Abstract
Liver abscesses (LA) resulting from bacterial infection in cattle pose a significant global challenge to the beef and dairy industries. Economic losses from liver discounts at slaughter and reduced animal performance drive the need for effective mitigation strategies. Tylosin phosphate supplementation is widely used to reduce LA occurrence, but concerns over antimicrobial overuse emphasize the urgency to explore alternative approaches. Understanding the microbial ecology of LA is crucial to this, and we hypothesized that a reduced timeframe of tylosin delivery would alter LA microbiomes. We conducted 16S rRNA sequencing to assess severe liver abscess bacteriomes in beef cattle supplemented with in-feed tylosin. Our findings revealed that shortening tylosin supplementation did not notably alter microbial communities. Additionally, our findings highlighted the significance of sample processing methods, showing differing communities in bulk purulent material and the capsule-adhered material. Fusobacterium or Bacteroides ASVs dominated LA, alongside probable opportunistic gut pathogens and other microbes. Moreover, we suggest that liver abscess size correlates with microbial community composition. These insights contribute to our understanding of factors impacting liver abscess microbial ecology and will be valuable in identifying antibiotic alternatives. They underscore the importance of exploring varied approaches to address LA while reducing reliance on in-feed antibiotics.
Keywords: 16 s, Bacteroides, cattle, Fusobacterium, Liver Abscess, microbiome
Hepatic abscess in cattle contain a low diversity microbial community dominated by Fusobacterium and Bacteroides.
Introduction
Liver abscesses (LA) in cattle result from bacterial infection of the hepatic parenchyma and present a significant economic and welfare challenge to the beef industry globally. In North America, LA severity is graded at slaughter using the Elanco scoring system (0 = healthy, A = one or more small abscesses surrounded by healthy liver tissue, A+ = one or more large abscesses surrounded by inflamed liver tissue; Elanco, USA). Liver abscess prevalence rose from 9.9% in 2010–2011 to 19.3% in 2016–17 (National Beef Quality Audit 2018), and can exceed 90% in some cohorts (Nagaraja and Lechtenberg 2007). Severe LA inhibit animal performance, causing a 5%–15% reduction in average daily gain and a 9.7% reduction in feed efficiency (Brink et al. 1990, Brown and Lawrence 2010, Rezac et al. 2014). In Canada, discounts due to LA cost the industry ∼ $61.2 million annually, but this is likely an underestimate as it does not account for losses incurred by reduced animal productivity (National Beef Quality Audit 2018).
Although LA can occur in all cattle, occurrence is most common in feedlot cattle receiving high-grain diets. It is hypothesized that the acidotic conditions created by rapid microbial fermentation of starch disrupts rumen and gut epithelial integrity, facilitating the translocation of opportunistic pathogens, including Fusobacterium necrophorum and Trueperella pyogenes to the liver via the portal circulation system (Pinnell and Morley 2022) The Gram-negative bacteria F. necrophorum has long been considered as the primary etiological agent of LA, and is recovered almost universally in culture-based studies of the LA microbiota (Nagaraja and Lechtenberg 2007, Pinnell and Morley 2022). Recent metataxonomic surveys have presented a considerable body of evidence that the microbial communities involved in LA are more complex than initially thought and that multiple Gram-positive bacteria likely play a role in LA etiology (Weinroth et al. 2019, Amachawadi et al. 2021, Fuerniss et al. 2022). Moreover, a recent study has proposed that LA microbiomes are almost universally dominated by either Fusobacterium or Bacteroides and can be classified into sub-types on this basis, challenging the long-held view of F. necrophorum as the primary etiological agent of LA (Pinnel et al. 2022). However, making equitable comparisons between LA microbiome studies is complicated by differences in the method of sample collection. Most studies have focused their examination of LA microbiomes on the purulent material of the abscess (Weinroth et al. 2017, Amachawadi et al. 2021, Stotz et al. 2021, Fuerniss et al. 2022, Pinnell et al. 2022), while one study also examined the microbial community associated with the abscess inner wall tissue (Amachawadi et al. 2021). Further research into the microbial processes driving LA formation is warranted to develop a comprehensive understanding of the microbial ecology of LA, which will aid in the identification of alternative methods for the prevention of LA in beef cattle.
At present, there is no ante-mortem diagnostic test for LA, and prevention relies mainly on in-feed administration of prophylactic antibiotics during the finishing period. The most widely used antibiotic for LA prevention is tylosin phosphate, a macrolide with broad-spectrum activity against Gram-positive bacteria (Shryock et al. 1998). Although tylosin is widely used in beef finishing systems throughout North America, macrolides are categorized as medically important antimicrobials (MIA) and are considered critical to human health by both Health Canada (Health Canada 2009) and the World Health Organization (WHO 2019). There are concerns that the use of MIA in food animals may contribute to antimicrobial resistance in human pathogens, which, coupled with the increased frequency of LA occurrence, highlights the importance of developing alternative practices to ameliorate LA in cattle. While alternative dietary formulations, feed supplements and vaccines have been evaluated for their efficacy in this regard, they have generally proven ineffective and have not been widely adopted by industry (Reinhardt and Hubbert 2015, Amachawadi and Nagaraja 2016).
We recently examined whether reductions in the amount of tylosin fed to beef cattle during the finishing period would increase the occurrence of LA and found that the duration of tylosin administration can be reduced by at least 25% without impacting LA frequency, animal health, performance, or carcass traits (Davedow et al. 2020). The present study builds on these findings by employing a 16S metataxonomic approach to evaluate the effect of a reduced timeframe of tylosin use on the microbiomes associated with both the purulent material and inner wall tissues of LA. Our data confirms previous observations of a sparse bacteriome in bovine LA, comprising ASVs of low prevalence across samples and dominated by either Fusobacterium or Bacteroides. We present evidence that microbial communities may vary between large and small abscesses, but that timing of tylosin administration has only a small impact on LA microbiota.
Materials and methods
Ethical statement
All animal procedures performed during the experiments described herein were reviewed and approved by the Animal Care Committees of Feedlot Health Management Services LTD (Okotoks, Alberta) and Lethbridge Research and Development Centre (AUP #:1642) in accordance with the guidelines set out by the Canadian Council on Animal Care, with informed consent from the animal owners.
Animal trial and sample collection
Details of experimental animal management have been published previously (Davedow et al. 2020). Briefly, a total of 7576 crossbred feedlot cattle, comprising both steers and heifers, were randomly assigned to one of three treatment groups: (i) tylosin fed throughout the feeding period (day 0–161; CTRL), (ii) tylosin fed during the first 78% of the feeding period (day 0–125; F-78), and (iii) tylosin fed during the last 75% of the feeding period (day 41–161; L-75). Each treatment group consisted of 10 pens, with an average of 253 animals per pen. Animals were adapted to a high-concentrate diet using a succession of four step-up diets over 20 days, prior to the feeding period. The finishing diet consisted of 85.8% concentrate, 11.5% roughage, and 2.8% supplement. The concentrate portion consisted of 70% corn with the remainder being tempered rolled barley / wheat. Tylosin phosphate (Tylosin 40, BioAgri Mix LP, Mitchell, ON) was administered to cattle at a rate of 11 ppm on a dry matter basis (as recommended for the prevention of LA in Canadian beef cattle (CFIA 2023) according to the treatment regimens described above. Monensin sodium was also included in diets at 33 ppm DM over the feeding period (Monensin Premix; Bio-Agri Mix LP, Mitchell, Ontario) again as recommended in Canadian feedlots (CFIA, 2023). Water and diets were provided ad libitum throughout the feeding period, with diets delivered twice daily.
After slaughter at a commercial abattoir, livers were obtained and evaluated by a certified CFIA inspector using the Elanco Liver Scoring System (0, A, A+). A subset of livers, ranging from 0 to 4 livers per pen within each treatment, was selected for further analysis. Specifically, severely abscessed livers with an A+ grade (n = 60) were chosen, with 6 out of 10 pens sampled for each treatment. A mobile laboratory facility was positioned near the abattoir's slaughter operations wing exit, and each selected liver was promptly transported in a sterile bag to the mobile lab immediately after retrieval. Prior to dissection, photographic records were taken to establish the appearance of the abscessed tissue for a subset of the samples (n = 34). Due to concerns over nucleic acid degradation, this step was not performed on all the livers. The categorization included (i) large—a single large abscess (>50 mm in diameter with purulent material), (ii) small—1–5 small abscesses with purulent material, (iii) small-multiple—more than five small abscesses on the liver with purulent material, or (iv) atypical—generally very large abscesses with solidified purulent/core mass, differing from typical purulent material. Tissue samples were collected aseptically. The liver surface was first disinfected with 70% ethanol, and a 2–3 cm3 piece of abscessed tissue, typically comprising half or a quarter of the abscess, including the abscess wall and purulent material (or the entire abscess in the case of smaller lesions), was collected using a sterile scalpel. Fresh gloves and scalpel blades were used for each sample. Using sterile forceps, the sample was carefully placed in the bottom of a sterile 50 ml falcon tube and immediately snap-frozen in liquid nitrogen. The samples were transported on dry ice to the laboratory and stored at −80°C awaiting molecular analysis.
Metagenomic DNA isolation
Prior to DNA isolation, each sample was processed aseptically to collect the bulk purulent material (BLP) and capsule-adhered (CAP) samples as follows. The BLP was collected by gently scooping or squeezing the material (pus) from already sliced (half or quarter) abscess using a sterile scalpel, or in the case of an intact abscess, cutting open the abscess in the middle and collecting the content. The inner wall tissue of the abscess capsule was scraped off and the scrapings were collected as the CAP sample.
DNA was isolated as previously described (Zaheer et al. 2018). Briefly, 325 mg of BLP/CAP was added to a sterile 2.0 ml safe-lock tube containing 0.3 g of 0.1 mm diameter and 0.1 g of 0.5 mm diameter zirconia beads with 1000 µL of resuspension buffer (600 mM NaCl, 120 mM Tris-HCl, 60 mM EDTA, 200 mM guanidine isothyocynate) and 5 µL of a 1 : 1 mix of β-mercaptoethanol: resuspension buffer. The tubes were mixed by inversion and 200 µL of a preheated (70°C) 10% aqueous solution of SDS was added. The samples were homogenised using an OMNI Bead Ruptor (Omni International, GA, USA) for 3 min at 5 m/s, followed by incubation at 70°C for 15 min with shaking at 300 r/m. Samples were then centrifuged at 16 000 × g for 5 min at 4 °C. The supernatant was retained, and the extraction process was repeated in its entirety using the remaining pellet. Both sample lysates were mixed with 200 µL of 10 M ammonium acetate, placed on ice for 10 min, and centrifuged at 4°C for 10 min at 16 000 × g. The supernatant was mixed with an equal volume of isopropanol, placed on ice for 30 min, and centrifuged at 4°C for 15 min at 16 000 × g. The nucleic acid pellet was washed with 70% ethanol and air-dried. Pellets from the same biological sample were dissolved in a total of 200 µL of TE buffer (10 mM Tris.HCl, 1 mM EDTA; pH 8.00) and combined. RNase and proteinase treatments were performed as previously described (Zaheer et al. 2018) followed by column purification using the QIAmp DNA Stool Minikit (QIAGEN Inc., Toronto, ON, Canada) as per the manufacturer's instructions. DNA quality was assessed using a Quant-iT™ PicoGreen kit (Thermo Fisher Scientific, Mississauga, ON, Canada). The purity of the DNA was determined by measuring the ratios of absorbance at 260/280 and 260/230 using a NanoDrop spectrophotometer (Thermo Fisher Scientific Waltham, MA, USA). Samples with a 260/280 ratio value of between 1.7 and 2.0 and a 260/230 ratio between 2.0 and 2.2 were regarded as acceptable for downstream analysis. Purified DNA was stored at −80°C until sequencing.
Library construction and sequencing
The primer pair 515F (5′-GTGCCAGCMGCCGCGGTAA-3′) and 806R (5′-GGACTACHVGGGTWTCTAAT-3′) targeting the V4 region of the 16S rRNA gene were used to examine bacterial communities in CAP and BLP samples (Caporaso et al. 2011). A 33 cycle PCR using 1 µL of a 1 : 10 dilution of genomic DNA and the Fast Start High Fidelity PCR System (Roche, Montreal, PQ) was conducted using the following conditions: 94°C for 2 min, followed by 33 cycles of 94°C for 30 s, 58°C for 30 s, and 72°C for 30 s, with a final elongation step at 72°C for 7 min. Fluidigm Corporation (San Francisco, CA, USA) barcodes were incorporated in a second PCR reaction using the following conditions: 95°C for 10 min, followed by 15 cycles of 95°C for 15 s, 60°C for 30 s, and 72°C for 1 min, followed by a final elongation step at 72°C for 3 min. After amplification, PCR products were assessed in a 2% agarose gel to confirm adequate amplification. All samples were quantified using the Quant-iT PicoGreen dsDNA Assay Kit and were pooled in equal proportions. Pooled samples were then purified using calibrated Ampure XP beads (Beckman Coulter, Mississauga, ON). Libraries were quantified using the Quant-iT PicoGreen dsDNA Assay Kit and the Kapa Illumina GA with Revised Primers-SYBR Fast Universal kit (Kapa Biosystems, Wilmington, MA, USA). The average fragment size was determined using a LabChip GX instrument (PerkinElmer, Waltham, MA, USA). Paired-end (2×250 bp) amplicon sequencing was performed at the Genome Quebec Innovation Centre (Montreal, Canada) using the Illumina MiSeq Reagent Kit v2 (500 cycle) following the manufacturer's guidelines, which incorporates a 20% PhiX spike-in.
Bioinformatic and statistical analysis
Sequence quality was assessed using FASTQC (Andrews 2010). Primer sequences were trimmed using BBDuk from the BBTools suite (Bushnell 2014). Data were processed using QIIME2 v.2022.8 (Bolyen et al. 2019). Both reads were truncated to 200 nt to facilitate high quality read merging and sequences were denoised into amplicon sequencing variants (ASVs) using DADA2 (Callahan et al. 2016), with MAFFT (Katoh and Standley 2013) used to generate a rooted phylogenetic tree. Amplicon sequencing variants were taxonomically classified using a Naïve Bayes classifier trained on the V4 hypervariable region of the 16S rRNA gene with the SILVA SSU 138.1 database (Quast et al. 2013). Taxonomic, phylogenetic, and raw ASV abundance data were exported for further analysis in R (v.4.0.1) using Phyloseq (McMurdie and Holmes 2013). To account for uneven library size, rarefaction curves were generated to identify an optimal point for subsampling prior to diversity analyses. Principle Coordinate Analysis (PCoA) plots were generated using both Unifrac and weighted Unifrac dissimilarity matrices (Lozupone and Knight 2005, Lozupone et al. 2011). PERMANOVA tests based on both distance metrics were used to identify differences in composition according to tylosin treatment, sample type, abscess type, and dominant genus rank using the ‘adonis2()’ function in Vegan (Oksanen et al. 2020). Beta-dispersion tests for the same distance metrics and experimental factors were also performed using Vegan. Alpha-diversity was measured using the Chao1 and Shannon indices on a per-sample basis, with Kruskal-Wallis and Dunn's post-hoc tests applied to identify statistically significant differences according to tylosin treatment regimen, abscess type, and sample type. Statistically significant differences for all diversity metrics were declared at P < 0.05. Prior to differential abundance (DA) testing, singleton ASVs were discarded and data were summarized at the phylum and genus levels. Differential abundance testing according to tylosin treatment, abscess type, and sample type was performed using parametric Wald tests implemented in DeSeq2 (Love et al. 2014), with animal included in the model. Raw, unnormalized count values were used as input for DeSeq2, as recommended by the authors (Love et al. 2014). Statistically significant differences were declared at an FDR-corrected P < 0.05. Average-linkage hierarchical clustering analysis was performed using Euclidean distances with the MicrobiotaProcess package in R (Xu et al. 2023) (Xu et al. 2023). Bar charts were generated using MicroViz (Barnett et al. 2021). All figures were prepared in R using ggplot2 (Wickham 2009).
Results
Liver abscess morphology
Of the 34 livers categorised by appearance, 13 had large abscesses, 8 had multiple small abscesses, and 9 had 1–2 small abscesses. The remaining 4 did not conform to any of these classifications and were denoted as atypical. Example images of each abscess type are presented in Supplementary Fig. S1, alongside an image of a healthy liver from the same animal cohort. Metadata corresponding to these livers are also provided as supplementary material.
Data Summary
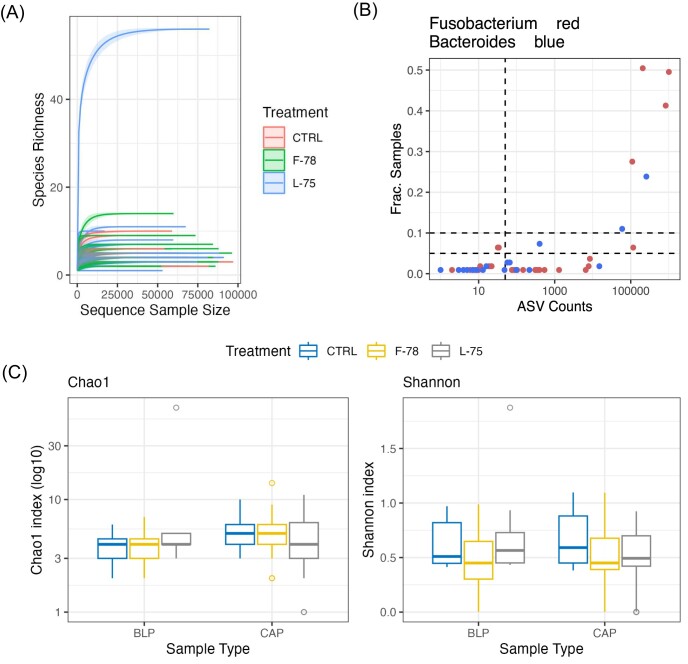
Amplicon sequencing of 16S rRNA libraries generated a total of 7 826 258 paired reads, ranging in number from 13 to 112 405 and with a median of 70 159.0 sequences per sample. An average of 89.71% ±8.22% of the input reads were successfully denoised by DADA2. Rarefaction analysis in 1000-read increments indicated that sequencing depth was sufficient (Supplementary Fig. S1A). Following the removal of unclassified ASVs, 242 remained which were allocated into 19 phyla and 82 genera. Microbial diversity and taxon prevalence was generally low, with just 6 phyla and 9 genera present in >5% of the samples. The ASVs allocated to the Fusobacterium and Bacteroides genera were the most prevalent across all the samples (Fig. 1B).
Figure 1.
(A) rarefaction curve in 1000-sequence increments with replacement. Lines represent individual samples and are colored according to treatment group (CTRL=control, F-78 = animals who received tylosin phosphate in the first 78% of the trial, L-75 = animals who received tylosin phosphate in the last 75% of the trial). (B) prevalence analysis at the ASV level. Fusobacterium and Bacteroides ASVs are highlighted in red and blue, respectively. Horizontal lines represent 5% and 10% ASV detection thresholds. The vertical line denotes 50 reads in total across all samples. The x-axis is provided on a logarithmic scale. Boxplots of (C) Chao1 and (D) Shannon diversity metrics across treatment groups and sample types. Chao1 values were log10 transformed prior to plotting. CAP = capsule-adhered purulent material; BLP = bulk purulent material
Alpha- and Beta-diversity results
Each sample was rarefied to 25 000 random reads prior to alpha- and beta-diversity testing, which excluded 2 samples (1 CAP sample, large abscess; 1 BLP sample, atypical abscess) from downstream analysis. Chao1 and Shannon diversity indices (values provided in Supplementary Data) were compared across groups using Kruskal-Wallis tests with Dunn's post-hoc tests where appropriate (Table 1). Chao1 and Shannon diversity indices were unaffected by the timing of tylosin phosphate administration (Fig. 1C and D; P > 0.05). Samples collected from steers were more diverse than those collected from heifers (Chao1 value: 5.29 vs 3.65; Shannon value: 0.61 vs 0.31; Table 1; P < 0.05), but as there were substantially more samples collected from steers (n = 46) than from heifers (n = 12) we interpret these findings of significance with caution. CAP samples had richer bacteriomes measured by Chao1 than BLP (Fig. 1C; P < 0.05). Bacteroides-dominated samples had higher Shannon diversity than Fusobacterium-dominated samples (Table 1; P < 0.05). Large abscesses had more diverse communities than single small abscesses (P < 0.05), but less diverse communities than multiple small abscesses (P < 0.05) using the Shannon index.
Table 1.
Alpha-diversity results
| Chao11 | Shannon | |||
|---|---|---|---|---|
| chi-squared | P-value | chi-squared | P-value | |
| Treatment | 0.05 | 0.976 | 4.95 | 0.084 |
| Dominant genus | 0.44 | 0.803 | 15.19 | 0.001 |
| Gender | 6.56 | 0.010 | 12.90 | <0.001 |
| Sample type | 6.87 | 0.009 | 0.09 | 0.760 |
| Abscess type | 1.23 | 0.745 | 9.15 | 0.027 |
Diversity indices were compared across groups using Kruskal-Wallis tests with Dunn's post-hoc tests where appropriate.
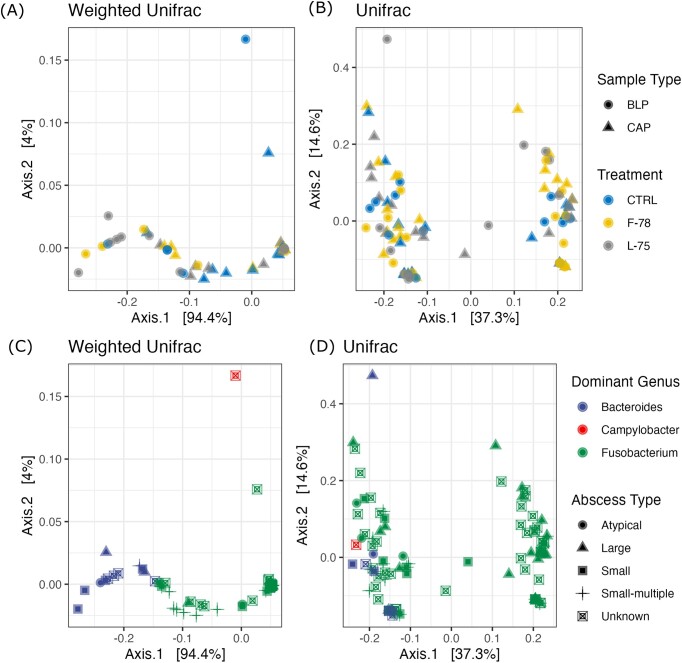
Beta-diversity was evaluated using PCoA plots (Fig. 2A–D) accompanied by PERMANOVA and beta-dispersion tests (Table 2), using both weighted and unweighted Unifrac distances. The timing of tylosin phosphate inclusion did not impact the composition of the LA microbiota using either test (Fig. 2A and B; P > 0.05). Tests using weighted Unifrac (Fig. 2A and C) indicated that CAP and BLP microbiotas differed, as well as those of Fusobacterium- and Bacteroides-dominated abscesses (P < 0.05). In contrast, classifying samples based on whether they originated from a liver with a single abscess, multiple abscesses or a very large abscess (abscess type) showed a trend toward differences in microbiome composition (Fig. 2D; P = 0.06, R2 = 0.06). Tests performed using the unweighted Unifrac distance matrix indicated significant differences in community composition according to both dominant genus and abscess type (Fig. 2B and D; P < 0.05). There were also statistically significant differences in beta dispersion according to dominant genus when tested using unweighted Unifrac distances (P < 0.05), and between sample types (CAP vs BLP) when tested using weighted Unifrac (P < 0.05).
Figure 2.
Principal Coordinate Analysis (PCoA) plots based on weighted (A, C) and unweighted (B, D) Unifrac distance matrices. Samples are categorized according to tylosin phosphate treatment group, sample type, abscess size, and their predominant genus. Treatment groups: CTRL=control, animals who received tylosin phosphate throughout the finishing period, F-78 = animals who received tylosin phosphate in the first 78% of the trial, L-75 = animals who received tylosin phosphate in the last 75% of the trial. CAP = capsule-adhered purulent material; BLP = bulk purulent material.
Table 2.
Beta-diversity results
| Unweighted Unifrac | Weighted Unifrac | |||||
|---|---|---|---|---|---|---|
| R21 | Perm. P-value2 | β-disp. P-value3 | R2 | Perm. P-value | β-disp. P-value | |
| Treatment | 0.02 | 0.75 | 0.66 | 0.01 | 0.56 | 0.453 |
| Sample type | 0.01 | 0.71 | 0.178 | 0.07 | <0.001 | 0.026 |
| Dominant genus | 0.06 | 0.01 | 0.016 | 0.55 | <0.001 | 0.537 |
| Abscess type | 0.12 | 0.01 | 0.514 | 0.06 | 0.06 | 0.136 |
| Gender | 0.01 | 0.54 | 0.414 | 0.01 | 0.21 | 0.034 |
Variation explainable by each factor.
PERMANOVA P-value.
β-dispersion test P-value.
Taxonomic composition of LA
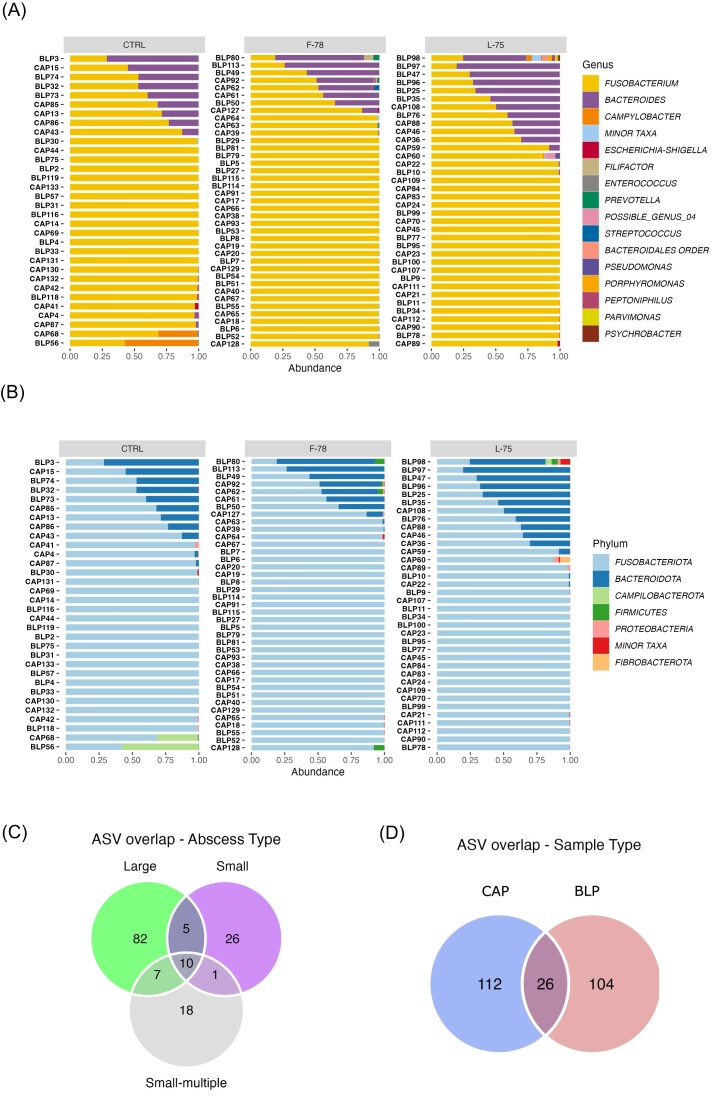
Fusobacteriota (85.93%) and Bacteroidota (12.56%) were the most prominent taxa detected in LA by mean relative abundance across all samples (Fig. 3A). Other phyla present in lower abundances included Campilobacterota (0.88%), Firmicutes (0.29%), Proteobacteria (0.24%), and Actinobacteria (0.05%). The mean abundances of all the remaining phyla were below 0.01%. There was considerable intersample variation in phylum abundance and prevalence as shown in Fig. 3A. All but 2 samples contained <10 genera. The 9 genera detected in >5% of the samples together accounted for 99.56% of all the sequences at the genus level. Fusobacterium (85.93%) was the most abundant overall and was the only ubiquitous genus present (Fig. 3B). Bacteroides (12.45%) was present in 59 of the 107 samples, with all remaining genera present in <50% of the samples. Campylobacter (0.87%) and Escherichia-Shigella (0.13%) were the other prominent genera, with the remainder all present at < 0.1% relative abundance (Fig. 3B).
Figure 3.
Bar charts of (A) phylum and (B) genus-level relative abundances across samples. Charts are organized according to tylosin treatment group. Sample type is denoted on the y-axis. For clarity, the 15 most abundant genera across all samples are presented, with the remainder grouped together as “Minor Taxa”. The corresponding phyla are denoted in the same way. CTRL=control, animals who received tylosin phosphate throughout the finishing period, F-78 = animals who received tylosin phosphate in the first 78% of the trial, L-75 = animals who received tylosin phosphate in the last 75% of the trial. The Venn diagrams display the overlap of unique ASVs between (C) abscess types = small; small-multiple; large, and (D) sample types = CAP = capsule-adhered purulent material; BLP = bulk purulent material.
To further evaluate relationship between abscess microbiomes, we identified the ASVs which were shared between abscesses categorized according to size (Fig. 3C) and between BLP and CAP samples (Fig. 3D). Large abscesses had more unique ASVs (82) than small (26) or small-multiple (18) abscesses with only 10 ASVs ubiquitous across all abscess types (Fig. 3C). These 10 ubiquitous ASVs accounted for >90% of the reads in the majority of the samples. There were 26 ASVs present in both BLP and CAP samples, with 112 unique to the CAP samples, and 104 to the BLP samples (Fig. 3D).
Impact of tylosin administration group on microbial composition
The impact of tylosin feeding regimen was evaluated at phylum and genus levels using DeSeq2. The timing of tylosin feeding did not have any effect on the abundance of Fusobacteriota (P > 0.05). Firmicutes were 3.16 times more abundant (log2 fold-change; log2FC) in F-78 samples than in the controls (P < 0.05), while the abundance of Bacteroidota was 2.7 times greater in the L-75 samples than in the controls (P < 0.05). No phyla exhibited significant differences in abundance between the L-75 and F-78 treatment groups (P < 0.05). When examined at genus level, F-78 samples tended to have greater abundances of Filifactor (1.67 log2 FC) and Acinetobacter (1.35 log2 FC) than the controls (P < 0.1). There were no significant differences in the abundances of any genera between the L-75 and control samples (P > 0.05). Acinetobacter was also more abundant in the F-78 samples than in the L-75 cohort (P < 0.05; log2FC = 1.65), while the higher abundance of Filifactor in the same group trended toward significance (P = 0.06; log2FC = 1.47). The lower abundance of Bacteroides in the F-78 samples compared to the L-75 samples showed a similar trend (P = 0.06; log2 FC = 2.49). Given the large variation in prevalence of genera across samples, we exercise some caution in interpreting these DA findings.
Differences between capsule-associated and purulent material microbiomes
The abundance of Fusobacteriota didn't differ between sample types (P > 0.05). The abundance of Proteobacteria was greater (2.13 log2 FC) in the CAP samples than in the BLP samples (P < 0.05). The CAP samples had higher proportions of both Acinetobacter (1.48 log2 FC) and Escherichia-Shigella (1.78 log2 FC) versus BLP (P < 0.05), while the higher abundances of Enterococcus (0.90 log2 FC) and Mycoplasma (0.89 log2 FC) showed a tendency to be significant (P < 0.1), but with low fold-change values.
Differences according to liver abscess morphology
Abscess type was the only factor that distinguished the microbial communities using both weighted and unweighted Unifrac metrics (Table 2; Fig. 2C and D). Samples collected from livers with one small abscess and multiple small abscesses had greater abundances of Bacteroidota than those from large abscesses (P < 0.05). Additionally, samples collected from livers with multiple small abscesses had higher abundances of Proteobacteria and Firmicutes compared to large abscesses (P < 0.05). The abundance of Fusobacteriota was not impacted by liver abscess morphology (P > 0.05). Small abscesses had more sequences assigned to Firmicutes than livers with multiple small abscesses (3.66 log2 fold change; P<0.05). At the genus level, large abscesses had lower abundances of Filifactor (−5.83 log2 FC) and Bacteroides (−4.62 log2 FC) compared to samples collected from livers with one small abscess (P < 0.05). The abundance of Bacteroides was also significantly lower when microbial profiles of large abscesses were compared to those of livers with multiple small abscesses (−4.44 log2 FC; P < 0.05). The abundance of Filifactor was significantly higher in samples generated from livers with one small abscess compared to those with multiple small abscesses (5.79 log2 FC; P < 0.05), but this taxa was only present in three samples overall.
Classification of LA samples according to genus predominance
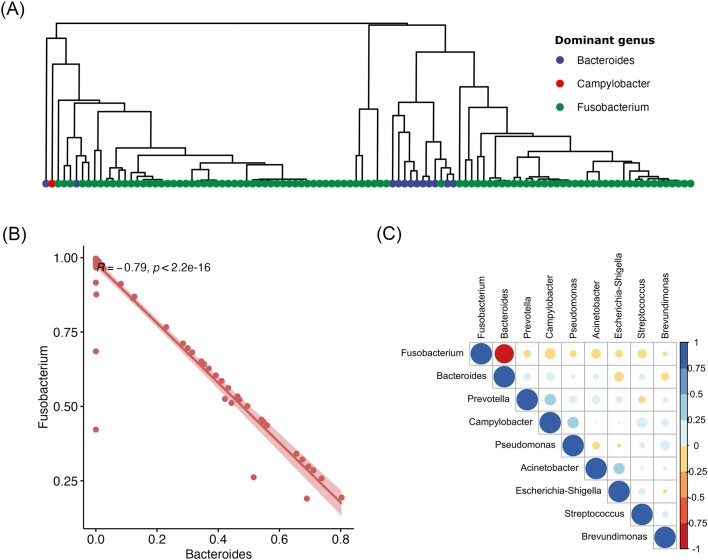
When samples were classified according to their most abundant genus (Fig. 4), Fusobacterium predominated in 94 of the 107 samples, followed by Bacteroides (12) and Campylobacter (1). Hierarchical clustering analysis using Euclidean distances showed clear clustering according to dominant genus classification (Fig. 4A), in agreement with the PCoA and PERMANOVA results. Spearman correlations showed a strong (R = −0.79) negative relationship between the relative abundances of Fusobacterium and Bacteroides (Fig. 4B). No other pairs of taxa exhibited strong correlations in either direction (Fig. 4C).
Figure 4.
(A) Hierarchical clustering dendrogram based on average-linkage based with Euclidean distances. Samples are highlighted according to dominant taxa. (B) Spearman correlation between Fusobacteria and Bacteroides relative abundances, with the R-value for the correlation provided as text alongside the P-value and (C) Spearman correlations between all genera present in more than 5% of the samples. R-values for each correlation are represented as a color gradient from −1 to 1.
Evaluation of Fusobacterium and Trueperella abundance profiles
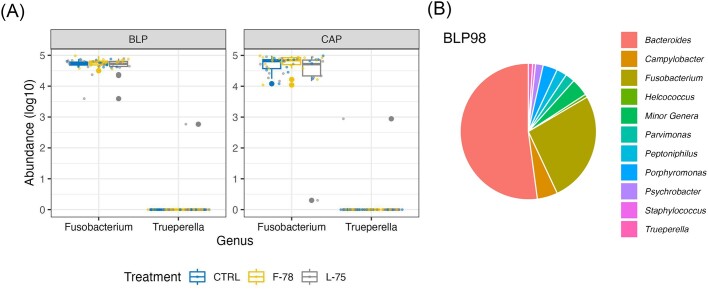
Of the 35 ASVs assigned to the genus Fusobacterium, 33 were classified as F. necrophorum, while the remainder had no species-level annotation. Fusobacterium ASVs were present in all samples (Fig. 5A), while Trueperella was detected in only one sample of BLP (BLP98; Fig. 5A), collected from a steer and contained within a large abscess. This sample also harbored a considerably richer and more diverse microbiota than any other sample, as evidenced by its rarefaction curve (Supplementary Fig. S1A) and taxonomic profile (Fig. 5B). It's Chao1 value was 55.5, while the average for all the other samples was 4.51 (Supplementary Material S2). A total of 45 genera were present in this sample, while the average number of genera detected in each of the other samples was 3.09. This was the most diverse sample in our dataset, and was dominated by Bacteroides (51.28% of sequences), followed by Fusobacterium (26.17%), Campylobacter (4.80%), Prophyromonas (3.38%), Peptoniphilus (2.41%), Parvinmonas (2.24%), Psychrobacter (1.78%), and Trueperella (1.00%) (Fig. 5B). An additional 16 genera had relative abundances of >0.1% in this sample.
Figure 5.
Boxplot showing the abundance of ASVs assigned to (A) Fusobacterium and Trueperella genera across treatment groups. Data were log-transformed for ease of presentation. (B) Pie-chart showing the relative abundance of genera detected in sample CAP-98. CTRL=control, animals who received tylosin phosphate throughout the finishing period, F-78 = animals who received tylosin phosphate in the first 78% of the trial, L-78 = animals who received tylosin phosphate in the last 75% of the trial. CAP 98 = Capsule-adhered purulent material sample number 98.
Discussion
Liver abscesses pose significant economic and welfare challenges in beef production systems, and a comprehensive understanding of the microbial basis of LA pathogenesis is key in the development of novel mitigation strategies. This study employed 16S rRNA sequencing to characterize the bulk-purulent material (BLP) and capsule-adhered (CAP) microbiota of LA. Our findings provide further information concerning the polymicrobial community inhabiting LA, and support previous studies that have documented the predominance of Fusobacteria and Bacteroides in LA (Stotz et al. 2021, Pinnell et al. 2022). We show that the microbial communities associated with BLP differ from that associated with the inner CAP of the abscess. LA morphology also appears to influence the composition of the liver abscess microbiome with changes in composition and diversity during abscess development.
Liver abscesses contain a low diversity microbiota with high individual variation, and are dominated by a small number of bacteria.
All but one of the samples in our dataset contained a low-diversity microbiota, with the majority containing <10 unique ASVs, and only one containing >20. This contradicts other studies; Weinroth et al. (2017) identified a far greater number of OTUs in their LA samples than we did, exceeding 10 000 in one sample. Rarefaction curves for all samples analyzed in this work reached a plateau at ∼25 000 reads, indicating that further sequencing would not have yielded additional ASVs in our dataset. Similarly, a more recent study of LA microbial ecology identified 4167 OTUs and 388 genera in samples from both abscessed and healthy livers (Stotz et al. 2021). The reason for these differences is hard to ascertain and may lie in the choice of sample type (i.e. combined sample type vs separate capsule tissue and purulent material), laboratory procedures (e.g. DNA extraction methods, sample processing), data analysis tools, different sampling depths, or a combination of the above. As more studies employ high-throughput sequencing to characterize LA microbiomes under different conditions, our understanding of the factors influencing these communities will improve.
Recent high-throughput sequencing studies have challenged the dogmatic view of F. necrophorum as the primary infectious agent in LA, with evidence emerging of a polymicrobial LA microbiome, which may originate from other regions of the gastrointestinal tract (GIT) (Weinroth et al. 2019, Fuerniss et al. 2022, Pinnell et al. 2022). The literature is inconsistent regarding the taxonomic composition of LA microbiomes using NGS. Our dataset was dominated by Fusobacteriota, with Bacteroidota a distant second, in agreement with recent studies that reported these as the predominant phyla in LA (Amachawadi et al. 2021, Stotz et al. 2021). Although Bacteroidota was the second most abundant phylum here, it was totally absent in almost 45% of the samples (Fig. 3A). This contrasts with another metataxonomic study of LA, which found that Bacteroidetes (since renamed Bacteroidota), and Proteobacteria were the dominant phyla, followed by Fusobacteria (Weinroth et al. 2017). Proteobacteria comprised just 0.24% of the reads on average in our study, and were found in ∼61% of the samples. Stotz et al. (2021) reported that Proteobacteria was the most abundant phyla present in healthy liver tissue, and the high abundance of Proteobacteria reported by Weinroth et al. (2017) compared to our and other studies may reflect amplification of microbes associated with healthy liver tissue alongside those of the abscess. This variation may also reflect systemic differences in experimental approaches used by each group.
Similar patterns were evident at genus level, where large intersample variation was also clear. Sequence count sparsity (i.e. the presence of a large number of distinct taxa, which are found only in a small number of samples, leading to a high amount of zeros in the taxonomic count matrix) is an inherent issue facing microbiome NGS studies (Pan 2021), and low prevalence of many taxa has been reported in other studies of LA (Pinnell and Morley 2022). While 82 genera were detected overall, just 9 were found in >5% of the samples and only 2 samples contained more than 10 unique genera. Additionally, the abundance ranges of the taxa across samples were large, particularly for the minor taxa. Recent studies have reported two distinct LA community subtypes in severely abscessed livers, a Fusobacteria-dominated subtype and a Bacteroidetes (Bacteroidota)-dominated subtype (Stotz et al. 2021, Pinnell et al. 2022). We ranked our data according to dominant genus, with Fusobacterium and Bacteroides being predominant in all but one of the samples. This partially contradicts Pinnell et al. (2022), who reported Porphyromonas as being the predominant genus in several of the Bacteroidetes-type abscesses, our data generally supports recent findings that many LA are dominated by bacteria other than Fusobacterium. PERMANOVA tests and accompanying R2 values showed that ranking according to dominant genus was substantially more discriminating of LA microbial communities than any other factor or combination of factors in our study. The proportions of Fusobacterium and Bacteroides were strongly negatively correlated, further underlining the strong inverse relationship between Fusobacterium and Bacteroides abundances. Interestingly, one sample was dominated by Campylobacter spp., a well-known human food pathogen known to reside in the ruminant gut (Stanley and Jones 2003, Schnur 2021). Campylobacter species have been implicated in liver abscess formation in humans, and have been reported as part of the LA microbiome in cattle (Pinnell et al. 2022). To our knowledge, this is the first example of a Campylobacter-dominated LA in cattle and suggests that bacteria other than Fusobacterium and Bacteroides have the potential to act as major infectious agents in bovine LA syndrome.
Escherichia-Shigella, Prevotella, Pseudomonas, Acinetobacter, and Brevundimonas were all present in a significant number of our samples. Escherichia-Shigella contains well-known pathogens like Escherichia coli, and was found in ∼38% of the samples and was more abundant in LA of tylosin-fed cattle than controls in a previous study (Amachawadi et al. 2021), suggesting that antimicrobial action of tylosin may make animals more susceptible to translocation of gut E. coli. Prevotella contains multiple important gut species, and while they have been previously identified in bovine LA, their role in pathogenesis is unknown (Pinnell et al. 2022). Many of these other prominent genera contain recognised opportunistic species. Pseudomonas are Proteobacteria which have been identified in LA using NGS (Amachawadi et al. 2021, Pinnell et al. 2022) and contain known pathogens (Juhas 2015). Acinetobacter are also Proteobacteria, and have been implicated in wound, bloodstream (Wareth et al. 2019) and liver infections in humans (Singh et al. 2016). This genus’ presence has also been reported in other studies of cattle LA microbiomes (Stotz et al. 2021, Pinnell et al. 2022) at varying proportions. Brevundimonas has not been previously noted as a major member of bovine LA microbiomes in cattle, but its capability as an opportunistic pathogen has been described (Ryan and Pembroke 2018). The high variation in prevalence and abundance of the microbes across samples supports the hypothesis that members of one or two genera act as primary infectors in LA syndrome, with the remaining constituents of LA microbiomes composed of opportunistic pathogens.
We detected additional taxa at high abundance in a very small number of samples, which is reflective of the high interindividual variation in the composition of LA microbiomes. For example, Enterococcus has been reported to play a role in pyogenic liver abscess syndrome in humans (Mücke et al. 2017), and was as abundant as 8.0% among 5 of the samples analyzed in the present study. Tylosin feeding increases AMR among fecal enterococci in cattle (Zaheer et al. 2013) and its high abundance in several samples here suggests that it may play a role in bovine LA pathogenesis. A companion study examining the impact of tylosin inclusion on the level of AMR genes in fecal enterococci collected during the same animal experiment found that their abundance linearly increased during the feeding period (Davedow et al. 2020). It is possible that enterococci may have translocated from either the rumen or hind-gut into the blood stream and established in the liver of these animals. Little is known about the role of the hind-gut microbiome in the development of LA and this could be an area for future study. Filifactor has also been previously linked to LA in cattle (Pinnell et al. 2022) and was found in two samples in our study. Filifactor belongs to the Firmicutes phylum and has been reported as being discriminant of Bacteroides-dominated LA (Pinnell et al. 2022), although another study reported its correlation with the abundance of Fusobacterium (Amachawadi and Nagaraja 2022).
The high intersample variation in the microbial communities that we observed is also exemplified in one sample that had a substantially richer microbial profile compared to the others (Fig. 1A). While dominated by Bacteroides and Fusobacterium, this BLP sample also many sequences assigned to other taxa. Among them, Parvimonas, Helcococcus, Prophyromonas, and Trueperella have all been previously reported in LA microbiomes (Weinroth et al. 2017, Amachawadi and Nagaraja 2022). The fact that they were only prominent (>1% abundance) in a single sample here again underlines the high degree of variability in LA microbiota even within the same animal cohort. We were surprised to find that this was the only sample that contained sequences assigned to Trueperella, a genus widely implicated in bovine LA etiology (Herrick et al. 2022, Pinnell and Morley 2022). This contrasts with recent NGS studies which have reported high abundances of Trueperella in LA (Stotz et al. 2021, Pinnell et al. 2022) but is in agreement with the results of Amachawadi et al. (2021) who also found this microbe to be present in a small number of samples. Another study reported that Trueperella was primarily isolated from samples collected from regions with temperate climates, exemplified by cooler climates and higher rainfall (Herrick et al. 2022). Our data does not reveal with certainty the source of these discrepancies, and the collection of samples from multiple environments will be necessary to generate a comprehensive picture of LA microbial ecology.
Sample and abscess type have more impact on LA microbiomes than tylosin feeding duration
Agreeing with previous studies (Amachawadi et al. 2021, Fuerniss et al. 2022), in-feed tylosin supplementation had only a marginal effect on the LA microbiome with no impact on global diversity metrics. Acinetobacter was more abundant in F-78 samples than in either L-75 and controls, even though controls received tylosin throughout the feeding period. Like most of the taxa present in Fusobacterium- and Bacteroides-dominated LA, Acinetobacter is a gram-negative opportunistic pathogen and is implicated in wound formation in the mammalian gut (Eliopoulos et al. 2008). Earlier research identified this bacteria as more prevalent in the LA of tylosin-fed cattle (Amachawadi et al. 2021). However, due to its low prevalence in our dataset, we cannot definitively infer its role in LA formation.
The lack of a consistent approach to sample collection (i.e. only purulent material vs whole abscess area) may have contributed to the variation in results from NGS-based LA microbiome studies to date. Here, we generated libraries from both inner wall capsule tissue (CAP) and BLP. Weighted Unifrac dissimilarities were different between sample types while unweighted Unifrac were not, indicating that even with a large number of ASVs unique to each sample type, it was differences in abundance rather than the presence/absence of taxa which drove the separation. Additionally, CAP samples were more diverse (as indicated by Chao1) and had greater abundances of Acinetobacter and Escherichia-Shigella, with the greater abundance of Enterococcus approaching significance. These CAP associated genera may be more metabolically active than those associated with the BLP (Amachawadi et al. 2021). While the differences between CAP and BLP microbiomes were relatively small overall, these findings point to potential variation in LA microbiomes between abscess regions, which should be considered in future studies.
We were surprised to find differences between abscess types (small vs multiple small vs large), which has not been previously documented to our knowledge. The separation was greater for the unweighted Unifrac distance (R2 = 0.12 vs 0.06 for weighted), indicating that this difference was likely driven by rare taxa (Lozupone and Knight 2005). The microbiomes of large abscesses were more diverse (Shannon index) and had considerably more ASVs than either of the other groups though this was likely driven by the single sample of high diversity. Additionally, they had significantly lower abundance of Bacteroides, indicating that major LA taxa vary between large and small abscesses. A microbiome inhabiting healthy liver tissue has also been reported recently, and many of the microbes found in abscessed tissue are also found in healthy liver tissue (Stotz et al. 2021). This intriguing finding suggests that opportunistic pathogens may be present in the liver tissues of cattle consuming high-grain diets, even in the absence of LA. The biological mechanisms that may favor these pathogens are unknown, and investigation of these potential stressors is warranted.
Conclusion
These data indicate that the microbiota of LA is sparsely populated by a wide range of mostly Gram-negative bacteria, and reflects the results of previous studies that showed LA are dominated by either Fusobacteria or Bacteroides. We acknowledge that in the absence of non-template control samples, we cannot totally exclude the possibility of external contamination. However, we took every practical precaution possible to avoid this, and have confidence in our findings. Very few taxa were highly prevalent across samples, and the large inter-sample variation in composition suggests that one or two bacterial taxa act as major infectors (primarily Bacteroides and Fusobacteria), while the remainder likely derive from the gastrointestinal tract and include opportunistic pathogens. Timing of tylosin administration did not have a significant global effect on the LA microbiome, but some shifts in individual taxon abundances were found. Additionally, differences in composition between the microbiomes of BLP and inner wall tissue, as well as between abscesses of divergent morphology, indicate that these are confounding factors that should be considered in future studies of LA microbiomes.
Supplementary Material
Acknowledements
Sequencing was conducted at the Centre d'expertise et de services Génome Québec (Montreal, QC). The authors acknowledge the staff from Feedlot Health Management Services for their assistance in collecting liver samples.
Contributor Information
Eóin O'Hara, Lethbridge Research and Development Centre, Agriculture and Agri-Food Canada, 5403 1st Ave S, Lethbridge, AB, T1J 4B1, Canada.
Rahat Zaheer, Lethbridge Research and Development Centre, Agriculture and Agri-Food Canada, 5403 1st Ave S, Lethbridge, AB, T1J 4B1, Canada.
Sara Andrés-Lasheras, Lethbridge Research and Development Centre, Agriculture and Agri-Food Canada, 5403 1st Ave S, Lethbridge, AB, T1J 4B1, Canada.
Tim A McAllister, Lethbridge Research and Development Centre, Agriculture and Agri-Food Canada, 5403 1st Ave S, Lethbridge, AB, T1J 4B1, Canada.
Robert J Gruninger, Lethbridge Research and Development Centre, Agriculture and Agri-Food Canada, 5403 1st Ave S, Lethbridge, AB, T1J 4B1, Canada.
Author contributions
Eóin O'Hara (Formal analysis, Visualization, Writing – original draft, Writing – review & editing), Rahat Zaheer (Conceptualization, Methodology, Writing – review & editing), Sara Andrés-Lasheras (Conceptualization, Methodology, Writing – review & editing), Tim A. McAllister (Conceptualization, Project administration, Resources, Writing – review & editing), and Robert J. Gruninger (Conceptualization, Methodology, Project administration, Resources, Writing – original draft, Writing – review & editing)
Conflicts of interest
The authors declare that this research was carried out in the absence of any conflicts of interest.
Funding
This work was supported by Agriculture and Agri-Food Canada, Genome Alberta (grant ID: MGLA to TAM) and Beef Cattle Research Council (grant ID: ANH.02.19 to RJG). The funders played no role in the design of the experiment or interpretation of the results.
Data Availability
The raw sequencing data have been deposited to SRA under BioProject number PRJNA98398.
References
- Amachawadi RG, Nagaraja TG. Liver abscesses in cattle: a review of incidence in Holsteins and of bacteriology and vaccine approaches to control in feedlot cattle. J Anim Sci. 2016;94:1620–32. 10.2527/jas.2015-0261. [DOI] [PubMed] [Google Scholar]
- Amachawadi RG, Nagaraja TG. Pathogenesis of liver abscesses in cattle. Vet Clin North Am Food Anim Pract. 2022;38:335–46. 10.1016/j.cvfa.2022.08.001. [DOI] [PubMed] [Google Scholar]
- Amachawadi RG, Tom WA, Hays MP et al. Bacterial community analysis of purulent material from liver abscesses of crossbred cattle and Holstein steers fed finishing diets with or without tylosin. J Anim Sci. 2021;99:skab076. 10.1093/jas/skab076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrews S. FastQC: a Quality Control Tool for High Throughput Sequence Data. 2010. http://www.bioinformatics.babraham.ac.uk/projects/fastqc/.
- Barnett DJm, Arts ICw, Penders J. microViz: an R package for microbiome data visualization and statistics. J Open Source Software. 2021;6:3201. 10.21105/joss.03201. [DOI] [Google Scholar]
- Bolyen E, Rideout JR, Dillon MR et al. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat Biotechnol. 2019;37:852–7. 10.1038/s41587-019-0209-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brink DR, Lowry SR, Stock RA et al. Severity of liver abscesses and efficiency of feed utilization of feedlot cattle. J Anim Sci. 1990;68:1201–7. 10.2527/1990.6851201x. [DOI] [PubMed] [Google Scholar]
- Brown TR, Lawrence TE. Association of liver abnormalities with carcass grading performance and value. J Anim Sci. 2010;88:4037–43. 10.2527/jas.2010-3219. [DOI] [PubMed] [Google Scholar]
- Bushnell B. BBMap: a Fast, Accurate, Splice-Aware Aligner (LBNL-7065E). Berkeley, CA (United States): Lawrence Berkeley National Lab. (LBNL), 2014. https://www.osti.gov/biblio/1241166-bbmap-fast-accurate-splice-aware-aligner. [Google Scholar]
- Callahan BJ, McMurdie PJ, Rosen MJ et al. DADA2: high-resolution sample inference from Illumina amplicon data. Nat Methods. 2016;13:581–3. 10.1038/nmeth.3869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caporaso JG, Lauber CL, Walters WA et al. Global patterns of 16S rRNA diversity at a depth of millions of sequences per sample. Proc Natl Acad Sci USA. 2011;108 Suppl 1:4516–22. 10.1073/pnas.1000080107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CFIA . Canadian Food Inspection Agency Index of Medicating Ingredients Approved by Livestock Species. 2023. Available online at: http://inspection.gc.ca/animals/feeds/medicating-ingredients/mib/livestock-species/eng/1522783196554/1522783196850#a4 (1 December 2023, date last accessed).
- Davedow T, Narvaez-Bravo C, Zaheer R et al. Investigation of a reduction in tylosin on the prevalence of liver abscesses and antimicrobial resistance in enterococci in feedlot cattle. Front Vet Sci. 2020;7. https://www.frontiersin.org/articles/10.3389/fvets.2020.00090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eliopoulos GM, Maragakis LL, Perl TM. Acinetobacter baumannii: epidemiology, antimicrobial resistance, and treatment options. Clin Infect Dis. 2008;46:1254–63. 10.1086/529198. [DOI] [PubMed] [Google Scholar]
- Fuerniss LK, Davis HE, Belk AD et al. Liver abscess microbiota of feedlot steers finished in natural and traditional management programs. J Anim Sci. 2022;100:skac252. 10.1093/jas/skac252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Health Canada . Categorization of Antimicrobial Drugs Based on Importance in Human Medicine. 2009. Available online at https://www.canada.ca/en/health-canada/services/drugs-health-products/veterinary-drugs/antimicrobial-resistance/categorization-antimicrobial-drugs-based-importance-human-medicine.html (1 December 2023, date last accessed).
- Herrick RT, Rogers CL, McEvers TJ et al. Exploratory observational quantification of liver abscess incidence, specific to region and cattle type, and their associations to viscera value and bacterial flora. Applied Animal Science. 2022;38:170–82. 10.15232/aas.2021-02228. [DOI] [Google Scholar]
- Juhas M. Pseudomonas aeruginosa essentials: an update on investigation of essential genes. Microbiology. 2015;161:2053–60. 10.1099/mic.0.000161. [DOI] [PubMed] [Google Scholar]
- Katoh K, Standley DM. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 2013;30:772–80. 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:Article 12. 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lozupone C, Knight R. UniFrac: a new phylogenetic method for comparing microbial communities. Appl Environ Microbiol. 2005;71:8228–35. 10.1128/AEM.71.12.8228-8235.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lozupone C, Lladser ME, Knights D et al. UniFrac: an effective distance metric for microbial community comparison. ISME J. 2011;5:169–72. 10.1038/ismej.2010.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMurdie PJ, Holmes S. phyloseq: an R package for reproducible interactive analysis and graphics of microbiome census data. PLoS One. 2013;8:e61217. 10.1371/journal.pone.0061217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mücke MM, Kessel J, Mücke VT et al. The role of Enterococcus spp. And multidrug-resistant bacteria causing pyogenic liver abscesses. BMC Infect Dis. 2017;17:450. 10.1186/s12879-017-2543-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagaraja TG, Lechtenberg KF. Liver abscesses in feedlot cattle. The Veterinary Clinics of North America Food Animal Practice. 2007;23:351–69., ix. 10.1016/j.cvfa.2007.05.002. [DOI] [PubMed] [Google Scholar]
- National Beef Quality Audit . National Beef Quality Audit. 2018. https://www.beefresearch.ca/files/pdf/NBQA-Carcass-Audit-Mar-27-2018-F.pdf [Google Scholar]
- Oksanen J, Blancet G, Friendly M et al. vegan-.ackage: community Ecology Package: ordination, Diversity and… in Vegan: community Ecology Package (2.5-7). 2020. https://rdrr.io/cran/vegan/man/vegan-package.html.
- Pan AY. Statistical analysis of microbiome data: the challenge of sparsity. Curr Opin Endocr Metab Res. 2021;19:35–40. 10.1016/j.coemr.2021.05.005. [DOI] [Google Scholar]
- Pinnell LJ, Morley PS. The microbial ecology of liver abscesses in cattle. Vet Clin North Am Food Anim Pract. 2022;38:367–81. 10.1016/j.cvfa.2022.08.004. [DOI] [PubMed] [Google Scholar]
- Pinnell LJ, Whitlow CW, Huebner KL et al. Not all liver abscesses are created equal: the impact of tylosin and antibiotic alternatives on bovine liver abscess microbial communities and a first look at bacteroidetes-dominated communities. Front Microbiol. 2022;13:882419. 10.3389/fmicb.2022.882419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quast C, Pruesse E, Yilmaz P et al. The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res. 2013;41:D590–6. 10.1093/nar/gks1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinhardt CD, Hubbert ME. Control of liver abscesses in feedlot cattle: a review. Appl Anim Sci. 2015;31:101–8. 10.15232/pas.2014-01364. [DOI] [Google Scholar]
- Rezac DJ, Thomson DU, Bartle SJ et al. Prevalence, severity, and relationships of lung lesions, liver abnormalities, and rumen health scores measured at slaughter in beef cattle. J Anim Sci. 2014;92:2595–602. 10.2527/jas.2013-7222. [DOI] [PubMed] [Google Scholar]
- Ryan MP, Pembroke JT. Brevundimonas spp: emerging global opportunistic pathogens. Virulence. 2018;9:480–93. 10.1080/21505594.2017.1419116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnur SE. Antimicrobial Alternatives to Control Pathogens Involved in Liver Abscesses, Food Safety and Mastitis in Cattle. 2021. https://krex.k-state.edu/handle/2097/41586.
- Shryock TR, Mortensen JE, Baumholtz M. The effects of macrolides on the expression of bacterial virulence mechanisms. J Antimicrob Chemother. 1998;41:505–12. 10.1093/jac/41.5.505. [DOI] [PubMed] [Google Scholar]
- Singh NP, Sagar T, Nirmal K et al. Pyogenic liver abscess caused by Acinetobacter lwoffii: a case report. J Clin Diagn Res. 2016;10:DD01–2. 10.7860/JCDR/2016/18256.7943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanley K, Jones K. Cattle and sheep farms as reservoirs of Campylobacter. J Appl Microbiol. 2003;94(s1):104–13. [DOI] [PubMed] [Google Scholar]
- Stotz MK, Henry DD, Crossland WL. Characterization of bacterial DNA identified in abscessed and non-abscessed bovine hepatic tissue at the time of harvest. J Anim Sci. 2021;99:skab280. 10.1093/jas/skab280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wareth G, Neubauer H, Sprague LD. Acinetobacter baumannii—a neglected pathogen in veterinary and environmental health in Germany. Vet Res Commun. 2019;43:1–6. 10.1007/s11259-018-9742-0. [DOI] [PubMed] [Google Scholar]
- Weinroth MD, Carlson CR, Martin JN et al. Rapid communication: 16S ribosomal ribonucleic acid characterization of liver abscesses in feedlot cattle from three states in the United States1. J Anim Sci. 2017;95:4520–5. 10.2527/jas2017.1743. [DOI] [PubMed] [Google Scholar]
- Weinroth MD, Martin JN, Doster E et al. Investigation of tylosin in feed of feedlot cattle and effects on liver abscess prevalence, and fecal and soil microbiomes and resistomes1. J Anim Sci. 2019;97:4567–78. 10.1093/jas/skz306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickham H. ggplot2: elegant Graphics for Data Analysis. New York, NY: Springer-Verlag, 2009. 10.1007/978-0-387-98141-3. [DOI] [Google Scholar]
- World Health Organization . Critically important antimicrobials for human medicine, 6th revision. Geneva, Switzerland; Licence: CC BY-NC-SA 3.0 IGO. 2019. Available at: https://apps.who.int/iris/bitstream/handle/10665/312266/9789241515528-eng.pdf. [Google Scholar]
- Xu S, Zhan L, Tang W et al. MicrobiotaProcess: a comprehensive R package for deep mining microbiome. Innovation. 2023;4:100388. 10.1016/j.xinn.2023.100388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaheer R, Cook S, Klima C et al. Effect of subtherapeutic vs. Therapeutic administration of macrolides on antimicrobial resistance in Mannheimia haemolytica and enterococci isolated from beef cattle. Front Microbiol. 2013;4. https://www.frontiersin.org/articles/10.3389/fmicb.2013.00133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaheer R, Noyes N, Ortega Polo R et al. Impact of sequencing depth on the characterization of the microbiome and resistome. Sci Rep. 2018;8:5890. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The raw sequencing data have been deposited to SRA under BioProject number PRJNA98398.