Abstract
The promyelocytic leukemia (PML) protein forms nuclear bodies which are relocated to the cytoplasm by the RNA virus lymphocytic choriomeningitis virus (LCMV). The viral Z protein directly binds to PML and can relocate the nuclear bodies. Others have observed that LCMV virions may contain ribosomes; hence, we investigated the effects of infection on the distribution of ribosomal P proteins (P0, P1, and P2) with PML as a reference point. We demonstrate an association of PML bodies with P proteins by indirect immunofluorescence and coimmunoprecipitation experiments, providing the first evidence of nucleic acid-binding proteins associated with PML bodies. We show that unlike PML, the P proteins are not redistributed upon infection. Immunofluorescence and coimmunoprecipitation studies indicate that the viral Z protein binds the nuclear, but not the cytoplasmic, fraction of P0. The nuclear fraction of P0 has been associated with translationally coupled DNA excision repair and with nonspecific endonuclease activity; thus, P0 may be involved in nucleic acid processing activities necessary for LCMV replication. During the infection process, PML, P1, and P2 are downregulated but P0 remains unchanged. Further, P0 is present in virions while PML is not, indicating some selectivity in the assembly of LCMV.
The promyelocytic leukemia (PML) protein is part of a large multiprotein nuclear complex referred to here as a PML nuclear body, which is distinct from small nuclear ribonucleoprotein particles and nucleoli (1, 10, 20, 26, 54). Several types of viral infection, oncogenic transformation, heat shock, and interferon treatment affect the morphology of these nuclear bodies. As a result of a chromosomal translocation, the PML protein is fused to retinoic acid receptor alpha in acute promyelocytic leukemia (8, 15, 19, 20, 31). In acute promyelocytic leukemia patients, the PML nuclear bodies are disrupted, forming a microparticulate pattern with the majority of PML found in the cytoplasm (10, 21, 51). PML bodies contain at least six other proteins, including SP100, an autoantigen of primary biliary cirrhosis (43); PIC1, a ubiquitin-like molecule (2); NDP55 (1); PML-associated factor (18); and PML (10, 21, 51).
The PML protein contains three cysteine-rich zinc-binding domains known as RING, B1 B box, and B2 B box. The domains are followed by a leucine coiled-coil region which forms a tripartite or RBCC motif (33). The RING domain is approximately 60 residues long and has been identified in over 80 other proteins, several of which are oncoproteins (5, 39). The RING and B-box motifs are thought to be involved in protein-protein interactions and do not appear to bind nucleic acids directly (5–7, 39). A loss of PML bodies in transiently transfected cells results from the mutation of any of these zinc-binding regions (6, 7, 22, 30).
Infection with lymphocytic choriomeningitis virus (LCMV) redistributes PML nuclear bodies to the cytoplasm (3). Arenaviruses, including LCMV, are carried by rodents and occasionally transmitted to primates, causing disease (29). Arenaviruses encode five different proteins: a nucleocapsid protein, two envelope glycoproteins, an RNA polymerase, and the Z protein (36, 37). The Z protein is a 90-residue protein containing a RING domain and a proline-rich region. The Z proteins are highly conserved among the arenaviridae (9) and are thought to function in genome synthesis (14). Z alone is sufficient to redistribute PML to the cytoplasm during infection and binds directly to PML (3).
We examined the effect of infection on ribosomal P proteins because arenavirus virions reportedly contain ribosomes (23). The association of PML and Z has been described and served as a standard for examining the association of the ribosomal P proteins with PML and Z. There are three P proteins which form part of the large ribosomal subunit in eukaryotes: P0, P1, and P2. The P1 and P2 proteins are homologous to the prokaryotic L7/L12 heterodimers which are anchored to the 60S ribosomal subunit by the P0 protein, a homolog of prokaryotic protein L10 (34, 46). P0 binds directly to 28S RNA, while P1 and P2 do not (48). P0, P1, and P2 associate with eukaryotic elongation factors EF1 and EF2 (45, 46), and the P proteins are essential for aminoacyl tRNA binding, EF2-dependent GTPase activity, and polypeptide synthesis (45, 46). P proteins have both a nuclear and a cytoplasmic distribution (52). In nonexponentially growing cells, the nuclear fraction is excluded from the nucleolus (52). The nuclear fraction of P protein has been shown to have an endonuclease activity and could function in translation-coupled base excision-repair (52).
Our studies indicate that while the distribution of PML is changed radically by LCMV infection, the distribution of P proteins is not. Intriguingly, the fraction of PML remaining in the nucleus after infection colocalizes and coimmunoprecipitates with P proteins. The nuclear portion of Z colocalizes with P proteins in infected cells. During infection, the levels of P0 protein remain unchanged, but those of P1, P2, and PML are substantially downregulated. Confocal microscopy and coimmunoprecipitation studies confirm that Z and P0 interact directly in the nuclear, but not the cytoplasmic, fraction. These data are consistent with the observations of others in terms of the differential distribution of P proteins’ functions between the nucleus and cytoplasm. Finally, we found P0 protein and not PML within LCMV virions, indicating selective LCMV assembly.
MATERIALS AND METHODS
Virus infection.
NIH 3T3 cells were grown on coverslips and infected with LCMV Armstrong at a multiplicity of 1 PFU per cell. At 70 or 90 h after infection, coverslips were washed in phosphate-buffered saline (PBS) and fixed in acetone for 5 min. Fixed cells were stained with hyperimmune guinea pig anti-LCMV serum to confirm infection. Guinea pig serum was heated for 1 h at 78°C to inactivate the virus. The serum was used at a 1:500 dilution with a 1:200 fluorescein isothiocyanate (FITC)-conjugated goat anti-guinea pig secondary antibody (Jackson Immunoresearch).
Immunofluorescence studies.
Our immunofluorescence methods were adapted from those of Harlow and Lane (16). Briefly, fixed cells were washed twice in PBS. Coverslips were then blocked in 10% newborn calf serum (NCS; Gibco) and washed twice in PBS. The first antibody (in 10% NCS–0.1% Tween 20) was then applied for 1 h at room temperature. Afterward, the cells were washed in PBS three times for 5 min each time. The second antibody (in 10% NCS–0.1% Tween 20) was applied for 30 min at room temperature. Coverslips were then washed once in PBS with 0.1% Tween 20, three times in PBS, and once in water. Each wash lasted 5 min. Coverslips were mounted in Vectoshield.
PML polyclonal antibody was used at a dilution of 1:200 (2, 6) and detected with either a 1:200 dilution of goat anti-rabbit FITC (Jackson Immunoresearch) or a 1:100 dilution of goat anti-rabbit Texas Red secondary antibody (Vector). The serum of a lupus patient with autoantibodies to the ribosomal P proteins, a kind gift of Eng Tan, was used at a dilution of 1:200. The P proteins were detected by using an FITC-conjugated donkey anti-human secondary antibody (Dakopatts). Fluorescence was observed by using a confocal laser microscope (Zeiss and Leica) with excitation recorded at 568 nm (red) or 488 nm (green). The two channels were recorded independently to avoid cross talk between the channels. The pinhole for the colocalization experiments was 18 to ensure that the FITC signal did not break into the red channel. Under these conditions, there was no breakthrough of the FITC signal into the red channel or vice versa. Each confocal microscope experiment was performed at least twice, and the number of cells in each sample exceeded 500. Images were overlaid in Photoshop and enlarged for the counting of the PML, P, and Z bodies found within each nucleus and recording of the percentage of bodies which colocalized. The total number of PML and P bodies counted in uninfected cells was 140, and for infected cells it was 98. For the infected cells, the total number of P and Z bodies was 116. The P-PML analysis involved nine uninfected cells and six infected cells. The P-Z analysis involved six infected cells. One can distinguish whether one or both antigens are present by overlaying confocal microscope images; nuclear bodies which contain only one antigen stain red or green, and nuclear bodies which contain both appear yellow.
Transient transfection and subcellular fractionation.
NIH 3T3 cells were transfected with lipofectamine as directed by the manufacturer (Gibco), with 1 μg of the appropriate construct. The mammalian expression construct containing Z was prepared as described previously (3). At 72 h after transfection, cells were fractionated as previously described (16, 41). In short, cells were grown in T80 flasks and rinsed in EDTA (Gibco). A solution of 0.25% trypsin–1 mM EDTA was added, and the mixture was incubated for 2 min. Ten percent fetal bovine serum in Dulbecco modified Eagle medium (Gibco) was added. The samples were moved to 50-ml Falcon tubes and centrifuged. The cells were resuspended in 5 ml of cold PBS (per flask), spun, and resuspended in buffer A (110 mM potassium acetate, 2 mM magnesium, 2 mM dithiothreitol [DTT], 10 mM HEPES [pH 7.3]). The cells were spun again and resuspended with protease inhibitors and 20 μM cytochalasin B in buffer B (10 mM potassium acetate, 2 mM magnesium acetate, 2 mM DTT, 5 mM HEPES [pH 7.3]). Protease inhibitors included leupeptin at 2 μg/ml, pepstatin A at 2 μg/ml, and aprotinin at 0.5%. The buffered cells were incubated on ice for 10 min. Cells were then disrupted by passage through 18-, 21-, and 23-gauge needles. The KCl concentration was adjusted to 100 mM, at which point an aliquot was saved and referred to as the total cell lysate.
The lysate was spun at 1,500 × g for 15 min at 4°C to yield a pellet and a supernatant designated the nuclear and cytoplasmic fractions. Additionally, the cytoplasmic fraction was spun at 100,000 × g to yield a pellet (P100) and a supernatant (S100) fraction. Both fractions were precleared before coimmunoprecipitation. The final EDTA concentration for the lysate was 5.0 mM. The nuclear pellets were resuspended in 1.0 ml of coimmunoprecipitation buffer (100 mM KCl, 5 mM EDTA, 1 mM DTT, 10 mM HEPES [pH 7.3]). Rabbit prebleed serum was added to both the cytoplasmic and nuclear lysates, and the mixtures were incubated for 1 h at 4°C. After being equilibrated in coimmunoprecipitation buffer, protein A-Sepharose was added to the lysates. The mixture was incubated while rotating for 30 min at 4°C. The mixtures were respun, and the resulting supernatants were moved to clean the tubes. Fresh protein A-Sepharose was then added, and the mixture was incubated for 30 min at 4°C. The resulting supernatants were moved to clean tubes and saved.
Coimmunoprecipitation studies.
Protein-protein interactions were demonstrated by coimmunoprecipitation assays. Cell lysates (described above) were mixed with rabbit anti-Z serum and bovine serum albumin for immunoprecipitations as described previously (3). After being incubated at 4°C for 1 h with protein A-Sepharose (Pharmacia), the samples were washed twice in coimmunoprecipitation buffer and 0.5% Triton X-100, boiled in a reducing buffer, and subjected to sodium dodecyl sulfate (SDS)–20% polyacrylamide gel electrophoresis (PAGE). Subsequently, samples were Western blotted onto Immobolin P membranes, and ECL (Amersham) was used to visualize the bound antibodies. The blots were probed with the P protein at a dilution of 1:1,000. As a control, the elongation initiation factor 4E (eIF-4E) monoclonal antibody (MAb) (Transduction Laboratories) was also used at 1:625 to determine if Z would precipitate it. Band intensities and areas were measured by using NIH Image v.1.5.8 software.
In separate coimmunoprecipitation experiments, a MAb specific to human PML, MAb 5E10, was covalently attached to protein A-Sepharose (Pharmacia) by cross-linking with dimethyl pimelimidate as previously described (24, 40). MAb 5E10 is specific for the human form of PML and has been extensively characterized previously (42). Briefly, protein A-Sepharose beads were washed twice in 0.2 M sodium borate (pH 9.0). Approximately 2 mg of antibody per ml of beads was mixed continuously for 2 h at room temperature (RT). Beads were washed with sodium borate solution, followed by 0.2 M triethanolamine (pH 8.5). An equal volume of 40 mM dimethyl pimelimidate in 0.2 M triethanolamine was added, the mixture was gently agitated for 1 h at RT, and spun, and the supernatant was removed. An equal volume of 0.2 M ethanolamine (pH 8.2) was added to the beads, and the mixture was incubated for 5 min at RT. The beads were washed with the sodium borate solution and stored at 4°C until use. The beads were rinsed in PBS prior to use.
Human PML was immunoprecipitated from human fibroblast 551 cells (ATCC CCL-110). The cells were washed twice in serum-free medium, trypsinized, and pelleted at 800 × g and RT for 5 min. Pellets were washed three times in medium prewarmed to 37°C. Cells were lysed in IPB buffer (150 mM NaCl, 20 mM Tris-HCl [pH 7.4], 1% Nonidet P-40 [NP-40], 100 μM phenylmethylsulfonyl fluoride, 5 μg each of chemostatin, leupeptin, pepstatin A, aprotinin, and antipain per ml). Cells were spun at 800 × g for 5 min at 4°C. Pellets were resuspended and spun at 13,000 × g for 20 min at 4°C, and supernatants were precleared as described by Harlow and Lane (15). Supernatants were collected and mixed with 20 μl of protein A-Sepharose plus mouse immunoglobulin for at least 30 min at 4°C and spun to remove beads. Protein A-MAb 5E10 beads were added to precleared supernatants and mixed for 2 h at 4°C. The beads were then washed three times with modified IPB buffer (0.1% deoxycholate, no NP-40). The beads were spun and collected, and an equal volume of reducing sample buffer was added in preparation for SDS-PAGE.
Time course of infection.
HeLa cells were grown in T75 flasks with one flask per time point (24, 48, 72, and 96 h after infection). Each T75 flask had approximately 8.4 × 106 HeLa cells that were uninfected or infected at a multiplicity of 1 PFU per cell and harvested as described below. Cells were washed in PBS, overlaid with 1 to 2 ml of lysis buffer (1% NP-40, 0.1% SDS, 100 μg of phenylmethylsulfonyl fluoride per ml, 10 μg of aprotinin per ml in PBS [pH 7.4]). Lysates were collected and sonicated for 30 s on the maximum setting. Extracts were spun at 14,000 × g at 4°C. No pellets were observed. Additional protease inhibitors were added, resulting in 200 μg of PMSF per ml and 20 μg of aprotonin per ml. These extracts were stored at −20°C. Extracts were boiled in a reducing loading buffer, subjected to SDS–20% PAGE, and blotted onto Immobilon P in accordance with standard methods (16). Each lane represents 1 × 105 to 2 × 105 cells.
Preparation of virions.
Virus was precipitated from the medium of 48-h-infected BHK cells by adding polyethylene glycol 8000 to 7% (wt/vol) at 4°C overnight. The precipitate was centrifuged at 6,000 × g for 45 min, and the pellet was resuspended in 1× TNE (10 mM Tris HCl [pH 8], 1 mM EDTA, 100 mM NaCl) at 2 ml of TNE per 100 ml of the original medium volume. Virus suspensions were further purified on Renografin 76 (Squibb, Princeton, N.J.). Gradients were formed in SW41 centrifuge tubes in 1-ml 50%, 2-ml 40%, and 3-ml 10% Renografin layers. Virus in 5 to 6 ml of TNE was layered on each gradient and spun at 35,000 rpm for 75 min in a Beckman SW41 rotor. The opalescent virus band was collected at the 40-to-50% Renografin interface, diluted threefold in TNE, pelleted for 1 h at 30,000 × g, and suspended in 1× TNE (100 μl per 500 ml of the original medium volume). The yield was approximately 1 mg or 1010 PFU of LCMV per liter of medium. A 2-μl volume of this suspension was boiled in Laemmli buffer for Western blotting.
RESULTS
P protein has a nuclear punctate and a diffuse cytoplasmic pattern.
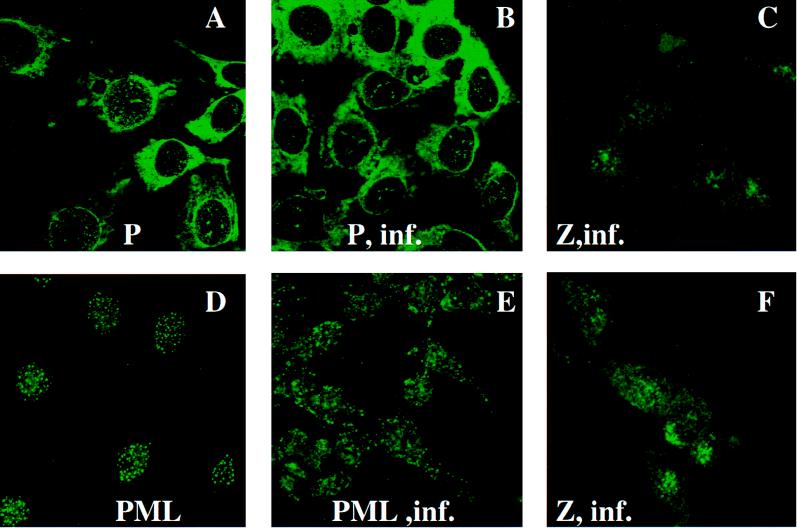
Confocal microscopy (Fig. 1) of cells stained with the P protein antibody (recognizing P0, P1, and P2 and characterized in references 11, 13, and 34) confirmed the cytoplasmic and nuclear distribution within NIH 3T3 cells that has been described previously (50, 52). The cytoplasmic fraction of the P proteins formed an intense, diffuse pattern, as expected, given its presence in the ribosome. The nuclear fraction was punctate. As shown in Fig. 1A and B, the nuclear portion was excluded from the nucleolus. Previous fractionation experiments indicated that P0 was in the nuclear, and not the nucleolar, fraction of quiescent cells (52).
FIG. 1.
Effect of LCMV infection on P proteins, PML, and Z in NIH 3T3 cells. Experiments were carried out as described in Materials and Methods. A and B, Cells stained with anti-P protein antibody in uninfected and infected (inf.) cells (90 h), respectively; D and E, Cells stained with the PML polyclonal antibody in uninfected and infected cells (90 h), respectively; C and F, staining for affinity-purified Z antibody in cells infected for 90 h. Magnifications: ×100 for A and B and ×40 for C to F with zooms of 3.3 (C), 2.6 (D), 2.8 (E), and 3.2 (F). Panels A and B represent single sections through the cell, whereas C, D, E, and F represent projections of several sections through the entire cell.
P0 subcellular distribution does not appear to change during infection by LCMV.
Figure 1B shows cells which have been stained with the P protein antibody after 90 h of LCMV infection. There appears to be no appreciable difference in the P proteins’ subcellular distribution before and after infection (Fig. 1A and B). The nuclear component is still present. For comparison, NIH 3T3 cells were stained with the PML antibody before and 90 h after infection with LCMV (Fig. 1D and E). PML’s normal nuclear distribution is shown in Fig. 1D, but Fig. 1E shows that the majority of PML bodies are relocated to the cytoplasm during infection, as we have demonstrated previously (3).
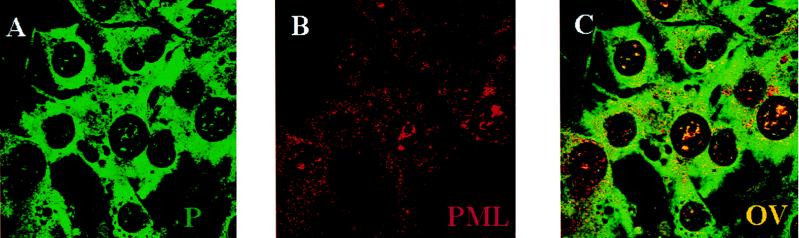
P proteins colocalize with PML in uninfected cells.
The nuclear punctate pattern of P protein is reminiscent of the pattern of PML bodies. To ascertain whether PML and P proteins colocalize, we undertook experiments using standard immunofluorescence and confocal microscopy. The anti-P protein staining indicates a punctate nuclear distribution with an intense, diffuse cytoplasmic pattern (Fig. 2A). Figure 2B shows the typical punctate nuclear staining observed for the polyclonal PML antibody in NIH 3T3 cells. Note that there is some cytoplasmic staining that has also been reported by other groups (12, 26, 44). Cells were analyzed to determine the percentages of the two proteins which colocalized. Approximately 60% ± 13% of the P proteins colocalized with PML; 77% ± 19% of PML colocalized with P proteins (see Materials and Methods). The majority of PML and P proteins found in the nucleus colocalized (Fig. 2C), with larger bodies tending to colocalize completely, leaving only small microbodies not colocalizing.
FIG. 2.
PML and P proteins colocalize in uninfected NIH 3T3 cells. Panels: A, cells stained with the P protein antibody (green); B, cells stained with the PML antibody (red); C, overlay (OV) (yellow). These confocal micrographs represent single slices through the plane of cells. Magnification, ×100. FITC was excited at 488 nm, and Texas Red was excited at 568 nm. The two channels were recorded independently.
Post-LCMV infection, the remaining PML nuclear component colocalizes with P proteins.
We ascertained whether PML and P proteins colocalize in infected cells (Fig. 3). In Fig. 3A, cells are stained with the anti-P protein antibody (green); in Fig. 3B, cells are stained with the PML antibody (red). Several cells no longer have many nuclear bodies, as expected, in Fig. 3B. Unlike PML in uninfected cells (Fig. 1D and 2B), there is significant staining in the cytoplasm. It is apparent that PML and P proteins still colocalize in those cells which retain PML in the nucleus (Fig. 3C). Approximately 61% ± 26% of P protein bodies colocalized with PML, and 95% ± 8% of PML bodies colocalized with the P proteins. The proportion of P proteins that colocalized with PML was essentially the same as in uninfected cells; however, the proportion of PML bodies colocalizing with P protein rose from 77% in uninfected cells to 95% in infected cells. This suggests that the subpopulation remaining in the nucleus was selected on the basis of protein-binding ability. It is interesting that although P proteins and PML colocalize in uninfected cells, the subcellular distribution of P proteins is not significantly affected by infection (Fig. 1A and B).
FIG. 3.
PML and P proteins colocalize in NIH 3T3 cells infected for 90 h with LCMV. Panels: A, cells stained with the P protein antibody (green); B, cells stained with the PML antibody (red); C, overlay (OV) (yellow). These confocal micrographs represent single slices through the plane of cells. Magnification, ×100. FITC was excited at 488 nm, and Texas Red was excited at 568 nm. The two channels were recorded independently.
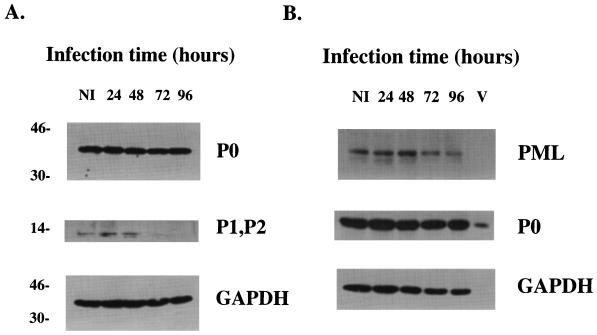
Expression of P1 and P2, but not that of P0, is altered during LCMV infection.
P protein levels during the infection process were monitored by using standard Western blotting techniques (see Materials and Methods). Extracts of infected or uninfected HeLa cells were probed with the P protein antibody (Fig. 4A) to reveal bands at the expected molecular sizes (34). The levels for P0 (molecular mass, approximately 37 kDa) were unchanged during the infection process. However, the P1 and P2 proteins (approximately 14 kDa) were substantially downregulated between 48 and 72 h after infection (Fig. 4A). There was also a drop in PML expression between 48 and 72 h (Fig. 4B). As a control, glyceraldehyde 3-phosphate dehydrogenase (GAPDH) levels were also monitored. These levels remained unchanged for the duration of the time course. The presence of various host cell proteins in the virions was also monitored. These data revealed that the P0 protein is found in virions (Fig. 4B). Neither GAPDH, a highly abundant cellular protein, nor PML was incorporated into virions.
FIG. 4.
The PML, P1, and P2 proteins are downregulated during infection, but the levels of P0 remain unchanged. Infection and Western blot analysis were carried out as described in Materials and Methods. NI, not infected. The levels of GAPDH were measured during the same time course as a control. In panel B, blots were prepared with virion preparations (lane V), as well as infected cells, as in panel A. The values to the left are molecular sizes in kilodaltons.
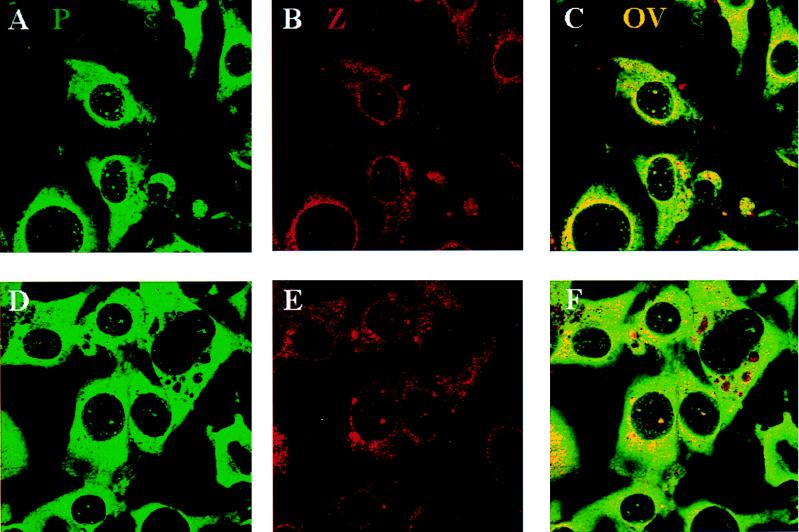
The Z protein colocalizes with P0, but only in the nucleus.
Cells were stained with the anti-P protein antibody (Fig. 5A and D) or with affinity-purified anti-Z antibody (Fig. 5B and E). Although the nuclear staining of the Z antibody is weak, it is clearly present. The Z and P proteins colocalize in the nucleus (Fig. 5C and F). The cytoplasmic pattern of P protein remains intense and diffuse; it does not concentrate with the Z bodies. Approximately 85% ± 17% of Z colocalized with P proteins, while only 38% ± 10% of P proteins colocalized with Z. P protein bodies that did not colocalize with Z tended to be smaller than the larger colocalizing bodies. Note that the same punctate nuclear pattern is evident for Z bodies in single-staining experiments (Fig. 1C and F), so that this nuclear and cytoplasmic punctate pattern is not a result of the signal from the P protein leaking into the red channel.
FIG. 5.
The viral Z protein and the P proteins colocalize in infected cells. Cells were infected for 90 h. Panels A and D show cells stained with the P protein antibody (green), panels B and E show cells stained with the affinity-purified Z antibody (red), and panels C and F show the overlay (OV) (yellow). These confocal micrographs represent single slices through the plane of cells. Magnification, ×100. FITC was excited at 488 nm, and Texas Red was excited at 568 nm. The two channels were recorded independently.
The Z nuclear fraction associates with P0 in cells transfected with Z.
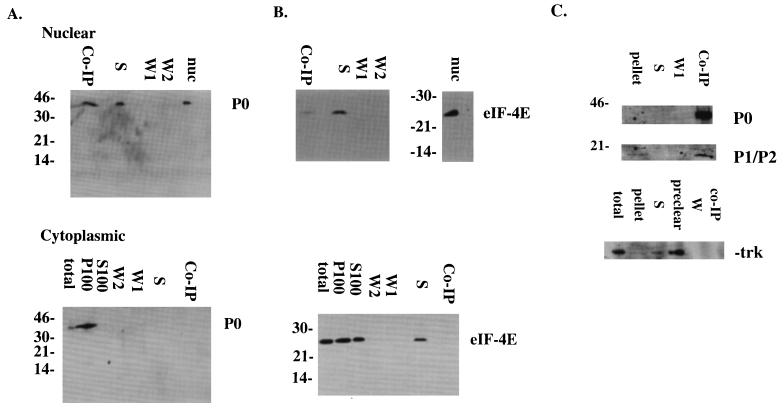
To determine whether the interaction between P and Z was direct, coimmunoprecipitation studies were conducted (Fig. 6A). The Z gene was transfected into NIH 3T3 cells that were subsequently fractionated into nuclear and cytoplasmic components. These fractions were then coimmunoprecipitated with the anti-Z antibody as described in Materials and Methods. The Z antibody precipitated P0 in the nuclear, but not the cytoplasmic, fraction (Fig. 6A). Band intensities were measured by using NIH Image v.1.5.8 software. This analysis indicated that approximately 50% of the P protein was precipitated after extensive washing (see Materials and Methods). This result is consistent with the ratio of P protein (approximately 38% ± 10%) observed colocalizing in the nuclei of infected cells (see above). The Z protein precipitated P0, but there was no indication of the P1 and P2 proteins. Further, the lack of colocalization in the cytoplasm was also consistent with our confocal studies (Fig. 5): P0 is found mainly in the ribosomal or P100 fraction, and trace amounts are found in the S100 fraction (34 and this study). The Z protein is found in the S100 fraction (3, 36). As expected, Z proteins did not coimmunoprecipitate P0 in the cytoplasm. It is possible that there was so little P0 in the S100 fraction that we could not detect it in the coimmunoprecipitation fraction. As a control, the ability of the Z protein to immunoprecipitate eIF-4E was ascertained. The samples for this experiment were identical to those used for the experiment in Fig. 6A. Z did not precipitate eIF-4E (25 kDa) in the cytoplasmic fraction, and only a trace amount was present in the nuclear fraction precipitate lane (Fig. 6B).
FIG. 6.
Direct interaction between the Z and P proteins in transfected cells (A). Cells were fractionated and coimmunoprecipitated with the Z antibody as described in Materials and Methods. In panel B, blots were probed with a control antibody, eIF-4E (140 kDa) to indicate that the coimmunoprecipitations were specific (see text for details). In panel C, cell lysates were coimmunoprecipitated with MAb 5E10 and probed with the P protein antibody as described in Materials and Methods. As a negative control, blots were probed with the trk antibody. Lanes: Co-IP, coimmunoprecipitated fraction; S, supernatant after coimmunoprecipitation; W1 and W2, washes one and two, respectively, after coimmunoprecipitation; nuc, nuclear fraction; total, total cell lysate. P100 and S100 are described in Materials and Methods. The values on the left are molecular sizes in kilodaltons.
PML and P proteins coimmunoprecipitate.
Human 551 fibroblasts were used to determine whether PML and P proteins directly interacted (Fig. 6C). PML coimmunoprecipitated P0, P1, and P2 from whole cell lysates. In Fig. 6C, the bands for P1 and P2 have coalesced; however, no P proteins are present in the supernatant lane. In our confocal microscopy studies, all of the P0 in the cells clearly did not colocalize with PML, especially as P0 is mainly cytoplasmic. Whole cell lysates were used in these coimmunoprecipitation experiments. Thus, this experiment does not reflect the normal compartmentalization within the cell. As a negative control, these blots were probed with an unrelated protein, trk (140 kDa; Santa Cruz), which did not coimmunoprecipitate with PML (Fig. 6C).
DISCUSSION
Little is known about arenavirus interactions with host cell molecules. Arenaviruses are known to replicate in the cytoplasm, but the viral Z gene product can be found in the nucleus in association with the host PML protein (3) and with ribosomal P proteins (this report). It has yet to be determined whether these associations are incidental to or essential for virus replication. Others have shown that cells enucleated prior to 12 h after infection cannot complete virus replication (reviewed in reference 35). Therefore, the host nucleus is essential for arenavirus replication and the PML and P proteins are reasonable candidates for the host cell nuclear components involved. The ribosomal P0 protein has nucleolytic activity (52) and could possibly function in viral RNA processing.
Arenaviruses are thought to contain ribosomes due to their appearance in electron micrographs and the ability of virus preparations to polymerize radioactive amino acids (23). We show here that ribosomal protein P0 is incorporated into the virion. However, the route of incorporation is unclear. It does not appear to be directly mediated through the Z protein for two reasons: (i) Z does not redistribute nuclear P0 upon infection, and (ii) Z and P0 do not appear to colocalize, coimmunoprecipitate, or cofractionate in the cytoplasm. The association of Z and P0 in the nucleus and the appearance of P0 in the virion appear to be separate events. Therefore, the incorporation of P0 into virions may be a result of interaction with other viral proteins.
LCMV infection results in the downregulation of P1 and P2, but not P0. There is also a less substantial decrease in PML levels. P1 and P2 downregulation is detectable at 48 to 72 h by immunofluorescence and Western analysis, and by 70 to 100 h, only P0 is detectable. The P1 and P2 proteins are required for active translation, as demonstrated in previous immunodepletion experiments (25, 49). P1- and P2-deficient ribosomes lose the abilities to bind EF1 and EF2 and to hydrolyze GTP (25, 28, 38). It is generally held that arenavirus infection does not perturb essential cell functions like protein synthesis, although 70- to 100-h-infected cell cultures do show decreased rates of cell division. Therefore, either the infection has an inhibitory effect on ribosome function or the virus supplies something to replace the functions of P1 and P2.
The interaction between P0 and Z is dependent on subcellular compartmentalization. The function of P0 is linked to its subcellular distribution; nuclear P0 has endonuclease and excision repair activities (52), whereas most of the cytoplasmic P0 is associated with ribosomes and translational activities (45–49). P0 is known to undergo phosphorylation, and therefore the nuclear and cytoplasmic fractions may differ in levels of phosphorylation or some other posttranslational modification. The PML/P0 bodies and the Z/P0 bodies are only observed in the nucleus, which must reflect the different functions associated with the nuclear and cytoplasmic fractions of P0.
Despite a wealth of information, the function of PML nuclear bodies remains enigmatic. This is the first report to link PML with ribosomal proteins in cell culture. Direct interactions between PML and two ribosome-associated proteins, EF1 and leucine zipper (L7), were observed in yeast two-hybrid studies (2) but not pursued because the cytoplasmic subcellular distribution of these components appeared to make them inappropriate partners for PML. However, more recent data, as well as the data presented here, indicate that the P proteins have a nuclear, as well as a cytoplasmic, distribution (50, 52). Another ribosomal protein, S3, has both a nuclear and a cytoplasmic distribution and is associated with DNA repair activity (53). PML interacts with at least three ribosomal proteins: P0, EF1, and L7. This association might explain why up to 20% of PML is found in the cytoplasm of normal cells (12, 44). Spatially, P0 and EF1 are located near each other on the ribosome (47). As ribosomal subunits are often exported to the cytoplasm in a partially assembled state, the associations in the cytoplasm probably reflect associations in the nucleus. Thus, PML nuclear bodies may be involved in some level of translational control in the cell.
The interaction of LCMV with host PML and P proteins may be related to its ability to establish chronic infection in both organisms and tissue cultures (35). Previously, it was established that PML is proapoptotic (4) and that both genetic disruption of PML (4) and LCMV infection reduce this activity (4). Recent reports suggest a connection between ribosomal proteins and oncogenesis. Several ribosomal proteins, including S3 and P0, are elevated in colorectal tumors and polyps (32). A fusion protein containing the L7a protein has been isolated from transformed cells and appears to be active in transformation (55). The cleavage of 28S RNA is thought to be an important step in the execution of apoptosis (17), and as P0 directly binds 28S RNA, it would be ideally placed to perform such a function. Thus, the interaction of PML with P proteins demonstrated here suggests a mechanism for PML’s proapoptotic activities: in uninfected cells, PML would interact with P0 and promote its proapoptotic function, and in infected cells, PML would relocate to the cytoplasm, no longer promoting P0 and resulting in reduced apoptosis.
In summary, we have established the interaction of the LCMV Z protein with another set of host proteins, the ribosomal P proteins in the nuclei of infected cells. We have shown that infection downregulates the ribosomal P1 and P2 proteins but not the P0 protein. Our data support claims that virions contain ribosomal components. Finally, we have demonstrated colocalization of PML and ribosomal proteins within complexes in the nuclei, but not in the cytoplasm, of uninfected cells.
ACKNOWLEDGMENTS
We thank Lynne Maillet-Frotten and W. G. Tatton for use of confocal microscopes. We are very grateful for the PML polyclonal antibody from K. Howe, M. N. Boddy, P. Freemont, and E. Solomon and for the lupus patient antiserum from Eng Tan. We thank L. Etkin and P. Freemont for their helpful conversations. We also thank I. Lukashevich for critical comments and Rhea MacDonald and Li Xia for technical assistance.
K.L.B.B. acknowledges financial support from the MRC of Canada (MT-13608). M.S.S. was supported by NIH grants RO1 AI32107 and R29 AI25522.
REFERENCES
- 1.Ascoli C A, Maul G G. Identification of a novel nuclear domain. J Cell Biol. 1991;112:785. doi: 10.1083/jcb.112.5.785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Boddy M N, Howe K, Etkin L D, Solomon E, Freemont P S. PIC1, a novel ubiquitin-like protein which interacts with the PML component of a multiprotein complex that is disrupted in acute promyelocytic leukaemia. Oncogene. 1996;13:971–982. [PubMed] [Google Scholar]
- 3.Borden K L B, CampbellDwyer E J, Salvato M S. An arenavirus RING (zinc-binding) protein binds the oncoprotein PML and relocates PML nuclear bodies to the cytoplasm. J Virol. 1998;72:758–766. doi: 10.1128/jvi.72.1.758-766.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Borden K L B, CampbellDwyer E J, Salvato M S. The promyelocytic leukemia protein PML has a pro-apoptotic activity mediated through its RING. FEBS Lett. 1997;418:30–34. doi: 10.1016/s0014-5793(97)01344-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Borden K L B, Freemont P S. The RING finger: an example of a sequence structure family. Curr Opin Struct Biol. 1996;6:395–401. doi: 10.1016/s0959-440x(96)80060-1. [DOI] [PubMed] [Google Scholar]
- 6.Borden K L B, Boddy M N, Lally J, O’Reilly N J, Martin S, Howe K, Solomon E, Freemont P S. The solution structure of the RING finger domain from the acute promyelocytic leukaemia proto-oncoprotein, pml. EMBO J. 1995;14:1532–1541. doi: 10.1002/j.1460-2075.1995.tb07139.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Borden K L B, Lally J M, Martin S R, O’Reilly N J, Solomon E, Freemont P S. In vivo and in vitro characterization of the B1 and B2 zinc-binding domains from the acute promyelocytic leukemia protein PML. Proc Natl Acad Sci USA. 1996;93:1601–1606. doi: 10.1073/pnas.93.4.1601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.de The H C, Lavau C A, Marchio C, Chrornr L, Degos L, Dejean A. The PML-RAR alpha fusion mRNA generated by t(15;17) translocation in acute promyelocytic leukemia encodes a functionally altered RAR. Cell. 1991;66:675–684. doi: 10.1016/0092-8674(91)90113-d. [DOI] [PubMed] [Google Scholar]
- 9.Djavani M, Lukashevich I S, Sanchez A, Nichol S T, Salvato M S. Completion of the Lassa fever virus sequence and identification of a RING finger open reading frame at the L RNA 5′ end. Virology. 1997;235:414–418. doi: 10.1006/viro.1997.8722. [DOI] [PubMed] [Google Scholar]
- 10.Dyck J A, Maul G G, Miller W H, Chen D J, Kakizuka A, Evans R M. A novel macromolecular structure is a target of the promyelocyte-retinoic acid receptor oncoprotein. Cell. 1994;76:333–343. doi: 10.1016/0092-8674(94)90340-9. [DOI] [PubMed] [Google Scholar]
- 11.Elkon K B, Parnassa A P, Foster C L. Lupus autoantibodies target ribosomal P proteins. J Exp Med. 1985;162:459–471. doi: 10.1084/jem.162.2.459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Flenghi L, Fagioli M, Tomassoni L, Pileri S, Gambacorta M, Pacini R, Grignani F, Casini T, Ferrucci P F, Martelli M F, Pelicci P G, Falini B. Characterization of a new monoclonal antibody (PG-M3) directed against the amino-terminal portion of the PML gene product: immuncytochemical evidence for high expression of PML proteins on activated macrophages, endothelial cells and epithelia. Blood. 1995;85:1871–1880. [PubMed] [Google Scholar]
- 13.Francoeur A, Peebles C L, Heckman K J, Lee J C, Tan E M. Identification of ribosomal protein autoantibodies. J Immunol. 1985;135:2378–2384. [PubMed] [Google Scholar]
- 14.Garcin D, Rochat S, Kolakofsky D. The Tacaribe arenavirus small zinc finger protein is required for both mRNA synthesis and genome replication. J Virol. 1993;67:807–812. doi: 10.1128/jvi.67.2.807-812.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Goddard A D, Borrow J, Freemont P S, Solomon E. Characterization of a zinc finger gene disrupted by the t(15:17) in acute promyelocytic leukemia. Science. 1991;254:1371–1373. doi: 10.1126/science.1720570. [DOI] [PubMed] [Google Scholar]
- 16.Harlow E, Lane D. Antibodies: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1988. [Google Scholar]
- 17.Houge G, Doskeland S O. Divergence towards a dead end? Cleavage of the divergent domains of ribosomal RNA in apoptosis. Experientia. 1996;52:963–967. doi: 10.1007/BF01920105. [DOI] [PubMed] [Google Scholar]
- 18.Ishov A M, Maul G G. The periphery of nuclear domain 10 (ND10) as site of DNA virus deposition. J Cell Biol. 1996;134:815–826. doi: 10.1083/jcb.134.4.815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kakizuka A, Miller W, Umensono K, Warrell R, Frankel S R, Murty V V, Dmitrovsky E, Evans R M. Chromosomal translocation t(15;17) in human acute promyelocytic leukemia fuses RARα with a novel putative transcription factor PML. Cell. 1991;66:663–674. doi: 10.1016/0092-8674(91)90112-c. [DOI] [PubMed] [Google Scholar]
- 20.Kastner P, Perez A, Lutz Y, Rochette-Egly C, Gaub B, Durnad M, Lanotte M, Berger R, Chambon P. Structure, localization and transcriptional properties of two classes of retinoic acid receptor alpha fusion proteins in acute promyelocytic leukemia (APL): structural similarities with a new family of oncoproteins. EMBO J. 1992;11:629–642. doi: 10.1002/j.1460-2075.1992.tb05095.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Koken M H M, Puvion-Dutilleul F, Guillemin M C, Viron V, Linares-Cruz G, Stuurman N, de Jong L, Szostecki C, Calvo F, Chomienne C, de The H. The t(15:17) translocation alters a nuclear body in a retinoic acid reversible fashion. EMBO J. 1994;13:1073–1083. doi: 10.1002/j.1460-2075.1994.tb06356.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Le X F, Yang P, Chang K S. Analysis of the growth and transformation suppressor domains of promyelocytic leukemia gene PML. J Biol Chem. 1996;271:130–135. doi: 10.1074/jbc.271.1.130. [DOI] [PubMed] [Google Scholar]
- 23.Leung W C, Rawls W E. Virion associated ribosomes are not required for the replication of Pichinde virus. Virology. 1977;81:17406. doi: 10.1016/0042-6822(77)90070-8. [DOI] [PubMed] [Google Scholar]
- 24.Ley S C, Verbi W, Pappin D J C, Druker B, Davies A A, Crumpton M J. Tyrosine phosphorylation of a tubulin in human T lymphocytes. Eur J Immunol. 1994;24:99–106. doi: 10.1002/eji.1830240116. [DOI] [PubMed] [Google Scholar]
- 25.MacConnell W P, Kaplan N P. The activity of the acidic phosphoproteins from the 80S rat liver ribosome. J Biol Chem. 1982;257:5395–5366. [PubMed] [Google Scholar]
- 26.Maul G G, Yu E, Ishov A M, Epstein A L. Nuclear domain 10 (ND10) associated proteins are present in nuclear bodies and redistribute to hundreds of nuclear sites after stress. J Cell Biochem. 1995;59:499–514. doi: 10.1002/jcb.240590410. [DOI] [PubMed] [Google Scholar]
- 27.Maul G G, Everett R D. The nuclear location of PML, a cellular member of the C3HC4 zinc-binding domain protein family, is rearranged during herpes simplex virus infection by the C3HC4 viral protein ICP0. J Gen Virol. 1994;75:1223–1233. doi: 10.1099/0022-1317-75-6-1223. [DOI] [PubMed] [Google Scholar]
- 28.Moller W, Slobin L I, Amons R, Richter D. Isolation and characterization of two acidic proteins of 60S ribosomes from Artemia salina cysts. Proc Natl Acad Sci USA. 1975;72:4744–4748. doi: 10.1073/pnas.72.12.4744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Montali R J, Connolly B M, Armstrong D L, Scanga C A, Holmes K V. Pathology and immunohistochemistry of callitrichid hepatitis, an emerging disease of captive new world primates caused by lymphocytic choriomeningitis virus. Am J Pathol. 1995;148:1441–1449. [PMC free article] [PubMed] [Google Scholar]
- 30.Mu Z-M, Chin K-V, Liu J-H, Lozano G, Chang K-S. PML, a growth suppressor disrupted in acute promyelocytic leukemia. Mol Cell Biol. 1994;14:6858–6867. doi: 10.1128/mcb.14.10.6858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pandolfi P P, Grignani F, Alcalay M, Mencarelli A, Biondi A, LoCoco F, Grignani F, Pellicci P G. Structure and origin of the acute promyelocytic leukemia myl/RARα cDNA and characterization of its retinoid-binding and transactivation properties. Oncogene. 1991;6:1285–1292. [PubMed] [Google Scholar]
- 32.Pogue-Geile K, Geiser J R, Shu M, Miller C, Wool I G, Meisler A I, Pipas J M. Ribosomal protein genes are overexpressed in colorectal cancer: isolation of a cDNA clone encoding the human S3 ribosomal protein. Mol Cell Biol. 1991;11:3842–3849. doi: 10.1128/mcb.11.8.3842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Reddy B A, Etkin L D, Freemont P S. A novel zinc finger coiled coil domain in a family of nuclear proteins. Trends Biochem Sci. 1992;17:344–345. doi: 10.1016/0968-0004(92)90308-v. [DOI] [PubMed] [Google Scholar]
- 34.Rich B E, Steitz J A. Human acidic ribosomal phosphoproteins P0, P1, and P2: analysis of cDNA clones, in vitro synthesis, and assembly. Mol Cell Biol. 1987;7:4065–4074. doi: 10.1128/mcb.7.11.4065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Salvato M S, Rai K S. Arenaviruses. In: Mahy B, editor. Topley and Wilson’s microbiology and microbial infections. 9th ed. London, United Kingdom: Arnold; 1996. pp. 629–650. [Google Scholar]
- 36.Salvato M S, Schweighofer K J, Burns J, Shimomaye E M. Biochemical and immunological evidence that the 11 kDa zinc-binding protein of lymphocytic choriomeningitis virus is a structural component of the virus. Virus Res. 1992;22:185–198. doi: 10.1016/0168-1702(92)90050-j. [DOI] [PubMed] [Google Scholar]
- 37.Salvato M S, Shimomaye E M. The completed sequence of lymphocytic choriomeningitis virus reveals a unique RNA structure and a gene for a zinc finger protein. Virology. 1989;173:1–10. doi: 10.1016/0042-6822(89)90216-x. [DOI] [PubMed] [Google Scholar]
- 38.Sanchez-Madrid F, Vidales F J, Ballesta J P G. Functional role of acidic ribosomal proteins. Interchangeability of proteins from bacterial and eukaryotic cells. Biochemistry. 1981;20:3263–3266. doi: 10.1021/bi00514a043. [DOI] [PubMed] [Google Scholar]
- 39.Saurin A J, Borden K L B, Boddy M N, Freemont P S. Does this have a familiar RING? Trends Biochem Sci. 1996;246:208–213. [PubMed] [Google Scholar]
- 40.Schneider C, Newman R A, Sutherland D R, Asser A, Greaves M F. A one-step purification of membrane proteins using a high efficiency immunomatrix. J Biol Chem. 1982;257:10766–10769. [PubMed] [Google Scholar]
- 41.Siomi M C, Zhang Y, Siomi H, Dreyfuss G. Specific sequences in the fragile Z syndrome protein FMR1 and the FXR proteins mediate their binding to 60S ribosomal subunits and the interactions among them. Mol Cell Biol. 1996;16:3825–3832. doi: 10.1128/mcb.16.7.3825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Stuurman N, De Graaf A, Floore A, Josso A, Humbel B, De Jong L, Van Driel R. A monoclonal antibody recognizing nuclear matrix associated nuclear bodies. J Cell Sci. 1992;101:773–784. doi: 10.1242/jcs.101.4.773. [DOI] [PubMed] [Google Scholar]
- 43.Szostecki J, Guldner H H, Netter H J, Will H. Isolation and characterization of cDNA encoding a human nuclear antigen predominantly recognized by autoantibodies from patients with primary biliary cirrhosis. J Immunol. 1990;145:4338–4347. [PubMed] [Google Scholar]
- 44.Terris B, Baldin V, Dubois S, Degott C, Flejou J-F, Henin D, Dejean A. PML nuclear bodies are general targets for inflammation and cell proliferation. Cancer Res. 1995;55:1590–1597. [PubMed] [Google Scholar]
- 45.Uchiumi T, Ogata K. Cross-linking study on localization of the binding site for elongation factor 1a on rat liver ribosomes. J Biol Chem. 1986;261:9668–9671. [PubMed] [Google Scholar]
- 46.Uchiumi T, Wahba A J, Traut R R. Topography and stoichiometry of acidic proteins in large ribosomal subunits from Artemia salina as determined by cross-linking. Proc Natl Acad Sci USA. 1987;84:5580–5584. doi: 10.1073/pnas.84.16.5580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Uchiumi T, Traut R R, Kominami R. Monoclonal antibodies against acidic phosphoproteins P0, P1, and P2 of eukaryotic ribosomes as functional probes. J Biol Chem. 1990;265:89–95. [PubMed] [Google Scholar]
- 48.Uchiumi T, Kominami R. Binding of mammalian ribosomal protein complex P0-P1-P2 and protein L12 to the GTPase associated domain of 28S ribosomal RNA and effect on the accessibility to anti-28S RNA autoantibody. J Biol Chem. 1997;272:3302–3308. doi: 10.1074/jbc.272.6.3302. [DOI] [PubMed] [Google Scholar]
- 49.Van Agthoven A J, Maassen J A, Moller W. Structure and phosphorylation of an acidic protein from 60S ribosomes and its involvement in elongation factor 2 GTP hydrolysis. Biochem Biophys Res Commun. 1977;77:989–998. doi: 10.1016/s0006-291x(77)80075-2. [DOI] [PubMed] [Google Scholar]
- 50.Vater C A, Bartle L M, Leszyk J D, Lambert J M, Goldmacher V S. Ricin A chains can be chemically cross-linked to the mammalian ribosomal proteins L9 and L10e. J Biol Chem. 1995;270:12933–12940. doi: 10.1074/jbc.270.21.12933. [DOI] [PubMed] [Google Scholar]
- 51.Weiss K, Rambaud S, Lavau C, Jansen J, Carvalho T, Carmo-Foneseca M, Lamond A, Dejean A. Retinoic acid regulates aberrant nuclear localization of PML-RARα in acute promyelocytic leukemia cells. Cell. 1994;76:345–356. doi: 10.1016/0092-8674(94)90341-7. [DOI] [PubMed] [Google Scholar]
- 52.Yacoub A, Kelley M R, Deutsch W A. Drosophila ribosomal protein P0 contains apurinic/apyrimidinic endonuclease activity. Nucleic Acids Res. 1996;24:4298–4303. doi: 10.1093/nar/24.21.4298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yacoub A, Auglri L, Kelley M R, Doetsch P, Deutsch W A. A Drosophila ribosomal protein contains 8-oxoguanine and abasic site DNA repair activities. EMBO J. 1996;15:2306–2312. [PMC free article] [PubMed] [Google Scholar]
- 54.Zhu Z, Cai W, Schaffer P A. Cooperativity among herpes simplex virus type 1 immediate early regulatory proteins: ICP4 and ICP27 affect the intracellular localization of ICP0. J Virol. 1994;68:3027–3040. doi: 10.1128/jvi.68.5.3027-3040.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ziemiecki A, Muller R G, Xiao-Chang F, Hynes N E, Kozma S. Oncogenic activation of the human trk protooncogene by recombination with the ribosomal large subunit protein L7a. EMBO J. 1990;9:191–196. doi: 10.1002/j.1460-2075.1990.tb08095.x. [DOI] [PMC free article] [PubMed] [Google Scholar]