentage of the value obtained with wild-type F protein (Fig. 3). As expected, in the absence of trypsin, Q114P directed the formation of syncytia at levels comparable to those of wild-type F protein, while expression of the cleavage-negative mutants did not result in significant syncytium formation. However, after addition of trypsin, all mutant proteins could direct syncytium formation, although to different extents. The reason that mutants K115G, R113G,K115G, and AF do not direct as extensive fusion as the wild-type F protein is unclear, since all mutants are cleaved to the same extent after trypsin addition.
FIG. 3.
Fusion activity of mutant proteins without and with trypsin activation. In duplicate, Cos cells were transfected with equal amounts of HN protein DNA and either wild-type (WT) F protein or mutant F protein DNAs. At 48 h posttransfection, one set of monolayers was incubated with trypsin as described in Materials and Methods. After 10 to 12 h of further incubation, the sizes of syncytia were measured as previously described (24). The values obtained are expressed as percentages of that obtained with the wild-type F protein DNA. Error bars indicate the variation in three independent experiments.
Addition of trypsin to the wild-type protein-expressing cells did not increase the fusion activity. This result was consistent with the results described above, which suggested that the F0 detected in wild-type transfected cells was intracellularly located.
Effect of mutant F protein on the cleavage of wild-type protein.
Like other glycoproteins, oligomerization of the F proteins occurs in the rough endoplasmic reticulum (RER) (18) prior to cleavage of F0 to F1+F2, which occurs in the trans-Golgi membranes (15). However, it is unclear if all subunits in an oligomer must be cleaved in concert or if individual members of the oligomer are cleaved independently. To explore this question, the extent of cleavage of a constant amount of wild-type protein in the presence of increasing expression of a mutant protein was determined (Fig. 4). Clearly, while the amount of F0 increased with increasing expression of mutant protein, the amount of F1 detected was unchanged. Thus, the presence of uncleaved fusion protein had no detectable effect on the cleavage of the wild-type protein.
FIG. 4.
Mutant F protein does not affect the cleavage of wild-type protein. Cos cells were transfected with wild-type F DNA (WT) or wild-type F DNA with increasing amounts of K115G. Radioactively labelled cell extracts, prepared as described in the legend to Fig. 2, were incubated with anti-Ftail antibody, and the resulting precipitated protein was electrophoresed in the presence of reducing agent. F0, uncleaved F protein; F1, the cleaved fusion protein. F2 is not resolved on this gel. Marker, infected-cell extract.
Wild-type and mutant proteins form heteroligomers.
One interpretation of the result shown in Fig. 4 is that individual members of an oligomer are cleaved independent of the cleavage of other members of an oligomer. If so, then it should be possible to demonstrate the presence of heterooligomers, or oligomers that contain both the cleaved and uncleaved forms of the protein. To explore the formation of heterooligomers, the Flag epitope was inserted at the carboxy terminus of the F protein sequence of the mutants AF and K115G. Addition of this epitope had no effect on the intracellular transport of the F protein (unpublished observations). Cells were transfected with either of these mutants as well as with wild-type F protein cDNA and the Flag-tagged mutant F protein was precipitated with anti-Flag antibody. If heteroligomers form, then precipitates should contain both the uncleaved F protein as well as the cleaved F protein derived from the wild-type protein gene. Indeed, precipitates derived from cells cotransfected with the wild-type protein as well as either of the mutant proteins contain both cleaved as well as uncleaved F protein (not shown). However, because of the stringent wash conditions required to visualize F protein above background material, the amount of F1 detected in the precipitate was low. Thus, cells were subjected to cross-linking, with the membrane-impermeable cross-linker DTSSP, to stabilize any complexes (Fig. 5).
FIG. 5.
Wild-type and mutant proteins form heterooligomers. Cells transfected with the DNAs indicated at the top of each panel were radioactively labeled with [35S]methionine and [35S]cysteine as described in the legend to Fig. 2. Intact cells were subjected to cross-linking with DTSSP as previously described (A). Cell lysates were incubated with cross-linker as described in Materials and Methods (B and C). Proteins present in extracts were precipitated with anti-Flag (A and B) or anti-Ftail (C), and the resulting precipitated proteins were electrophoresed in the presence of reducing agent. Marker, infected-cell extract. The DNAs used are indicated at the top of each lane. F0, uncleaved F protein; F1, cleaved F protein.
Cross-linking was accomplished with intact cells (Fig. 5A) as well as cell lysates (Fig. 5B). In both cases, precipitation of protein with the anti-Flag antibody resulted in precipitation of not only the Flag-tagged F0 protein but also the F1 protein, which must be derived from the wild-type protein, since little F1 is detected after transfection of the mutant DNAs alone. The results were similar whether or not the HN protein was also expressed. Figure 5C shows precipitation of all F proteins from the total cell extracts with anti-Ftail antibody and shows that the wild-type protein was expressed.
Fusion activity is proportional to expression of the wild-type protein.
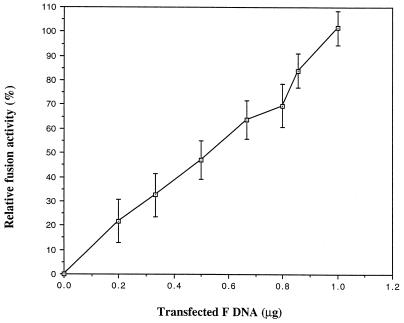
Because the effect of mutant proteins on the activities of wild-type proteins often provides insight into the function of the wild-type protein (8), it was of interest to determine the effects of cleavage mutant proteins on the fusion activity of wild-type protein. To explore this question, it was first necessary to determine if syncytium size was proportional to the amount of expression of fusion-active F protein. To address this question, cells were transfected with various amounts of wild-type F protein cDNA, and the amount of F protein expression at each DNA concentration was determined by immunoprecipitation from cell extracts (Fig. 6). Clearly the amount of fusion protein detected increased linearly with the amount of DNA used for transfection. Next, the syncytium size at each DNA concentration was measured, and, as shown in Fig. 7, the size of syncytia at 48 h posttransfection was directly proportional to the amount of F protein DNA. Thus, syncytium size can be used to measure the effects of mutants on wild-type activities.
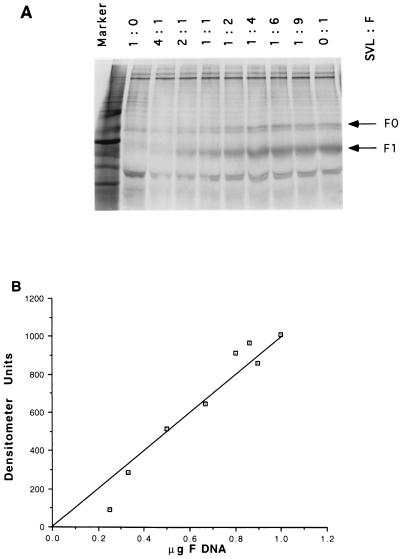
FIG. 6.
F protein expression as a function of amount of transfected DNA. Cos cells were transfected with vector (SVL) or increasing amounts of wild-type F DNA (WT). Vector DNA (pSVL) was added to each transfection such that the total DNA was at 1 μg. The ratios of wild-type and pSVL DNAs are shown at the top of panel A. The amounts of F protein cDNA used in each transfection are shown on the x axis in panel B. Transfected cells were radioactively labeled with [35S]methionine and [35S]cysteine for 4 h at approximately 48 h posttransfection and then subjected to a nonradioactive chase for 4 h. Cell extracts were precipitated with anti-Ftail antibody, and the resulting precipitated proteins were resolved on 10% polyacrylamide gels in the presence of reducing agent (β-mercaptoethanol [A]). (B) Quantitation by a densitometer of the total F protein present in each lane.
FIG. 7.
Fusion activity is proportional to the level of F protein expression. The sizes of syncytia in Cos cells, transfected as described in the legend to Fig. 4, were determined at 48 h posttransfection as described in Materials and Methods and are shown versus the amount of DNA used in the transfection. Error bars indicate the actual variation in three independent experiments.
Effect of mutant proteins on wild-type fusion activity.
The effect of cleavage mutants on the activity of the wild-type protein was determined by cotransfection of Cos cells with different ratios of wild-type and mutant DNAs. Mutants K115G and R113G,K115G were selected, since little cleavage was observed with these mutants, yet trypsin digestion resulted in significant fusion, suggesting that the conformation of these mutants was least affected by mutation. The effect of these mutants on the fusion activity of wild type assayed by syncytium size and the results were identical. The results obtained with K115G are shown in Fig. 8. Fusion activity is expressed as a percentage of wild-type activity as described in Materials and Methods. Both mutants significantly inhibited the activity of the wild-type protein. At equimolar ratios of wild-type and mutant cDNAs, the fusion activity was approximately 12% that observed with wild-type F alone (Fig. 8). Thus, the mutant proteins exert a dominant negative phenotype. The dominant negative phenotype of the cleavage mutants suggested that the active form of the F protein is an oligomer and that inclusion of a mutant protein in an oligomer inhibits its activity. Figure 8 also shows predicted inhibition curves calculated for dimers, trimers, tetramers, hexomers, and nine-mers (see Discussion).
FIG. 8.
Predicted and measured fusion activities of wild-type F protein in the presence of various amounts of mutant proteins. To determine the fusion activity of wild-type protein in the presence of different amounts of mutant protein, cells were cotransfected with 1.5 μg of HN protein DNA, and different ratios of wild-type and K115G DNA were expressed as the fraction that is wild-type DNA. The syncytium sizes were determined at 48 h posttransfection and are shown as data points with error bars (experimental curve). Error bars show the actual variation in three experiments. For each ratio of wild-type and mutant DNAs, the predicted amount of fusion was determined by calculating the probability of a fully wild-type oligomer (see text). The different curves show the calculations for dimers, trimers, tetramers, heptamers, and nine-mers at different ratios of wild-type and mutant DNAs.
DISCUSSION
The results presented in this paper characterize the effect of coexpression of two forms of the NDV F protein, the protein with a furin sequence, termed wild-type here, and a mutant protein without such a site. Clearly the synthesis and stability of the wild-type protein as well as those of the mutant protein were unaffected. Since the proteins form oligomers in the RER (18) and cleavage occurs in the trans-Golgi membranes (15), it seemed possible that a wild-type protein that oligomerized with a mutant protein in the RER might not be a substrate for the furin protease if the protease recognized only an oligomeric substrate. However, the presence of the mutant protein had no effect on the cleavage of the wild-type protein. This result could indicate that cleavage of a wild-type polypeptide occurs regardless of the sequence at the cleavage site of other members of the oligomer. Alternatively, it was possible that the sequence in the cleavage site somehow dictated oligomerization specificity. This possibility was not likely, however, since cross-linking experiments suggested complexes can form between mutant F0 and the wild-type F1.
The presence of the mutant F protein did, however, inhibit the fusion activity as measured by syncytium formation. A dominant negative phenotype of a mutant protein can be interpreted to indicate that a protein functions as an oligomer (8). Indeed, the mature F protein has been found to exist as an oligomer, although there is some disagreement as to the number of subunits in the oligomer. It has been argued that the F protein forms a dimer or tetramer (4, 23). Recently, however, size determinations on sucrose gradients and cross-linking experiments have suggested that the NDV F protein and the SV5 F protein both form trimers (17, 18).
That the mutant F protein inhibits the activity of the wild-type protein suggests that inclusion of mutant protein in a heterooligomer inhibits the activity of the oligomer. This inhibition may take several forms. First, it is possible that the only active oligomer is composed of entirely wild-type subunits and that heterooligomers have no activity. In this case, the mutation is strictly a dominant negative mutation. Alternatively, it is possible that some heterooligomers have partial activity. In this case, the activity measured in cotransfections with mutant and wild-type DNAs would be a combination of the activity of fully wild-type oligomers as well as full or partial activity of some heterooligomers. In this case, a mutant is not dominant.
The results presented above showed that the expression of F protein was proportional to the input DNA and that the expression levels of wild-type and mutant DNAs were comparable for a given amount of DNA. Furthermore, the cleavage mutants had no effect on cleavage of the wild-type protein, and heterooligomers likely form. Thus if one assumes that the formation of oligomers is totally random, i.e., that one to three amino acid changes in the cleavage site had no effect on oligomer formation, then the concentration of various forms of homo- and heterooligomers can be calculated for each ratio of wild-type and mutant DNA according to simple statistical considerations. Calculations can be accomplished with the formula
 |
where T is the total population of F molecules, w is the fraction of wild-type molecules, m is the fraction of mutant molecules, n is the number of subunits in the oligomer, and Ai = n!/(i!) · (n − i!). The first term of the equation represents the concentration of fully wild-type oligomers, the last term represents the concentration of fully mutant oligomers, and the middle term represents all combinations of heterooligomers. Thus, for a trimer, the total population is represented by the formula
 |
If the concentration of the wild type is 50% of the total, then the fraction of fully wild-type oligomers would be 12.5%, while the fraction of trimers containing two wild-type molecules would be 37.5%, the fraction of oligomers containing only one wild-type molecule would be 37.5%, and the fraction of molecules containing only mutant protein would be 12.5%.
The percent of the population at each ratio of wild-type and mutant DNA composed of only wild-type subunits can be calculated with the first term of the equation. The results for dimers (n = 2), trimers (n = 3), and tetramers (n = 4), as well as larger complexes (n = 6 and n = 9) are shown in Fig. 8. It was shown above that the fusion activity was directly proportional to the input DNA; therefore, the activity at different ratios of wild-type and mutant DNA was compared to that of these predicted populations. Interestingly, the experimental values obtained most closely coincided with the predicted concentrations of wild-type oligomer if the oligomer were a trimer. This result indicates that the active form of the F protein is likely larger than a dimer. The simplest interpretation of these results is that the active form of F is a trimer and that the active form contains only wild-type subunits. The data are not consistent with the idea that a trimer with some mutant subunits has partial activity. If such were the case, the observed activity at each ratio of DNA would be larger and would fall closer to the dimer curve or above.
An alternative explanation for the observed results is that the active form of the F protein is larger than a trimer, such as a heptamer or a nine-mer. However, if such were the case, then subunits containing only some wild-type subunits would have to have partial activity to account for the results obtained. That is, some of the population represented by the middle term of the equation would have to have partial activity.
The question of the number of oligomers required for fusion has been addressed in the influenza virus system by several approaches, all of which suggest that several trimers must function together during the fusion process (5, 7, 29). Thus, the results obtained in this study were unexpected. However, our results do not preclude the possibility that multiple trimers in a large complex function in the fusion process mediated by the paramyxovirus F protein. As noted above, it is possible that complexes of trimers with some inactive monomers may have partial activity. However, it should be noted that our attempts to generate a theoretical curve for a hexomer or a nine-mer that fit the experimental data assuming various partial activities of heterooligomers were not successful.
Alternatively, an uncleaved F0 may not interfere with a step in the fusion process involving multiple oligomers. Studies in a number of systems have proposed that the fusion process involves multiple steps, including aggregation and activation of fusion proteins, close approach of target and attack membranes, hemifusion, pore formation, and pore expansion (7, 13). How fusion proteins mediate each of these steps is not well understood. Perhaps an uncleaved fusion protein is functional in some steps but not others. If this were the case, the assay described above may measure the active unit as a trimer and not a larger complex.
In summary, it was shown that coexpression of two forms of the NDV F protein, one with a furin site and one without, has no effect on the cleavage of the F protein with a furin site, although the cleaved and uncleaved forms of the fusion protein can form heterooligomers. These results argue that furin recognizes and cleaves individual subunits of the fusion protein oligomer. Furthermore, at various ratios of expression of mutant and wild-type protein, results were obtained that are most consistent with the notion that the size of the functionally active NDV F protein in syncytium formation is a trimer, although partial activity of a larger oligomer containing wild-type and mutant protein cannot be excluded.
ACKNOWLEDGMENT
This work was supported by grant AI30572 from the National Institutes of Health.
REFERENCES
- 1.Bagai S, Lamb R A. Quantitative measurement of paramyxovirus fusion: differences in requirements of glycoproteins between simian virus 5 and human parainfluenza virus 3 or Newcastle disease virus. J Virol. 1995;69:6712–6719. doi: 10.1128/jvi.69.11.6712-6719.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Blumenthal R, Sarkar K P, Durell S, Howard D E, Morris S J. Dilation of the influenza hemagglutinin fusion pore revealed by the kinetics of individual cell-cell fusion events. J Cell Biol. 1996;135:63–71. doi: 10.1083/jcb.135.1.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Campbell M J. Lipofection reagents prepared by a simple ethanol injection technique. BioTechniques. 1995;18:1027–1032. [PubMed] [Google Scholar]
- 4.Collins P, Mottet G. Posttranslational processing and oligomerization of the fusion glycoproteins of human respiratory syncytial virus. J Gen Virol. 1991;72:3095–3101. doi: 10.1099/0022-1317-72-12-3095. [DOI] [PubMed] [Google Scholar]
- 5.Danieli T, Pelletier S L, Henis Y I, White J M. Membrane fusion mediated by the influenza virus hemagglutinin requires the concerted action of at least three hemagglutinin trimers. J Cell Biol. 1996;133:559–569. doi: 10.1083/jcb.133.3.559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Freed E O, Delwart E L, Buchschacher J G L, Panganiban A T. A mutation in the human immunodeficiency virus type 1 transmembrane glycoprotein gp41 dominantly interferes with fusion and infectivity. Proc Natl Acad Sci USA. 1992;89:70–74. doi: 10.1073/pnas.89.1.70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hernandez L D, Hoffman L R, Wolfsberg T G, White J M. Virus-cell and cell-cell fusion. Annu Rev Cell Biol. 1996;12:627–661. doi: 10.1146/annurev.cellbio.12.1.627. [DOI] [PubMed] [Google Scholar]
- 8.Herskowitz I. Functional inactivation of genes by dominant negative mutations. Nature. 1987;329:219–222. doi: 10.1038/329219a0. [DOI] [PubMed] [Google Scholar]
- 9.Lamb R A. Paramyxovirus fusion: a hypothesis of changes. Virology. 1993;197:1–11. doi: 10.1006/viro.1993.1561. [DOI] [PubMed] [Google Scholar]
- 10.Lamb R A, Kolakofsky D. Paramyxoviridae: the viruses and their replication. In: Fields B N, Knipe D M, Howley P M, editors. Fields virology. 3rd ed. Vol. 1. Philadelphia, Pa: Lippincott-Raven; 1996. pp. 1177–1206. [Google Scholar]
- 11.McGinnes L, Sergel T, Morrison T G. Mutations in the transmembrane domain of the HN protein of Newcastle disease virus affect the structure and activity of the protein. Virology. 1993;196:101–110. doi: 10.1006/viro.1993.1458. [DOI] [PubMed] [Google Scholar]
- 12.McGinnes L W, Morrison T G. The role of the individual cysteine residues in the formation of the mature, antigenic HN protein of Newcastle disease virus. Virology. 1994;200:470–483. doi: 10.1006/viro.1994.1210. [DOI] [PubMed] [Google Scholar]
- 13.Monck J R, Fernandez J M. The exocytotic fusion pore. J Cell Biol. 1992;119:1395–1404. doi: 10.1083/jcb.119.6.1395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Morrison T, McQuain C, Sergel T, McGinnes L, Reitter J. The role of the amino terminus of F1 of the Newcastle disease virus fusion protein in cleavage and fusion. Virology. 1993;193:997–1000. doi: 10.1006/viro.1993.1214. [DOI] [PubMed] [Google Scholar]
- 15.Morrison T, Ward L J, Semerjian A. Intracellular processing of the Newcastle disease virus fusion glycoprotein. J Virol. 1985;53:851–857. doi: 10.1128/jvi.53.3.851-857.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nagai Y, Klenk H D. Activation of precursors to both glycoproteins of Newcastle disease virus by proteolytic cleavage. Virology. 1977;77:125–134. doi: 10.1016/0042-6822(77)90412-3. [DOI] [PubMed] [Google Scholar]
- 17.Reitter J N, Sergel T, Morrison T G. Mutational analysis of the leucine zipper motif in the Newcastle disease virus fusion protein. J Virol. 1995;69:5995–6004. doi: 10.1128/jvi.69.10.5995-6004.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Russell R, Paterson R G, Lamb R A. Studies with crosslinking reagents on the oligomeric form of the paramyxovirus fusion protein. Virology. 1994;199:160–168. doi: 10.1006/viro.1994.1108. [DOI] [PubMed] [Google Scholar]
- 19.Sakaguchi T, Toyoda T, Gotoh B, Inocencio N M, Kuma K, Miyata T, Nagai Y. Newcastle disease virus evolution. I. Multiple lineages defined by sequence variability of the hemagglutinin-neuraminidase gene. Virology. 1989;169:260–272. doi: 10.1016/0042-6822(89)90151-7. [DOI] [PubMed] [Google Scholar]
- 20.Sakaguchi T, Tu Q, Pinto L H, Lamb R A. The active oligomeric state of the minimalistic influenza M2 ion channel is a tetramer. Proc Natl Acad Sci USA. 1997;94:5000–5005. doi: 10.1073/pnas.94.10.5000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Scheid A, Choppin P W. Identification of biological activities of paramyxovirus glycoproteins. Activation of cell fusion, hemolysis, and infectivity by proteolytic cleavage of an inactive protein of Sendai virus. Virology. 1974;57:470–490. doi: 10.1016/0042-6822(74)90187-1. [DOI] [PubMed] [Google Scholar]
- 22.Scheid A, Choppin P W. Two disulfide-linked polypeptide chains constitute active F protein of paramyxoviruses. Virology. 1978;80:51–66. doi: 10.1016/0042-6822(77)90380-4. [DOI] [PubMed] [Google Scholar]
- 23.Sechoy O, Philippot J R, Bienvenue A. F protein-F protein interaction within the Sendai virus identified by native bonding of chemical cross-linking. J Biol Chem. 1987;262:11519–11523. [PubMed] [Google Scholar]
- 24.Sergel T, McGinnes L W, Morrison T G. The fusion promotion activity of the NDV HN protein does not correlate with neuraminidase activity. Virology. 1993;196:831–834. doi: 10.1006/viro.1993.1541. [DOI] [PubMed] [Google Scholar]
- 25.Sergel T, McGinnes L W, Peeples M E, Morrison T G. The attachment function of the Newcastle disease virus hemagglutinin-neuraminidase protein can be separated from fusion promotion by mutation. Virology. 1993;193:717–726. doi: 10.1006/viro.1993.1180. [DOI] [PubMed] [Google Scholar]
- 26.Sergel T, Morrison T. Mutations in the cytoplasmic domain of the fusion glycoprotein of Newcastle disease virus depress syncytia formation. Virology. 1995;210:264–272. doi: 10.1006/viro.1995.1343. [DOI] [PubMed] [Google Scholar]
- 27.Sergel-Germano T, McQuain C, Morrison T. Mutations in the fusion peptide and heptad repeat regions of the Newcastle disease virus fusion protein block fusion. J Virol. 1994;68:7654–7658. doi: 10.1128/jvi.68.11.7654-7658.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang C, Raghu G, Morrison T, Peeples M E. Intracellular processing of the paramyxovirus F protein: critical role of the predicted amphipathic alpha helix adjacent to the fusion domain. J Virol. 1992;66:4161–4169. doi: 10.1128/jvi.66.7.4161-4169.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.White J M. Membrane fusion. Science. 1992;258:917–924. doi: 10.1126/science.1439803. [DOI] [PubMed] [Google Scholar]