Abstract
Hepatitis C virus (HCV) is a leading cause of chronic hepatitis in the world. The study of HCV has been hampered by the low level of viral particles in infected individuals, the inability to propagate efficiently the virus in cultured cells, and the lack of a convenient animal model. Due to these obstacles, neither the structure of the virus nor the prerequisites for its assembly have been clearly defined. In this report, we describe a model for the production and purification of HCV-like particles in insect cells using a recombinant baculovirus containing the cDNA of the HCV structural proteins. In insect cells, expressed HCV structural proteins assembled into enveloped viruslike particles (40 to 60 nm in diameter) in large cytoplasmic cisternae, presumably derived from the endoplasmic reticulum. Biophysical characterization of viruslike particles by CsCl and sucrose gradient centrifugation revealed biophysical properties similar to those of putative virions isolated from infected humans. The results suggested that HCV core and envelope proteins without p7 were sufficient for viral particle formation. Analysis of particle-associated nucleic acids demonstrated that HCV RNAs were selectively incorporated into the particles over non-HCV transcripts. The synthesis of HCV-like particles in insect cells may provide an important tool to determine the structural requirements for HCV particle assembly as well as to study viral genome encapsidation and virus-host interactions. The described system may also represent a potential approach toward vaccine development.
Hepatitis C virus (HCV) is a major causative agent of posttransfusion and community-acquired hepatitis in the world (2, 23, 26). The majority of HCV-infected individuals develop chronic hepatitis progressing eventually to liver cirrhosis and hepatocellular carcinoma (48). Neither an effective treatment for chronic HCV infection nor a vaccine to prevent HCV infection is available at the present time (19, 28).
HCV is a member of the Flaviviridae family (44). The virion contains a positive-stranded RNA genome of 9.5 kb. The genome consists of a highly conserved 5′ noncoding region (35) followed by a long open reading frame of 9,030 to 9,099 nucleotides (nt) that is translated into a single polyprotein of 3,010 to 3,030 amino acids (16, 35). Initiation of translation occurs by a mechanism of internal ribosomal entry requiring the 5′ untranslated region (UTR) and a short stretch of HCV coding sequences (43). Processing of the polyprotein occurs with a combination of host and viral proteases. The HCV structural proteins comprise the nucleocapsid or core protein (C) and the two envelope glycoproteins, E1 and E2 (for a review, see reference 39). The cleavage of structural proteins from the polyprotein is catalyzed by a host signal peptidase (16, 30), whereas polyprotein cleavage in the nonstructural region requires HCV-encoded proteases (11). An additional cleavage product in the coding region of the structural proteins was recently identified as p7 (30, 41). Although the characterization of the viral genome organization has been described in detail (35), analysis of the structural features of HCV has been hampered by the inability to propagate the virus efficiently in cultured cells. The levels of viral particles present in infected patient plasma or liver tissues are very low, making it difficult to visualize the virus. In analogy to other members of the Flaviviridae, the genome organization of HCV suggests a viral structure consisting of a nucleocapsid or core protein and a viral genome coated by a lipid envelope containing envelope glycoproteins E1 and E2. Transmission studies with chimpanzees, the only reliable animal model for HCV, have provided evidence that HCV is inactivated by chloroform, indicating that it contains lipids and therefore is probably enveloped (9). Filtration studies have estimated the virion particle size at a diameter of 30 to 60 nm (15).
The baculovirus-insect cell expression system has been applied successfully for the synthesis of viral capsids for viruses of various families (10, 24, 49) but not for the enveloped RNA viruses of the Flaviviridae family. The baculovirus-insect cell expression system has two features which make it attractive for HCV protein expression. First, eukaryotic insect cells are known to carry out a number of co- or posttranslational modifications, including fatty acid acetylation and glycosylation, similar to mammalian cells (33). Second, in contrast to many mammalian cell expression systems, the baculovirus expression system allows high-level synthesis of heterologous proteins (33). We therefore rationalized that the baculovirus system may be able to direct the synthesis of HCV-like particles in insect cells.
(This work was presented in part at the 47th Annual Meeting of the American Association for the Study of Liver Diseases, 8 to 12 November 1996, Chicago, Ill., and the 4th International Meeting on Hepatitis C Virus and Related Viruses, 6 to 10 March 1997, Kyoto, Japan.)
MATERIALS AND METHODS
Baculovirus constructs and insect cell cultures.
For the construction of recombinant baculoviruses, a recently described baculovirus expression system was applied (Bac-to-Bac; Gibco BRL, Gaithersburg, Md.) (32). The cDNA for the HCV structural proteins, cloned from a Japanese patient with chronic hepatitis (HCV-J strain, genotype 1b), was used to generate the recombinant baculovirus BVHCV.S. pFastBacHCV.S was generated by subcloning an EcoRI-Tth111I fragment (nt 259 to 2819) of pCMV980 (23) into the EcoRI and SpeI sites of pFastBac. The Tth111I and SpeI sites were blunt ended with the Klenow fragment before ligation. An in-frame stop codon is present in the vector sequence close to the 3′ end of the subcloned cDNA. pFastBacHCV.Sp7− was generated by PCR with the following primers: 5′ GAGACAGACGTGCCTGCTACTTAG CAACACGCG 3′ (sense nt 1918 to 1951) and 5′ TCGAAAGCTTAGGCCT CAGCCTGGGCTATCAGC 3′ (antisense nt 2567 to 2543). A stop codon and a HindIII site were introduced at the 3′ end of the p7 protein coding region. The NotI-HindIII digestion product of the PCR fragment was subcloned into NotI-HindIII site (multiple cloning site) of pFastBacHCV.S. pFastBacHIVgp160 containing the cDNA for the human immunodeficiency virus (HIV) glycoprotein precursor gp160 was generated by subcloning a HindIII-NotI fragment (nt 1 to 2481) of pSyngp160 (kindly provided by Brian Seed, Department of Genetics, Massachusetts General Hospital, Boston) into the StuI and NotI sites of pFast Bac. The HindIII site was blunt ended with the Klenow fragment before ligation. The correct sequences of the pFastBacHCV.S, pFastBacHCV.Sp7−, and pFast BacHIVgp160 constructs were confirmed by restriction enzyme digestions and DNA sequencing. pFastBacGUS (Gibco BRL) containing the coding sequence of the enzyme β-glucuronidase (GUS) was used to generate the control baculovirus BVGUS.
Recombinant baculoviruses were generated as described previously (32), identified by immunofluorescence and immunoblotting of transfected Spodoptera frugiperda Sf9 insect cells with specific antibodies, and amplified by subsequent rounds of Sf9 cell infection until a final titer of 5 × 107 PFU/ml was achieved. Sf9 insect cells were maintained in spinner or monolayer cultures at 28°C in Sf-900 II serum-free medium (Gibco BRL). For all protein expression experiments, Sf9 cells in mid-log growth in monolayer cultures were infected with a multiplicity of infection (MOI) of 1 to 10. Infection of insect cells with BVGUS served as a negative control in all experiments.
Anti-HCV antibodies.
The monoclonal anti-core, anti-E1, and anti-E2(G/H) mouse antibodies (7) as well as the polyclonal anti-E2(9284) rabbit antibody (29, 42) were described previously. Additional monoclonal anti-E1 and anti-E2 antibodies were obtained from Johnson Lau (University of Florida, Gainesville). Human serum containing antibodies against HCV was obtained from two patients with chronic hepatitis C and high-titer anti-HCV antibodies. The patients were serologically negative for hepatitis B virus (HBV), hepatitis A virus, and HIV.
Immunofluorescence of HCV proteins.
At 96 h postinfection, BVHCV.S- and BVGUS-infected Sf9 cells were fixed in 7% paraformaldehyde (fixative B, described below), dehydrated, embedded, and sectioned as described below (electron microscopy). Semithin sections (0.5 to 1 μm) were incubated with anti-HCV (serum from HCV-infected individuals as described above; diluted 1:200 in 1% bovine serum albumin [BSA]–phosphate-buffered saline [PBS]), anti-core (diluted 1:200 in 1% BSA–PBS), anti-E1 (diluted 1:100 in 1% BSA–PBS), or anti-E2(9284) (diluted 1:200 in 1% BSA–PBS) antibody or 1% BSA in PBS followed by fluorescein isothiocyanate-conjugated anti-human (for serum), anti-mouse (for anti-core and anti-E1), or anti-rabbit (for anti-E2) antibody (all from Jackson Laboratories, West Grove, Pa; diluted 1:500 in 1% BSA–PBS) each for 30 min at room temperature. Between steps, plates were rinsed three times with PBS.
Immunoblotting and immunoprecipitation of HCV proteins.
For immunoblot analysis, Sf9 cells infected with BVHCV.S, BVHCV.Sp7−, and BVGUS were lysed with a buffer containing 1% sodium deoxycholate, 0.1% sodium dodecyl sulfate (SDS), 1% Triton X-100, 10 mM Tris, and 140 mM NaCl (pH 8.0), whereas for immunoprecipitation analysis, the lysis buffer consisted of 0.1% Nonidet P-40 (NP-40), 50 mM Tris, 50 mM NaCl, and 5 mM EDTA (pH 7.5). All lysis buffers contained 1 mM phenylmethylsulfonyl fluoride (PMSF), 2 μg of aprotinin per ml, and 2 μg of leupeptin per ml. The cell lysate was cleared of cell debris and nuclei by low-speed centrifugation (15 min at 15,000 × g and 4°C). For immunoblotting, a fraction of the supernatant (containing 50 μg of protein) was subjected to 12% polyacrylamide gel electrophoresis (PAGE). For immunoprecipitation, 400 μl of the cleared supernatant of either BVHCV.S- or BVGUS-infected cells was incubated with 1 μl of anti-E2(9284) antibody for 16 h at 4°C and then with 50 μl of protein A-Sepharose 4B-Cl beads (Pharmacia Biotech Inc., San Francisco, Calif.) for 1 h at room temperature with mixing. The beads were washed repeatedly, and the bound proteins were released and denatured by heating for 5 min at 95°C in SDS sample buffer (13). The immunoprecipitated proteins were analyzed by electrophoresis on a 12% polyacrylamide gel. After gel transfer to polyvinylidene difluoride membranes (Westran; Schleicher & Schuell, Keene, N.H.), the blots were probed with anti-core (diluted 1:2,000), anti-E1 (diluted 1:2,000), or anti-E2(G/H) (diluted 1:1,000) antibody followed by horseradish peroxidase-conjugated anti-mouse immunoglobulin G antibody (diluted 1:4,000; Amersham Corp., Arlington Heights, Ill.), with subsequent chemiluminescence detection (ECL; Amersham). For analysis of the expression of the HCV structural proteins in mammalian cells, BSC-1 cells (grown in a monolayer in modified Eagle medium–2% fetal calf serum) were infected at an MOI of 10 with wild-type vaccinia or a vaccinia virus (vvHCV.S) containing the same HCV structural cDNA as the baculovirus BVHCV.S (provided by E. V. Schmidt, Massachusetts General Hospital, Boston). At 8 h postinfection, expression of the HCV structural proteins was analyzed as described above.
Electron microscopy.
Sf9 cells infected with BVHCV.S, BVHCV.Sp7−, and BVGUS were washed with PBS and fixed in various solutions. For morphological studies, the fixative buffer consisted of 1.25% paraformaldehyde, 2.5% glutaraldehyde in 0.03% picric acid, and 0.05 M cacodylate buffer at pH 7.4 (fixative A) (21). For immunodecoration, the cells were fixed in 7% paraformaldehyde–0.25 M sucrose in 0.03% picric acid–0.05 M cacodylate buffer at pH 7.4 (fixative B). The cells were scraped from the cell culture dishes with a razor blade, pelleted in an Eppendorf desktop centrifuge for 10 min at 18,000 × g, and postfixed with 1% osmium tetroxide in 0.05 M cacodylate buffer for 15 min. The pellets were washed in 0.1 M maleate buffer (pH 5.0), treated with 1% uranyl acetate (pH 5.0) for 30 min, washed with maleate buffer, dehydrated in a graded series of ethanol solutions followed by propylene oxide, embedded in a mixture of Epon 812 and Araldite, and polymerized at 40°C for 3 days. Thin sections were stained with saturated uranyl acetate diluted to 50% with acetone and then with lead citrate for electron microscopy examination. Prior to immunogold labeling of thin sections, plastic-embedded cells in semithin sections (0.5 to 1 μm) were mounted on glass slides and stained with anti-HCV-positive human serum, anti-E1, or anti-E2(9284) antibody as described above. For immunogold labeling, ultrathin sections collected on nickel grids were etched with saturated NaIO4. After being washed with PBS, the grids were incubated with 3% BSA in PBS for 30 min. The grids were then incubated for 1 h with either anti-HCV (HCV patient serum; diluted 1:100 in 1% BSA–PBS), anti-E1 (diluted 1:50 in 1% BSA–PBS), or anti-E2 ([polyclonal anti-E2(9284) rabbit; diluted 1:50 in 1% BSA–PBS] antibody or 1% BSA–PBS only. After five washes with PBS, samples were incubated with protein A coupled to 10-nm gold particles (Jan Schlott Laboratories, Utrecht, The Netherlands) in PBS (dilution 1:200) and rinsed five times with PBS. After counterstaining with uranyl acetate and lead citrate, samples were examined with a transmission electron microscope (JEOL 1200 EX) operated at 80 kV. For electron microscopy of partially purified viruslike particles, sucrose gradient fractions were pooled, diluted 1:10 with PBS, and subjected to ultracentrifugation (Beckman SW55 rotor; 40,000 rpm, 2 h, 4°C). The pellet was fixed in fixative A or B and subjected to the same processing as that described above.
Purification of HCV-like particles.
At 96 h postinfection, insect cells infected with BVHCV.S or BVHCV.Sp7− (approximately 5 × 107 cells, grown in suspension) were lysed in 50 mM Tris–50 mM NaCl–0.5 mM EDTA (pH 7.5) with 1 mM PMSF, 2 μg of aprotinin per ml, and 2 μg of leupeptin per ml. In some experiments, 0.1% NP-40 was added to the lysis buffer, and sonication of the lysate was performed. The lysate was homogenized and subjected to low-speed centrifugation (15 min at 4°C and 15,000 × g), and the supernatant was pelleted over a 30% (wt/vol) sucrose (in 20 mM Tris–150 mM NaCl [pH 7.4]) cushion (6 h at 4°C and 150,000 × g). The pellet, containing the HCV-like particles, was resuspended in 50 mM Tris–100 mM NaCl (pH 7.4) or PBS, homogenized, and subjected to a second sucrose or CsCl equilibrium gradient centrifugation. For sucrose equilibrium gradient centrifugation, the resuspended pellet was layered onto a 20 to 60% (wt/wt) sucrose (in 50 mM Tris–100 mM NaCl [pH 7.4]) gradient and centrifuged for 22 h at 4°C and 150,000 × g (17). Ten 0.5-ml fractions were collected from the top and analyzed by SDS–12% PAGE. For CsCl equilibrium gradient centrifugation, 0.5 ml of the resuspended pellet was mixed with 4.5 ml of PBS containing 0.5 mM PMSF and 1.67 g of CsCl (33% [wt/wt] in PBS) and centrifuged for 72 h at 4°C and 300,000 × g (14). After centrifugation, 10 0.5-ml fractions were collected from the top, extensively dialyzed against PBS at 4°C, and analyzed by SDS–12% PAGE. After gel transfer, the blots were probed with anti-core, anti-E1, or anti-E2(G/H) antibody and horseradish peroxidase-labeled anti-mouse antibody as described above. For sucrose sedimentation velocity centrifugation, insect cell lysates were layered onto a 10 to 60% (wt/wt) sucrose (in 50 mM Tris–100 mM NaCl [pH 7.4]) gradient and centrifuged for 2.5 h at 4°C and 200,000 × g. Ten fractions were collected from the top and analyzed for HCV structural proteins as described above. Empty HBV surface particles and HBV core particles were subjected to the same sucrose gradient velocity centrifugation and used as sedimentation markers.
HBV surface and core particles were isolated from the cell culture medium and cytosol of hepatoma cell lines transfected with a replication-competent HBV DNA construct (1, 3, 14). Sedimentation of empty HBV surface particles was analyzed by hepatitis B surface antigen detection in sucrose gradient fractions with a hepatitis B surface antigen-specific radioimmunoassay (Ausria II; Abbott Laboratories, Abbott Park, Ill.). Sedimentation of HBV core particles was analyzed by immunoblotting of sucrose gradient fractions with an anti-HBV core-specific antibody (Dako Corp., Carpinteria, Calif.).
Analysis of HCV-like particle-associated nucleic acids.
Insect cells grown in monolayers (5 × 106 cells) were infected with various combinations of BVGUS, BVHCV.S, and BVHIVgp160. At 96 h postinfection, total RNA was purified by the guanidium isothiocyanate-acid-phenol method (5), and a total of 20 μg of RNA was subjected to Northern blot analysis as described below. To analyze HCV particle-incorporated RNA, HCV-like particles were isolated either by immunoprecipitation with a polyclonal rabbit anti-E2(9284) antibody or by pelleting of insect cell lysates over a 30% sucrose cushion and subsequent immunoprecipitation of the resuspended pellet with a polyclonal rabbit anti-E2(9284) antibody (immunoprecipitation buffer: 50 mM Tris, 100 mM NaCl, 0.5% digitonin [pH 7.4]) as described above. After digestion of the precipitated particles with Staphylococcus aureus nuclease (Boehringer Mannheim Biochemicals, Indianapolis, Ind.) at a concentration of 24 μg/ml and RNase A (Sigma, St. Louis, Mo.) at a concentration of 50 μg/ml for 2 h at 37°C (in 20 mM Tris [pH 8.8], 5 mM CaCl2, 2 mM NaCl) to eliminate any nonincorporated nucleic acids (3, 14), particle-associated RNA was isolated by the guanidium isothiocyanate-acid-phenol method (5) and subjected to Northern blot analysis with [α-32P]dCTP-labeled HCV (nt 259 to 2819)-, GUS-, or HIV gp160-specific cDNA probes (equal cDNA counts per minute per nanogram in each probe).
RESULTS
Expression and interaction of HCV structural proteins in insect cells.
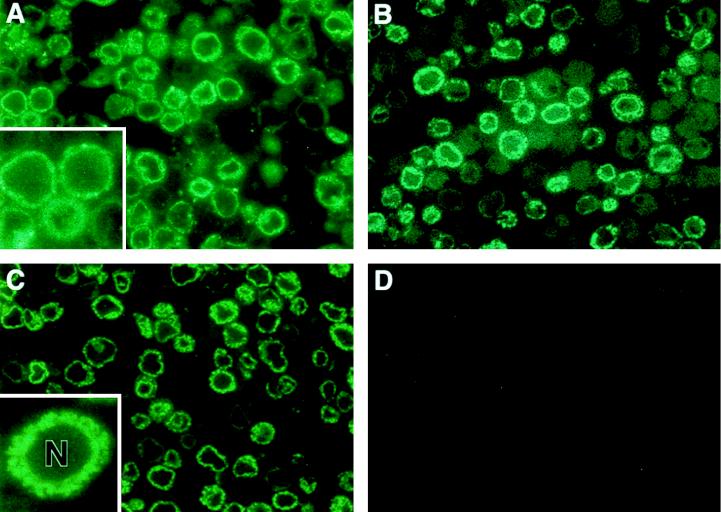
Recombinant baculovirus BVHCV.S, containing the coding sequences for the core, E1, E2, and p7 proteins and 21 amino acids of the NS2 protein (HCV nt 259 to 2819; Fig. 1), directed the production of HCV structural proteins in insect cells, as demonstrated by immunofluorescence analysis of infected insect cells with anti-HCV antibodies (Fig. 2) and immunoblotting of insect cell lysates (Fig. 3). Immunofluorescence analysis of semithin sections of BVHCV.S-infected insect cells with anti-HCV antibodies demonstrated that the expression of HCV structural proteins was confined to the cytoplasm (Fig. 2A to C). Cytoplasmic staining with clusters of immunoreactivity was observed with serum from an HCV-infected individual with high-titer anti-HCV antibodies (Fig. 2A) and specific antibodies against E1 (Fig. 2B), E2 (Fig. 2C), or the core (data not shown). The anti-HCV antibodies used in this study did not display any cross-reactivity against insect cell or baculovirus proteins (Fig. 2D).
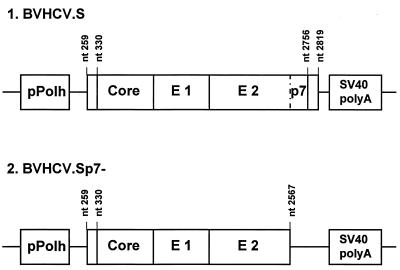
FIG. 1.
Map depicting segments of the HCV genome in the recombinant baculoviruses expressing HCV structural proteins. The BVHCV.S construct contains a short stretch of the 5′ UTR as well as the coding sequences for the core, E1, E2, and p7 proteins and 21 amino acids of the NS2 protein. The polyprotein open reading frame begins at nt 330, and the junction of E2-p7 and NS2 is at nt 2756. BVHCV.Sp7− contains the same 5′ UTR, core, E1, and E2 coding sequences as BVHCV.S but not p7 and the amino-terminal part of the NS2 protein. pPolh, baculovirus polyhedrin promoter; SV40, simian virus 40.
FIG. 2.
Immunofluorescence of HCV structural proteins expressed in insect cells. (A, B, and C) Insect cells were infected with the recombinant baculovirus BVHCV.S containing the cDNA for the HCV structural proteins at an MOI of 10. At 96 h postinfection, the cells were fixed and processed as described in Materials and Methods. Protein expression was analyzed with serum obtained from an HCV-infected human (A), anti-E1 antibody (B), or anti-E2 antibody (C) as described in Materials and Methods. The insets show a higher magnification of the stained insect cells. Immunostaining was confined to the cytosol, whereas the nucleus (N) remained unstained. (D) Insect cells infected with the control baculovirus BVGUS containing the cDNA for GUS were subjected to staining with a mixture of the same serum and antibodies as those shown in panels A, B, and C.
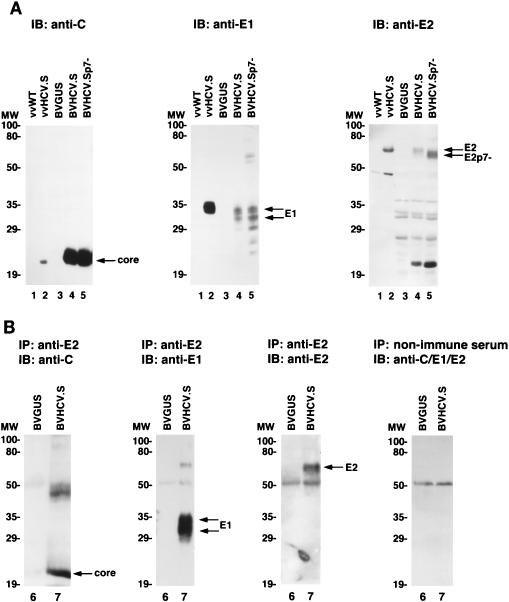
FIG. 3.
(A) Expression of HCV structural proteins in insect and mammalian cells. Sf9 insect cells were infected either with control baculovirus BVGUS (lane 3) or with the HCV expression baculoviruses BVHCV.S (lane 4) and BVHCV.Sp7− (lane 5). At 96 h postinfection, the cells were lysed as described in Materials and Methods. To compare the expression of HCV structural proteins between insect and mammalian cells, BSC-1 cells were infected with a wild-type vaccinia virus (vvWT) (lane 1) or a vaccinia virus (vvHCV.S) containing the same cDNA for the HCV structural proteins as baculovirus BVHCV.S (lane 2). At 8 h postinfection, the cells were lysed as described in Materials and Methods. Sf9 and BSC-1 cell lysates were then subjected to SDS-PAGE and immunoblotting (IB) with anti-core (left panel), anti-E1 (middle panel), or anti-E2 (right panel) antibodies. Molecular masses (in kilodaltons) of protein molecular weight (MW) markers are indicated on the left; HCV-specific proteins are indicated on the right. (B) Coimmunoprecipitation of HCV structural proteins in insect cells. Insect cells were infected with BVGUS (lane 6) or BVHCV.S (lane 7). At 96 h postinfection, the cells were lysed and subjected to immunoprecipitation (IP) with an anti-E2 antibody or nonimmune serum as indicated. The immunoprecipitated proteins were subjected to SDS-PAGE and immunoblotting (IB) with anti-core (C), anti-E1, or anti-E2 antibodies as indicated; the control immunoprecipitate with nonimmune serum was probed with a mixture of antibodies to core, E1, and E2 proteins. The exposure times for the probed blot were adjusted to visualize the proteins and do not represent a quantitation of the precipitated proteins.
Analysis of insect cell lysates by SDS-PAGE and immunoblotting with monoclonal antibodies against the core, E1, and E2 proteins revealed appropriate posttranslational processing of the HCV structural proteins. As shown in Fig. 3A, the core protein had an apparent molecular mass of 22 kDa, the E1 protein was present in various glycosylated forms and had a molecular mass of 30 to 35 kDa, and the E2 protein had a molecular mass of approximately 66 kDa. The size of the core protein was identical to that of the core protein expressed in a mammalian tissue culture system (Fig. 3A), whereas the sizes of the E1 and E2 proteins were similar to the sizes of envelope proteins expressed in mammalian cells (E1, 34 to 36 kDa; E2, 68 kDa; Fig. 3A). These data suggested similar posttranslational processing of HCV structural proteins in insect and mammalian expression systems (39). The variations in the sizes of the envelope proteins probably reflected differences in glycosylation between the two protein expression systems. This finding has been noted by other investigators (34) and is not unexpected, since glycosylation in insect and mammalian cells has been shown to be distinctly different (33). The difference in glycosylation may have resulted in different signal intensities on the immunoblot, thereby explaining the apparently higher levels of E1 and E2 relative to the core in mammalian cells than in insect cells (Fig. 3A). On the other hand, it is possible that aberrant translational termination or accelerated degradation of a particular protein species may have been responsible for this difference between mammalian and insect cells. As shown in Fig. 3B, the E2 protein was coimmunoprecipitated with the E1 and core proteins. The interaction of the E2 protein with the E1 and core proteins was also confirmed by metabolic labeling and coimmunoprecipitation (data not shown). These results extend the findings of recent studies which demonstrated interactions of the E1 and E2 proteins (7) as well as the core and E1 proteins (31).
HCV-like particle assembly occurs in cytoplasmic vesicles.
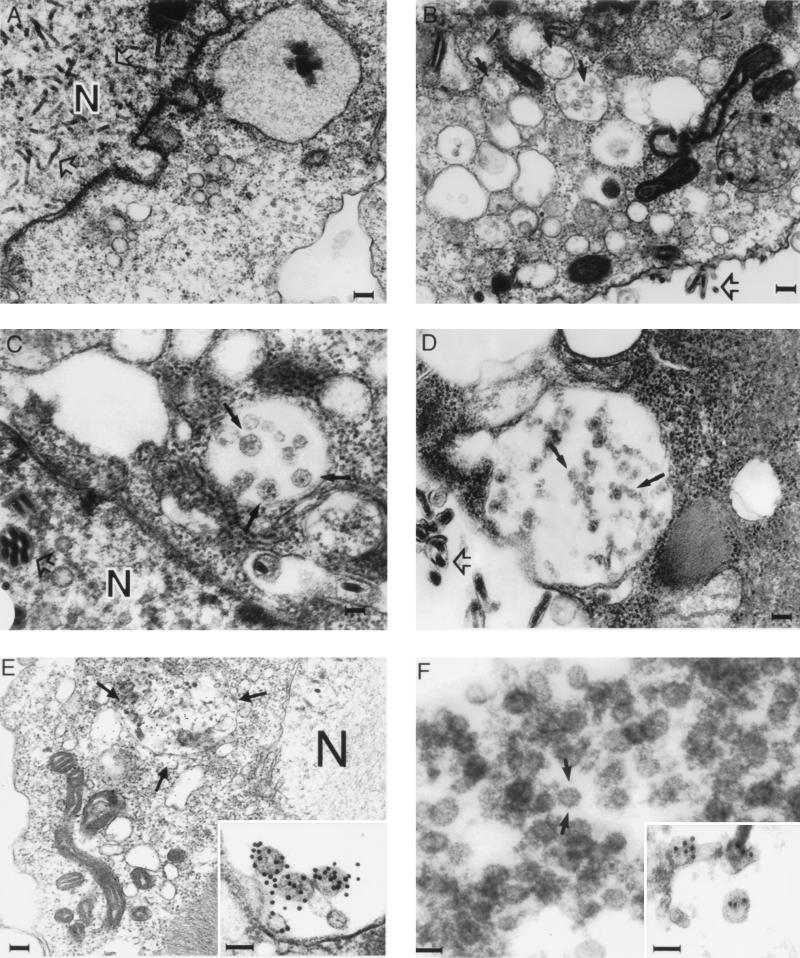
Transmission electron microscopy of BVHCV.S-infected cells revealed abundant viruslike particles in cytoplasmic vesicles or vacuoles, presumably derived from the endoplasmic reticulum (ER) of the insect cells (Fig. 4B, C, and E). These particles, 40 to 60 nm in diameter, were polymorphic in appearance and had an envelope consisting of a membrane (Fig. 4B, C, and E); many of them had unevenly distributed electron-dense structures suggestive of possible nucleocapsids. Visualization of these particles was only possible after osmium treatment, which stains membranes. These features resemble the previously described structure and morphology of pestiviruses in infected cells (4, 12, 44). The particles formed predominantly in cytoplasmic vesicles, giving the appearance of virion transport through the ER secretory pathway of the cells. These particle-containing structures were not observed in insect cells infected with a control baculovirus expressing GUS (BVGUS; Fig. 4A) or uninfected insect cells (data not shown), suggesting that they were not related to baculovirus protein expression and replication. In addition to these vesicles containing viruslike particles, floccular membranous materials with irregular structures of 20 to 100 nm were clustered in large vacuoles in the cytoplasm (Fig. 4B). These diversely polymorphic structures also contained membranous envelopes, but most of them demonstrated no viruslike structures.
FIG. 4.
Electron microscopy of HCV-like particles in insect cells infected with BVHCV.S or BVHCV.Sp7−. Insect cells in monolayer cultures were infected with BVGUS, BVHCV.S, or BVHCV.Sp7−. At 96 h postinfection, the cells were fixed and processed for electron microscopy as described in Materials and Methods. (A) Insect cells infected with the control baculovirus BVGUS. Electron microscopy demonstrates abundant replicative forms of baculovirus (open arrows) in the nucleus (N). No HCV-like particles can be seen in the cytoplasm. Bar, 120 nm. (B) Insect cells infected with BVHCV.S. Electron microscopy demonstrates numerous enveloped, viruslike particles 40 to 60 nm in diameter (closed arrows) in vacuoles. In addition, the synthesis of baculovirus is visualized (open arrow). Bar, 120 nm. (C) Higher magnification of viruslike particles (closed arrows) seen in BVHCV.S-infected cells. Bar, 40 nm. (D) Insect cells infected with BVHCV.Sp7−. Electron microscopy demonstrates viruslike particle formation in a large cytoplasmic vacuole similar to that seen in BVHCV.S-infected cells (solid arrows). Baculovirus particles (open arrows) can be distinguished easily from the HCV-like particles in both panels C and D. Bar, 100 nm. (E) Immunogold labeling of viruslike particles (arrows) similar to those seen in panels B, C, and D with serum from an HCV-infected individual. Immunostaining was confined to the cytoplasmic cisternae and ER, whereas the N remained unstained. Bar, 120 nm. The inset shows a higher magnification of the viruslike particles labeled with an anti-E2 antibody. Bar, 50 nm. (F) Electron microscopy of viruslike particles (arrows) partially purified by sucrose gradient centrifugation. Bar, 50 nm. The inset shows labeling of partially purified particles with an anti-E2 antibody. Bar, 50 nm.
The viruslike particles were immunostained specifically with human anti-HCV (Fig. 4E), anti-E2 (Fig. 4E, inset), and anti-E1 (data not shown) antibodies, suggesting that the viruslike particles were derived from HCV structural proteins. In addition to strong immunostaining of the particles, labeling of the ER and the vesicular membranous complexes mentioned above was also present. No nuclear staining was observed with any of the antibodies. The immunolabeling was highly specific for the indicated structures. No labeling of any cellular or baculovirus structures was seen in BVGUS-infected (Fig. 4A) or noninfected insect cells, and staining was not present in samples not incubated with primary antibodies (data not shown). Since examination of the spent medium at various times was negative for HCV-like particles but abundantly positive for baculoviruses, the viruslike particles appeared not to be released or secreted into the culture medium.
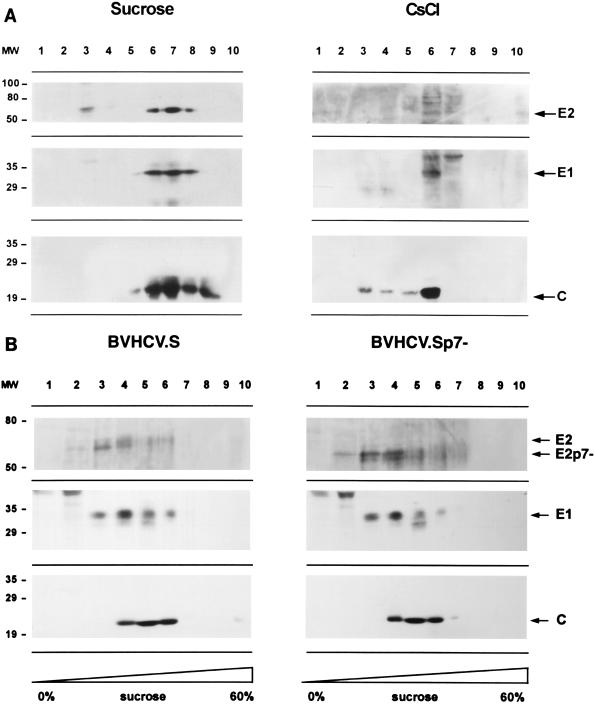
Purification of HCV-like particles by sucrose and CsCl gradient centrifugation.
In order to purify the viruslike particles, lysates of baculovirus-infected insect cells were subjected to sucrose or CsCl equilibrium gradient centrifugation. In both sucrose and CsCl equilibrium gradients, the HCV-like particles banded in specific fractions (Fig. 5A), confirming the assembly of viruslike particles. The densities of the fractions demonstrating immunoreactivity for HCV-like particles were 1.14 to 1.18 g/cm3 in sucrose equilibrium gradients (fractions 6 to 8 in Fig. 5A) and 1.14 to 1.16 g/cm3 in CsCl equilibrium gradients (fraction 6 in Fig. 5A). Colocalization of the HCV structural proteins in specific sucrose fractions was confirmed by coimmunoprecipitation of the HCV structural proteins. To confirm that the colocalization of the HCV structural proteins in the gradient fractions did not represent randomly assembled protein aggregates, sucrose gradient fractions showing immunoreactivity for HCV structural proteins were examined with transmission electron microscopy. Like the structures seen within the BVHCV.S-infected insect cells, 40- to 60-nm enveloped viruslike particles were present (Fig. 4F) and were labeled specifically with an anti-E2 antibody (Fig. 4F, inset).
FIG. 5.
Purification of HCV-like particles by sucrose and CsCl gradient centrifugation. (A) Sucrose and CsCl equilibrium centrifugation. At 96 h postinfection, insect cells infected with BVHCV.S were lysed and subjected to low-speed centrifugation, and the supernatant was pelleted over a 30% sucrose cushion. The pellet containing the viruslike particles was resuspended and subjected to a second sucrose or CsCl equilibrium gradient centrifugation as described in Materials and Methods. Ten fractions were collected from the top and analyzed by SDS-PAGE and immunoblotting with anti-core (C), anti-E1, or anti-E2 antibodies. Molecular masses (in kilodaltons) of protein molecular weight (MW) markers are indicated on the left; HCV-specific proteins are indicated on the right. (B) Sucrose sedimentation velocity centrifugation. Lysates of insect cells infected with BVHCV.S and BVHCV.Sp7− were layered onto a 10 to 60% sucrose gradient and centrifuged for 2.5 h at 4°C and 200,000 × g. Ten fractions were collected from the top and analyzed for HCV structural proteins as described above.
To further analyze the biophysical properties of the viruslike particles, insect cell lysates were subjected to sucrose sedimentation velocity centrifugation (Fig. 5B). Immunoblotting of the sucrose fractions revealed colocalization of the HCV structural proteins in high-sedimentation fractions (fractions 4 to 6), indicating the presence of viruslike particles. Fractions 2 and 3 contained E1 and E2 only, probably representing E1 and E2 complexes as assembly intermediates prior to interaction with the core. To estimate the sedimentation coefficient of the HCV-like particles, we determined the percentage of sucrose in the particle-containing sucrose gradient fractions and applied the approximation tables of McEwen (37). The sedimentation coefficient S20,Ω can be approximated by the Svedberg equation, including the parameters of the applied angular velocity Ω, centrifugation time and temperature, percentage or density of sucrose in which the particle is sedimenting, and the density of the sedimenting particle (37). The validity of this approximation under the experimental conditions was confirmed by sedimentation analysis of HBV surface and core particles as standards. Empty HBV surface particles with a sedimentation coefficient of 39S to 54S and a particle density of 1.16 g/cm3 (18) sedimented to fraction 3 of the sucrose velocity gradient, and HBV core particles with a sedimentation coefficient of 124S and a particle density of 1.30 g/cm3 (18) sedimented to fractions 5 and 6 of the sucrose velocity gradient. Using the approximation of McEwen (37) and an assumed HCV-like particle density of 1.14 to 1.18 g/cm3 (from sucrose and CsCl equilibrium gradients; Fig. 5A), we estimated the sedimentation coefficient S20,w of HCV-like particles to be between 100S and 230S (peak at approximately 160S). The sedimentation coefficient S20,w of HCV-like particles was different from that of similarly sedimenting HBV core particles because of the difference in particle densities. The S value for HCV-like particles is within the range of reported sedimentation coefficients for other members of the Flaviviridae family (44).
Since the visualized viral particles were present predominantly in membrane-enclosed cytoplasmic vesicles, various cell lysis conditions were studied to increase the yield of partially purified particles. Although the qualitative findings did not change, the quantity of purified particles was enhanced significantly by the addition of a low concentration of a mild nonionic detergent (0.1% NP-40 or 0.5% digitonin) to the lysis buffer. The well-known detergent effect of the de-envelopment of viruses has been shown to be a time- and concentration-dependent process. Since the majority of the detergent in the cell lysate was bound in micellar aggregates with cellular proteins and lipids, the effective detergent concentration, being much lower than the original concentration, resulted in the release of viruslike particles from the cytoplasmic vesicles without affecting its envelope. In contrast, higher concentrations of detergent in the lysis buffer (>0.5% NP-40) resulted in the disruption of the core-envelope interactions, as indicated by a lack of their coimmunoprecipitation (data not shown). On the other hand, the sucrose gradient-purified particles were highly sensitive to detergent treatment: incubation of partially purified particles with 0.1% NP-40 disrupted the cosedimentation of HCV structural proteins in the sucrose gradient and eliminated particular viruslike structures, as seen by electron microscopy in Fig. 4F.
HCV protein p7 is not required for viral assembly in insect cells.
In order to study whether HCV protein p7 is required for viral assembly, a recombinant baculovirus containing a deletion of the p7 protein (BVHCV.Sp7−) was generated. This construct contained the same 5′ UTR, core, E1, and E2 coding sequences as BVHCV.S but not p7 (Fig. 1). Infection of insect cells with BVHCV.Sp7− resulted in levels of core and E1 protein expression similar to those seen in BVHCV.S-infected cells. In contrast, the molecular mass of the E2 protein was approximately 7 kDa lower in insect cells infected with the construct BVHCV.Sp7− than in insect cells infected with the construct BVHCV.S (Fig. 3A). These data indicate that p7 is not cleaved efficiently from the E2-p7 polyprotein in BVHCV.S-infected insect cells and are consistent with observations in mammalian expression systems (30, 41). Electron microscopy of insect cells infected with BVHCV.Sp7− demonstrated particle assembly (Fig. 4D) similar to that seen in insect cells infected with BVHCV.S (Fig. 4B and C). We next studied the effect of the p7 deletion on the sedimentation profile of putative HCV-like particles in sucrose gradients. In both sucrose velocity centrifugation (Fig. 5B) and sucrose equilibrium centrifugation (data not shown), viruslike particles derived from BVHCV.Sp7− demonstrated a sedimentation pattern similar to that of BVHCV.S-derived viruslike particles. These data suggest that the HCV p7 protein is not required for the assembly of HCV-like particles in insect cells, although we cannot rule out the possibility that p7 may affect particle assembly in a subtle way.
HCV-like particles incorporate HCV RNAs.
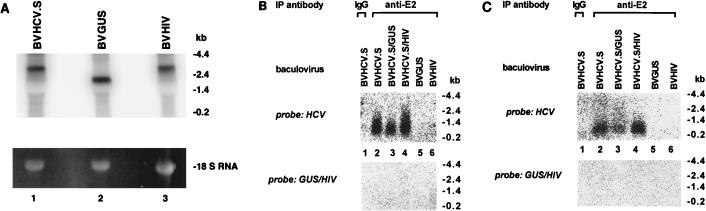
Since the HCV-like particles sedimented at a density typical of nucleic acid-containing particles, we reasoned that these particles might contain nucleic acids. Although the HCV cDNA used in our study contained only a partial genome expressing the structural proteins, it is conceivable that RNA transcribed from this partial genome could have been incorporated into the HCV-like particles. To analyze whether the viruslike particles contained nucleic acids, HCV-like particles were purified by immunoprecipitation with an anti-E2 antibody (Fig. 6B) or by sucrose gradient centrifugation followed by immunoprecipitation (Fig. 6C). After extensive digestion with S. aureus nuclease and RNase A, nuclease-resistant nucleic acids were purified from the immunopurified viral particles and subjected to Northern blot analysis. The HCV-like particles incorporated short HCV transcripts, as indicated by hybridization of the RNA with an HCV-specific probe (Fig. 6B and C). The signal was specific for RNA because treatment of the purified nucleic acids from the viruslike particles with RNase eliminated all of the signal, whereas DNase treatment had no effect. The incorporated HCV RNA appeared to be of degraded forms, as evidenced by the smear of RNA species at the low molecular size, whereas the cDNA transcript of BVHCV.S was about 2.8 kb (Fig. 6A). In order to distinguish between preferential encapsidation and random, nonspecific incorporation of HCV RNA by the viruslike particles, insect cells were coinfected with BVHCV.S and BVGUS (containing the cDNA for GUS) or BVHIV (containing the cDNA for the HIV glycoprotein precursor gp160). Northern blot analysis of total cellular RNA demonstrated similar levels of HCV, GUS, and HIV gp160 transcripts (Fig. 6A). Analysis of RNA encapsidated into the HCV-like particles of the coinfected insect cells revealed the absence of GUS or HIV gp160 transcripts (Fig. 6B and C), suggesting preferential incorporation into the HCV-like particles of HCV transcripts over non-HCV transcripts.
FIG. 6.
(A) Analysis of total RNA of insect cells infected with BVHCV.S, BVGUS, or a baculovirus containing the cDNA for the HIV glycoprotein precursor gp160 (BVHIV). At 96 h postinfection, total RNA was isolated as described in Materials and Methods and subjected to Northern blot analysis with an HCV (lane 1)-, GUS (lane 2)-, or HIV gp160 (lane 3)-specific cDNA probe. Ethidium bromide staining of the 18S RNA was used as a control for RNA loading. (B and C) Encapsidation of HCV RNA into HCV-like particles. In order to distinguish between preferential encapsidation and random, nonspecific incorporation of HCV RNA by the viruslike particles, insect cells were coinfected with BVHCV.S, BVGUS, and BVHIV. HCV-like particles were isolated by immunoprecipitation (IP) (B) or sucrose gradient centrifugation followed by immunoprecipitation (IP) (C) as described in Materials and Methods. Particle-associated RNA was purified as described in Materials and Methods and analyzed by Northern blotting with an HCV-specific cDNA probe (HCV nt 259 to 2819; upper blots) or a GUS- and HIV gp160-specific cDNA probe (lower blots). IgG, immunoglobulin G.
DISCUSSION
In this study, we presented several lines of evidence that HCV structural proteins expressed in recombinant baculovirus-infected insect cells appeared to assemble into viruslike structures with a lipid bilayer envelope. First, viruslike particles were visualized specifically in insect cells infected with BVHCV.S, and their morphology was similar to that of the putative HCV particles detected in cytoplasmic vesicles of an HCV-infected chimpanzee liver (45), an HCV-infected human B-cell line (45), and HCV cDNA-transfected HeLa cells (40). Second, the envelope of the particles, presumably containing properly assembled E1 and E2, was labeled specifically by anti-E1, anti-E2, and anti-HCV-infected human serum containing high titers of anti-HCV antibodies. The same anti-E2 antibody has been shown to bind HCV virions isolated from human serum (42). Third, biophysical characterization of purified HCV-like particles by CsCl and sucrose gradient centrifugation revealed densities similar to those of putative HCV virions visualized by electron microscopy (1.14 to 1.16 g/cm3 [22]) or demonstrated by detection of HCV genomes (1.03 to 1.20 g/cm3 [47], 1.10 to 1.16 g/cm3 [46], or 1.08 g/cm3 [38]) by sucrose equilibrium centrifugation of HCV-infected human plasma. Fourth, the particles could be partially purified and exhibited morphology and immunoreactivity to anti-HCV antibodies similar to those of the particles observed within cells. Fifth, the partially purified particles contained HCV-specific RNA.
Although the current knowledge of HCV virion morphology is limited and the biophysical characterization of putative virions is highly variable in different systems, our data suggest that the morphology and biophysical properties of HCV-like particles synthesized in insect cells are similar to the features described for putative virions isolated from HCV-infected humans. The lack of an efficient tissue culture system for HCV propagation and the difficulty of HCV virion detection in infected humans or chimpanzees other than by PCR precluded a direct side-by-side comparison of the HCV-like particles with authentic virions at the present time.
Prior studies demonstrated the expression of HCV structural proteins in a baculovirus-insect cell system but did not report HCV-like particle assembly (20, 27, 34, 36). The system used in the present study differed from those used in the other studies in that our expression construct contained part of the 5′ UTR and the HCV cDNA was obtained from a different patient source (23). It is possible but unlikely that the 5′ UTR included in the expression construct may be required for appropriate initiation of protein translation and polyprotein processing as a prerequisite for viral assembly (43). Since a recent study (25) elegantly demonstrated that only a minority of circulating viral genomes represent infectious genomes, it is possible that the cDNA sequences used in other baculovirus expression studies were unable to form viral particles due to mutations in domains critical for viral assembly. Furthermore, we found that the optimal processing of infected cells for electron microscopy was crucial for visualization of the viruslike particles. Preservation of cellular and viral structures with an optimal initial fixative buffer and a short period of osmium treatment were important parameters for visualization of the viruslike particles.
The described system may allow the determination of structural requirements for HCV particle assembly. As a first step toward that goal, we demonstrated that HCV protein p7 is not required for particle assembly in insect cells. The function of protein p7, which is conserved among the pestiviruses and HCV, is unknown. Its genomic localization between the structural and nonstructural proteins of the pestiviruses and HCV and the presence of two species of E2 proteins, with and without p7, have led to the suggestion that p7 may play a role in glycoprotein maturation or virus morphogenesis (8, 30, 41). However, no functional data have been presented so far to support this hypothesis. For pestiviruses, it has been shown that the p7 protein is not a major structural component of the virion (8). Our functional analysis demonstrates that the p7 protein does not seem to be necessary for viral assembly in insect cells, although we cannot completely exclude the possibility of p7 affecting viral assembly at the ultrastructural level.
Analysis of particle-associated nucleic acids demonstrated that HCV-like particles contained short HCV RNAs and that these RNAs were selectively incorporated into the particles over non-HCV transcripts. The HCV RNA transcripts generated in this study may contain sufficient cis-acting information (encapsidation signal) interacting specifically with viral structural proteins for encapsidation. However, since the incorporated transcripts were substantially degraded compared to the 2.8-kb transcript of the HCV cDNA, the incorporation more likely represents a cis-dominant, nonspecific effect of the transcripts interacting with the structural proteins; i.e., the physical proximity of the transcripts with their translated proteins conferred a preferential interaction. During such a process, cytosolic RNases may have access to the transcripts, resulting in partial degradation. Further studies are under way to elucidate the mechanism of the observed RNA encapsidation.
The synthesis of HCV-like particles in insect cells is potentially an important tool for studying viral assembly and virus-cell interactions and for the development of an HCV vaccine. In the latter case, efforts to generate recombinant HCV subunit vaccines have been directed at the expression of portions of individual structural proteins in soluble form (6). This approach has met with marginal success, partly as a result of the nonnative forms of the expressed viral proteins. In contrast to the proteins in recombinant subunit vaccines, the HCV proteins in HCV-like particles presumably are presented in a native, virionlike conformation and may therefore be superior in eliciting protective humoral and cellular immune responses. Since HCV-like particles synthesized in insect cells are derived from partial viral genomes without the nonstructural genes required for viral replication, they are noninfectious and therefore represent excellent candidates for an HCV vaccine. Studies are under way to determine the immunogenicity of the HCV-like particles as a potential vaccine.
ACKNOWLEDGMENTS
We thank Kunitada Shimotohno for generously providing construct pCVM980, Brian Seed for kindly providing plasmid pSyngp160, Richard Lesniewski for the gift of polyclonal rabbit anti-E2(9284) antibody, Johnson Lau for monoclonal mouse anti-E1 and anti-E2 antibodies, Emmett V. Schmidt for providing recombinant vaccinia virus vvHCV.S, and Bernard Moss for the gift of wild-type vaccinia virus vvWT. We also thank Harry B. Greenberg, Stephen M. Feinstone, and Jay H. Hoofnagle for helpful discussions. Excellent technical assistance by Hucheng Bei, John Vergalla, Louise Trakimus, and Maria Ericsson is gratefully appreciated.
This work was supported in part by a postdoctoral fellowship grant from the Deutsche Forschungsgemeinschaft (Ba 1417/1-1) to T.F.B. and by grants from the National Institutes of Health to T.F.B. (VF-DK-14361) and T.J.L. (DK-01952 and CA-54525). S.I. was supported by the Harvard Digestive Disease Center. D.T.W. was a recipient of a Glaxo Institute for Digestive Health scientific research award and was supported by NIH grants T30DK38707 and AI95-012.
REFERENCES
- 1.Acs G, Sells M A, Purcell R H, Price P, Engle R, Shapiro M, Popper H. Hepatitis B virus produced by transfected HepG2 cells causes hepatitis in chimpanzees. Proc Natl Acad Sci USA. 1987;84:4641–4644. doi: 10.1073/pnas.84.13.4641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alter H J, Purcell R H, Shih J W, Melpolder J C, Houghton M, Choo Q-L, Kuo G. Detection of antibody to hepatitis C virus in prospectively followed transfusion recipients with acute and chronic non-A, non-B hepatitis. N Engl J Med. 1989;321:1494–1500. doi: 10.1056/NEJM198911303212202. [DOI] [PubMed] [Google Scholar]
- 3.Baumert T F, Rogers S A, Hasegawa K, Liang T J. Two core promoter mutations identified in a hepatitis B virus strain associated with fulminant hepatitis result in enhanced viral replication. J Clin Invest. 1996;98:2268–2276. doi: 10.1172/JCI119037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bielefeldt Ohmann H, Bloch B. Electron microscopic studies of bovine viral diarrhea virus in tissues of diseased calves and in cell cultures. Arch Virol. 1981;71:57–74. doi: 10.1007/BF01315175. [DOI] [PubMed] [Google Scholar]
- 5.Chomczynski P N, Sacchi N. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem. 1987;162:156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- 6.Choo Q-L, Kuo G, Ralston R, Weiner A J, Chien D, Van Nest G, Han J, Berger K, Thudium K, Kuo C, Kansopon J, McFarland J, Tabrizi A, Ching K, Moss B, Cummins L B, Houghton M, Muchmore E. Vaccination of chimpanzees against infection by the hepatitis C virus. Proc Natl Acad Sci USA. 1994;91:1294–1298. doi: 10.1073/pnas.91.4.1294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dubuisson J, Hsu H H, Cheung R C, Greenberg H B, Russell D R, Rice C M. Formation and intracellular localization of hepatitis C virus envelope glycoprotein complexes expressed by recombinant vaccinia and sindbis viruses. J Virol. 1994;68:6147–6160. doi: 10.1128/jvi.68.10.6147-6160.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Elbers K, Tautz N, Becher P, Stoll D, Rümenapf T, Thiel H-J. Processing of the pestivirus E2-NS2 region: identification of proteins p7 and E2p7. J Virol. 1996;70:4131–4135. doi: 10.1128/jvi.70.6.4131-4135.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Feinstone S M, Mihalik K B, Kamimura T, Alter H J, London W T, Purcell R H. Inactivation of hepatitis B viruses and non-A, non-B hepatitis by chloroform. Infect Immun. 1983;41:816–821. doi: 10.1128/iai.41.2.816-821.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gheysen D, Jacobs E, De Foresta F, Thiriart C, Francotte M, Thines D, De Wilde M. Assembly and release of HIV-1 precursor pr55gag virus-like particles from recombinant baculovirus-infected insect cells. Cell. 1989;59:103–112. doi: 10.1016/0092-8674(89)90873-8. [DOI] [PubMed] [Google Scholar]
- 11.Grakoui A, Wychowski C, Lin C, Feinstone S M, Rice C M. Expression and identification of hepatitis C virus polyprotein cleavage products. J Virol. 1993;67:1385–1395. doi: 10.1128/jvi.67.3.1385-1395.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gray E W, Nettleton P F. The ultrastructure of cell cultures infected with border disease and bovine virus diarrhoea viruses. J Gen Virol. 1987;68:2339–2346. doi: 10.1099/0022-1317-68-9-2339. [DOI] [PubMed] [Google Scholar]
- 13.Harlow E, Lane D. Antibodies. A laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1988. [Google Scholar]
- 14.Hasegawa K, Huang J, Rogers S A, Blum H E, Liang T J. Enhanced replication of a hepatitis B virus mutant associated with an epidemic of fulminant hepatitis. J Virol. 1994;68:1651–1659. doi: 10.1128/jvi.68.3.1651-1659.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.He L E, Alling D, Popkin D, Shapiro M, Alter H J, Purcell R H. Determining the size of non-A, non-B hepatitis virus by filtration. J Infect Dis. 1987;156:636–640. doi: 10.1093/infdis/156.4.636. [DOI] [PubMed] [Google Scholar]
- 16.Hijikata M, Kato N, Ootsuyama Y, Nakagawa M, Shimotohno K. Gene mapping of the putative structural region of the hepatitis C virus genome by in vitro processing analysis. Proc Natl Acad Sci USA. 1991;88:5547–5551. doi: 10.1073/pnas.88.13.5547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hijikata M, Shimuzu Y K, Kato H, Iwamoto A, Shih J W, Alter H J, Purcell R H, Yoshikura H. Equilibrium centrifugation studies of hepatitis C virus: evidence for circulating immune complexes. J Virol. 1993;67:1953–1958. doi: 10.1128/jvi.67.4.1953-1958.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hollinger F B. Hepatitis B virus. In: Fields B N, Knipe D M, Howley P M, editors. Fields virology. 3rd ed. Philadelphia, Pa: Lippincott-Raven; 1996. pp. 2738–2808. [Google Scholar]
- 19.Hoofnagle J H, Di Bisceglie A M. The treatment of chronic viral hepatitis. N Engl J Med. 1997;336:347–356. doi: 10.1056/NEJM199701303360507. [DOI] [PubMed] [Google Scholar]
- 20.Hsu H, Donets M, Greenberg H B, Feinstone S M. Characterization of hepatitis C virus structural proteins with a recombinant baculovirus expression system. Hepatology. 1993;17:763–771. [PubMed] [Google Scholar]
- 21.Ito S, Karnovsky M J. Formaldehyde-glutaraldehyde fixatives containing trinitro compounds. J Cell Biol. 1968;39:168A. [Google Scholar]
- 22.Kaito M, Watanabe S, Tsukiyama-Kohara K, Yamaguchi K, Kobayashi Y, Konishi M, Yokoi M, Ishida S, Suzuki S, Kohara M. Hepatitis C virus particle detected by immunoelectron microscopic study. J Gen Virol. 1994;75:1755–1760. doi: 10.1099/0022-1317-75-7-1755. [DOI] [PubMed] [Google Scholar]
- 23.Kato N, Hijikata M, Ootsuyama Y, Nakagawa M, Ohkoshi S, Sugimura T, Shimotohno K. Molecular cloning of the human hepatitis C virus genome from Japanese patients with non-A, non-B hepatitis. Proc Natl Acad Sci USA. 1990;87:9524–9528. doi: 10.1073/pnas.87.24.9524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kirnbauer R, Booy F, Cheng N, Lowy D R, Schiller J T. Papillomavirus L1 major capsid protein self-assembles into virus-like particles that are highly immunogenic. Proc Natl Acad Sci USA. 1992;89:12180–12184. doi: 10.1073/pnas.89.24.12180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kolykhalov A A, Agapov E V, Blight K J, Mihalik K, Feinstone S M, Rice C M. Transmission of hepatitis C by intrahepatic inoculation with transcribed RNA. Science. 1997;277:570–574. doi: 10.1126/science.277.5325.570. [DOI] [PubMed] [Google Scholar]
- 26.Kuo G, Choo Q-L, Alter H J, Gitnick G L, Redeker A G, Purcell R H, Miyamura T, Dienstag J L, Alter M J, Stevens C E, Tegtmeier G E, Bonino F, Colombo M, Lee W-S, Kuo C, Berger K, Shuster R J, Overby L R, Bradley D W, Houghton M. An assay for circulating antibodies to a major etiologic virus of human non-A, non-B hepatitis. Science. 1989;244:362–364. doi: 10.1126/science.2496467. [DOI] [PubMed] [Google Scholar]
- 27.Lanford R E, Notvall L, Chavez D, White R, Frenzel G, Simonsen C, Kim J. Analysis of hepatitis C virus capsid, E1, and E2/NS1 proteins expressed in insect cells. Virology. 1993;197:225–235. doi: 10.1006/viro.1993.1583. [DOI] [PubMed] [Google Scholar]
- 28.Lemon S M, Thomas D L. Vaccines to prevent viral hepatitis. N Engl J Med. 1997;336:197–203. doi: 10.1056/NEJM199701163360307. [DOI] [PubMed] [Google Scholar]
- 29.Lesniewski R, Okasinski G, Carrick R, Van Sant C, Desai S, Johnson R, Scheffel J, Moore B, Mushahwar I. Antibody to hepatitis C virus second envelope (HCV-E2) glycoprotein: a new marker of HCV infection closely associated with viremia. J Med Virol. 1995;45:415–422. doi: 10.1002/jmv.1890450411. [DOI] [PubMed] [Google Scholar]
- 30.Lin C, Lindenbach B D, Pragai B, McCourt D W, Rice C M. Processing of the hepatitis C E2-NS2 region: identification of p7 and two distinct E2-specific products with different C termini. J Virol. 1994;68:5063–5073. doi: 10.1128/jvi.68.8.5063-5073.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lo S-Y, Selby M S, Ou J-H. Interaction between hepatitis C virus core protein and E1 envelope protein. J Virol. 1996;70:5177–5182. doi: 10.1128/jvi.70.8.5177-5182.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Luckow V A, Lee S C, Barry G F, Olins P O. Efficient generation of infectious recombinant baculoviruses by site-specific transposon-mediated insertions of foreign genes into a baculovirus genome propagated in Escherichia coli. J Virol. 1993;67:4566–4579. doi: 10.1128/jvi.67.8.4566-4579.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Luckow V A, Summers M D. Trends in the development of baculovirus expression vectors. Bio/Technology. 1988;6:47–55. [Google Scholar]
- 34.Matsuura Y, Harada S, Susuki R, Watanabe Y, Inoue Y, Saito I, Miyamura T. Expression of processed envelope protein of hepatitis C virus in mammalian and insect cells. J Virol. 1992;66:1425–1431. doi: 10.1128/jvi.66.3.1425-1431.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Matsuura Y, Miyamura T. The molecular biology of hepatitis C virus. Semin Virol. 1993;4:297–304. [Google Scholar]
- 36.Matsuura Y, Suzuki T, Suzuki R, Sato M, Aizaki H, Saito I, Miyamura T. Processing of E1 and E2 glycoproteins of hepatitis C virus expressed in mammalian and insect cells. Virology. 1994;205:141–150. doi: 10.1006/viro.1994.1629. [DOI] [PubMed] [Google Scholar]
- 37.McEwen C R. Tables for estimating sedimentation through linear concentration gradients of sucrose solution. Anal Biochem. 1967;20:114–149. doi: 10.1016/0003-2697(67)90271-0. [DOI] [PubMed] [Google Scholar]
- 38.Miyamoto H, Okamoto H, Sato K, Tanaka T, Mishiro S. Extraordinarily low density of hepatitis C virus estimated by sucrose gradient centrifugation and the polymerase chain reaction. J Gen Virol. 1992;73:715–718. doi: 10.1099/0022-1317-73-3-715. [DOI] [PubMed] [Google Scholar]
- 39.Miyamura T, Matsuura Y. Structural proteins of hepatitis C virus. Trends Microbiol. 1993;1:229–231. doi: 10.1016/0966-842x(93)90137-g. [DOI] [PubMed] [Google Scholar]
- 40.Mizuno M, Yamada G, Tanaka T, Shimotohno K, Takatani M, Tsuji T. Virion-like structures in HeLa G cells transfected with the full-length sequence of the hepatitis C virus genome. Gastroenterology. 1995;109:1933–1940. doi: 10.1016/0016-5085(95)90761-0. [DOI] [PubMed] [Google Scholar]
- 41.Mizushima H, Hijikata M, Asabe S-I, Hirota M, Kimura K, Shimotohno K. Two hepatitis C virus glycoprotein E2 products with different C termini. J Virol. 1994;68:6215–6222. doi: 10.1128/jvi.68.10.6215-6222.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Prince A M, Huima-Byron T, Parker T S, Levine D M. Visualization of hepatitis C virions and putative defective interfering particles isolated from low-density lipoproteins. J Viral Hepatitis. 1996;3:11–17. doi: 10.1111/j.1365-2893.1996.tb00075.x. [DOI] [PubMed] [Google Scholar]
- 43.Reynolds J E, Kaminski A, Kettinen H J, Grace K, Clarke B E, Carrol A R, Rowlands D J, Jackson R J. Unique features of internal initation of hepatitis C virus RNA translation. EMBO J. 1995;14:6010–6020. doi: 10.1002/j.1460-2075.1995.tb00289.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rice C M. Flaviviridae: the viruses and their replication. In: Fields B N, Knipe D M, Howley P M, editors. Fields virology. 3rd ed. Philadelphia, Pa: Lippincott-Raven; 1996. pp. 931–959. [Google Scholar]
- 45.Shimuzu Y K, Feinstone S M, Kohara M, Purcell R, Yoshikura H. Hepatitis C virus: detection of intracellular virus particles by electron microscopy. Hepatology. 1996;23:205–209. doi: 10.1002/hep.510230202. [DOI] [PubMed] [Google Scholar]
- 46.Shindo M, Di Bisceglie A M, Akatsuka T, Fong T-L, Scaglione L, Donets M, Hoofnagle J H, Feinstone S M. The physical state of the negative strand of hepatitis C virus RNA in serum of patients with chronic hepatitis C. Proc Natl Acad Sci USA. 1994;91:8719–8723. doi: 10.1073/pnas.91.18.8719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Thomssen R, Bonk S, Propfe C, Heermann K-H, Koechel H G, Uy A. Association of hepatitis C virus in human sera with beta-lipoprotein. Med Microbiol Immunol. 1992;181:293–300. doi: 10.1007/BF00198849. [DOI] [PubMed] [Google Scholar]
- 48.Tong M J, El-Farra N S, Reikes A R, Co R L. Clinical outcomes after transfusion-associated hepatitis C. N Engl J Med. 1995;332:1463–1466. doi: 10.1056/NEJM199506013322202. [DOI] [PubMed] [Google Scholar]
- 49.Zeng C Q-Y, Wentz M J, Cohen J, Estes M K, Ramig R F. Characterization and replicase activity of double-layered and single-layered rotavirus-like particles expressed from baculovirus recombinants. J Virol. 1996;70:2736–2742. doi: 10.1128/jvi.70.5.2736-2742.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]