Abstract
The versatility of cellular metabolism in converting various substrates to products inspires sustainable alternatives to conventional chemical processes. Metabolism can be engineered to maximize the yield, rate, and titer of product generation. However, the numerous combinations of substrate, product, and organism make metabolic engineering projects difficult to navigate. A perfect trifecta of substrate, product, and organism is prerequisite for an environmentally and economically sustainable metabolic engineering endeavor. As a step toward this endeavor, we propose a reverse engineering strategy that starts with product selection, followed by substrate and organism pairing. While a large bioproduct space has been explored, the top ten compounds have been synthesized mainly using glucose and model organisms. Unconventional feedstocks (e.g., hemicellulosic sugars and CO2) and non-model organisms are increasingly gaining traction for advanced bioproduct synthesis due to their specialized metabolic modes. Judicious selection of the substrate-organism-product combination will illuminate the untapped territory of sustainable metabolic engineering.
Introduction
Over the past two centuries, humanity has relied primarily on fossil resources for power generation and chemical production. The one-way process involving the extraction and combustion of fossil resources has led to an ever-increasing accumulation of non-biodegradable and gas waste. A shift toward utilization of renewable feedstocks and sustainable processes is needed. Biotechnology and metabolic engineering allow us to harness a complex network of highly specific biochemical reactions to produce advanced bioproducts, from fuels to materials to agrochemicals to medicine. By engineering microorganisms, we can increase access to these products in a sustainable manner.
A perfect trifecta of substrate, product, and organism is necessary for bioproduct synthesis that maximizes economic and environmental benefits. An integral process of utilizing more abundant and metabolically efficient substrates, synthesizing valuable products, and selecting the organism with specialized metabolic capability would lead to a quantum leap in biotechnology. In this review, we explore a reverse engineering approach to metabolic engineering, starting from selecting a commercially valuable product and an environmentally sustainable substrate to harnessing an organism excelling at bridging the gap between the substrate and the product (Fig. 1). Focusing initially on product-substrate pairings would allow researchers to ponder on pairs of ambitious real-world problems to tackle and invent “two birds one stone” solutions.
Figure 1. Proposed reverse metabolic engineering approach for green chemistry alternatives.
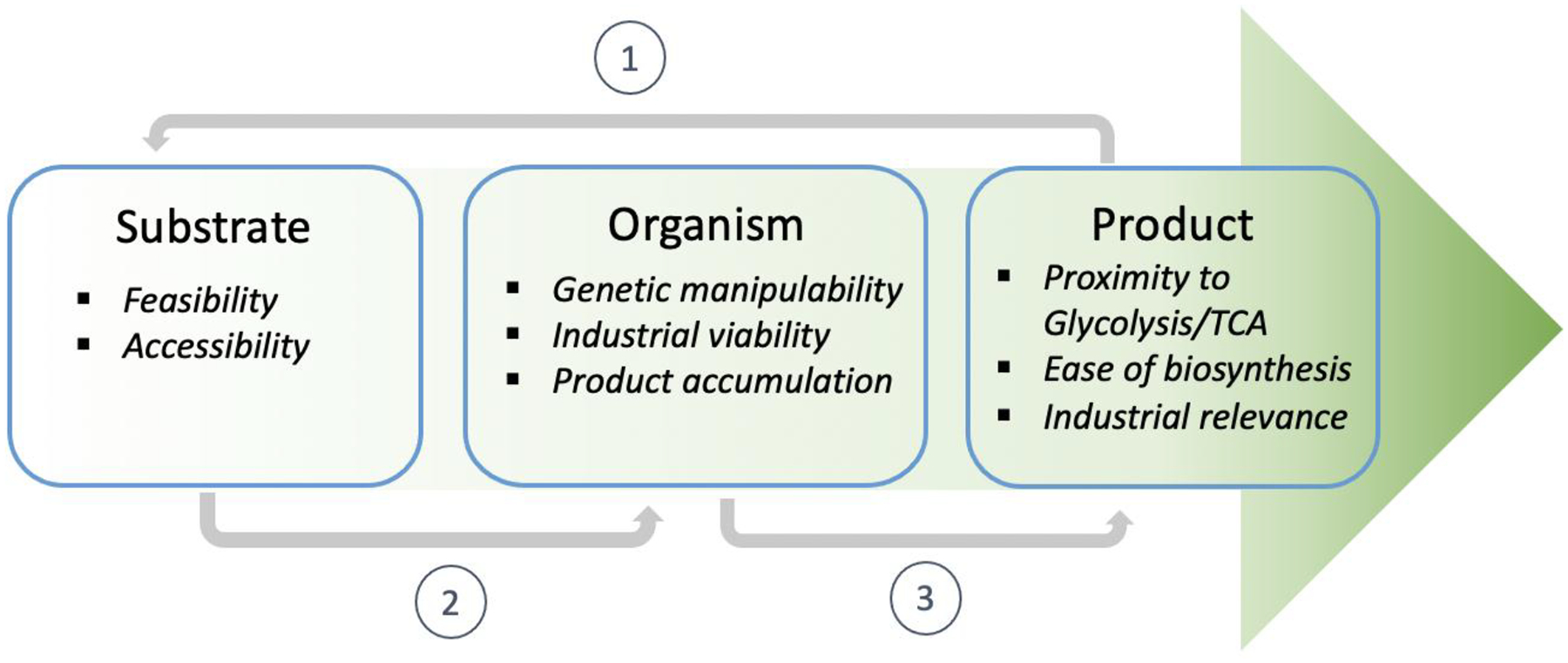
Suitable bioprocesses will maximize synergy between a substrate, organism, and product trifecta. We propose selecting (1) a product and a reasonable desired starting substrate, (2) an organism which can either naturally, or via metabolic engineering, grow on this substrate and (3) engineer the organism to bridge the gap between the substrate and product.
Products
What products are we making?
To explore the product scope of the metabolic engineering landscape, we searched through literature published between 1990 and 2019 using the Web of Science database. “Metabolic engineering” and “synthetic biology” were used as keywords to search for relevant research articles. We then extracted the abstract of the resulting articles and selected only those that reported a titer. All prefiltered abstracts were then manually screened for the microorganism that was used and the product that was synthesized. Articles related to in vitro biocatalysis, co-cultures/consortia, protein production, and biomass production were removed. Following this approach, we compiled a dataset consisting of 566 unique small-molecule products that have been reported. The top ten products account for ~25% of the dataset, suggesting that a large degree of diversity is established for microbial synthesis of chemical compounds (Fig. 2a). Many of the top ten most commonly produced compounds are platform chemicals that are one or two steps removed from glycolysis and the TCA cycle. The compounds proximal to central carbon metabolism lends themselves to biodegradation[1]. To examine any trends in different types of chemicals, we categorized our dataset into commodity chemicals (chemicals produced in bulk for large global markets), specialty chemicals (chemicals used in specific industries such as flavors, cosmetics, cosmetics, and adhesives), fuels, natural products (secondary metabolites including terpenes, polyketides, phenylpropanoids, and alkaloids), and amino acids (standard, nonstandard, and non-proteinogenic). Specialty chemicals were the largest class of products from microbial synthesis (Fig. 2b). The proportion of specialty chemicals is at 29.5%, natural products at 23.4%, commodity chemicals at 21.6%, fuels at 15.8%, and amino acids at 9.7%. While there is no majority within the specific categories, chemicals that are generally considered low value mass production (commodity, specialty, and fuels) make up the majority of bio-produced chemicals. While overall microbial bioproduct synthesis endeavors rapidly increased starting 2010, the proportion of entries between the different categories remains relatively similar (Fig. 2c). This product distribution hinted at the challenges of producing high-margin compounds.
Figure 2. Most produced products ranked by the number of associated entries in the dataset.
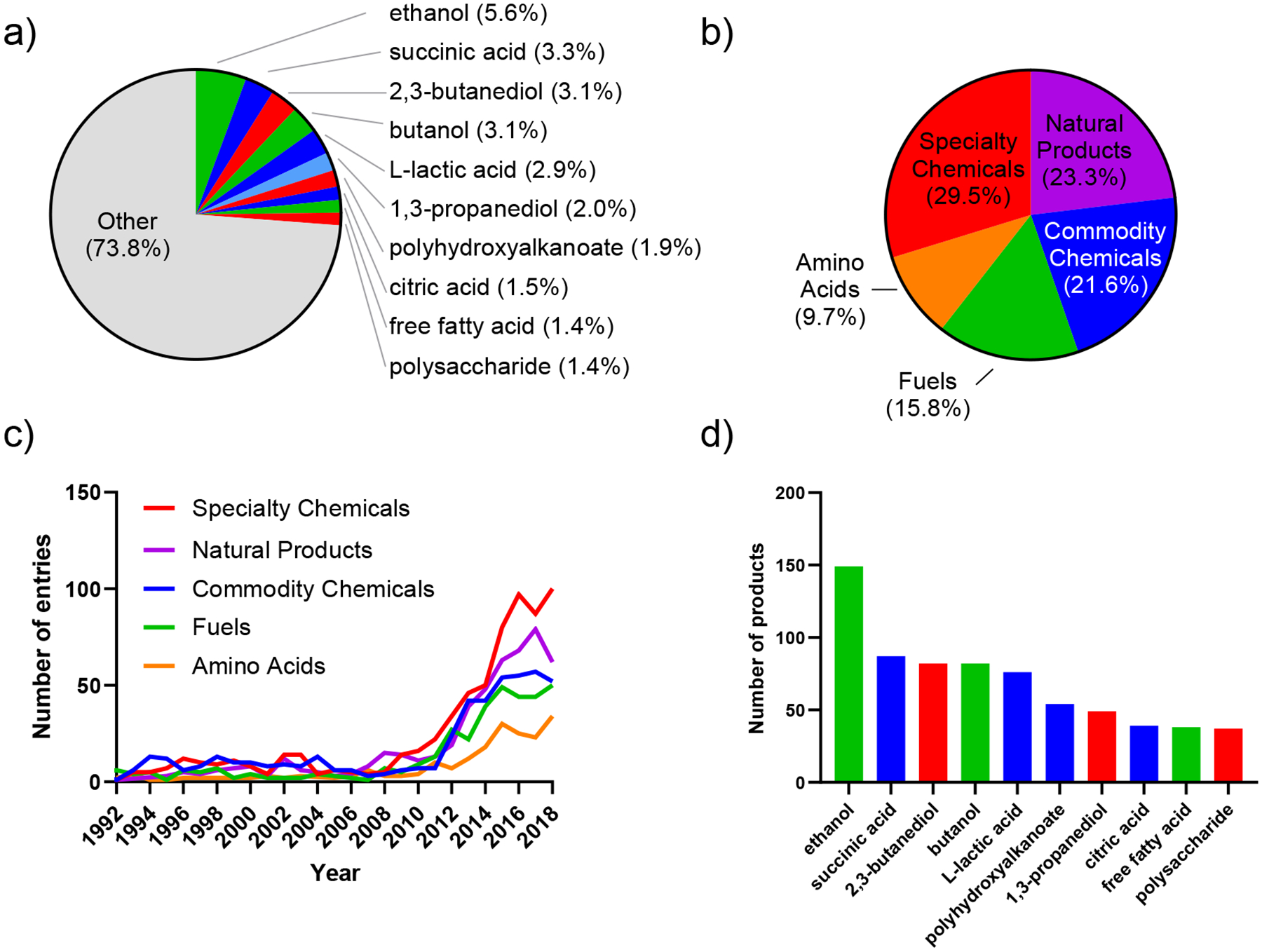
(a) Number of entries associated with each product relative to the total number of entries in the dataset (2,644). Only the top ten are shown. (b) Distribution of the number of entries associated with the five major classes of products according to the dataset. (c) Five major classes over time. Data shown from 1992–2018. (d) Absolute number of entries associated with the top ten products. Database is available on our lab website (https://parklab.ucla.edu/resources.html/).
Top high-volume, low-margin chemicals
While engineering metabolic pathways to generate mass-produced chemicals is relatively straightforward, optimizing the pathways to achieve economically relevant production metrics is challenging and a key area of current research. Since commodity chemicals as well as some fuels and specialty chemicals are typically within a few reaction steps from the TCA cycle and glycolysis, metabolic engineering efforts are focused on downregulating competing pathways while maintaining sufficient growth. While a low-cost feedstock is an important consideration, production of bulk commodities also requires high titer (g/L) for ease of downstream process, high productivity (g/L/hr) for tractable capital expenses, and high yield (g of product/g of substrate) for lowering operating costs.
The top three microbially produced high-volume, low-margin chemicals are ethanol, succinic acid, and 2,3-butanediol (Fig. 2d). Ethanol is used as a fuel and fuel additive as well as in medicine and synthetic chemistry[2,3]. Furthermore, it can be readily derivatized into a variety of commodity chemicals, such as ethylene and ethyl acetate[4]. Ethanol is derived from pyruvate, the end product of glycolysis. Ethanol synthesis pathway is endogenous to many organisms, including the model organisms E. coli and S. cerevisiae[5]. Thus, engineering strategies include overexpressing the endogenous or more efficient heterologous biosynthetic genes, increasing tolerance to ethanol or inhibitors of its synthesis, and increasing or changing substrate preference of the microorganism [6–8]. Non-model organisms may already have these advantages, and thus can also be used for ethanol bioproduction but will require different engineering strategies such as accelerating uptake of alternative substrates or increasing growth[9]. Succinic acid is used as a precursor to 1,4-butanediol, making it important in many industrial fields for solvents and reagents in chemical synthesis and polymers[10], though research has been done for direct biosynthesis of 1,4-butanediol[11]. Being a metabolite in the TCA cycle, it requires no additional enzymes for its biosynthesis. Instead, engineering efforts include removing byproduct formation pathways, enhancing catalytic activity of TCA enzymes, and optimizing energy availability and NADH/NAD+ ratios[12]. 2,3-butanediol can be derivatized to a variety of polyesters, polyurethanes, building blocks for use in chemical industry and cosmetics[13]. Although 2,3-butanediol is derivatized from pyruvate through several reduction steps, not all microorganisms have an endogenous biosynthetic route toward it. This pathway is absent in E. coli and inefficient in wild-type S. cerevisiae[14], which would necessitate incorporating heterologous biosynthetic enzymes. Other strategies include downregulating competing ethanol production and engineering acetoin racemase to produce enantiomerically pure 2,3-butanediol[15].
Top high-margin, low-volume chemicals
Microbial production of high-margin, low-volume chemicals such as natural products for food, agriculture, or pharmaceutical industries poses a different challenge than bulk commodity production because natural products are derived from often long, heterologous secondary metabolic pathways[16]. Natural products in endogenous producers are often produced in low quantities and sometimes only in certain environments[17,18]. Many engineering efforts look to increase the production of the compound in a model organism with more synthetic biology tools available for strain engineering. Thus, the primary focus for natural product synthesis is on incorporating a functional pathway into a host organism. Challenges arise due to issues related to cofactor compatibility, enzyme efficiency in heterologous hosts, and altered energetic demand[19,20]. Though their production metrics (i.e., yield, productivity, and titer) may be lower than that of bulk commodities, this problem is offset by their high-value nature as complex chemicals that are challenging to synthesize. Terpenoids and phenylpropanoids make up the majority of natural products produced (40.7% and 28.6%, respectively), while polyketides and alkaloids account for 19.2% and 11.5% (Fig. 3a). This distribution may be due to the prevalence of more structurally simple terpenoids and phenylpropanoids, while polyketides and alkaloids are often more functionalized. For the more structurally complex and functionalized natural products, metabolic engineering endeavors often reside in the proof-of-concept stage, resulting in fewer publications.
Figure 3. Most produced natural products and categories ranked by the number of associated entries in the dataset.
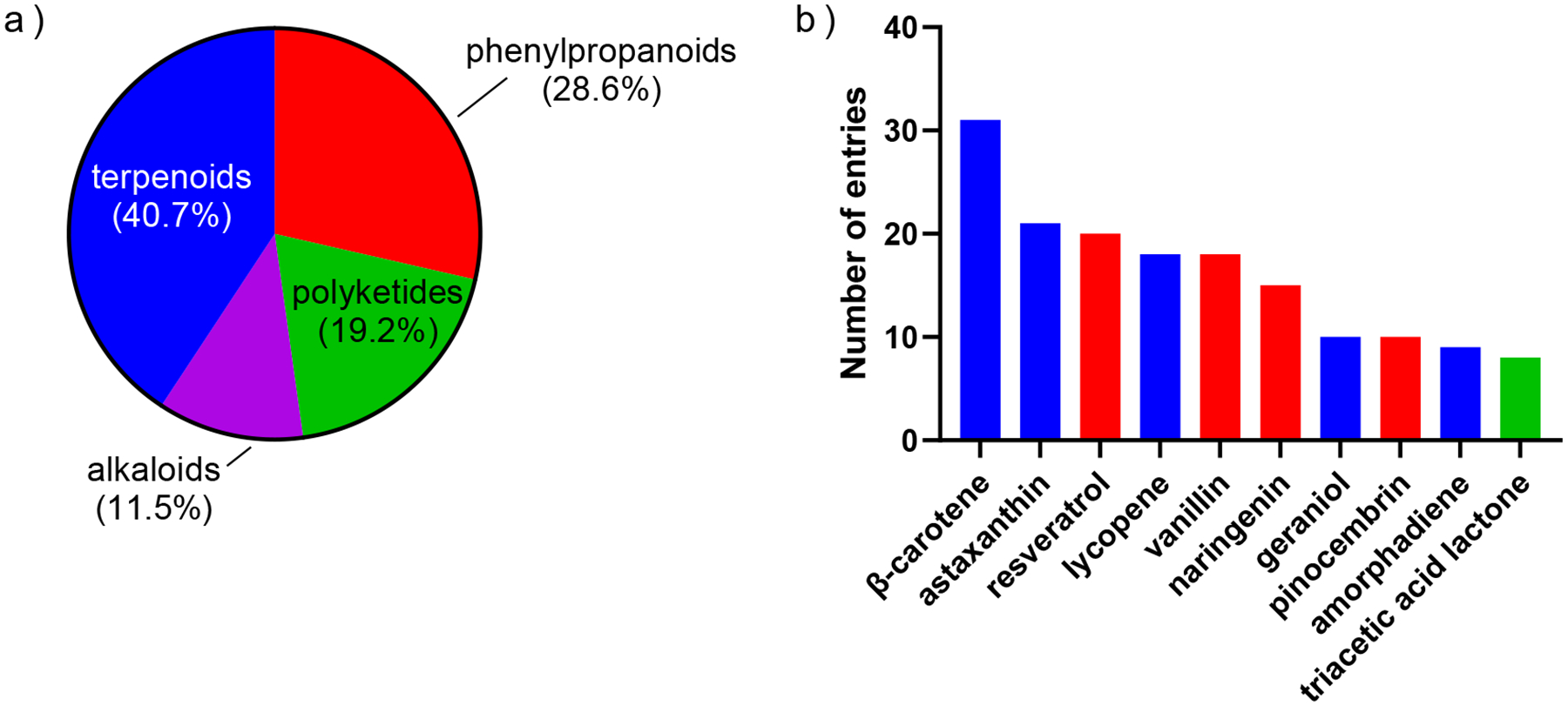
(a) Distribution of the number of entries associated with the four major classes of natural products according to the dataset. (b) Absolute number of entries associated with the top ten natural products.
The three most produced natural products are β-carotene, astaxanthin, and resveratrol (Fig. 3b). β-Carotene is a terpenoid mainly used as a nutritional supplement (as provitamin A) and a food coloring agent[21]. Astaxanthin is a terpenoid commonly used in agriculture and aquaculture as a coloring agent and as a dietary supplement for humans[22]. Common strategies for increasing terpenoid production are incorporating the biosynthetic pathway in a model organism, introducing additional pathways for increased supply of terpene carbon backbones (isopentenyl pyrophosphate and dimethylallyl pyrophosphate), ATP, and NADPH[23–25]. Phenylpropanoid-derived resveratrol is a phytoalexin and antioxidant used a dietary supplement for potential health benefits[26]. Its titer can be increased by tethering biosynthetic enzymes together for metabolite channeling and downregulating competing pathways[27]. The accumulated knowledge of natural product biosynthesis so far lays a solid foundation for future specialty and fine chemicals.
What products can we be making?
With the introduction and advancement of new technologies, new and diverse compounds can be made. Using computer-assisted pathway modeling and rational enzyme engineering, biosynthetic routes can be implemented for new-to-nature chemicals. With improved genome mining and synthetic biology tools, more biosynthetic gene clusters can be discovered and expressed in novel hosts for production of more complex natural products. Microbial synthesis of new-to-nature bulk commodities and complex natural products comes with unique challenges and benefits.
New-to-nature bulk commodities and specialty chemicals
While there are many examples of microbially produced bulk commodity chemicals and fuels, a variety of non-natural chemicals can theoretically be synthesized de novo by derivatization of the intermediates of glycolysis and the TCA cycle. Due to the prevalence of carboxylic acid, ketone, and alcohol functionality in primary metabolites, many synthetic building blocks and solvents have potential to be produced completely through metabolic engineering, reducing the amount of downstream processing. For de novo biosynthesis of non-natural bulk commodities, the difficulty is developing novel synthetic routes and novel enzyme activity to produce a noncanonical metabolite. However, with computer-assisted tools[28,29], moving away from petroleum-based synthetic routes to bio-based ones is a promising future direction. As each bulk commodity poses distinct opportunities for future engineering, understanding the unique challenge associated with each chemical is important for efficient biosynthesis.
Adipic acid is a dicarboxylic acid and is used as a precursor for polymers and plasticizers, most notably nylon[30]. Adipic acid represents a new-to-nature chemical with various noncanonical pathways established in the literature[31]. The synthetic enzymatic routes to adipic acid generally start with the condensation of acetyl-CoA and succinyl-CoA and utilize various reduction steps to yield the final product[32]. Further research can be done to establish biosynthesis in microorganisms with higher precursor availability or a more efficient synthetic route towards adipic acid. 1-octanol is an important primary alcohol in chemical industry, which is often esterified for use in various flavorings and fragrances and is a precursor for polymers and surfactants[33]. As a straight chain alcohol, microbial production requires reduction of octanoyl-ACP or octanoyl-CoA. With a straightforward noncanonical metabolic route, improving 1-octanol synthesis requires the engineering of enzymes for higher specificity and activity. A recent paper by Lozada et al. demonstrates the viability of identifying and optimizing acyl-CoA reductases and synthetases[34]. Levulinic acid is a keto acid that is often used as a platform chemical, being synthesized into polymers, pharmaceuticals, and fragrances[35]. There is no record of microbial production, and thus levulinic acid represents a chemical where computational tools to develop novel routes can be utilized. A recent paper by Vila-Santa et al. provided a systematic framework for constructing synthetic routes in microorganisms and demonstrated its applicability for levulinic acid[36]. In this paper, the optimal route used the natural 3-oxoadipic acid as the precursor, followed by two decarboxylase steps and one methylketone synthase step. Future work can implement and compare the in vivo viability of the various proposed routes. While all three bulk commodities discussed here can potentially be synthesized by derivatizing central carbon metabolites, future metabolic engineering endeavors may result in more efficient microbial production.
Pharmaceutical and agrochemical natural products
The unique benefit of metabolism is its ability to synthesize complex natural products with high specificity. Considering the most commonly produced natural products, one can notice the lack of functionalization, which adds challenging reaction steps to carbon backbone synthesis (Table 1). Furthermore, engineering of entire biosynthetic pathways of complex natural products remains a daunting task with the sheer number of enzymes, though better tools for gene integration and protein engineering would facilitate the creation of microbial strains capable of producing potent natural products. Although many natural products have elucidated biosynthetic pathways, one must consider the length, precursors, and complexity of these routes. For example, strong heterologous oxygenase expression is often difficult to accomplish, as these enzymes can be heavily dependent on correct cellular localization and energy supply[37]. These new advances can help identify and overcome the challenges of de novo biosynthesis.
Table 1. List of commonly made chemicals and potential chemicals to be made.
Pathway length denotes the number of enzymatic steps from the nearest intermediates of central carbon metabolism, where → denotes <5 steps, →→ denotes 5–10 steps, →→→ denotes 10–20 steps, and →→→→ denotes >20 steps. For value, $ denotes <$3/kg, $ $ denotes $3–9/kg, $ $ $ denotes $10–500/kg, $ $ $ $ denotes >$500/kg (prices from Alibaba, Sigma-Aldrich, and Biosynth). For market size, • denotes data not available or <$100 million, •• denotes $100–199 million, ••• denotes $200 million-1 billion, and •••• denotes >$1 billion (data from GrandView Research, Future Market Insights, and StrategyHelix).
| Product | Category | Pathway Length | # of Oxygenases | Value | Market Size |
|---|---|---|---|---|---|
| Ethanol | Fuels | → | 0 | $ | •••• |
| Succinic acid | Bulk commodities | → | 0 | $ | ••• |
| 2,3-butanediol | Fuels | → | 0 | $ | •• |
| β-carotene | Natural products | →→→ | 0 | $ $ $ | ••• |
| Astaxanthin | Natural products | →→→ | 0 | $ $ $ | ••• |
| Resveratrol | Natural products | →→→ | 0 | $ $ $ | •• |
| Adipic acid | Bulk commodities | →→ | 0 | $ | •••• |
| 1-octanol | Fuels | →→ | 0 | $ $ | •••• |
| Levulinic acid | Bulk commodities | → | 0 | $ | • |
| Avermectins | Natural products | →→→→ | 1 | $ $ $ $ | ••• |
| Nodulisporic acids | Natural products | →→→ | 3 | $ $ $ $ | • |
| Pyrrolnitrin | Natural products | →→→→ | 1 | $ $ $ $ | • |
Avermectins are a group of related polyketides found in Streptomyces avermitilis that have potent insecticidal and anthelmintic bioactivity[38]. They are 16-membered macrocyclic lactones that are produced by a Type I polyketide synthase, before further modification and glycosylation. Avermectins thus represent a typical glycosylated macrolide polyketide, posing the challenge of requiring a large and diverse array of enzymes for biosynthesis. To date, the only metabolic engineering efforts have been in S. avermitilis through engineering regulators and increasing precursor supply[39,40]. Thus, research into heterologous expression of the polyketide synthase or tailoring and glycosylation enzymes for semisynthesis or de novo biosynthesis would provide a novel approach. Nodulisporic acid is an indole diterpene natural product derived from Hypoxylon pulicicidum that has potential in pharmaceutical and agricultural industries, with selective toxicity to insects over mammals[41]. As its biosynthesis requires prenylation of an indole ring[42], nodulsporic acid represents a natural product where efficient convergent biosynthesis of two classes of metabolites is needed. No metabolic engineering efforts have been identified, though the recent elucidation of its biosynthesis will hopefully spur investigation into heterologous expression of the involved oxygenases, transferases, and cyclase. Pyrrolnitrin is a halogenated tryptophan-derived alkaloid with potent antifungal activity that was found in various Pseudomonas species[43]. Its four-step biosynthesis from tryptophan consists of two halogenases, a synthase, and an oxygenase[44,45]. Although its biosynthesis is short compared to other natural products, pyrrolnitrin represents a natural product with difficult biocatalytic steps that require enzyme engineering for efficient microbial production. Heterologous expression of the biosynthetic genes has been done[46], but high titers have yet to be reached. The three complex natural products encompass different natural product categories and have unique challenges to overcome in metabolic engineering endeavors.
Substrates
The criteria for an ideal substrate for green chemistry can be grouped under two main ideas: feasibility and accessibility. For specific products, certain substrates will be inherently more efficient and/or bioenergetically favorable. Therefore, the relevant existing or engineered pathways for production will play an important role in process feasibility and substrate selection. Before considering a substrate, either natural pathways to metabolize it should be known or plans for engineered pathways should be well underway. Novel substrates can be used by integrating pathways from one organism into another, or by pre-treating the substrate to convert it to other more well-known starting points for biochemical reactions. Related to the ideas of accessibility, is the idea of scale. To make an impact, the substrate should be abundant and made at scale. To be considered accessible, it should be inexpensive, renewable and/or sourced from waste, not compete with food sources, and not involve complex or energy intensive pre-treatment processes.
Sugars
Different families of organisms utilize different pathways to metabolize nutrients and transform them to biomass and bioproducts. Familiarity with the relevant natural pathways and organisms which possess these pathways is essential to picking an initial starting substrate. The most widely used substrates are sugars such as glucose and sucrose[47–49]. Glucose is catabolized in the glycolysis pathway where it is converted to two molecules of pyruvate as well as cellular energy ATP and NADH. Sucrose is cleaved by invertase and split into fructose and glucose. Sugars are simple and reliable substrates, with abundant natural pathways and organisms to work with.
Unfortunately, the use of sugars directly competes with food security, and the common sources of sugar are non-renewable due to its cultivation being water intensive. Furthermore, sucrose and glucose are not inexpensive starting materials, creating a need for alternate substrates. Plant cell wall and storage polysaccharides can be broken down into monosaccharides. Starch is a storage polysaccharide found in cereal grains and consists of multiple glucose units and is an abundant renewable raw material[50]. Plant detritus, such as lignocellulose, which is the inedible woody parts, represents a large renewable feedstock. Lignocellulose is composed of cellulose, hemicellulose, and lignin. Cellulose is a polymer of glucose, hemicellulose a polymer of various sugars, and lignin has a phenolic backbone[51]. Thus, a spectrum of sugars can be extracted from lignocellulose and used as substrates for fermentation[52]; however, the complex pretreatment and toxic lignin byproducts make it a challenging starting substrate[53–55]. Strategies have been developed to improve strain resistance to these stressors[56,57].
Non-sugar substrates
Alternatively, gluconeogenesis can be used to convert non-sugar substrates (e.g., lactate, glycerol, amino acids, acetate, etc.) to glucose. Among these substrates, acetate is particularly interesting as it can be produced from CO/CO2 and renewably generated H2 with high efficiency[58]. Acetate can be viewed as an accessible and scalable liquid phase intermediate, circumventing the challenges associated with the utilization of renewable gaseous substrates (CO, CO2, H2) by microbes. Numerous studies have demonstrated the feasibility of using acetate as the starting substrate of fermentation[59]. Chen et al. engineered E. coli metabolism to synthesize bioplastics polyhydroxyalkanoates (PHA) from acetate rather than glucose[60]. Bioplastics can be produced from glucose, but the high environmental and economic cost of glucose-derived plastics make them a poor competitor to replace petroleum-based plastics. Their engineering approach included the overexpression of the existing phosphotransacetylase/acetate kinase pathway to improve acetate assimilation, and they further engineered the strain to produce poly-3-hydroxybutyrate (P3HB), poly(3-hydroxybutyrate-co-4-hydroxybutyrate) (P3HB4HB), and poly(3-hydroxybutyrate-co-3-hydroxyvalerate) (PHBV).
The use of acetate is particularly advantageous for fatty acid, terpenoid, and polyketide production. Acetyl-CoA is the initial substrate of their biosynthetic pathways, and it can be generated from acetate without loss of carbon in fewer enzymatic steps than from glucose, which goes through glycolysis and decarboxylation for acetyl-CoA generation. However, the slower microbial growth on acetate is often a drawback, and NADPH generation using acetate is also a major challenge[61]. One way to overcome this issue is to introduce secondary substrates that can be dedicated to generating the limiting factors[62,63].
One-carbon substrates
Use of waste one-carbon (C1) substrates (CO2, CO, formate, methanol, methane) is particularly industrially relevant, since these represent a large percentage of pollutants and greenhouse gases that the industrialized society emits. Both formate and methanol can be produced electrochemically from CO2 with high efficiency and renewable energy. Kim et al. have used a four-step modular engineering approach to develop a linear synthetic pathway, the reductive glycine pathway (rGlyP) which supports E. coli growth on formate or methanol[64]. Their short-term laboratory evolution experiment resulted in cells with improved growth on formate with the promise of further improvement. Attractive alternatives include starting from organisms (e.g., methylotrophs and acetogens) that natively utilize C1 substrates to grow[65,66]. To get the best out of these 1C-utilizing organisms, autotrophic or near-autotrophic growth is desired from the perspective of carbon yield, which often comes at the cost of lower titer and productivity.
Potential and challenges for waste substrate utilization
An attractive benefit of bioprocesses is the ability to convert industrial waste and pollutants into value-added products with fewer separation steps. Engineering approaches such as directed evolution, rational design and modular engineering of novel pathways can be used to reprogram organisms to produce biomass from waste carbon, nitrogen, and hydrogen sourced from the chemical industry[67]. A variety of waste sources can be used to derive the starting sugars, starches, non-carbohydrate precursors and C1 substrates discussed above. 1.3 billion tons of food waste (FW) is produced globally each year[68]. FW is difficult to dispose of safely without polluting groundwater and emitting toxic gases[69]. FW is rich in organic matter which can be converted to value-added products by microbes. CO2 and CO are pollutants emitted by the chemical, steel, energy, and agriculture industries that can serve as carbon substrates. Steel mill flue gas contains CO2, CO and H2 which can be used as feedstock for gas fermentation[70–75].
While waste substrates sound exciting on paper, it can be inherently difficult to source these at the scale needed to transform the industry. For example, the technology to convert waste cooking oil (WCO) to jet diesel is well established; however, the poor recovery rate and lack of infrastructure to properly separate WCO on the restaurant scale makes commercialization difficult[76]. The necessary pretreatment and separation steps required for upcycling waste compounds is also a major challenge.
Processes that utilize a combination of multiple waste sources and abundant renewable sources will be most industrially relevant. More complex processes, such as bioelectrochemical processes[77,78], biorefineries, or co-culturing systems can be combined with metabolic engineering to help us utilize challenging substrates and combine waste streams. Biorefineries improve substrate accessibility by converting biomass from multiple sources into a variety of sugars and then to a range of chemical products via microbial fermentation[79].
Organisms
What organisms have we been using?
Using our compiled database, we explored the variety of organisms used over the last 30 years. The prokaryotic bacteria E. coli and the eukaryotic yeast S. cerevisiae were the organism of choice for nearly half of all the metabolic engineering products made (Fig. 4a). These are model organisms with sequenced genomes and a plethora of available synthetic biology tools[80–82], and their prevalence in academia is paralleled in industry as well[83]. None of the top ten most used organisms are classically recognized as prolific natural product producing organisms (Fig. 4b), perhaps resulting in more bulk commodities being made over natural products. Looking at the history of organism usage, E. coli has always been the most extensively used chassis for metabolic engineering, with its usage increasing more than other organisms during the 2010s (Fig 4c). Furthermore, S. cerevisiae has always been the second most utilized organism, as the eukaryotic model organism counterpart to E. coli, allowing for more complex proteins and higher industrial potential[84]. As new chassis are increasingly utilized with the growth of the metabolic engineering field, the dominance of E. coli has been on the decline since mid-2010s. The rankings of organisms based on the number of unique products mirrored the rankings of most used organisms in the literature with the top five of these rankings being identical (Fig 4d). Unsurprisingly, these model organisms boast high product diversity. The number of unique products seems to reflect the engineerability of the organism.
Figure 4. Most used organisms ranked by the number of associated entries in the dataset.
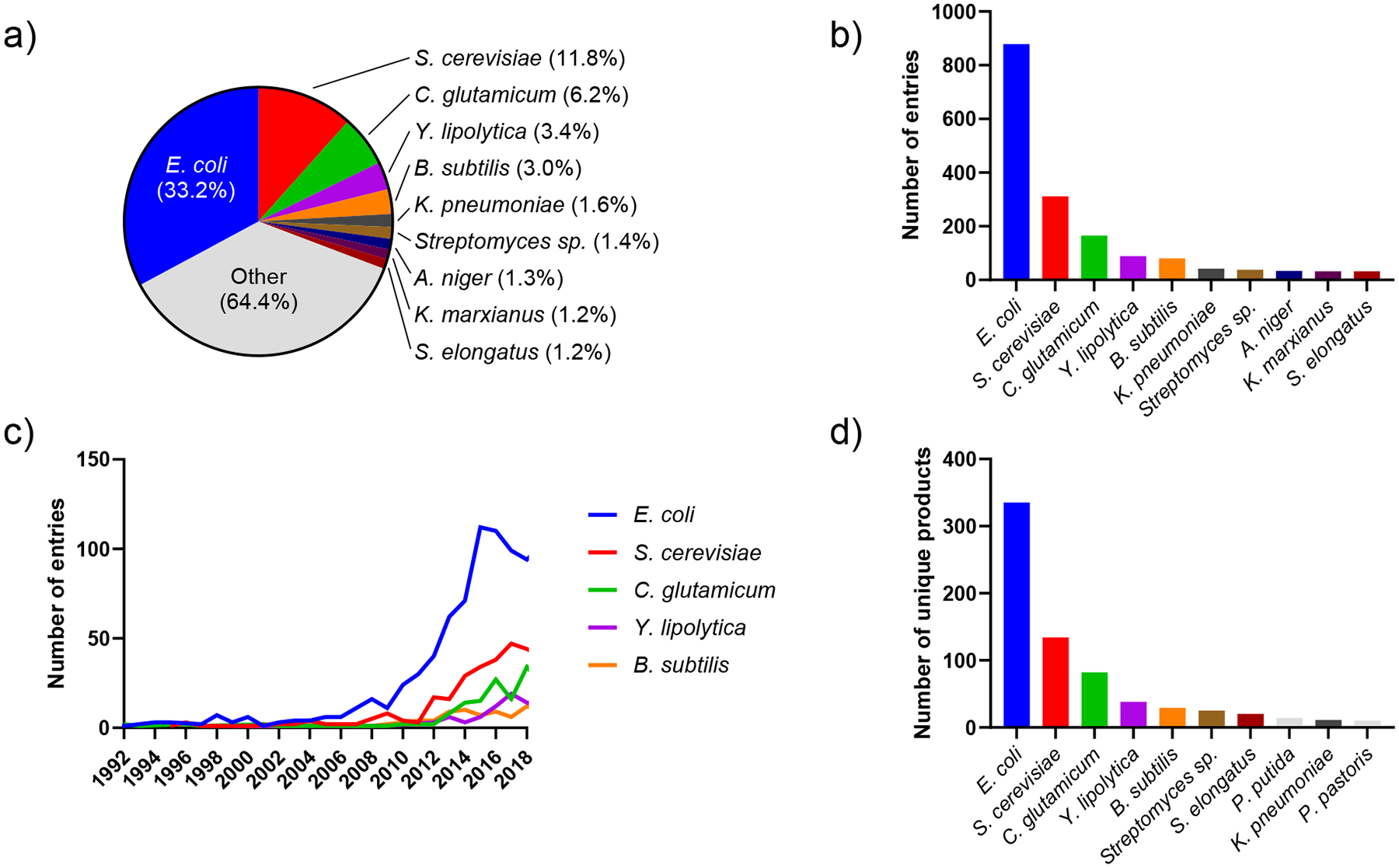
(a) Number of entries associated with each strain relative to the total number of entries in the dataset (2,632). Only the top ten are labelled. (b) Absolute number of entries associated with each strain. (c) Top five most used organisms with number of entries from 1992 to 2018. (d) Top ten most used organisms ranked by the number of unique products.
What considerations go into choosing an organism?
In metabolic engineering, the organism bridges the gap between the starting substrate and the target product. Careful organism selection can alleviate engineering challenges and facilitate industrial implementation, and thus several considerations are warranted. Genetic manipulability allows easier engineering of strains, as many model organisms have large synthetic biology toolboxes. Some organisms such as autotrophic and cellulolytic organisms naturally utilize more desirable substrates. Differences in growth and basal metabolic rate can impact viability, as heterotrophs often grow faster than autotrophs. Growth conditions such as pH, temperature, osmolarity, and nutrient requirements are also important considerations[85]. For example, low temperature yeast fermentation may reduce energy costs while high temperature culturing of thermophiles in warmer regions may reduce potential for contamination[86]. Some organisms are more amenable to product accumulation, such as increased ethanol tolerance in S. cerevisiae and sequestration of hydrophobic compounds to lipid droplets in Yarrowia lipolytica[87,88]. Thus, there is no one-size-fits-all organism, and the advantages and the disadvantages of different biological systems should be carefully weighed for chassis selection.
Model organism advantages
Model organisms for metabolic engineering, namely E. coli and S. cerevisiae, are robust and inexpensive chassis for bioproduction. Their well understood biology, fast growth rates, and availability of different strains and tools propel the large proportion of metabolic engineering projects. They have the benefit of a wide range of available in silico and in vivo tools, highly annotated genomes, and numerous previous efforts to reference[80,81,89,90]. These recent advancements in systems biology, synthetic biology, and pathway engineering make engineering these model organisms more streamlined[91]. With these more sophisticated tools, model organisms have been engineered to make a wide variety of bulk commodities, fuels, and natural products, as over the last twenty years publications about E. coli and S. cerevisiae have covered over 300 and nearly 150 products, respectively (Fig 4d).
Using these model organisms also makes bioprocesses more implementable at a large industry scale since their fermentation conditions are already well established[92]. For example, yeast fermentation plants for ethanol have been implemented in various locations of Brazil and the US[93,94]. Many of industrial fermentation plants make bulk commodities, though industrial precedents for these organisms allow for product scope expansion, as different products will not require major changes in equipment. Combined with their understood physiology and molecular genetics, model organisms are well-suited for scaling up fermentation processes in industry.
Non-model organism advantages
The variety of non-model organisms offers the potential for discovering more beneficial physiological or metabolic traits over model organisms, providing a unique advantage for utilization of unconventional substrates and synthesis of complex natural products. Although the substrate of choice is often glucose for model organisms (without further engineering), the use of other organisms readily unlocks access to a wide range of substrates that are more renewable, such as Clostridium cellulolyticum being able to degrade cellulose or Cupriavidus necator being able to fix CO2[95–97]. Thus, by utilizing the endogenous metabolism of a non-model organism, one can bypass engineering steps to confer alternative substrate utilization and focus on improving product synthesis. Furthermore, certain non-model organisms have inherent metabolic capacity to make advanced product at high metrics (i.e., titer, yield, and productivity). For example, Y. lipolytica is increasingly used as a host for lipophilic terpenoids and carotenoids. Its strong mevalonate pathway flux, large capacity for lipophilic compound storage, and ability to grow to high culture densities make it a better organism for these specific compounds over model organisms[98].
Non-model organisms can endogenously produce various compounds of interest. For example, 2,3-butanediol is natively produced in the generally recognized as safe microorganism Klebsiella oxytoca, where engineering efforts can focus on increasing precursor supply rather than heterologous expression[99]. Less genetic modification provides an advantage in long-term fermentation as it does not require heterologous enzymes and potential expression issues. Episomal-based model organism strains can be unstable over the course of fermentation, as even in minimal or selective media they can lose their plasmid[100]. Thus, finding a non-model organism that is inherently capable of utilizing an environmentally and economically sensible substrate and synthesizing a desirable product and has analogous physiology to a model organism can benefit biotechnology.
Co-culturing systems
Co-cultures allow us to harness synergistic physiology and metabolism of multiple microbes. Co-culturing systems can relieve the metabolic burden from one single organism and accelerate conversion of complex raw materials, as long as both organisms can co-exist[69,101]. Each organism in a co-culture can catabolize a different part of a heterogenous complex substrate. Food waste is a complex combination of organic substrates that is difficult to convert with a single organism. An artificial microbial consortium (AMC) of Bacillus amyloliquefacie and Y. lipolytica has been used for the efficient conversion of FW to lipopeptides[69]. The AMC takes advantage of Y. lipolytica being able to convert oily waste into fatty acids and B. amyloliquefacie to convert starchy waste and fatty acid into lipopeptides. Thus, judicious selection of multiple organisms allows for collective utilization of environmentally and economically sensible substrates and synthesis of desirable products.
Conclusion
With broad spectra of substrates, products, and organisms in nature, careful consideration including technoeconomic analysis of the combinations of these three components of metabolic engineering can lead to a quantum leap in biotechnology by eliciting synergies. With the wide variety of bioproducts that can be synthesized, one must consider the compound’s metabolic demands, the available enzymes and pathways, and how many heterologous or difficult enzymatic steps are required. Different substrates are routed to different metabolic pathways, providing biochemical precursors and cellular energy for bioproduct synthesis and cell proliferation differently. Organisms can help bridge the gap between product and substrate, which can be chosen through consideration of its genetic manipulability, physiology, and metabolism. All three components must be compatible and collectively optimized for sustainable metabolic engineering and biotechnology.
Highlights.
Bulk commodities constitute the majority of microbially produced compounds, but the advancement of metabolic engineering offers the opportunity to produce complex natural products and new-to-nature chemicals.
Non-carbohydrate substrates, one-carbon substrates, and waste chemicals can be upcycled into value-added products by leveraging their utility in efficiently supplying cognate chemical building blocks and energy.
While model organisms dominate the metabolic engineering landscape, non-model organisms confer beneficial metabolic and physiological capabilities for select substrate-product pairs.
Acknowledgements
The authors would like to thank the members of the Park lab, the UCLA Molecular Instrumentation Center, and the UCLA Metabolomics Center for helpful discussion. This work was supported by the National Institute of General Medical Sciences of the National Institutes of Health under Award Number R35GM143127, the National Science Foundation Research Traineeship in Integrated Urban Solutions for Food, Energy, and Water Management (NSF-INFEWS) under Award Number DGE-1735325, the BioPACIFIC Materials Innovation Platform of the National Science Foundation under Award Number DMR-1933487, and the California NanoSystems Institute. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health or the National Science Foundation.
References
• Of special interest
•• Of outstanding interest
- 1.García-Depraect O, Bordel S, Lebrero R, Santos-Beneit F, Börner RA, Börner T, Muñoz R: Inspired by nature: Microbial production, degradation and valorization of biodegradable bioplastics for life-cycle-engineered products. Biotechnol Adv 2021, 53:107772. [DOI] [PubMed] [Google Scholar]
- 2.Pohorecky LA, Brick J: Pharmacology of ethanol. Pharmacol Therapeut 1988, 36:335–427. [DOI] [PubMed] [Google Scholar]
- 3.Logsdon JE: Kirk‐Othmer Encyclopedia of Chemical Technology. 2013, doi: 10.1002/0471238961.0520080112150719.a01.pub2. [DOI] [Google Scholar]
- 4.Dagle RA, Winkelman AD, Ramasamy KK, Dagle VL, Weber RS: Ethanol as a Renewable Building Block for Fuels and Chemicals. Ind Eng Chem Res 2020, 59:4843–4853. [Google Scholar]
- 5.Zaldivar J, Nielsen J, Olsson L: Fuel ethanol production from lignocellulose: a challenge for metabolic engineering and process integration. Appl Microbiol Biotechnol 2001, 56:17–34. [DOI] [PubMed] [Google Scholar]
- 6.Ingram LO, Gomez PF, Lai X, Moniruzzaman M, Wood BE, Yomano LP, York SW: Metabolic engineering of bacteria for ethanol production. Biotechnol Bioeng 1998, 58:204–214. [DOI] [PubMed] [Google Scholar]
- 7.López JM, Duran L, Avalos JL: Physiological limitations and opportunities in microbial metabolic engineering. Nat Rev Microbiol 2022, 20:35–48. [DOI] [PubMed] [Google Scholar]
- 8.Woo JE, Jang Y-S: Metabolic engineering of microorganisms for the production of ethanol and butanol from oxides of carbon. Appl Microbiol Biotechnol 2019, 103:8283–8292. [DOI] [PubMed] [Google Scholar]
- 9.Choi KR, Jiao S, Lee SY: Metabolic engineering strategies toward production of biofuels. Curr Opin Chem Biol 2020, 59:1–14. [DOI] [PubMed] [Google Scholar]
- 10.Nghiem NP, Kleff S, Schwegmann S: Succinic Acid: Technology Development and Commercialization. Ferment 2017, 3:26. [Google Scholar]
- 11.Yim H, Haselbeck R, Niu W, Pujol-Baxley C, Burgard A, Boldt J, Khandurina J, Trawick JD, Osterhout RE, Stephen R, et al. : Metabolic engineering of Escherichia coli for direct production of 1,4-butanediol. Nat Chem Biol 2011, 7:445–452. [DOI] [PubMed] [Google Scholar]
- 12.Vuoristo KS, Mars AE, Sanders JPM, Eggink G, Weusthuis RA: Metabolic Engineering of TCA Cycle for Production of Chemicals. Trends Biotechnol 2016, 34:191–197. [DOI] [PubMed] [Google Scholar]
- 13.Celińska E, Grajek W: Biotechnological production of 2,3-butanediol—Current state and prospects. Biotechnol Adv 2009, 27:715–725. [DOI] [PubMed] [Google Scholar]
- 14.Ng C, Jung M, Lee J, Oh M-K: Production of 2,3-butanediol in Saccharomyces cerevisiae by in silico aided metabolic engineering. Microb Cell Fact 2012, 11:68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Maina S, Prabhu AA, Vivek N, Vlysidis A, Koutinas A, Kumar V: Prospects on bio-based 2,3-butanediol and acetoin production: Recent progress and advances. Biotechnol Adv 2022, 54:107783. [DOI] [PubMed] [Google Scholar]
- 16.Zhang J, Hansen LG, Gudich O, Viehrig K, Lassen LMM, Schrübbers L, Adhikari KB, Rubaszka P, Carrasquer-Alvarez E, Chen L, et al. : A microbial supply chain for production of the anti-cancer drug vinblastine. Nature 2022, doi: 10.1038/s41586-022-05157-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yang D, Park SY, Park YS, Eun H, Lee SY: Metabolic Engineering of Escherichia coli for Natural Product Biosynthesis. Trends Biotechnol 2020, 38:745–765. [DOI] [PubMed] [Google Scholar]
- 18.Krivoruchko A, Nielsen J: Production of natural products through metabolic engineering of Saccharomyces cerevisiae. Curr Opin Biotech 2015, 35:7–15. [DOI] [PubMed] [Google Scholar]
- 19.Li Y, Li S, Thodey K, Trenchard I, Cravens A, Smolke CD: Complete biosynthesis of noscapine and halogenated alkaloids in yeast. Proc National Acad Sci 2018, 115:E3922–E3931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chemler JA, Fowler ZL, McHugh KP, Koffas MAG: Improving NADPH availability for natural product biosynthesis in Escherichia coli by metabolic engineering. Metab Eng 2010, 12:96–104. [DOI] [PubMed] [Google Scholar]
- 21.Mayne ST: Beta‐carotene, carotenoids, and disease prevention in humans. Faseb J 1996, 10:690–701. [PubMed] [Google Scholar]
- 22.Naguib YMA: Antioxidant Activities of Astaxanthin and Related Carotenoids. J Agr Food Chem 2000, 48:1150–1154. [DOI] [PubMed] [Google Scholar]
- 23.Wang L, Liu Z, Jiang H, Mao X: Biotechnology advances in β-carotene production by microorganisms. Trends Food Sci Tech 2021, 111:322–332. [Google Scholar]
- 24.Wan X, Zhou X-R, Moncalian G, Su L, Chen W-C, Zhu H-Z, Chen D, Gong Y-M, Huang F-H, Deng Q-C: Reprogramming microorganisms for the biosynthesis of astaxanthin via metabolic engineering. Prog Lipid Res 2021, 81:101083. [DOI] [PubMed] [Google Scholar]
- ••25.Ma Y, Liu N, Greisen P, Li J, Qiao K, Huang S, Stephanopoulos G: Removal of lycopene substrate inhibition enables high carotenoid productivity in Yarrowia lipolytica. Nat Commun 2022, 13:572. [DOI] [PMC free article] [PubMed] [Google Scholar]; Authors increased lycopene production by diverting flux away from inhibitor production while maintaining product formation. This work demonstrates the importance of considering metabolic regulation while designing engineered organisms with complex pathways
- 26.Frémont L: Biological effects of resveratrol. Life Sci 2000, 66:663–673. [DOI] [PubMed] [Google Scholar]
- 27.Halls C, Yu O: Potential for metabolic engineering of resveratrol biosynthesis. Trends Biotechnol 2008, 26:77–81. [DOI] [PubMed] [Google Scholar]
- 28.Richter F, Leaver-Fay A, Khare SD, Bjelic S, Baker D: De Novo Enzyme Design Using Rosetta3. Plos One 2011, 6:e19230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Song X, Dong M, Liu M: PyMiner: A method for metabolic pathway design based on the uniform similarity of substrate-product pairs and conditional search. Plos One 2022, 17:e0266783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Castellan A, Bart JCJ, Cavallaro S: Industrial production and use of adipic acid. Catal Today 1991, 9:237–254. [Google Scholar]
- 31.Kruyer NS, Peralta-Yahya P: Metabolic engineering strategies to bio-adipic acid production. Curr Opin Biotech 2017, 45:136–143. [DOI] [PubMed] [Google Scholar]
- 32.Skoog E, Shin JH, Saez-Jimenez V, Mapelli V, Olsson L: Biobased adipic acid – The challenge of developing the production host. Biotechnology Advances 2018, 36:2248–2263. [DOI] [PubMed] [Google Scholar]
- 33.Kim Y, Im HB, Jung UH, Park JC, Youn MH, Jeong H-D, Lee D-W, Rhim GB, Chun DH, Lee KB, et al. : Production of linear α-olefin 1-octene via dehydration of 1-octanol over Al2O3 catalyst. Fuel 2019, 256:115957. [Google Scholar]
- 34.Lozada NJH, Simmons TR, Xu K, Jindra MA, Pfleger BF: Production of 1-octanol in Escherichia coli by a high flux thioesterase route. Metab Eng 2020, 61:352–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Klingler FD, Ebertz W: Ullmann’s Encyclopedia of Industrial Chemistry. 2013, doi: 10.1002/14356007.a18_313. [DOI] [Google Scholar]
- ••36.Prather KLJ, Vila-Santa A, Islam MA, Ferreira FC, Mira NP: Prospecting Biochemical Pathways to Implement Microbe-Based Production of the New-to-Nature Platform Chemical Levulinic Acid. ACS Synth Biol 2021, 10:724–736. [DOI] [PubMed] [Google Scholar]; Authors created a systematic framework for in silico design and evaluation of new-to-nature biochemical pathways, demonstrated for de novo production of levulinic acid. The applied use of available computational tools should contribute to design and implementation of novel biosynthetic routes.
- 37.Hu B, Zhao X, Wang E, Zhou J, Li J, Chen J, Du G: Efficient heterologous expression of cytochrome P450 enzymes in microorganisms for the biosynthesis of natural products. Crit Rev Biotechnol 2023, 43:227–241. [DOI] [PubMed] [Google Scholar]
- 38.Fisher MH: Recent advances in avermectin research. Pure Appl Chem 1990, 62:1231–1240. [Google Scholar]
- 39.Robertsen HL, Weber T, Kim HU, Lee SY: Toward Systems Metabolic Engineering of Streptomycetes for Secondary Metabolites Production. Biotechnol J 2018, 13:1700465. [DOI] [PubMed] [Google Scholar]
- 40.Guo J, Zhang X, Chen Z, Wen Y, Li J: Two Adjacent and Similar TetR Family Transcriptional Regulator Genes, SAV577 and SAV576, Co-Regulate Avermectin Production in Streptomyces avermitilis. Plos One 2014, 9:e99224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Meinke P, Smith M, Shoop W: Nodulisporic Acid: Its Chemistry and Biology. Curr Top Med Chem 2002, 2:655–674. [DOI] [PubMed] [Google Scholar]
- •42.Richardson AT, Cameron RC, Stevenson LJ, Singh AJ, Lukito Y, Berry D, Nicholson MJ, Parker EJ: Biosynthesis of Nodulisporic Acids: A Multifunctional Monooxygenase Delivers a Complex and Highly Branched Array. Angew Chem Int Ed 2022, 61:e202213364. [DOI] [PMC free article] [PubMed] [Google Scholar]; Authors characterized a single promiscuous monooxygenase, which serves as a pivotal enzyme for nodulisporic acid congeners. This study opens potential for heterologous production.
- 43.Arima K, Imanaka H, Kousaka M, Fukuta A, Tamura G: Pyrrolnitrin, a New Antibiotic Substance, Produced by Pseudomonas. Agr Biol Chem Tokyo 2014, 28:575–576. [Google Scholar]
- 44.Kirner S, Hammer PE, Hill DS, Altmann A, Fischer I, Weislo LJ, Lanahan M, van Pée K-H, Ligon JM: Functions Encoded by Pyrrolnitrin Biosynthetic Genes from Pseudomonas fluorescens. J Bacteriol 1998, 180:1939–1943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.van Pée K-H, Ligon JM: Biosynthesis of pyrrolnitrin and other phenylpyrrole derivatives by bacteria. Nat Prod Rep 2000, 17:157–164. [DOI] [PubMed] [Google Scholar]
- 46.Hill DS, Stein JI, Torkewitz NR, Morse AM, Howell CR, Pachlatko JP, Becker JO, Ligon JM: Cloning of Genes Involved in the Synthesis of Pyrrolnitrin from Pseudomonas fluorescens and Role of Pyrrolnitrin Synthesis in Biological Control of Plant Disease. Appl Environ Microb 1994, 60:78–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Guo X, Li Z, Wang X, Wang J, Chala J, Lu Y, Zhang H: De novo phenol bioproduction from glucose using biosensor‐assisted microbial coculture engineering. Biotechnol Bioeng 2019, 116:3349–3359. [DOI] [PubMed] [Google Scholar]
- 48.Zhou M, Li Y, Che H, Sun Y, wang S, Zhan Y, Cai D, Chen S: Metabolic Engineering of Bacillus licheniformis for the Bioproduction of Nicotinamide Riboside from Nicotinamide and Glucose. ACS Sustain Chem Eng 2023, 11:6201–6210. [Google Scholar]
- 49.Mohamed ET, Mundhada H, Landberg J, Cann I, Mackie RI, Nielsen AT, Herrgård MJ, Feist AM: Generation of an E. coli platform strain for improved sucrose utilization using adaptive laboratory evolution. Microb Cell Factories 2019, 18:116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tian C, Yang J, Li Y, Zhang T, Li J, Ren C, Men Y, Chen P, You C, Sun Y, et al. : Artificially designed routes for the conversion of starch to value-added mannosyl compounds through coupling in vitro and in vivo metabolic engineering strategies. Metab Eng 2020, 61:215–224. [DOI] [PubMed] [Google Scholar]
- 51.Sanderson K: Lignocellulose: A chewy problem. Nature 2011, 474:S12–S14. [DOI] [PubMed] [Google Scholar]
- 52.Boboescu I-Z, Damay J, Chang JKW, Beigbeder J-B, Duret X, Beauchemin S, Lalonde O, Lavoie J-M: Ethanol production from residual lignocellulosic fibers generated through the steam treatment of whole sorghum biomass. Bioresource Technol 2019, 292:121975. [DOI] [PubMed] [Google Scholar]
- 53.Palmqvist E, Hahn-Hägerdal B: Fermentation of lignocellulosic hydrolysates. II: inhibitors and mechanisms of inhibition. Bioresource Technol 2000, 74:25–33. [Google Scholar]
- 54.Kabel MA, Bos G, Zeevalking J, Voragen AGJ, Schols HA: Effect of pretreatment severity on xylan solubility and enzymatic breakdown of the remaining cellulose from wheat straw. Bioresource Technol 2007, 98:2034–2042. [DOI] [PubMed] [Google Scholar]
- 55.MODIG T, LIDÉN G, TAHERZADEH MJ: Inhibition effects of furfural on alcohol dehydrogenase, aldehyde dehydrogenase and pyruvate dehydrogenase. Biochem J 2002, 363:769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Li W-C, Zhu J-Q, Zhao X, Qin L, Xu T, Zhou X, Li X, Li B-Z, Yuan Y-J: Improving co-fermentation of glucose and xylose by adaptive evolution of engineering xylose-fermenting Saccharomyces cerevisiae and different fermentation strategies. Renew Energ 2019, 139:1176–1183. [Google Scholar]
- 57.Qin L, Dong S, Yu J, Ning X, Xu K, Zhang S-J, Xu L, Li B-Z, Li J, Yuan Y-J, et al. : Stress-driven dynamic regulation of multiple tolerance genes improves robustness and productive capacity of Saccharomyces cerevisiae in industrial lignocellulose fermentation. Metab Eng 2020, 61:160–170. [DOI] [PubMed] [Google Scholar]
- 58.Yoneda N, Kusano S, Yasui M, Pujado P, Wilcher S: Recent advances in processes and catalysts for the production of acetic acid. Appl Catal Gen 2001, 221:253–265. [Google Scholar]
- •59.Perez CMT, Watanabe K, Okamura Y, Nakashimada Y, Aki T: Metabolite Profile Analysis of Aurantiochytrium limacinum SR21 Grown on Acetate-based Medium for Lipid Fermentation. J Oleo Sci 2019, 68:ess19020. [DOI] [PubMed] [Google Scholar]; Authors explored the use of acetate as a substrate for Aurantiochytrium limacium SR21 for fatty acid production. Acetate assimilation rewires metabolism. Growth assays, fatty acid yields and metabolomics studies were used to identify targets to improve fatty acid productivity.
- 60.Chen J, Li W, Zhang Z-Z, Tan T-W, Li Z-J: Metabolic engineering of Escherichia coli for the synthesis of polyhydroxyalkanoates using acetate as a main carbon source. Microb Cell Fact 2018, 17:102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Liu N, Qiao K, Stephanopoulos G: 13C Metabolic Flux Analysis of acetate conversion to lipids by Yarrowia lipolytica. Metab Eng 2016, 38:86–97. [DOI] [PubMed] [Google Scholar]
- ••62.Chen L, Yan W, Qian X, Chen M, Zhang X, Xin F, Zhang W, Jiang M, Ochsenreither K: Increased Lipid Production in Yarrowia lipolytica from Acetate through Metabolic Engineering and Cosubstrate Fermentation. Acs Synth Biol 2021, 10:3129–3138. [DOI] [PubMed] [Google Scholar]; Acetyl-CoA synthetase, acetyl-CoA carboxylase and fatty acid synthetase were overexpressed in Y. lipolytica to produce lipids from acetate. A cosubstrate fed-batch fermentation process was also explored. This demonstrates how the synergy between substrate, organism and product selection as well as more advanced process design choices such as cosubstrate utilization can lead to economic and efficient processes.
- 63.Park JO, Liu N, Holinski KM, Emerson DF, Qiao K, Islam MA, Stephanopoulos G, Woolston BM, Xu J, Lazar Z, et al. : Synergistic substrate cofeeding stimulates reductive metabolism. Nat Metab [date unknown], 1:643–651. [DOI] [PubMed] [Google Scholar]
- 64.Kim S, Lindner SN, Aslan S, Yishai O, Wenk S, Schann K, Bar-Even A: Growth of E. coli on formate and methanol via the reductive glycine pathway. Nat Chem Biol 2020, 16:538–545. [DOI] [PubMed] [Google Scholar]
- 65.Nguyen AD, Park JY, Hwang IY, Hamilton R, Kalyuzhnaya MG, Kim D, Lee EY: Genome-scale evaluation of core one-carbon metabolism in gammaproteobacterial methanotrophs grown on methane and methanol. Metab Eng 2020, 57:1–12. [DOI] [PubMed] [Google Scholar]
- 66.Wiechmann A, Trifunović D, Klein S, Müller V: Homologous production, one-step purification, and proof of Na+ transport by the Rnf complex from Acetobacterium woodii, a model for acetogenic conversion of C1 substrates to biofuels. Biotechnol Biofuels 2020, 13:208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- •67.Lu H, Diaz DJ, Czarnecki NJ, Zhu C, Kim W, Shroff R, Acosta DJ, Alexander BR, Cole HO, Zhang Y, et al. : Machine learning-aided engineering of hydrolases for PET depolymerization. Nature 2022, 604:662–667. [DOI] [PubMed] [Google Scholar]; Authors used a machine learning algorithm to engineer a poly(ethylene terephthalate) (PET) hydrolase with five mutations, which can depolymerize a variety of plastics.
- 68.Uekert T, Dorchies F, Pichler CM, Reisner E: Photoreforming of food waste into value-added products over visible-light-absorbing catalysts. Green Chem 2020, 22:3262–3271. [Google Scholar]
- ••69.Bai S, Qiao B, Hou Z-J, Gao G-R, Cao C-Y, Cheng J-S, Yuan Y-J: Mutualistic microbial community of Bacillus amyloliquefaciens and recombinant Yarrowia lipolytica co-produced lipopeptides and fatty acids from food waste. Chemosphere 2023, 310:136864. [DOI] [PubMed] [Google Scholar]; Authors designed an artificial microbial consortium to use complex food waste for precursors of valuable chemicals. Stable population ratios of Bacillus amyloliquefaciens and recombinant Yarrowia lipolytica were used to convert starch and fat into fatty acids and lipopetides.
- 70.Kato J, Takemura K, Kato S, Fujii T, Wada K, Iwasaki Y, Aoi Y, Matsushika A, Murakami K, Nakashimada Y: Metabolic engineering of Moorella thermoacetica for thermophilic bioconversion of gaseous substrates to a volatile chemical. Amb Express 2021, 11:59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Liew FE, Nogle R, Abdalla T, Rasor BJ, Canter C, Jensen RO, Wang L, Strutz J, Chirania P, Tissera SD, et al. : Carbon-negative production of acetone and isopropanol by gas fermentation at industrial pilot scale. Nat Biotechnol 2022, 40:335–344. [DOI] [PubMed] [Google Scholar]
- 72.Bae J, Song Y, Lee H, Shin J, Jin S, Kang S, Cho B-K: Valorization of C1 gases to value-added chemicals using acetogenic biocatalysts. Chem Eng J 2022, 428:131325. [Google Scholar]
- 73.Debabov VG: Acetogens: Biochemistry, Bioenergetics, Genetics, and Biotechnological Potential. Microbiology 2021, 90:273–297. [Google Scholar]
- 74.Pavan M, Reinmets K, Garg S, Mueller AP, Marcellin E, Köpke M, Valgepea K: Advances in systems metabolic engineering of autotrophic carbon oxide-fixing biocatalysts towards a circular economy. Metab Eng 2022, 71:117–141. [DOI] [PubMed] [Google Scholar]
- 75.Köpke M, Simpson SD: Pollution to products: recycling of ‘above ground’ carbon by gas fermentation. Curr Opin Biotechnol 2020, 65:180–189. [DOI] [PubMed] [Google Scholar]
- 76.Goh BHH, Chong CT, Ge Y, Ong HC, Ng J-H, Tian B, Ashokkumar V, Lim S, Seljak T, Józsa V: Progress in utilisation of waste cooking oil for sustainable biodiesel and biojet fuel production. Energ Convers Manage 2020, 223:113296. [Google Scholar]
- 77.Szydlowski L, Lan TCT, Shibata N, Goryanin I: Metabolic engineering of a novel strain of electrogenic bacterium Arcobacter butzleri to create a platform for single analyte detection using a microbial fuel cell. Enzyme Microb Tech 2020, 139:109564. [DOI] [PubMed] [Google Scholar]
- 78.Vellingiri A, Song YE, Munussami G, Kim C, Park C, Jeon B, Lee S, Kim JR: Overexpression of c‐type cytochrome, CymA in Shewanella oneidensis MR‐1 for enhanced bioelectricity generation and cell growth in a microbial fuel cell. J Chem Technology Biotechnology 2019, 94:2115–2122. [Google Scholar]
- 79.Baptista SL, Costa CE, Cunha JT, Soares PO, Domingues L: Metabolic engineering of Saccharomyces cerevisiae for the production of top value chemicals from biorefinery carbohydrates. Biotechnol Adv 2021, 47:107697. [DOI] [PubMed] [Google Scholar]
- 80.Keseler IM, Collado-Vides J, Gama-Castro S, Ingraham J, Paley S, Paulsen IT, Peralta-Gil M, Karp PD: EcoCyc: a comprehensive database resource for Escherichia coli. Nucleic Acids Res 2005, 33:D334–D337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Botstein D, Chervitz SA, Cherry M: Yeast as a Model Organism. Science 1997, 277:1259–1260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Becker J, Wittmann C: Industrial Biotechnology. 2018, doi: 10.1002/9783527807796.ch6. [DOI] [Google Scholar]
- 83.Buschke N, Schäfer R, Becker J, Wittmann C: Metabolic engineering of industrial platform microorganisms for biorefinery applications – Optimization of substrate spectrum and process robustness by rational and evolutive strategies. Bioresource Technol 2013, 135:544–554. [DOI] [PubMed] [Google Scholar]
- 84.Pompon D, Louerat B, Bronine A, Urban P: [6] Yeast expression of animal and plant P450s in optimized redox environments. Methods Enzymol 1996, 272:51–64. [DOI] [PubMed] [Google Scholar]
- 85.Wu Z-Y, Sun W, Shen Y, Pratas J, Suthers PF, Hsieh P-H, Dwaraknath S, Rabinowitz JD, Maranas CD, Shao Z, et al. : Metabolic engineering of low-pH-tolerant non-model yeast, Issatchenkia orientalis, for production of citramalate. Metab Eng Commun 2023, 16:e00220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Shaw AJ, Podkaminer KK, Desai SG, Bardsley JS, Rogers SR, Thorne PG, Hogsett DA, Lynd LR: Metabolic engineering of a thermophilic bacterium to produce ethanol at high yield. Proc National Acad Sci 2008, 105:13769–13774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Casey GP, Ingledew WMM: Ethanol Tolerance in Yeasts. Crc Cr Rev Microbiol 1986, 13:219–280. [DOI] [PubMed] [Google Scholar]
- 88.Beopoulos A, Chardot T, Nicaud J-M: Yarrowia lipolytica: A model and a tool to understand the mechanisms implicated in lipid accumulation. Biochimie 2009, 91:692–696. [DOI] [PubMed] [Google Scholar]
- 89.Mienda BS, Dräger A: Computational Methods in Synthetic Biology. Methods Mol Biology 2020, 2189:217–229. [DOI] [PubMed] [Google Scholar]
- 90.Chen B, Lee HL, Heng YC, Chua N, Teo WS, Choi WJ, Leong SSJ, Foo JL, Chang MW: Synthetic biology toolkits and applications in Saccharomyces cerevisiae. Biotechnol Adv 2018, 36:1870–1881. [DOI] [PubMed] [Google Scholar]
- 91.Chae TU, Choi SY, Kim JW, Ko Y-S, Lee SY: Recent advances in systems metabolic engineering tools and strategies. Curr Opin Biotech 2017, 47:67–82. [DOI] [PubMed] [Google Scholar]
- 92.Ferrer-Miralles N, Domingo-Espín J, Corchero JL, Vázquez E, Villaverde A: Microbial factories for recombinant pharmaceuticals. Microb Cell Fact 2009, 8:17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Basso LC, Amorim HVD, Oliveira AJD, Lopes ML: Yeast selection for fuel ethanol production in Brazil. Fems Yeast Res 2008, 8:1155–1163. [DOI] [PubMed] [Google Scholar]
- 94.Kramer D: Whatever happened to cellulosic ethanol? Phys Today 2022, 75:22–24. [Google Scholar]
- 95.Tao X, Xu T, Kempher ML, Liu J, Zhou J: Precise promoter integration improves cellulose bioconversion and thermotolerance in Clostridium cellulolyticum. Metab Eng 2020, 60:110–118. [DOI] [PubMed] [Google Scholar]
- •96.Tao X, Morgan JS, Liu J, Kempher ML, Xu T, Zhou J: Target integration of an exogenous β-glucosidase enhances cellulose degradation and ethanol production in Clostridium cellulolyticum. Bioresour Technol 2023, 376:128849. [DOI] [PubMed] [Google Scholar]; Authors increased endogenous cellulose-degrading ability of non-model organism Clostridium cellulolyticum, demonstrating utilization of CRISPR-Cas9 and identification of an integration site for heterologous expression.
- 97.Panich J, Fong B, Singer SW: Metabolic Engineering of Cupriavidus necator H16 for Sustainable Biofuels from CO2. Trends Biotechnol 2021, 39:412–424. [DOI] [PubMed] [Google Scholar]
- 98.Zhu Q, Jackson EN: Metabolic engineering of Yarrowia lipolytica for industrial applications. Curr Opin Biotech 2015, 36:65–72. [DOI] [PubMed] [Google Scholar]
- 99.Park JM, Song H, Lee HJ, Seung D: Genome-scale reconstruction and in silico analysis of Klebsiella oxytoca for 2,3-butanediol production. Microb Cell Fact 2013, 12:20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Hahn-Hägerdal B, Karhumaa K, Larsson CU, Gorwa-Grauslund M, Görgens J, Zyl WH van: Role of cultivation media in the development of yeast strains for large scale industrial use. Microb Cell Fact 2005, 4:31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Lyu X, Zhao G, Ng KR, Mark R, Chen WN: Metabolic Engineering of Saccharomyces cerevisiae for De Novo Production of Kaempferol. J Agr Food Chem 2019, 67:5596–5606. [DOI] [PubMed] [Google Scholar]


