Summary
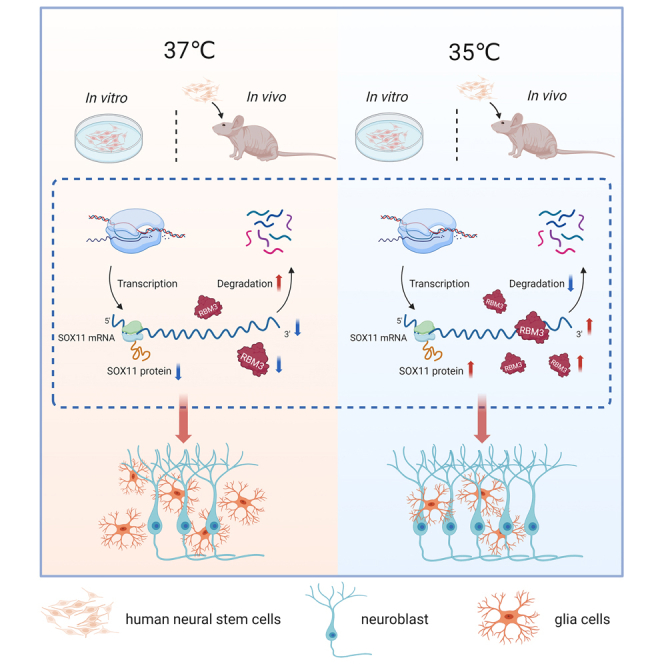
Both therapeutic hypothermia and neural stem cells (NSCs) transplantation have shown promise in neuroprotection and neural repair after brain injury. However, the effects of therapeutic hypothermia on neuronal differentiation of NSCs are not elucidated. In this study, we aimed to investigate whether mild hypothermia promoted neuronal differentiation in cultured and transplanted human NSCs (hNSCs). A significant increase in neuronal differentiation rate of hNSCs was found when exposed to 35°C, from 33% to 45% in vitro and from 7% to 15% in vivo. Additionally, single-cell RNA sequencing identified upregulation of RNA-binding motif protein 3 (RBM3) in neuroblast at 35°C, which stabilized the SRY-box transcription factor 11 (SOX11) mRNA and increased its protein expression, leading to an increase in neuronal differentiation of hNSCs. In conclusion, our study highlights that mild hypothermia at 35°C enhances hNSCs-induced neurogenesis through the novel RBM3-SOX11 signaling pathway, and provides a potential treatment strategy in brain disorders.
Subject areas: Neuroscience, Cell biology, Stem cells research
Graphical abstract
Highlights
-
•
Mild hypothermia of 35°C promotes cultured hNSCs to differentiate into neurons
-
•
RBM3-SOX11 signaling pathway regulates the neuronal differentiation of hNSCs
-
•
RBM3 binds to the 3′UTR of SOX11 mRNA and stabilizes SOX11 mRNA
-
•
Mild hypothermia promotes neuronal differentiation of transplanted hNSCs in vivo
Neuroscience; Cell biology; Stem cells research
Introduction
The potential use of stem or progenitor cells to reduce brain damage or promote regeneration is a promising strategy that has been tested in different central nervous system disorders and traumatic brain diseases.1,2,3 Transplantation of neural stem cells (NSCs) into the injured brain represents a significant method in neural repair, involving neuronal regeneration and improving neurological function.4,5,6,7,8 However, the immune and inflammatory microenvironment, as well as reduced trophic support, often induce apoptotic responses in grafted NSCs after brain damage.9,10,11 Thus, for more effective transplantation, NSCs are usually induced into neurons.6 Unfortunately, the protocol based on mitogen removal and exposure to fetal bovine serum or cytokines results in less than 20% of β-tubulin III positive neurons.12 Although, several methods have been discovered to induce the differentiation of neurons, including NRC-interacting factor 1 (NIF-1),13 gallic acid,14 murine suppressor/enhancer lin12-like (mSEL-1L),15 NOP2/Sun RNA methyltransferase 2 (NSUN2),16 and SUMOylation,17 a simple and effective method for promoting neuronal differentiation is still needed.
Both laboratory experiments and multi-center prospective clinical trials have demonstrated the neuroprotective effects of therapeutic hypothermia (ranging from 33°C to 35°C, also known as mild hypothermia) after brain injury,18,19,20,21,22,23,24,25 which is achieved by reducing neuron energy demands, glutamate release, and reactive oxygen species formation, as well as regulating inflammatory and apoptotic factor expression.26,27,28,29 Mild hypothermia could promote the long-term survival of newborn cells in the dentate gyrus after traumatic brain injury by reducing a pro-apoptotic microenvironment.30 Additionally, mild hypothermia combined with NSCs transplantation for hypoxic-ischemic encephalopathy is achieved through anti-inflammatory and anti-apoptotic mechanisms.31 However, the direct impact of mild hypothermia on NSCs differentiation remains unclear. It has been reported that 30°C hypothermia affects neurogenesis in embryonic mice during pregnancy and development.32 Moderate hypothermia at 32°C can inhibit the differentiation and proliferation of marrow-derived mesenchymal stem cells33 and MEB5 mouse NSC.34 However, these temperatures are not suitable for clinical treatment of patients with brain injuries, as such hypothermia can lead to potential side effects. Therefore, it is important to thoroughly investigate the effects of mild hypothermia on neuronal differentiation. Additionally, it is also necessary to determine the precise temperature range and duration of hypothermia that provide optimal benefits for NSCs.
In the present study, we aimed to explore the effects of therapeutic mild hypothermia on human NSCs (hNSCs)-induced neuronal differentiation with different temperatures and elucidate the underlying molecular mechanism. We expect to seek a novel, efficient, and practical approach to enhance neurogenesis in neural repair and regeneration.
Results
Mild hypothermia of 35°C promoted the neuronal differentiation of cultured hNSCs
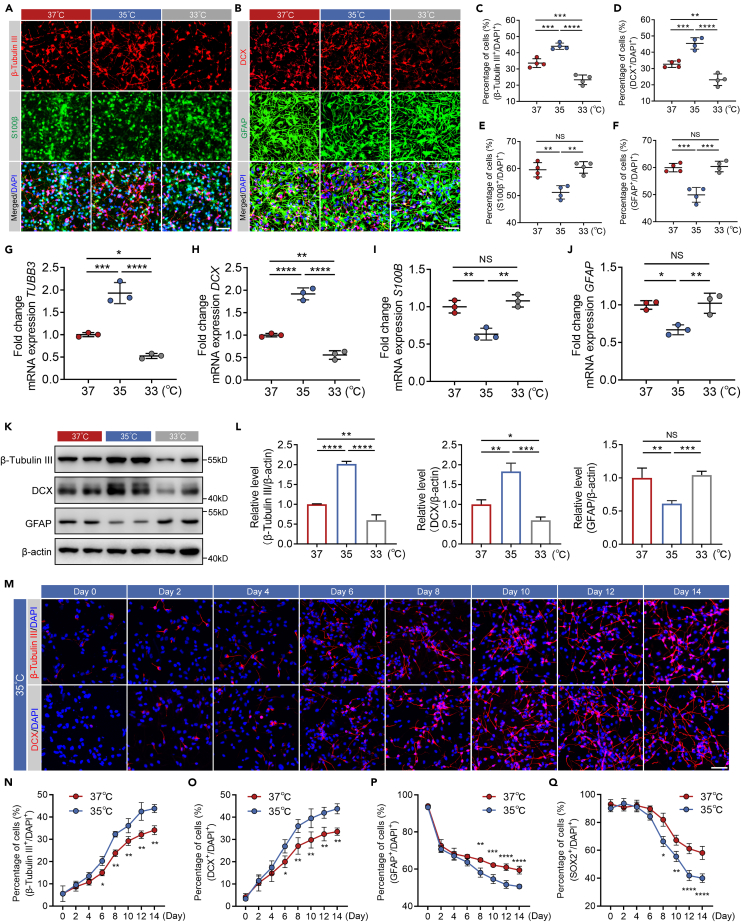
Moderate hypothermia at 32°C can inhibit the differentiation and proliferation of marrow-derived mesenchymal stem cells33 and MEB5 mouse NSC.34 However, it was still unknown whether mild hypothermia affected the differentiation of hNSCs, and whether different temperatures had varying effects. Besides, in order to accurately represent human conditions, we used embryo-derived hNSCs in our study. To answer whether mild hypothermia could promote neuronal differentiation of hNSCs or not, we first investigated the effect of mild hypothermia on neuronal differentiation of cultured hNSCs compared with normothermia (37°C) treatment. A mild hypothermia treatment of 33°C and 35°C was chosen based on the results of clinical studies.18 Cultured hNSCs were incubated with neural differentiation medium under mild hypothermia and normothermia for 14 days, and the expression of β-tubulin III and DCX (neuron markers), and S100β and GFAP (glial cell markers) was examined by immunofluorescence (Figures 1A and 1B). In comparison with 37°C-treatment, the percentage of β-tubulin III or DCX positive cells was significantly increased at 35°C (Figures 1C and 1D). Specifically, ∼45% of hNSCs differentiated into neurons at 35°C compared to ∼33% at 37°C (Figures 1C and 1D). In other words, cooling down to 35°C promoted about an additional 12% of hNSCs to differentiate into neurons. In contrast, compared with 37°C-treatment, the percentage of S100β or GFAP positive cells was significantly decreased at 35°C (Figures 1E and 1F). Surprisingly, although 33°C belongs to mild hypothermia as well, the percentage of β-tubulin III or DCX positive cells was decreased and the percentage of S100β or GFAP positive cells did not change at 33°C compared with 37°C-treatment (Figures 1A–1F). The mRNA and protein expression levels of β-tubulin III, DCX and GFAP shown by the qRT-PCR and western blotting were consistent with the microscopic data (Figures 1G–1L). These results confirm that mild hypothermia of 35°C is capable of promoting neuronal differentiation of cultured hNSCs, whereas 33°C has the opposite effect. It suggests that the mild hypothermia of 35°C, but not 33°C, induces neurogenesis of cultured hNSCs.
Figure 1.
Mild hypothermia of 35°C promoted the neuronal differentiation of cultured hNSCs
(A and B) Representative immunofluorescence images showing β-tubulin III, DCX, S100β, and GFAP positive differentiated hNSCs at Day 14 with 37°C-, 35°C-, and 33°C-treatments.
(C–F) The percentage of β-tubulin III, DCX, S100β, and GFAP positive cells in (A) and (B).
(G–J) qRT-PCR analysis showing the relative mRNA expression of TUBB3, DCX, S100B, and GFAP of cultured hNSCs at Day 14 with 37°C-, 35°C-, and 33°C-treatments.
(K) Western blots showing the protein expression of β-tubulin III, DCX, and GFAP of cultured hNSCs at Day 14 with 37°C-, 35°C-, and 33°C-treatments. β-actin was used as the loading control.
(L) The quantification of western blots of (K). Normalized β-tubulin III, DCX, and GFAP to corresponding loading control were summarized for three independent trials.
(M) Representative immunofluorescence images showing β-tubulin III and DCX positive cells from Day 0 to Day 14 every two days with 35°C-treatment.
(N–Q) The percentage of β-tubulin III, DCX, GFAP, and SOX2 positive cells from Day 0 to Day 14 every two days with 35°C- and 37°C-treatment. All data presented as mean ± SD. One-way ANOVA tests were used in (C–J) and (L). Two-way ANOVA tests were used in (N–Q). NS, not significant; ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001. Scale bars represent 50 μm in (A), (B), and (M).
See also Figures S1 and S2.
Understanding the timeline of hNSCs differentiation at 35°C is critical for evaluating the neurogenesis effect of mild hypothermia of 35°C on cultured hNSCs. Therefore, the expression of β-tubulin III, DCX, and GFAP was examined by immunofluorescence every two days from Day 0 to Day 14 after neuronal induction at 35°C and 37°C (Figures 1M and S1A–S1C). The longitudinal immunofluorescence revealed a statistically significant effect of 35°C-treatment over time. There was an increase in the percentage of β-tubulin III or DCX positive cells from Day 0 until Day 6 (Figures 1N and 1O) and a decrease in the percentage of GFAP positive cells from Day 0 until Day 8 (Figure 1P). The different signal kinetics and relative signal changes between 35°C and 37°C continued until 14 days. These data support that mild hypothermia of 35°C induces neuronal differentiation of hNSCs after 6-day incubation. Besides, we also examined the expression of SOX2, an NSCs marker, and found that SOX2 positive cells gradually decreased during the initial 4 days (Figures S1B and S1C). A significant reduction in the percentage of SOX2 positive cells treated with 35°C was observed until Day 8 (Figure 1Q), which once more illustrates that the differentiation of hNSCs can be facilitated by mild hypothermia of 35°C.
We then explored the proliferation of cultured hNSCs at 35°C or 37°C by EdU staining. As shown in Figures S2A, S2B, and S2E, the percentage of EdU positive cells at 35°C was significantly decreased compared with 37°C from Day 8 to Day 14, showing that mild hypothermia of 35°C repressed the proliferation of cultured hNSCs incubated with neural differentiation medium. However, the percentage of EdU+DCX+ cells to EdU+ cells at 35°C was significantly increased from Day 6 to Day 14, while the percentage of EdU+GFAP+ cells EdU+ cells at 35°C was significantly decreased from Day 8 to Day 14, which further supports that mild hypothermia of 35°C promotes neurogenesis of cultured hNSCs (Figures S2A–S2D).
RBM3 was identified as a potential regulator in neuronal differentiation of hNSCs
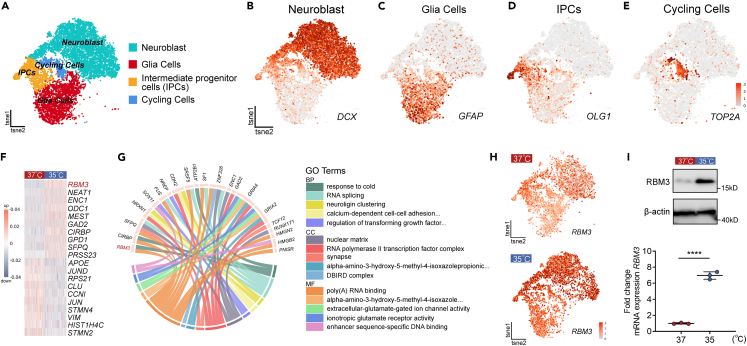
In order to elucidate why mild hypothermia of 35°C could promote neurogenesis of cultured hNSCs, we performed single-cell RNA sequencing (scRNA-seq) to reveal the molecular mechanism. Firstly, we conducted scRNA-seq of hNSCs at Day 0 and the analysis of specific marker genes revealed that most cells at Day 0 were PAX6 or NES positive radical glia cells and TJP1 positive neuroepithelial cells, which meant the hNSCs were pluripotent35,36 (Figure S3). Then, cultured hNSCs which underwent neuronal induction for 14 days at 37°C served as controls and were compared to hNSCs with the same culture conditions at 35°C. Using Drop-seq, we sequenced 6,238 and 6,751 cells at 35°C and 37°C, respectively. A single-cell digital gene expression matrix was generated and then projected into two dimensions using t-distributed stochastic neighbor embedding (t-SNE) to clarify cell clusters. As shown in Figure 2A, four clusters of cells with similar gene expression patterns were identified. We resolved the cell type identities based on the cluster-specific gene signatures and recovered the four clusters as known cell types including neuroblast, glia cells, intermediate progenitor cells (IPCs), and cycling cells. We then validated each cluster by DCX for neuroblast, GFAP for glia cells, OLG1 for IPCs, and TOP2A for cycling cells (Figures 2B–2E).
Figure 2.
RBM3 was identified as a potential regulator in neuronal differentiation of hNSCs by mild hypothermia
(A) The t-SNE plot of single-cell RNA sequencing showing individual cell types of differentiated hNSCs at Day 14 based on maker genes expression, including neuroblast, glia cells, intermediate progenitor cells and cycling cells.
(B–E) t-SNE plots highlighting cell types by marker genes expression: neuroblast (DCX), glia cells (GFAP), intermediate progenitor cells (OLIG1), and cycling cells (TOP2A).
(F) Heatmap of top 10 and last 10 differentially expressed genes (DEGs) between 35°C and 37°C in neuroblast clusters. Each row represents a gene, and each column represents a cell.
(G) Circos plot showing significant upregulated genes and GO analysis. The bottom half of circos plot shows the top 5 GO terms including BP, MF, CC and the top half shows the DEGs of the depicted GO enrichment analysis.
(H) t-SNE plots highlighting the RBM3 expression in differentiated hNSCs at Day 14 with 35°C- and 37°C-treatments. (I) Western blots and qRT-PCR analysis showing the protein and mRNA expressions of RBM3 in 35°C- and 37°C-treatments. β-actin was used as the loading control. All data presented as mean ± SD. Student’s t test was used in (I). ∗∗∗∗p < 0.0001.
To reveal potential mechanisms underlying neuronal differentiation observed at distinct temperature, we included only the neuroblast cluster for further analysis. As shown in Figure 2F and Table S1, heatmap depicted the top 10 upregulated and downregulated genes of neuroblast cluster at 35°C compared with 37°C-treatment, which indicated that these differentially expressed genes were likely to play an important role in regulating neuronal differentiation of hNSCs. To be able to assess subtle transcriptomic alteration, we performed gene ontology (GO) enrichment analysis of all significantly differentially regulated genes (Table S2). In many cases, single genes are often assigned to many ontological terms and therefore the complex relationships between genes and GO-terms were visualized using circos plots including logFC values and gene symbols. Based on the circos plots of GO analysis, it was found that “response to cold” was one of the top five upregulated biological process (BP) terms in the 35°C-treated neuroblast clusters, and RBM3, CIRBP, and ATP2B1 were the genes differentially expressed within “response to cold” (Figure 2G; Table S2). Combined with the heatmap in Figure 2F, RBM3 is the gene that was most significantly upregulated in 35°C-cultured hNSCs, and we believed that RBM3 might be the key regulator in neuronal differentiation at 35°C. RBM3 is a cold-inducible mRNA binding protein that regulates global protein synthesis both physiologically and mild hypothermically.37 It is widely expressed in human cells and brain tissue and increased on cooling, and thus has been suggested as a biomarker in therapeutic hypothermia.37 Pharmacologically inducing RBM3 expression represents a new possibility for neuroprotection in the absence of hypothermia.38 Consistent with previous reports, RBM3 was expressed in almost 4 clusters, but the upregulation of RBM3 was most noticeable in neuroblast cluster at 35°C (Figure 2H). It was confirmed that the protein and mRNA levels of RBM3 were significantly increased after 14-day neuronal induction of hNSCs at 35°C by western blotting and qRT-PCR analysis (Figure 2I). In conclusion, these data demonstrate that RBM3 probably take part in promoting neurogenesis of cultured hNSCs by mild hypothermia of 35°C.
RBM3 promoted neuronal differentiation of hNSCs under mild hypothermia of 35°C
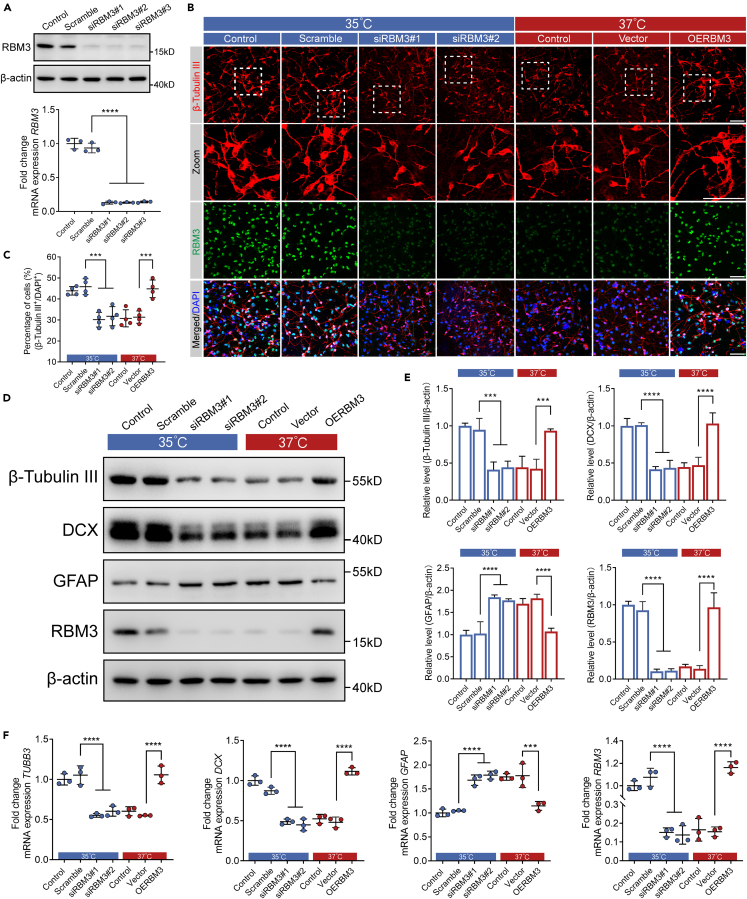
To further confirm the role of RBM3 in neuronal differentiation of cultured hNSCs at 35°C, we knocked down endogenous RBM3 expression by RNAi strategy in hNSCs and determined whether loss of RBM3 would affect neuronal differentiation of cultured hNSCs at 35°C. Using RBM3-specific siRNAs (Table S3), we achieved a more than 80% knockdown of RBM3 protein expression in hNSCs incubated at 35°C for 4 days (Figure 3A). As determined by qRT-PCR and immunofluorescence analysis, the knockdown effectiveness of RBM3 mRNA and protein expression were consistent with the results of western blotting (Figures 3A and 3B). Wild-type, scrambled, and RBM3 knocked-down hNSCs were incubated with neural differentiation medium at 35°C and 37°C for 14 days, and the expression of β-tubulin III was examined by immunofluorescence. Consistent with the aforementioned results, 35°C significantly increased the percentage of β-tubulin III positive cells in wild-type hNSCs, as well as scrambled hNSCs, compared with wild-type hNSCs at 37°C (Figures 3B and 3C). As expected, 35°C lost its effect on the percentage of β-tubulin III positive cells after knockdown of RBM3 in comparison with scrambled hNSCs incubated at 35°C, which indicated that RBM3 played an important role in increasing neuronal differentiation of cultured hNSCs by mild hypothermia of 35°C (Figures 3B and 3C). Furthermore, the knockdown of RBM3 at 35°C resulted in no significant difference of β-tubulin III as well as DCX protein and mRNA expression, compared with wild-type hNSCs at 37°C (Figures 3B–3F). To further validate the role of RBM3, we overexpressed RBM3 in hNSCs at 37°C using lentivirus. After 14-day neuronal induction at 37°C, overexpression of RBM3 significantly increased the percentage of β-tubulin III positive cells compared with hNSCs infected with vector-lentivirus, which demonstrated that neuronal differentiation of cultured hNSCs could be facilitated by RBM3 (Figures 3B and 3C). The increased protein and mRNA levels of β-tubulin III and DCX also supported the aforementioned results (Figures 3D–3F). Besides, according to western blotting and qRT-PCR analysis, we found that overexpression of RBM3 significantly inhibited the GFAP expression at 37°C, while knockdown of RBM3 increased GFAP expression at 35°C, which suggested that RBM3 took part in regulating glial differentiation of cultured hNSCs as well (Figures 3D–3F). In these experiments, RBM3 is shown to be an important regulator in the promotion of neuronal differentiation by mild hypothermia of 35°C in cultured hNSCs.
Figure 3.
RBM3 promoted neuronal differentiation of cultured hNSCs
(A) Western blots and qRT-PCR analysis showing the protein and mRNA expression of RBM3 after scramble or RBM3 siRNA transfection. β-actin was used as the loading control.
(B) Representative immunofluorescence images of β-tubulin III positive cells and RBM3 expression at Day 14. (Left 4 panels) hNSCs were transfected with scramble or RBM3 siRNA with 35°C-treatment. (Right 3 panels) hNSCs were transducted with vector or RBM3 by lentivirus with 37°C-treatment.
(C) The percentage of β-tubulin III positive cells in (B).
(D) Western blot showing β-tubulin III, DCX, GFAP, and RBM3 protein expression at Day 14. (Left 4 lanes) hNSCs were transfected with scramble or RBM3 siRNA with 35°C-treatment. (Right 3 lanes) hNSCs were transducted with vector or RBM3 by lentivirus with 37°C-treatment. β-actin was used as the loading control.
(E) The quantification of western blots of (D). Normalized β-tubulin III, DCX, GFAP, and RBM3 to corresponding loading control were summarized for three independent trials.
(F) qRT-PCR analysis showing the mRNA level of TUBB3, DCX, GFAP, and RBM3 at Day 14. (Left 4 columns) hNSCs were transfected with scramble or RBM3 siRNA with 35°C-treatment. (Right 3 columns) hNSCs were transducted with vector or RBM3 by lentivirus with 37°C-treatment. All data presented as mean ± SD. One-way ANOVA tests were used in (A), (C), (E) and (F). ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001. Scale bars represent 50 μm in (B).
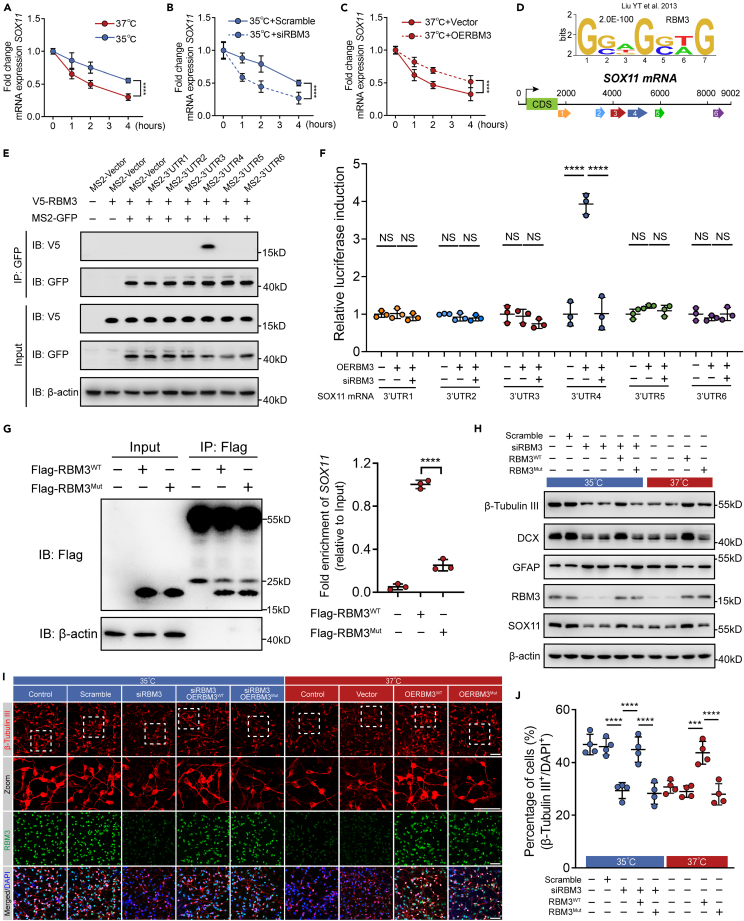
RBM3-SOX11 signaling pathway regulates neuronal differentiation of hNSCs during mild hypothermia of 35°C
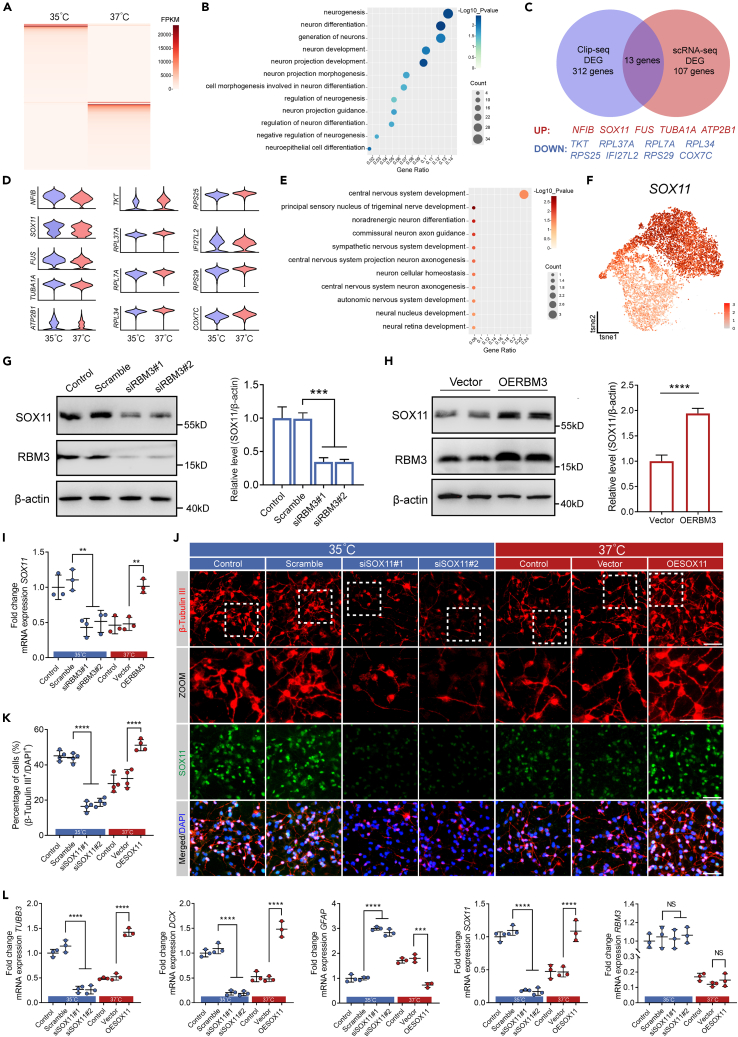
Upon identifying RBM3 as an RNA-binding protein, we performed crosslinking immunoprecipitation RNA sequencing (CLIP-seq) analysis to isolate RNA molecules bound to RBM3 using differentiated hNSCs cultured in neural differentiation medium at 35°C and 37°C. Our investigation revealed 312 differently expressed mRNAs bound to RBM3 in hNSCs between 35°C and 37°C (Figure 4A; Table S4). Furthermore, we identified 166 upregulated and 146 downregulated mRNAs at 35°C compared to at 37°C, along with 160 mRNAs exclusive to the 35°C-treatment and 140 mRNAs exclusive to the 37°C-treatment (Figure 4A; Table S4). We then executed GO enrichment analysis on the mRNA bound to RBM3, pinpointing 12 enriched BP terms related to neuron differentiation and neurogenesis (Figure 4B; Table S5). A subsequent comparison of the 312 expressed mRNAs from CLIP-seq with the 107 differentially expressed genes from scRNA-seq revealed only 13 common mRNA molecules, including upregulated NFIB, SOX11, FUS, TUBA1A, and ATP2B1, and downregulated TKT, RPL37A, RPL34, RPS25, IFI27L2, RPS29, and COX7C at 35°C compared to at 37°C (Figure 4C). The scRNA-seq violin plots depicted the expression levels of these 13 mRNAs in differentiated hNSCs at Day 14 (Figure 4D). We conducted GO enrichment analysis on these 13 mRNAs, and found that three mRNAs were associated with neuronal differentiation (Figure 4E; Table S6). Upon through examination of the gene distribution and expression, we proposed SOX11 as a potential target of RBM3 (Figure 4F).
Figure 4.
SOX11 was regulated by RBM3 and promoted neuronal differentiation of cultured hNSCs
(A) Heatmap of CLIP-seq showing differentially RBM3-bound mRNA with 35°C- and 37°C-treatment.
(B) GO enrichment analysis showing 12 enriched BP terms related to neuron differentiation and neurogenesis on differentially RBM3-bound mRNA.
(C) Venn diagram showing 13 differentially expressed genes in both CLIP-seq and scRNA-seq.
(D) Violin maps of the 13 differentially expressed genes in scRNA-seq.
(E) GO enrichment analysis on the 13 differentially expressed genes.
(F) t-SNE plot highlighting the SOX11 expression in cultured hNSCs at Day 14.
(G) (Left) Western blots analysis showing the protein expression of SOX11 and RBM3 at Day 14 after scramble or RBM3 siRNA transfection. β-actin was used as the loading control. (Right) Normalized SOX11 to corresponding loading control were summarized for three independent trials.
(H) (Left) Western blots analysis showing the protein expression of SOX11 and RBM3 at Day 14 after vector or RBM3 overexpression by lentivirus. β-actin was used as the loading control. (Right) Normalized SOX11 to corresponding loading control were summarized for three independent trials.
(I) qRT-PCR analysis showing the mRNA level of SOX11 in cultured hNSCs at Day 14. (Left 4 columns) hNSCs were transfected with scramble or RBM3 siRNA with 35°C-treatment. (Right 3 columns) hNSCs were transducted with vector or RBM3 by lentivirus with 37°C-treatment.
(J) Representative immunofluorescence images of β-tubulin III positive cells and SOX11 expression at Day 14. (Left 4 panels) hNSCs were transfected with scramble or SOX11 siRNA with 35°C-treatment. (Right 3 panels) hNSCs were transducted with vector or SOX11 by lentivirus with 37°C-treatment.
(K) The percentage of β-tubulin III positive cells in (J).
(L) qRT-PCR analysis showing the mRNA level of TUBB3, DCX, GFAP, SOX11, and RBM3 at Day 14. (Left 4 columns) hNSCs were transfected with scramble or SOX11 siRNA with 35°C-treatment. (Right 3 columns) hNSCs were transducted with vector or SOX11 by lentivirus with 37°C-treatment. All data presented as mean ± SD. One-way ANOVA tests were used in (G), (I), (K) and (L). Student’s t test was used in (H). NS, not significant; ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001. Scale bars represent 50 μm in (J).
To validate that endogenous RBM3 regulated SOX11 expression in hNSCs at 35°C, the protein and mRNA expression of SOX11 were examined in RBM3 knocked-down and scrambled hNSCs after neuronal induction for 14 days by western blotting and qRT-PCR analysis. As shown in Figures 4G and 4I, largely deleted endogenous RBM3 significantly decreased the protein and mRNA levels of SOX11 at 35°C. Besides, the protein and mRNA levels of SOX11 were also increased in cultured hNSCs with overexpression of RBM3 at 37°C using lentivirus (Figures 4H and 4I). These data indicate that RBM3 regulates SOX11 expression in cultured hNSCs under mild hypothermia of 35°C.
To verify the role of SOX11 in neuronal differentiation of cultured hNSCs regulated by RBM3 at 35°C, we knocked down endogenous SOX11 expression by SOX11-specific siRNAs. As expected, more than 80% SOX11 was deleted by SOX11-siRNAs in hNSCs incubated at 35°C for 4 days (Figure S4A). Wild-type, SOX11 knocked-down and scrambled hNSCs were incubated with neural differentiation medium at 35°C and 37°C for 14 days, and the expression of β-tubulin III was examined by immunofluorescence. Similarly, the percentage of β-tubulin III positive cells in wild-type hNSCs as well as scrambled hNSCs was significantly increased at 35°C (Figures 4J and 4K). The knockdown of SOX11 led to significant reduction of the percentage of β-tubulin III positive cells at 35°C compared with scrambled hNSCs at 35°C, showing that SOX11 regulated neurogenesis of cultured hNSCs under mild hypothermia of 35°C (Figures 4J and 4K). Meanwhile, hNSCs were incubated with neural differentiation medium at 37°C for 14 days after infection with SOX11-lentivirus. Compared to hNSCs infected with vector-lentivirus, the percentage of β-tubulin III positive cells was significantly increased in hNSCs overexpressed with SOX11 (Figures 4J and 4K). Nevertheless, the RBM3 was increased in hNSCs at 35°C in comparison with hNSCs at 37°C, while no significant change in RBM3 expression was observed when SOX11 was knocked down or overexpressed (Figures S4B, S4C, and 4L). The β-tubulin III and DCX protein and mRNA expression were consistent with the aforementioned immunofluorescence results (Figures S4B, S4C, and 4L). Similar to the function of RBM3, at protein and mRNA level, overexpression of SOX11 significantly inhibited the GFAP expression at 37°C, while knockdown of SOX11 increased GFAP expression at 35°C, which indicated that SOX11 also regulated glial differentiation of cultured hNSCs (Figures S4B, S4C, and 4L). In conclusion, RBM3-SOX11 axis plays an important role in facilitating neurogenesis by mild hypothermia of 35°C in cultured hNSCs.
To further validate the significance of RBM3-SOX11 axis, we then elucidated the association between RBM3 and SOX11. Based on the transcripts versus RNA-binding proteome analysis, SOX11 might be bound to RBM3 through the PF00076 domain predicted by Pfam (Figure S5A).
RBPs play key roles in post-transcriptional processing of RNAs, including pre-mRNA splicing, stabilizing/destabilizing mRNA, mRNA localization, turnover, polyadenylation, translational control, nuclear export,37 etc. As an RBP, RBM3 was reported to modulate mRNA stability in cold stress. Therefore, we firstly examined the half-life of SOX11 mRNA in cultured hNSCs with loss or overexpression of RBM3 at distinct temperature. Cultured hNSCs were incubated with neural differentiation medium at 35°C and 37°C for 14 days, and then treated with actinomycin D (Act. D) that was widely used in mRNA stability assays to inhibit the synthesis of new mRNA. SOX11 mRNA levels were measured after 0, 1, 2, and 4 h of Act. D incubation by qRT-PCR analysis, and the half-life of SOX11 mRNA in 35°C-cultured cells was increased significantly compared with 37°C-cultured cells, showing that mild hypothermia inhibited SOX11 mRNA degradation as expected (Figure 5A). Next, we verified the role of RBM3 in regulating the half-life of SOX11 mRNA. The knockdown of RBM3 decreased the half-life of SOX11 mRNA at 35°C (Figure 5B). Similarly, hNSCs infected with RBM3- or vector-lentivirus were incubated at 37°C for neural differentiation and treated with Act. D, and the overexpression of RBM3 increased the half-life of SOX11 mRNA (Figure 5C). These data suggest that mild hypothermia of 35°C increases SOX11 expression through stabilizing SOX11 mRNA by elevating RBM3 expression.
Figure 5.
RBM3 regulated SOX11 mRNA stability via binding to the 3′UTR of SOX11 mRNA
(A) qRT-PCR analysis showing the relative mRNA expression of SOX11 of cultured hNSCs after actinomycin D incubation at 35°C or 37°C. Data for each time point were collected from three independent trials.
(B) qRT-PCR analysis showing the relative mRNA expression of SOX11 of cultured hNSCs transfected with scramble or RBM3 siRNA after actinomycin D incubation at 35°C. Data for each time point were collected from three independent trials.
(C) qRT-PCR analysis showing the relative mRNA expression of SOX11 of cultured hNSCs expressing vector or RBM3 after actinomycin D incubation at 37°C. Data for each time point were collected from three independent trials.
(D) The SOX11 mRNA 3′UTR sequence that was divided into six parts containing RBM3 recognition motifs.
(E) Western blots analysis showing the expression of RNA pull-downed RBM3 of HEK-293T cells transfected with MS2-3′UTR1-6 plasmids under overexpression of RBM3. β-actin was used as the loading control.
(F) Dual luciferase reporter assay showing the luciferase activity of HEK-293T cells transfected with pmirGLO-3′UTR1-6 plasmids under overexpression or knockdown of RBM3.
(G) (Left) Western blots analysis showing the expression of immunoprecipitated RBM3 in cultured hNSCs expressing RBM3WT or RBM3 mutant (RBM3Mut) at Day 14. β-actin was used as the loading control. (Right) qRT-PCR analysis showing the mRNA level of SOX11 bound to RBM3 in cultured hNSCs expressing RBM3WT or RBM3Mut.
(H) Western blots analysis showing β-tubulin III, DCX, GFAP, RBM3, and SOX11 protein expression at Day 14. (Left 6 lanes) hNSCs were transfected with scramble, RBM3 siRNA, RBM3 siRNA + RBM3WT, or RBM3 siRNA + RBM3Mut with 35°C-treatment. (Right 4 lanes) hNSCs were transducted with vector, RBM3WT, or RBM3Mut by lentivirus with 37°C-treatment. β-actin was used as the loading control.
(I) Representative immunofluorescence images of β-tubulin III positive cells and RBM3 expression at Day 14. (Left 5 panels) hNSCs were transfected with scramble, RBM3 siRNA, RBM3 siRNA + RBM3WT or RBM3 siRNA + RBM3Mut with 35°C-treatment. (Right 4 panels) hNSCs were transducted with vector, RBM3WT or RBM3Mut by lentivirus with 37°C-treatment.
(J) The percentage of β-tubulin III positive cells in (I). All data presented as mean ± SD. Two-way ANOVA tests were used in (A–C). One-way ANOVA tests were used in (F), (G), and (J). NS, not significant; ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001. Scale bars represent 50 μm in (I).
See also Figure S5.
It is common for RBPs to bind to the mRNA 3′UTR for regulating mRNA half-life. As reported in previous study,39 we found several potential RBM3 binding sequences in the 3′UTR of SOX11 mRNA. Considering that sequences were located in six different parts of the 3′UTR of SOX11 mRNA, we named these six parts 3′UTR1–6 (Figure 5D). To determine the binding sequence, an MS2-tagged RNA pull-down assay was performed in HEK-293T cells expressing MS2-3′UTR1-6 with or without V5-RBM3 and MCP-GFP plasmids. The binding of RBM3 with 3′UTR4 of SOX11 mRNA was detected (Figure 5E). Conversely, to examine whether the RBM3 was associated with 3′UTR4 of SOX11 mRNA, dual luciferase reporter assay was performed in HEK-293T cells transfected with pmirGLO-3′UTR1-6 plasmids under overexpression or knockdown of RBM3. The results showed that overexpression of RBM3 increased the luciferase activity driven by 3′UTR4 but not 3′UTR1-3 and 3′UTR5-6 (Figure 5F). The overexpression of RBM3 failed to do the same after transfection of RBM3-siRNA (Figure 5F). Collectively, RBM3 stabilizes SOX11 mRNA by binding to the 3′UTR of SOX11 mRNA. To further confirm the association between RBM3 and SOX11 mRNA, we examined SOX11 expression by mutating the RNA recognition motif (RRM) domain, N-terminal 84 residue, of RBM3 (RBM3Mut). According to the interactions at the RNA-RRM interface, Ser2 (S2), Arg47 (R47), and Gln76 (Q76) which were interacted with the head, middle, and tail of the recognized RNA were mutated to alanine (A) in RBM3Mut (Figure S5B). Next, CLIP-qRT-PCR was performed from hNSCs expressing Flag-RBM3WT or Flag-RBM3Mut at Day 14. Based on the same efficiency of immunoprecipitation, RBM3WT but not RBM3Mut increased the level of SOX11 mRNA bound to RBM3 (Figure 5G). In summary, RBM3 stabilizes SOX11 mRNA by binding to its mRNA.
Lastly, we investigated whether the association between RBM3 and SOX11 mRNA took part in the neuronal differentiation of cultured hNSCs at 35°C. Cultured hNSCs who’s endogenous RBM3 were knocked down by RBM3-siRNA were infected with RBM3WT- or RBM3Mut-lentivirus, and underwent neuronal induction for 14 days at 35°C. The percentage of β-tubulin III positive cells in hNSCs expressing RBM3Mut was significantly decreased compared with hNSCs expressing RBM3WT, and was similar to the RBM3 knocked-down cells (Figures 5I and 5J). Then cultured hNSCs overexpressed with RBM3WT or RBM3Mut were incubated with neural differentiation medium for 14 days at 37°C. The results also showed the loss of function in promoting neuronal differentiation of RBM3Mut (Figures 5I and 5J). The β-tubulin III and DCX protein levels were consistent with the microscopic images (Figure 5H). The change of GFAP protein level acquired by western blotting also supported that RBM3Mut lost its control of the regulation of glial differentiation in cultured hNSCs (Figure 5H). These data suggest that the interaction between RBM3 and SOX11 mRNA plays a key role in the neuronal differentiation of cultured hNSCs under mild hypothermia of 35°C.
To sum up, RBM3-SOX11 signaling pathway regulates neuronal differentiation of hNSCs during mild hypothermia of 35°C.
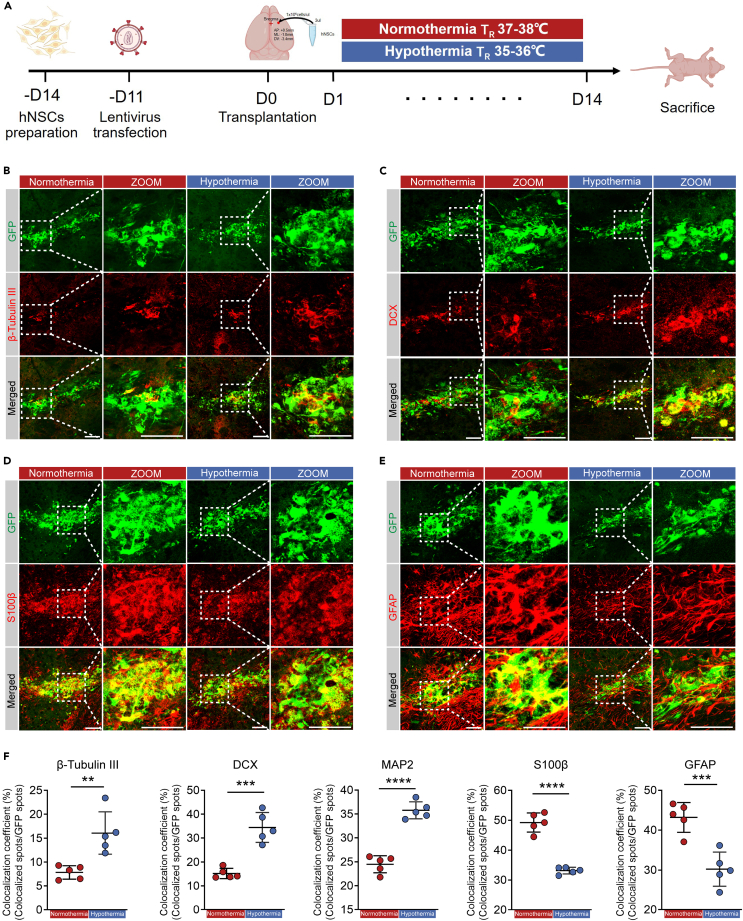
Mild hypothermia of 35°C promoted neuronal differentiation of transplanted hNSCs in vivo
To explore the functional significance of mild hypothermia of 35°C in promoting neuronal differentiation of transplanted hNSCs in mice, nude mice were transplanted with GFP-labeled hNSCs into striatum, and underwent 14-day normothermia or mild hypothermia (35°C) treatment for 2 h every day under rectal temperature monitoring (Figures 6A, S6A, and S6B). Specifically, for precise control of brain temperature, the mouse rectal and brain temperatures were measured. Consistent with previous reports, the mouse rectal temperature was about 0.9°C higher than brain temperature (Figure S6C), which meant that the rectal temperature of mice during normothermia or mild hypothermia was maintained at around 38°C or 36°C, respectively (Figure S6D). After 14 days normothermia or mild hypothermia treatment, nude mice were sacrificed, and the expression of β-tubulin III, DCX, MAP2, S100β, and GFAP in the striatum was examined by immunofluorescence to analyze the differentiation of transplanted hNSCs (Figures 6B–6E and S6E). In comparison with normothermia treatment, the colocalization coefficients of β-tubulin III, DCX, and MAP2 positive cells with GFP positive hNSCs were significantly increased after mild hypothermia treatment, indicating that mild hypothermia could facilitate the differentiation of transplanted hNSCs into neuron (Figure 6F). As expected, the glial differentiation of transplanted hNSCs was inhibited after mild hypothermia treatment (Figure 6F). In conclusion, neuronal differentiation of transplanted hNSCs could be promoted by mild hypothermia treatment.
Figure 6.
Mild hypothermia of 35℃ promoted transplanted hNSCs differentiation into neurons in vivo
(A) The timeline of the hNSCs transplantation into mouse striatum with normothermia or mild hypothermia treatments.
(B–E) Representative immunofluorescence images of β-tubulin III, DCX, S100β, and GFAP positive GFP-labeled cells at Day 14 with normothermia or mild hypothermia treatments.
(F) The colocalization coefficient of β-tubulin III, DCX, MAP2, S100β, and GFAP positive GFP-labeled cells in (B–E) and Figure S6E. All data presented as mean ± SD. Student’s t tests were used in (F). ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001. Scale bars represent 50 μm in (B–E).
See also Figure S6.
Discussion
Therapeutic hypothermia is considered to protect the brain from secondary injury after primary brain injury, while there is controversial information regarding whether hypothermia facilitates or represses injury-related NSCs proliferation and death in clinical sessions.40,41,42,43 Severe neuronal damage from brain injury diseases, such as traumatic brain injury (TBI) or neurodegenerative disorders, highlights the paramount significance of promoting neural regeneration and repair to improve neurological function.44,45 However, the regenerative capacity of neurons is notably limited, primarily due to the scarcity of intrinsic NSCs and their restricted differentiation potential, even after NSCs transplantation, which inevitably hinder its clinical application.12,46,47 Currently, there are a total of 34 registered studies on the transplantation of NSCs in the ClinicalTrials.gov database (https://clinicaltrials.gov/). Recent studies have identified several factors involved in modulating the neuronal differentiation of NSCs, including NIF-1,13 gallic acid,14 murine suppressor/enhancer lin12-like (mSEL-1L),15 NOP2/Sun RNA methyltransferase 2 (NSUN2),16 and SUMOylation.17 Nevertheless, the mechanism of combining hypothermia therapy and hNSCs transplantation remains unknown.
In the study, we used the hNSCs obtained from cortical brain tissue of aborted embryos at 6–8 weeks of gestational age to do the further research. Most hNSCs were radial glia cells and neuroepithelial cells, which were pluripotent.36 Here, we have demonstrated that the neuronal differentiation rate of hNSCs cultured at 35°C was significantly increased both in vitro and in vivo. Mechanistically, RBM3 was upregulated, which stabilized SOX11 mRNA and increased its expression. This novel finding might be of potential in promoting neural repair in hNSCs-based treatment after brain injury or in neuro-degenerative disorders. And consistent with previous studies, a further hypothermia at 33°C injured the cultured hNSCs which had no increase in neuronal differentiation rate in vitro.
RBM3 has previously been identified to promote the proliferation and suppress the apoptotic of NSCs in vitro.34,48,49 This protein not only protects neural stem/progenitor cells (NSPCs) from apoptosis induced by hypoxic ischemia, but also promotes NSPC proliferation and neuronal differentiation.1,49 However, its cellular location is unknown. We pinpointed RBM3 as a key regulator located in neuroblast via scRNA-seq in cultured hNSCs under mild hypothermia compared to normothermia. Furthermore, our study revealed 35°C led to an increase in the expression of RBM3, which in turn resulted in the stabilization of SOX11 mRNA, thereby facilitating the differentiation of hNSCs into neurons.
SOX11 plays a vital role in regulating various processes, including neural progenitor cell (NPC) proliferation,50 neuronal migration,51 and differentiation processes.52 Notably, the targeted deletion of SOX11 in NPCs significantly impairs neurogenesis within the adult hippocampus.1 However, the relationship between RBM3 and SOX11 remains unclear. In this study, we employed a combination of molecular methods, such as actinomycin D chase assay, binding site prediction, RNA pulldown, luciferase reporter assays, CLIP-seq, and mutation of interaction sequences. Our findings demonstrate that RBM3 is upregulated at 35°C, which in turn stabilizes SOX11 mRNA, increases SOX11 expression, and enhances neurogenesis in cultured and transplanted hNSCs. Therefore, we establish the critical role of the RBM3-SOX11 signaling pathway in regulating neurogenesis of hNSCs under mild hypothermia. Consequently, developing a small compound to stabilize the RBM3 protein presents a promising strategy for enhancing neural repair.
Although the research did not address how hypothermia increases RBM3 expression, recent studies have explored the related mechanism. Studies have shown that hypothermia enhanced the HNRNPH1 binding to the G-rich sequence in RBM3 mRNA, which prevented the expression of toxic exons of RBM3 mRNA and inhibited the mRNA degradation. Then it promoted the RBM3 expression.53 Besides, in addition to regulating SOX11 mRNA stability, RBM3 could also bind to Yes-associated protein 1 (Yap1) mRNA, inhibiting its degradation and promoting neuron generation.32
In conclusion, our findings reveal that mild hypothermia at 35°C promotes the neurogenesis of hNSCs through the RBM3-SOX11 signaling pathway. This study opens avenues for further research and suggests that targeting RBM3 could be a promising approach in neural repair.
Limitations of the study
It is important to note some limitations of this study. We did not investigate the specific effects of mild hypothermia on the transplantation of hNSCs in animal models of brain diseases, such as the controlled cortical impact (CCI) or transient middle cerebral artery occlusion (tMACO) models. Besides, hypothermia may affect the surrounding brain tissue to produce chemistry signals, such as fibroblast growth factors (FGFs), insulin-like growth factors (IGFs), sonic hedgehog (Shh), retinoic acid, bone morphogenic proteins (BMPs), Wnts, notch ligands, and purines, to affect the differentiation process of transplanted hNSCs.54 Finally, the functions of the newly generated neurons and their integration into neuronal circuits remain areas for future exploration.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| β-Tubulin III | Abcam | Cat# ab78078; RRID: AB_2256751 |
| β-Tubulin III | Abcam | Cat# ab52623; RRID: AB_869991 |
| β-Tubulin III | Sigma | Cat# T8660; RRID: AB_477590 |
| DCX | Abcam | Cat# ab18723; RRID: AB_732011 |
| S100β | Abcam | Cat# ab52642; RRID: AB_882426 |
| GFAP | Cell Signaling Technology | Cat# 3670; RRID: AB_561049 |
| GFAP | Cell Signaling Technology | Cat# 12389; RRID: AB_2631098 |
| SOX2 | Cell Signaling Technology | Cat# 3579s; RRID: AB_2195767 |
| RBM3 | Proteintech | Cat# 14363-1-AP; RRID: AB_2269266 |
| SOX11 | Millipore | Cat# HPA000536; RRID: AB_1080060 |
| SOX11 | Abmart | Cat# T55823 |
| β-actin | Cell Signaling Technology | Cat# 4970; RRID: AB_2223172 |
| GFP | Abcam | Cat# ab13970; RRID: AB_300798 |
| V5 | Abmart | Cat# M20052 |
| Flag | Abways | Cat# AB0008; RRID: AB_2943672 |
| MAP2 | Abcam | Cat# ab5392; RRID: AB_2138153 |
| Anti-Rabbit 488 | Cell Signaling Technology | Cat# 4412; RRID: AB_1904025 |
| Anti-Rabbit 594 | Cell Signaling Technology | Cat# 8889; RRID: AB_2716249 |
| Anti-Mouse 488 | Cell Signaling Technology | Cat# 4408; RRID: AB_10694704 |
| Anti-Mouse 594 | Cell Signaling Technology | Cat# 8890; RRID: AB_2714182 |
| Anti-Chicken 647 IgY | Invitrogen | Cat# A-21449; RRID: AB_2535866 |
| HRP-linked anti-Rabbit IgG (H + L) | Beyotime | Cat# A0208; RRID: AB_2892644 |
| HRP-linked anti-Mouse IgG (H + L) | Beyotime | Cat# A0216; RRID: AB_2860575 |
| HRP-linked anti-Chicken IgG (H + L) | Abcam | Cat# ab6877; RRID: AB_955465 |
| Bacterial and virus strains | ||
| DH5α | Shanghai Sangon Biotechnology Co., Ltd | B528413 |
| pLenti-EF1-EGFP-P2A-Puro | OBIO Technology (Shanghai) Co., Ltd | N/A |
| Chemicals, peptides, and recombinant proteins | ||
| Basic fibroblast growth factor | Gibco | PHG0367 |
| Epidermal growth factor | Proteintech | HZ-1326 |
| Critical commercial assays | ||
| DMEM/F12 | Gibco | 10565018 |
| B27 supplement | Gibco | 17504044 |
| Accutase | Gibco | A1110501 |
| Lipofectamine 2000 | Invitrogen | 11668019 |
| neurobasal medium | Gibco | 21103049 |
| L-GlutaMax | Gibco | 35050079 |
| polybrene | Biosharp | Bl628A |
| 4-Thiouridine | Sigma | T4509 |
| DTT | ThermoFisher | R0862 |
| RNase inhibitor | Sangon Biotech | B600478 |
| EDTA-Free Protease Inhibitor Cocktail | Bimake | B14001 |
| TRIzol | Vazyme | R401-01 |
| RNase 1 | Thermo Scientific | EN0601 |
| Protease K | Sangon Biotech | A610451 |
| RIPA buffer | Beyotime | P0013B |
| SDS-PAGE Gel | Vazyme | E304/305 |
| BCA protein assay kit | Sangon | C503021 |
| ECL | Millipore | WBULS0500 |
| HiScript III 1st Strand cDNA Synthesis Kit | Vazyme | R312-01 |
| ChamQ Universal SYBR qRT-PCR Master Mix | Vazyme | Q711-02 |
| Actinomycin D | MedChemExpress | HY-17559 |
| EdU assays kit | EpiZyme | CX004 |
| Dual-Luciferase Reporter Assay Kit | Yeasen | 11402ES60 |
| DAPI | Ysasen | 40727ES10 |
| Deposited data | ||
| Single-cell RNA sequencing data | This paper | GEO: GSE241889 |
| CLIP-RNA sequencing data | This paper | GEO: GSE241548 |
| Original, unprocessed data | This paper | Mendeley Data: https://doi.org/10.17632/7527hjwbnv.1 |
| Experimental models: Cell lines | ||
| Human neural stem cells | Shanghai Angecon Biotechnology Co., Ltd | N/A |
| HEK293T | Cell Bank/Stem Cell Bank, Chinese Academy of Sciences (Shanghai, China) | CSTR:19375.09.3101HUMSCSP5209 |
| Experimental models: Organisms/strains | ||
| BALB/C nude mice | Experimental Animal Center of Ren Ji Hospital | N/A |
| Oligonucleotides | ||
| siRNA oligo sequence | This study, see Table S3 | N/A |
| Primers for qRT-PCR | This study, see Table S7 | N/A |
| Recombinant DNA | ||
| pMS2 | Shanghai Jiao Tong University | N/A |
| pMS2-GST | Shanghai Jiao Tong University | N/A |
| pmirGLO | Beijing Tsingke Biotechnology Co., Ltd | N/A |
| pCDH-CMV-RBM3-EF1-copGFP | Beijing Tsingke Biotechnology Co., Ltd | N/A |
| pLVX-RBM3-3Flag-Puro | Beijing Tsingke Biotechnology Co., Ltd | N/A |
| pCDH-CMV-RBM3mut-EF1-copGFP | Beijing Tsingke Biotechnology Co., Ltd | N/A |
| pLVX-RBM3mut-3Flag-Puro | Beijing Tsingke Biotechnology Co., Ltd | N/A |
| pLX304-CMV-RBM3-V5 | Shanghai Jiao Tong University | N/A |
| pCDH-CMV-SOX11-EF1-copGFP | Beijing Tsingke Biotechnology Co., Ltd | N/A |
| pLVX-SOX11-3Flag-Puro | Beijing Tsingke Biotechnology Co., Ltd | N/A |
| Software and algorithms | ||
| Prism 8 | GraphPad | https://www.graphpad.com/ |
| ImageJ | ImageJ | https://imagej.net/software/imagej/ |
| catRAPID omics v2.0 | catRAPID | http://service.tartaglialab.com/page/catrapid_omics2_group |
| Biorender | Biorender | https://biorender.com/ |
| Other | ||
| Temperature-controlled blanket | CWE Inc | TC-1000 |
| Stereotactic apparatus | RWD | N/A |
| Micro syringe pump | Harvard Apparatus | Pump 11 Elite |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Junfeng Feng (fengjfmail@163.com).
Materials availability
Plasmids generated in this study are available from the lead contact upon reasonable request. This study did not generate new unique reagents.
Date and code availability
-
•
Single-cell RNA-seq data and CLIP-RNA-seq data have been deposited at GEO and are publicly available as of the date of publication. Accession numbers are listed in the key resources table. Original images have been deposited at Mendeley Data and are publicly available as of the date of publication.
-
•
This paper does not report original code.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
Experimental model and study participant details
Male BALB/c nude mice, aged 6 to 8 weeks and weighing an average of 20–25 g, were obtained from the Experimental Animal Center of Ren Ji Hospital in Shanghai, China. These mice were housed in a controlled environment at the Experimental Animal Center of Ren Ji Hospital, ensuring specific pathogen-free conditions, a 12-h light-dark cycle, and access to food and water ad libitum. To eliminate bias, randomization and allocation concealment techniques were employed to allocate mice to different experimental groups. The surgeries and outcome assessments were conducted by investigators who were blinded to the group assignments, thereby ensuring impartiality and minimizing the potential for bias during data analysis. All experiments were approved by the Ren Ji Hospital Institutional Animal Care and Use Committee (RJ 2022-0820) and performed in accordance with the Institutional Guide for the Care and Use of Laboratory Animals.
The hNSCs utilized in this study were obtained from cortical brain tissue of aborted embryos at 6-8 weeks of gestational age using microdissection microscopes and ophthalmic forceps. To ensure cell homogeneity, all batches of cells used came from the same individual. Ethical approval was obtained for this research. First, cell lines were established and expanded. The cells were then frozen for long-term preservation. Before use, the cells were thawed and cell identification was conducted using flow cytometry. Then the cells were prepared for further experiments after identification.
Method details
Cells isolation and cultures
The hNSCs were procured from Shanghai Angecon Biotechnology Co., Ltd. The hNSCs were firstly cultured in a proliferation medium. Upon receipt from Shanghai Angecon Biotechnology Co., Ltd, the hNSCs suspension was centrifuged at 300 g for 5 min. The supernatant was discarded, and the cells were resuspended and seeded into T25 low-attached flasks with 4 mL expansion medium. The expansion medium contained DMEM/F12 (Gibco, 10565018), 2% B27 supplement (Gibco, 17504044), 20 ng/mL of basic fibroblast growth factor (bFGF) (Gibco, PHG0367), 20 ng/mL of epidermal growth factor (EGF) (Proteintech, HZ-1326), and 1% penicillin-streptomycin to promote growth. The hNSCs grew as neurospheres, and one flask was supplemented with 1 mL expansion medium every two days during hNSCs expansion. After the hNSCs had expanded for about 7–10 days and the neurosphere diameters reached approximately 100 μm, the cells were collected and passed through digestion with Accutase (Gibco, A1110501) at 37°C for 15 min. Subsequently, 1×106 cells were seeded into a new T25 low-attached flask for another generation. All hNSCs utilized in this study were within passages 3–4.
Plasmids and RNA interference
V5-RBM3 was procured from Shanghai Jiao Tong University School of Medicine. To obtain lentivirus plasmids, the complete RBM3 WT, RBM3 mutation, and SOX11 sequences were cloned into pCDH-CMV-MCS-EF1-copGFP and pLVX-MCS-3Flag-Puro vectors. The pMS2-Vector and pMS2-GST plasmids were provided by Prof. Yong Li from Shanghai Jiao Tong University School of Medicine in Shanghai, China. The RBM3 and SOX11 siRNA duplexes were synthesized by Tsingke Biotechnology in Beijing, China, and the oligo sequences were listed in Table S3. The siRNA oligos were then transfected into hNSCs cells at Day 0 and Day 7 with a final concentration of 50 nM using Lipofectamine 2000 (Invitrogen, 11668019) in accordance with the manufacturer’s protocol.
Neuronal induction and transfection of hNSCs
For the process of neuronal induction, the neurospheres were dissociated into a single-cell suspension and plated on coverslips that had been previously coated with 0.05 mg/ml poly-L-lysine at a density of 2×104 cells/cm2. Subsequently, the cells were cultured in expansion medium for 48 h, to allow for adequate recovery from passage. Once the recovery period had elapsed, the cultured medium was changed to a neural differentiation medium, which comprised of neurobasal medium (Gibco, 21103049), 2% B27 supplement, 1% penicillin/streptomycin, and 1% L-GlutaMax (Gibco, 35050079). Following 14 days of culture, the cells were collected for immunofluorescence, qRT-PCR and western blotting analysis. During the transfection of cells with siRNA and lentivirus plasmids, the digested cells were plated on coverslips coated with poly-L-lysine and cultured in expansion medium for 36 h. Then, a mixture of siRNA with Lipofectamine 2000 was added to the medium, at a final concentration of 50 nM. Additionally, the lentivirus medium was added to the culture medium, with the supplementation of 10 μg/mL polybrene (Biosharp, Bl628A). Following 8 h of culture, the medium was changed to the neural differentiation medium.
Crosslinking immunoprecipitation (CLIP)
The hNSCs were treated with 4-Thiouridine (Sigma, T4509) at a final concentration of 100 μM, 24 h prior to cells collection. Subsequently, the hNSCs cultured in a 6 cm plate for 14 days of differentiation were carefully washed twice with ice-cold PBS and subjected to cross-linkage using 4000 J of UV irradiation for three times. Following this process, the cells were gently scraped off using 1 mL PBS and centrifuged at 300 g × 5 min, after which they were re-suspended in 300 μL of complete IP cell lysis buffer (20 μM of Tris-HCl pH 7.5, 10 μM of KCl, 5 μM of MgCl2, 0.5% NP-40, 1 μM of DTT (ThermoFisher, R0862), 40 units/ml of RNase inhibitor (Sangon Biotech, B600478), and EDTA-Free Protease Inhibitor Cocktail (Bimake, B14001)). Besides, after centrifugation, 100 μL of scraped cells were added with TRIzol (Vazyme, R401-01) labeled as “Input” for RNA analysis. The cells were then incubated on ice for 15 min and centrifuged at 12000 g for 15 min at 4°C. After centrifugation, 30 μL of cell lysate supernatant was transferred to a new tube labeled as “Input” for protein analysis. The remaining supernatant was then treated with or without RNase 1 (Thermo Scientific, EN0601), at a final concentration of 1 unit/μl, for 10 min at room temperature. Subsequently, the lysate supernatant was mixed with 10 μL of antibody-coupling agarose beads and incubated overnight at 4°C with rotation. The next day, the samples were centrifuged at 3000 g for 5 min and the supernatant was carefully discarded. The samples were then washed thrice with 1 mL ice-cold PBS. After the third centrifugation, the beads were resuspended with 200 μL IP buffer, and 20 μL of the sample were taken for protein testing. Subsequently, Protease K (Sangon Biotech, A610451) was added to a concentration of 100 μg/mL and incubated at 55°C for 2 h. Finally, 1 mL TRIzol was added to one sample to extract RNA for further sequence and qRT-PCR analysis.
MS2-tagged RNA pull-down assay
The plasmid pMS2 boasts a pcDNA3 scaffold encompassing 24 repeats of MS2 sequences situated between the EcoRI and EcoRV restriction sites. The chosen SOX11 mRNA 3′UTR complementary DNA segments were conjugated to the upstream region of the MS2 repeats by employing the cloning sites HindIII and EcoRI. Subsequently, mammalian HEK-293T cells were cultivated to achieve 80% confluence within a 6 cm2 culture dish. In the next step, 4.5 μg of p (SOX11 3′UTR cDNA)-MS2 or pMS2 vacant vector, in conjunction with 1.5 μg of pMS2-GST, was transfected into the prepared HEK-293T cells. After 24 hours, the cellular medium was replaced, and the V5-RBM3 or vector plasmid was introduced into the HEK-293T cells. Following an additional 24-h incubation, the cells were harvested to conduct the RNA pull-down assay.
Transfected cells expressing MS2 and RBM3 plasmids were cultured in a 6 cm2 plate, then gently rinsed with ice-cold PBS before being dislodged in 1 mL PBS. The cell suspension was then centrifuged at 300 g for 5 min and resuspended in 500 μL of complete NP-40 cell lysis buffer (composed of 20 mM Tris-HCl pH 7.5, 100 mM KCl, 5 mM MgCl2, and 0.5% NP-40, supplemented with 1 mM DTT, 40 units/ml RNase inhibitor, and EDTA-Free Protease Inhibitor Cocktail). Following incubation on ice for 10 min, the cell lysates were subjected to centrifugation at 10000 g for 30 min at 4°C. The supernatant was collected and 50 μL of the cell lysate was separated into a new tube labeled “Input” for subsequent protein analysis. The remaining cell lysate supernatant was mixed with 10 μL of antibody-coupled agarose beads and allowed to incubate with rotation for 2 h at 4°C. The samples were then centrifuged at 2000 g for 2 min at 4°C and washed three times with NP-40 lysis buffer. Subsequently, 20 units of RNase-free DNase I (Yeasen, 10325ES80) were added to 100 μL of NP-40 lysis buffer and incubated for 15 min at 37°C. After incubation, 700 μL of NT2 buffer (composed of 50 mM Tris-HCl pH 7.5, 150 mM NaCl, 1 mM MgCl2, and 0.05% NP-40) was added and the samples were centrifuged at 2000 g for 2 min at 4°C. The supernatant was discarded, and the samples were resuspended in 1×loading buffer for further protein analysis.
Immunofluorescence
hNSCs were fixed with 4% paraformaldehyde (PFA) for 30 min at room temperature, followed by washing with PBS. The samples were permeabilized with 1% Triton X-100 (PBS) and blocked with 10% donkey serum (PBS) for 1 h. Primary antibodies were then applied and allowed to incubate overnight at 4°C. The next day, after washing with PBS, the samples were incubated with secondary antibodies conjugated with Alexa Fluor 488, 594, or 647 and 4′,6-diamidino-2-phenylindole (DAPI) diluted at 1:1000 in 10% donkey serum (PBS) for 1 h at room temperature. The samples were then visualized using a confocal fluorescence microscope or fluorescence microscope to capture high-resolution images.
Western blotting
Cultured cells were harvested and lysed in RIPA buffer (Bioytime, P0013B). The protein concentration was determined using the BCA protein assay kit (Sangon, C503021), and an equal amount of protein was loaded and separated using SDS-PAGE Bis-Tris Protein Gel (Vazyme, E304/305) with SDS running buffer. The proteins were then electrophoretically transferred to a PVDF membrane using Transfer Buffer with 20% methanol. The membranes were blocked with 5% skim milk (TBS-T) for 1 h at room temperature and incubated with primary antibodies overnight at 4°C. After washing with TBS-T three times for 5 min, secondary antibodies conjugated to horseradish peroxidase (HRP) were incubated for 1 h at room temperature. Following three additional washing steps, signal development was performed using an enhanced chemiluminescence (ECL) detection kit (Millipore, WBULS0500).
RNA extraction and real-time reverse transcription (qRT) PCR
Total RNAs from samples were extracted with TRIzol according to manufacture instruction. 1μg of total RNA was reversed to cDNA using the HiScript III 1st Strand cDNA Synthesis Kit (Vazyme, R312-01). qRT-PCR was performed using the ChamQ Universal SYBR qRT-PCR Master Mix (Vazyme, Q711-02) in a Roche LightCycler 480 II. All reactions were performed according to the manufactured protocol. Primer sequences are listed in Table S7 and were synthesized. Results were normalized to β-actin.
RNA half-life test
As delineated in recent research,32 cultured cells were treated with 5 μg/mL Actinomycin D (MedChemExpress, HY-17559) to inhibit mRNA transcription. Following the treatment, the cells were harvested at 0, 1, 2, and 4-h intervals, and total mRNA was extracted using TRIzol reagent following the manufacturer’s protocol. The isolated mRNA was then converted into cDNA via RNA reverse-transcription kit. qRT-PCR analysis was subsequently performed according to previously established procedures, and human SOX11 mRNA levels were quantified and normalized using β-actin. The mRNA expression level of SOX11 at the initial time point was defined as 1.
EdU assays
To assess hNSCs proliferation, an EdU assays kit was procured from EpiZyme Life Technologies (CX004). EdU was initially introduced into the cell culture medium at a final concentration of 10 μM. After 24 h, the hNSCs were collected for further immunofluorescence analysis. Following immunofluorescence, 500 μL of click additive buffer (composed of 430 μL click reaction buffer, 20 μL CuSO4, 1 μL 647 azide, and 50 μL click additive solution) was added to one 6-well plate. The samples were then incubated for 30 min at 37°C and subsequently washed three times with PBS containing 3% BSA. Finally, the samples were stored in PBS for subsequent fluorescence testing.
Dual luciferase reporter assays
Dual luciferase reporter assays were carried out in HEK-293T cells. The six SOX11 mRNA 3′UTR sequences were incorporated into pmirGLO reporter, and the resultant gene expression reporter plasmids were subsequently transfected into HEK-293T cells under different conditions. After 48 h, the expression of the luciferase was assessed using the Dual-Luciferase Reporter Assay Kit (Yeasen, 11402ES60).
GFP+ hNSCs preparation
In order to label transplanted hNSCs, we used lentivirus expressing EGFP to infect hNSCs to distinguish endogenous NSCs from transplanted hNSCs. Initially, 2×106 newly derived hNSCs were cultured in a T75 low-attachment flask with proliferation medium. After 3 days of incubation, lentivirus carrying the GFP vector with 10 μL/mL polybrene were introduced into one T75 flask. Following an 8-h incubation, the cells were transferred to new proliferation medium. After 4 days, the majority of the cells were identified as GFP-positive. Subsequently, GFP+hNSCs from one T75 flask were sub-cultured into two new low-attachment flasks and continued to be cultured for 7 days. At the end of the incubation period, the GFP+ hNSCs were dissociated into a single-cell suspension and diluted with PBS to a concentration of 1×105 cells/μL for further transplantation.
Transplantation of GFP+hNSCs into nude mice
For cell transplantation, mice were placed under 2–5% isoflurane anesthesia at stereotaxic instrument. Then hNSCs (1×105 cells/μL, 3 μL) were stereotaxic injected into right striatum at the speed of 0.6 μL/min into the dorsal striatum (AP+0.5, ML-1.8, DV-3.4 from bregma) with the aid of stereotactic apparatus and electrical pump to drive the syringe.
Normothermia and hypothermia treatment to mice
Nude mice that underwent hNSCs transplantation were randomly divided into two groups: normothermia group and hypothermia group. After the transplantation procedure, the mice were allowed to recover from surgery. Beginning on the second day after transplantation, the mice received normothermia or hypothermia treatment for 2 h each day over the course of 14 days. All mice were placed on a temperature-controlled blanket (CWE Inc, TC-1000) under 2–5% isoflurane anesthesia. Rectal temperature was measured using a detector. The rectal temperature of the normothermia group mice was controlled within the range of 37°C–38°C, while the rectal temperature of the hypothermia group mice was controlled within the range of 35°C–36°C, achieved by regulating the temperature of the blanket. Following the 2-h treatment period, the mice were removed from isoflurane and the temperature-controlled blanket, and allowed to recover to their normal body temperature. After 14 days treatment, mice were sacrificed with pentobarbital sodium injection and to do the further experiments.
Quantification and statistical analysis
All statistical analyses were performed using GraphPad Prism 8 software (GraphPad Software, Inc., USA). The RNA-Protein binding site was predicted by catRAPID omics v2.0 (http://service.tartaglialab.com/page/catrapid_omics2_group). All data are presented as mean ± SD. Statistical tests were employed by One- and two-way ANOVA and Student’s t tests and number of independent culture replicates or animals/groups are stated in figures and legends. Alpha levels were set as ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001 and ∗∗∗∗p < 0.0001.
To evaluate the differentiation ratio of transplanted hNSCs in vivo with different temperature treatments, colocalization coefficients related to GFP spots were calculated. Merged immunofluorescence pictures were imported into ImageJ. Then the threshold was automatically set to remove the background. Then the plugin coloc 2 was used to evaluate the Manders’ Colocalization Coefficients (MCC). For two probes, denoted as red (R) and green (G), two different MCC values are derived, M1, the fraction of R in compartments containing G and M2, the fraction of G in compartments containing R. These coefficients are simply calculated as: M1 = where Ri,colocal = Ri if Gi > 0 and Ri,colocal = 0 if Gi = 0 and M2 = where Gi,colocal = Gi if Ri > 0 and Gi,colocal = 0 if Ri = 022. In the research, M2 values were further performed statistical analysis.
Acknowledgments
This study was supported by grants from National Natural Science Foundation of China (82071358, 81771317 to J.F.; 81971825 to J.J.), Program of Shanghai Academic Research Leader (21XD1422400 to J.F.), Shanghai Municipal Education Commission-Gaofeng Clinical Medicine Support (02.101005.001.30.30A to J.F.), and Project of Shanghai Medical And Health Development Foundation (20224Z0012 to J.F.).
Author contributions
Conceptualization: J.F. and W.W.; investigation: Y.M., Z.H., J.W., and Z.M.; formal analysis: Q.Z. and J.H.; resources: X.Z.; data curation: Y.M., P.Z., Q.L., and Q.Z.; writing – original draft: Y.M. and P.Z.; writing – review & editing: W.W. and J.F.; supervision: W.W., J.J., and J.F.
Declaration of interests
The authors declare no competing interests.
Published: March 6, 2024
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2024.109435.
Contributor Information
Weiji Weng, Email: zigzagweng@163.com.
Jiyao Jiang, Email: jiyaojiang@126.com.
Junfeng Feng, Email: fengjfmail@163.com.
Supplemental information
References
- 1.Mu L., Berti L., Masserdotti G., Covic M., Michaelidis T.M., Doberauer K., Merz K., Rehfeld F., Haslinger A., Wegner M., et al. SoxC transcription factors are required for neuronal differentiation in adult hippocampal neurogenesis. J. Neurosci. 2012;32:3067–3080. doi: 10.1523/jneurosci.4679-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Abu-El-Rub E., Khasawneh R.R., Almahasneh F.A., Aloud B.M., Zegallai H.M. The Molecular and Functional Changes of Neural Stem Cells in Alzheimer's Disease: Can They be Reinvigorated to Conduct Neurogenesis. Curr. Stem Cell Res. Ther. 2023;18:580–594. doi: 10.2174/1574888x17666220831105257. [DOI] [PubMed] [Google Scholar]
- 3.Zhang Z., Zheng X., Liu Y., Luan Y., Wang L., Zhao L., Zhang J., Tian Y., Lu H., Chen X., Liu Y. Activation of metabotropic glutamate receptor 4 regulates proliferation and neural differentiation in neural stem/progenitor cells of the rat subventricular zone and increases phosphatase and tensin homolog protein expression. J. Neurochem. 2021;156:465–480. doi: 10.1111/jnc.14984. [DOI] [PubMed] [Google Scholar]
- 4.Carr M.J., Toma J.S., Johnston A.P.W., Steadman P.E., Yuzwa S.A., Mahmud N., Frankland P.W., Kaplan D.R., Miller F.D. Mesenchymal Precursor Cells in Adult Nerves Contribute to Mammalian Tissue Repair and Regeneration. Cell Stem Cell. 2019;24:240–256.e9. doi: 10.1016/j.stem.2018.10.024. [DOI] [PubMed] [Google Scholar]
- 5.Gantner C.W., de Luzy I.R., Kauhausen J.A., Moriarty N., Niclis J.C., Bye C.R., Penna V., Hunt C.P.J., Ermine C.M., Pouton C.W., et al. Viral Delivery of GDNF Promotes Functional Integration of Human Stem Cell Grafts in Parkinson's Disease. Cell Stem Cell. 2020;26:511–526.e5. doi: 10.1016/j.stem.2020.01.010. [DOI] [PubMed] [Google Scholar]
- 6.Chen Y., Xiong M., Dong Y., Haberman A., Cao J., Liu H., Zhou W., Zhang S.C. Chemical Control of Grafted Human PSC-Derived Neurons in a Mouse Model of Parkinson's Disease. Cell Stem Cell. 2016;18:817–826. doi: 10.1016/j.stem.2016.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hu Z., Gajavelli S., Spurlock M.S., Mahavadi A., Quesada L.S., Gajavelli G.R., Andreoni C.B., Di L., Janecki J., Lee S.W., et al. Human neural stem cell transplant location-dependent neuroprotection and motor deficit amelioration in rats with penetrating traumatic brain injury. J. Trauma Acute Care Surg. 2020;88:477–485. doi: 10.1097/ta.0000000000002510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Badner A., Reinhardt E.K., Nguyen T.V., Midani N., Marshall A.T., Lepe C.A., Echeverria K., Lepe J.J., Torrecampo V., Bertan S.H., et al. Freshly Thawed Cryobanked Human Neural Stem Cells Engraft within Endogenous Neurogenic Niches and Restore Cognitive Function after Chronic Traumatic Brain Injury. J. Neurotrauma. 2021;38:2731–2746. doi: 10.1089/neu.2021.0045. [DOI] [PubMed] [Google Scholar]
- 9.Lei X., Hu Q., Ge H., Zhang X., Ru X., Chen Y., Hu R., Feng H., Deng J., Huang Y., Li W. A redox-reactive delivery system via neural stem cell nanoencapsulation enhances white matter regeneration in intracerebral hemorrhage mice. Bioeng Transl Med. 2023;8:e10451. doi: 10.1002/btm2.10451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Upadhya R., Madhu L.N., Attaluri S., Gitaí D.L.G., Pinson M.R., Kodali M., Shetty G., Zanirati G., Kumar S., Shuai B., et al. Extracellular vesicles from human iPSC-derived neural stem cells: miRNA and protein signatures, and anti-inflammatory and neurogenic properties. J. Extracell. Vesicles. 2020;9:1809064. doi: 10.1080/20013078.2020.1809064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yan Y., Jin B., Zhang X., Li H., Zhou W., Zhang D. Effects of neural stem cells in combination with nerve neurotrophic factor 3 gene modified olfactory ensheathing cell co-transplants on functional recovery after traumatic brain injury in the adult rats. Chin J Exp Surg. 2020;37:71–73. doi: 10.3760/cma.j.issn.1001-9030.2020.01.021. [DOI] [Google Scholar]
- 12.Conti L., Cattaneo E. Neural stem cell systems: physiological players or in vitro entities? Nat. Rev. Neurosci. 2010;11:176–187. doi: 10.1038/nrn2761. [DOI] [PubMed] [Google Scholar]
- 13.Chen G., Li X., Zhu H., Wu H., He D., Shi L., Wei F., Liu X., Chen N., Liu S. Transplanting neurofibromatosis-1 gene knockout neural stem cells improve functional recovery in rats with spinal cord injury by enhancing the mTORC2 pathway. Exp. Mol. Med. 2022;54:1766–1777. doi: 10.1038/s12276-022-00850-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jiang J., Hai J., Liu W., Luo Y., Chen K., Xin Y., Pan J., Hu Y., Gao Q., Xiao F., Luo H. Gallic Acid Induces Neural Stem Cell Differentiation into Neurons and Proliferation through the MAPK/ERK Pathway. J. Agric. Food Chem. 2021;69:12456–12464. doi: 10.1021/acs.jafc.1c04011. [DOI] [PubMed] [Google Scholar]
- 15.Cardano M., Diaferia G.R., Conti L., Baronchelli S., Sessa A., Broccoli V., Barbieri A., De Blasio P., Biunno I. mSEL-1L deficiency affects vasculogenesis and neural stem cell lineage commitment. J. Cell. Physiol. 2018;233:3152–3163. doi: 10.1002/jcp.26153. [DOI] [PubMed] [Google Scholar]
- 16.Flores J.V., Cordero-Espinoza L., Oeztuerk-Winder F., Andersson-Rolf A., Selmi T., Blanco S., Tailor J., Dietmann S., Frye M. Cytosine-5 RNA Methylation Regulates Neural Stem Cell Differentiation and Motility. Stem Cell Rep. 2017;8:112–124. doi: 10.1016/j.stemcr.2016.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bernstock J.D., Peruzzotti-Jametti L., Leonardi T., Vicario N., Ye D., Lee Y.J., Maric D., Johnson K.R., Mou Y., Van Den Bosch A., et al. SUMOylation promotes survival and integration of neural stem cell grafts in ischemic stroke. EBioMedicine. 2019;42:214–224. doi: 10.1016/j.ebiom.2019.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hui J., Feng J., Tu Y., Zhang W., Zhong C., Liu M., Wang Y., Long L., Chen L., Liu J., et al. Safety and efficacy of long-term mild hypothermia for severe traumatic brain injury with refractory intracranial hypertension (LTH-1): A multicenter randomized controlled trial. EClinicalMedicine. 2021;32:100732. doi: 10.1016/j.eclinm.2021.100732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tisherman S.A., Rodriguez A., Safar P. Therapeutic hypothermia in traumatology. Surg. Clin. 1999;79:1269–1289. doi: 10.1016/s0039-6109(05)70077-3. [DOI] [PubMed] [Google Scholar]
- 20.Adelson P.D., Wisniewski S.R., Beca J., Brown S.D., Bell M., Muizelaar J.P., Okada P., Beers S.R., Balasubramani G.K., Hirtz D., Paediatric Traumatic Brain Injury Consortium Comparison of hypothermia and normothermia after severe traumatic brain injury in children (Cool Kids): a phase 3, randomised controlled trial. Lancet. Neurol. 2013;12:546–553. doi: 10.1016/s1474-4422(13)70077-2. [DOI] [PubMed] [Google Scholar]
- 21.Inoue A., Hifumi T., Kuroda Y., Nishimoto N., Kawakita K., Yamashita S., Oda Y., Dohi K., Kobata H., Suehiro E., Maekawa T., Brain Hypothermia B-HYPO Study Group in Japan Mild decrease in heart rate during early phase of targeted temperature management following tachycardia on admission is associated with unfavorable neurological outcomes after severe traumatic brain injury: a post hoc analysis of a multicenter randomized controlled trial. Crit. Care. 2018;22:352. doi: 10.1186/s13054-018-2276-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wu X., Tao Y., Marsons L., Dee P., Yu D., Guan Y., Zhou X. The effectiveness of early prophylactic hypothermia in adult patients with traumatic brain injury: A systematic review and meta-analysis. Aust. Crit. Care. 2021;34:83–91. doi: 10.1016/j.aucc.2020.05.005. [DOI] [PubMed] [Google Scholar]
- 23.Madden L.K., Hill M., May T.L., Human T., Guanci M.M., Jacobi J., Moreda M.V., Badjatia N. The Implementation of Targeted Temperature Management: An Evidence-Based Guideline from the Neurocritical Care Society. Neurocrit Care. 2017;27:468–487. doi: 10.1007/s12028-017-0469-5. [DOI] [PubMed] [Google Scholar]
- 24.Quine E.J., Murray L., Trapani T., Cooper D.J. Thromboelastography to Assess Coagulopathy in Traumatic Brain Injury Patients Undergoing Therapeutic Hypothermia. Ther. Hypothermia Temp. Manag. 2021;11:53–57. doi: 10.1089/ther.2020.0025. [DOI] [PubMed] [Google Scholar]
- 25.Chandrasekaran P.N., Dezfulian C., Polderman K.H. What is the right temperature to cool post-cardiac arrest patients? Crit. Care. 2015;19:406. doi: 10.1186/s13054-015-1134-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang F., Dong H., Lv T., Jin K., Jin Y., Zhang X., Jiang J. Moderate hypothermia inhibits microglial activation after traumatic brain injury by modulating autophagy/apoptosis and the MyD88-dependent TLR4 signaling pathway. J. Neuroinflammation. 2018;15:273. doi: 10.1186/s12974-018-1315-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liu T., Zhao D.X., Cui H., Chen L., Bao Y.H., Wang Y., Jiang J.Y. Therapeutic hypothermia attenuates tissue damage and cytokine expression after traumatic brain injury by inhibiting necroptosis in the rat. Sci. Rep. 2016;6:24547. doi: 10.1038/srep24547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jiang J.Y., Liang Y.M., Luo Q.Z., Zhu C. Effect of mild hypothermia on brain dialysate lactate after fluid percussion brain injury in rodents. Neurosurgery. 2004;54:713–717. doi: 10.1227/01.neu.0000109535.58429.49. discussion 717-718. [DOI] [PubMed] [Google Scholar]
- 29.Wu T.C., Grotta J.C. Hypothermia for acute ischaemic stroke. Lancet. Neurol. 2013;12:275–284. doi: 10.1016/s1474-4422(13)70013-9. [DOI] [PubMed] [Google Scholar]
- 30.Chen C., Ma T.Z., Wang L.N., Wang J.J., Tu Y., Zhao M.L., Zhang S., Sun H.T., Li X.H. Mild hypothermia facilitates the long-term survival of newborn cells in the dentate gyrus after traumatic brain injury by diminishing a pro-apoptotic microenvironment. Neuroscience. 2016;335:114–121. doi: 10.1016/j.neuroscience.2016.08.038. [DOI] [PubMed] [Google Scholar]
- 31.Wang L., Jiang F., Li Q., He X., Ma J. Mild hypothermia combined with neural stem cell transplantation for hypoxic-ischemic encephalopathy: neuroprotective effects of combined therapy. Neural. Regen. Res. 2014;9:1745–1752. doi: 10.4103/1673-5374.143417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Xia W., Su L., Jiao J. Cold-induced protein RBM3 orchestrates neurogenesis via modulating Yap mRNA stability in cold stress. J. Cell Biol. 2018;217:3464–3479. doi: 10.1083/jcb.201801143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liu X., Ren W., Jiang Z., Su Z., Ma X., Li Y., Jiang R., Zhang J., Yang X. Hypothermia inhibits the proliferation of bone marrow-derived mesenchymal stem cells and increases tolerance to hypoxia by enhancing SUMOylation. Int. J. Mol. Med. 2017;40:1631–1638. doi: 10.3892/ijmm.2017.3167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Saito K., Fukuda N., Matsumoto T., Iribe Y., Tsunemi A., Kazama T., Yoshida-Noro C., Hayashi N. Moderate low temperature preserves the stemness of neural stem cells and suppresses apoptosis of the cells via activation of the cold-inducible RNA binding protein. Brain Res. 2010;1358:20–29. doi: 10.1016/j.brainres.2010.08.048. [DOI] [PubMed] [Google Scholar]
- 35.Eze U.C., Bhaduri A., Haeussler M., Nowakowski T.J., Kriegstein A.R. Single-cell atlas of early human brain development highlights heterogeneity of human neuroepithelial cells and early radial glia. Nat. Neurosci. 2021;24:584–594. doi: 10.1038/s41593-020-00794-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Akula S.K., Exposito-Alonso D., Walsh C.A. Shaping the brain: The emergence of cortical structure and folding. Dev. Cell. 2023;58:2836–2849. doi: 10.1016/j.devcel.2023.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhu X., Bührer C., Wellmann S. Cold-inducible proteins CIRP and RBM3, a unique couple with activities far beyond the cold. Cell. Mol. Life Sci. 2016;73:3839–3859. doi: 10.1007/s00018-016-2253-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Peretti D., Smith H.L., Verity N., Humoud I., de Weerd L., Swinden D.P., Hayes J., Mallucci G.R. TrkB signaling regulates the cold-shock protein RBM3-mediated neuroprotection. Life Sci. Alliance. 2021;4:e202000884. doi: 10.26508/lsa.202000884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liu Y., Hu W., Murakawa Y., Yin J., Wang G., Landthaler M., Yan J. Cold-induced RNA-binding proteins regulate circadian gene expression by controlling alternative polyadenylation. Sci. Rep. 2013;3:2054. doi: 10.1038/srep02054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Staykov D., Wagner I., Volbers B., Doerfler A., Schwab S., Kollmar R. Mild prolonged hypothermia for large intracerebral hemorrhage. Neurocrit Care. 2013;18:178–183. doi: 10.1007/s12028-012-9762-5. [DOI] [PubMed] [Google Scholar]
- 41.Volbers B., Herrmann S., Willfarth W., Lücking H., Kloska S.P., Doerfler A., Huttner H.B., Kuramatsu J.B., Schwab S., Staykov D. Impact of Hypothermia Initiation and Duration on Perihemorrhagic Edema Evolution After Intracerebral Hemorrhage. Stroke. 2016;47:2249–2255. doi: 10.1161/strokeaha.116.013486. [DOI] [PubMed] [Google Scholar]
- 42.Su X., Zheng K., Ma Q., Huang J., He X., Chen G., Wang W., Su F., Tang H., Wu H., Tong S. Effect of local mild hypothermia on regional cerebral blood flow in patients with acute intracerebral hemorrhage assessed by 99mTc-ECD SPECT imaging. J. Xray. Sci. Technol. 2015;23:101–109. doi: 10.3233/xst-140473. [DOI] [PubMed] [Google Scholar]
- 43.Zhao J., Mao Q., Qian Z., Zhu J., Qu Z., Wang C. Effect of mild hypothermia on expression of inflammatory factors in surrounding tissue after minimally invasive hematoma evacuation in the treatment of hypertensive intracerebral hemorrhage. Exp. Ther. Med. 2018;15:4906–4910. doi: 10.3892/etm.2018.6014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wohl S.G., Reh T.A. miR-124-9-9∗ potentiates Ascl1-induced reprogramming of cultured Müller glia. Glia. 2016;64:743–762. doi: 10.1002/glia.22958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hong X., Jian Y., Ding S., Zhou J., Zheng X., Zhang H., Zhou B., Zhuang C., Wan J., Tong X. Kir4.1 channel activation in NG2 glia contributes to remyelination in ischemic stroke. EBioMedicine. 2023;87:104406. doi: 10.1016/j.ebiom.2022.104406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bergmann O., Frisén J. Neuroscience. Why adults need new brain cells. Science. 2013;340:695–696. doi: 10.1126/science.1237976. [DOI] [PubMed] [Google Scholar]
- 47.Schneider J., Weigel J., Wittmann M.T., Svehla P., Ehrt S., Zheng F., Elmzzahi T., Karpf J., Paniagua-Herranz L., Basak O., et al. Astrogenesis in the murine dentate gyrus is a life-long and dynamic process. EMBO J. 2022;41:e110409. doi: 10.15252/embj.2021110409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhang Q., Wang Y.Z., Zhang W., Chen X., Wang J., Chen J., Luo W. Involvement of Cold Inducible RNA-Binding Protein in Severe Hypoxia-Induced Growth Arrest of Neural Stem Cells In Vitro. Mol. Neurobiol. 2017;54:2143–2153. doi: 10.1007/s12035-016-9761-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhu X., Yan J., Bregere C., Zelmer A., Goerne T., Kapfhammer J.P., Guzman R., Wellmann S. RBM3 promotes neurogenesis in a niche-dependent manner via IMP2-IGF2 signaling pathway after hypoxic-ischemic brain injury. Nat. Commun. 2019;10:3983. doi: 10.1038/s41467-019-11870-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ling K.H., Hewitt C.A., Beissbarth T., Hyde L., Banerjee K., Cheah P.S., Cannon P.Z., Hahn C.N., Thomas P.Q., Smyth G.K., et al. Molecular networks involved in mouse cerebral corticogenesis and spatio-temporal regulation of Sox4 and Sox11 novel antisense transcripts revealed by transcriptome profiling. Genome Biol. 2009;10:R104. doi: 10.1186/gb-2009-10-10-r104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Song K., Lin Z., Cao L., Lu B., Chen Y., Zhang S., Lu J., Xu H. Sox11b regulates the migration and fate determination of Müller glia-derived progenitors during retina regeneration in zebrafish. Neural. Regen. Res. 2023;18:445–450. doi: 10.4103/1673-5374.346550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chiang S.Y., Wu H.C., Lin S.Y., Chen H.Y., Wang C.F., Yeh N.H., Shih J.H., Huang Y.S., Kuo H.C., Chou S.J., Chen R.H. Usp11 controls cortical neurogenesis and neuronal migration through Sox11 stabilization. Sci. Adv. 2021;7:eabc6093. doi: 10.1126/sciadv.abc6093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lin J.Q., Khuperkar D., Pavlou S., Makarchuk S., Patikas N., Lee F.C., Zbiegly J.M., Kang J., Field S.F., Bailey D.M., et al. HNRNPH1 regulates the neuroprotective cold-shock protein RBM3 expression through poison exon exclusion. EMBO J. 2023;42:e113168. doi: 10.15252/embj.2022113168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Taverna E., Götz M., Huttner W.B. The cell biology of neurogenesis: toward an understanding of the development and evolution of the neocortex. Annu. Rev. Cell Dev. Biol. 2014;30:465–502. doi: 10.1146/annurev-cellbio-101011-155801. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.