Abstract
To define the molecular features characteristic of the early stages of infection of lymphocytes with human herpesvirus 6 (HHV-6) variant A or B, we studied the temporal regulation of expression of selected sets of viral genes. Thus, U42, U94, U89-U90, U73, and U39 are α genes since their transcripts (i) were made in the presence of inhibitors of protein synthesis and (ii) were detected 3 h after infection of untreated cells. U41, U53, U31, and U19 are β genes since their expression is inhibited by cycloheximide but not by phosphonoacetate, an inhibitor of DNA synthesis. U100 is a γ gene since its spliced transcript encoding the structural glycoprotein gp82/105 was first detected 16 h after infection of untreated cells but could not be detected in cells treated with phosphonoacetate. HHV-6 variants differ in the transcription patterns of their genes. U16-U17 originates a splice transcript and is regulated as α in HHV-6B and as β in HHV-6A. U91 generates two transcripts, amplified as 476- and 374-bp PCR fragments. The 476-bp fragment is α in HHV-6A-infected cells but β in HHV-6B-infected cells. Conversely, the 374-bp fragment is β in HHV-6A-infected cells and α in HHV-6B-infected cells. Furthermore, the spliced product of U18-U19-U20 (526 bp) is β in HHV-6A-infected cells, but only a partially spliced form (1.9 kb) was detected at late stages of infection in HHV-6B. HHV-6 transcription was also studied in nonproductive lymphoid cells, and the same transcription pattern detected during lytic infection was observed. Also, HHV-6 variants maintain the differences in U91, U16-17, and U18-U19-U20. We conclude that, as expected from the sequencing data, gene expression is generally similar in HHV-6 variants. However, transcription of selected genes in HHV-6A and HHV-6B differs with respect to temporal regulation and splicing pattern. Furthermore, the identification of viral functions expressed during the different stages of lytic replication suggests that reverse transcription-PCR for HHV-6 genes is a useful diagnostic approach to differentiate between latent and productive HHV-6 infection.
Human herpesvirus 6 (HHV-6) is widespread in the human population. Primary infection usually takes place in the first years of life, causing exanthem subitum (ES) (37). Severe or fatal involvement of HHV-6 in pneumonitis, hepatitis, and encephalitis has also been described, although this is less common (3, 4, 18, 28).
The virus establishes a latent infection (20), and spontaneous reactivation of HHV-6 is a rare event in the healthy adult population. In contrast, the virus is frequently isolated from patients with acquired or drug-induced immunodeficiency, such as human immunodeficiency virus-infected individuals, transplant recipients, and patients with autoimmune diseases. In these cases, viral reactivation has been associated with significant pathologic findings (8, 9, 12, 17, 26).
HHV-6 isolates form two groups, HHV-6A and HHV-6B. These groups are genetically related but show consistent differences in biological, immunological, and molecular properties (reviewed in reference 14). For example, (i) specific T-cell lines support the growth of HHV-6A but not of HHV-6B, and vice versa; (ii) monoclonal antibodies can recognize variant specific epitopes; and (iii) each variant displays a characteristic DNA restriction pattern. These differences are highly conserved, and until now no chimeric form between HHV-6A and HHV-6B has been described. HHV-6 variants also differ with respect to the pattern of disease with which they are associated. Thus, HHV-6B, but not HHV-6A, is frequently isolated from children with ES or febrile disease (29). It has been suggested that HHV-6 is more virulent than HHV-6B in that it has a more pronounced effect on suppression of bone marrow function (10). The molecular mechanisms responsible for the biological differences between HHV-6A and HHV-6B have yet to be identified.
Herpesvirus gene transcription and expression are tightly and coordinately regulated during the infectious cycle: immediate-early (IE) proteins, encoded by α-genes, activate β-genes, which in turn switch on the synthesis of late products encoded by γ-genes. β-Genes can be further differentiated into two groups (β1 and β2), according to their temporal appearance. γ-Genes also form two groups differentiated on the basis of their independence (γ1) or dependence (γ2) on viral DNA synthesis for their expression. The regulatory cascade has been extensively studied in cells infected with herpes simplex virus (HSV) and, more recently, in those infected with human cytomegalovirus (HCMV). Both HHV-6 and HCMV belong to the Betaherpesvirinae subfamily of the herpesvirus family as determined by colinearity of gene clusters and homology of their proteins (19, 21). It is reasonable to assume that HHV-6 gene transcription is coordinately expressed in a cascade fashion, but no experimental evidence is available and specific studies have yet to be reported.
Elucidation of the pattern of HHV-6 gene regulation would be expected to affect the current knowledge of HHV-6 molecular biology, provide the basis for identification of viral gene expression during latent infection and the various stages of productive infection, and provide useful markers to diagnose the state of viral infection in clinical infections with these viruses.
In this report, we describe the results of analyses of the temporal patterns of regulation of transcription of HHV-6 in untreated infected cells and in cells infected and maintained in the presence of cycloheximide (CEX) or emetine, inhibitors of protein synthesis, or phosphonoacetate (PAA), an inhibitor of viral DNA replication (13). In these studies, viral transcripts were detected by reverse transcription-PCR (RT-PCR). The objectives of the studies were to determine the kinetic class of selected viral genes with the aim of identifying the pattern of transcription of the HHV-6A and HHV-6B DNAs in productive and latent infection.
MATERIALS AND METHODS
Virus and cells.
Strain U1102 of HHV-6A (16) was grown and analyzed in the JJhan T-cell line. Strain CV of HHV-6B (15) was grown and analyzed in the SupT1 T-cell line. Both cell lines were grown in suspension at 37°C in RPMI 1640 supplemented with 10% fetal calf serum. Cell-free viral inocula were obtained by pelletting 1 liter of HHV-6-infected cell cultures exhibiting complete cytopathic effects (CPE). Infected cells, resuspended in 2 ml of 100% fetal calf serum supplemented with RNase (Boehringer; 50 μg/ml), were disrupted by four cycles of freezing in liquid nitrogen and thawing at 37°C. The resulting inoculum was completely free of living cells, as checked by microscopic observation and cultivation, and was also analyzed by RT-PCR (both for β-actin and for the panel of viral mRNAs) to ensure that RNA was completely absent. Infection was performed by adding the viral inoculum to 107 cells/ml. After 1 h of adsorption at 37°C, the cells were diluted with fresh medium to a final concentration of 5 × 105 cells/ml. For cultures which were treated with drugs, the cells were mixed with the appropriate concentrations of the drug 1 h before infection. Adsorption of virus to cells, dilution, and incubation took place in the continuous presence of the drug.
To identify α-genes, the cells were infected in the presence of CEX at 200 μg/ml. The experiments were repeated with another inhibitor of protein synthesis, emetine (Sigma), at 50 μg/ml. Aliquots of 5 × 106 cells were collected 3, 6, and 8 h after infection for RNA extraction. To identify β-genes, cells were infected in the presence of PAA (Sigma), an inhibitor of viral DNA replication, at 500 μg/ml. The cells were harvested 8, 16, 24, and 36 h after infection. Control cells were infected under similar conditions, but no drug was added to the medium. The cells were washed in phosphate-buffered saline and immediately frozen at −80°C until used for RNA extraction.
RNA purification and RT.
Total RNA was extracted from cells harvested at each time point and purified with Tripure isolation reagent (Boheringer); DNA contamination was eliminated by three cycles of digestion with 20 U of RNase-free DNase (Boheringer) at room temperature for 30 minutes in MgSO4-acetate buffer. RNA was purified by three phenol-chloroform extractions and recovered by ethanol precipitation. After a 75% ethanol rinse, the RNA pellet was resuspended in water treated with diethylpyrocarbonate and stored at −80°C with the addition of 40 U of RNase inhibitor (Amersham). The complete absence of DNA contaminants was confirmed by PCR, amplifying 200 ng of total RNA with human β-actin primers (38) and two different sets of primers designed to amplify the HHV-6 U31 and U94 genes (Table 1).
TABLE 1.
HHV-6 primers used in this study
| HHV-6 ORF | Primer sequence (3′-5′) | Amplimer size (bp)a
|
|
|---|---|---|---|
| cDNA | DNA | ||
| U16-U17 | AGAACTGCAAATCGTTCCG | 315b | 401 |
| CGTAGAACAGAAGACCGGC | |||
| U18-U19-U20 | TGATGAAGTGCCTATGGTGATT | 526b | 2,064 |
| TAACATCGCAAGGTTGATCAG | |||
| U19 | ACCGAACACTGTCGTATCGT | 685 | 685 |
| ATCGTCAACTGGTCAAGTGC | |||
| U31 | GATCCGACGCCTACAAACAC | 831 | 831 |
| CGGTGTTACACAGCATGAACTCTC | |||
| U39 | ACTGGACACAGTAGTCACCG | 478 | 478 |
| AGACATTGAGGTATTGGATGC | |||
| U41 | GTGAAAACTACGATTCAGGC | 264 | 264 |
| TTTCGGAACATTGTTGAGC | |||
| U42 | ACGATGGACATGGCTTGTTG | 525 | 525 |
| ACCTTACAACGGAGACGCC | |||
| U53 | CTGCGTTGCGCAGATACAG | 652 | 652 |
| CGTTGTAGCTGTCGCAGACTC | |||
| U73 | CAACTTCACTAACGGAGAGG | 319 | 319 |
| AAGTTGAGAGGCATGCGG | |||
| U89-U90c | ACAGCTACCGTCCCGATCC | 682 | 789 |
| GACATCTCTTTGTTGTGTGCC | |||
| ACAGCTACCATCCCGATCC | —d | 810e | |
| GACATCTCTTTGTTGTGTGCC | |||
| U91 | CAGAACAGTCAGGTATTTTCCCC | 374 | 476 |
| GGCGCTGAAGCATGTAAGCA | |||
| U94 | CATCGCATACGTCTCCCAG | 450 | 450 |
| TCTCTAACGTGTCCGTGCC | |||
| U100 | TGATCGGCCCCGACTCATC | 703 | 1,045 |
| GTTGCTCCCGAAAGCGCC | |||
Random primer first-strand cDNA synthesis from 1 μg of total RNA was carried out with the cDNA cycle kit (Invitrogen) as recommended by the manufacturer, with random hexamer primers. The cDNAs were purified by phenol-chloroform extractions and ethanol precipitated. After being rinsed with 75% ethanol, the cDNAs were stored at −80°C. Due care was taken to avoid sample-to-sample contamination: different rooms were used for RNA extraction, PCR setup, and gel analyses, all pipette tips had filters for aerosol protection, samples were intersped with blank reaction mixtures, etc. To assay whether cDNAs from different samples were retrotranscribed with similar efficiencies, 10,000-fold dilutions of cDNAs were analyzed by PCR for the detection of the human β-actin gene.
PCR analysis.
We analyzed 13 HHV-6 genes. All the primers were derived by us from the published HHV-6 sequence (19), with the exception of the primers for U31, which were described by Aubin et al. (5), and the primers for U73 and U41, which were developed by Rapp and Pellett (29a). The primer sequences, as well as the expected size of amplified fragments resulting from DNA and cDNA, are shown in Table 1. All primer sets amplified the DNA of both HHV-6 variants, with the exception of U89-U90, for which separate sets of primers had to be developed.
PCRs were done in the presence of 400 nM primers (500 nM for β-actin and U31), 1.5 mM MgCl2 (3 mM for U41, U39, and U16-U17; 2.5 mM for U31), 200 mM deoxynucleoside triphosphates, and 1 U of AmpliTaq DNA polymerase (Perkin Elmer) in the buffer supplied by the manufacturer.
After an initial denaturation of 5 min at 94°C, a thermal cycle of 1 min at 94°C, 1 min at 55°C (63°C for U91 and U42; 68°C for U100; 58°C for U89-U90; 60°C for U31), and 1 min at 72°C (with a 3-s increase at each new cycle) was repeated 35 times. The amplification products were run on an agarose gel (1 to 2%, according to the expected fragment size) and visualized under UV illumination after ethidium bromide staining.
Sequencing.
After electrophoresis, the amplified fragments were purified with Gene Clean II (Bio 101) and were sequenced with the FMOL sequencing system (Promega). Reaction products were separated on a 6% polyacrylamide gel, with 8 M urea and Tris-borate buffer. The gels were dried on filter paper and exposed to autoradiographic film. The sequences, obtained with both sense and antisense primers, were manually determined. Open reading frames were determined with the DNA Strider 1.0 software.
RESULTS
Temporal regulation of HHV-6 transcription.
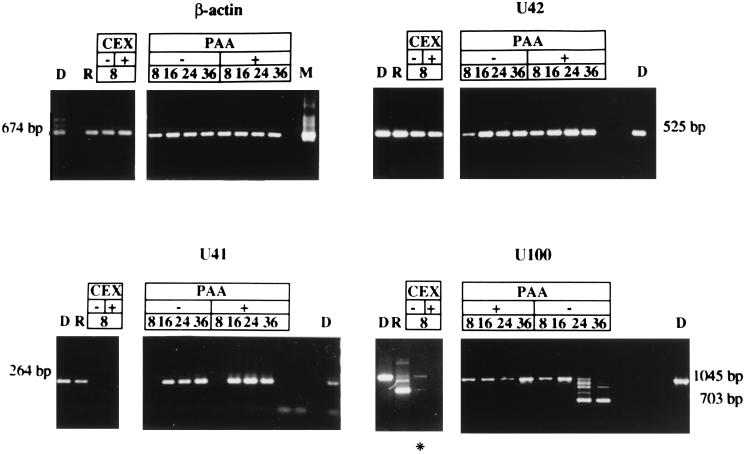
In this series of experiments, cells were infected with HHV-6A or HHV-6B and maintained in the presence or absence of CEX or emetine (inhibitors of protein synthesis) and PAA (inhibitor of viral DNA replication) to identify the temporal class of viral transcripts. The concentrations of drugs compatible with cell viability but effective in inhibiting viral protein or DNA synthesis, respectively, had been determined in preliminary experiments. All the results were verified in at least two independent experiments. To minimize experimental variations, RNA extracted from infected cells was retrotranscribed after priming with random hexamers, and therefore the resulting cDNAs could be analyzed for the whole panel of HHV-6 genes. In addition, the same samples were retrotranscribed in different experiments, and equivalent results were always obtained. Moreover, aliquots of the viral inoculum were carefully analyzed by rtPCR, to ensure that the cDNAs detected originated from newly synthesized mRNAs and not from transcripts present in the inoculum. The samples were also checked for the absence of viral DNA: samples from all time points were analyzed by PCR, without retrotranscription, for the presence of residual HHV-6 DNA, and were retrotranscribed only when found negative. No contaminant DNA was present in the samples, as shown in Fig. 1 (U41, 8 h postinfection [p.i.]).
FIG. 1.
Agarose gels stained with ethidium bromide showing the results of RT-PCR for human β-actin and HHV-6 U42 (an α-gene), U41 (a β-gene), and U100 (a γ-gene). RNA was extracted at the indicated time (hours) after infection from cells infected with HHV-6 in the presence or absence of CEX and PAA. D, PCR on DNA from HHV-6 infected cells, extracted 4 days p.i., at complete CPE; R, RT-PCR on RNA extracted from HHV-6-infected cells at complete CPE; M, human DNA extracted from mock-infected cells. The asterisk marks a sample yielding a faint band, difficult to reproduce in print. The sizes of amplified fragments are shown. No contaminant DNA was detected in the samples (U41, 8 h after infection).
The efficiency of retrotranscription was checked by amplification of the human β-actin cDNA, as shown in Fig. 1, after a 1:10,000 dilution. Samples from all time points were amplified with similar efficiencies, excluding the possibility of marked differences in retrotranscription for different samples. Furthermore, the presence of CEX and PAA did not affect the mRNA levels of a housekeeping gene such as the β-actin gene.
Since the primers for PCR amplification were derived from HHV-6A sequence, we checked whether they could also amplify HHV-6B DNA. Similar efficiencies were obtained, with the exception of U89-U90, for which an HHV-6B-specific set of primers had to be developed (Table 1).
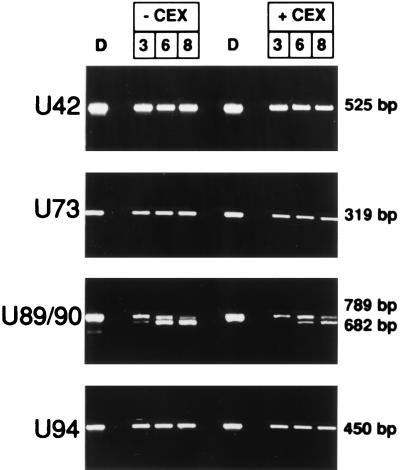
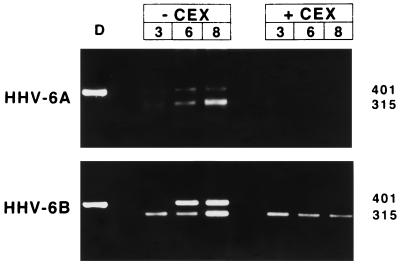
The genes analyzed in our study are shown in Table 2; they include genes with predicted regulatory functions, genes encoding proteins which orchestrate viral replication, and genes encoding structural proteins. The α-genes which included U42, U89-U90, U94, U39, and U73, were expressed as early as 3 h after infection even in the presence of CEX (Fig. 2). The results obtained with CEX and emetine are identical (data not shown). A typical example of an α-gene whose expression is not affected by CEX or PAA is U42 (Fig. 1).
TABLE 2.
Temporal mapping of HHV-6 transcripts
| ORF | Function | Transcriptional class in:
|
||
|---|---|---|---|---|
| HHV-6A | HHV-6B | Herpesviruses | ||
| U39 | gB | α | α | γ1 |
| U42 | Conserved transactivator | α | α | α/β |
| U73 | Origin binding protein | α | α | β |
| U89-U90 | IE-A (Kpn) | α | α | α |
| U94 | Rep | α | α | |
| U16-U17 | IE-B single splice, US22 family | β | α | α |
| U91 | IE-A (antisense) nonspliced | α | β | |
| IE-A (antisense) single splice | β | α | ||
| U18-U20 | IE-B | β (526bp) | 1.9kb | α |
| U19 | IE-B | β | β | α |
| U31 | Large tegument protein | β | β | γ |
| U41 | Major DNA binding protein | β | β | β |
| U53 | Protease | β | β | γ |
| U100 | gp82/105 | γ | NDa | |
ND, not determined.
FIG. 2.
Agarose gel stained with ethidium bromide showing the results of RT-PCR for HHV-6 α-genes (U42, U73, U89-U90, and U94). RNA was extracted 3, 6, or 8 h after infection from cells infected in the presence of absence of CEX. D, PCR on DNA from HHV-6 infected cells, extracted 4 days p.i., at complete CPE. The sizes of the amplified fragments are shown.
U19, U31, U41, and U53 were regulated as β-genes. Transcripts of these genes were detected at 8 h after infection. They were inhibited by CEX but unaffected by the addition of PAA. The example of U41 is shown in Fig. 1.
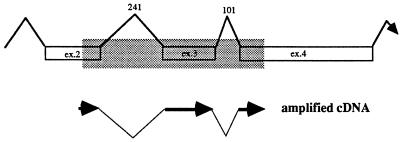
Only one gene in our panel, U100, encoding gp82/105, was regulated as a γ-gene. The cDNA of HHV-6A U100 was identified by Pfeiffer et al. (27) as a multiply spliced product of a gene containing 12 exons. The primers used by us amplified a 1,045-bp fragment of the DNA, encompassing part of exon 2, exon 3, and part of exon 4, and detected both unspliced and spliced mRNAs (Fig. 3). Low levels of unspliced mRNA were detected at 8 h after infection but could not be detected in cells infected and maintained in the presence of CEX (Fig. 1). Spliced RNA was detected at late times after infection and was detected 24 h after infection. The unspliced form gradually disappeared, and at the time the cells exhibited complete CPE, 4 days p.i., only spliced RNAs were detected. In the presence of PAA, no spliced RNAs could be detected, suggesting that splicing of U100 RNA was regulated as or by a γ-gene function.
FIG. 3.
Schematic representation of part of U100 (exons 2 to 4) from HHV-6A (27), containing two introns of 241 and 101 bases. The PCR amplified both a genomic fragment of 1,045 bp, highlighted in the shaded area, and a multiply spliced cDNA of 703 bp.
Transcription of HHV-6A and HHV-6B IE-A region.
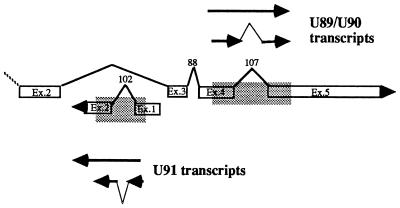
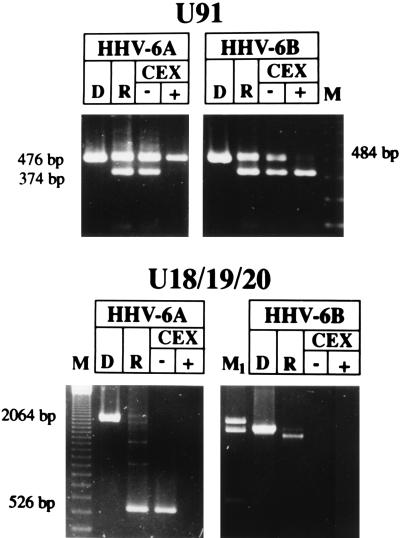
The patterns of expression of the genes listed above are similar in HHV-6A- and HHV-6B-infected cells. However, other genes exhibited temporal patterns specific for HHV-6A or HHV-6B. U91 and U89-U90 are encoded by the HHV-6 IE region homologous to the major IE locus of HCMV. The domain of this region relevant to our studies is represented in Fig. 4. Schiewe et al. (31) described two spliced transcripts derived from U89-U90 in the published HHV-6 sequence and identified them as α-genes on the basis of the observation that they are expressed in the presence of CEX. In addition, they reported the presence of another spliced mRNA, derived from U91 mapping on the complementary strand. Our results confirm that U89-U90 of both HHV-6A and HHV-6B are α-genes and that similar splicing events take place in the homologous regions of HHV-6A and HHV-6B. However, the pattern of transcription of U91 differed in cells infected with these two viruses. Our assay amplified part of exons 1 and 2 and detected both unspliced and spliced transcripts (Fig. 4). The results, shown in Fig. 5, indicate that the unspliced sequence (476 bases) of HHV-6A was detected in cells infected and maintained in the presence of CEX whereas the spliced form (374 bases) was detected only in untreated, infected cells, suggesting that the splicing requires the intervention of a function expressed earlier in infection. Interestingly, the same transcripts exhibited a different pattern of synthesis in cells infected with HHV-6B. In fact, the spliced form was maintained in the presence of CEX whereas the unspliced transcript (484 bases) was not detected (Fig. 5). Therefore, in HHV-6B the shorter transcript is α and the longer is transcribed later in infection.
FIG. 4.
Schematic representation of the portion of U89-U90 and U91 from HHV-6A (31) amplified by PCR. The amplified DNA fragment is shown within the shaded area. U89-U90 amplified cDNAs were 789 bp (unspliced) and 682 bp (spliced); the U91 amplified cDNAs were 476 and 374 bp, respectively.
FIG. 5.
Agarose gels stained with ethidium bromide showing the cDNA amplified by RT-PCR for U91 and U18-U19-U20. RNA was extracted 8 h p.i. from cells infected with HHV-6A (U1102) or HHV-6B (CV) in the presence or absence of CEX. D, PCR on DNA from HHV-6-infected cells, extracted 4 days p.i., at complete CPE. R, RT-PCR on RNA extracted from HHV-6-infected cells at complete CPE; M, 123-bp ladder molecular size marker; M1, lambda HindIII molecular size marker, showing the 2,322-, 2,027-, and 564-bp fragments. The sizes of the amplified fragments are shown.
Transcription of the HHV-6A and HHV-6B IE-B region.
The IE-B region, homologous to HCMV IE, encodes two sets of spliced genes. U16 and U17 correspond to the EFLF-2 and EFLF-1 open reading frames (ORFs) originally described by Nicholas and Martin (25). Our analysis confirms that this region yields a spliced RNA transcript hypothesized earlier (25). In HHV-6A-infected cells, the transcript was regulated as a β-gene function: it appeared early in the course of infection, being detected by 3 h p.i., but was absent in cells treated with inhibitors of protein synthesis (Fig. 6). However, this transcript has a different regulation in HHV-6B: the spliced product is unaffected by the presence of either CEX or emetine and therefore is an α-gene function (Fig. 6).
FIG. 6.
Agarose gel stained with ethidium bromide showing the results of RT-PCR for HHV-6 U16-U17. RNA was extracted 3, 6, or 8 h p.i. from cells infected in the presence or absence of CEX. D, PCR on DNA from HHV-6 infected cells, extracted 4 days p.i. at the time the infected cells exhibited complete CPE.
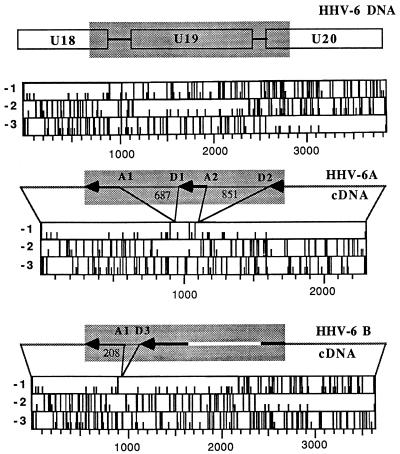
The other group of spliced genes is U18, U19, and U20. We detected a transcript originating from U19, presumably regulated as a β-gene function in both variants (Table 2). However, this region also encodes multiply spliced transcripts, and some spliced forms of U18-U19-U20 differ in cells infected with HHV-6A or HHV-6B. Nicholas and Martin (25) suggested that the EJFL6, EJFL4, and EJFL3 ORFs, renamed U18, U19, and U20 in the published HHV-6 sequence (19), were joined as result of multiple splice events, and mRNA species were proposed on the basis of putative splice donor and acceptor sites. We designed PCR primers that permitted the amplification of a 2,064-bp DNA sequence comprising part of U18, all of U19, and part of U20 (Fig. 7). Analyses of HHV-6A-infected cells showed that a spliced mRNA was detected 8 h after infection. This mRNA was detected in infected cells maintained in the presence of PAA (data not shown) but not in cultures infected and maintained in the presence of CEX (Fig. 5). The amplified product of HHV-6A cDNA was completely sequenced and showed a perfect correspondence to the published sequence (19). The amplified fragment from cDNA was 526 bp long, and the splice pattern is shown in Fig. 7. Splice donor and splice acceptor sequences were identified at the intron boundaries (Table 3). Two splicing sites were present (donor sites D1 and D2 and acceptor sites A1 and A2), originating two introns of 687 and 851 bases. According to the analysis of the coding regions, this transcript has the potential of synthesizing two proteins, one corresponding to the complete U18 and the other corresponding to U20 partially deleted in the carboxy terminus (Fig. 7).
FIG. 7.
Schematic representation of the portion of U18-U19-U20 amplified by our PCR assay. The amplified DNA fragment is shown within the shaded area. The sequenced cDNAs of HHV-6A (U1102) and HHV-6B (CV) are indicated by thick lines, and the white line indicates the undetermined sequence. Spliced regions and their sizes are indicated. The size of exon D1/A2 is 170 bp. The locations of splice acceptor (A) and splice donor (D) sites are shown. The corresponding ORFs are shown. Full bars indicate stop codons, and small bars indicate start codons.
TABLE 3.
Intron splice junctions detected in cDNA for U18/U19/U20
| HHV-6 variant | Intron | Donor sitea | Acceptor sitea |
|---|---|---|---|
| HHV-6A | 1 | D1: GGCA(AG)/(GT)GTGT | A1: CCAC(AG)/AGACGC |
| HHV-6B | 1 | D3: GCTG(AG)/(GT)AGGT | A1: CCAC(AG)/AGATGC |
| HHV-6A | 2 | D2: CAAA(AG)/(GT)CCGT | A2: TGAC(AG)/(G)TGTAT |
| Consensus sequence | N(AG)/(GT)AAGT | TTNC(AG)/G |
Point mutation in HHV-6B is shown in boldface type. Parentheses indicate nucleotides within the splice junctions which show 100% identity to the consensus sequence.
The cDNA from HHV-6B-infected cells yielded a significantly larger amplimer, which differed from that of HHV-6A-infected cells both for size, approximately 1.9 kb, and for the time of its appearance, being detected only 24 h p.i. (Fig. 5). We should stress that preliminary experiments had shown that the PCR had similar efficiencies in amplifying the transcripts of both HHV-6A and HHV-6B and therefore the disparity of results cannot be attributed to the amplification procedure. HHV-6B cDNA, as well as DNA, were partially sequenced, and only one intron was detected. The splice donor and acceptor sites are given in Table 3. The splice acceptor site A1, also present in HHV-6A, was maintained. However, the corresponding donor site (D3) was different from the site in HHV-6A, being located only 214 bp upstream on the HHV-6B DNA sequence (Fig. 7). Analysis of the coding regions from HHV-6B DNA showed that this transcript has the potential of synthesizing three complete ORFs, corresponding to U18, U19, and U20 (Fig. 7). Therefore, U19, spliced out in the final transcript of HHV-6A, is instead transcribed in HHV-6B.
Interestingly, HHV-6A DNA contains an identical splice donor site (GCTGAG/GTAGGT) in a position corresponding to the D3 site in HHV-6B (19), but it does not give rise to introns. Likewise, HHV-6B DNA also contains typical splice donor sites (GGGACG/GTGTGT and CAAAAC/GTCCGT) in the same position as D1 and D2, respectively, with some point mutations (underlined) not affecting the consensus sequence, which do not originate introns.
HHV-6 transcription in nonproductive cells.
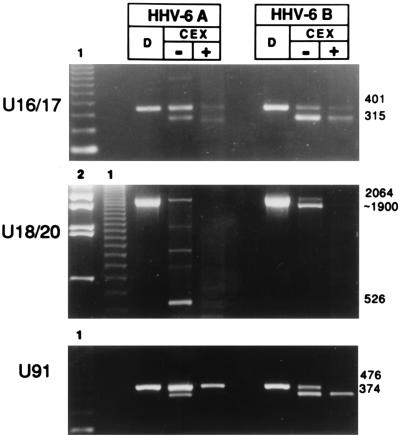
To ensure that the differences detected between HHV-6A and HHV-6B were not due to the different cell lines used, the experiments were repeated but SupT1 cells were infected with HHV-6A (U1102) and JJhan cells were infected with HHV-6B (CV). As expected (14), viral growth did not occur. Nevertheless, transcription took place with the same pattern as in lytically infected cells (data not shown). The results reported in Fig. 8 show that for nonproductive infection, HHV-6 variants also maintain the three main differences already described for U91, for U16-U17, and for the multiply spliced transcript of U18-U19-U20. The larger transcript from U91 is temporally regulated as α in HHV-6A and as β in HHV-6B, while the supposedly spliced form is β in HHV-6A and α in HHV-6B. The U16-U17 splice is regulated as α in HHV-6B and as β in HHV-6A. Finally, the β spliced transcript from U18-U20, revealed as a 526-bp fragment by the RT-PCR assay, was present only in HHV-6A, whereas HHV-6B showed only the larger, 1.9-kb fragment (Fig. 8).
FIG. 8.
Agarose gels stained with ethidium bromide showing RT-PCR of mRNA transcribed during nonproductive HHV-6 infection of lymphoid cells. SupT1 cells were infected with HHV-6A (U1102), and JJhan cells were infected with HHV-6B (CV), in the presence or absence of CEX. The PCR products of amplification upon DNA are shown (D). Sizes are shown in base pairs. The sizes shown for U91 are derived from HHV-6A; the respective sizes in HHV-6B are 484 and 388 bp. Molecular weight markers are indicated by 1 (123-bp ladder) and 2 (λ DNA digested with BstE).
DISCUSSION
HHV-6 was discovered in 1986 (30), the existence of two distinct groups (HHV-6A and HHV-6B) was described in 1991 (32), but the knowledge of its molecular biology has experienced slow progresses and is still limited. The nucleotide sequence of HHV-6A was published only in 1995 (19), the complete sequence of HHV-6B is still not available, and studies on the transcription of viral genes have been restricted to ORFs homologous to IE genes of HCMV. We describe here for the first time the temporal regulation of transcription of a selected set of HHV-6 genes. These studies were undertaken for two practical reasons: (i) the temporal mapping of HHV-6 transcripts is a necessary step to identify viral genes which may be expressed during latency, following reactivation, and during the early stages of infection; and (ii) the different biological behaviors of HHV-6A and HHV-6B could reflect differences in gene expression, potentially revealed by a comparative analysis of their transcription.
Our results show that HHV-6 transcription follows a typical herpesvirus cascade pattern in which α-, β-, and γ-genes are coordinately and sequentially transcribed. The kinetic classes of genes such as U42, U89-U90, and U41 correspond to those of their homologous counterparts in the other HHVs (Table 2), but other HHV-6 genes are transcribed earlier than in other members of the family; this is the case for U31, U39, U53, and U73. It should be noted that several factors may complicate the temporal analysis of transcriptional classes. The presence of a specific mRNA does not necessarily imply protein expression, and transcripts may accumulate before protein synthesis is observed. Furthermore, regardless of when it begins, transcription continues throughout the infection and mRNA levels are significantly higher at late times p.i. For example, mRNA from U39 is unaffected by the presence or absence of CEX, but its levels are lower at 8 h p.i. than at later times (data not shown), suggesting that a promoter may be active at a low level throughout the reproductive cycle or that its activity could be enhanced at specific times during infection. The sensitive approach we used, RT-PCR, does not allow us to determine the relative abundance of viral transcripts and could result in an overestimate of the amounts of mRNAs from scarcely transcribed genes. Furthermore, different members of the herpesvirus family express homologous genes at different times during the lytic cycle. Such is the case, for example, for a conserved herpesvirus transactivator gene: in HSV, ICP27 is clearly an α-gene product, but the counterpart of this gene in HCMV, UL69, is a β-gene, expressed at 7 h after infection and not detected under IE conditions (39). The HHV-6 homolog U42 is regulated as an α-gene and in this respect resembles the HSV counterpart more closely than the HCMV counterpart.
Several genes were transcribed under IE conditions (Table 2) in that their mRNA was present at 3 h after infection, CEX and emetine did not inhibit their synthesis, and the mRNA was also detected at later times p.i.
In this report, we show that U94, which does not have counterparts in the other herpesviruses, is regulated as an α-gene. It has been reported that the product of U94, a homolog of the Rep protein of adeno-associated parvovirus type 2, has transactivating potential and can negatively regulate several heterologous promoters, such as the HIV long terminal repeat and the ras promoter (2). The finding that U94 was expressed under IE conditions supports the notion that it is a regulatory gene which may play an important role in the viral replicative cycle, either by regulating viral activity or by turning off host cell functions.
The only γ-gene revealed by our study is U100: the mRNA was transcribed in its unspliced form under β-gene conditions, but the spliced product was detected only 16 h p.i. and was inhibited by PAA (Fig. 1), showing that HHV-6 controls gp82/105 production by temporally regulated splicing mechanisms. An indirect observation is that the HHV-6 replicative cycle is longer than 16 h, since at earlier times the viral envelope would not be complete.
Transcription from the IE-B region shows a distinctive pattern for each variant. One difference resides in U16-U17. The final splice product is present in both variants, but it is α in HHV-6B and β in HHV-6A (Fig. 6). Therefore, HHV-6 variants modulate transcription from this region differently. Other variant-specific differences were observed in the remaining portion of IE-B. Nicholas and Martin proposed a speculative mRNA structure for the U18-U19-U20 region (25), on the basis of DNA sequence and by comparison with the splicing pattern in the HCMV homologous region. Our results demonstrate that transcription from this region is complex. In fact, both variants show the presence of a distinct β-gene transcript originating from U19, but a multiply spliced transcript is also found. In HHV-6A the cDNA amplified in our assay is larger than expected (25), since a putative 118-bp intron is not spliced out and a splice junction is present within an intron, resulting in an additional 170-bp exon (Fig. 7). Interestingly, HHV-6B shows a different cDNA structure. The size of the amplified cDNA in HHV-6B is about 1.9 kb; sequence analysis showed that one splice donor/acceptor site is missing (A2/D2, Fig. 7) and that the D1 site, still present in HHV-6B, is not recognized and instead is substituted by a donor site (D3) closer to A1. This pattern reflects a different coding potential in the spliced transcript detected for each variant (Fig. 7). Theoretically, the HHV-6A mRNA has the ability to transcribe a complete product from U18 and a partially deleted protein from U20. Instead, the HHV-6B transcript can perform synthesis for all three ORFs, including U19. Furthermore, the cDNA in HHV-6B appeared later during infection than did that in HHV-6A (24 and 8 h p.i., respectively). Considering the late appearance and large size, it is possible that this 1.9-kb cDNA represents a low-abundance intermediate species, yielding a final transcript(s) not detected by the PCR primers, possibly due to a different splice pattern between HHV-6 variants. By analogy to the homologous UL36-38 region in HCMV, it is also possible to propose that these transcripts function as polycistronic messengers, encoding distinct proteins from nonoverlapping ORFs (35). Each variant showed its characteristic splice pattern in both cell lines used (Fig. 8), strengthening the idea that the observed differences are variant specific. The different splice patterns detected within U18-U19-U20 are not due to differences in the sequence of HHV-6 variants, because splice sites D1 and D3 are present both in HHV-6A and HHV-6B. Therefore, it is possible that differences reside in spliceosome formation or in the splicing machinery.
Another difference in transcription between HHV-6 variants was observed for U91. The 476- and 374-base transcripts generated by U91 show opposite temporal regulations in HHV-6 variants (Table 2; Fig. 5). Schiewe et al. (31), on the basis of sequence data on cDNA, suggested that the transcript from this region is spliced. However, in our experiments with HHV-6B, the 374-bp fragment corresponding to the spliced mRNA appeared earlier than the 476-bp amplimer corresponding to the full-length molecule, from which it should originate. The results are compatible with the hypothesis that U91 transcription is regulated by two promoters, temporally regulated as different functions in HHV-6 variants: the promoter yielding the 476-base transcript is α in HHV-6A and β in HHV-6B, while the 374-base mRNA is transcribed from a β promoter in HHV-6A and from an α promoter in HHV-6B. It is also possible that, similarly to the situation already described for U16-U17, different splicing mechanisms take place in HHV-6 variants. The observation that transcription of HHV-6 variants differ in this region is supported by sequence data, showing that the homology between IE regions is approximately 75 to 85% for the DNA sequence and 62 to 70% for the amino acid sequence, whereas in other genomic regions the sequence homology is often above 90% (11, 36).
The differences observed were not due to the cell lines, since all three variant-specific transcription patterns were also detected when HHV-6 strains infected cell lines that did not support viral growth (Fig. 8). Incidentally, these results show that nonproductive lymphoid cells are susceptible to infection and support active transcription; however, the lack of viral growth is indicative of restricted or abortive infection.
Although both HHV-6 variants have a predominant tropism for CD4+ cells, HHV-6B requires cell maturation associated with the expression of the CD3 antigen (34) but HHV-6A seems to prefer less differentiated CD4+ T cells (23). The two variants show distinct characteristics of growth in continuous cell lines. Variant B, unlike HHV-6A, does not propagate in JJhan and HSB-2 cell lines (1, 40) but grows in other T cells, such as SupT1, Molt-3, and MT-4, which are not susceptible to HHV-6A productive infection (1, 6). NK cells are more permissive to HHV-6A than to HHV-6B infection and replication (22). Finally, both variants can infect the monkey Macaca nemestrina, but only HHV-6B, and not HHV-6A, infects and replicates in peripheral blood mononuclear cells of Macaca mulatta (24). It has also been suggested that HHV-6 variants may have a different disease association or a different pathogenic potential, and, indeed, HHV-6B is frequently isolated from children with ES whereas HHV-6A is isolated mostly from immunosuppressed individuals. The biological and pathological implications of these differences between HHV-6 variants are still controversial, but the present results, showing that at least three viral genes expressed during the early phases of replication have distinct features in each variant, provide a molecular basis for the different biological behaviors of HHV-6 variants.
Several observations support the notion that HHV-6 can be an opportunistic pathogen in immunosuppressed patients (33), and viral infection or reactivation in transplant recipients may result in graft-versus-host disease, interstitial pneumonitis, and encephalitis. In these cases, an early diagnosis of viral infection is important to permit prompt antiviral treatment, and PCR for HHV-6 DNA is often an obligate diagnostic choice. However, the mere detection of viral genomes is not sufficient to discriminate between latent and acute infection, and even quantitative PCR is not suitable to distinguish between a low-level productive infection and an increased load of latently infected cells in immunocompromised patients (7). Such a distinction might be important, since even low levels of productive infection by HHV-6 may cause clinical disease (7, 10). Our results, identifying the viral functions associated with the different phases of lytic infection, provide the necessary conditions for proposing RT-PCR as a useful diagnostic approach to the differentiation between latent and productive infection by HHV-6.
ACKNOWLEDGMENTS
This work was supported by grants from Ministero della Sanità (Istituto Superiore Sanità, AIDS Project), from BiomedII (European Community), from Associazione Italiana per la Ricerca sul Cancro (AIRC), and from MURST.
REFERENCES
- 1.Ablashi D V, Balachandran N, Josephs S F, Hung C L, Krueger G R F, Kramarsky B, Salahuddin S Z, Gallo R C. Genomic polymorphism, growth properties, and immunologic variations in human herpesvirus-6 isolates. Virology. 1991;184:545–552. doi: 10.1016/0042-6822(91)90424-a. [DOI] [PubMed] [Google Scholar]
- 2.Araujo J C, Doniger J, Kashanchi F, Hermonat P L, Thompson J, Rosenthal L J. Human herpesvirus 6A ts suppresses both transformation by H-ras and transcription by the H-ras and human immunodeficiency virus type 1 promoters. J Virol. 1995;69:4933–4940. doi: 10.1128/jvi.69.8.4933-4940.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Asano Y, Yoshikawa T, Kajita Y, Ogura R, Suga S, Yazaki T, Nakashima T, Yamada A, Kurata T. Fatal encephalitis/encephalopathy in primary human herpesvirus-6 infection. Arch Dis Child. 1992;67:1484–1485. doi: 10.1136/adc.67.12.1484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Asano Y, Yoshikawa T, Suga S, Yazaki T, Kondo K, Yamanishi K. Fatal fulminant hepatitis in an infant with human herpesvirus-6 infection. Lancet. 1990;i:862–863. doi: 10.1016/0140-6736(90)90983-c. [DOI] [PubMed] [Google Scholar]
- 5.Aubin J T, Collandre H, Candotti D, Ingrand D, Rouzioux C, Burgard M, Richard S, Huraux J M, Agut H. Several groups among human herpesvirus 6 strains can be distinguished by Southern blotting and polymerase chain reaction. J Clin Microbiol. 1991;29:367–372. doi: 10.1128/jcm.29.2.367-372.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Black J B, Sanderlin K C, Goldsmith C S, Gary H E, Lopez C, Pellett P E. Growth properties of human herpesvirus 6 strain Z29. J Virol Methods. 1989;26:133–145. doi: 10.1016/0166-0934(89)90143-2. [DOI] [PubMed] [Google Scholar]
- 7.Carrigan D R. Human herpesvirus-6 and bone marrow transplantation. Blood. 1995;85:294–295. [PubMed] [Google Scholar]
- 8.Carrigan D R, Drobyski W R, Russler S K, Tapper M A, Knox K K, Ash R C. Interstitial pneumonitis associated with human herpesvirus-6 infection after marrow transplantation. Lancet. 1991;338:147–149. doi: 10.1016/0140-6736(91)90137-e. [DOI] [PubMed] [Google Scholar]
- 9.Carrigan D R, Knox K K. Human herpesvirus 6 (HHV-6) isolation from bone marrow: HHV-6-associated bone marrow suppression in bone marrow transplant patients. Blood. 1994;84:3307–3310. [PubMed] [Google Scholar]
- 10.Carrigan D R, Knox K K. Bone marrow suppression by human herpesvirus-6: comparison of the A and B variants of the virus. Blood. 1995;86:835–836. [PubMed] [Google Scholar]
- 11.Chou S, Marousek G I. Analysis of interstrain variation in a putative immediate-early region of human herpesvirus 6 DNA and definition of variant-specific sequences. Virology. 1994;198:370–376. doi: 10.1006/viro.1994.1044. [DOI] [PubMed] [Google Scholar]
- 12.Cone R W, Hackman R C, Huang M L, Bowden R A, Meyers J D, Metcalf M, Zeh J, Ashley R, Corey L. Human herpesvirus 6 in lung tissue from patients with pneumonitis after bone marrow transplantation. N Engl J Med. 1993;329:156–161. doi: 10.1056/NEJM199307153290302. [DOI] [PubMed] [Google Scholar]
- 13.Di Luca D, Katsafanas G, Schirmer E, Frenkel N. The replication of viral and cellular DNA in human herpesvirus 6 infected cells. Virology. 1990;175:199–210. doi: 10.1016/0042-6822(90)90200-b. [DOI] [PubMed] [Google Scholar]
- 14.Di Luca D, Mirandola P, Ravaioli T, Bigoni B, Cassai E. Distribution of HHV-6 variants in human tissues. Infect Agents Dis. 1996;5:203–214. [PubMed] [Google Scholar]
- 15.Di Luca D, Mirandola P, Secchiero P, Cermelli C, Aleotti A, Bovenzi P, Portolani M, Cassai E. Characterization of human herpesvirus 6 strains isolated from patients with exanthem subitum with or without cutaneous rash. J Infect Dis. 1992;166:689. doi: 10.1093/infdis/166.3.689. [DOI] [PubMed] [Google Scholar]
- 16.Downing R G, Sewankambo N, Serwadda D, Honess R W, Crawford D, Jarrett R, Griffin B E. Isolation of human lymphotropic herpesviruses from Uganda. Lancet. 1987;ii:390. doi: 10.1016/s0140-6736(87)92403-2. [DOI] [PubMed] [Google Scholar]
- 17.Drobyski W R, Dunne W M, Burd E M, Knox K K, Ash R C, Horowitz M M, Flomenberg N, Carrigan D R. Human herpesvirus-6 (HHV-6) infection in allogeneic bone marrow transplant recipients: evidence of a marrow-suppressive role for HHV-6 in vivo. J Infect Dis. 1993;167:735–739. doi: 10.1093/infdis/167.3.735. [DOI] [PubMed] [Google Scholar]
- 18.Drobyski W R, Knox K K, Majewski D, Carrigan D R. Brief report: fatal encephalitis due to variant B human herpesvirus-6 infection in a bone marrow-transplant recipient. N Engl J Med. 1994;330:1356–1360. doi: 10.1056/NEJM199405123301905. [DOI] [PubMed] [Google Scholar]
- 19.Gompels U A, Nicholas J, Lawrence G, Jones M, Thomson B J, Martin M E D, Efstathiou S, Craxton M, Macaulay H A. The DNA sequence of human herpesvirus-6: structure, coding content, and genome evolution. Virology. 1995;209:29–51. doi: 10.1006/viro.1995.1228. [DOI] [PubMed] [Google Scholar]
- 20.Kondo K, Kondo T, Okuno T, Takahashi M, Yamanishi K. Latent human herpesvirus 6 infection of human monocytes/macrophages. J Gen Virol. 1991;72:1401–1408. doi: 10.1099/0022-1317-72-6-1401. [DOI] [PubMed] [Google Scholar]
- 21.Lawrence G L, Chee M, Craxton M A, Gompels U A, Honess R W, Barrell B G. Human herpesvirus 6 is closely related to human cytomegalovirus. J Virol. 1990;64:287–299. doi: 10.1128/jvi.64.1.287-299.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lusso P, Gallo R C. Human herpesvirus 6 in AIDS. Immunol Today. 1995;16:67–71. doi: 10.1016/0167-5699(95)80090-5. [DOI] [PubMed] [Google Scholar]
- 23.Lusso P, Malnati M, De Maria A, Balotta C, De Rocco S E, Markham P D, Gallo R C. Productive infection of CD4+ and CD8+ mature human T cell populations and clones by human herpesvirus 6. Transcriptional down-regulation of CD3. J Immunol. 1991;147:685–691. [PubMed] [Google Scholar]
- 24.Lusso P, Secchiero P, Crowley R W. In vitro susceptibility of Macaca nemestrina to human herpesvirus 6: a potential animal model of coinfection with primate immunodeficiency viruses. AIDS Res Hum Retroviruses. 1994;10:181–187. doi: 10.1089/aid.1994.10.181. [DOI] [PubMed] [Google Scholar]
- 25.Nicholas J, Martin M E. Nucleotide sequence analysis of a 38.5-kilobase-pair region of the genome of human herpesvirus 6 encoding human cytomegalovirus immediate-early gene homologs and transactivating functions. J Virol. 1994;68:597–610. doi: 10.1128/jvi.68.2.597-610.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Okuno T, Higashi K, Shiraki K, Yamanishi K, Takahashi M, Kokado Y, Ishibashi M, Takahara S, Sonoda T, Tanaka K. Human herpesvirus 6 infection in renal transplantation. Transplantation. 1990;49:519–522. doi: 10.1097/00007890-199003000-00009. [DOI] [PubMed] [Google Scholar]
- 27.Pfeiffer B, Thomson B, Chandran B. Identification and characterization of a cDNA derived from multiple splicing that encodes envelope glycoprotein gp105 of human herpesvirus 6. J Virol. 1995;69:3490–3500. doi: 10.1128/jvi.69.6.3490-3500.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Prezioso P J, Cangiarella J, Lee M, Nuovo G J, Borkowsky W, Orlow S J, Greco M A. Fatal disseminated infection with human herpesvirus-6. J Pediatr. 1992;120:921–923. doi: 10.1016/s0022-3476(05)81962-6. [DOI] [PubMed] [Google Scholar]
- 29.Pruksananonda P, Hall C B, Insel R A, McIntyre K, Pellett P E, Long C E, Schnabel K C, Pincus P H, Stamey F R, Dambaugh T R, Stewart J A. Primary human herpesvirus 6 infection in young children. N Engl J Med. 1992;326:1445–1450. doi: 10.1056/NEJM199205283262201. [DOI] [PubMed] [Google Scholar]
- 29a.Rapp, J., and P. Pellett. Personal communication.
- 30.Salahuddin S Z, Ablashi D V, Markham P D, Josephs S F, Sturznegger S, Kaplan M, Haleigan G, Biberfeld P, Wong-Staal F, Kramarsky B, Gallo R C. Isolation of a new virus, HBLV, in patients with lymphoproliferative disorders. Science. 1986;234:596–601. doi: 10.1126/science.2876520. [DOI] [PubMed] [Google Scholar]
- 31.Schiewe U, Neipel F, Schreiner D, Fleckenstein B. Structure and transcription of an immediate-early region in the human herpesvirus 6 genome. J Virol. 1994;68:2978–2985. doi: 10.1128/jvi.68.5.2978-2985.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schirmer E C, Wyatt L S, Yamanishi K, Rodriguez W J, Frenkel N. Differentiation between two distinct classes of viruses now classified as human herpesvirus 6. Proc Natl Acad Sci USA. 1991;88:5922–5926. doi: 10.1073/pnas.88.13.5922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Singh N, Carrigan D R. Human herpesvirus-6 in transplantation: an emerging pathogen. Ann Intern Med. 1996;124:1065–1071. doi: 10.7326/0003-4819-124-12-199606150-00007. [DOI] [PubMed] [Google Scholar]
- 34.Takahashi K, Sonoda S, Higashi K, Kondo T, Takahashi H, Takahashi M, Yamanishi K. Predominant CD4 T-lymphocyte tropism of human herpesvirus 6-related virus. J Virol. 1989;63:3161–3162. doi: 10.1128/jvi.63.7.3161-3163.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tenney D S, Colberg-Poley A M. Expression of the human cytomegalovirus UL 36-38 immediate early region during permissive infection. Virology. 1991;182:199–210. doi: 10.1016/0042-6822(91)90663-v. [DOI] [PubMed] [Google Scholar]
- 36.Yamamoto M, Black J B, Stewart J A, Lopez C, Pellett P E. Identification of a nucleocapsid protein as a specific serological marker of human herpesvirus 6 infection. J Clin Microbiol. 1990;28:1957–1962. doi: 10.1128/jcm.28.9.1957-1962.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yamanishi K, Okuno T, Shiraki K, Takahashi M, Kondo T, Asano Y, Kurata T. Identification of human herpesvirus-6 as a causal agent for exanthem subitum. Lancet. 1988;i:1065–1067. doi: 10.1016/s0140-6736(88)91893-4. [DOI] [PubMed] [Google Scholar]
- 38.Walthr W, Stein U, Eder C. RNA analysis using miniprep RNA in reverse transcription PCR. BioTechniques. 1994;17:674–675. [PubMed] [Google Scholar]
- 39.Winkler M, Rice S A, Stamminger T. UL69 of human cytomegalovirus, an open reading frame with homology to ICP27 of herpes simplex virus, encodes a transactivator of gene expression. J Virol. 1994;68:3943–3954. doi: 10.1128/jvi.68.6.3943-3954.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wyatt L S, Balachandran N, Frenkel N. Variations in the replication and antigenic properties of human herpesvirus 6 strains. J Infect Dis. 1990;162:852–857. doi: 10.1093/infdis/162.4.852. [DOI] [PubMed] [Google Scholar]