Abstract
Core protein is the major component of the core particle (nucleocapsid) of human hepatitis B virus. Core particles and core proteins are involved in a number of important functions in the replication cycle of the virus, including RNA packaging, DNA synthesis, and recognition of viral envelope proteins. Core protein is a phosphoprotein with most, if not all, of the phosphorylation on C-terminal serine residues. In this study, we identified a serine kinase activity from the ribosome-associated protein fraction of cytoplasm that could specifically bind and phosphorylate the C-terminal portion of recombinant core protein. This kinase is referred to as core-associated kinase (CAK). CAK could be inhibited by the kinase inhibitors heparin and manganese ions but not by spermidine, DRB, H89, or H7, indicating that CAK is distinct from protein kinase A and protein kinase C. CAK could be partially purified by heparin-Sepharose CL-6B and phosphocellulose P11 columns. By using a far-Western assay, three specific proteins, of 46, 35, and 13 kDa, were shown to interact with the C-terminal part of the core protein. These three proteins were present only in the eluted fractions that contains the CAK activity. An in-gel kinase assay showed that a 46-kDa kinase in the same fraction could bind and phosphorylate the C-terminal part of the recombinant core protein. These results indicate that this 46-kDa kinase is most probably CAK. A similar 46-kDa kinase, which exhibits the same profile of sensitivity to kinase inhibitors as that of CAK, is present in both purified intracellular core particles and extracellular 42-nm virions, suggesting that CAK is a candidate for the core particle-associated kinase.
Human hepatitis B virus (HBV) is a small, enveloped DNA virus. The 42-nm virion is composed of a 27-nm core particle (nucleocapsid) and an envelope. The core particle is involved in a number of important functions in the replication cycle of the virus, including RNA packaging, DNA synthesis, and recognition of viral envelope proteins. The first step of core particle assembly is the formation of dimers from monomeric core protein (32). The subsequent assembly into a core particle is combined with the specific packaging of the pregenomic RNA, which is mediated by the viral polymerase (2, 12). The 21-kDa core protein can be detected in the nucleus, the cytoplasm, or both within infected hepatocytes (22). Its N-terminal 144 amino acid residues can direct the assembly of the core particle (7, 10). In contrast, the protamine-like carboxyl terminus, from residues 150 to 185 (HBV subtype adw) or 183 (ayw), is dispensable for particle assembly but mediates interactions between the core protein and the nucleic acid. This region contains four blocks of arginine residues (amino acids 150 to 152, 159 to 161, 166 to 169, and 174 to 177) and three overlapping SPRRR motifs (amino acids 157 to 161, 164 to 168, and 172 to 176). The first block of arginine residues drives RNA binding, while the other three blocks of arginine residues are required for binding of viral DNA (10, 23, 29).
In addition to core proteins, polymerase, and pregenomic RNA, a protein kinase is present in the core particle. This enzyme activity, termed core particle-associated kinase, was initially identified in 1980 by the ability of virions to incorporate radioactive phosphate into core proteins upon incubation with labeled ATP in vitro (1). It was capable of phosphorylating the serine residues at the C-terminal SPRRR motifs of core proteins in core particles, which are exposed to the inside of core particles (8, 21, 26). This kinase was also found in core particles of duck hepatitis B virus, woodchuck hepatitis virus, and ground squirrel hepatitis virus (6, 9, 24). Core particle-associated kinase is apparently of cellular origin, since core particles contain an encapsidated kinase when produced in insect cells in the absence of other viral gene products (18). Since phosphorylation occurs in a region which is important for the packaging of pregenomic RNA and the synthesis of viral DNA, phosphorylation has been implicated in modulation of the ability of core protein to interact with viral genome in vivo (10, 21). The evolutionary conservation of core particle-associated kinase activity among these hepadnaviruses and its possible phosphorylation effect on genome replication suggest that this cellular kinase plays important role(s) in the viral life cycle. The identity and characteristics of this core particle-associated kinase remain unknown.
Core protein is a phosphoprotein in vivo. Most, if not all, of the amino acid residues for phosphorylation in the core protein are the serine residues at the C-terminal SPRRR motifs (19, 25). The serine residues of three SPRRR motifs can serve as acceptor sites for phosphorylation with equal efficiency. The nuclear localization of the core protein appears to be regulated in a cell cycle-dependent manner and correlates with its phosphorylation status (28). Phosphorylation has been proposed to negatively regulate the nuclear localization (19).
In this paper, we identify a serine kinase activity that can specifically bind and phosphorylate the C-terminal part of recombinant core protein. This kinase is designated core-associated kinase (CAK). By using an in-gel kinase assay, we show that a 46-kDa kinase which coeluted with the CAK activity can bind and phosphorylate the C-terminal part of the recombinant core protein. A similar 46-kDa kinase which exhibits the same profile of sensitivity to inhibitors as that of CAK is present in both purified core particles and 42-nm secreted virions.
MATERIALS AND METHODS
Plasmid construction.
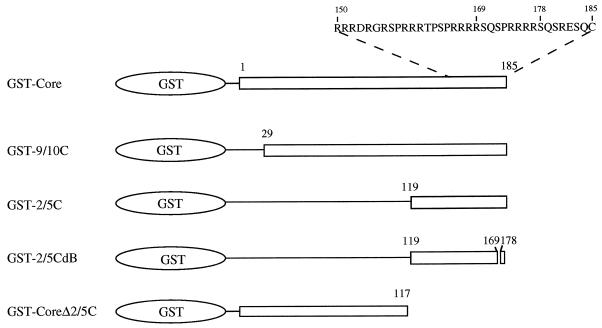
Plasmids GST-Core, GST-9/10C, and GST-2/5C contain sequences encoding the entire length, the segment from amino acids 29 to 185, and the segment from amino acids 119 to 185, respectively, of the HBV core protein on the pGEX-3X vector. Plasmid GST-2/5CdB is derived from GST-2/5C and contains an internal deletion corresponding to amino acids 170 to 177 of the core protein in GST-2/5C. Plasmid GST-CoreΔ2/5C is derived from GST-Core and contains the segment encoding amino acids 1 to 117. Plasmid pHBV3.6 consists of more than a unit length of the HBV genome with the sequence from nucleotides 1636 to 1990 (30).
In vitro binding and kinase reaction.
All glutathione S-transferase (GST) fusion proteins were expressed in bacterial strain RR1. Bacterial cultures (50 ml) was grown to mid-log phase and then induced with 1 mM isopropyl-β-d-galactoside (IPTG). The culture was then grown for another 3 h. The cells were pelleted and lysed at 4°C for 15 min in 5 ml of phosphate-buffered saline (PBS) (170 mM NaCl, 3 mM KCl, 10 mM Na2PO4, 2 mM KH2PO4) containing 1% Triton X-100. All subsequent steps were carried out at 4°C. The lysates were broken in a French press and centrifuged at 18,000 × g for 30 min. The supernatant was filtered through a 0.45-μm-pore-size filter (Millipore).
Approximately 1 μg of GST fusion protein in 1 ml of PBS was incubated with 40 μl of glutathione-Sepharose beads (50% slurry) preequilibrated in PBS containing 0.5% Triton X-100. The beads were pelleted at 200 × g for 2 min and washed three times with PBS containing 0.5% Triton X-100. The beads were then incubated with ribosome-associated proteins (RAP) (see below) for 30 min at 4°C with rocking, collected after centrifugation at 200 × g for 2 min, and washed four times with kinase buffer (20 mM Tris-HCl [pH 7.5], 10 mM MgCl2, 100 mM NaCl). The kinase assay was performed at room temperature for 20 min with 50 μl of a reaction mixture containing 20 μl of beads, 10 μl of 5× kinase buffer, and 50 μCi of [γ-32P]ATP (5,000 Ci/mmol; Amersham). After the kinase reaction, the beads were washed four times with PBS containing 0.5% Triton X-100, and the adsorbed proteins, including kinase-treated products, were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) (12% polyacrylamide) and visualized by autoradiography.
For kinase inhibitor experiments, inhibitors such as H7 [1-(5-isoquinolinesulfonyl)-2-methylpiperazine 2 HCl] (100 μM, Sigma), H89 [N-(2-[p-bromocinnamylamino]-ethyl)-5-isoquinolinesulfoamide 2 HCl] (100 μM; Sigma), DRB (5,6-dichloro-1-β-d-ribofuranosylbenzimidazole) (6 μM; BioMol), heparin (25 U/ml; Sigma), spermidine (500 μM; Sigma), and manganese (5 mM) were added individually to kinase reaction mixtures (3, 5, 11, 31). For phosphorylation of the core protein by protein kinase A (PKA) and protein kinase C (PKC), 1 μl of PKA (100,000 U/ml; Promega) and 1 μl of PKC (100,000 U/ml; Promega), respectively, were incubated with GST-2/5C.
Preparation of ribosome-associated proteins and purification of CAK.
Ribosomes and ribosome-associated proteins were prepared essentially as described by Lin (20) and Spedding (27). HuH-7 cells or minced porcine liver tissues were washed with 0.25 M sucrose in TKM buffer (20 mM Tris-HCl [pH 7.6], 50 mM KCl, 12.5 mM MgCl2) three times, homogenized with 9 volumes of the same buffer with 13 strokes, and centrifuged at 8,000 × g for 15 min. The supernatant was then filtered through glass wool fabric. Triton X-100 was added to the filtrate to a final concentration of 1%. After centrifugation at 8,000 × g for 15 min, the supernatant was centrifuged in an SW41 rotor (Beckman) at 280,000 × g for 2.5 h and 4°C on a 1 M sucrose cushion in TKM buffer. The pellet (80S ribosome) was stored at −70°C. The 80S ribosome pellet was further washed with TKM buffer containing 0.5 M KCl and centrifuged at 280,000 × g for 2.5 h at 4°C. The supernatant, containing RAP, was dialyzed against TKM buffer and stored at −70°C.
For further purification of CAK, 20 mg of RAP in TKM buffer was loaded onto a heparin-Sepharose CL-6B (Pharmacia) column (10 mm by 10 cm) equilibrated with TKM buffer and eluted stepwise at a flow rate of 1 ml/min with the same buffer containing 0.05, 0.1, 0.2, 0.3, 0.4, 0.5, 0.6, or 0.8 M KCl. Each fraction was pooled, dialyzed against TKM buffer, concentrated about 50-fold, and tested for CAK activity. For an additional round of purification, 200 μg of proteins from the combined 0.3 and 0.4 M fractions from the heparin-Sepharose CL-6B column was loaded onto a phosphocellulose P11 column (Whatman) and eluted under the same condition as for the heparin-Sepharose CL-6B column.
Analysis of phosphoamino acids.
The GST-Core protein and its deletion proteins which were labeled in the in vitro kinase reaction and resolved by SDS-PAGE were excised from the gel and partially hydrolyzed in 5.7 M HCl at 110°C for 1 h. The phosphoamino acids were separated on thin-layer cellulose plates in two dimensions, the first dimension in pH 1.9 buffer (formic acid-acetic acid-water, 1:3.55:40.77) and the second dimension in pH 3.5 buffer (glacial acetic acid-pyridine-water, 10:1:189). The migration of cold phosphoamino acid standards was determined by ninhydrin staining, while that of radioactive amino acids was detected by autoradiography.
Far-Western analysis of associated proteins.
Samples (20 μg) of proteins from the 0.2 and 0.4 M fractions from the phosphocellulose P11 column were resolved by SDS-PAGE (12% polyacrylamide). After being transferred to nitrocellulose filters in transfer buffer (39 mM glycine, 48 mM Tris-HCl, 0.037% SDS, 20% methanol), proteins on filters were renatured for 24 h at 4°C in renaturation buffer (10 mM HEPES [pH 7.5], 10 mM MgCl2, 0.1 mM EDTA, 1 mM dithiothreitol [DTT], 100 mM KCl, 10% glycerol, 5% nonfat milk) and then washed with binding buffer (20 mM HEPES [pH 7.5], 10 mM MgCl2, 0.25 mM EDTA, 0.5 mM DTT, 100 mM KCl, 0.25% nonfat milk) three times. The filters were then incubated with 100 μg of GST fusion protein in binding buffer for 1 h at 4°C and washed with TNT buffer (10 mM Tris-HCl [pH 7.5], 150 mM NaCl, 0.05% Tween 20) three times. These filters with bound GST fusion protein were incubated with anti-GST antibody in TNT buffer for 1 h at room temperature, washed with TNT buffer three times, incubated with horseradish peroxidase (HRP)-conjugated secondary antibody (Promega) for 1 h at room temperature, and washed with TNT buffer. Finally, these treated filters were incubated at room temperature in the 4-chloro-1-naphthol developing solution. The multitude of proteins present in the 0.2 and 0.4 M fractions was visualized by using the silver stain method.
In-gel kinase assay.
Our in-gel kinase assay was modified from previously reported procedures (4, 13). Briefly, 20-μg samples of proteins from fractions eluted with 0.2 and 0.4 M salt were resolved on by SDS-PAGE (12% polyacrylamide) containing 2 mg of GST-2/5C protein per ml. The gel was washed four times with 200 ml of 40 mM HEPES (pH 7.6) buffer and twice with the same buffer containing 25% 2-propanol over a period of 4 to 6 h. Proteins in the gel were denatured in denaturation buffer (7 M guanidine-HCl, 50 mM Tris-HCl [pH 7.5], 2 mM EDTA, 10 mM DTT) for 1 h at room temperature and renatured in renaturation kinase buffer (25 mM HEPES [pH 7.9], 12.5 mM MgCl2, 100 mM KCl, 0.1 mM EDTA, 0.1% Nonidet P-40) for 8 h at room temperature. Proteins in the gel were then subjected to kinase reactions for 1 h at room temperature in renaturation kinase buffer containing 250 μCi of [γ-32P]ATP (5,000 Ci/mmol; Amersham). The gel was then washed with 40 mM HEPES (pH 7.4) buffer six times. A 20-g portion of Sephadex G-50 resin, which could bind free [γ-32P]ATP, was included in the buffer during the last two washes. The gel was washed for an additional 3 h in 40 mM HEPES (pH 7.6) buffer containing 1% sodium pyrophosphate. Then the gel was fixed in 250 ml of fixation solution (10% 2-propanol, 5% acetic acid, 1% sodium pyrophosphate) for 1 h, vacuum dried, and autoradiographed.
Preparation of intracellular core particles and extracellular 42-nm virions.
Human hepatoma HuH-7 cells were transiently transfected with plasmid pHBV3.6, which contains more than a unit length of the HBV genome and is capable of viral replication and 42-nm virion assembly (30). For purifying intracellular core particles, transfectants were pelleted at 6,000 × g for 10 min. The cell pellet was then treated for 2 h at 4°C with PBS containing 0.5% Triton X-100 and centrifuged at 10,000 × g for 15 min. Intracellular core particles were immunoprecipitated from the supernatant with human anti-core protein A–Sepharose beads and washed extensively with PBS buffer containing 0.5% Nonidet P-40. To purify extracellular 42-nm virions, the culture medium of the transfectants was centrifuged at 20,000 × g for 30 min at 4°C. The supernatant was then centrifuged in a Ti 55.2 rotor (Beckman) at 227,000 × g for 80 min at 4°C. The pellet was resuspended in TNE buffer (50 mM Tris-HCl [pH 7.5], 150 mM NaCl, 1 mM EDTA), immunoprecipitated with anti-surface protein A–Sepharose beads, and washed extensively with TNE buffer.
The immunoprecipitated core particles were pretreated with proteinase K in 50 μl of TNE containing 1, 5, 25, 50, 100, or 200 ng of proteinase K at 37°C for 20 min (2).
Core particle-associated kinase activity.
The assays of core particle-associated kinase activity for both purified core particles and virions were performed by adding 10 μl of kinase buffer followed by 50 μCi of [γ-32P]ATP (5,000 Ci/mmol) and then incubating the mixtures at room temperature for 10 min. For virions, additional Triton X-100 was added to the kinase buffer to a final concentration of 0.5%. Phosphorylated core proteins were visualized by autoradiography after SDS-PAGE. Other conditions were as described for the in vitro kinase assay. For kinase inhibitor experiments, kinase inhibitors were added before [γ-32P]ATP.
Endogenous DNA polymerase activity.
To assay the endogenous DNA polymerase activity of intracellular core particles and extracellular virions (16), samples were incubated in polymerase assay buffer (50 mM Tris-HCl [pH 7.5], 40 mM NH4Cl, 5 mM MgCl2, 0.5% Nonidet P-40, 0.2% 2-mercaptoethanol, 50 μM each TTP, dGTP, and dCTP, 100 μCi of [α-32P]dATP [3,000 Ci/mmol]) at 37°C for 2 h, dATP was added to 50 μM, and the mixtures were further incubated for 1.5 h. DNA bound to core particles was digested with Staphylococcus aureus nuclease at a final concentration of 15 U/ml for 1 h at 37°C. The core particles were subsequently digested with proteinase K at 0.5 mg/ml in the presence of 1% SDS for 2 h at 37°C. The samples were then extracted several times with equal volumes of chloroform. Yeast tRNA was added to each sample to a final concentration of 1 mg/ml, and the mixture was passed through a Sephadex G-50 column and precipitated with ethanol. The precipitate was dissolved in TE buffer (10 mM Tris-HCl [pH 8.0], 1 mM EDTA), electrophoresed in a 1% agarose gel, and autoradiographed.
RESULTS
Phosphorylation of the core protein by a kinase present in RAP.
The core protein of human HBV is phosphorylated by PKA and PKC (14) (see Fig. 4). We have recently detected a kinase activity in the RAP (17). To test whether the core protein can be phosphorylated by this kinase, a GST-Core fusion protein construct expressing full-length HBV core protein was made (Fig. 1). The GST-Core fusion protein was purified with glutathione-Sepharose beads and analyzed by SDS-PAGE (Fig. 2A). The GST-Core fusion protein on the beads was incubated with RAP purified from a differentiated human hepatoma cell line HuH-7. After extensive washing, the bound complex was incubated with [γ-32P]ATP in an in vitro kinase assay and the product was analyzed by SDS-PAGE. As shown in Fig. 2B, phosphorylation of the GST-Core protein (lane 2) but not GST alone (lane 1) was detected. Without prior incubation with RAP, no phosphorylation of the GST-Core protein was detected (data not shown). This result indicates that the core protein can interact with a kinase present in RAP and serve as its substrate. We refer to this kinase activity as CAK.
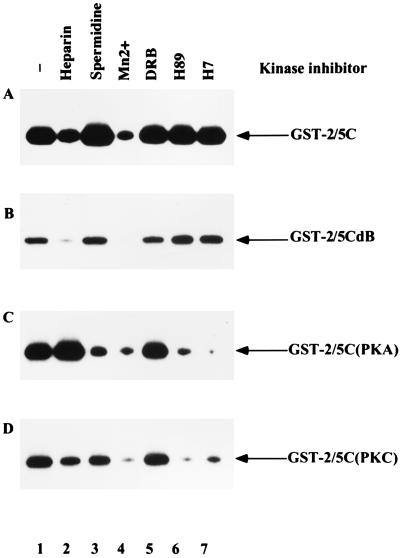
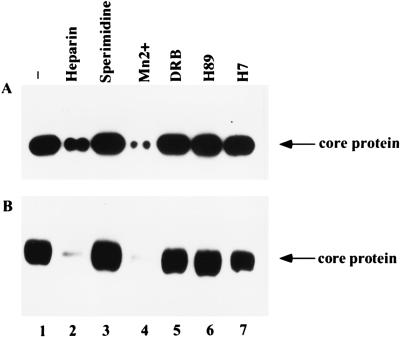
FIG. 4.
Sensitivity of CAK to different kinase inhibitors. Beads with GST-2/5C (A) or GST-2/5CdB (B) were incubated with RAP. After extensive washing, the sensitivity of CAK, adsorbed onto beads coated with GST fusion protein, to different kinase inhibitors was tested. As a control, beads with GST-2/5C were incubated with purified PKA (C) or PKC (D). The inhibitors used in the assay were 25 U of heparin per ml (lane 2), 500 mM spermidine (lane 3), 5 mM manganese (lane 4), 6 mM DRB (lane 5), 100 mM H89 (lane 6), and 100 mM H7 (lane 7). Lane 1 is no inhibitor control.
FIG. 1.
Depiction of wild-type and deletion mutant GST-Core fusion proteins. The core protein has 185 (adw subtype) amino acids. GST-Core contains the full-length core protein. Deletion mutants GST-9/10C and GST-2/5C contain the segments from amino acids 29 to 185 and 119 to 185 of core protein, respectively. GST-2/5CdB is derived from GST-2/5C and contains an internal deletion from amino acids 170 to 177 of the core protein in GST-2/5C. GST-CoreΔ2/5C is derived from GST-Core and contains the segment from amino acids 1 to 117 of the core protein.
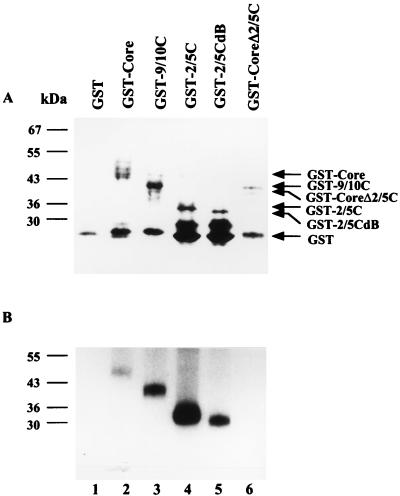
FIG. 2.
Phosphorylation of wild-type and deletion mutant GST-Core proteins by CAK. (A) Expression of GST fusion proteins. The glutathione-Sepharose beads were incubated with GST fusion proteins, extensively washed, and then analyzed by SDS-PAGE (12% polyacrylamide) followed by Coomassie brilliant blue staining. (B) Phosphorylation of GST fusion proteins by CAK. Beads with the same amount of GST fusion proteins as in panel A were incubated with RAP of HuH-7 cells, extensively washed, and subjected to an in vitro kinase assay. The products were analyzed by SDS-PAGE (12% polyacrylamide). Other experimental details are as described in Materials and Methods. Lanes: 1, GST; 2, GST-Core; 3, GST-9/10C; 4, GST-2/5C; 5, GST-2/5CdB; 6, GST-CoreΔ2/5C.
Minimal sequence of the core protein sufficient for phosphorylation by CAK.
Two core protein deletion mutants were constructed to determine the minimal sequence of the core protein which could interact with and be phosphorylated by CAK. GST-9/10C and GST-2/5C contained segments of core protein from amino acids 29 to 185 and from 119 to 185, respectively (Fig. 1). Both GST-9/10C and GST-2/5C proteins bound to glutathione-Sepharose beads could interact with CAK and could also be phosphorylated by CAK (Fig. 2, lanes 3 and 4). We then created an internal deletion from amino acids 170 to 177 of the C-terminal portion of the core protein in the GST-2/5C construct, GST-2/5CdB, by taking advantage of an internal BglII site. Phosphorylation of GST-2/5CdB was also detected (lane 5). However, GST-CoreΔ2/5C, which contained the segment of the core protein from amino acids 1 to 117, could not be phosphorylated by CAK (lane 6). Therefore, the region from amino acids 119 to 185 of the core protein is sufficient for interaction with and phosphorylation by CAK.
Phosphorylation of the core protein by CAK at serine residues.
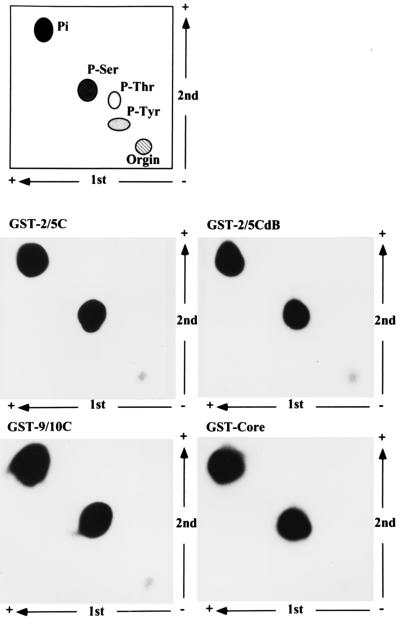
GST-Core, GST-9/10C, GST-2/5C, and GST-2/5CdB were labeled by in vitro kinase reactions with CAK. Phosphorylated GST fusion proteins were eluted from SDS-PAGE gels and subjected to acid hydrolysis. The products were resolved on a two-dimensional thin-layer cellulose plate (bottom four panels of Fig. 3). The migration of unlabeled phosphoserine, phosphothreonine, and phosphotyrosine controls was determined by ninhydrin staining (top panel of Fig. 3). The labeled amino acid from GST-Core, GST-9/10C, GST-2/5C and GST-2/5CdB migrated exclusively with phosphoserine.
FIG. 3.
Phosphorylation of core protein by CAK on serine residue(s). GST-Core, GST-9/10C, GST-2/5C, and GST-2/5CdB labeled by an in vitro kinase assay were eluted from 12% polyacrylamide SDS-PAGE gels and subjected to hydrolysis. Phosphoamino acid analysis was performed in two dimensions as described in Materials and Methods and shown in the bottom four panels. The migration of phosphoamino acid standards visualized by ninhydrin staining is shown in the top panel.
Sensitivity of CAK to a variety of kinase inhibitors.
A variety of kinase inhibitors were tested for their ability to inhibit the phosphorylation of GST-2/5C (Fig. 4A) and GST-2/5CdB (Fig. 4B) by CAK. Heparin, manganese ions, and spermidine are inhibitors of S6 kinase (5). DRB, H89, and H7 are inhibitors of casein kinase II, PKA, and PKC, respectively (3, 11, 31). Inhibitors were added during the kinase assays, and the phosphorylation of GST-2/5C and GST-2/5CdB was measured. As controls, PKA (Fig. 4C) and PKC (Fig. 4D) were incubated with GST-2/5C, which served as a substrate. Our results showed that phosphorylation of both GST-2/5C and GST-2/5CdB by CAK could be inhibited by heparin and manganese ions but not by spermidine, DRB, H89, or H7. CAK appears to be distinct from PKA or PKC since they exhibit different sensitivities to the panel of inhibitors.
Partial purification of CAK.
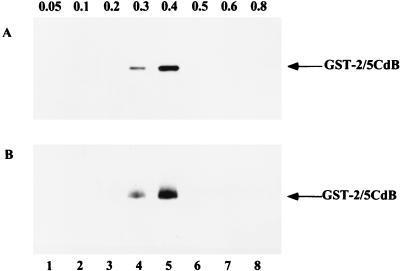
To purify CAK, RAP was fractionated by ion-exchange chromatography by being loaded onto a heparin-Sepharose CL-6B column and step eluted with buffers containing increasing concentrations of KCl. As shown in Fig. 5A, CAK activity was first detected in the fraction with 0.3 M KCl and was completely eluted with buffer containing 0.4 M KCl. When the combined 0.3 and 0.4 M KCl fractions were subjected to a second round of ion-exchange chromatography on a phosphocellulose P11 column, CAK activity was detected in both 0.3 and 0.4 M KCl fractions, as shown in Fig. 5B.
FIG. 5.
Partial purification of CAK. (A) A 20-mg portion of RAP was loaded on a heparin-Sepharose column and eluted with TKM buffer containing increasing concentrations of KCl, from 0.05 to 0.8 M. Each fraction was collected, dialyzed, concentrated, and analyzed for CAK activity. CAK activity was measured by phosphorylation of bead-bound GST-2/5CdB as in Fig. 4. (B) After dialysis, the pooled 0.3 and 0.4 M fractions from heparin-Sepharose column chromatography were loaded onto a phosphocellulose P11 column and eluted with TKM buffer containing 0.05 to 0.8 M KCl. Each fraction was collected, dialyzed, concentrated, and analyzed for CAK activity.
Identification of proteins interacting with core protein.
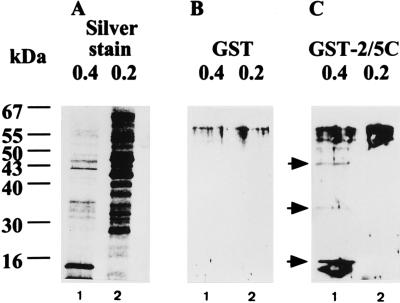
To identify CAK, far-Western assays were performed to search for proteins that could interact with the C-terminal portion of the core protein. The CAK-containing fraction (eluted with 0.4 M KCl; Fig. 6, lanes 1) and CAK-free fraction (eluted with 0.2 M KCl, lane 2) from the P11 column chromatography were subjected to SDS-PAGE, blotted onto nitrocellulose filters, and renatured. These filters were then incubated with GST (Fig. 6B) and GST-2/5C (Fig. 6C), respectively. Proteins that could interact with the GST-2/5C protein were subsequently visualized with antiserum against GST. GST-2/5C, compared with GST, bound to three specific proteins, of 46, 35, and 13 kDa, which were present in the 0.4 M fraction but not in the 0.2 M fraction. These three proteins were present only in the fraction containing CAK activity. This suggested that they were candidates for CAK. The silver-staining pattern of proteins in two fractions is shown in Fig. 6A.
FIG. 6.
Detection of proteins binding to the C-terminal portion of core protein by a far-Western assay. (B and C) Proteins (20 μg) from 0.4 M KCl (lane 1) and 0.2 M (lane 2) KCl eluted fractions from phosphocellulose P11 column chromatography were separated by SDS-PAGE (12% polyacrylamide) and transferred to nitrocellulose filters. After renaturation, these filters were blotted with GST (B) and GST-2/5C (C) first and then incubated with a secondary anti-GST, HRP-conjugated antibody. Proteins capable of binding to GST fusion proteins were visualized by the addition of HRP substrates. (A) Silver-staining pattern of proteins present in each fraction.
CAK is most probably a 46-kDa protein.
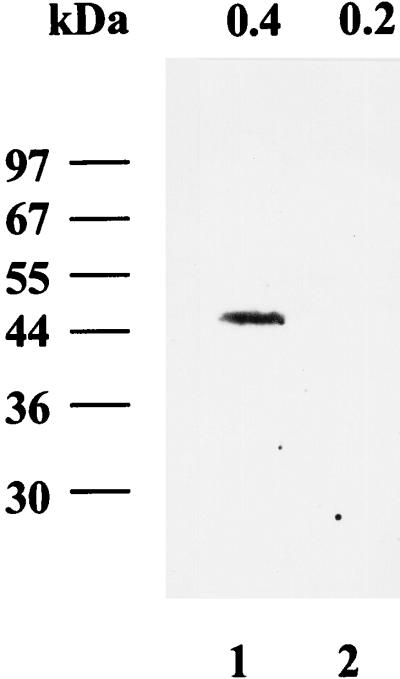
In-gel kinase assays with GST-2/5C as the substrate were performed to determine which protein(s) possessed kinase activity and therefore was likely to be CAK. 0.4 M (Fig. 7, lane 1) and 0.2 M (lane 2) KCl eluted fractions from P11 column chromatography were subjected to SDS-PAGE containing the GST-2/5C protein. Proteins in the gel were denatured, renatured, and then incubated with [γ-32P]ATP under conditions optimized for kinase activity. The gel was then extensively washed, dried, and autoradiographed. As shown in Fig. 7, a 46-kDa phosphorylated signal was present in the 0.4 M KCl fraction (lane 1) but not in the 0.2 M KCl fraction (lane 2). No signal was detected when GST was used as the substrate (data not shown). This result demonstrated that a 46-kDa protein could phosphorylate the C-terminal portion of the core protein (GST-2/5C) and was present only in the fraction where CAK activity was present.
FIG. 7.
Detection of the CAK by an in-gel kinase assay. Proteins (20 μg) from 0.4 M KCl (lane 1) and 0.2 M KCl (lane 2) eluted fractions were separated by SDS-PAGE (12% polyacrylamide) with GST-2/5C protein as the substrate. After denaturation and renaturation, proteins in the gel were incubated with [γ-32P]ATP and tested for kinase activity. After being washed, the protein(s) containing the kinase activity was visualized by autoradiography.
The observations that (i) a 46-kDa protein can bind to the GST-2/5C protein, (ii) a 46-kDa protein can phosphorylate the GST-2/5C protein, and (iii) this 46-kDa protein is present only in the fraction possessing CAK activity indicate that this 46-kDa protein is most probably CAK.
CAK as a candidate for the core particle-associated kinase.
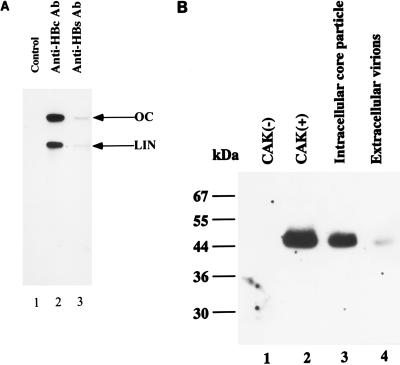
The ability of CAK to bind and phosphorylate the C-terminal portion of the core protein raised the possibility that CAK was the core particle-associated kinase. To address this possibility, we transiently transfected pHBV3.6, which was able to produce intracellular core particles and extracellular 42-nm virions, in the human hepatoma HuH-7 cell line (30). Intracellular core particles (Fig. 8A, lane 2) were isolated with immunoprecipitation from cell lysates by anti-core (anti-HBc) antibody. The extracellular virions (lane 3) were collected with immunoprecipitation from medium by anti-surface (anti-HBs) antibody. The integrity of these core particles and secreted virions was verified by endogenous DNA polymerase assays. These assays measured the incorporation of radioactively labeled deoxynucleotides into incomplete viral genomes by an endogenous reverse transcriptase enclosed in core particles. Both open-circular and linear forms of viral DNA products were detected. This result demonstrated that intracellular core particles and extracellular virions could be produced and harvested in this transient-transfection system.
FIG. 8.
Comigration of the core particle-associated kinase with CAK in an in-gel kinase assay. (A) Production of intracellular core particles and extracellular secreted virions as analyzed by endogenous DNA polymerase activity. Plasmid pHBV3.6 was transfected into HuH-7 cells. The cell lysate and medium were collected 4 days after the transient transfection. Intracellular core particles were prepared by immunoprecipitation of the cell lysate with anti-HBc antibody (Ab) (lane 2). Extracellular virions were collected by immunoprecipitation of medium with anti-HBs antibody (lane 3). The endogenous DNA polymerase activity was then measured as described in Materials and Methods. OC, open circular; LIN, linear. (B) Proteins from the 0.2 M KCl eluted CAK-negative fraction (lane 1), the 0.4 M KCl eluted CAK-positive fraction (lane 2), immunoprecipitated intracellular core particles (lane 3), and extracellular virions (lane 4) were separated by SDS-PAGE (12% polyacrylamide) with GST-2/5C protein as the substrate. An in-gel kinase assay was performed as described in the legend to Fig. 7.
An in-gel kinase assay of intracellular core particles was performed to characterize the core particle-associated kinase. Intracellular core particles were subjected to SDS-PAGE with GST-2/5C protein as the substrate, followed by an in-gel kinase reaction as described above. A 0.4 M KCl CAK-positive fraction (Fig. 8B, lane 2) and a 0.2 M KCl CAK-negative fraction (lane 1) were included as controls. The intracellular core particles contained a 46-kDa kinase, the same molecular mass as CAK, that could phosphorylate the C-terminal portion of core protein (lane 3).
The core particle-associated kinase could also be detected by phosphorylation of the core protein after incubation of the core particles with [γ-32P]ATP (Fig. 9A, lane 1). When core particles were incubated with different inhibitors before the addition of [γ-32P]ATP, core particle-associated kinase appeared to exhibit the same profile of sensitivity toward different inhibitors as CAK did (compare Fig. 9A with Fig. 4A).
FIG. 9.
Sensitivity of core particle-associated kinase to different kinase inhibitors. The activity of the core particle-associated kinase from intracellular core particles either without the proteinase K pretreatment (A) or with the proteinase K (25 ng) pretreatment (B) was assayed by the incorporation of [γ-32P]ATP into core protein. The kinase assay was performed in the absence (lane 1) or presence of kinase inhibitors: 25 U of heparin per ml (lane 2), 500 mM spermidine (lane 3), 5 mM manganese (lane 4), 6 mM DRB (lane 5), 100 mM H89 (lane 6), and 100 mM H7 (lane 7).
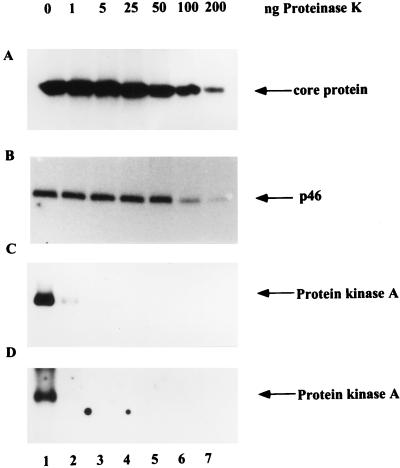
To show that the 46-kDa kinase was truly encapsidated rather than simply adsorbed on the outside of core particles, a protease protection experiment was performed (2). First, isolated intracellular core particles were treated with increasing concentrations of proteinase K to degrade exposed protein. Core particle-associated kinase was then detected by an in-gel kinase assay (Fig. 10B) and by phosphorylation of the core protein after incubation of core particles with [γ-32P]ATP (Fig. 10A). Solubilized PKA, which could phosphorylate GST-2/5C in the in-gel kinase assay (Fig. 10D) and could also autophosphorylate after incubation with [γ-32P]ATP (Fig. 10C), was used as a control. The kinase activity in intracellular core particles could withstand the treatment with 25 ng of proteinase K and showed a gradual decline at higher concentrations (Fig. 10A and B). In contrast, the activity of PKA in solution was completely abolished at 1 ng of proteinase K (Fig. 10C and D). Thus, the 46-kDa kinase was indeed located inside the core particle. In line with the above observation, the core particle-associated kinase in intracellular core particles after treatment with 25 ng of proteinase K still exhibited the same profile of sensitivity toward different inhibitors as did the one without treatment (compare Fig. 9A and B).
FIG. 10.
The activity of core particle-associated kinase is resistant to proteinase K digestion. Purified intracellular core particles (A and B) and solubilized PKA (C and D) were treated with increasing concentrations of proteinase K (1, 5, 25, 50, 100, and 200 ng) and assayed by incorporation of [γ-32P]ATP into core protein (A) and autophosphorylation of PKA (C) or by an in-gel kinase assay with GST-2/5C as the substrate, as described in the legend to Fig. 7 (B and D).
To demonstrate that this 46-kDa kinase is associated with extracellular virions, virions were subjected to SDS-PAGE with GST-2/5C protein as the substrate and an in-gel kinase reaction was performed. The virions indeed contained a 46-kDa kinase that could phosphorylate the C-terminal portion of the core protein (Fig. 8B, lane 4). The core particle-associated kinase activity present in secreted virions could be demonstrated by the incorporation of radioactive labels into core proteins of permeabilized virions after 0.5% Triton X-100 treatment. This core particle-associated kinase from isolated virions exhibited the same profile of sensitivity toward different inhibitors as that of CAK (data not shown).
The above evidence indicates that CAK is a candidate for core particle-associated kinase.
DISCUSSION
In this paper, we present evidence that the C-terminal portion of the core protein can be phosphorylated at its serine residues by a cellular kinase termed CAK. As shown by the in-gel kinase assay, CAK is apparently a 46-kDa protein. The activity of CAK was discovered during our study of another serine kinase in ribosome-associated proteins of cytoplasm (17). Our preliminary study showed that CAK is localized predominantly in RAP of cytoplasm; no CAK activity is detected in the nucleus, mitochondria, endoplasmic reticulum, Golgi complex, or lysosome (data not shown). Our observation that deletion of the last SPRRR motif led to the reduction of the phosphorylation of the C-terminal portion of core protein by CAK is consistent with a recent report that serine residues of three SPRRR motifs are phosphorylated with equal efficiency in vivo (19).
Although PKA and PKC can also phosphorylate serine residues at the C-terminal portion of core protein (14) (Fig. 4), CAK has a profile of sensitivity toward kinase inhibitors quite distinct from those of PKA and PKC. Although it has also been reported that the core protein can be phosphorylated in a cell cycle-dependent manner, the nature and localization of the responsible kinase have not been characterized (19, 28). The possibility that CAK is involved in this cell cycle-dependent phosphorylation of core protein cannot be excluded.
A 46-kDa protein can bind and phosphorylate the GST-2/5C protein in the in-gel kinase experiment. Besides, it is present only in the eluted fraction possessing CAK activity. These results indicated that this 46-kDa protein is most probably CAK. On the other hand, three specific proteins, of 46, 35, and 13 kDa, could interact with the C-terminal part of the core protein in the far-Western experiment. These three proteins were present only in the eluted fraction that contained the CAK activity. These suggested that the 46-kDa protein detected in the far-Western experiment is most probably the 46-kDa CAK detected in the in-gel kinase experiment. The 35- and 13-kDa proteins can also interact with the C-terminal part of the core protein. Whether they form a complex with the 46-kDa CAK requires further study. Since the 46-kDa CAK alone has the kinase activity, neither the 35- nor the 13-kDa protein is required for the kinase activity of the 46-kDa CAK. Our studies (see below) showed that the 46-kDa CAK may be present in the core particles. Whether the 35- and 13-kDa proteins are also present in the core particles needs further study.
A serine kinase, which phosphorylates amino acid residues in the arginine-rich domain in the C-terminal part of the core protein, is present in core particles of human HBV, woodchuck hepatitis virus, and ground squirrel hepatitis virus (1, 6, 9, 24). This encapsidated kinase is apparently of cellular origin (18); its characteristics remain largely unknown. Our study indicates that CAK, a 46-kDa serine kinase found in RAP, is a candidate for the core particle-associated kinase. This conclusion is drawn based upon the following: (i) both CAK and core particle-associated kinase can bind and phosphorylate the C-terminal portion of core protein; (ii) both CAK and core particle-associated kinase are serine kinases; (iii) in the in-gel kinase assay, CAK comigrated with a 46-kDa kinase activity present in both core particles and virions; (iv) this 46-kDa protein is resistant to protease treatment and therefore is encapsidated within core particles; and (v) both CAK and kinase present in core particles and virions display the same profile of sensitivity toward a panel of kinase inhibitors. An earlier report excluded the possibility that PKA or cdc kinases were the core particle-associated kinases (15). A subsequent study showed by immunoblotting that PKC might be present in liver-derived core particles (14). Our study showed, however, that both CAK and core particle-associated kinase were distinct from PKC based upon their molecular weights and sensitivity to kinase inhibitors. The reason for this discrepancy is not clear.
The function of the core particle-associated kinase is unknown. Since the phosphorylation occurs within a region which is important for pregenome encapsidation and conversion of viral RNA pregenome to DNA (10), this kinase may have essential functions in the viral life cycle. Our finding that a 46-kDa kinase is a candidate for the core particle-associated kinase is an important step toward understanding the role of this kinase in the HBV life cycle.
ACKNOWLEDGMENTS
We are grateful to members of Alan Lin’s laboratory (National Yang-Ming University) for technical guidance during the preparation of ribosome and ribosome-associated proteins. We are indebted to Shiuh-Wen Luoh for stimulating suggestions and discussions in the course of these experiments and for critical reading of and invaluable comments on the manuscript. We thank Suh-Der Tsen (National Yang-Ming University) for improving the manuscript.
This study was supported by research grants NSC84-2331-B-010-005MH, NSC85-2331-B-010-027, and NSC86-2314-B-010-036 from the National Science Council, Republic of China.
REFERENCES
- 1.Albin C, Robinson W S. Protein kinase activity in hepatitis B virus. J Virol. 1980;34:297–302. doi: 10.1128/jvi.34.1.297-302.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bartenschlager R, Junker-Niepmann M, Schaller H. The P gene product of hepatitis B virus is required as a structural component for genomic RNA encapsidation. J Virol. 1990;64:5324–5332. doi: 10.1128/jvi.64.11.5324-5332.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chijiwa T, Mishima A, Hagiwara M, Sano M, Hayoshi K, Inoue T, Naito K, Toshioka T, Hidaka H. Inhibition of forskolin-induced neurite outgrowth and protein phosphorylation by a newly synthesized selective inhibitor of cyclic AMP-dependent protein kinase, N-[2-(p-bromocinnamylamino)-ethyl]-5-isoquinolinesulfoamide (H-89), of PC12D pheochromocytoma. J Biol Chem. 1990;265:5262–5272. [PubMed] [Google Scholar]
- 4.Diskstein R, Ruppert S, Tjian R. TAFII-250 is a bipartite protein kinase that phosphorylates the basal transcription factor RAP74. Cell. 1996;84:781–790. doi: 10.1016/s0092-8674(00)81055-7. [DOI] [PubMed] [Google Scholar]
- 5.Erikson E, Maller J L. Purification and characterization of a protein kinase from Xenopus eggs highly specific for ribosomal protein S6. J Biol Chem. 1986;261:350–355. [PubMed] [Google Scholar]
- 6.Feitelson M A, Marion P L, Robinson W S. Core particles of hepatitis B virus and ground squirrel hepatitis virus. J Virol. 1982;43:741–748. doi: 10.1128/jvi.43.2.741-748.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gallina A, Bonelli F, Zentilin L, Rindi G, Muttini M, Milanesi G. A recombinant hepatitis B core antigen polypeptide with the protamine-like domain deleted self-assembles into capsid particles but fails to bind nucleic acids. J Virol. 1989;63:4645–4652. doi: 10.1128/jvi.63.11.4645-4652.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gerlich W H, Goldmann U, Muller R, Stibbe W, Wolff W. Specificity and localization of the hepatitis B virus-associated protein kinase. J Virol. 1982;42:761–766. doi: 10.1128/jvi.42.3.761-766.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hantz O, Fourel I, Buendia B, Baginski I, Trepo C. Specificity of the woodchuck hepatitis virus-associated protein kinase. In: Vyas G N, et al., editors. Viral hepatitis and liver disease. New York, N.Y: Alan R. Liss, Inc.; 1988. pp. 471–475. [Google Scholar]
- 10.Hatton T, Zhou S, Standring D N. RNA- and DNA-binding activities in hepatitis B virus capsid protein: a model for their roles in viral replication. J Virol. 1992;66:5232–5241. doi: 10.1128/jvi.66.9.5232-5241.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hidaka H, Inagaki M, Kawamoto S, Sasaki Y. Isoquinolinesulfonamide, novel and potent inhibitors of cyclic nucleotide dependent protein kinase and protein kinase C. Biochemistry. 1984;23:5036–5041. doi: 10.1021/bi00316a032. [DOI] [PubMed] [Google Scholar]
- 12.Hirsch R C, Lavine J E, Chang L-J, Varmus H E, Ganem D. Polymerase gene products of hepatitis B viruses are required for genomic RNA packaging as well as for reverse transcription. Nature (London) 1990;344:552–555. doi: 10.1038/344552a0. [DOI] [PubMed] [Google Scholar]
- 13.Hutchcroft J E, Anostario M, Harrison M L, Geahlen R L. Renaturation and assay of protein kinases after electrophoresis in sodium dodecyl sulfate-polyacrylamide gels. Methods Enzymol. 1991;200:417–423. doi: 10.1016/0076-6879(91)00157-r. [DOI] [PubMed] [Google Scholar]
- 14.Kann M, Gerlich W H. Effect of core protein phosphorylation by protein kinase C on encapsidation of RNA within core particles of hepatitis B virus. J Virol. 1994;68:7993–8000. doi: 10.1128/jvi.68.12.7993-8000.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kann, M., R. Thomssen, H. G. Kochel, and W. H. Gerlich. 1993. Characterization of the endogenous protein kinase activity of the hepatitis B virus. Arch. Virol. 8(Suppl.):52–62. [DOI] [PubMed]
- 16.Kaplan P, Greenman R, Gerin J, Purcell R, Robinson W. DNA polymerase associated hepatitis B antigen. J Virol. 1973;12:995–1005. doi: 10.1128/jvi.12.5.995-1005.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kau J-H. A candidate core particle-associated kinase interacts with both terminal protein domain of polymerase and core protein of hepatitis B virus. Ph.D. thesis. Taipei, Taiwan: National Yang-Ming University; 1997. [Google Scholar]
- 18.Lanford R E, Notvall L. Expression of hepatitis B virus core and precore antigens in insect cells and characterization of core-associated kinase activity. Virology. 1990;176:222–233. doi: 10.1016/0042-6822(90)90247-o. [DOI] [PubMed] [Google Scholar]
- 19.Liao W, Ou J H. Phosphorylation and nuclear localization of the hepatitis B virus core protein: significance of serine in the three repeated SPRRR motifs. J Virol. 1995;69:1025–1029. doi: 10.1128/jvi.69.2.1025-1029.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lin A. Localization of surface peptide from ribosomal protein L7 on 80S ribosome by biotinylation. FEBS Lett. 1991;287:121–124. doi: 10.1016/0014-5793(91)80030-7. [DOI] [PubMed] [Google Scholar]
- 21.Machida A, Ohnuma H, Tsuda F, Yoshikawa A, Hoshi Y, Tanaka T, Kishimoto S, Akahane Y, Miyakawa Y, Mayumi M. Phosphorylation in the carboxyl-terminal domain of the capsid protein of hepatitis B virus: evaluation with a monoclonal antibody. J Virol. 1991;65:6024–6030. doi: 10.1128/jvi.65.11.6024-6030.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mondelli M, Tedder R S, Ferns B, Pontisso P, Realdi G, Alberti A. Differential distribution of hepatitis B core and e antigens in hepatocytes: analysis by monoclonal antibodies. Hepatology. 1986;6:199–204. doi: 10.1002/hep.1840060208. [DOI] [PubMed] [Google Scholar]
- 23.Nassal M. The arginine-rich domain of the hepatitis B virus core protein is required for pregenome encapsidation and productive viral positive-strand DNA synthesis but not for virus assembly. J Virol. 1992;66:4107–4116. doi: 10.1128/jvi.66.7.4107-4116.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pugh J, Zweidler A, Summers J. Characterization of the major duck hepatitis B virus core particle protein. J Virol. 1989;63:1371–1376. doi: 10.1128/jvi.63.3.1371-1376.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Roossinck M J, Siddiqui A. In vivo phosphorylation and protein analysis of hepatitis B virus core antigen. J Virol. 1987;61:955–961. doi: 10.1128/jvi.61.4.955-961.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Seifer M, Standring D N. A protease-sensitive hinge linking the two domains of the hepatitis B virus core protein is exposed on the viral capsid surface. J Virol. 1994;68:5548–5555. doi: 10.1128/jvi.68.9.5548-5555.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Spedding C. Ribosomes and proteins synthesis. A practical approach. Oxford: IRL Press; 1990. pp. 9–12. [Google Scholar]
- 28.Yeh C T, Wong S W, Fung Y K, Ou J H. Cell cycle regulation of nuclear localization of hepatitis B virus core protein. Proc Natl Acad Sci USA. 1993;90:6459–6463. doi: 10.1073/pnas.90.14.6459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yu M, Summers J. A domain of the hepadnaviral capsid protein specifically required for DNA maturation and virus assembly. J Virol. 1991;65:2511–2517. doi: 10.1128/jvi.65.5.2511-2517.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yuh C H, Chang Y L, Ting L P. Transcriptional regulation of precore and pregenomic RNAs of hepatitis B virus. J Virol. 1992;66:4073–4084. doi: 10.1128/jvi.66.7.4073-4084.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zandomeni R, Zandomeni M C, Shugar D, Weinmann R. Casein kinase type II is involved in the inhibition by 5,6-dichloro-1-β-d-ribofuranosylbenzimidazole of specific RNA polymerase II transcription. J Biol Chem. 1986;261:3414–3419. [PubMed] [Google Scholar]
- 32.Zhou S, Standring D N. Hepatitis B virus capsid particles are assembled from core-protein dimer precursors. Proc Natl Acad Sci USA. 1992;89:10046–10050. doi: 10.1073/pnas.89.21.10046. [DOI] [PMC free article] [PubMed] [Google Scholar]