Summary
Background
An intronic GAA repeat expansion in FGF14 was recently identified as a cause of GAA-FGF14 ataxia. We aimed to characterise the frequency and phenotypic profile of GAA-FGF14 ataxia in a large Chinese ataxia cohort.
Methods
A total of 1216 patients that included 399 typical late-onset cerebellar ataxia (LOCA), 290 early-onset cerebellar ataxia (EOCA), and 527 multiple system atrophy with predominant cerebellar ataxia (MSA-c) were enrolled. Long-range and repeat-primed PCR were performed to screen for GAA expansions in FGF14. Targeted long-read and whole-genome sequencing were performed to determine repeat size and sequence configuration. A multi-modal study including clinical assessment, MRI, and neurofilament light chain was conducted for disease assessment.
Findings
17 GAA-FGF14 positive patients with a (GAA)≥250 expansion (12 patients with a GAA-pure expansion, five patients with a (GAA)≥250-[(GAA)n (GCA)m]z expansion) and two possible patients with biallelic (GAA)202/222 alleles were identified. The clinical phenotypes of the 19 positive and possible positive cases covered LOCA phenotype, EOCA phenotype and MSA-c phenotype. Five of six patients with EOCA phenotype were found to have another genetic disorder. The NfL levels of patients with EOCA and MSA-c phenotypes were significantly higher than patients with LOCA phenotype and age-matched controls (p < 0.001). NfL levels of pre-ataxic GAA-FGF14 positive individuals were lower than pre-ataxic SCA3 (p < 0.001) and similar to controls.
Interpretation
The frequency of GAA-FGF14 expansion in a large Chinese LOCA cohort was low (1.3%). Biallelic (GAA)202/222 alleles and co-occurrence with other acquired or hereditary diseases may contribute to phenotypic variation and different progression.
Funding
This study was funded by the National Key R&D Program of China (2021YFA0805200 to H.J.), the National Natural Science Foundation of China (81974176 and 82171254 to H.J.; 82371272 to Z.C.; 82301628 to L.W.; 82301438 to Z.L.; 82201411 to L.H.), the Innovation Research Group Project of Natural Science Foundation of Hunan Province (2020JJ1008 to H.J.), the Key Research and Development Program of Hunan Province (2020SK2064 to H.J.), the Innovative Research and Development Program of Development and Reform Commission of Hunan Province to H.J., the Natural Science Foundation of Hunan Province (2024JJ3050 to H.J.; 2022JJ20094 and 2021JJ40974 to Z.C.; 2022JJ40783 to L.H.; 2022JJ40703 to Z.L.), the Project Program of National Clinical Research Center for Geriatric Disorders (Xiangya Hospital, 2020LNJJ12 to H.J.), the Central South University Research Programme of Advanced Interdisciplinary Study (2023QYJC010 to H.J.) and the Science and Technology Innovation Program of Hunan Province (2022RC1027 to Z.C.). D.P. holds a Fellowship award from the Canadian Institutes of Health Research (CIHR).
Keywords: GAA-FGF14 ataxia, GAA-impure expansion, Co-occurrence, Long-read sequencing, Biallelic (GAA)n allele
Research in context.
Evidence before this study
An intronic GAA repeat expansion ≥250 repeat units in the FGF14 gene was recently shown to cause GAA-FGF14 ataxia. The genetic and phenotypic spectrum of GAA-FGF14 ataxia needs to be fully delineated. We searched PubMed up to Oct 23, 2023, for the following terms without language restriction: “GAA-FGF14 ataxia” or “spinocerebellar ataxia 27B” or “late-onset ataxia” or “adult-onset cerebellar ataxia”. Currently, GAA-FGF14 ataxia was identified as one of the most common causes of late-onset cerebellar ataxia (LOCA) in non-Asian ataxia population, and shared an overlap with cerebellar ataxia, neuropathy, and vestibular areflexia syndrome (CANVAS). This search yielded no report on genetic and phenotypic spectrum of GAA-FGF14 ataxia in Chinese cohort.
Added value of this study
We screened the FGF14 GAA expansion in a large Chinese ataxia cohort of 1216 patients that included 399 typical late-onset cerebellar ataxia (LOCA), 290 early-onset cerebellar ataxia (EOCA), and 527 multiple system atrophy with predominant cerebellar ataxia (MSA-c). The frequency of GAA-FGF14 expansion in a large Chinese LOCA cohort was lower than in non-Asian ataxia populations. The findings included: 1) expansion could arise from alleles of 200–249 repeat units during intergenerational transmission; 2) biallelic (GAA)202/222 alleles may also be associated with disease; 3) GAA-FGF14 expansion co-occurrent with other acquired or hereditary diseases may contribute to complicated clinical spectrum and progression of GAA-FGF14 positive cases with EOCA and MSA-c phenotypes.
Implications of all the available evidence
Our study provides a comprehensive genetic assessments of a large Chinese ataxia cohort and expands the understanding of clinical heterogeneity of GAA-FGF14 ataxia. Our finding highlights further genetic assessment is warranted to explore unsolved causes responsible for disease phenotype when the phenotype is not compatible with typical GAA-FGF14 ataxia. GAA-FGF14 expansion might be a modulator of disease onset in other ataxias.
Introduction
Hereditary ataxias (HA) are a group of neurodegenerative disorders characterised by a progressive cerebellar syndrome with high clinical and genetic heterogeneity.1,2 Short tandem repeats (STRs) in coding or non-coding regions of causative genes are a major cause of HA, such as CAG expansions for polyglutamine spinocerebellar ataxia (SCA), GAA expansion for Friedreich ataxia (FRDA), and AAGGG expansion for cerebellar ataxia, neuropathy, and vestibular areflexia syndrome (CANVAS).2,3 As noted by functional genomics analysis from 100,000 Genomes Project, childhood- and adult-onset hereditary ataxia may share a common clinical spectrum rather than being distinct entities and the diagnostic rate may increase by removing the disease onset partition and modified STRs screening strategy,4 suggesting that the disease heterogeneity of repeat expansion ataxia may be underestimated.
Recently, an autosomal dominant (GAA)n repeat expansions in intron 1 of the fibroblast growth factor 14 (FGF14) gene was found to cause GAA-FGF14 ataxia, one of the most common causes of late-onset cerebellar ataxia (LOCA).5, 6, 7, 8, 9 A recent natural history study showed that GAA-FGF14 ataxia typically present as a late-onset slowly progressive pancerebellar syndrome, with frequent episodic features and cerebellar oculomotor signs.7 Another study has also observed that GAA-FGF14 ataxia may be a cause of cerebellar ataxia with polyneuropathy and/or bilateral vestibulopathy, suggesting an overlap between GAA-FGF14 ataxia and RFC1-related CANVAS.10 Considering the heterogeneity of repeat expansion ataxia, whether there are other clinical symptoms and a broader phenotypic spectrum in carriers of an FGF14 GAA expansion and how frequent the GAA-FGF14 ataxia is in an Asian ataxia cohort require further investigation.
In this study, we screened the FGF14 GAA expansion in a Chinese ataxia cohort of 1216 patients, including patients with typical LOCA, early-onset cerebellar ataxia (EOCA) and multiple system atrophy with predominant cerebellar ataxia (MSA-c), to gain insight of the genotype and phenotypic landscape of GAA-FGF14 ataxia in China.
Methods
Participants
A total of 1216 index cases, including 399 patients with typical LOCA (age onset >30 years old), 290 patients with EOCA (age onset ≤30 years old) and 527 patients with MSA-c were enrolled from 2013 to 2022 in the Department of Neurology, Xiangya Hospital, Central South University. To be included in this study, patients with typical LOCA and EOCA needed to have progressive cerebellar ataxia and exclusion of acquired etiologies (such as toxic causes, acute injury, stroke, infection, immune and neoplasm).11 In typical LOCA cohort, 228 probands with autosomal dominant cerebellar ataxia (ADCA) and 171 patients with sporadic ataxia were included, while 162 probands with ADCA and 128 patients with sporadic ataxia were enrolled in EOCA cohort. The patients with MSA-c were diagnosed based on the second consensus statement of diagnostic criteria for MSA.12 The common genetic causes for ataxia including (CAG)n/(GAA)n expansion for SCA1, 2, 3, 6, 7 and FRDA were excluded in all patients with typical LOCA, MSA-c, and most patients with EOCA, except 63 probands with ADCA and 30 patients with sporadic ataxia in EOCA cohort without (CAG)n/(GAA)n expansion screen.
Ethics
The study was approved by the Ethics Committee of Xiangya Hospital, Central South University in China (Reference number: 202310206) and all participants gave written informed consent in accordance with the Declaration of Helsinki.
Clinical assessments
All patients underwent a thorough neurological examination performed by at least two experienced neurologists. The overall disease severity was measured with the Scale for Assessment and Rating of Ataxia (SARA) for patients with familial and sporadic ataxia,13 and with the Unified Multiple System Atrophy Rating Scale (UMSARS) for patients with MSA-c.14 The Spinocerebellar Degeneration Functional Score (SDFS) scale was used to evaluate the functional disability of all patients.15 Brain magnetic resonance imaging (MRI) data were collected on patients with GAA expansion when available. For patients carrying a GAA expansion in FGF14, plasma neurofilament light (pNfL) levels were measured with the single molecule array (Simoa) technique on the automated Simoa HD-X platform (Quanterix, MA), as described previously.16,17 These positive cases were classified as pre-ataxic stage with SARA < 3 and ataxic stage with SARA ≥ 3.18,19 In addition, 30 patients with SCA3 (including 15 pre-ataxic and 15 ataxic), 15 GAA-FGF14-negative patients with MSA-c, and 15 age-matched healthy individuals were enrolled as controls for pNfL testing.
Long-range PCR and repeat-primed PCR
The genomic DNA was extracted by a standard phenol–chloroform method. The screens for FGF14 repeat locus were performed as described previously.20 Briefly, long-range PCR was used to amplify the intronic FGF14 repeat locus. The repeat sizes were determined by capillary electrophoresis and agarose-gel electrophoresis of fluorescent long-range PCR amplification products. Next, bidirectional repeat-primed PCRs (RP-PCR) were performed and a sawtooth pattern in RP-PCR electropherograms generally suggested the presence of a GAA repeat expansion. Expansions of ≥250 GAA repeat units were considered pathogenic (alleles of 250–300 units are incompletely/variably penetrant whereas larger alleles are considered fully penetrant).5,6
Targeted long-read sequencing
Targeted long-read sequencing (LRS) was conducted on positive candidates with sufficient DNA available to measure the repeat size precisely and to detect interruptions within the expansions.21 Other tandem repeats related to neurological diseases were also included and the list of targeted genes is provided in Supplementary Table S1. Sequencing libraries were constructed using SMRTbell Express Template Kit 2.0 (Pacific Biosciences) according to the manufacturer's protocol. The libraries were then loaded onto a PacBio sequencing flow cell and ran on the PacBio Sequel IIe sequencing platform. Subsequently, the PacBio subreads were transformed into HiFi reads using circular consensus sequencing (CCS) with default parameters.22 Following sequencing, the aligned HiFi reads were mapped to the human genome 19 (hg19) using minimap2 (version 2.24).23 RepeatHMM was run at the known pathogenic repeats from STRipy database.24 Expansions were further visualised using IGV (version 2.16.2).
Whole-genome sequencing
Whole-genome sequencing (WGS) was performed on positive patients for the detection of other causal variants. Sequencing libraries were prepared according to the DNBSEQ-T7 PCR-free library construction protocol. The circularization was performed to produce single-stranded cyclised products which were then replicated through rolling cycle amplification (RCA) to produce DNA nanoballs (DNBs). Qualified DNBs were then loaded into patterned nanoarrays and sequenced through DNBSEQ-T7 platform. The WGS data were aligned to the reference genome hg38 by the BWA software (version 0.7.17) and the BAM products were sorted through the Samtools software (version 1.16). The GATK HaplotypeCaller (version 4.0.4.0) was used for the variants calling and the SnpEff (version 4.3) was used for variants annotation. The allele frequencies for each variant in our dataset and publicly available databases, including dbSNP, 1KGP, gnomAD and TOPmed were calculated. Variants with an allele frequency greater than 1% were then excluded. The ClinVar and HGMD (Human Gene Mutation Database) were utilised for the pathogenic classification of variants.25
Statistics
All statistical analyses were performed with SPSS software (version 25.0). Quantitative variables were presented as median (range) or mean ± SD and categorical variables were shown as absolute numbers (percentages). Analysis of covariance (ANCOVA) and Wilcoxon rank sum test were used for comparisons of quantitative variables, and Pearson's chi-square test was utilised for comparisons of categorical variables between groups. Pearson correlation was conducted between NfL levels and age in normal distribution. Spearman correlation was conducted between GAA repeat sizes and age at onset in non-normal distribution. A value of p < 0.05 was considered statistically significant. Sex/gender was self-reported by study participants for the analysis of intergenerational instability and parental transmission.
Role of funders
This study was funded by the National Key R&D Program of China, the National Natural Science Foundation of China, the Innovation Research Group Project of Natural Science Foundation of Hunan Province, the Key Research and Development Program of Hunan Province, the Innovative Research and Development Program of Development and Reform Commission of Hunan Province, the Natural Science Foundation of Hunan Province, the Project Program of National Clinical Research Center for Geriatric Disorders, the Central South University Research Programme of Advanced Interdisciplinary Study and the Science and Technology Innovation Program of Hunan Province, and the Canadian Institutes of Health Research (CIHR). The funders had no role in study design, data collection, data analysis, data interpretation, or writing of the report.
Results
Distribution of GAA-FGF14 expansion
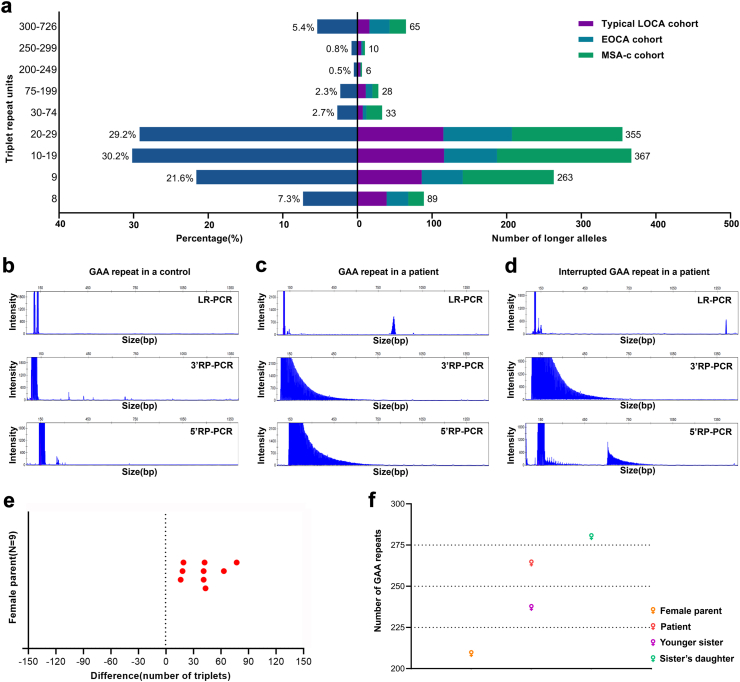
In total, 1216 index cases, including 399 patients from the typical LOCA cohort, 290 patients from the EOCA cohort, and 527 patients from the MSA-c cohort, were screened for the FGF14 GAA expansion by long-range PCR and RP-PCR (Fig. 1). Of the 1216 patients, 93.8% (n = 1141) carried an allele between 8 and 234 GAA repeat units (Fig. 2a and b). Interestingly, two siblings in family 1 from the typical LOCA cohort (P-1 and P-2) were found to be compound heterozygous for pure (GAA)202 and (GAA)222 alleles by target LRS, both presenting a slowly progressive cerebellar ataxia with gait ataxia, upper limb ataxia and dysarthria at age 40. Co-segregation analysis in other family members revealed that (GAA)202 and (GAA)222 alleles might not be pathogenic, but compound heterozygosity for (GAA)202/222 alleles may be associated with typical GAA-FGF14 ataxia (Fig. 5a, Supplementary Figure S1, Supplementary Table S2).
Fig. 1.
Workflow of GAA-FGF14 expansion screen in typical LOCA, EOCA, and MSA-c cohorts. ADCA, autosomal dominant cerebellar ataxia; EOCA, early-onset cerebellar ataxia; LOCA, late-onset cerebellar ataxia; MSA-c, multiple system atrophy with predominant cerebellar ataxia.
Fig. 2.
Genetic distribution, long-range PCR, repeat-primed PCR, and genetic instability of the GAA-FGF14 expansion in the cohort. (a) The distribution in terms of percentage and quantity of longer triplet repeat units in FGF14 gene in typical LOCA, EOCA, and MSA-c cohorts. Long-range PCR and repeat-primed PCR results for control with normal GAA repeats (b), patient with (GAA)≥250 expansion (c) and patient with (GAA)≥250 expansion with interruption (d). (e) The repeated length variation of (GAA)≥250 expansion transmission during nine different maternal meiotic events. (f) The transmission of GAA-FGF14 expansion across two generations in a family.
Fig. 5.
Pedigree chart and co-occurrence screening of GAA-FGF14 positive cases. (a) 13 pedigrees of positive GAA-FGF14 positive cases. The content in parentheses represented the age of onset and the age at latest visit in hospital, respectively. Patients from Family 4 and Family 5 were identified to have (GAA)≥250-[(GAA)n (GCA) m]z expansions in the FGF14 gene. (b)In family 2, the proband (P-3) was identified to have a heterozygous variant of c.1801G > A in COL1A2 gene by Sanger sequencing. (c)In family 3, a co-segregation analysis by Sanger sequencing revealed that both parents had a heterozygous variant of c.544A > G in NEU1 gene. (d) In family 4, the proband (P-6) was identified with (CAG)78 expansions in the ATXN3 gene. (e) In family 5, both the proband (P-8) and her younger sister (P-9) were identified to have (CAG)50 expansions in the ATXN1 gene.
Among the 1216 index cases, the remaining 75 probands were found to carry an FGF14 GAA expansion of at least 250 triplet repeats (21 cases with typical LOCA, 29 cases with EOCA and 25 cases with MSA-c) (Fig.2a, c and d). Among them, four patients with typical LOCA (P-12, P-14, P-15 and P-16), three patients with EOCA (P-3, P-4 and P-11) and three patients with MSA-c (P-17, P-18 and P-19) carried an uninterrupted GAA-pure expansions ranging from 250 to 302 repeats were identified. Among the other 65 patients carrying an interrupted non-GAA-pure expansion, 56 underwent subsequent target LRS and nine were excluded due to insufficient DNA quantity or quality. The target LRS results showed that 14 patients with typical LOCA, 21 patients with EOCA and 21 patients with MSA-c carried a [(GAA)n (GCA)m]z expansion, with the total trinucleotide repeats ranging from 296 to 726 repeats and GCA repeats ranging from 41 to 273 repeats (Supplementary Figure S1). Among them, two probands with EOCA (P-6 and P-8) carried a (GAA)≥250-[(GAA)n (GCA)m]z expansion which co-segregated with disease (Fig. 5a, Supplementary Table S2). The other 54 patients (14 typical LOCA, 19 EOCA and 21 MSA-c) carried a [(GAA)n (GCA)m]z expansion not containing a (GAA)≥250 expansion that did not co-segregate with disease phenotype. The [(GAA)n (GCA)m]z expansion without (GAA)≥250 expansion was also present in our in-house control database (3/150), suggesting that such expansion with less than 250 uninterrupted GAA-pure repeats may be polymorphic rather than pathogenic, as previously suggested.26 Thus, 12 positive cases were identified in the 75 probands. Additional screening on family members of the 12 positive cases detected five more patients with GAA-FGF14 ataxia (P-5, P-7, P-9, P-10 and P-13).
In total, 17 positive patients with a (GAA)≥250 expansion and two possible positive patients with biallelic (GAA)202/222 alleles were identified, including seven from typical LOCA cohort, nine from EOCA cohort and three from MSA-c cohort. The frequency of index GAA-FGF14 positive patients was 1.3% in the typical LOCA cohort.
Genetic instability of GAA expansion in FGF14
Five of eight dominant ataxia index cases (62.5%) were maternally inherited. Expansion ranging from 16 to 77 repeats was observed during nine maternal transmissions of expanded (GAA)≥250 alleles (Fig. 2e). Notably, the alleles expanded from (GAA)200-249 to (GAA)≥250 upon maternal transmission across two generations (Fig. 2f).
Phenotypic profile
Clinical spectrum and subgroup analysis
19 positive and possible positive patients were classified into LOCA subgroup (n = 9), EOCA subgroup (n = 6) and MSA-c subgroup (n = 4) based on their clinical phenotype. In LOCA subgroup, six were from the typical LOCA cohort and three were from EOCA families who presented late-onset slowly progressive cerebellar ataxia. Their clinical features were similar to those observed in the European GAA-FGF14 ataxia cohort (Table 1). The remaining 10 cases showed atypical GAA-FGF14 ataxia including six with EOCA phenotype (all from the EOCA cohort) and four with MSA-c phenotype (one from the typical LOCA cohort and three from the MSA-c cohort) (Fig. 4a–c, Supplementary Table S3). Interestingly, in family 10, the family members presented a slowly progressive LOCA phenotype but the proband (P-16) exhibited MSA-c phenotype, suggesting a possible secondary MSA-c in the proband.7 Thus, the proband from family 10 was classified into the MSA-c subgroup.
Table 1.
The comparison of the clinical features of GAA-FGF14 positive cases with LOCA from Chinese and other populations.
| Participants |
GAA-FGF14 ataxia cases |
|||
|---|---|---|---|---|
| Population | Chinese (n = 9)a | German (n = 48)b French-Canadian (n = 68)b Australian (n = 3)b, Indian (n = 3)b |
European (n = 50)c | European (n = 17)d |
| Sex | ||||
| Male | 3 (33%) | 62 (51%) | 25 (50%) | 13 (76%) |
| Female | 6 (67%) | 60 (49%) | 25 (50%) | 4 (24%) |
| GAA count of the larger allele | 286 (222–383) | NA | 346 (252–583) | 343 (258–637) |
| Age at onset/years | 47 (40–60) | 55 ± 13e | 61 (37–78) | 63 (28–78) |
| Duration/years | 10 (1–31) | NA | 9 (0.4–25.7) | 14 (4–24) |
| Episodic symptoms | 6/9 (67%) | 56/121 (46%) | 6/47 (13%) | 10/17 (59%) |
| Ataxia features | ||||
| Downbeat nystagmus | 4/7 (57%) | 50/119 (42%) | NA | 10/17 (59%) |
| Gaze-evoked horizontal nystagmus | 7/7 (100%) | 65/119 (55%) | NA | NA |
| Dysarthria | 4/9 (44%) | 63/118 (53%) | 25/42 (60%) | 2/17 (12%) |
| Upper limb ataxia | 4/9 (44%) | NA | 26/43 (60%) | 12/17 (71%) |
| Rombergism | 7/9 (78%) | NA | 27/40 (68%) | 8/15 (53%) |
| Gait ataxia | 9/9 (100%) | 113/118 (96%) | 41/43 (95%) | 17/17 (100%) |
| Additional features | ||||
| Visual impairment | 2/7 (29%) | 57/120 (48%) | 16/33 (48%) | 9/16 (56%) |
| Urinary urgency | 0/7 (0%) | NA | 11/40 (28%) | 8/14 (57%) |
| Dysesthesia | 1/8 (13%) | NA | 1/31 (3%) | NA |
| Cerebellar atrophy on MRI | 6/6 (100%) | 67/91 (74%) | 28/29 (97%) | 13/17 (76%) |
Quantitative variables were presented as median and range.
Abbreviations: NA, no available; MRI, magnetic resonance imaging.
All patients with LOCA phenotype were included.
Pellerin et al., NEJM 2023.
Wilke et al., Brain 2023.
Pellerin et al., JNNP 2023.
The quantitative variables was presented as mean and standard deviation.
Fig. 4.
Clinical assessments of patients with LOCA, EOCA and MSA-c phenotypes. Brain MRI revealed that mild cerebellar atrophy in the patient with LOCA phenotype (P-15) (a), slight cerebellar atrophy in the patient with EOCA phenotype (P-6) (b), and olivopontocerebellar atrophy in the patient with MSA-c phenotype (P-18) (c). (d) The evolution of Spinocerebellar Ataxia Functional Index (SDFS) in the patients with LOCA, EOCA, and MSA-c phenotypes. X-axis: disease duration; Y-axis: SDFS score. (e) Age at onset of Chinese and European LOCA groups. The European GAA-FGF14 ataxia cohort exhibited a significantly higher age at onset compared to patients with LOCA phenotypes (p = 0.006). Each bar represented the median and range for independent group (∗∗p < 0.01, Wilcoxon rank sum test). (f) NfL levels of GAA-FGF14 positive cases in ataxic (LOCA, EOCA, and MSA-c phenotypes) and pre-ataxic stage, as well as controls with age. (g) NfL levels of patients with LOCA, EOCA and MSA-c phenotypes, patients with SCA3, GAA-FGF14 negative patients with MSA-c and controls. The patients with EOCA and MSA-c phenotypes exhibited similar NfL level with patients with SCA3 and GAA-FGF14 negative patients with MSA-c, but significantly higher NfL levels than LOCA phenotype and controls after age correction (p < 0.001, Bonferroni corrected). Each bar represented the mean ± SD for independent group (∗∗p < 0.01, ∗∗∗p < 0.001, ANCOVA). (h) NfL levels of pre-ataxic GAA-FGF14 positive individuals, pre-ataxic patients with SCA3 and controls. Pre-ataxic GAA-FGF14 positive individuals exhibited similar NfL level with the controls but significantly lower than pre-ataxic patients with SCA3 after age correction (p < 0.001, Bonferroni corrected). Each bar represented the mean ± SD for independent group (∗∗∗p < 0.001, ANCOVA).
The median of GAA repeats for the LOCA, EOCA and MSA-c subgroups were 294 (ranging from 250 to 357 GAA repeats), 335 (ranging from 277 to 416 GAA repeats) and 267 (ranging from 264 to 275 GAA repeats), respectively (Supplementary Table S3). The EOCA subgroup exhibited significantly higher numbers of GAA repeats compared to the MSA-c subgroup (p = 0.029, Wilcoxon rank sum test), while no significant difference was observed in other pairwise comparisons.
The age at onset, extracerebellar involvements, and disease progression differed between LOCA, EOCA, and MSA-c subgroups. Compared to a European GAA-FGF14 ataxia cohort of median age at onset of 61 years (37–78 years),7 the median age at disease onset was 47 years (40–60 years) in our LOCA subgroup (p = 0.006, Wilcoxon rank sum test) (Fig. 4e). The most frequent permanent ataxia features were gait ataxia (9/9, 100%) and gaze-evoked horizontal nystagmus (7/7, 100%) in all patients, while six patients experienced episodic symptoms including vertigo or dizziness (4/9, 44%), episodic ataxia (3/7, 43%) (Fig. 3). Overall, patients with LOCA phenotype presented a mild ataxia with independent ambulation despite a median disease duration of 10 years (Fig. 4d). Additionally, we found no correlation between GAA expansion and age at onset (p = 0.691, spearman correlation) (Supplementary Figure S2).
Fig. 3.
Clinical features among patients with LOCA, EOCA, and MSA-c phenotypes. Numbers in brackets indicated the number of affected patients over the total number of patients assessed for this feature.
The median age at onset in the EOCA subgroup was 20.5 years (12–35 years). The patients with EOCA phenotype showed permanent gait ataxia and dysarthria (6/6, 100%), but no episodic symptom. Action tremor was observed in one patient (1/6, 16.7%) (Fig. 3). The median duration was 10 years to walking aid-dependent stage, and 12 years to wheelchair-dependent stage (Fig. 4d).
Four patients presented an MSA-c phenotype (two possible and two probable) with brainstem atrophy and hot cross bun sign observed on MRI. Overall, autonomic dysfunction was detected in the MSA-c subgroup (4/4, 100%), but was not common in the EOCA and LOCA subgroups (Fig. 3). The patients in the MSA-c subgroup experienced the most rapid progression with a median of 3.3 years from onset to walking aid-dependent stage and 4 years from onset to wheelchair-dependent stage (Fig. 4d). We further compared the main clinical features between patients with MSA-c phenotype (n = 4) and age-matched GAA-FGF14 negative patients with MSA-c (n = 297) in our cohort, and found no significant difference in UMSARS Ⅰ (p = 0.257, Wilcoxon rank sum test) and UMSARS Ⅱ (p = 0.294, Wilcoxon rank sum test) after correction for disease duration. However, a trend toward slower disease progression in patients with MSA-c phenotype was observed compared to the GAA-FGF14 negative MSA-c group (p = 0.054, Wilcoxon rank sum test) (Table 2).
Table 2.
The clinical measures of GAA-FGF14 positive and negative patients with MSA-c.
| GAA-FGF14 positive MSA-c (n = 4) | GAA-FGF14 negative MSA-c (n = 297) | p value | |
|---|---|---|---|
| Sex | 0.708a | ||
| Male | 2 (50%) | 176 (59%) | |
| Female | 2 (50%) | 121 (41%) | |
| Age at onset/years | 55.5 (48–68) | 53 (36–71) | 0.457b |
| Duration/years | 3 (3–3) | 2 (0.5–8) | 0.088b |
| UMSARS Ⅰ score | 13.5 (7–25) | 16 (5–44) | 0.257c |
| UMSARS Ⅱ score | 13.5 (9–21) | 15 (5–48) | 0.294c |
| Rate of disease progression | 9.5 (5.7–14) | 16 (2.4–67) | 0.054b |
Quantitative variables were presented as median and range.
Pearson's chi-square test.
Wilcoxon rank sum test.
Analysis of covariance (ANCOVA). .
Exploration of clinical heterogeneity
To explore the possibility that other genetic modifiers modify the clinical spectrum of GAA-FGF14 positive and possible positive cases, we retrospectively performed WGS combined with targeted LRS on 19 patients with GAA expansion to screen for other single nucleotide variants (SNVs) and STRs.
In the LOCA and MSA-c subgroups, no causative or potentially disease-causing SNV or STR was identified in any of the 13 patients. Remarkably, co-occurrence with other genetic diseases was found in five of six patients in the EOCA subgroup (83%), including SCA1, SCA3, sialidosis type 1, and osteogenesis imperfecta type I. In general, co-occurrence accounted for the majority of the phenotypic variation from typical GAA-FGF14 ataxia manifestation. The remaining patient with EOCA phenotype (P-11) from family 6 exhibited a pure GAA expansion of 302 repeats with no causative or potentially pathogenic genetic mutations identified. His ataxia symptoms appeared from the age of 12 years and progressed into frequent falls within one year with a SARA score of 10.
Co-occurrence with other genetic bases
In family 2, the proband (P-3) developed ataxia at age 17, with a history of short stature, scoliosis, and limb fractures. The increase of cross-sectional SARA scores with disease duration was 1.3 points/year when he was 23. He was found to carry an FGF14 (GAA)277 expansion. Additionally, WGS showed a c.1801G > A (p.G601S) variant in COL1A2 gene responsible for osteogenesis imperfecta type I, a dominant connective tissue disorder characterised mainly by bone fragility (Fig. 5a and b, Supplementary Table S3).
In family 3, the proband (P-4) came from a consanguineous Chinese family. He presented with early-onset ataxia, myoclonus, action tremor and seizure since the age of 20, with longitudinal increase of 1.1 SARA points/year. However, the mother (P-5) of the index case presented with a slowly progressive ataxic syndrome suggestive of GAA-FGF14 ataxia at the age of 57, without myoclonus and seizure, with cross-sectional increase of 0.6 SARA points/year. The proband and his mother were found to carry an FGF14 (GAA)302 and (GAA)286 expansion, respectively. However, a common homozygous c.544A > G (p.S182G) variant in NEU1 gene was identified in the proband. Co-segregation analysis revealed that both parents harbored a heterozygous c.544A > G variant in NEU1 gene, suggesting that the proband fulfilled the genetic diagnosis of sialidosis type 1 (Fig. 5a and c, Supplementary Table S3). The proband had a phenotype typical of sialidosis type 1, whereas his mother exhibited a phenotype typical of GAA-FGF14 ataxia.
In family 4, the proband (P-6) developed gait ataxia at age 21 which progressed rapidly as exemplified by the total SARA score of 6 after two years. In addition to carrying an FGF14 (GAA)416-[(GAA)n (GCA)m]z expansion, she was found to carry an ATXN3 (CAG)78 expansion causing SCA3 (Fig. 5a and d, Supplementary Table S3). Remarkably, the age at onset of the proband (P-6) was earlier than the expected age at onset (21 years vs expected 28 years) based on the size of the ATXN3 (CAG)78 expansion, as reported previously.27 Her mother carried an FGF14 (GAA)397-[(GAA)n (GCA)m]z expansion and was asymptomatic at age of 43, while her grandmother carried a (GAA)297-[(GAA)n (GCA) m]z expansion and developed gait ataxia at age 57 with a slow increase of 0.6 SARA points/year (Supplementary Table S3). However, her father, an obligate ATXN3 expansion carrier, exhibited progressive ataxia at age 21 and walking aid-dependent at 26, and died at 35 without genetic diagnosis.
In family 5, the proband (P-8) and her younger sister (P-9) experienced similar disease progression characterised by rapidly progressive gait imbalance. The disease onset in the proband and her younger sister were 30 and 35, respectively. The increase of cross-sectional SARA scores with disease duration was 1.7 points/year when the proband was 40. Her younger sister presented cross-sectional increase of SARA by 2.5 points/year when she underwent clinical assessment at age 39. Notably, the proband's mother developed gait ataxia at age 60 and progressed slowly with a SARA score of 4 when she was 80 (20 years of disease duration). However, both the proband and her younger sister died at age of 53 due to pulmonary infection and respiratory failure. The index case, her younger sister, and her mother were all found to carry an FGF14 (GAA)349-390-[(GAA)n (GCA) m]z expansion. In addition, both the proband and her younger sister, but not their mother, were found to carry a (CAG)50 expansion in the ATXN1 gene causing SCA1. The expected age at onset based on the size of the ATXN1 (CAG)50 expansion is 35 years,28 which is similar to that of P-9 (35 years) but later than that of P-8 (30 years), who was also found to carry a longer FGF14 GAA expansion (Fig. 5a and e, Supplementary Table S3).
NfL level distribution in GAA-FGF14 ataxia, SCA3, and MSA-c
As NfL was considered as an objective biomarker for disease severity in degenerative ataxia, we compared NfL levels of GAA-FGF14 positive patients to patients with SCA3 and GAA-FGF14 negative patients with MSA-c. We measured the pNfL in 16 GAA-FGF14 positive individuals (including six patients with LOCA phenotype, two patients with EOCA phenotype, three patients with MSA-c phenotype, and 5 pre-ataxic), 30 patients with SCA3 (including 15 ataxic and 15 pre-ataxic), and 15 GAA-FGF14 negative patients with MSA-c.
Plasma NfL levels increased with age in controls (10.7 ± 4.7 pg/ml) (p = 0.001, r = 0.761, pearson correlation). Compared to controls, patients with EOCA and MSA-c phenotypes showed a tendency for higher NfL levels, while LOCA and controls presented comparable NfL levels when age got adjusted (Fig. 4f, Supplementary Table S3).
The NfL levels in patients with atypical GAA-FGF14 ataxia (EOCA and MSA-c phenotypes: 41.6 ± 27.5 pg/ml) were significantly higher than that of patients with LOCA phenotype (21.8 ± 11.8 pg/ml) and controls (10.7 ± 4.7 pg/ml) after age correction (p < 0.001, Bonferroni corrected, ANCOVA). Furthermore, the patients with atypical GAA-FGF14 ataxia had similar NfL levels to patients with SCA3 (29.9 ± 4.6 pg/ml) and GAA-FGF14 negative patients with MSA-c (37.8 ± 9.4 pg/ml) (p > 0.05, Bonferroni corrected, ANCOVA) (Fig. 4g, Supplementary Table S3).
After age adjustment, we also found that NfL levels of pre-ataxic GAA-FGF14 positive individuals (6.5 ± 3.2 pg/ml) were lower than that of individuals with pre-ataxic SCA3 (19.1 ± 7.4 pg/ml) (p < 0.001, Bonferroni corrected, ANCOVA). No significant difference of NfL level between pre-ataxic GAA-FGF14 ataxia and controls was observed (p > 0.05, Bonferroni corrected, ANCOVA) (Fig. 4h, Supplementary Table S3).
Discussion
In this study, we explored the genetic and clinical spectrum of GAA-FGF14 ataxia in China and found a frequency of FGF14 GAA expansion of 1.3% in a large Chinese LOCA cohort, which is lower than what has previously been reported in non-Asian ataxia populations.5,7 We also observed that LOCA, EOCA, and MSA-c phenotypes of GAA-FGF14 positive cases differed in terms of age at onset, clinical features, and disease progression due to different genetic bases and detected new findings: 1) expansion could arise from alleles of 200–249 repeat units during intergenerational transmission; 2) biallelic (GAA)202/222 alleles may also be associated with disease; 3) GAA-FGF14 expansion co-occurrent with other acquired or hereditary diseases may contribute to complicated clinical spectrum and progression of GAA-FGF14 positive cases with EOCA and MSA-c phenotypes. Overall, our study in the Chinese population will help to better understand the genotype–phenotype correlations of GAA-FGF14 ataxia.
In our cohort, 6.2% patients carried an FGF14 expansion of at least 250 triplet repeat units. The repeat configuration of the expansions consisted of the GAA motif and the GAA-GCA motif. GAA-GCA expansions were identified in 5.3% of patients while GAA expansions were identified in 0.9% of (0.7% for [GAA]250-299 expansions and 0.2% for [GAA]≥300 expansions).
Notably, the sequence of the GAA-GCA expansions was characterised by GAA expansion followed by various [(GAA)n (GCA)m]z patterns. Although the GAA-GCA motif was more frequent, co-segregation analysis in patients with expanded GAA-GCA repeats revealed that the pathogenicity of such expansions was determined by the size of consecutive uninterrupted GAA repeats rather than the size of the GAA-GCA repeats.
Interestingly, biallelic (GAA)202/222 expansions were observed in one family and co-segregated with disease in both affected individuals exhibiting typical GAA-FGF14 ataxia. Other known mutations were excluded by WGS and targeted LRS in affected relatives from this family, suggesting that biallelic GAA alleles of less than 250 repeats might be associated with disease. Further studies including large-scale case–control series and functional studies are needed to clarify whether such biallelic alleles are pathogenic.
Our results confirm that GAA repeat expansions tend to expand further upon maternal transmission.5,29 We also document an expansion through meiosis from a non-pathological range (200–249 repeats) to a pathological range (GAA repeats greater than 250) upon maternal transmission. Till now, the FGF14-GAA expansion in the female germline and contraction in the male germline are noted, which is similar to FRDA and other noncoding repeat expansion disorders.30 Additional studies looking at the probability of expansion/contraction based on parental sex, the impact of GAA repeat length on instability, and the influence of the common 5’ flanking variant29 are warranted.
Clinically, in addition to the LOCA phenotype previously reported in GAA-FGF14 positive cases, we observed EOCA and MSA-c phenotypes in GAA-FGF14 positive Chinese patients. In GAA-FGF14 positive cases, subgroup analysis showed that patients with LOCA phenotype presented with late onset and mild ataxia, while MSA-c phenotype was associated with the most rapid progression and poorest prognosis, and EOCA phenotype showed early onset and intermediate disease severity. Specifically, the absence of dysarthria and downbeat nystagmus, which was considered typical in non-Asian patients with GAA-FGF14 ataxia, was found in our Chinese cohort.5,7,10 Notably, as a hallmark of typical GAA-FGF14 ataxia, episodic symptoms were common in individuals with LOCA rather than cases with EOCA.31 Similar to a recently reported European GAA-FGF14 ataxia cohort, the NfL level of patients with LOCA phenotype was similar to controls, but significantly lower than that of patients with EOCA and MSA-c phenotypes. Thus, the three phenotypically different subgroups presented various clinical features and NfL levels. We assume that co-occurrence of second-hit diseases (acquired or hereditary) may contribute to clinical phenotypic variation of our GAA-FGF14 positive cases, while the elevated NfL levels in the EOCA subgroup is likely caused by the second cause of ataxia.
Co-occurrence with other genetic diseases were identified in 5/6 of GAA-FGF14 positive cases with EOCA phenotype, all of which presented with early onset and rapidly progressive ataxia. The finding that SCA1, SCA3, sialidosis type 1, and osteogenesis imperfecta type I may co-occur with an FGF14 GAA expansion in patients with EOCA phenotype highlights the importance of searching for other genetic diseases in patients with a phenotype considered atypical for GAA-FGF14 ataxia. Thus, when the phenotype is not compatible with typical GAA-FGF14 ataxia, further genetic assessment is warranted to explore unsolved causes responsible for disease phenotype. Although we found patients with EOCA carrying FGF14 GAA expansion together with SCA1 or SCA3, the common SCA subtypes need to be screened first before FGF14 GAA expansion test. Then atypical GAA-FGF14 ataxia need to be further tested for uncommon subtypes of inherited ataxia. Notably, the probands carrying an ATXN1 or ATXN3 expansion and a GAA-FGF14 expansion showed an earlier disease onset than that expected based on the size of their ATXN1 or ATXN3 expansion, suggesting that the GAA-FGF14 expansion might be a modulator of disease onset in other ataxias. The validation experiments will be conducted in large SCA1 and SCA3 cohort for next step.
Notably, the genome-wide screen failed to identify other causative mutations in patients with an MSA-c phenotype. Previous reports indicated that GAA-FGF14 ataxia differs from the MSA-c type.7 Here, expansions within the incompletely penetrant range (264–275 GAA repeats) were observed in our GAA-FGF14 positive patients with MSA-c, suggesting it insufficiently causes MSA-c phenotype. Further, as reported in European cohorts, atypical disease severity or progression in GAA-FGF14 ataxia may be attributed to another concomitant disease.7 The possibility that the MSA-c phenotype developed as an independently comorbid neurodegeneration may occur and complicate the clinical spectrum of GAA-FGF14 positive cases. In our cohort, the clinical features did not significantly differ between GAA-FGF14 positive patients with MSA-c and age-matched GAA-FGF14 negative patients with MSA-c, but a tendency for slower disease progression was observed in the GAA-FGF14 positive patients with MSA-c.
Some limitations in our study need to be considered. Due to our limited positive cases, larger sample size of positive cases is required to comprehensively and systematically characterise clinical features of GAA-FGF14 ataxia. Whether GAA-FGF14 expansions within the incompletely penetrant range is a modifier or a pathogenic mutation in patients with ataxia may be uncovered in follow-up assessments longitudinally.
In conclusion, our study provides a comprehensive genetic assessment of a large Chinese ataxia cohort and expand the understanding of clinical heterogeneity of GAA-FGF14 ataxia. The frequency of GAA expansion in Chinese patients was lower than in non-Asian ataxia populations. We also showed that biallelic (GAA)202/222 alleles of less than 250 repeats and co-occurrence with other genetic or diseases (especially in cases with an atypical phenotype), may contribute to disease onset and phenotypic variation, which shed light on disease heterogeneity of clinical spectrum and progression. The future investigations to examine the role of GAA-FGF14 expansion combined with other unknown modifiers may help to better delineate the disease trajectory of GAA-FGF14 ataxia and improve efficacy of individualised clinical management.
Contributors
R.O., Z.C., L.W., and H.J. contributed to the conception and design of the study; R.O., Z.C., L.W., Z.L., J.H., Q.J., C.W., L.P., H.P., L.H., R.Q., J.W., J.G., L.S., B.T., and H.J. contributed to the acquisition and analysis of data; R.O., Z.C., D.P., L.W., B.B., M.C.D., S.Z., and H.J. contributed to the drafting a significant portion of the manuscript and figures. All authors read and approved the final version of the manuscript. R.O., Z.C., L.W., and H.J. confirmed that they had full access to all the data in the study and accept responsibility to submit for publication.
Data sharing statement
De-identified data and a data dictionary generated during the study are available from the corresponding author on reasonable request. Please send data access requests of proposal to the corresponding author. The requests must be approved by the appropriate ethics boards and data custodians.
Declaration of interests
S.Z. has received consultancy honoraria from Neurogene, Aeglea BioTherapeutics, Applied Therapeutics, and is an unpaid officer of the TGP foundation, all unrelated to the present manuscript. The other authors report no conflicts of interest.
Acknowledgements
We thank all of the participants for their involvement in this study. This study was funded by the National Key R&D Program of China (2021YFA0805200 to H.J.), the National Natural Science Foundation of China (81974176 and 82171254 to H.J.; 82371272 to Z.C.; 82301628 to L.W.; 82301438 to Z.L.; 82201411 to L.H.), the Innovation Research Group Project of Natural Science Foundation of Hunan Province (2020JJ1008 to H.J.), the Key Research and Development Program of Hunan Province (2020SK2064 to H.J.), the Innovative Research and Development Program of Development and Reform Commission of Hunan Province to H.J., the Natural Science Foundation of Hunan Province (2024JJ3050 to H.J.; 2022JJ20094 and 2021JJ40974 to Z.C.; 2022JJ40783 to L.H.; 2022JJ40703 to Z.L.),the Project Program of National Clinical Research Center for Geriatric Disorders (Xiangya Hospital, 2020LNJJ12 to H.J.), the Central South University Research Programme of Advanced Interdisciplinary Study (2023QYJC010 to H.J.) and the Science and Technology Innovation Program of Hunan Province (2022RC1027 to Z.C.). D.P. holds a Fellowship award from the Canadian Institutes of Health Research (CIHR).
Footnotes
Supplementary data related to this article can be found at https://doi.org/10.1016/j.ebiom.2024.105077.
Contributor Information
Zhao Chen, Email: czcf98@csu.edu.cn, czcf98@126.com.
Hong Jiang, Email: jianghong73868@csu.edu.cn, jianghong73868@126.com.
Appendix A. Supplementary data
References
- 1.Jayadev S., Bird T.D. Hereditary ataxias: overview. Genet Med. 2013;15(9):673–683. doi: 10.1038/gim.2013.28. [DOI] [PubMed] [Google Scholar]
- 2.Klockgether T., Mariotti C., Paulson H.L. Spinocerebellar ataxia. Nat Rev Dis Primers. 2019;5(1):24. doi: 10.1038/s41572-019-0074-3. [DOI] [PubMed] [Google Scholar]
- 3.Cortese A., Simone R., Sullivan R., et al. Biallelic expansion of an intronic repeat in RFC1 is a common cause of late-onset ataxia. Nat Genet. 2019;51(4):649–658. doi: 10.1038/s41588-019-0372-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen Z., Tucci A., Cipriani V., et al. Functional genomics provide key insights to improve the diagnostic yield of hereditary ataxia. Brain. 2023;146(7):2869–2884. doi: 10.1093/brain/awad009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pellerin D., Danzi M.C., Wilke C., et al. Deep intronic FGF14 GAA repeat expansion in late-onset cerebellar ataxia. N Engl J Med. 2023;388(2):128–141. doi: 10.1056/NEJMoa2207406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rafehi H., Read J., Szmulewicz D.J., et al. An intronic GAA repeat expansion in FGF14 causes the autosomal-dominant adult-onset ataxia SCA27B/ATX-FGF14. Am J Hum Genet. 2023;110(6):1018. doi: 10.1016/j.ajhg.2023.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wilke C., Pellerin D., Mengel D., et al. GAA-FGF14 ataxia (SCA27B): phenotypic profile, natural history progression and 4-aminopyridine treatment response. Brain. 2023;146:4144. doi: 10.1093/brain/awad157. [DOI] [PubMed] [Google Scholar]
- 8.Wirth T., Clément G., Delvallée C., et al. Natural history and phenotypic spectrum of GAA-FGF14 sporadic late-onset cerebellar ataxia (SCA27B) Mov Disord. 2023;38:1950. doi: 10.1002/mds.29560. [DOI] [PubMed] [Google Scholar]
- 9.Hengel H., Pellerin D., Wilke C., et al. As frequent as polyglutamine spinocerebellar ataxias: SCA27B in a large German autosomal dominant ataxia cohort. Mov Disord. 2023;38(8):1557–1558. doi: 10.1002/mds.29559. [DOI] [PubMed] [Google Scholar]
- 10.Pellerin D., Wilke C., Traschütz A., et al. Intronic FGF14 GAA repeat expansions are a common cause of ataxia syndromes with neuropathy and bilateral vestibulopathy. J Neurol Neurosurg Psychiatry. 2023;95:175. doi: 10.1136/jnnp-2023-331490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Klockgether T. Sporadic ataxia with adult onset: classification and diagnostic criteria. Lancet Neurol. 2010;9(1):94–104. doi: 10.1016/S1474-4422(09)70305-9. [DOI] [PubMed] [Google Scholar]
- 12.Gilman S., Wenning G.K., Low P.A., et al. Second consensus statement on the diagnosis of multiple system atrophy. Neurology. 2008;71(9):670–676. doi: 10.1212/01.wnl.0000324625.00404.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schmitz-Hübsch T., du Montcel S.T., Baliko L., et al. Scale for the assessment and rating of ataxia: development of a new clinical scale. Neurology. 2006;66(11):1717–1720. doi: 10.1212/01.wnl.0000219042.60538.92. [DOI] [PubMed] [Google Scholar]
- 14.Wenning G.K., Tison F., Seppi K., et al. Development and validation of the unified multiple system atrophy rating scale (UMSARS) Mov Disord. 2004;19(12):1391–1402. doi: 10.1002/mds.20255. [DOI] [PubMed] [Google Scholar]
- 15.Anheim M., Monga B., Fleury M., et al. Ataxia with oculomotor apraxia type 2: clinical, biological and genotype/phenotype correlation study of a cohort of 90 patients. Brain. 2009;132(Pt 10):2688–2698. doi: 10.1093/brain/awp211. [DOI] [PubMed] [Google Scholar]
- 16.Peng Y., Zhang Y., Chen Z., et al. Association of serum neurofilament light and disease severity in patients with spinocerebellar ataxia type 3. Neurology. 2020;95(22):e2977–e2987. doi: 10.1212/WNL.0000000000010671. [DOI] [PubMed] [Google Scholar]
- 17.Montroull L.E., Danelon V., Cragnolini A.B., Masco D.H. Loss of TrkB signaling due to status epilepticus induces a proBDNF-dependent cell death. Front Cell Neurosci. 2019;13:4. doi: 10.3389/fncel.2019.00004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Maas R.P., van Gaalen J., Klockgether T., van de Warrenburg B.P. The preclinical stage of spinocerebellar ataxias. Neurology. 2015;85(1):96–103. doi: 10.1212/WNL.0000000000001711. [DOI] [PubMed] [Google Scholar]
- 19.Chen Z., Liao G., Wan N., et al. Synaptic loss in spinocerebellar ataxia type 3 revealed by SV2A positron emission tomography. Mov Disord. 2023;38(6):978–989. doi: 10.1002/mds.29395. [DOI] [PubMed] [Google Scholar]
- 20.Bonnet C., Pellerin D., Roth V., et al. Optimized testing strategy for the diagnosis of GAA-FGF14 ataxia/spinocerebellar ataxia 27B. Sci Rep. 2023;13(1):9737. doi: 10.1038/s41598-023-36654-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chintalaphani S.R., Pineda S.S., Deveson I.W., Kumar K.R. An update on the neurological short tandem repeat expansion disorders and the emergence of long-read sequencing diagnostics. Acta Neuropathol Commun. 2021;9(1):98. doi: 10.1186/s40478-021-01201-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wenger A.M., Peluso P., Rowell W.J., et al. Accurate circular consensus long-read sequencing improves variant detection and assembly of a human genome. Nat Biotechnol. 2019;37(10):1155–1162. doi: 10.1038/s41587-019-0217-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li H. New strategies to improve minimap2 alignment accuracy. Bioinformatics. 2021;37(23):4572–4574. doi: 10.1093/bioinformatics/btab705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li J.Y., Liu J., Manaph N.P.A., Bobrovskaya L., Zhou X.F. ProBDNF inhibits proliferation, migration and differentiation of mouse neural stem cells. Brain Res. 2017;1668:46–55. doi: 10.1016/j.brainres.2017.05.013. [DOI] [PubMed] [Google Scholar]
- 25.Cao Y., Li L., Xu M., et al. The ChinaMAP analytics of deep whole genome sequences in 10,588 individuals. Cell Res. 2020;30(9):717–731. doi: 10.1038/s41422-020-0322-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pellerin D., Iruzubieta P., Tekgül Ş., et al. Non-GAA repeat expansions in FGF14 are likely not pathogenic-reply to: "shaking up ataxia: FGF14 and RFC1 repeat expansions in affected and unaffected members of a Chilean family". Mov Disord. 2023;38(8):1575–1577. doi: 10.1002/mds.29552. [DOI] [PubMed] [Google Scholar]
- 27.Chen Z., Zheng C., Long Z., et al. (CAG)n loci as genetic modifiers of age-at-onset in patients with Machado-Joseph disease from mainland China. Brain. 2016;139(Pt 8):e41. doi: 10.1093/brain/aww087. [DOI] [PubMed] [Google Scholar]
- 28.Wang P., Chen Z., Peng Y., et al. (CAG)(n) loci as genetic modifiers of age at onset in patients with spinocerebellar ataxia type 1 from mainland China. Eur J Neurol. 2019;26(8):1130–1136. doi: 10.1111/ene.13954. [DOI] [PubMed] [Google Scholar]
- 29.Pellerin D., Gobbo G.D., Couse M., et al. A common flanking variant is associated with enhanced meiotic stability of the FGF14 -SCA27B locus. bioRxiv. 2023 doi: 10.1101/2023.05.11.540430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Depienne C., Mandel J.L. 30 years of repeat expansion disorders: what have we learned and what are the remaining challenges? Am J Hum Genet. 2021;108(5):764–785. doi: 10.1016/j.ajhg.2021.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ashton C., Indelicato E., Pellerin D., et al. Spinocerebellar ataxia 27B: episodic symptoms and acetazolamide response in 34 patients. Brain Commun. 2023;5(5):fcad239. doi: 10.1093/braincomms/fcad239. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.