Abstract
Objectives
Immunotherapies targeting natural killer (NK) cell receptors have shown promise against leukaemia. Unfortunately, cancer immunosuppressive mechanisms that alter NK cell phenotype prevent such approaches from being successful. The study utilises advanced cytometry to examine how cancer immunosuppressive pathways affect NK cell phenotypic changes in clinical samples.
Methods
In this study, we conducted a high‐dimensional examination of the cell surface expression of 16 NK cell receptors in paediatric patients with acute myeloid leukaemia and acute lymphoblastic leukaemia, as well as in samples of non‐age matched adult peripheral blood (APB) and umbilical cord blood (UCB). An unsupervised analysis was carried out in order to identify NK cell populations present in paediatric leukaemias.
Results
We observed that leukaemia NK cells clustered together with UCB NK cells and expressed relatively higher levels of the NKG2A receptor compared to APB NK cells. In addition, CD56dimCD16+CD57− NK cells lacking NKG2A expression were mainly absent in paediatric leukaemia patients. However, CD56br NK cell populations expressing high levels of NKG2A were highly represented in paediatric leukaemia patients. NKG2A expression on leukaemia NK cells was found to be positively correlated with the expression of its ligand, suggesting that the NKG2A‐HLA‐E interaction may play a role in modifying NK cell responses to leukaemia cells.
Conclusion
We provide an in‐depth analysis of NK cell populations in paediatric leukaemia patients. These results support the development of immunotherapies targeting immunosuppressive receptors, such as NKG2A, to enhance innate immunity against paediatric leukaemia.
Keywords: AML, B‐ALL, NK cells, NKG2A
We provide an in‐depth unsupervised immunophenotyping of NK cell populations in paediatric leukaemia patients. Our findings lend support to the development of immunotherapies that target immunosuppressive receptors, such as NKG2A, in order to boost innate immunity against paediatric leukaemia.
Introduction
Acute leukaemia is the most prevalent paediatric cancer. 1 There are two subtypes of paediatric leukaemia: acute lymphoblastic leukaemia (ALL) and acute myeloid leukaemia (AML). 2 , 3 Overall survival rates exceed 90% in paediatric ALL patients, but only 60–70% in paediatric AML patients. 4 Therefore, novel approaches are urgently needed to prevent relapse and overcome the long‐term toxicities of current therapies.
NK cells are innate lymphocytes that constitute promising therapeutic targets against acute leukaemias. 5 NK cell function is tightly controlled by the balance of activating and inhibitory signals received from germline‐encoded NK cell receptors. 6 Upon binding to their ligands on the surface of tumor cells, activating receptors such as NKG2D, NKp30, NKp44 and DNAM‐1 are known to trigger NK cell cytotoxic functions, while receptors such as NKG2A, KIRs, TIGIT and TIM‐3 are putative suppressors of NK cell tumoricidal activity. 7 Most studies divide human NK cells into two subsets, which are CD56brCD16− and CD56dimCD16+ NK cells. 8 However, the stochastic expression of NK cell receptors results in broad NK cell phenotypic diversity, with mass‐spectrometry analyses identifying over 100 000 NK cell populations in the blood of healthy donors. 9 Furthermore, a single‐cell RNA sequencing study revealed higher phenotypic heterogeneity among human bone marrow and blood NK cells, and they identified a transitional NK cell population between CD56br and CD56dim NK cells. 10
NK cell‐based immunotherapies are safe and have demonstrated promising results in clinical trials after their therapeutic allogeneic infusion against leukaemia 11 , 12 , 13 , 14 and solid cancers. 15 , 16 The blockade of NK cell receptor NKG2A via anti‐NKG2A antibodies or through genetic engineering has been proposed as a strategy to boost NK cell anti‐tumor immunity. 17 , 18 An alternative approach is the crosslinking of activating receptors such as NKG2D on NK cells with antigens highly expressed on tumor cells. As an example of a therapeutic approach, NKG2D bispecific antibodies (BiKE) were shown to successfully induce NK cell‐mediated killing of multiple myeloma cells. 19 The efficacy of immunotherapies targeting NK cell receptors might be impaired by tumor‐immunosuppressive mechanisms that have been shown to alter NK cell receptor expression profiles. 20 , 21 An extensive characterisation of these profiles is a prerequisite to the development of receptor‐targeting therapies. Previous studies using flow cytometry have reported low expression of the activating receptors NKp46, NKp30, NKp44 22 , 23 and DNAM‐1, 24 while the inhibitory receptors NKG2A, 25 PD‐1, 26 TIM‐326 and TIGIT 24 were found to be up‐regulated in adult AML patients. Alterations in NK cell receptor profiles have been associated with poor clinical outcomes in adult AML 22 , 25 , 26 and B‐ALL. 27 In comparison to AML, B‐ALL has received less attention in the study of NK cells. It was shown that NK cells in paediatric B‐ALL displayed high expression of NKp46, but their expression of NKG2D, NKp30, NKp44 and DNAM‐1 was similar to those of healthy donors. 27 , 28
Here, we applied high‐dimensional flow cytometry to provide in‐depth characterisation of NK cell receptor expression profiles in AML and ALL. Our findings revealed that CD56dimCD16+ NK cells characterised by low surface protein levels of NKG2A were poorly represented in paediatric leukaemia compared to non‐age match adult healthy blood samples. By contrast, CD56brCD57−KIRs− NK cells expressing high levels of NKG2A were abundant in leukaemia samples. Our results strengthen the rationale for targeting NKG2A in paediatric leukaemia patients.
Results
NK cells from paediatric leukaemia patients resemble those from UCB
We developed a 20‐colour flow cytometry receptor panel (Supplementary table 1) for the analysis of NK cell receptors in the peripheral blood (PB) and the matched bone marrow (BM) of paediatric patients diagnosed with AML or B‐ALL (Table 1). NK cells were gated as cells expressing CD56 and/or CD16 after pre‐gating on live CD45+CD33−CD19−CD3− lymphocytes (Supplementary figure 1). In agreement with published studies, 29 , 30 we observed significantly lower percentages of NK cells in the blood and BM of paediatric leukaemia AML and B‐ALL patients compared with healthy donor (HD) umbilical cord blood (UCB) and HD adult peripheral blood (APB; Figure 1a).
Table 1.
Paediatric leukaemia patient's characteristics
| Characteristic | AML | B‐ALL |
|---|---|---|
| Patient number | n = 15 | n = 12 |
| Age at diagnosis (years, median, range) | 10.66 (4.03–17.36) | 11.64 (2.31–17.13) |
| Relapse, No. (%) | 4 (27%) | 1 (8%) |
| In CR1 | 11 (73%) | 11 (92%) |
| In CR2 | 4 (27%) | 1 (8%) |
| Procedure type | ||
| Diagnostic | 15 (100%) |
12 (100%) |
| CMV results | ||
| CMV negative | 7 (47%) | 9 (75%) |
| IgG reactive | 6 (40%) | 0 (0%) |
| IgM non‐reactive | 2 (13%) | 1 (8%) |
| Unavailable | 0 (0%) | 2 (17%) |
AML, acute myeloid leukaemia; B‐ALL, B‐cell acute lymphoblastic leukaemia; CMV, cytomegalovirus; CR, complete remission.
Figure 1.
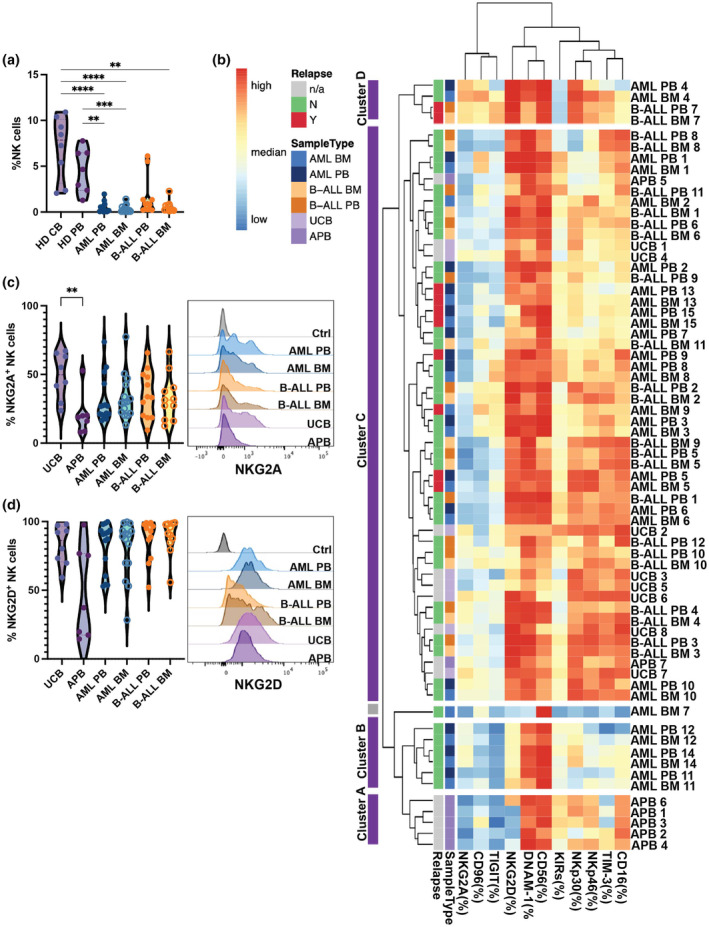
Low frequencies of NK cells in paediatric leukaemia patients. (a) Percentages of total NK cells in healthy donor umbilical cord blood (UCB; n = 8), healthy donor adult peripheral blood (APB; n = 7), peripheral blood (PB) and bone marrow (BM) of patients with acute myeloid leukaemia (AML; n = 15) or B‐cell acute lymphoblastic leukaemia (B‐ALL; BM: n = 11, PB: n = 12). (b) Hierarchical clustering heatmap display percentage of indicated markers of total NK cells from each donor. Purple clusters are NK cell clusters from different patients. The grey cluster is the non‐NK cell artefact population. (c) and (d) Representative flow cytometry histograms (right) and bar graphs (left) showing the surface expression of NKG2A and NKG2D on NK cells from different donors Data are shown as mean ± sem, data were tested using a Kruskal–Wallis test with Dunn's multiple comparisons test. **P < 0.01, ***P < 0.001, ****P < 0.0001.
Hierarchical clustering of the samples revealed four clusters, with most APB samples clustering apart from leukaemia and UCB samples (Figure 1b). Interestingly, BM and PB NK cells from a given patient clustered together, suggesting that NK cell phenotypic diversity was mostly driven by donor variability rather than organ location. Of note, BM AML sample #7 was considered as an artefact because these cells did not harbour any additional NK markers other than CD56. Cluster A regrouped five out of seven of the HD APB samples. Compared with the other clusters, Cluster A expressed relatively low levels of NKG2A, NKG2D and TIGIT. Cluster B regrouped PB and BM samples from three out of 15 AML patients. Like Cluster A, Cluster B was CD56bright and TIGIT−. However, compared with Cluster A, Cluster B expressed higher levels of NKG2A and NKG2D but lower levels of NKp46, KIRs and CD16. Cluster C was the largest cluster, regrouping most AML and B‐ALL samples together with all UCB samples. Compared to Clusters A and B, Cluster C had higher expression of NKG2D, NKp30, NKp46 and TIM‐3. Cluster D regrouped NK cells from one AML and one B‐ALL patient. Like Cluster C, Cluster D expressed high levels of NKG2D. Moreover, Cluster D expressed higher levels of NKG2A, CD96 and NKp30 than all the other clusters (Figure 1b).
We observed that the frequencies of NKG2D+ and NKG2A+ NK cells in leukaemia patients were comparable to those observed in UCB and higher than those observed in APB, despite not reaching statistical significance (Figure 1c and d). Moreover, cytomegalovirus (CMV) infection has been shown to increase NKG2C+/NKG2A− NK cells. 31 , 32 In AML patients, we compared the frequencies of NKG2A+ NK cells between those who were CMV negative, IgG reactive and IgM non‐reactive AML patients. Although there was no significant difference between the groups, AML patients who did not get CMV infection tended to have somewhat more NKG2A+ NK cells (Supplementary figure 2).
Interestingly, increased percentages of TIGIT+ NK cells in both AML and B‐ALL samples, and higher TIM‐3+ NK cells in B‐ALL samples; however, significant differences were only observed between B‐ALL BM and APB samples (Supplementary figure 3a and b). Higher DNAM‐1+ NK cells was also observed in B‐ALL PB samples compared to UCB samples (Supplementary figure 3c). Although there was expression in detectable levels, no significant difference in percentages of NK cells expressing KIRs, NKp30, CD96 and NKp46 was observed between leukaemia samples and HD samples (Supplementary figure 3d–g). We detected lower to no expression of CTLA‐4, LAG‐3, NKp44 and PD‐1 on NK cells (not shown in the paper). Taken together, these data indicate that the receptor expression profiles of NK cells in paediatric leukaemia patients resemble more those of UCB NK cells than those of APB NK cells, and they are characterised by relatively high expression levels of NKG2A and NKG2D.
In AML, NK cell expression of the NKG2A receptor correlates with the expression levels of their respective ligands on tumor cells
According to the cancer immunoediting process, 33 high expression of a given activating receptor on NK cells would lead to the selection of tumor cells expressing low levels of the corresponding ligand. 34 We applied two flow cytometry panels (Supplementary table 2 and 3) to evaluate the expression of ligands for NK cell receptors on leukaemia tumor cells, where the gating strategy for leukaemia tumor cells is shown in (Supplementary figures 4a and b, 5a and 6a and b). In AML, we observed positive correlations between the expression of NKG2A and its ligand HLA‐E on AML tumor cells (Figure 2a). To investigate whether the expression of NKG2A provides prognostic value, we analysed the gene expression and clinical data from the TARGET AML. We found that paediatric AML patients with high expression of KLRC1 (gene encoding NKG2A) had a worse prognosis compared to those with low gene expression (Figure 2b). Furthermore, paediatric AML patients with high expression of HLA‐E had a worse prognosis (Figure 2c). NKG2A expression on NK cells was shown in earlier research to be downregulated when NK cells matured from newborns to adults, and CMV infection affected the frequencies of NKG2A− NK cells. 32 Consequently, we used multivariable correlation analysis to look into any potential relationships between the age of patients and the expression of HLA‐E on tumors and NKG2A on NK cells. We verified that both the expression of NKG2A and HLA‐E did not correlate with the patient's age in both blood and BM samples (Figure 2d, e). Additionally, we used multiple regression analysis to see whether the CMV status affected the correlation between NKG2A and HLA‐E (Supplementary table 4). Given that the coefficient for CMV results had a P‐value greater than 0.05, the CMV results had no effect on the NKG2A‐HLA‐E connection.
Figure 2.
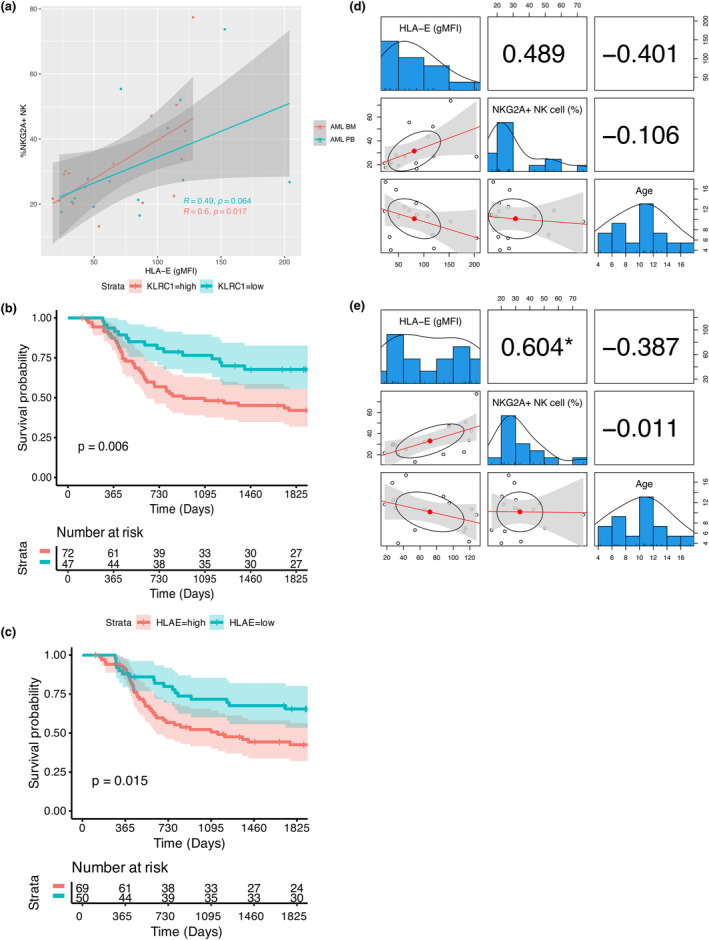
Correlation analysis of receptor‐ligand for AML patients. (a) Correlation between the NK cell receptor NKG2A expression and its corresponding ligand HLA‐E expression on AML tumors. The Pearson correlation coefficients and corresponding P‐value is displayed in the plot. (b, c) Kaplan–Meier 5‐year survival analysis of paediatric AML patients stratified by KLRC1 (gene encoding NKG2A) or HLA‐E expression levels from TARGET AML project. A Log‐rank test was performed. A P‐value of < 0.5 is significant. (d, e) Correlation analysis among the NK cell receptor NKG2A expression and its corresponding ligand HLA‐E expression on AML tumors, and age of the patients. * indicates a P‐value of < 0.5 is significant. An absence of * indicates a P‐value of > 0.05 is not significant.
NKG2D expression on NK cells was also positively correlated with its ligands, MICA and MICB, on AML tumor cells (Supplementary figure 7a). We also found that paediatric AML patients with high expression of KLRK1 (the gene encoding NKG2D) in the TARGET AML database had a poorer prognosis than those with low gene expression of KLRK1 (Supplementary figure 8a). When we divided the patients based on the NK cell score and KLRK1 expression level, we discovered no significant differences (Supplementary figure 8b). This was achieved using an NK cell score that predicted NK cell frequency. Nevertheless, MICA and MICB expression levels in paediatric AML patients do not indicate changes in the prognosis (Supplementary figure 8c and d). The observed correlation between NKG2D and MICA/B, contrary to expectations of immune evasion, suggests that mechanisms beyond mere expression levels may impair the activating signals of NKG2D. Additionally, we did not observe any significant correlation for other receptor‐ligand pairs analysed, including TIGIT‐CD112, TIGIT‐CD155, DNAM‐1‐CD155, DNAM‐1‐CD112, CD96‐CD155 and KIRs‐HLA‐ABC (Supplementary figure 7b–g). No significant correlations were observed in B‐ALL (Supplementary figure 9a–h). Taken together, our results suggest that the receptor‐ligand interaction involving NKG2A‐HLA‐E may contribute to the co‐regulation of NK cell responses in paediatric AML and the high expression of genes encoding NKG2A in paediatric leukaemia samples is associated with unfavorable outcomes.
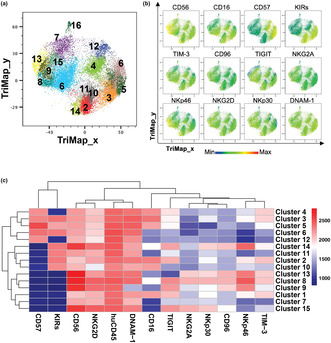
High‐dimensional analysis identifies 15 distinct NK cell populations
Phenograph analysis 35 of NK cells in leukaemia patients and HD samples identified 16 clusters (Figure 3a) and each marker expression of NK cells were demonstrated in Figure 3b. Cluster 16 was mostly composed of cells from AML sample #7 which, as discussed earlier, are likely to be gating artefacts. Therefore, only clusters 1 to 15 were further considered for analysis (Figure 3c).
Figure 3.
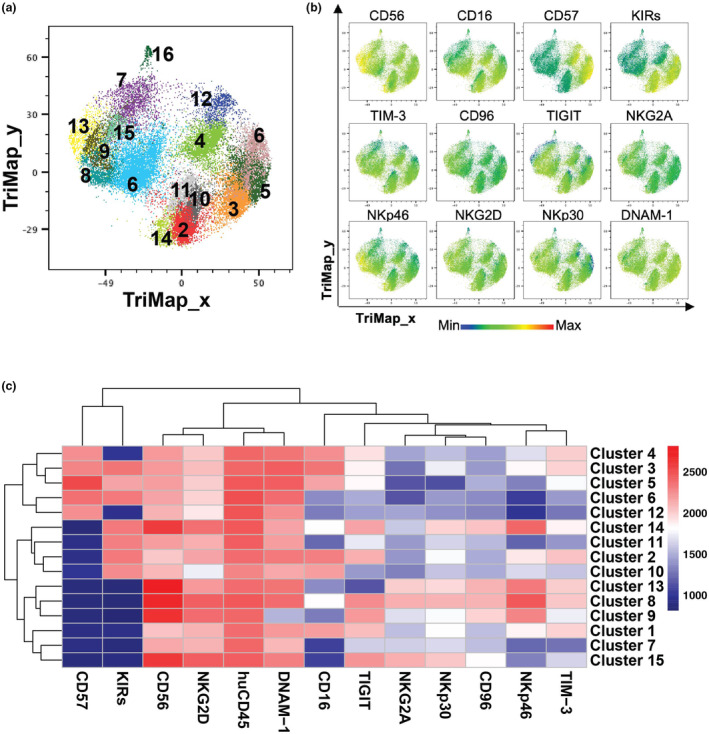
Fifteen NK cell populations identified in paediatric leukaemia patients. (a) TriMap plots of 15 Phenograph‐identified NK cells from healthy donor umbilical cord blood (UCB; n = 8), healthy donor adult peripheral blood (APB; n = 7), peripheral blood (PB) and bone marrow (BM) of patients with acute myeloid leukaemia (AML; n = 15) or B‐cell acute lymphoblastic leukaemia (B‐ALL; BM: n = 11, PB: n = 12). (b) TriMap plots of total NK cell expression of the indicated markers. (c) Heatmap displays the median fluorescence intensity (MFI) of the indicated markers for each NK cell cluster identified by Phenograph.
CD56brNKG2A+ are speculated to be an immature NK cell subset, 36 which were identified as Clusters 8, 13 and 15, which together represented 8.7% of all NK cells (Table 2). Clusters 2, 3, 5 and 10 corresponded to conventional CD56dimCD16+ cytotoxic NK cells 36 and made up for 36.4% of all NK cells (Table 2). Among these, clusters 3 and 5 were identified as terminally matured NK cells because of their expression of CD57. 37 Moreover, Clusters 4, 6 and 12 corresponded to terminally matured NK cells lacking CD16 and/or KIR expression, which are atypical NK cell populations identified in a previous study 38 ; they represented 19.3% of NK cells. Cluster 1 corresponded to CD56dimCD16+ NK cells that were negative for both NKG2A and KIR and might represent maturation intermediates between CD56brKIR− and CD56dimKIR+ NK cells. Clusters 7 and 11 were identified as CD56dimCD16− NK cells that were negative for CD57 expression, representing < 10% of NK cells. Cluster 9 was the only subset lacking DNAM‐1 expression. Finally, cluster 14 was characterised by a CD56brNKp46brCD16loKIR+ phenotype. Both clusters 9 and 14 were relatively small subsets and represented 4.4% and 2% of NK cells respectively (Table 2).
Table 2.
NK cell populations identified by Phenograph in paediatric leukaemia samples and healthy cord blood and adult peripheral blood samples
| Population | Additional population characteristics | % of total NK cells | Interpretations | |
|---|---|---|---|---|
| CD56brNKG2A+ | 8 | CD16loNKp46brCD57−KIRs−NKG2D+ | 8.70 | Immature NK cells |
| 13 | CD16−NKp46brCD57−KIRs−NKG2D+ | |||
| 15 | CD16−NKp46−CD57−KIRs−NKG2D+ | |||
| CD56dimCD16+KIRs+ | 2 | CD57−NKG2A− NKG2D+ | 12.10 | Conventional cytotoxic NK cells |
| 10 | CD57+NKG2A− NKG2Dlo | 4.00 | ||
| 3 | CD57+NKG2A− NKG2D+ | 20.3 | ||
| 5 | CD57+NKG2A− NKG2D+ | |||
| CD56dim CD57+ lacking CD16 and/or KIRs expression | 4 | CD16+KIRs− NKG2A− | 10.60 | Unconventional terminally differentiated NK cells |
| 6 | CD16−KIRs+ NKG2A− | 6.05 | ||
| 12 | CD16−KIRs− NKG2A− | 2.65 | ||
| CD56dimCD16+KIRs− | 1 | CD56dimCD16+CD57−KIRs− NKG2A− | 18.9 | Possible maturation intermediates between CD56brKIR− and CD56dimKIR+ NK cells |
| CD56dimCD16− | 7 | CD57−KIRs− NKG2A−TIGIT− | 5.78 | Possible maturation intermediates between CD56brCD16− and CD56dimCD16+NK cells |
| 11 | CD57−KIRs+ NKG2A−TIGIT− | 3.80 | ||
| CD56brDNAM‐1− | 9 | CD16−KIR−NKG2Alo NKG2D+ | 4.44 | Population highly presented in patient's BM and UCB |
| CD56brKIR+ | 14 | CD16loCD57− NKG2Alo NKG2D+ | 1.97 | Potentially activated NK cells that have shed CD16 expression |
CD56dimCD16+NKG2A− NK cells are poorly represented in paediatric leukaemia samples
Cluster 10, which was composed of CD56dimCD16+ NK cells, represented less than 1.5% of total NK cells in paediatric leukaemia samples while it accounted for 15.7% and 7.8% of APB and UCB NK cells respectively (Figure 4a and b; Supplementary figure 10a and b). These cells were KIR+, but negative for CD57 (Figure 4a), suggesting that they were mature but not terminally differentiated. 8 These cells were the NK cell subset expressing the lower levels of NKG2D and negative for NKG2A (Figure 3c). We noted that Clusters 3 and 5 were very similar to Cluster 10, as they shared a CD56dimCD16+KIR+NKp46loNKp30loNKG2A− phenotype. However, Clusters 3 and 5 were both positive for CD57 and expressed TIGIT and TIM‐3. Clusters 6 and 12, two other CD57+ terminally differentiated subsets resembling clusters 3 and 5 but lacking CD16, were also more represented in AML samples (13.4% PB NK cells and 17.6% BM NK cells in AML), but not in B‐ALL samples. Of note, clusters 3, 5, 6 and 12 were characterised by CD56dimCD57+ NK cells, and they represented a similar proportion of NK cells in both B‐ALL samples and APB (Figure 4b). Taken together, these data indicate a reduced representation of CD56dimCD16+CD57−NKG2A− NK cells (cluster 10) in paediatric leukaemia samples compared to UCB and APB samples.
Figure 4.
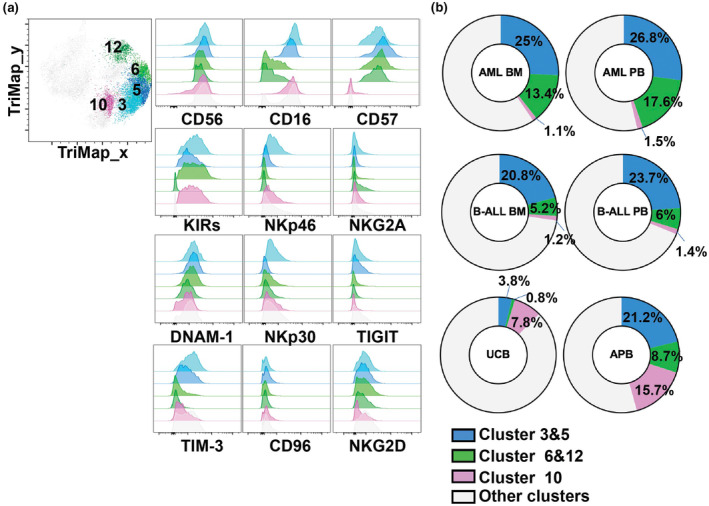
NK cell clusters differentially represented in leukaemia patients. (a) TriMap plots of NK cell clusters 3, 5, 6, 12 and flow cytometry histograms showing the NK cell cluster expression of indicated markers. (b) Donut plots showing percentage of NK cell clusters 3, 5, 6, 12 from healthy donor umbilical cord blood (UCB; n = 8), healthy donor adult peripheral blood (APB; n = 7), peripheral blood (PB) and bone marrow (BM) of patients with acute myeloid leukaemia (AML; n = 15) or B‐cell acute lymphoblastic leukaemia (B‐ALL; BM, n = 11; PB, n = 12).
CD56br NK cell populations expressing high levels of NKG2A are over‐represented in leukaemia samples
CD56br populations (i.e. clusters 8, 9, 13, 14 and 15) were more represented in leukaemia and UCB samples compared to APB (Supplementary figure 10a and b). All these subsets expressed relatively high levels of NKG2D (Figures 5a and 5c). Clusters 8, 13 and 15 might correspond to immature CD56brCD16lo/− NK cells characterised by high levels of NKG2A and a lack of KIR and CD57 expression. 39 Immature CD56br NK cells were more represented in UCB and paediatric leukaemia samples, comprising approximately 8% total NK cells in AML, 11% in B‐ALL, 9% in UCB, but only 3% in APB (Figure 5a and b). We further identified two CD56br NK cell populations that we qualified as “atypical” because of their relatively lower expression of NKG2A expression typically seen on the CD56br subset. 39 Atypical CD56br NK cells encompassed clusters 9 and 14, which were also more represented in paediatric leukaemia patients (Figure 5b). As previously mentioned, cluster 9 was characterised by its lack of DNAM‐1 expression. Interestingly, this population of DNAM‐1− NK cells was mostly found in the BM of leukaemia patients and UCB. By contrast, cluster 14 comprised CD16+ and CD16− sub‐populations (Figure 5c), and this cluster was uniformly positive for KIRs, which are known to be expressed on mature NK cells. 8 Therefore, in contrast to the four other CD56br clusters, cluster 14 NK cells are likely to be a mature NK cell population with up‐regulated CD56 upon activation. 40 , 41 This result suggests that CD56br NK cell populations expressing intermediate to high levels of NKG2A are more prevalent in paediatric leukaemia samples.
Figure 5.
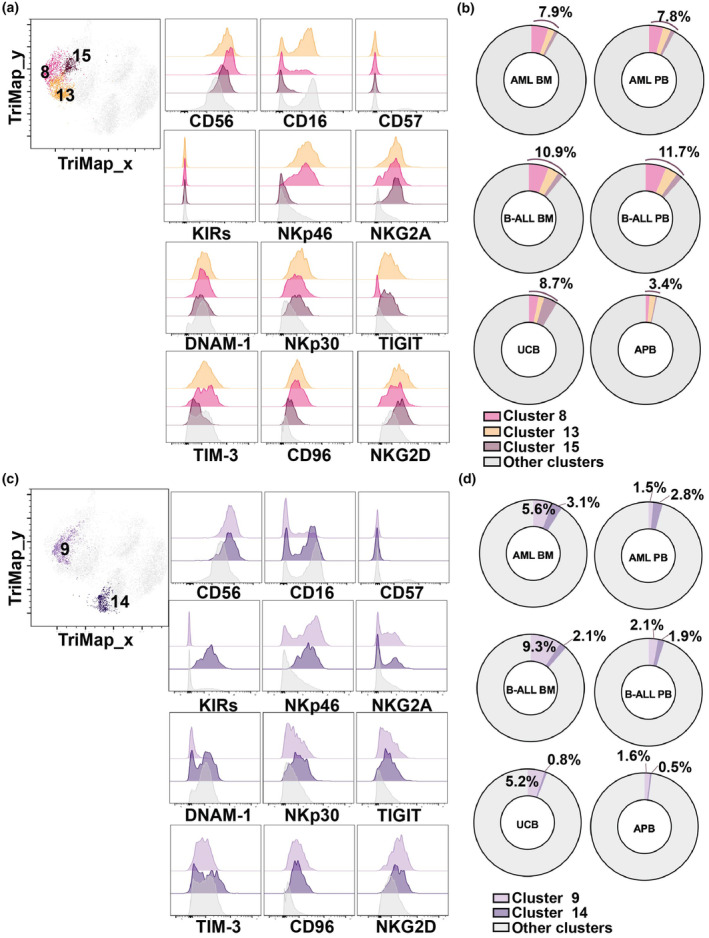
(a) TriMap plots of NK cell clusters 8, 13, 15 and flow cytometry histograms showing the NK cell cluster expression of indicated markers. (b) Donut plots showing percentage of NK cell clusters 8, 13, 15 from healthy donor umbilical cord blood (UCB; n = 8), healthy donor adult peripheral blood (APB; n = 7), peripheral blood (PB) and bone marrow (BM) of patients with acute myeloid leukaemia (AML; n = 15) or B‐cell acute lymphoblastic leukaemia (B‐ALL) CD56br NK cell clusters differentially represented in leukaemia patients. (c) TriMap plots of NK cell clusters 9, 14 and flow cytometry histograms showing the NK cell cluster expression of indicated markers. (d) Donut plots showing percentage of NK cell clusters 9, 14 from healthy donor umbilical cord blood (UCB; n = 8), healthy donor adult peripheral blood (APB; n = 7), peripheral blood (PB) and bone marrow (BM) of patients with acute myeloid leukaemia (AML; n = 15) or B‐cell acute lymphoblastic leukaemia (B‐ALL; BM, n = 11; PB, n = 12).
Discussion
In‐depth characterisation of NK cell phenotypic alterations in paediatric leukaemia may shed light on leukaemia cell immune evasion from NK cells and contribute to the establishment of more effective NK cell‐based targeted therapeutic approaches for paediatric leukaemia patients. Our results revealed the NK cell phenotypic similarity of paediatric leukaemia and UCB, characterised by relatively higher surface protein expression levels of NKG2D and NKG2A compared to APB. We found that CD56br NK cell populations expressing high levels of NKG2A were more represented in paediatric acute leukaemia compared with HD samples. Furthermore, high expression of genes encoding NKG2A in paediatric leukaemia patients is correlated with a poor prognosis. Collectively, these results highlight the prognostic and therapeutic potential of NKG2A receptors in paediatric leukaemia.
In this study, we analysed BM and blood samples from paediatric patients along with two sources of HD blood: UCB and APB. Unfortunately, we were unable to access age‐matched BM and blood samples from HD, and for this reason, we cannot determine whether differences between leukaemia and HD are caused by the tumor or are because of age differences. UCB have been previously reported to express high levels of NKG2A, which has been associated with NK cells' immature development stages. 38 It is possible that the high expression of NKG2A and NKG2D in paediatric leukaemia patients reflects their young age. Interestingly, in AML, we observed significant correlations between the expression of NKG2A on NK cells and their respective ligand expression on tumor cells, suggesting that immunoediting occurred along this immunological axis. Therefore, these findings imply that the elevated expression levels of NKG2A in AML patients may be a result of the tumor microenvironment. Additionally, although CMV infection has been shown to expand NKG2C+/NKG2A− NK cells, 31 , 32 the positive correlation between NKG2A and HLA‐E expression does not appear to be influenced by CMV status, according to the multiple regression analysis. While NKG2C characterisation of leukaemia NK cell is missing in our study, further investigation is needed to fully capture all the NK cell phenotypic changes in leukaemia patients.
Our hierarchical clustering analysis grouped most APB samples together, while leukaemia samples were spread across three clusters. One cluster of AML samples (cluster B) was grouped with the APB cluster, and these two clusters were characterised by low expression of TIGIT, CD96 and TIM‐3. Interestingly, none of the patients regrouped in this AML cluster experienced relapse. Moreover, this cluster of AML samples expressed lower levels of NKG2A, TIGIT, CD96 and TIM‐3 compared to two other clusters containing leukaemia and UCB samples. In light of our findings, we hypothesise that low levels of immune suppressors on NK cells may be a protective factor for paediatric AML patients.
We found a distinct composition of NK cell subsets in paediatric leukaemia patients, with a notable reduction of conventional mature NK cells (CD56dimCD16+CD57−) and an increase in CD56brCD57− immature NK cells as compared to APB. Both paediatric AML and B‐ALL had lower representations of the conventional cytotoxic CD56dimCD16+CD57− NK cell population with low NKG2A expression. While AML samples displayed a slightly higher proportion of terminally differentiated unconventional CD56dimCD57+ NK cells (cluster 3 and 5) that expressed intermediate to high levels of NKG2A, TIM‐3 and TIGIT than APB and UCB samples. This phenomenon was not observed in B‐ALL samples. To the best of our understanding, we are the first to report this phenomenon. Given that NKG2A, TIM‐3 and TIGIT are checkpoint molecules that have been shown to limit NK cell immune responses in cancers, 18 , 42 , 43 , 44 the higher proportion of CD56dimCD57+ NK cells expressing these immune checkpoints in AML may explain why patients with AML often have a worse prognosis. 4 Moreover, AML patients have a higher proportion of CD56dimCD16dim NK cells than UCB and APB, but this also does not occur in B‐ALL patients. CD56dimCD16dim NK cells have previously been studied; they degranulated more than CD56dimCD16bright NK cells but less than CD56dimCD16− NK cells, 45 perhaps representing a subgroup of NK cells with intermediate functional features. The function of CD56dimCD16dim NK cells should be investigated further to assess whether they contribute to making AML worse than ALL.
Additionally, paediatric leukaemia patients had higher proportions of immature NK cell populations that were CD56brCD57−NKG2A+. Our findings align with a prior study that found an increased frequency of NKG2A+CD57− immature NK cells and decreased NKG2A−CD57+ mature NK cells in paediatric ALL compared to healthy donors. 46 This high level of NKG2A may make it more difficult for NK cells to recognise and eliminate malignant cells, promoting disease progression and enhancing immune evasion. Furthermore, the observed positive correlation between NKG2A expression on NK cells and the presence of its cognate receptor, HLA‐E, on AML tumor cells might suggest that AML tumor cells upregulate HLA‐E to avoid NK cell‐mediated immune surveillance. The positive correlation between NKG2A and HLA‐E has also been observed in intratumor hepatocellular carcinoma (HCC) tissues, and the NKG2A+ NK cells from HCC patients exhibited reduced IFNγ production, suggesting high NKG2A expression contributed to NK cell dysfunction. 47 Previous research on the NKG2A‐HLA‐E interaction revealed that it mediated the immune evasion of pancreatic ductal adenocarcinoma cells from NK cell immune surveillance and inhibiting the NKG2A‐HLA‐E interaction prevented tumor metastasis by restoring NK cell function. 42 Therefore, blocking NKG2A may be promising for enhancing NK cell antileukaemic activity.
Among the 15 identified NK cell populations, we identified NK cells that are differentially presented between paediatric leukaemias and healthy APB. In particular, cluster 14 (CD56brKIR+ NK cell cluster) has greater abundance in paediatric leukaemias; cells of this cluster were atypical CD56brNKG2D+NKG2AloNK cells with low CD16 expression. Their KIR+NKG2Alo phenotype suggests that they are mature, 8 and they are NKp46brCD16lo, which might be an indication of activation. 48 The low CD16 expression in CD56brKIR+ NK cell cluster may potentially be attributed to the downregulation of CD16 following activation by leukaemic cells. 49 The shedding of CD16 can limit NK cell antibody‐dependent cell cytotoxicity (ADCC). 49 A recent study found that NK cells from paediatric ALL patients displayed a significant reduction in their ability to sustain ADCC. 46 The overrepresentation of NK cells with low CD16 levels in paediatric leukaemia patients implies that ADCC activity may be less robust. This finding is significant because it implies that leukaemia patients may have an increase in NK cells with impaired ADCC capacity because of CD16 shedding, and NK cells with low CD16 levels may be unable to carry out efficient ADCC‐mediated tumor cell eradication.
In conclusion, our study demonstrates a pivotal role for NKG2A expression on NK cells as a prognostic marker significantly correlated with poor clinical outcomes in paediatric leukaemia patients. Moreover, given the high expression levels of NKG2A in patients, targeting NKG2A could pave the way for developing immunotherapies aimed at boosting NK cell‐mediated antileukaemic responses. The precise mechanisms underlying how NKG2A impacts NK cells in paediatric leukaemia patients have yet to be determined. However, our findings support the development of paediatric leukaemia immunotherapies that target the NKG2A receptor.
Methods
Patient samples
Peripheral blood (PB) and matched bone marrow‐derived aspirates (BM) were collected from patients with newly diagnosed AML (n = 15) and B‐ALL (BM, n = 11; PB = 12), which were provided by Queensland Children's Tumor Bank (Brisbane, Australia). Adult peripheral blood (APB) samples were collected from healthy individuals (n = 7). Umbilical cord blood (UCB; n = 8) was provided by the Queensland Cord Blood Bank at the Mater (Brisbane, Australia). All human sample donors gave written informed consent to sample acquisition following the Declaration of Helsinki, and the study has been approved by the Mater Human Research Ethics Committee (HREC/13/MHS/83, 22 310/1407AP and 1586 M) and ratified by the UQ HREC.
APB mononuclear cells (MNCs) were cryopreserved in 90% heat‐inactivated foetal calf serum (FBS; Gibco, Waltham, USA) and 10% dimethyl sulfoxide (DMSO; Sigma‐Aldrich, St. Louis, US). Cord blood MNCs were cryopreserved using 90% cord blood plasma and 10% DMSO solution. Cells were frozen at a maximum concentration of 50 × 106 cells mL−1 in 1‐mL cryovials. MNCs derived from AML and B‐ALL patient BM and PB samples were collected in EDTA vacutainer collection tubes and cryopreserved in 10% DMSO, 20% FBS and RPMI 1640 medium (Gibco). Cryovials were placed in NALGENE Mr.Frosty at −80°C for 12–24 h and subsequently relocated into long‐term −196°C liquid nitrogen storage.
Sample preparation
The isolation of cells from APB and UCB samples was conducted under sterile conditions at room temperature according to previously established protocol. 38 The blood was first diluted with an equal volume of PBS, resulting in a total volume of 35 mL. A 50‐mL conical tube was then prepared by underlaying 15 mL of Ficoll‐Paque Plus blood cell density gradient medium (Cytivia, Massachusetts, USA). To maintain the integrity of the density gradient, the tubes were centrifuged at 500 g for 20 min without any interruptions. Subsequently, the mononuclear cells located at the plasma/Ficoll interface were carefully collected using a transfer pipette, and the cells were washed twice with PBS.
Thawing of cryopreserved APB MNCs and CB MNCs samples was performed by quickly swirling the cryovial in a 37°C water bath until partially thawed, followed by transferring the cells into chilled complete RPMI‐1640 medium: RPMI‐1640 medium containing HEPES (10 mm) (Gibco), GlutaMAX‐I (2 mm) (Gibco), FBS (10%) (Gibco), MEM non‐essential amino acids (0.1 mm) (Gibco), sodium pyruvate (1 mm) (Gibco), penicillin–streptomycin (100 U mL−1) (Gibco), β‐Mercaptoethanol (50 μm) (Sigma‐Aldrich). The cells were gently added to the medium and rinsed to maximise cell recovery. The cells were then pelleted, resuspended in complete RPMI‐1640 medium with DNase I (Roche Diagnostics, Mannheim, Germany, 12.5 μg mL−1), and incubated at 37°C for 15 min. The DNase I reaction was stopped by adding complete RPMI‐1640 medium, and the cells were washed and resuspended. Cells from each sample were transferred to a 96‐well plate for flow cytometry panel staining.
For AML and B‐ALL samples, a stepwise thawing process was employed to preserve cell viability. Thawed cells were transferred to a room temperature RPMI‐1640 medium with 60% FBS, ensuring thorough washing of the cryovial. After centrifugation, the cells were sequentially resuspended in RPMI‐1640 medium with 60% FBS and RPMI medium with 40% FBS. Each resuspension step was followed by a brief 3‐min rest period. Then, the cells were centrifuged, the supernatant was aspirated, and they were resuspended in complete RPMI‐1640 medium. Cells were then transferred to a 96‐well plate for flow cytometry panel staining. The stepwise decrement in FBS concentration during thawing was implemented to minimise cell viability loss.
Flow cytometry
Cells were stained with 10% mouse serum (Sigma‐Aldrich), 10% rat serum (Sigma‐Aldrich) and 10% MACS buffer (Miltenyi Biotec, Bergisch Gladbach, Germany) at a 1:1:1 ratio for 10 min at 4°C to block the non‐specific binding, as described in a previous study. 38 Cells were then washed with PBS and performed LiveDead staining with Fixable Viability Stain 575 V (BD Biosciences, San Jose, USA) or Zombie Aqua™ Fixable Viability Kit (Biolegend, San Deigo, USA) for 10 min at 4°C in the dark. Cells were subsequently washed with MACS buffer and stained with other cell surface antibodies for 15–20 min at 4°C. The expression of NK cell receptors and immune checkpoints on NK cells were analysed using a 21‐colour flow cytometry panel (Supplementary table 1) and the expression of the corresponding ligands on the tumor cells was analysed using a 20‐colour flow cytometry panel (Supplementary tables 2 and 3). The cells were then washed and resuspended in MACS buffer and acquired on a FACSymphony A5 instrument (BD Biosciences).
Flow cytometry analysis
Flow cytometry data were analysed using FlowJo v.10.9.0 (BD Biosciences) and FlowJo plugins (https://flowjo.com/exchange/#/). Samples from one UCB donor were used as reference control samples that were recorded along with each batch of samples, and the FlowJo plugin CytoNorm was performed to remove the unwanted batch effect by using the reference control samples. For high dimensional analysis, NK cells from each donor were downsampled and concatenated, which contained about 500 NK cell events per sample. One BM sample from B‐ALL was excluded in this study as it only had 14 cell counts for NK cells. TriMap and PhenoGraph were performed for the NK cell concatenated file to conduct dimensionally reduction analysis. TriMap was conducted using CD56, CD16, CD57, KIRs, TIM‐3, CD96, TIGIT, NKG2A, NKp46, NKG2D, NKp30, DNAM‐1, CTLA‐4, PD‐1, NKp44, LAG‐3 and using the default setting: distance function = Euclidean and nearest neighbours = 10. PhenoGraph was also conducted using the same markers and the default number of nearest neighbours (K = 30). Hierarchical clustering heatmap in Figure 1b displays the percentage of indicated markers of total NK cells from each donor made by using the R package pheatmap.
Bioinformatics analyses: TARGET database analysis
RNA‐seq expression data and clinical characteristics for the bone marrow (BM) samples collected at the diagnosis stage of paediatric AML patients were collected from the TARGET database (Target‐AML database, n = 119) (https://www.cancer.gov/ccg/research/genome‐sequencing/target/using‐target‐data). The optimal cut‐off for the single factor in the survival analysis was determined using the ‘surv_cutpoint’ function with the ‘maxstat’ method. The analysis was performed using the ‘survival’ package in R, and the resulting Kaplan–Meier (KM) curves were plotted using the ‘survminer’ package. To generate the NK score for survival analysis, a NK signature curation given by Cursons et al. 50 has been used to generate the NK score using the singscore as described in the previous publication.
Statistical analysis
Statistical data were analysed using GraphPad Prism 9. Comparisons between groups were performed using a Kruskal–Wallis test, followed by a Dunn's multiple comparisons test. The correlation analysis between receptor expression on NK cells and their corresponding ligands on tumor cells was analysed using Pearson's correlation test. The data are presented as mean ± sem, and the significant cut‐off was 0.05. * P < 0.05, ** P < 0.01, *** P < 0.001, **** P < 0.0001.
Author contributions
Cui Tu: Conceptualization; data curation; formal analysis; investigation; visualization; writing – original draft; writing – review and editing. Irina Buckle: Investigation. Ingrid Leal Rojas: Investigation. Gustavo Rodrigues Rossi: Writing – original draft; writing – review and editing. Kristen Radford: Resources. David P Sester: Conceptualization. Andrew S Moore: Resources. Camille Guillerey: Conceptualization; resources; supervision; writing – original draft; writing – review and editing. Fernando Souza‐Fonseca‐Guimaraes: Writing – original draft; writing – review and editing.
Conflict of interest
The authors have no conflict of interest to declare.
Supporting information
Supporting Information
Acknowledgments
This work was supported by the Mater Foundation, Brisbane, Australia and by an Innovator Grant from the Children’ Hospital Foundation (Queensland) attributed to Camille Guillerey (RPC00037). We acknowledge financial support provided by the Translational Research Institute (TRI) by way of access to the Symphony Reagent Support Fund. Camille Guillerey is supported by a Discovery Early Career Research Award (DECRA) from the Australian Research Council (ARC) (DE210101144). The Guimaraes Laboratory is funded by a US Department of Defense—Breast Cancer Research Program—Breakthrough Award Level 1 (#BC200025), a grant (#2019485) awarded through the Medical Research Future Fund, an Ian Frazer Centre for Children's Immunotherapy Research—Immunotherapy Research Grant (IF0022021), and by a Metro South Health Research Support Scheme—Co‐Funded Collaboration Grant (#RSS_2023_085). Cui Tu is funded by a UQ Research Training Program PhD Scholarship. We acknowledge the support of the Queensland Children's Tumor Bank, which is funded by the Children's Hospital Foundation (Queensland). Open access publishing was facilitated by The University of Queensland, as part of the Wiley ‐ The University of Queensland agreement via the Council of Australian University Librarians.
Contributor Information
Cui Tu, Email: cui.tu@uq.net.au.
Fernando Souza‐Fonseca‐Guimaraes, Email: f.guimaraes@uq.edu.au.
Data availability statement
The patient data that support the findings of this study are available on request from the corresponding author. The data are not publicly available because of privacy or ethical restrictions. Raw transcriptomics data used in this study are freely available as TARGET Datasets (https://www.cancer.gov/ccg/research/genome‐sequencing/target/using‐target‐data).
References
- 1. Mezei G, Sudan M, Izraeli S, Kheifets L. Epidemiology of childhood leukemia in the presence and absence of down syndrome. Cancer Epidemiol 2014; 38: 479–489. [DOI] [PubMed] [Google Scholar]
- 2. Björk‐Eriksson T, Boström M, Bryngelsson I‐L et al. Mortality among pediatric patients with acute lymphoblastic leukemia in Sweden from 1988 to 2017. JAMA Netw Open 2022; 5: e2243857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. He J‐R, Yu Y, Fang F et al. Evaluation of maternal infection during pregnancy and childhood leukemia among offspring in Denmark. JAMA Netw Open 2023; 6: e230133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Ward E, DeSantis C, Robbins A, Kohler B, Jemal A. Childhood and adolescent cancer statistics, 2014. CA Cancer J Clin 2014; 64: 83–103. [DOI] [PubMed] [Google Scholar]
- 5. Myers JA, Miller JS. Exploring the NK cell platform for cancer immunotherapy. Nat Rev Clin Oncol 2021; 18: 85–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Guillerey C. NK cells in the tumor microenvironment. Adv Exp Med Biol 2020; 1273: 69–90. [DOI] [PubMed] [Google Scholar]
- 7. Wong JKM, Dolcetti R, Rhee H, Simpson F, Souza‐Fonseca‐Guimaraes F. Weaponizing natural killer cells for solid cancer immunotherapy. Trends Cancer 2023; 9: 111–121. [DOI] [PubMed] [Google Scholar]
- 8. Luetke‐Eversloh M, Killig M, Romagnani C. Signatures of human NK cell development and terminal differentiation. Front Immunol 2013; 4: 499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Horowitz A, Strauss‐Albee DM, Leipold M et al. Genetic and environmental determinants of human NK cell diversity revealed by mass cytometry. Sci Transl Med 2013; 5: 208ra145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Yang C, Siebert JR, Burns R et al. Heterogeneity of human bone marrow and blood natural killer cells defined by single‐cell transcriptome. Nat Commun 2019; 10: 3931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Liu E, Tong Y, Dotti G et al. Cord blood NK cells engineered to express IL‐15 and a CD19‐targeted CAR show long‐term persistence and potent antitumor activity. Leukemia 2018; 32: 520–531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Bednarski JJ, Zimmerman C, Berrien‐Elliott MM et al. Donor memory‐like NK cells persist and induce remissions in pediatric patients with relapsed AML after transplant. Blood 2022; 139: 1670–1683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Tang X, Yang L, Li Z et al. First‐in‐man clinical trial of CAR NK‐92 cells: Safety test of CD33‐CAR NK‐92 cells in patients with relapsed and refractory acute myeloid leukemia. Am J Cancer Res 2018; 8: 1083–1089. [PMC free article] [PubMed] [Google Scholar]
- 14. Marin D, Li Y, Basar R et al. Safety, efficacy and determinants of response of allogeneic CD19‐specific CAR‐NK cells in CD19+ B cell tumors: A phase 1/2 trial. Nat Med 2024. 10.1038/s41591-023-02785-8. Online ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Lee SC, Shimasaki N, Lim JSJ et al. Phase I trial of expanded, activated autologous NK‐cell infusions with Trastuzumab in patients with HER2‐positive cancers. Clin Cancer Res 2020; 26: 4494–4502. [DOI] [PubMed] [Google Scholar]
- 16. Khatua S, Cooper LJN, Sandberg DI et al. Phase I study of intraventricular infusions of autologous ex vivo expanded NK cells in children with recurrent medulloblastoma and ependymoma. Neuro‐Oncology 2020; 22: 1214–1225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Andre P, Denis C, Soulas C et al. Anti‐NKG2A mAb is a checkpoint inhibitor that promotes anti‐tumor immunity by unleashing both T and NK cells. Cell 2018; 175: 1731–1743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kamiya T, Seow SV, Wong D, Robinson M, Campana D. Blocking expression of inhibitory receptor NKG2A overcomes tumor resistance to NK cells. J Clin Invest 2019; 129: 2094–2106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Chan WK, Kang S, Youssef Y et al. A CS1‐NKG2D bispecific antibody collectively activates cytolytic immune cells against multiple myeloma. Cancer Immunol Res 2018; 6: 776–787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Lion E, Willemen Y, Berneman ZN, Van Tendeloo VF, Smits EL. Natural killer cell immune escape in acute myeloid leukemia. Leukemia 2012; 26: 2019–2026. [DOI] [PubMed] [Google Scholar]
- 21. Kaweme NM, Zhou F. Optimizing NK cell‐based immunotherapy in myeloid leukemia: Abrogating an immunosuppressive microenvironment. Front Immunol 2021; 12: 683381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Costello RT, Sivori S, Marcenaro E et al. Defective expression and function of natural killer cell‐triggering receptors in patients with acute myeloid leukemia. Blood 2002; 99: 3661–3667. [DOI] [PubMed] [Google Scholar]
- 23. Fauriat C, Just‐Landi S, Mallet F et al. Deficient expression of NCR in NK cells from acute myeloid leukemia: Evolution during leukemia treatment and impact of leukemia cells in NCRdull phenotype induction. Blood 2007; 109: 323–330. [DOI] [PubMed] [Google Scholar]
- 24. Valhondo I, Hassouneh F, Lopez‐Sejas N et al. Characterization of the DNAM‐1, TIGIT and TACTILE Axis on circulating NK, NKT‐like and T cell subsets in patients with acute myeloid leukemia. Cancers (Basel) 2020; 12: 2171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Stringaris K, Sekine T, Khoder A et al. Leukemia‐induced phenotypic and functional defects in natural killer cells predict failure to achieve remission in acute myeloid leukemia. Haematologica 2014; 99: 836–847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Liu G, Zhang Q, Yang J et al. Increased TIGIT expressing NK cells with dysfunctional phenotype in AML patients correlated with poor prognosis. Cancer Immunol Immunother 2022; 71: 277–287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Duault C, Kumar A, Taghi Khani A et al. Activated natural killer cells predict poor clinical prognosis in high‐risk B‐ and T‐cell acute lymphoblastic leukemia. Blood 2021; 138: 1465–1480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Rouce RH, Shaim H, Sekine T et al. The TGF‐beta/SMAD pathway is an important mechanism for NK cell immune evasion in childhood B‐acute lymphoblastic leukemia. Leukemia 2016; 30: 800–811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Chretien AS, Devillier R, Granjeaud S et al. High‐dimensional mass cytometry analysis of NK cell alterations in AML identifies a subgroup with adverse clinical outcome. Proc Natl Acad Sci USA 2021; 118: e2020459118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Valenzuela‐Vazquez L, Nunez‐Enriquez JC, Sanchez‐Herrera J et al. Functional characterization of NK cells in Mexican pediatric patients with acute lymphoblastic leukemia: Report from the Mexican Interinstitutional Group for the Identification of the causes of childhood leukemia. PLoS One 2020; 15: e0227314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Bigley AB, Rezvani K, Shah N et al. Latent cytomegalovirus infection enhances anti‐tumour cytotoxicity through accumulation of NKG2C+ NK cells in healthy humans. Clin Exp Immunol 2016; 185: 239–251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Manser AR, Uhrberg M. Age‐related changes in natural killer cell repertoires: Impact on NK cell function and immune surveillance. Cancer Immunol Immunother 2016; 65: 417–426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Dunn GP, Bruce AT, Ikeda H, Old LJ, Schreiber RD. Cancer immunoediting: From immunosurveillance to tumor escape. Nat Immunol 2002; 3: 991–998. [DOI] [PubMed] [Google Scholar]
- 34. Guillerey C, Smyth MJ. NK cells and cancer immunoediting. Curr Top Microbiol Immunol 2016; 395: 115–145. [DOI] [PubMed] [Google Scholar]
- 35. Levine JH, Simonds EF, Bendall SC et al. Data‐driven phenotypic dissection of AML reveals progenitor‐like cells that correlate with prognosis. Cell 2015; 162: 184–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Juelke K, Killig M, Luetke‐Eversloh M et al. CD62L expression identifies a unique subset of polyfunctional CD56dim NK cells. Blood 2010; 116: 1299–1307. [DOI] [PubMed] [Google Scholar]
- 37. Lopez‐Verges S, Milush JM, Pandey S et al. CD57 defines a functionally distinct population of mature NK cells in the human CD56dimCD16+ NK‐cell subset. Blood 2010; 116: 3865–3874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Buckle I, Johnson A, Rojas IL et al. High dimensional analysis reveals distinct NK cell subsets but conserved response to stimulation in umbilical cord blood and adult peripheral blood. Eur J Immunol 2023; 53: e2250118. [DOI] [PubMed] [Google Scholar]
- 39. Freud AG, Caligiuri MA. Human natural killer cell development. Immunol Rev 2006; 214: 56–72. [DOI] [PubMed] [Google Scholar]
- 40. Vukicevic M, Chalandon Y, Helg C et al. CD56bright NK cells after hematopoietic stem cell transplantation are activated mature NK cells that expand in patients with low numbers of T cells. Eur J Immunol 2010; 40: 3246–3254. [DOI] [PubMed] [Google Scholar]
- 41. Loza MJ, Perussia B. The IL‐12 signature: NK cell terminal CD56+high stage and effector functions. J Immunol 2004; 172: 88–96. [DOI] [PubMed] [Google Scholar]
- 42. Liu X, Song J, Zhang H et al. Immune checkpoint HLA‐E:CD94‐NKG2A mediates evasion of circulating tumor cells from NK cell surveillance. Cancer Cell 2023; 41: 272–287. [DOI] [PubMed] [Google Scholar]
- 43. Yu L, Liu X, Wang X et al. TIGIT+ TIM‐3+ NK cells are correlated with NK cell exhaustion and disease progression in patients with hepatitis B virus‐related hepatocellular carcinoma. Onco Targets Ther 2021; 10: 1942673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Maas RJ, Hoogstad‐van Evert JS, Van der Meer JM et al. TIGIT blockade enhances functionality of peritoneal NK cells with altered expression of DNAM‐1/TIGIT/CD96 checkpoint molecules in ovarian cancer. Onco Targets Ther 2020; 9: 1843247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Amand M, Iserentant G, Poli A et al. Human CD56dimCD16dim cells As an individualized natural killer cell subset. Front Immunol 2017; 8: 699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Valenzuela‐Vazquez L, Nunez‐Enriquez JC, Sanchez‐Herrera J et al. NK cells with decreased expression of multiple activating receptors is a dominant phenotype in pediatric patients with acute lymphoblastic leukemia. Front Oncol 2022; 12: 1023510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Sun C, Xu J, Huang Q et al. High NKG2A expression contributes to NK cell exhaustion and predicts a poor prognosis of patients with liver cancer. Onco Targets Ther 2017; 6: e1264562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Hadad U, Thauland TJ, Martinez OM, Butte MJ, Porgador A, Krams SM. NKp46 clusters at the immune synapse and regulates NK cell polarization. Front Immunol 2015; 6: 495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Romee R, Foley B, Lenvik T et al. NK cell CD16 surface expression and function is regulated by a disintegrin and metalloprotease‐17 (ADAM17). Blood 2013; 121: 3599–3608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Cursons J, Souza‐Fonseca‐Guimaraes F, Foroutan M et al. A gene signature predicting natural killer cell infiltration and improved survival in melanoma patients. Cancer Immunol Res 2019; 7: 1162–1174. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting Information
Data Availability Statement
The patient data that support the findings of this study are available on request from the corresponding author. The data are not publicly available because of privacy or ethical restrictions. Raw transcriptomics data used in this study are freely available as TARGET Datasets (https://www.cancer.gov/ccg/research/genome‐sequencing/target/using‐target‐data).


