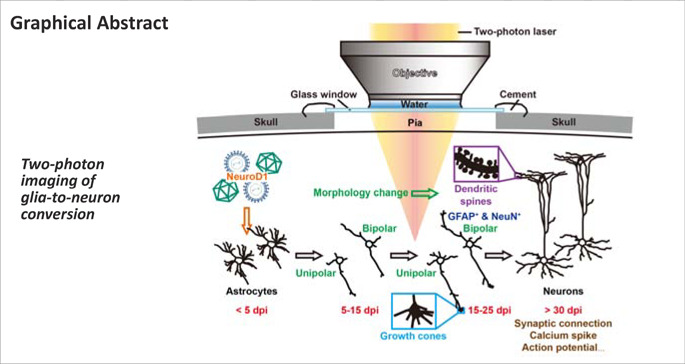
Keywords: astrocyte-to-neuron conversion, Ca2+ imaging, direct lineage conversion, glia, astrocyte; in vivo reprogramming, lineage-tracing mice, NeuroD1, neuron, two-photon imaging
Abstract
Over the past decade, a growing number of studies have reported transcription factor-based in situ reprogramming that can directly convert endogenous glial cells into functional neurons as an alternative approach for neuroregeneration in the adult mammalian central nervous system. However, many questions remain regarding how a terminally differentiated glial cell can transform into a delicate neuron that forms part of the intricate brain circuitry. In addition, concerns have recently been raised around the absence of astrocyte-to-neuron conversion in astrocytic lineage-tracing mice. In this study, we employed repetitive two-photon imaging to continuously capture the in situ astrocyte-to-neuron conversion process following ectopic expression of the neural transcription factor NeuroD1 in both proliferating reactive astrocytes and lineage-traced astrocytes in the mouse cortex. Time-lapse imaging over several weeks revealed the step-by-step transition from a typical astrocyte with numerous short, tapered branches to a typical neuron with a few long neurites and dynamic growth cones that actively explored the local environment. In addition, these lineage-converting cells were able to migrate radially or tangentially to relocate to suitable positions. Furthermore, two-photon Ca2+ imaging and patch-clamp recordings confirmed that the newly generated neurons exhibited synchronous calcium signals, repetitive action potentials, and spontaneous synaptic responses, suggesting that they had made functional synaptic connections within local neural circuits. In conclusion, we directly visualized the step-by-step lineage conversion process from astrocytes to functional neurons in vivo and unambiguously demonstrated that adult mammalian brains are highly plastic with respect to their potential for neuroregeneration and neural circuit reconstruction.
Introduction
Progressive neurodegeneration and neuronal loss are common features of many neurodegenerative diseases, such as Alzheimer's disease and Parkinson's disease. The possibility of replacing degenerated neurons with new, functional neurons to treat these devastating disorders has been studied extensively (Goldman, 2016). However, adult neurogenesis is limited to a few restricted areas in mammalian brains and is particularly sparse in human brains (Boldrini et al., 2018; Sorrells et al., 2018). Transplantation of external stem cells with the potential to generate new neurons has shown some promise in treating specific neurological disorders (Schweitzer et al., 2020; Tao et al., 2021), but challenges such as immunorejection and possible tumorigenesis limit the widespread use of this approach as a therapeutic intervention (Mollinari et al., 2018; Sugaya and Vaidya, 2018).
An alternative to replacing lost neurons is to induce direct glia-to-neuron conversion in vivo via ectopic expression of transcription factors such as neuronal differentiation 1 (NeuroD1; Guo et al., 2014; Matsuda et al., 2019; Wang et al., 2022; Xu et al., 2023), SRY (sex-determining region Y)-box 2 (Sox2; Niu et al., 2013; Heinrich et al., 2014; Su et al., 2014), neurogenin 2 (Ngn2; Grande et al., 2013), distal-less homeobox 2 (Dlx2; Lentini et al., 2021), and achaete-scute homolog 1 (Ascl1; Liu et al., 2015; Torper et al., 2015), or knockdown of RNA-binding protein polypyrimidine tract binding protein 1 (PTBP1; Qian et al., 2020; Jiao et al., 2022). These direct trans-differentiation approaches could potentially be used therapeutically to treat many neurodegenerative disorders and central nervous system (CNS) injuries (Li and Chen, 2016; Barker et al., 2018; Lei et al., 2019; Qian et al., 2021; Qian and Fu, 2021; Bocchi et al., 2022). On the other hand, because these studies were largely carried out in fixed tissues collected from different animal models, many questions remain regarding how a terminally differentiated glial cell can directly transform into a neuronal cell within the intricate three-dimensional structure of the brain. For instance, it remains unclear what the time course is of glia-to-neuron conversion, how a newly generated neuron finds its targets and migrates within the brain, and how a newly converted neuron interacts with its surrounding environment and integrates into pre-existing neural networks.
To address these questions, we employed two-photon live imaging (Fu et al., 2012; Ma et al., 2016; Pilz et al., 2018) to visualize the cell conversion process in the mouse cortex over the course of several weeks, using blood vessels as landmarks to keep track of the cells of interest. Ultimately, we were able to observe the conversion of both reactive astrocytes infected with retrovirus expressing the transcription factor NeuroD1 and lineage-traced astrocytes in Aldh1l1-CreERT2 mice infected with AAV9 GFAP::NeuroD1 into neurons. Our findings unambiguously demonstrate that astrocytes can be directly converted into neurons, which may help resolve the concerns around in situ glia-to-neuron conversion in the CNS raised by a few recent studies reporting a lack of conversion in astrocytic lineage-tracing mice (Wang et al., 2021; Hoang et al., 2022).
Methods
Animals
To avoid the complexity caused by the estrus cycle and the higher cost of female mice, only male mice were used in this study. Approximately 80 6- to 8-week-old male wild-type (C57BL/6J) mice (weight ~21 g, specific pathogen-free) were purchased from Experimental Animal Center of Guangdong Province (Guangzhou, China, animal license No. SYXK (Yue) 2022-0002). B6N.FVB-Tg(Aldh1l1-cre/ERT2)1 Khakh/J or Aldh1l1-CreERT2 transgenic mice (JAX stock# 031008) and B6.Cg-Gt(ROSA)26Sortm14(CAG-tdTomato)Hze/J or Ai14 knock-in mice (JAX stock #007914) were purchased from The Jackson Laboratory (Bar Harbor, ME, USA). All animals were kept under standard laboratory conditions (temperature: 20–24°C, humidity: 50–70%) with a 12/12-hour light/dark cycle and were supplied with standard mouse chow (Guangdong Medical Laboratory Animal Center) and distilled water. All animal procedures were performed according to Animal Research Reporting of In Vivo Experiments (ARRIVE) guidelines (Percie du Sert et al., 2020) and were approved by the Laboratory Animal Ethics Committee at Jinan University (approval No. IACUC-20180330-06) on June 29, 2018.
Viruses
Retroviral vectors and single-stranded adeno-associated viruses (AAVs) were used in this study. The CAG::GFP and CAG::NeuroD1-P2A-GFP retroviruses were generated in our laboratory. The titer of viral particles ranged from 1 × 107 to 1 × 108 viral genomes (vg)/mL. AAV serotype 9 (AAV9) GFAP::GFP, AAV9 GFAP (CMVe)::NeuroD1, and AAV9 GFAP1.6::NeuroD1-P2A-GFP were produced by PackGene® Biotech, LLC (Guangzhou, China). Quantitative PCR was used to validate AAV genome titer. The titer used in this study was 5 × 1011 genome copies (gc)/mL. AAV5 hSyn::GCaMP6s was a gift from Dr. Li Zhang's laboratory at Jinan University.
Thinned-skull cranial window preparation and viral injection
Brain surgery was performed on 4- to 8-week-old mice to prepare them for viral injection. The mice were anesthetized by intraperitoneal injection of 20 mL/kg 1.25% Avertin (Sigma, T48402, St. Louis, MO, USA). Eye ointment (Guangzhou Baiyunshan Pharmaceutical Co., Ltd., Guangzhou, China) was applied to cover and protect the eyes, and the fur was removed from most of the head by shaving. An incision was made in the scalp, and the connective tissue was removed to expose the skull. A small amount of cyanoacrylate glue (New Century Dental, Shanghai, China) was placed around the edge of a head-holding adaptor to hold the skull to an immobilization device (Beijing Xing Lin Biotechnology Co., Ltd., Beijing, China). Then, a drop of artificial cerebrospinal fluid (ACSF) was placed on the surface of the exposed skull for lubrication, and a high-speed microdrill (Microdrill 78001, RWD Life Science, Shenzhen, China) was used to thin a circular area of skull over the mouse neocortex under a dissecting microscope (Carl Zeiss Stemi 305, Oberkochen, Germany). After removing the majority of the spongy bone, skull thinning was continued with a microsurgical blade to obtain a very thin, smooth cranial window. Next, we performed viral injection close to the thinned-skull cranial window using stereotaxic instruments (Model 940, KOPF Instruments, Tujunga, CA, USA), an ultra microsyringe pump (UMP3 UltraMicropPump, World Precision Instruments, Sarasota, FL, USA), and a microprocessor-based controller (Micro4, World Precision Instruments). The injection volume and flow rate were 1 μL and 0.12 μL/min, respectively. After viral injection, the needle was kept in place for about 10 minutes and then slowly withdrawn.
Open-skull cranial window preparation
One day after viral injection, open-skull cranial windows were created in a vertical laminar flow clean bench (SW-CJ-1FD, AIRTECH System Co., Ltd., Hsinchu, Taiwan, China). The mice were anesthetized with Avertin. The head was shaved, and the scalp was washed with iodine tincture and ethanol. A stainless steel head-holding adaptor and cyanoacrylate glue (Beijing Xing Lin Biotechnology Co., Ltd.) were used to immobilize the head. A high-speed microdrill (Microdrill 78001, RWD Life Science, Shenzhen, China) was used to thin the circumference of an area on the skull surface (2–3 mm diameter) until a piece of skull could be lifted up and removed. Immediately afterward, the hole in the skull was covered with a coverslip (Warner Instruments, CS-3R, Hamden, CT, USA) that was attached with Vetbond Tissue Adhesive (3M, 1469SB, St. Paul, MN, USA) and sealed with dental acrylic (Changshu Shang Chi Dental Material Co., Ltd., Changshu, Jiangsu, China). To avoid inducing an inflammatory response, we administered dexamethasone (~2 mg/kg body weight, Rui Cheng Ke Long, Ruicheng, Shanxi, China) by intramuscular injection. The mice were allowed to recover for 7–10 days prior to imaging to improve imaging quality.
In vivo two-photon live imaging
A two-photon microscope (Zeiss LM780, Oberkochen, Germany) equipped with a Ti:Sapphire laser source (120 fs width pulses, 90 MHz repetition rate, Coherent Inc., Santa Clara, CA USA) was used to track individual cells. Each mouse was immobilized on an in vivo imaging platform (Beijing Xing Lin Biotechnology Co., Ltd.) during the long-term, repetitive imaging process. The excitation wavelength was set to 920 nm. Imaging was achieved using a 20× water-immersion objective (N.A. 1.0, Carl Zeiss). Each image was 106.27 μm × 106.27 μm or 212.55 μm × 212.55 μm. The image depth was about 100–700 μm from the dura (layer II–V of the cortex).
Tissue preparation and immunofluorescence staining
Animals were anesthetized with 20 mL/kg 1.25% Avertin and then perfused with normal saline solution (0.9% NaCl), followed by 4% paraformaldehyde (Sigma-Aldrich, St. Louis, MO, USA) to fix the brain. The brains were dissected out and post-fixed in 4% paraformaldehyde overnight at 4°C, and then dehydrated by sequentially soaking in 10%, 20%, and 30% sucrose solutions (Sigma-Aldrich) before being sliced into 20- to 30-μm sections using a cryostat (CryoStar NX50, Thermo Fisher Scientific, Waltham, MA, USA). The coronal brain sections were first pretreated in phosphate-buffered saline (PBS, pH 7.4) for 10 minutes three times, followed by incubation in 5% normal donkey serum (Gibco, Beverly, MA, USA), 3% bovine serum albumin (Sigma-Aldrich), and 0.2% Triton X-100 in PBS for 1 hour. Then, the sections were incubated with primary antibodies diluted in 5% normal donkey serum, 3% bovine serum albumin, and 0.2% Triton X-100 in PBS overnight at 4°C. Next, the samples were washed with 0.2% PBST (0.2% Tween-20 in PBS) and incubated with 4′,6-diamidino-2-phenylindole (dihydrochloride, DAPI) and appropriate secondary antibodies conjugated to Alexa Fluor 488, Alexa Fluor 555, or Alexa Fluor 647 (all 1:1000) for 2 hours at room temperature (~20°C), then washed again with PBST. All information regarding the primary antibodies and secondary antibodies used is shown in Table 1. Finally, the coverslips were mounted onto glass slides with fluorescence mounting medium (VECTASHIELD, Vector Laboratories, Inc., Burlingame, CA, USA). The samples were imaged with a confocal microscope (Zeiss, LSM880) and analyzed with ZEN 2.3 blue edition (Zeiss, LSM880) and ImageJ 1.8.0 (National Institutes of Health, Bethesda, MD, USA) (Schneider et al., 2012).
Table 1.
The information of primary and secondary antibodies
| Host species | Manufacturer | Catalog number | RRID | |
|---|---|---|---|---|
| Primary antibody | ||||
| Monoclonal anti-NeuN | Rabbit | Abcam | ab177487 | AB_2532109 |
| Monoclonal anti-GFAP | Mouse | Sigma | G3893 | AB_477010 |
| Polyclonal anti-GFP | Chicken | Abcam | ab13970 | AB_300798 |
| Monoclonal anti-NeuroD1 | Mouse | Abcam | ab60704 | AB_943491 |
| Polyclonal anti-MAP2 | Chicken | Abcam | ab5392 | AB_2138153 |
| Monoclonal anti-Ctip2 | Rat | Abcam | ab18465 | AB_2064130 |
| Monoclonal anti-Cux1 | Mouse | Abcam | ab54583 | AB_2917999 |
| Polyclonal anti-synapsin1 | Rabbit | Sigma | S193 | AB_261457 |
| Secondary antibody | ||||
| Goat anti-chicken Alexa Fluor 488 | Goat | Invitrogen | A11039 | AB_2534096 |
| Donkey anti-rat Alexa Fluor 488 | Donkey | Invitrogen | A21208 | AB_2535794 |
| Donkey anti-mouse Alexa Fluor 488 | Donkey | Invitrogen | A21202 | AB_141607 |
| Donkey anti-rabbit Alexa Fluor 488 | Donkey | Invitrogen | A21206 | AB_2535792 |
| Donkey anti-rat Alexa Fluor 555 | Donkey | Jackson Immunoresearch | 712-165-150 | AB_2340666 |
| Donkey anti-mouse Alexa Fluor 555 | Donkey | Invitrogen | A31570 | AB_2536180 |
| Donkey anti-rabbit Alexa Fluor 555 | Donkey | Invitrogen | A31572 | AB_162543 |
| Goat anti-chicken Alexa Fluor 647 | Goat | Invitrogen | A21449 | AB_2535866 |
| Donkey anti-mouse Alexa Fluor 647 | Donkey | Invitrogen | A31571 | AB_162542 |
| Donkey anti-rabbit Alexa Fluor 647 | Donkey | Invitrogen | A31573 | AB_2536183 |
| Goat anti-rat Alexa Fluor 647 | Goat | Invitrogen | A21247 | AB_141778 |
Ctip2: COUP-TF-interacting protein 2; Cux1: cut-like homeobox 1; GFAP: glial fibrillary acidic protein; GFP: green fluorescent protein; MAP2: microtubule association protein-2; NeuroD1: neuronal differentiation 1.
The following primary antibodies were used: monoclonal anti-NeuN (1:1000), monoclonal anti-glial fibrillary acidic protein (1:1000), polyclonal anti-green fluorescent protein (1:1000), monoclonal anti-NeuroD1 (1:1000), polyclonal anti-microtubule associated protein 2 (1:2000), monoclonal anti-Ctip2 (1:1000), monoclonal anti-Cux1 (1:500), and polyclonal anti-synapsin1 (1:1000). All information regarding the primary antibodies used is shown in Table 1.
Antibodies and reagents
4-Hydroxytamoxifen (Sigma-Aldrich, Cat# 68392-35-8) was dissolved in ethanol (20 mg/mL) and added to the culture medium at a final concentration of 1 μM. After 24 hours, the culture medium was replaced with normal medium that did not contain 4-hydroxytamoxifen. The antibody information was shown in Table 1.
Sholl analysis
To quantify neurite outgrowth and arborization, we used the Sholl Analysis Plugin designed by the Ghosh lab (Cambridge, MA, USA). Briefly, a series of concentric rings with a diameter difference of 5 or 10 μm radiating from the center of the neuronal soma was overlaid on each microscopy image. Then, the number of neuronal dendritic branches that intersected each of the concentric rings was counted.
Two-photon calcium imaging
We performed two-photon calcium imaging in astrocytic lineage-tracing Aldh1l1-CreERT2 x Ai14 mouse 30 days after AAV9 GFAP (CMVe)::NeuroD1 and AAV5 hSyn::GCaMP6s injection. The mice were immobilized in an in vivo imaging platform (Beijing Xing Lin Biotechnology Co., Ltd.) and dark-adapted for 30 min before imaging. To investigate whether the astrocyte-converted neurons were functionally integrated into the local brain circuitry, we imaged the tdTomato+ neuronal cells infected with AAV5 hSyn::GCaMP6s. The excitation wavelength was set to 920 nm, and images were obtained using a 20× water-immersion objective (N.A. 1.0, Carl Zeiss). The images were 106.27 μm × 106.27 μm, and the sample frequency was set to 7 Hz. The image depth was about 100 μm from the dura (layer II of the cortex).
Patch-clamp recording
Cortical slices were prepared about 1 month after virus injection. The mouse brains were cut into 300-μm-thick coronal slices using a Leica vibratome (Leica VT1000S, Wetzlar, Germany) and ice-cold cutting solution (containing 75 mM sucrose, 85 mM NaCl, 2.5 mM KCl, 0.5 mM CaCl2, 4 mM MgCl2, 24 mM NaHCO3, 1.25 mM NaH2PO4, and 25 mM glucose), and the slices were placed in ACSF containing 126 mM NaCl, 2.5 mM KCl, 1.25 mM NaH2PO4, 26 mM NaHCO3, 2 mM MgCl2, 2 mM CaCl2, and 10 mM glucose. The slices immersed in ACSF were continuously bubbled with 95% O2 and 5% CO2, first at 33°C for 15 minutes, and then at room temperature. Whole-cell recordings were performed using a pipette solution containing 135 mM K-gluconate, 10 mM KCl, 5 mM Na-phosphocreatine, 10 mM HEPES, 2 mM EGTA, 4 mM MgATP, and 0.5 mM Na2GTP (pH 7.3, adjusted with KOH, 290 mOsm/L). The pipette resistance was 3–8 MΩ. The holding potential for the voltage-clamp experiments was –70 mV. Data were collected using pClamp 10.4 software (Molecular Devices, Palo Alto, CA, USA), sampled at 10 kHz and filtered at 1 kHz, and then analyzed with Clampfit software (Molecular Devices).
Statistical analysis
Data are shown as mean ± standard deviation (SD), with only a few exceptions. All statistical analyses were performed in a double-blinded manner. Two-way analysis of variance followed by Sidak's post hoc test or one-way analysis of variance followed by Tukey's post hoc test or two-way analysis of variance followed by Bonferroni post hoc multiple comparison test were applied using GraphPad Prism (version 7.0.0 for Windows, GraphPad Software, Boston, MA, USA, www.graphpad.com).
Results
The step-by-step conversion of proliferating reactive glial cells into neurons in the adult mouse neocortex observed by two-photon live imaging
While many studies have used immunostaining to show that overexpression of neural transcription factors in glial cells can convert them into neurons (Grande et al., 2013; Niu et al., 2013; Guo et al., 2014; Heinrich et al., 2014; Su et al., 2014; Liu et al., 2015; Torper et al., 2015; Gascón et al., 2016; Matsuda et al., 2019; Qian et al., 2020; Lentini et al., 2021; Xiang et al., 2021), the precise conversion process has yet to be observed using a more direct imaging approach. To investigate the in situ glia-to-neuron conversion process, we visualized glial cells overexpressing the transcription factor NeuroD1 (Guo et al., 2014) in the mouse somatosensory cortex for several weeks using two-photon live imaging. All of the two-photon images were acquired through a permanent open-skull cranial window (Figure 1A) (Holtmaat et al., 2009). In order to keep track of the cells of interest during consecutive imaging sessions, blood vessels on the surface of the mouse cortex were used as landmarks to relocate previously imaged areas (Figure 1A) (Holtmaat et al., 2009; Yang et al., 2010). Because retroviruses only express target genes in dividing cells like proliferating glial cells, but not non-dividing cells such as neurons (Katz et al., 2005), we used the retroviral vector CAG::NeuroD1-P2A-GFP to achieve glial cell-specific expression of NeuroD1 in the mouse cortex. The dividing glial cells transfected with the CAG::NeuroD1-P2A-GFP retrovirus in the mouse cortex were actually quite rare (< 1%), consistent with a previous report of only a small population of dividing glial cells in adult mouse brains (Ge and Jia, 2016). Since retroviral expression of NeuroD1 under the chicken beta-actin (CAG) promoter was very strong, we observed a rapid conversion process that occurred over 7–9 days. The control retrovirus CAG::GFP infected dividing glial cells as expected, and virtually all GFP-expressing glial cells retained normal glial morphology throughout the imaging period (Figure 1B). In contrast, many glial cells infected with the CAG::NeuroD1-P2A-GFP retrovirus exhibited rapid morphological changes during the first 3–9 days post injection (dpi) (Figure 1C and D). More specifically, a NeuroD1-overexpressing glial cell would retract some of its glial processes and extend one, or occasionally two, long axon-like neurites to adopt a neuron-like morphology within 7–9 days (Figure 1C and D). Following two-photon live imaging, brain samples were fixed and immunostained to confirm that many of those neuron-like cells were indeed NeuN+ neurons by 10–12 dpi (Figure 1E and F). Moreover, quantification of the retrovirus-infected cells from the two-photon microscopy images revealed that ~40% of the NeuroD1-GFP-expressing cells displayed neuronal morphology with long neurites by 5–7 dpi, and >70% by 7–9 dpi, while only a small percentage retained glial morphology (Figure 1G, n = 10 animals, Ftime (2, 36) = 77.41, P < 0.0001, Fgroup (1, 18) = 139.1, P < 0.0001). Lastly, quantification of the retrovirus-infected cells immunostained with an anti-NeuN antibody after two-photon imaging showed that ~30% of the NeuroD1-GFP-expressing glial cells had converted into NeuN+ neurons by 5–7 dpi, and ~60% by 7–9 dpi (Figure 1H, n = 10 animals, Ftime (2, 36) = 50.25, P < 0.0001, Fgroup (1, 18) = 116, P < 0.0001). These results suggest that at least 60% of the dividing glial cells transfected with the retrovirus CAG::NeuroD1-P2A-GFP eventually converted directly into NeuN+ neurons in adult mouse brains. Thus, the in situ glia-to-neuron conversion process was observed in real time in this study, and our results demonstrate unambiguously that proliferating reactive glial cells can be directly converted into neurons by NeuroD1 overexpression in the adult mouse cortex.
Figure 1.
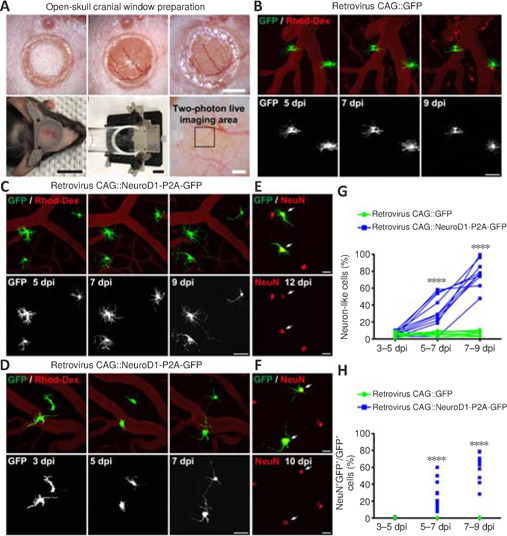
Two-photon imaging of the direct conversion of proliferating glial cells to neurons following retrovirus-mediated NeuroD1 overexpression in the adult mouse cortex.
(A) Permanent cranial window preparation, and design of the head immobilization device. Blood vessels on the cortical surface were used as landmarks (black box) to relocate previously imaged areas. Scale bars: 1 mm, 1 cm, 2 cm, 500 μm (from upper left to lower right). (B) Two-photon time-lapse images illustrating the lack of major morphological or positional changes of the control glial cells infected with the CAG::GFP retrovirus. Blood vessels were labeled with rhodamine-conjugated dextran (Rhod-Dex). Scale bars: 50 μm. (C, D) Two-photon time-lapse images illustrating rapid glia-to-neuron conversion induced by the CAG::NeuroD1-P2A-GFP retrovirus within approximately 1 week of infection. Note that, by 7–9 dpi, some converted neurons had already retracted many short lamellate branches and extended long, curved axons. During the same period, some converting cells began migrating away from their original positions (D). Blood vessels were labeled with rhodamine-conjugated dextran. Scale bars: 50 μm. (E, F) Following two-photon live imaging, the mouse brains were fixed and immunostained with the neuronal marker NeuN. Some of the morphologically neuron-like cells shown in C and D were indeed NeuN-positive by 10–12 dpi, confirming their neuronal identities. Arrows indicate NeuN+ cells. Scale bars: 20 μm. (G) Quantification of the percentage of morphologically neuron-like cells at different time points after retroviral infection. The percentage increased continuously over 5–9 dpi with NeuroD1 overexpression, while the percentage stayed unchanged, at ~5%, with GFP overexpression alone. Two-way analysis of variance followed by Sidak's post hoc test, n = 10 animals, ****P < 0.0001, Ftime (2, 36) = 77.41, Fcolumn factor (1, 18) = 139.1. (H) Quantification of the percentage of NeuN+ GFP+ cells among all GFP+ cells at different time points after retroviral infection. The percentage increased continuously over 5–9 dpi with NeuroD1 overexpression, while virtually no NeuN+ GFP+ cells were apparent with GFP overexpression alone. Two-way analysis of variance followed by Sidak's post hoc test, n = 10 animals, ****P < 0.0001, Ftime (2, 36) = 50.25, Fcolumn factor (1, 18) = 116. dpi: Day(s) post infection; GFP: green fluorescent protein; NeuroD1: neuronal differentiation 1.
To validate that proliferating astrocytes, a major subtype of proliferating glial cells involved in gliosis (Buffo et al., 2008), can be directly converted into neurons by NeuroD1 overexpression, we examined NeuroD1-induced astrocyte-to-neuron conversion in cultured proliferating astrocytes isolated from astrocytic lineage-tracing mice generated by crossing Aldh1l1-CreERT2 mice (Srinivasan et al., 2016) with Rosa-CAG-LSL-tdTomato (Ai14) mice. In the presence of 4-hydroxytamoxifen in the culture medium, the lineage conversion process took place even more rapidly in vitro than in the mouse brain, with some lineage-traced astrocytes already became tdTomato+ neuron-like cells with extended neurites at 4 dpi (Figure 2A). In fact, many tdTomato+ astrocytes had been converted into NeuN+ and MAP2+ (neuron-specific cytoskeletal protein microtubule-associated protein 2) neurons after retroviral expression of NeuroD1 by 6–8 dpi (Figure 2B), suggesting that many of the lineage-converting reactive glia cells we observed in the mouse cortex are proliferating reactive astrocytes.
Figure 2.
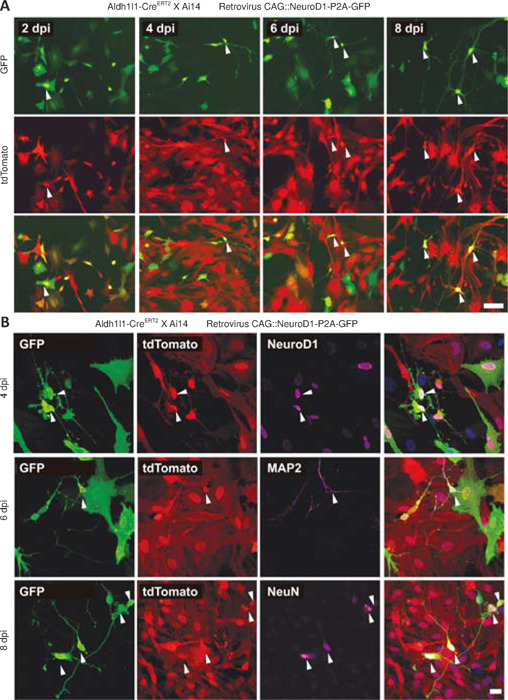
Direct in vitro conversion of astrocytes isolated from astrocytic lineage–tracing mice to neurons following retrovirus-mediated NeuroD1 overexpression.
(A) Representative fluorescence images illustrating rapid astrocyte-to-neuron conversion in cultured lineage-traced astrocytes induced by infection with the CAG::NeuroD1-P2A-GFP retrovirus from 2–8 dpi after the addition of 4-hydroxytamoxifen to the culture medium. Note that more and more retrovirus-infected tdTomato+ astrocytes gradually adopted neuron-like morphology with extended neurites over time (arrowhead). Scale bar: 50 μm. (B) Representative confocal images showing immunostaining for NeuroD1, MAP2, and NeuN (magenta, Alexa Fluor 647) in tdTomato+ astrocytic lineage-tracing cells overexpressing NeuroD1. Note the expression of NeuroD1 in tdTomato+ lineage-tracing cells at 4 dpi and the expression of the neuron-specific markers MAP2 and NeuN in newly converted tdTomato+ cells at 6–8 dpi, confirming their neuronal identities (arrowhead). Scale bar: 20 μm. dpi: Day(s) post infection; GFP: green fluorescent protein; MAP2: microtubule association protein-2; NeuroD1: neuronal differentiation 1.
The step-by-step conversion of lineage-traced astrocytes into neurons in adult mouse neocortex observed by two-photon live imaging
Little astrocyte proliferation occurs in healthy or mildly damaged brains (Sofroniew and Vinters, 2010), so retroviral vectors can only infect a very small portion of astrocytes. In order to monitor the in situ astrocyte-to-neuron conversion process in the vast majority of astrocytes in the mouse cortex, we utilized astrocytic lineage-tracing mice created by crossing Aldh1l1-CreERT2 mice with Ai14 mice. After injecting tamoxifen to induce Cre-mediated recombination, around 90–95% of astrocytes were labeled with tdTomato for lineage-tracing purpose (Additional Figure 1 (2.2MB, tif) ). Then, we employed AAV9 GFAP(CMVe)::NeuroD1 to overexpress NeuroD1 in astrocytes in the mouse cortex under the control of the astrocyte-specific promoter GFAP with a CMV enhancer (CMVe). In the vicinity of the AAV injection sites (~2 mm3), around 75% of the tdTomato+ lineage-traced astrocytes were transfected with AAV9 GFAP::GFP or AAV9 GFAP(CMVe)::NeuroD1, as demonstrated by immunostaining for GFP and NeuroD1 (Additional Figure 2 (3.1MB, tif) ). Two-photon microscopy showed that the tdTomato-traced astrocytes infected with AAV9 GFAP::GFP exhibited few changes in morphology or position over 4 weeks of imaging, consistent with the typical lack of astrocyte migration in the adult mouse brain (Figure 3A; Tsai et al., 2012). AAV-mediated NeuroD1 expression induced a gradual transformation of tdTomato-traced astrocytes from astrocytic morphology to neuronal morphology, with distinct long neurites from 7–27 dpi (Figure 3B). Most NeuroD1-overexpressing tdTomato+ astrocytes initially displayed typical stellate morphology (Figure 3B, 7 dpi), but some quickly lost fine processes and started to sprout neurite-like processes ~2 weeks after infection (Figure 3B, 13–25 dpi). Importantly, these lineage-traced astrocytes never retracted all of their processes, but rather retained several major processes that directly elongated into neurites during conversion (Figure 3B, arrowheads). After two-photon imaging, we performed immunostaining to verify the identity of the tdTomato+ cells in the mouse cortex with or without NeuroD1 infection (Figure 3C and Additional Figure 3 (4.7MB, tif) ). As expected, the tdTomato+ cells in the contralateral hemisphere of the brain without NeuroD1 overexpression were all NeuN- astrocytes (Additional Figure 3A, C (4.7MB, tif) and E (4.7MB, tif) ). In contrast, in the AAV9 GFAP(CMVe)::NeuroD1-injected hemisphere, many tdTomato+ cells had acquired neuronal morphology and stained positive for NeuN by 30 dpi (Figure 3C (4.7MB, tif) and Additional Figure 3B (4.7MB, tif) , D (4.7MB, tif) , F (4.7MB, tif) ), suggesting that these cells were originally astrocytes that had been converted into neurons. Interestingly, immunostaining also showed AAV9 GFAP1.6::NeuroD1-GFP-infected transitional-stage cells with both neuronal and astrocytic properties that stained positively for both GFAP and NeuN (Additional Figure 4 (2.6MB, tif) ). Moreover, many AAV9 GFAP1.6::NeuroD1-GFP-infected GFP+ cells expressed another neuron-specific cytoskeletal protein, MAP2, in their dendrites and cell bodies at 60 dpi (Additional Figure 5A (3.9MB, tif) ). Some of these newly converted neurons acquired an upper cortical layer identity (layers II–IV, cortical neuron marker Cux1+), while others acquired a deeper cortical layer identity (layers V–VI, cortical neuron marker Ctip2+) at 60 dpi (Additional Figure 5B (3.9MB, tif) ). Furthermore, quantification of the lineage-traced astrocytes from the two-photon images revealed that > 6% more tdTomato+ astrocytes in the NeuroD1-expressing hemisphere had adopted neuronal morphology compared with the control hemisphere by 19–21 dpi, and > 10% more by 25–27 dpi (Figure 3D, n = 10 animals, Ftime (3, 54) = 134.3, P < 0.0001, Fgroup (1, 18) = 132.2, P < 0.0001). In addition, quantification of the lineage-traced astrocytes immunostained with a NeuN antibody after two-photon imaging showed that consistently more tdTomato+ astrocytes in the NeuroD1-expressing hemisphere had been converted into NeuN+ neurons compared with the control hemisphere (Figure 3E, n = 10 animals, Ftime (3, 54) = 152.6, P < 0.0001, Fgroup (1, 18) = 78.14, P < 0.0001). A simple calculation indicated that approximately 15% of the lineage-traced astrocytes transfected with AAV9 GFAP(CMVe)::NeuroD1 would eventually be directly converted into NeuN+ neurons in adult astrocytic lineage-tracing mouse brains. Collectively, these results show that lineage-traced astrocytes in the adult mouse cortex can be directly reprogrammed into neurons by 4 weeks of NeuroD1 overexpression, with clear transitional stages (gradually losing glial morphology while adopting neuronal morphology), as observed by continuous two-photon live imaging.
Figure 3.
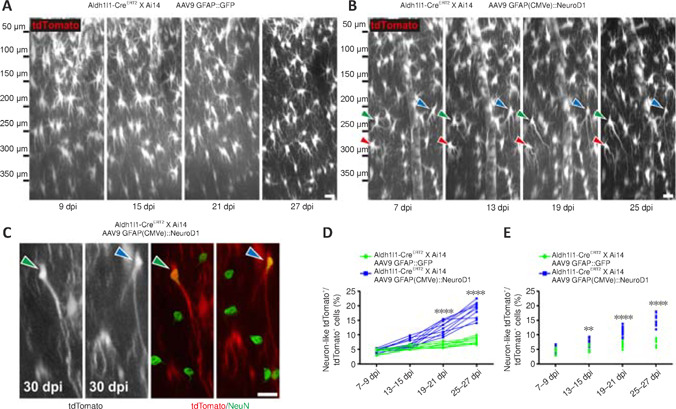
Two-photon imaging of the direct conversion of lineage-traced astrocytes into neurons following AAV-mediated NeuroD1 overexpression in adult lineage-tracing mice.
(A) Two-photon time-lapse images illustrating the lack of morphological or positional changes of tdTomato+ astrocytes in Aldh1l1-CreERT2 X Ai14 mice infected with AAV9 GFAP::GFP. Original Z-stack images were resliced along the X–Z plane to obtain a lateral view of the mouse cortex (pia/top, ventricle/bottom). Scale bar: 20 μm. (B) Two-photon time-lapse images illustrating astrocyte-to-neuron conversion of some tdTomato+ lineage-traced astrocytes (each colored arrowhead points to a specific converting cell) induced by AAV9 GFAP(CMVe)::NeuroD1 in Aldh1l1-CreERT2 X Ai14 mice within 4 weeks of infection. By 19–25 dpi, some converted neurons had retracted many short lamellate branches and projected long, thin axons toward the lateral ventricle. Note that the cells indicated by arrowheads were not the only ones undergoing lineage conversion in these images. Original Z-stack images were resliced along the X–Z plane to obtain a lateral view of the mouse cortex (pia/top, ventricle/bottom). Scale bar: 20 μm. (C) Following two-photon imaging, the mouse brains were fixed and immunostained with the neuronal marker NeuN. Some of the morphologically neuron-like lineage-traced tdTomato+ cells shown in B (green/blue arrowheads) were NeuN+ (green, Alexa Fluor 488) by 30 dpi, confirming their neuronal identities. Scale bar: 20 μm. (D) Quantification of the percentage of morphologically neuron-like tdTomato+ cells among all lineage-traced tdTomato+ cells at different time points after AAV infection. The percentage increased continuously over 19–27 dpi with NeuroD1 overexpression, while the percentage remained statistically unchanged at ~6% with GFP overexpression alone. Two-way analysis of variance followed by Sidak's post hoc test, n = 10 animals, ****P < 0.0001, Ftime (3, 54) = 134.3, Fcolumn factor (1, 18) = 132.2. (E) Quantification of the percentage of NeuN+ tdTomato+ cells among all lineage-traced tdTomato+ cells at different time points after AAV infection. The percentage increased continuously over 13–27 dpi with NeuroD1 overexpression, while the percentage remained statistically unchanged at ~5% with GFP overexpression alone. Two-way analysis of variance followed by Sidak's post hoc test, n = 10 animals, **P < 0.01, ****P < 0.0001, Ftime (3, 54) = 152.6, Fcolumn factor (1, 18) = 78.14. dpi: Day(s) post infection; GFAP: glial fibrillary acidic protein; GFP: green fluorescent protein; NeuroD1: neuronal differentiation 1.
Growth cone dynamics, cellular migration, structural maturation, and integration of the glia-converted neurons revealed by two-photon live imaging
After infection with the retroviral vector CAG::NeuroD1-P2A-GFP, we often observed growth cones at the distal end of newly elongated neurites (Figure 4A, white boxes), reminiscent of newly generated neurons during early brain development. Two-photon images of the newly converted neurons taken every 12 hours suggested that these growth cones were highly dynamic, as they exhibited both extension and retraction (Figure 4A, insets on the right). These observations indicate that the newly converted neurons in the adult mouse cortex extended growth cones to actively explore their local environment. More excitingly, during long-term two-photon imaging, we serendipitously observed the migration of converting cells in the mouse cortex. While endogenous astrocytes in the adult mouse brain rarely migrate due to the formation of gap junctions among adjacent astrocytes, some cells infected with the CAG::NeuroD1-P2A-GFP retrovirus displayed clear migration during time-lapse imaging (Figure 4B). As shown in Figure 4B, the NeuroD1-GFP-expressing cells at different time points were pseudo-colored to track their relative positions, and the images were then overlaid. The arrowheads in the figure indicate a NeuroD1-GFP-expressing cell that exhibited tangential somal translocation between 4 and 8 dpi (Figure 4B). Similarly, when we constructed vertical stacks of two-photon images of NeuroD1-GFP-expressing cells, we found that some of the converting cells also exhibited vertical somal translocation (Figure 4C). Quantification of the migration velocity demonstrated that the converting cells expressing NeuroD1 migrated ~5–8 μm per day, while the non-converting glial cells remained in place during the same time period (Figure 4D, n = 10 animals, Ftreatment (3, 36) = 72.59, P < 0.0001). Together, these data suggest that, like neuronal differentiation and migration during early brain development, the astrocyte-converted neurons can also migrate within the mouse cortex, possibly to find suitable target locations.
Figure 4.
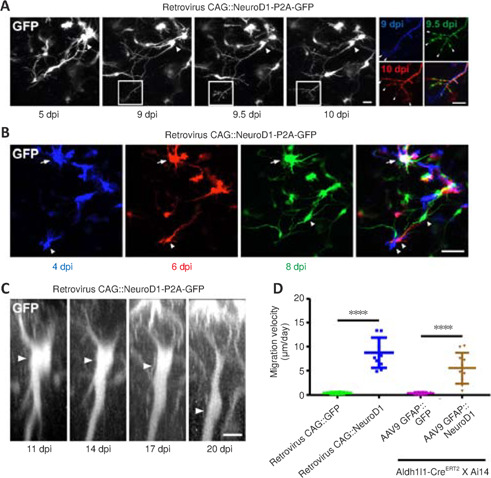
Dynamic growth cones and somal migration during astrocyte-to-neuron conversion, as observed by two-photon live imaging.
(A) Two-photon time-lapse images illustrating a newly converted neuron (arrowhead) extending a long axon-like neurite with multiple growth cones (white box). Note the retraction and extension of the growth cones (arrowheads) within 24 hours (right insets). The same group of growth cones in the white boxes at 9, 9.5, and 10 dpi is pseudocolored blue, green, and red, respectively, and then overlaid to highlight growth cone dynamics over time. Scale bars: 20 μm, 10 μm (insets). (B) Two-photon time-lapse images showing tangential migration of a lineage-converting cell (arrowhead). The same group of cells infected with the CAG::NeuroD1-P2A-GFP retrovirus at 4, 6, and 8 dpi was pseudocolored blue, red, and green, respectively, and then overlaid to highlight tangential somal translocation over time. A typical non-migrating cell is indicated with an arrow. Scale bar: 50 μm. (C) Two-photon time-lapse images showing vertical migration of a lineage-converting cell after AAV9 GFAP1.6::NeuroD1-P2A-GFP infection. Note the vertical somal translocation (arrowhead) from 11–20 dpi. Original Z-stack images were resliced along the X–Z plane to obtain a lateral view of the mouse cortex (pia/top, ventricle/bottom). Scale bar: 10 μm. (D) Quantification of cell migration velocity after viral infection. Note that the proliferating cells and lineage-traced cells overexpressing NeuroD1 migrated at average velocities of ~8 and ~6 μm/day, respectively. Meanwhile, the proliferating or lineage-traced astrocytes overexpressing GFP alone did not migrate at all. One-way analysis of variance followed by Tukey's post hoc test, mean ± SD, n = 10 animals, ****P < 0.0001, Ftreatment (3, 36) = 72.59. dpi: Day(s) post infection; GFAP: glial fibrillary acidic protein; GFP: green fluorescent protein; NeuroD1: neuronal differentiation 1.
To determine how fast the neurons induced to convert from glial cells by in situ reprogramming sent out neurites within a pre-existing neural circuit, we examined the time course of neurite outgrowth during astrocyte-to-neuron conversion in the mouse cortex via two-photon live imaging. Figure 5A illustrates a number of traced examples showing the major branches of cells infected with AAV9 GFAP::GFP (red), AAV9 GFAP1.6::NeuroD1-P2A-GFP (blue), or the CAG::NeuroD1-P2A-GFP retrovirus (green) at different time points following viral injection. Astrocytes expressing GFP alone projected many major branches that were relatively short; in contrast, converting cells expressing NeuroD1-GFP showed fewer branches, with one or two very long processes (Figure 5A). To quantify the major branch patterns of the converting cells, we employed Sholl analysis to track the number of major branches at different distances from the center of the soma. Endogenous astrocytes displayed ~10 major branches within a radius of 20 µm. In contrast, AAV9 GFAP1.6::NeuroD1-P2A-GFP-infected cells projected ~6 major branches at 7–9 dpi, which dropped further to < 3 at 19–21 dpi. Along with the retraction of astrocytic processes, the converting cells also sent out a few long branches that kept growing longer after 2–3 weeks of NeuroD1 overexpression (Figure 5B, n = 20 cells, Fgroup (3, 76) = 6.59, P = 0.0006). Similarly, proliferating cells infected with the CAG::NeuroD1-P2A-GFP retrovirus had extended their neurites significantly inside the mouse cortex after ~1 week of NeuroD1 overexpression (Figure 5C, n = 20 cells, Fgroup (2, 57) = 8.16, P = 0.0257). Based on the average neurite length data we collected over a defined period, the average neurite outgrowth speed of the converting cells within a well-established mouse brain circuit was estimated to be around 4 to 40 µm/day. In addition, many newly converted neurons generated an increasing number of dendritic spines after ~4–8 weeks of NeuroD1 overexpression (Figure 5D and E). The spine density was about 0.6 spine per µm for newly converted neurons originating from proliferating glial cells by 26 dpi, and about 1 spine per µm for newly converted neurons originating from lineage-traced astrocytes by 60 dpi (Figure 5F, n = 10 animals, Ftreatment (3, 36) = 33.86, P < 0.0001).
Figure 5.
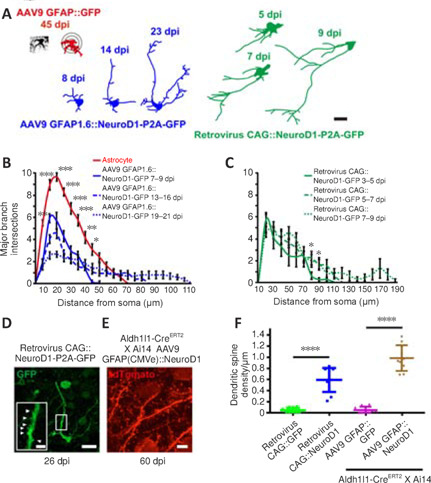
Neurite outgrowth and dendritic spine development during astrocyte-to-neuron conversion, as observed by two-photon live imaging.
(A) Representative tracings used for Sholl analysis showing the major branches of an AAV9 GFAP::GFP–infected astrocyte (red), a converting cell infected with the CAG::NeuroD1-P2A-GFP retrovirus (green), and an AAV9 GFAP1.6::NeuroD1-P2A-GFP-infected converting cell (blue) at different time points after infection. Note that the processes of NeuroD1-expressing cells adopted a neuron-like morphology over time. Scale bar: 20 μm. (B) Sholl analysis of GFP- (red) or NeuroD1- (blue) overexpressing cells at different time points after AAV infection. Note that many short branches retracted, while a couple of processes grew much longer over time with NeuroD1 overexpression. Two-way repeated-measures repeated-measures analysis of variance with Bonferroni post hoc test, mean ± SEM, n = 20 cells from 5 animals, ***P = 0.0006, Fcolumn factor (3, 76) = 6.59. *P < 0.05; **P < 0.01; ***P < 0.001. (C) Sholl analysis of NeuroD1-overexpressing cells at different time points after retrovirus infection. Note that a couple of processes grew much longer over time with NeuroD1 overexpression. Two-way repeated-measures analysis of variance with Bonferroni post hoc test, mean ± SEM, n = 20 cells from 4 animals, *P = 0.0257, Fcolumn factor (2, 57) = 8.16. *P < 0.05. (D, E) Representative immunostaining images following two-photon live imaging showing many dendritic spines in neurons newly converted from NeuroD1-expressing proliferating astrocytes at 26 dpi (D, arrowheads) or lineage-traced astrocytes at 60 dpi (E). Scale bars: 20 μm, 5 μm (D, inset). (F) Quantification of the dendritic spine density after viral infection. Note that the proliferating cells and lineage-traced cells overexpressing NeuroD1 grew ~0.6 and ~1.0 dendritic spine per μm dendritic shaft length, respectively. Meanwhile, the dendritic spines of proliferating or lineage-traced astrocytes overexpressing GFP alone barely grew. One-way analysis of variance followed by Tukey's post hoc test, mean ± SD, n = 10 animals, ****P < 0.0001, Ftreatment (3, 36) = 33.86. dpi: Day(s) post infection; GFAP: glial fibrillary acidic protein; GFP: green fluorescent protein; NeuroD1: neuronal differentiation 1.
Functional integration of astrocyte-converted neurons into pre-existing neural circuits, as revealed by two-photon Ca2+ imaging and patch-clamp recording
Using astrocytic lineage-tracing mice (Aldh1l1-CreERT2 X Ai14), we further investigated the functional relationship between astrocyte-converted neurons and pre-existing neurons through two-photon Ca2+ imaging with ultra-sensitive protein calcium sensor green fluorescent protein-calmodulin fusion protein 6 sensitive (GCaMP6s) (Chen et al., 2013). We first injected AAV5 GFAP::GCaMP6s and AAV9 GFAP(CMVe)::NeuroD1 into the neocortex of astrocytic lineage-tracing mice. At 5 dpi, nearly all of the tdTomato+ cells were still astrocytes, and their Ca2+ traces showed typical astrocytic Ca2+ spikes, with slow rising and decaying phases of several seconds' duration (Figure 6A). Then, we injected AAV5 hSyn::GCaMP6s and AAV9 GFAP(CMVe)::NeuroD1 together into the brain of the lineage-tracing mice. At 30 dpi, we found that some tdTomato+ cells exhibiting spontaneous somatic Ca2+ spikes very similar to those seen in the endogenous neurons (Figure 6B and C). More interestingly, during two-photon time-lapse imaging, we observed that some tdTomato+ astrocyte-converted neurons displayed somatic Ca2+ spikes that were synchronized with those of endogenous neurons (Figure 6D and E), suggesting that these astrocyte-converted neurons had successfully integrated into local neural circuits. Quantification of the rising and decaying time of somatic Ca2+ transients demonstrated much shorter rise-to-peak time and decay time in converted neurons (trise-to-peak ≈ 200 ms, tdecay ≈ 2 seconds) than in resident astrocytes (trise-to-peak ≈ 3.5 seconds, tdecay ≈ 8 seconds, Figure 6F), which is consistent with somatic Ca2+ signals in astrocytes generally occurring at a distinctively slower rate than neuronal Ca2+ events (Wang et al., 2006). In fact, the timescales of somatic Ca2+ transients in converted neurons were indistinguishable from those in endogenous neurons (Figure 6F, n = 10 animals, Ftreatment (5, 54) = 168.4, P < 0.0001), indicating that the newly converted neurons exhibited the same intracellular Ca2+ release features as mature neurons. Taken together, the results from two-photon Ca2+ imaging of astrocytic lineage-tracing mice provide further proof that lineage-traced astrocytes can be converted into functional neurons and then contribute to synchronized neural circuit activity.
Figure 6.
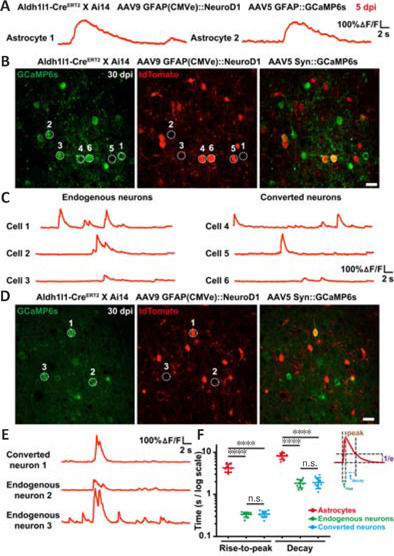
Functional integration of newly converted neurons into pre-existing neural circuits, as revealed by two-photon Ca2+ imaging in astrocytic lineage-tracing mice.
(A) Representative GCaMP6s traces (in ΔF/F) illustrating spontaneous Ca2+ spikes in tdTomato+ lineage-traced astrocytes (astrocytes 1 and 2) at 5 dpi. Note that the typical widths of the Ca2+ spikes of these astrocytes were significantly broader than those of the neurons. Scale bars: 100% ΔF/F, 2 seconds. (B) Representative frames from two-photon time-lapse Ca2+ imaging of tdTomato+ converted and tdTomato-endogenous neurons with GCaMP6s in Aldh1l1-CreERT2 X Ai14 mice at 30 dpi. Note that all the tdTomato- and tdTomato+ cells we observed expressed GCaMP6s under the control of the neuron-specific synapsin promoter, indicating their neuronal identities. Scale bar: 20 μm. (C) Representative GCaMP6s traces (in ΔF/F) showing spontaneous Ca2+ spikes in endogenous neurons (cells 1–3) and converted neurons (cells 4–6). Note that the amplitude and width of the Ca2+ spikes of the converted neurons were similar to those of the endogenous neurons, suggesting these newly converted neurons were functionally mature at this stage. Scale bars: 100% ΔF/F, 2 seconds. (D) Representative frames from two-photon time-lapse Ca2+ imaging of tdTomato+ converted and tdTomato-endogenous neurons with GCaMP6s in Aldh1l1-CreERT2 X Ai14 mice at 30 dpi. Note that converted neuron 1 and endogenous neurons 2 and 3 were in close vicinity to each other, perhaps within the same neural circuits. Scale bar: 20 μm. (E) Representative GCaMP6s traces (in ΔF/F) showing synchronized Ca2+ spikes among the newly converted (cell 1) and endogenous (cells 2, 3) neurons. These highly synchronized Ca2+ spikes suggest that some newly generated neurons made effective synaptic connections with surrounding neurons, and thus were functionally integrated into pre-existing neural circuits. Scale bars: 100% ΔF/F, 2 seconds. (F) Quantification of the rise and decay time of somatic Ca2+ transients. Note the much shorter rise-to-peak and decay time in converted neurons (trise-to-peak ≈ 200 ms, tdecay ≈ 2 seconds) compared with astrocytes (trise-to-peak ≈ 3.5 seconds, tdecay ≈ 8 seconds). The rise-to-peak time and decay time are defined as the time required to rise to 100% peak value and to decrease to ~36.8% (1/e) peak value. One-way analysis of variance followed by Tukey's post hoc test, mean ± SD, n = 10 animals, ****P < 0.0001, Ftreatment (5, 54) = 168.4. dpi: Day(s) post infection; GCaMP6s: green fluorescent protein-calmodulin fusion protein 6 sensitive; GFAP: glial fibrillary acidic protein; NeuroD1: neuronal differentiation 1.
As an alternative approach to investigate the functionality of astrocyte-converted neurons, we tested the electrophysiological properties of the newly converted neurons in cortical slices at 30 dpi using patch-clamp recording (Figure 7A). As expected, AAV9 GFAP::GFP-infected cells did not exhibit any action potentials (Figure 7B), voltage-gated sodium currents (Figure 7C), or spontaneous synaptic responses (Figure 7D), consistent with the properties of typical astrocytes (n = 11). In contrast, many AAV9 GFAP1.6::NeuroD1-GFP-infected cells exhibited repetitive action potentials (Figure 7E), large voltage-gated sodium currents (Figure 7F), and spontaneous synaptic responses (Figure 7G), consistent with the properties of typical neurons (n = 22). Moreover, immunostaining with the synaptic marker synapsin1 at 60 dpi showed presynaptic puncta juxtaposed with the dendritic shafts and dendritic spines of NeuroD1-GFP+ cells (Figure 7H), confirming that the astrocyte-converted neurons did become functionally incorporated into an established neural network in the mouse cortex. Collectively, our two-photon Ca2+ imaging and patch-clamp recording results demonstrate that astrocytes can be directly reprogrammed into fully functional neurons that can integrate into pre-existing neural circuits, and unambiguously show that adult mammalian brains are highly plastic in terms of neuroregeneration and neural circuit remodeling.
Figure 7.
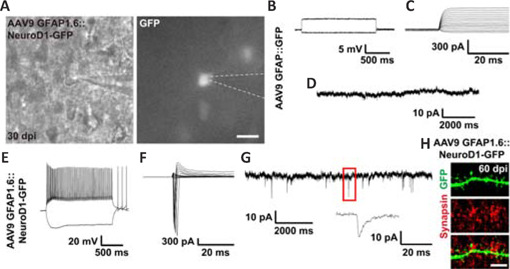
Functional synaptic connections formed by the astrocyte-converted neurons, as revealed by electrophysiological recordings.
(A) Representative phase contrast image (left) and fluorescent image (right, GFP signal) during brain slice recordings. The dashed lines outline the tip of the recording pipette. Scale bar: 20 μm. (B–D) Representative traces from an astrocyte infected with the control AAV9 GFAP::GFP (30 dpi). No action potential or synaptic responses were recorded. (E–G) Representative traces from a cell infected with AAV9 GFAP1.6::NeuroD1-GFP at 30 dpi, displaying repetitive action potentials (E), large Na+ currents (F), and spontaneous synaptic events (G). (H) Astrocyte-converted neurons (green) displayed dendritic spines that were juxtaposed with the synaptic marker synapsin (red, Alexa Fluor 555) at 60 dpi, suggesting functional synapse formation between newly converted neurons and local neural circuits. Scale bar: 5 μm. dpi: Day(s) post infection; GFAP: glial fibrillary acidic protein; GFP: green fluorescent protein; NeuroD1: neuronal differentiation 1.
Discussion
Over the past couple of years, a few recent studies and comments have questioned whether in vivo direct reprogramming of glia into neurons in the CNS is possible (Blackshaw and Sanes, 2021; Qian et al., 2021; Wang et al., 2021; Hoang et al., 2022). Thus, more direct evidence of intermediate stages between glial cells and neurons during the lineage conversion process is needed. In the current study, we used two-photon live imaging technology to provide unambiguous evidence of the step-by-step conversion process from proliferating or lineage-traced astrocytes to functional neurons in the neocortex of live animals, through direct visualization. We observed that both reactive astrocytes infected with a retrovirus expressing transcription factor NeuroD1 and lineage-traced astrocytes in Aldh1l1-CreERT2 mice infected with AAV9 GFAP::NeuroD1 gradually retracted numerous fine processes and extended one or two major branches, eventually adopting neuronal morphology within 4 weeks. Interestingly, the elongated neurites displayed dynamic growth cones that actively explored the local microenvironment. Additionally, some converting cells migrated tangentially or vertically within the mouse cortex. Moreover, many newly converted neurons exhibited synchronous calcium signals and neuronal electrophysiological properties, suggesting functional synaptic connections and local circuit integration.
Stages in the astrocyte-to-neuron transition process captured by repetitive two-photon live imaging
During the in situ glia-to-neuron conversion process, the converting cells presumably go through several consecutive transitional stages in which they gradually lose typical glial morphology and properties while progressively adopting neuronal morphology and properties. The most striking result from our study is the two-photon live cell images capturing these step-by-step cell fate transitional stages in the brains of living animals (Svoboda and Yasuda, 2006; Holtmaat et al., 2009; Yang et al., 2010). The continuous images of the direct astrocyte-to-neuron conversion events we provide here may help address some doubts that have been expressed about in situ direct reprogramming in the CNS (Blackshaw and Sanes, 2021; Qian et al., 2021; Wang et al., 2021; Hoang et al., 2022). The astrocyte-to-neuron conversion process started with the retraction of fine astrocytic processes, while some major astrocytic branches directly transformed into long neurites (future axons or dendrites). Such direct conversion of astrocytic processes into neuronal processes suggests that local neurons and astrocytes may share certain commonalities, which is supported by a recent study reporting that astrocytes are more likely to be converted into local neuron types during in vivo reprogramming (Herrero-Navarro et al., 2021). Interestingly, some of the converting cells exhibited both long neurite-like processes and short blood vessel-wrapping astrocytic processes in the early stages of conversion. This suggests that the newly converted neurons obtain nutrients from the blood supply directly before relying on neighboring astrocytes to supply nutrients once the astrocytic endfeet retract from the blood vessels. Furthermore, dynamic growth cones were observed at the tips of some of the long processes of the converting cells. This is reminiscent of developing embryonic neurons, suggesting that these newly generated neurons were actively exploring their local environment to find potential target areas (Vitriol and Zheng, 2012; Gomez and Letourneau, 2014; Goodhill et al., 2015). The discovery of active growth cones in converted neurons also provides direct evidence supporting previous reports that, after neuronal conversion, the newly generated neurons can project their axons to distal target areas, possibly by following the axonal pathway laid out by their neighboring neurons during early brain development (Chen et al., 2020; Wu et al., 2020; Zheng et al., 2022).
Migration of lineage-converting cells through the intricate brain network
Another unexpected finding from our study was the discovery of somal migration of the lineage-converting cells in the adult mouse brain. This is again reminiscent of newly generated neurons during early brain development (Huang, 2009; Tan and Shi, 2013; Barber and Pierani, 2016). More interestingly, we found that some lineage-converting cells migrated either tangentially or radially from superficial to deep layers of the cortex, which is opposite of the migration path along radial glia in an inside-out manner that occurs during early cortical development (Tan and Shi, 2013). The precise molecular mechanisms underlying such tangential and radial migration during astrocyte-to-neuron conversion should be explored in more detail in the future. Nevertheless, such migration of the lineage-converting cells indicates that there are sufficient local cues within the adult mouse cortex to interact with and guide the newly generated neurons to suitable positions. In fact, this type of neuronal migration has been observed during adult neurogenesis in the mouse subventricular zone and hippocampal subgranular zone, suggesting that it could also occur in adult mammalian brains (Mu et al., 2010; Ming and Song, 2011; Sun et al., 2015; Lim and Alvarez-Buylla, 2016; Gage, 2019).
In contrast to the lineage-converting cells that actively navigated through the local environment to arrive at the correct location for the newly generated neurons, many studies have shown that astrocytes generally do not migrate in mammalian brains or spinal cords, possibly due to the formation of intercellular gap junctions (Hatton et al., 1993; Tsai et al., 2012; Bardehle et al., 2013). The sharp contrast in migration capability between the endogenous astrocytes and the newly converted neurons provides further evidence that the astrocytes were converted into neurons in the adult mouse brains.
Functional integration of the newly converted neurons into the brain network
Ca2+ transients are a distinct feature that can distinguish astrocytes from neurons. In general, Ca2+ signals in astrocyte somata are much slower than neuronal Ca2+ events (Stobart et al., 2018; Lia et al., 2021). Compared with typical fast-rising (~180 ms) and rapidly recovering (~1.5 seconds) neuronal Ca2+ kinetics, astrocytic Ca2+ signals show a much slower onset (~3 seconds) and longer duration of recovery (> 15 seconds) (Wang et al., 2006; Wang et al., 2009; Deneux et al., 2016). Using astrocytic lineage-tracing mice, two-photon Ca2+ imaging of the tdTomato+ lineage-traced astrocytes with GCaMP6s in the mouse cortex identified many newly converted neurons that exhibited typical neuronal Ca2+ kinetics, indicating functional maturation of these converted neurons. More strikingly, two-photon time-lapse imaging revealed synchronized Ca2+ spikes between astrocyte-converted neurons and pre-existing neurons, suggesting that these newly converted neurons had formed functional synapses and been effectively incorporated into local neural networks. In addition to the two-photon Ca2+ imaging results, electrophysiological recordings of the newly astrocyte-converted neurons in cortical slices also showed repetitive action potentials, large voltage-gated sodium currents, and spontaneous synaptic responses, suggesting that these newly generated neurons and the endogenous neurons can make de novo synaptic connections with each other. Such functional connections have been reported by our group, as well as several other groups (Niu et al., 2013; Guo et al., 2014; Liu et al., 2015, 2020; Chen et al., 2020; Wu et al., 2020; Zheng et al., 2022). The formation of functional synapses between newly generated and pre-existing mature neurons indicates that the astrocyte-converted neurons were successfully incorporated into well-established brain circuits.
Like all other studies, this study had some limitations. First, the AAV we used to overexpress NeuroD1 in the tdTomato+ lineage-traced astrocytes does not have a GFP tag, so we are not able to directly visualize glia-to-neuron conversion of Aldh1l1-CreERT2-traced astrocytes with double fluorescence. In the future, we plan to use dual-color fluorescence two-photon imaging to better demonstrate the direct conversion process of lineage-traced astrocytes. Second, the acute inflammatory response due to preparing the open-skull cranial window makes it extremely challenging to take high quality two-photon images before 3 dpi, so we were unable to image the original state of the AAV-transfected glial cells and directly visualize the lineage-traced astrocytes from day 0. We are currently working on addressing this technical challenge to provide even clearer evidence for the morphologic transition from glial cells to neurons.
In conclusion, this study demonstrates unambiguously that astrocytes can be directly reprogrammed into neurons in the mouse cortex; newly generated neurons can send out active growth cones to explore the local environment; their neuronal soma can migrate to new positions inside the brain; and astrocyte-converted neurons can make functional synaptic connections with the pre-existing neural circuits. These findings indicate that adult mammalian brains are highly plastic in terms of neuroregeneration and suggest that in situ direct glia-to-neuron conversion is a potentially promising approach for neural circuit reconstruction.
Additional files:
Additional Figure 1 (2.2MB, tif) : The majority of astrocytes in Aldh1l1-CreERT2 X Ai14 mice were labeled by tdTomato for lineage-tracing purpose.
The majority of astrocytes in Aldh1l1-CreERT2 X Ai14 mice were labeled by tdTomato for lineage-tracing purpose.
(A) Representative images showing immunostaining against tdTomato (red), neuronal marker NeuN (green, Alexa Fluor 488) and astrocytic marker GFAP (green, Alexa Fluor 488) in the Aldh1l1-CreERT2 X Ai14 mouse cortex. Note the vast majority of GFAP+ astrocytes in Aldh1l1-CreERT2 X Ai14 mouse brains were tdTomato+. Scale bar: 20 μm. (B) Quantification of the percentage of tdTomato+ cells among NeuN+ or GFAP+ cells in Aldh1l1-CreERT2 X Ai14 mouse cortex. Student's t-test. All values are presented as mean ± SD. n = 5 per group. dpi: Day(s) post infection; GFAP: glial fibrillary acidic protein; NeuroD1: neuronal differentiation 1.
Additional Figure 2 (3.1MB, tif) : AAV infection and ectopic gene overexpression in Aldh1l1-CreERT2 X Ai14 mouse brains shown by GFP/NeuroD1 immunostaining.
AAV infection and ectopic gene overexpression in Aldh1l1-CreERT2 X Ai14 mouse brains shown by GFP/NeuroD1 immunostaining.
(A) Representative images showing GFP and tdTomato immunostaining in the Aldh1l1-CreERT2 X Ai14 mouse cortex. Note that most of the lineage-traced astrocytes overexpressed GFP 5 days after AAV9 GFAP::GFP injection. Scale bar: 20 μm. (B) Quantification of the percentage of GFP+ cells among tdTomato+ cells in Aldh1l1-CreERT2 X Ai14 mouse cortex. The value is presented as mean ± SD. n = 5 per group. (C) Representative images showing immunostaining against tdTomato (red) and neural transcription factor NeuroD1 (green, infrared pseudo-color, Alexa Fluor 647) in the Aldh1l1-CreERT2 X Ai14 mouse cortex. Note that the endogenous level of NeuroD1 in astrocytes is extremely low (undetectable by immunostaining), while most of the lineage-traced astrocytes overexpressed NeuroD1 5 days after AAV9 GFAP (CMVe)::NeuroD1 injection. Scale bar: 20 μm. (D) Quantification of the percentage of NeuroD1+ cells among tdTomato+ cells in Aldh1l1-CreERT2 X Ai14 mouse cortex. Student's t-test. All values are presented as mean ± SD. n = 5 per group. AAV: Adeno-associated virus; dpi: day(s) post infection; GFAP: glial fibrillary acidic protein; GFP: green fluorescent protein; NeuroD1: neuronal differentiation 1.
Additional Figure 3 (4.7MB, tif) : Lineage conversion from tdTomato+ astrocytic lineage-tracing cells into mature neurons by NeuroD1.
Lineage conversion from tdTomato+ astrocytic lineage-tracing cells into mature neurons by NeuroD1.
(A, B) Representative images illustrating the tdTomato+ cells in the astrocytic lineage-tracing mouse cortex without (A) or with NeuroD1-infection (B, 30 dpi). Many of the tdTomato+ astrocytes adopt neuronal morphology 30 days after NeuroD1 overexpression, while all the tdTomato+ astrocytes keep astrocytic morphology on the contralateral side of the brain without NeuroD1 overexpression. The white boxes show the positions of the ROIs in C and D. Scale bar: 100 μm. (C, D) Enlarged images showing that the tdTomato+ cells in contralateral cortex without viral injection were immuno-negative for neuronal marker NeuN (C, arrowheads), while the tdTomato+ cells in NeuroD1-infected cortex were immuno-positive for NeuN (D, arrowheads, 30 dpi, green, Alexa Fluor 488). Scale bar: 20 μm. (E, F) Enlarged images showing that the tdTomato+ cells in contralateral side without viral injection were immune-negative for NeuN (E, arrowheads, green, Alexa Fluor 488), and the tdTomato+ cells in NeuroD1-infected side were immune-negative for GFAP (F, arrowheads, 30 dpi, green, Alexa Fluor 488). Scale bar: 20 μm. dpi: Day(s) post infection; GFAP: glial fibrillary acidic protein; NeuroD1: neuronal differentiation 1; ROI: region of interest.
Additional Figure 4 (2.6MB, tif) : Transitional stage cell with both neuronal and astrocytic properties in the mouse cortex after NeuroD1 overexpression.
Transitional stage cell with both neuronal and astrocytic properties in the mouse cortex after NeuroD1 overexpression.
(A) Following two-photon live imaging, the mouse brain was fixed and immunostained with both neuronal marker NeuN and astrocytic marker GFAP. The AAV NeuroD1-GFP-infected cell was found NeuN (blue, Alexa Fluor 647) and GFAP (red, Alexa Fluor 555) double positive, indicating its transitional stage during lineage conversion. White arrowhead: GFAP+ NeuN- cell; red arrowhead: GFAP- NeuN+ cell; yellow arrowhead: GFAP+ NeuN+ cell. (B) Orthographic projection of A showing colocalization of GFP with NeuN and GFAP. Scale bars: 20 μm. dpi: day(s) post infection; GFAP: glial fibrillary acidic protein; GFP: green fluorescent protein; NeuroD1: neuronal differentiation 1.
Additional Figure 5 (3.9MB, tif) : Cell identities verified with post-fixation immunostaining after two-photon live imaging.
Cell identities verified with post-fixation immunostaining after two-photon live imaging.
(A) NeuroD1-converted neurons (AAV9 GFAP1.6::NeuroD1-GFP) were immunopositive for mature neuronal marker MAP2 (red, Alexa Fluor 555) at 60 dpi. (B) Some NeuroD1-converted neurons were immunopositive for upper cortical layer neuronal marker Cux1 (layers II-IV, red, Alexa Fluor 555), while others were immunopositive for deeper cortical layer neuronal marker Ctip2 (layers V-VI, red, Alexa Fluor 555) at 60 dpi. White arrowheads: GFP+ Cux1+/Ctip2+ cells. Scale bars: 20 μm. Ctip2: COUP-TF-interacting protein 2; Cux1: cut-like homeobox 1; dpi: day(s) post infection; GFAP: glial fibrillary acidic protein; GFP: green fluorescent protein; MAP2: microtubule association protein-2; NeuroD1: neuronal differentiation 1.
Acknowledgments:
We would like to thank Xiao-Peng Luo (Guangdong-Hong Kong-Macau Institute of CNS Regeneration (GHMICR), Jinan University), Dr. Qing-Song Wang (Guangdong-Hong Kong-Macau Institute of CNS Regeneration (GHMICR), Jinan University), and Long-Jiao Ge (Kunming University of Science and Technology) for virus packaging. We would also like to thank Dr. Li Zhang (Guangdong-Hong Kong-Macau Institute of CNS Regeneration (GHMICR), Jinan University) for the generous gift of AAV9 hSyn::GCaMP6s.
Funding Statement
Funding: This study was supported by the National Natural Science Foundation of China, No. 31970906 (to WLei); the Natural Science Foundation of Guangdong Province, No. 2020A1515011079 (to WLei); Key Technologies R&D Program of Guangdong Province, No. 2018B030332001 (to GC); Science and Technology Projects of Guangzhou, No. 202206060002 (to GC); the Youth Science Program of the National Natural Science Foundation of China, No. 32100793 (to ZX); the Pearl River Innovation and Entrepreneurship Team, No. 2021ZT09Y552; and Yi-Liang Liu Endowment Fund from Jinan University Education Development Foundation.
Footnotes
Conflicts of interest: GC is a co-founder of NeuExcell Therapeutics Inc. The other authors declare no conflict of interest.
Data availability statement: All data relevant to the study are included in the article or uploaded as Additional files.
C-Editor: Zhao M; S-Editors: Yu J, Li CH; L-Editors: Crow E, Yu J, Song LP; T-Editor: Jia Y
References
- Barber M, Pierani A. Tangential migration of glutamatergic neurons and cortical patterning during development: Lessons from Cajal-Retzius cells. Dev Neurobiol. 2016;76:847–881. doi: 10.1002/dneu.22363. [DOI] [PubMed] [Google Scholar]
- Bardehle S, Krüger M, Buggenthin F, Schwausch J, Ninkovic J, Clevers H, Snippert HJ, Theis FJ, Meyer-Luehmann M, Bechmann I, Dimou L, Götz M. Live imaging of astrocyte responses to acute injury reveals selective juxtavascular proliferation. Nat Neurosci. 2013;16:580–586. doi: 10.1038/nn.3371. [DOI] [PubMed] [Google Scholar]
- Barker RA, Götz M, Parmar M. New approaches for brain repair-from rescue to reprogramming. Nature. 2018;557:329–334. doi: 10.1038/s41586-018-0087-1. [DOI] [PubMed] [Google Scholar]
- Blackshaw S, Sanes JR. Turning lead into gold: reprogramming retinal cells to cure blindness. J Clin Invest. 2021;131:e146134. doi: 10.1172/JCI146134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bocchi R, Masserdotti G, Götz M. Direct neuronal reprogramming: fast forward from new concepts toward therapeutic approaches. Neuron. 2022;110:366–393. doi: 10.1016/j.neuron.2021.11.023. [DOI] [PubMed] [Google Scholar]
- Boldrini M, Fulmore CA, Tartt AN, Simeon LR, Pavlova I, Poposka V, Rosoklija GB, Stankov A, Arango V, Dwork AJ, Hen R, Mann JJ. Human hippocampal neurogenesis persists throughout aging. Cell Stem Cell. 2018;22:589–599.e5. doi: 10.1016/j.stem.2018.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buffo A, Rite I, Tripathi P, Lepier A, Colak D, Horn AP, Mori T, Götz M. Origin and progeny of reactive gliosis: A source of multipotent cells in the injured brain. Proc Natl Acad Sci U S A. 2008;105:3581–3586. doi: 10.1073/pnas.0709002105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen TW, Wardill TJ, Sun Y, Pulver SR, Renninger SL, Baohan A, Schreiter ER, Kerr RA, Orger MB, Jayaraman V, Looger LL, Svoboda K, Kim DS. Ultrasensitive fluorescent proteins for imaging neuronal activity. Nature. 2013;499:295–300. doi: 10.1038/nature12354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen YC, Ma NX, Pei ZF, Wu Z, Do-Monte FH, Keefe S, Yellin E, Chen MS, Yin JC, Lee G, Minier-Toribio A, Hu Y, Bai YT, Lee K, Quirk GJ, Chen G. A NeuroD1 AAV-Based Gene Therapy for Functional Brain Repair after Ischemic Injury through In Vivo Astrocyte-to-Neuron Conversion. Mol Ther. 2020;28:217–234. doi: 10.1016/j.ymthe.2019.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deneux T, Kaszas A, Szalay G, Katona G, Lakner T, Grinvald A, Rózsa B, Vanzetta I. Accurate spike estimation from noisy calcium signals for ultrafast three-dimensional imaging of large neuronal populations in vivo. Nat Commun. 2016;7:12190. doi: 10.1038/ncomms12190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu M, Yu X, Lu J, Zuo Y. Repetitive motor learning induces coordinated formation of clustered dendritic spines in vivo. Nature. 2012;483:92–95. doi: 10.1038/nature10844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gage FH. Adult neurogenesis in mammals. Science. 2019;364:827–828. doi: 10.1126/science.aav6885. [DOI] [PubMed] [Google Scholar]
- Gascón S, Murenu E, Masserdotti G, Ortega F, Russo GL, Petrik D, Deshpande A, Heinrich C, Karow M, Robertson SP, Schroeder T, Beckers J, Irmler M, Berndt C, Angeli JP, Conrad M, Berninger B, Götz M. Identification and successful negotiation of a metabolic checkpoint in direct neuronal reprogramming. Cell Stem Cell. 2016;18:396–409. doi: 10.1016/j.stem.2015.12.003. [DOI] [PubMed] [Google Scholar]
- Ge WP, Jia JM. Local production of astrocytes in the cerebral cortex. Neuroscience. 2016;323:3–9. doi: 10.1016/j.neuroscience.2015.08.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldman SA. Stem and progenitor cell-based therapy of the central nervous system: hopes, hype, and wishful thinking. Cell Stem Cell. 2016;18:174–188. doi: 10.1016/j.stem.2016.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez TM, Letourneau PC. Actin dynamics in growth cone motility and navigation. J Neurochem. 2014;129:221–234. doi: 10.1111/jnc.12506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodhill GJ, Faville RA, Sutherland DJ, Bicknell BA, Thompson AW, Pujic Z, Sun B, Kita EM, Scott EK. The dynamics of growth cone morphology. BMC Biol. 2015;13:10. doi: 10.1186/s12915-015-0115-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grande A, Sumiyoshi K, López-Juárez A, Howard J, Sakthivel B, Aronow B, Campbell K, Nakafuku M. Environmental impact on direct neuronal reprogramming in vivo in the adult brain. Nat Commun. 2013;4:2373. doi: 10.1038/ncomms3373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Z, Zhang L, Wu Z, Chen Y, Wang F, Chen G. In vivo direct reprogramming of reactive glial cells into functional neurons after brain injury and in an Alzheimer's disease model. Cell Stem Cell. 2014;14:188–202. doi: 10.1016/j.stem.2013.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatton JD, Finkelstein JP, U HS. Native astrocytes do not migrate de novo or after local trauma. Glia. 1993;9:18–24. doi: 10.1002/glia.440090104. [DOI] [PubMed] [Google Scholar]
- Heinrich C, Bergami M, Gascón S, Lepier A, Viganò F, Dimou L, Sutor B, Berninger B, Götz M. Sox2-mediated conversion of NG2 glia into induced neurons in the injured adult cerebral cortex. Stem Cell Reports. 2014;3:1000–1014. doi: 10.1016/j.stemcr.2014.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrero-Navarro Á, Puche-Aroca L, Moreno-Juan V, Sempere-Ferràndez A, Espinosa A, Susín R, Torres-Masjoan L, Leyva-Díaz E, Karow M, Figueres-Oñate M, López-Mascaraque L, López-Atalaya JP, Berninger B, López-Bendito G. Astrocytes and neurons share region-specific transcriptional signatures that confer regional identity to neuronal reprogramming. Sci Adv. 2021;7:eabe8978. doi: 10.1126/sciadv.abe8978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoang T, Kim DW, Appel H, Pannullo NA, Leavey P, Ozawa M, Zheng S, Yu M, Peachey NS, Blackshaw S. Genetic loss of function of Ptbp1 does not induce glia-to-neuron conversion in retina. Cell Rep. 2022;39:110849. doi: 10.1016/j.celrep.2022.110849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holtmaat A, Bonhoeffer T, Chow DK, Chuckowree J, De Paola V, Hofer SB, Hübener M, Keck T, Knott G, Lee WC, Mostany R, Mrsic-Flogel TD, Nedivi E, Portera-Cailliau C, Svoboda K, Trachtenberg JT, Wilbrecht L. Long-term, high-resolution imaging in the mouse neocortex through a chronic cranial window. Nat Protoc. 2009;4:1128–1144. doi: 10.1038/nprot.2009.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Z. Molecular regulation of neuronal migration during neocortical development. Mol Cell Neurosci. 2009;42:11–22. doi: 10.1016/j.mcn.2009.06.003. [DOI] [PubMed] [Google Scholar]
- Jiao Y, Sun YT, Chen NF, Zhou LN, Guan X, Wang JY, Wei WJ, Han C, Jiang XL, Wang YC, Zou W, Liu J. Human umbilical cord-derived mesenchymal stem cells promote repair of neonatal brain injury caused by hypoxia/ischemia in rats. Neural Regen Res. 2022;17:2518–2525. doi: 10.4103/1673-5374.339002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katz RA, Greger JG, Skalka AM. Effects of cell cycle status on early events in retroviral replication. J Cell Biochem. 2005;94:880–889. doi: 10.1002/jcb.20358. [DOI] [PubMed] [Google Scholar]
- Lei W, Li W, Ge L, Chen G. Non-engineered and engineered adult neurogenesis in mammalian brains. Front Neurosci. 2019;13:131. doi: 10.3389/fnins.2019.00131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lentini C, d'Orange M, Marichal N, Trottmann MM, Vignoles R, Foucault L, Verrier C, Massera C, Raineteau O, Conzelmann KK, Rival-Gervier S, Depaulis A, Berninger B, Heinrich C. Reprogramming reactive glia into interneurons reduces chronic seizure activity in a mouse model of mesial temporal lobe epilepsy. Cell Stem Cell. 2021;28:2104–2121.e10. doi: 10.1016/j.stem.2021.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Chen G. In Vivo Reprogramming for CNS repair: regenerating neurons from endogenous glial cells. Neuron. 2016;91:728–738. doi: 10.1016/j.neuron.2016.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lia A, Henriques VJ, Zonta M, Chiavegato A, Carmignoto G, Gómez-Gonzalo M, Losi G. Calcium signals in astrocyte microdomains, a decade of great advances. Front Cell Neurosci. 2021;15:673433. doi: 10.3389/fncel.2021.673433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim DA, Alvarez-Buylla A. The Adult Ventricular-Subventricular Zone (V-SVZ) and Olfactory Bulb (OB) Neurogenesis. Cold Spring Harb Perspect Biol. 2016;8:a018820. doi: 10.1101/cshperspect.a018820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu MH, Li W, Zheng JJ, Xu YG, He Q, Chen G. Differential neuronal reprogramming induced by NeuroD1 from astrocytes in grey matter versus white matter. Neural Regen Res. 2020;15:342–351. doi: 10.4103/1673-5374.265185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Miao Q, Yuan J, Han S, Zhang P, Li S, Rao Z, Zhao W, Ye Q, Geng J, Zhang X, Cheng L. Ascl1 converts dorsal midbrain astrocytes into functional neurons in vivo. J Neurosci. 2015;35:9336–9355. doi: 10.1523/JNEUROSCI.3975-14.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma L, Qiao Q, Tsai JW, Yang G, Li W, Gan WB. Experience-dependent plasticity of dendritic spines of layer 2/3 pyramidal neurons in the mouse cortex. Dev Neurobiol. 2016;76:277–286. doi: 10.1002/dneu.22313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuda T, Irie T, Katsurabayashi S, Hayashi Y, Nagai T, Hamazaki N, Adefuin AMD, Miura F, Ito T, Kimura H, Shirahige K, Takeda T, Iwasaki K, Imamura T, Nakashima K. Pioneer factor NeuroD1 rearranges transcriptional and epigenetic profiles to execute microglia-neuron conversion. Neuron. 2019;101:472–485.e7. doi: 10.1016/j.neuron.2018.12.010. [DOI] [PubMed] [Google Scholar]
- Ming GL, Song H. Adult neurogenesis in the mammalian brain: significant answers and significant questions. Neuron. 2011;70:687–702. doi: 10.1016/j.neuron.2011.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mollinari C, Zhao J, Lupacchini L, Garaci E, Merlo D, Pei G. Transdifferentiation: a new promise for neurodegenerative diseases. Cell Death Dis. 2018;9:830. doi: 10.1038/s41419-018-0891-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mu Y, Lee SW, Gage FH. Signaling in adult neurogenesis. Curr Opin Neurobiol. 2010;20:416–423. doi: 10.1016/j.conb.2010.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niu W, Zang T, Zou Y, Fang S, Smith DK, Bachoo R, Zhang CL. In vivo reprogramming of astrocytes to neuroblasts in the adult brain. Nat Cell Biol. 2013;15:1164–1175. doi: 10.1038/ncb2843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Percie du Sert N, Hurst V, Ahluwalia A, Alam S, Avey MT, Baker M, Browne WJ, Clark A, Cuthill IC, Dirnagl U, Emerson M, Garner P, Holgate ST, Howells DW, Karp NA, Lazic SE, Lidster K, MacCallum CJ, Macleod M, Pearl EJ, et al. The ARRIVE guidelines 2.0: Updated guidelines for reporting animal research. PLoS Biol. 2020;18:e3000410. doi: 10.1371/journal.pbio.3000410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pilz GA, Bottes S, Betizeau M, Jörg DJ, Carta S, April S, Simons BD, Helmchen F, Jessberger S. Live imaging of neurogenesis in the adult mouse hippocampus. Science. 2018;359:658–662. doi: 10.1126/science.aao5056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian C, Dong B, Wang XY, Zhou FQ. In vivo glial trans-differentiation for neuronal replacement and functional recovery in central nervous system. FEBS J. 2021;288:4773–4785. doi: 10.1111/febs.15681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian H, Kang X, Hu J, Zhang D, Liang Z, Meng F, Zhang X, Xue Y, Maimon R, Dowdy SF, Devaraj NK, Zhou Z, Mobley WC, Cleveland DW, Fu XD. Reversing a model of Parkinson's disease with in situ converted nigral neurons. Nature. 2020;582:550–556. doi: 10.1038/s41586-020-2388-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian H, Fu XD. Brain repair by cell replacement via in situ neuronal reprogramming. Annu Rev Genet. 2021;55:45–69. doi: 10.1146/annurev-genet-071719-023616. [DOI] [PubMed] [Google Scholar]
- Schneider CA, Rasband WS, Eliceiri KW. NIH Image to ImageJ: 25 years of image analysis. Nat Methods. 2012;9:671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schweitzer JS, Song B, Herrington TM, Park TY, Lee N, Ko S, Jeon J, Cha Y, Kim K, Li Q, Henchcliffe C, Kaplitt M, Neff C, Rapalino O, Seo H, Lee IH, Kim J, Kim T, Petsko GA, Ritz J, et al. Personalized iPSC-derived dopamine progenitor cells for Parkinson's disease. N Engl J Med. 2020;382:1926–1932. doi: 10.1056/NEJMoa1915872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sofroniew MV, Vinters HV. Astrocytes: biology and pathology. Acta Neuropathol. 2010;119:7–35. doi: 10.1007/s00401-009-0619-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorrells SF, Paredes MF, Cebrian-Silla A, Sandoval K, Qi D, Kelley KW, James D, Mayer S, Chang J, Auguste KI, Chang EF, Gutierrez AJ, Kriegstein AR, Mathern GW, Oldham MC, Huang EJ, Garcia-Verdugo JM, Yang Z, Alvarez-Buylla A. Human hippocampal neurogenesis drops sharply in children to undetectable levels in adults. Nature. 2018;555:377–381. doi: 10.1038/nature25975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srinivasan R, Lu TY, Chai H, Xu J, Huang BS, Golshani P, Coppola G, Khakh BS. new transgenic mouse lines for selectively targeting astrocytes and studying calcium signals in astrocyte processes in situ and in vivo. Neuron. 2016;92:1181–1195. doi: 10.1016/j.neuron.2016.11.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stobart JL, Ferrari KD, Barrett MJP, Glück C, Stobart MJ, Zuend M, Weber B. Cortical circuit activity evokes rapid astrocyte calcium signals on a similar timescale to neurons. Neuron. 2018;98:726–735.e4. doi: 10.1016/j.neuron.2018.03.050. [DOI] [PubMed] [Google Scholar]
- Su Z, Niu W, Liu ML, Zou Y, Zhang CL. In vivo conversion of astrocytes to neurons in the injured adult spinal cord. Nat Commun. 2014;5:3338. doi: 10.1038/ncomms4338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugaya K, Vaidya M. Stem cell therapies for neurodegenerative diseases. Adv Exp Med Biol. 2018;1056:61–84. doi: 10.1007/978-3-319-74470-4_5. [DOI] [PubMed] [Google Scholar]
- Sun GJ, Zhou Y, Stadel RP, Moss J, Yong JH, Ito S, Kawasaki NK, Phan AT, Oh JH, Modak N, Reed RR, Toni N, Song H, Ming GL. Tangential migration of neuronal precursors of glutamatergic neurons in the adult mammalian brain. Proc Natl Acad Sci U S A. 2015;112:9484–9489. doi: 10.1073/pnas.1508545112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svoboda K, Yasuda R. Principles of two-photon excitation microscopy and its applications to neuroscience. Neuron. 2006;50:823–839. doi: 10.1016/j.neuron.2006.05.019. [DOI] [PubMed] [Google Scholar]
- Tan X, Shi SH. Neocortical neurogenesis and neuronal migration. Wiley Interdiscip Rev Dev Biol. 2013;2:443–459. doi: 10.1002/wdev.88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao Y, Vermilyea SC, Zammit M, Lu J, Olsen M, Metzger JM, Yao L, Chen Y, Phillips S, Holden JE, Bondarenko V, Block WF, Barnhart TE, Schultz-Darken N, Brunner K, Simmons H, Christian BT, Emborg ME, Zhang SC. Autologous transplant therapy alleviates motor and depressive behaviors in parkinsonian monkeys. Nat Med. 2021;27:632–639. doi: 10.1038/s41591-021-01257-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torper O, Ottosson DR, Pereira M, Lau S, Cardoso T, Grealish S, Parmar M. In vivo reprogramming of striatal NG2 glia into functional neurons that integrate into local host circuitry. Cell Rep. 2015;12:474–481. doi: 10.1016/j.celrep.2015.06.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai HH, Li H, Fuentealba LC, Molofsky AV, Taveira-Marques R, Zhuang H, Tenney A, Murnen AT, Fancy SP, Merkle F, Kessaris N, Alvarez-Buylla A, Richardson WD, Rowitch DH. Regional astrocyte allocation regulates CNS synaptogenesis and repair. Science. 2012;337:358–362. doi: 10.1126/science.1222381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vitriol EA, Zheng JQ. Growth cone travel in space and time: the cellular ensemble of cytoskeleton, adhesion, and membrane. Neuron. 2012;73:1068–1081. doi: 10.1016/j.neuron.2012.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang LL, Serrano C, Zhong X, Ma S, Zou Y, Zhang CL. Revisiting astrocyte to neuron conversion with lineage tracing in vivo. Cell. 2021;184:5465–5481.e16. doi: 10.1016/j.cell.2021.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang T, Liao JC, Wang X, Wang QS, Wan KY, Yang YY, He Q, Zhang JX, Chen G, Li W. Unexpected BrdU inhibition on astrocyte-to-neuron conversion. Neural Regen Res. 2022;17:1526–1534. doi: 10.4103/1673-5374.325747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Lou N, Xu Q, Tian GF, Peng WG, Han X, Kang J, Takano T, Nedergaard M. Astrocytic Ca2+ signaling evoked by sensory stimulation in vivo. Nat Neurosci. 2006;9:816–823. doi: 10.1038/nn1703. [DOI] [PubMed] [Google Scholar]
- Wang X, Takano T, Nedergaard M. Astrocytic calcium signaling: mechanism and implications for functional brain imaging. Methods Mol Biol. 2009;489:93–109. doi: 10.1007/978-1-59745-543-5_5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Z, Parry M, Hou XY, Liu MH, Wang H, Cain R, Pei ZF, Chen YC, Guo ZY, Abhijeet S, Chen G. Gene therapy conversion of striatal astrocytes into GABAergic neurons in mouse models of Huntington's disease. Nat Commun. 2020;11:1105. doi: 10.1038/s41467-020-14855-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang Z, Xu L, Liu M, Wang Q, Li W, Lei W, Chen G. Lineage tracing of direct astrocyte-to-neuron conversion in the mouse cortex. Neural Regen Res. 2021;16:750–756. doi: 10.4103/1673-5374.295925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu D, Zhong LT, Cheng HY, Wang ZQ, Chen XM, Feng AY, Chen WY, Chen G, Xu Y. Overexpressing NeuroD1 reprograms Müller cells into various types of retinal neurons. Neural Regen Res. 2023;18:1124–1131. doi: 10.4103/1673-5374.355818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang G, Pan F, Parkhurst CN, Grutzendler J, Gan WB. Thinned-skull cranial window technique for long-term imaging of the cortex in live mice. Nat Protoc. 2010;5:201–208. doi: 10.1038/nprot.2009.222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng J, Li T, Qi S, Qin B, Yu J, Chen G. Neuroregenerative gene therapy to treat temporal lobe epilepsy in a rat model. Prog Neurobiol. 2022;208:102198. doi: 10.1016/j.pneurobio.2021.102198. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The majority of astrocytes in Aldh1l1-CreERT2 X Ai14 mice were labeled by tdTomato for lineage-tracing purpose.
(A) Representative images showing immunostaining against tdTomato (red), neuronal marker NeuN (green, Alexa Fluor 488) and astrocytic marker GFAP (green, Alexa Fluor 488) in the Aldh1l1-CreERT2 X Ai14 mouse cortex. Note the vast majority of GFAP+ astrocytes in Aldh1l1-CreERT2 X Ai14 mouse brains were tdTomato+. Scale bar: 20 μm. (B) Quantification of the percentage of tdTomato+ cells among NeuN+ or GFAP+ cells in Aldh1l1-CreERT2 X Ai14 mouse cortex. Student's t-test. All values are presented as mean ± SD. n = 5 per group. dpi: Day(s) post infection; GFAP: glial fibrillary acidic protein; NeuroD1: neuronal differentiation 1.
AAV infection and ectopic gene overexpression in Aldh1l1-CreERT2 X Ai14 mouse brains shown by GFP/NeuroD1 immunostaining.
(A) Representative images showing GFP and tdTomato immunostaining in the Aldh1l1-CreERT2 X Ai14 mouse cortex. Note that most of the lineage-traced astrocytes overexpressed GFP 5 days after AAV9 GFAP::GFP injection. Scale bar: 20 μm. (B) Quantification of the percentage of GFP+ cells among tdTomato+ cells in Aldh1l1-CreERT2 X Ai14 mouse cortex. The value is presented as mean ± SD. n = 5 per group. (C) Representative images showing immunostaining against tdTomato (red) and neural transcription factor NeuroD1 (green, infrared pseudo-color, Alexa Fluor 647) in the Aldh1l1-CreERT2 X Ai14 mouse cortex. Note that the endogenous level of NeuroD1 in astrocytes is extremely low (undetectable by immunostaining), while most of the lineage-traced astrocytes overexpressed NeuroD1 5 days after AAV9 GFAP (CMVe)::NeuroD1 injection. Scale bar: 20 μm. (D) Quantification of the percentage of NeuroD1+ cells among tdTomato+ cells in Aldh1l1-CreERT2 X Ai14 mouse cortex. Student's t-test. All values are presented as mean ± SD. n = 5 per group. AAV: Adeno-associated virus; dpi: day(s) post infection; GFAP: glial fibrillary acidic protein; GFP: green fluorescent protein; NeuroD1: neuronal differentiation 1.
Lineage conversion from tdTomato+ astrocytic lineage-tracing cells into mature neurons by NeuroD1.
(A, B) Representative images illustrating the tdTomato+ cells in the astrocytic lineage-tracing mouse cortex without (A) or with NeuroD1-infection (B, 30 dpi). Many of the tdTomato+ astrocytes adopt neuronal morphology 30 days after NeuroD1 overexpression, while all the tdTomato+ astrocytes keep astrocytic morphology on the contralateral side of the brain without NeuroD1 overexpression. The white boxes show the positions of the ROIs in C and D. Scale bar: 100 μm. (C, D) Enlarged images showing that the tdTomato+ cells in contralateral cortex without viral injection were immuno-negative for neuronal marker NeuN (C, arrowheads), while the tdTomato+ cells in NeuroD1-infected cortex were immuno-positive for NeuN (D, arrowheads, 30 dpi, green, Alexa Fluor 488). Scale bar: 20 μm. (E, F) Enlarged images showing that the tdTomato+ cells in contralateral side without viral injection were immune-negative for NeuN (E, arrowheads, green, Alexa Fluor 488), and the tdTomato+ cells in NeuroD1-infected side were immune-negative for GFAP (F, arrowheads, 30 dpi, green, Alexa Fluor 488). Scale bar: 20 μm. dpi: Day(s) post infection; GFAP: glial fibrillary acidic protein; NeuroD1: neuronal differentiation 1; ROI: region of interest.
Transitional stage cell with both neuronal and astrocytic properties in the mouse cortex after NeuroD1 overexpression.
(A) Following two-photon live imaging, the mouse brain was fixed and immunostained with both neuronal marker NeuN and astrocytic marker GFAP. The AAV NeuroD1-GFP-infected cell was found NeuN (blue, Alexa Fluor 647) and GFAP (red, Alexa Fluor 555) double positive, indicating its transitional stage during lineage conversion. White arrowhead: GFAP+ NeuN- cell; red arrowhead: GFAP- NeuN+ cell; yellow arrowhead: GFAP+ NeuN+ cell. (B) Orthographic projection of A showing colocalization of GFP with NeuN and GFAP. Scale bars: 20 μm. dpi: day(s) post infection; GFAP: glial fibrillary acidic protein; GFP: green fluorescent protein; NeuroD1: neuronal differentiation 1.
Cell identities verified with post-fixation immunostaining after two-photon live imaging.
(A) NeuroD1-converted neurons (AAV9 GFAP1.6::NeuroD1-GFP) were immunopositive for mature neuronal marker MAP2 (red, Alexa Fluor 555) at 60 dpi. (B) Some NeuroD1-converted neurons were immunopositive for upper cortical layer neuronal marker Cux1 (layers II-IV, red, Alexa Fluor 555), while others were immunopositive for deeper cortical layer neuronal marker Ctip2 (layers V-VI, red, Alexa Fluor 555) at 60 dpi. White arrowheads: GFP+ Cux1+/Ctip2+ cells. Scale bars: 20 μm. Ctip2: COUP-TF-interacting protein 2; Cux1: cut-like homeobox 1; dpi: day(s) post infection; GFAP: glial fibrillary acidic protein; GFP: green fluorescent protein; MAP2: microtubule association protein-2; NeuroD1: neuronal differentiation 1.


