Abstract
Following cell surface receptor binding and membrane fusion, human immunodeficiency virus (HIV) virion cores are released in the cytoplasm. Incoming viral proteins represent potential targets for cytosolic proteases. We show that treatment of target cells with the proteasome inhibitors MG132 and lactacystin increased the efficiency of HIV infection. Proteasome inhibitors were active at the early steps of the viral cycle. Incoming p24Gag proteins accumulated in the cytosol, and larger amounts of proviral DNA were synthesized. In vitro, purified 20S proteasome degraded HIV virion components. Thus, degradation of incoming viral proteins by the proteasome represents an early intracellular defense against infection.
The early phases of the human immunodeficiency virus (HIV) life cycle include receptor-specific binding of virions to target cells and fusion of viral and cell membranes (9, 14, 40). Virion cores are then released into the cytoplasm, where uncoating events take place and reverse transcription is performed (2, 27, 41). The resulting proviral DNA is present in large nucleoprotein complexes, termed preintegration complexes, containing the viral integrase and reverse transcriptase enzymes and nucleocapsid (NC), matrix (MA), and Vpr proteins (5, 15, 16, 26). Translocation of preintegration complexes into the nucleus is associated with structural changes, including a reduction in size and altered protein composition (26). The proviral DNA integrates into the host DNA to complete the infection cycle.
How the cell participates in or, just the opposite, protects itself against the viral aggression is poorly documented. Viral proteins newly released in the cytoplasm may be considered as abnormal components and attacked by cytoplasmic proteases. The proteasome is the main proteolytic complex operating in the cytosol and the nucleus. It is involved in many biological and degradative processes, ensuring the removal of misfolded or ubiquitinated proteins. It controls the cell cycle and generates antigenic peptides presented by major histocompatibility complex class I molecules (8, 12, 24, 36). Two forms of the proteasome exist in the cell. The 20S (700-kDa) proteasome contains multiple peptidase activities, and the 26S (2,000-kDa) proteasome, which degrades ubiquitinated proteins, contains an additional 19S regulatory complex, including ATPases and components necessary for binding protein substrates (8, 12, 24, 36). The yeast 20S proteasome crystal structure has been recently resolved (22).
In this report, we examined the role of the proteasome on the early steps of HIV replication cycle. The proteasome inhibitors MG132 (33) and lactacystin (17) dramatically increased the efficiency of infection, whereas calpain inhibitors were uneffective. In the presence of proteasome inhibitors, incoming p24Gag proteins accumulated in the cytosol of target cells and larger amounts of proviral DNA were synthesized. In vitro, purified 20S proteasome degraded HIV virion components. Our data strongly suggest that the proteasome acts as an early intracellular defense against infection by degrading incoming viral proteins.
MATERIALS AND METHODS
Viruses, cells, and reagents.
HIV-1 (NL43 strain) and HIVΔenv(VSV [for vesicular stomatitis virus]) were produced and infections were performed as described previously (32, 38). The HIV-HSA reporter virus, which encodes the CD24 marker, was a kind gift of N. Landau (Aaron Diamond AIDS Research Center, New York, N.Y.) and was used as described previously (23). P4 cells (HeLa CD4+ long terminal repeat (LTR)-lacZ) (7) were grown in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% fetal calf serum. CEM, Jurkat, and HUT78 human T-cell lines were grown in RPMI 1640 medium supplemented with 10% fetal calf serum. MG132 was a kind gift of F. Baleux (Institut Pasteur, Paris, France) (37). Lactacystin was obtained from BioMol, and calpain inhibitors I and II were obtained from Boehringer Mannheim.
Measurement of HIV infection in P4 cells.
A total of 104 P4 cells per well in 96-well plates were cultured for 24 h before infection with 200 μl of viral supernatants (in triplicate) in the absence or in the presence of the protease inhibitors. After the indicated period of time, cells were washed to remove unbound virus and inhibitors. After 24 h, β-galactosidase activity was measured. Cells were lysed in 100 μl of a mixture of 60 mM Na2HPO4, 40 mM NaH2PO4, 10 mM KCl, 10 mM MgSO4, 2.5 mM EDTA, 50 mM β-mercaptoethanol, and 0.125% Nonidet P-40. Lysates were mixed with 100 μl of 80 mM NaPO4 (pH 7.4), 10 mM MgCl2, 50 mM β-mercaptoethanol, and 6 mM chlorophenol red-β-galactopyranoside monosodium salt (CPRG) and incubated at 37°C before measurement of the optical density at 540 nm. Values are means of triplicate determinations, and variations for each point were less than 10%. In situ staining for β-galactosidase activity was performed as described previously (7).
Analysis of viral DNA synthesis.
A total of 12 × 106 P4 cells were exposed to the indicated amounts of HIV-1 in the presence or absence of MG132 (25 μM) for 5 h. Seventeen hours later, low-molecular-weight DNA was prepared by Hirt extraction, EcoRI digested, and analyzed by Southern blotting with a 1.9-kb fragment from the pol region of pNL43 as a probe, as described previously (38).
Measurement of amounts of cytosolic p24Gag protein.
A total of 107 P4 cells were exposed to HIV-1 (1 μg of p24Gag) for 1 h at 37°C in the absence or in the presence of MG132 (50 μM) or lactacystin (40 μM). Cells were then washed and incubated at 37°C for 0, 3, or 7 h in medium containing the indicated proteasome inhibitors. Cytosolic fractions were prepared as described previously (28). Briefly, to remove virus adsorbed at the cell surface, cells were incubated for 10 min on ice with 1 ml of pronase (7 mg/ml, freshly prepared in DMEM with 20 mM HEPES). Cells were resuspended with 1 ml of ice-cold DMEM containing 10% fetal calf serum, washed in ice-cold phosphate-buffered saline, and resuspended in 2 ml of swelling buffer (10 mM Tris-HCl [pH 8], 10 mM KCl, 1 mM EDTA) for 15 min at 4°C. Cells were then subjected to mechanical disruption by 15 strokes of a 7-ml β-pestles Dounce homogenizer. Nuclei and unbroken cells were pelleted at 3,000 rpm for 3 min at 4°C. The resulting postnuclear supernatant was spun at 60,000 rpm in a TL-100 Beckman centrifugator for 10 min at 4°C to separate the membrane and vesicle-rich pellet from the cytosolic supernatant. The supernatant was adjusted to 0.5% Triton X-100 and analyzed for p24Gag content by enzyme-linked immunosorbent assay (Dupont).
Analysis of LTR activity.
P4 cells were seeded at 8 × 104 cells per well of a 24-well plate 24 h before transfection by the Ca-phosphate coprecipitation technique. Cells were transfected with 1 μg of the pLTRX-Luc plasmid, containing the luciferase reporter gene driven by the HIV-1 long terminal repeat (LTR) (39), and with the indicated amounts of the pCMV-Tat plasmid, containing the tat gene driven by the immediate-early cytomegalovirus promoter (39). Twenty-four hours after transfection, cells were incubated, when stated, with MG132 (50 μM) for 1 h, washed, and lysed 5 or 17 h later as previously described (39). Luciferase activity contained in cytoplasmic extracts was measured with a luminometer (Lumat LB9501; Bertold). Results are expressed as relative luciferase units (RLU) per microgram of cellular protein and were obtained by subtracting background signal (from untransfected cells) from each value and dividing this value by the amount of cytoplasmic protein contained in the sample. Experiments were performed in triplicate, and variations for each point were less than 10%.
RESULTS AND DISCUSSION
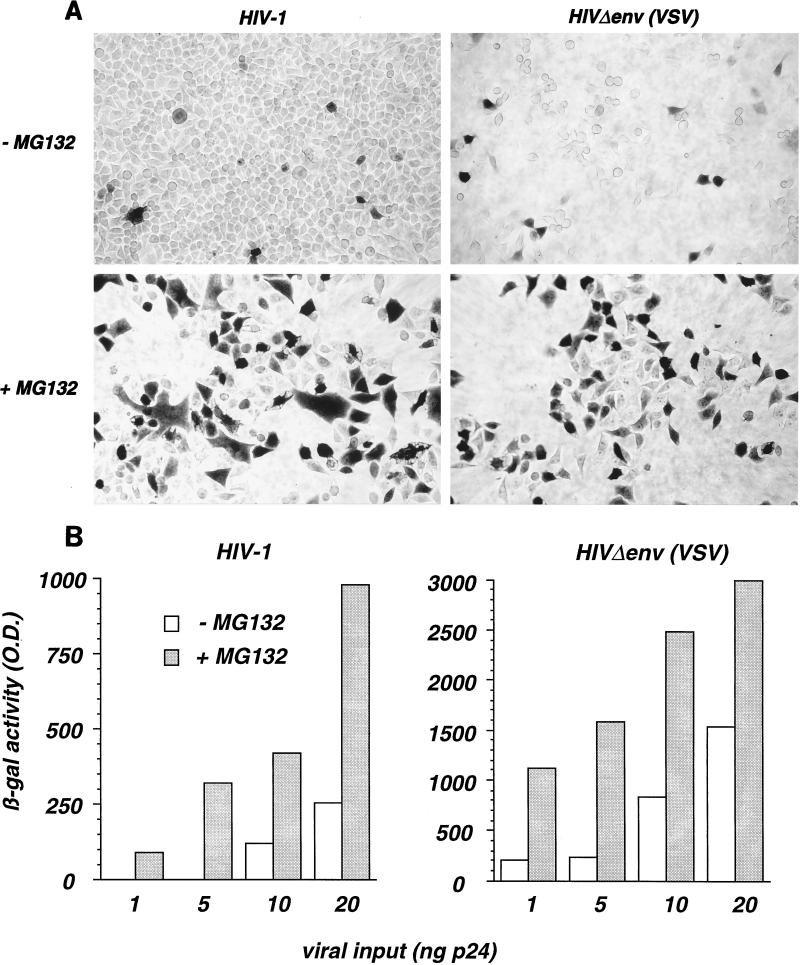
The peptide-aldehyde MG132 is a potent and reversible inhibitor of the chymotryptic-like activity of the proteasome (33). We examined the effects of MG132 on HIV-1 infection. P4 indicator cells are HeLa CD4+ cells carrying an integrated lacZ gene driven by the HIV-1 LTR and inducible by the viral transactivator Tat (7). HIV-infected P4 cells produce Tat and can be detected with high specificity by an in situ β-galactosidase assay (7). P4 cells were exposed to HIV-1, in the absence or presence of MG132 (25 μM), for 5 h. The proportion of β-galactosidase-positive cells, revealed 24 h after viral exposure, was much higher in the presence of MG132 (Fig. 1A, left panels). Infections were also performed with HIV-1 particles coated with the VSV-G envelope glycoprotein [HIVΔenv(VSV)]. VSV-G binds membrane phospholipids, allowing virus entry independently of the CD4 and chemokine receptors (6, 29, 31). Infection of P4 cells with HIVΔenv(VSV) was also increased by MG132 (Fig. 1A, right panels). Thus, the stimulation of HIV infection by the proteasome inhibitor is independent of gp120/gp41-mediated binding and entry.
FIG. 1.
The proteasome inhibitor MG132 increases HIV infection. (A) P4 indicator cells (HeLa CD4+ LTR-lacZ+) were incubated with HIV-1 (left panels) or with HIVΔenv(VSV), a defective HIV-1 strain with a deletion in the env gene and pseudotyped with the VSV-G envelope, in the absence and in the presence of MG132 (25 μM). Five hours later, virus and MG132 were removed. After 24 h, HIV-infected cells were revealed by in situ staining for β-galactosidase (β-gal) activity. (B) The effect of MG132 is independent of the viral input. HIV infection was quantified in P4 cells in a β-galactosidase based colorimetric assay. P4 cells were infected with the indicated amounts of HIV-1 (left panel) or HIVΔenv(VSV) (right panel) with or without MG132 (50 μM) for 1 h. After 24 h, cells were lysed, and β-galactosidase activity (optical density [O.D.] units) in cell extracts was measured. Values are means of triplicates. Variation between each point of triplicate determinations was below 10%. Data are representative of three independent experiments.
Since proteasome inhibitors affect the cell cycle, the effect of MG132 on HIV infection was examined in P4 cells previously arrested at the G1/S phase by aphidicolin treatment. Equivalent increases in β-galactosidase activity by MG132 were observed in arrested and nonarrested cells, indicating that the effect on HIV infection did not result from a modification of cell cycling (not shown).
MG132 activity on HIV infection was quantified by using a β-galactosidase-based colorimetric assay. P4 cells were infected with increasing amounts of HIV-1 or HIVΔenv(VSV), with or without MG132 (50 μM), for 1 h. β-Galactosidase activity, measured 24 h later, was three- to eightfold higher in the presence of MG132, independently of the viral input (Fig. 1B). The MG132 effect is known to be reversible, and a normal proteasomal activity is recovered 30 min after removal of the compound (12, 33). We then performed a 1-h pulse incubation with MG132 90 min before virus exposure. HIV infection was not increased under this condition (not shown). Thus, the peptide-aldehyde must be present at the same time as the viral proteins in order to increase infection. Altogether, these experiments strongly suggest that MG132 acts at an early step of the viral cycle.
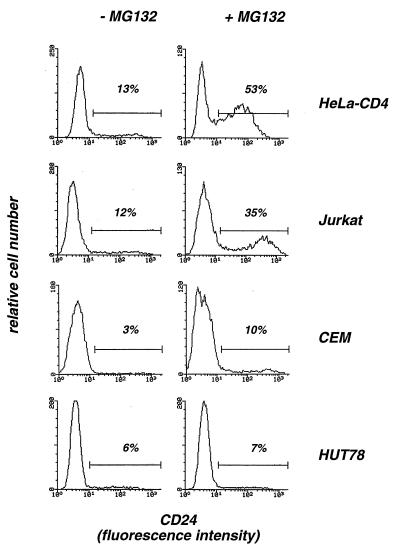
We next investigated whether MG132 increases HIV replication in lymphoid cell lines. Proteasome inhibitors appeared highly toxic in certain cell types, such as thymocytes and leukemic HL60 and U937 cells (13, 20, 25). Toxicity probably results from a modulation of the cell death program (13, 20, 25). Proteasome inhibitors were highly toxic in peripheral blood mononuclear cells (cell viability of <20% 24 h after a 1-h pulse treatment with MG132 at 50 μM), thus impairing study of primary T cells (not shown). MG132 (at 50 μM) was not toxic in P4 cells and in the T-lymphoid Jurkat and HUT78 cells (cell viability >90%, 24 h after a 1-h pulse treatment). In contrast, high proportions of dead cells were observed after treatment of T-lymphoid CEM and monocytic U937 cells (cell viabilities of 50 and 10%, respectively). With the aim of analyzing the effect of MG132 on a single round of viral infection, experiments were performed with a VSV-G pseudotype of the env-defective HIV-HSA reporter virus, which encodes the CD24 cell surface marker protein (23). Infected cells could be detected by measuring CD24 expression by flow cytometry. Susceptibility to HIV-HSA(VSV) infection varied from one cell line to another (Fig. 2). Twenty-four hours after a 1-h exposure to the virus (corresponding to 50 ng of p24), HeLa-CD4 and Jurkat cells showed 13 and 12% CD24+ cells, respectively. CEM and HUT78 were less susceptible (3 and 6% of CD24+ cells, respectively). Inhibition of the proteasome by MG132 induced a three- to fourfold increase in HIV infection in HeLa-CD4, Jurkat and CEM cells, but was ineffective in HUT78 cells. These experiments indicate that MG132 increases HIV replication in T-cell lines, although HUT78 cells appeared to be resistant to the activity of the compound. This resistance might be the consequence of a peculiar physiology of the proteasome or of impaired penetration of the drug in these cells.
FIG. 2.
Analysis of HIV-1 infection in the presence of MG132 on various cell lines. HeLa-CD4 cells and the Jurkat, CEM, and HUT78 T-lymphoid cell lines were infected with the HIV-HSA reporter virus pseudotyped with the VSV-G envelope, which contains, in the place of nef, the gene coding for HSA (CD24). Following integration and proviral expression, cells synthesize CD24, which can be detected at the cell surface. Cell lines were infected with HIV-HSA(VSV) (50 ng of p24Gag) for 1 h, with or without MG132 (50 μM). Infection was revealed 24 h later by staining with anti-CD24 monoclonal antibodies and flow cytometry analysis of gated living cells. Data are representative of three independent experiments.
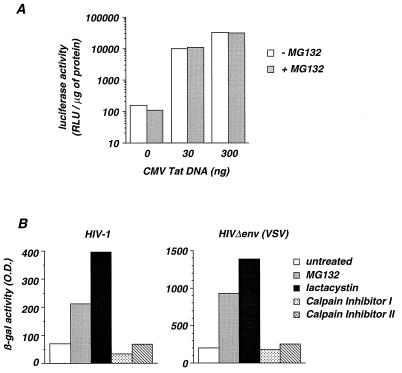
We then examined whether MG132 could act at a late step of the viral cycle, by increasing Tat-mediated transactivation of the HIV-1 LTR. P4 cells were transfected with the pLTRX-Luc plasmid, containing the luciferase reporter gene driven by the HIV-1 LTR, along with variable amounts of a Tat expression vector (pCMV-Tat). After 24 h, cells were incubated for 1 h in the absence or in the presence of MG132, and luciferase activity was measured 5 h later (Fig. 3A). In the absence of MG132, the LTR promoter yielded a baseline luciferase activity of 158 RLU, which rose to 9,963 and 31,644 RLU upon transfection of 30 and 300 ng of pCMV-Tat DNA, respectively. We did not observe any significant differences in both basal and Tat-induced luciferase activities when MG132 was included in the experiment (Fig. 3A). Similar results were obtained when MG132 was pulse-added for 1 h 17 h before measurement of luciferase activity (not shown). Therefore, inhibition of the proteasome does not significantly affect basal and Tat-induced transcriptional activities of the LTR.
FIG. 3.
Analysis of the effect of various protease inhibitors on HIV infection and on LTR activity. (A) Effect of MG132 on HIV-1 LTR activity. P4 cells were transfected with 1 μg of pLTRX-Luc and the indicated amounts of pCMV-Tat DNA. After 24 h, cells were pulse-incubated for 1 h with or without MG132 (50 μM), and 5 h later, luciferase activities in cell extracts were measured. Results are expressed as RLU per microgram of cellular protein. Values are means of triplicate determinations, and variation between each point was below 10%. (B) P4 cells were infected with HIV-1 (left panel) or HIVΔenv(VSV) (right panel) in the presence of the proteasome inhibitors MG132 (50 μM) or lactacystin (40 μM) or with calpain inhibitor I (50 μM) or calpain inhibitor II (50 μM) for 1 h and washed. Infections were revealed 24 h later by measuring β-galactosidase activity (optical density [O.D.] units). Data are representative of three independent experiments.
MG132 predominantly inhibits the proteasome but also affects, although with less efficiency, the cysteine proteases calpain and cathepsin B (33). We examined whether the inhibition of these cysteine proteases affects HIV infection. The calpain inhibitors I and II efficiently inhibit calpain and cathepsin B but have little effect on the proteasome (33, 36). In contrast, lactacystin is a specific and irreversible inhibitor of the chymotrypsin-like and trypsin-like activities of the proteasome (17). HIV infection was increased by MG132 and lactacystin (four- and sevenfold, respectively), whereas calpain inhibitors I and II had no effect (Fig. 3B). Therefore, inhibition of the proteasome and not of other cellular proteases increased HIV infection in P4 cells.
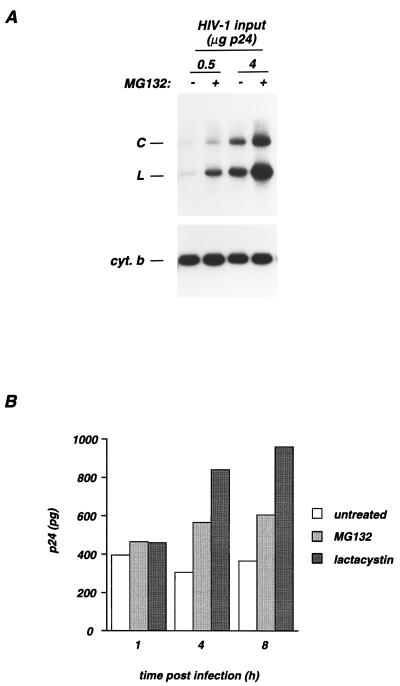
Since proteasome inhibitors affect HIV infection at early steps of the viral cycle, we monitored newly reverse-transcribed proviral DNA. P4 cells were incubated with HIV-1 for 5 h, with or without MG132. Nonintegrated viral DNA was extracted 17 h later and analyzed by Southern blotting (Fig. 4A). Samples were normalized for equal amounts of low-molecular-weight DNA by hybridization with the mitochondrial gene which codes for cytochrome b (Fig. 4A, lower panel). An HIV probe revealed viral linear DNA and one-LTR-circle (C) DNA (38) (Fig. 4A, upper panel). No signal was detected when target cells were treated with zidovudine, indicating that the hybridizing DNA was actually de novo synthesized during the infection period (not shown). In the absence of MG132, the intensity of the signal correlated with the amount of virus input, showing that the assay was conducted in its linear phase (Fig. 4A). Whatever the virus input (0.5 or 4 μg of p24), MG132 induced a fourfold increase in the amounts of proviral DNA. Viral DNA molecules undergoing circularization after their transport to the nucleus were used as a marker to monitor the nuclear import of preintegrative complexes (4). The ratios of circular to total viral DNA were equivalent with or without MG132. Thus, the proteasome inhibitor significantly increased proviral DNA synthesis and did not affect subsequent nuclear import of preintegration complexes.
FIG. 4.
MG132 induces an accumulation of proviral DNA and of cytosolic p24Gag proteins in target cells. (A) Synthesis of proviral DNA in newly infected cells. P4 cells were infected by a 5-h incubation period with HIV at the indicated amounts of p24Gag in the absence or in the presence of MG132 (25 μM). Seventeen hours later, low-molecular-weight DNA was extracted and analyzed by Southern blotting with an HIV probe (upper panel). Digestion with EcoRI produced diagnostic fragments with sizes of 5.7 and 9.1 kb from linear DNA (L) and DNA containing one LTR circle (C), respectively. Since variations in the yield of cellular low-molecular-weight DNA were observed, samples were normalized by hybridization with the mitochondrial gene coding for cytochrome b (cyt. b) (lower panel). (B) Accumulation of cytosolic p24Gag proteins. P4 cells were incubated with HIV-1 (1 μg of p24) in the absence (untreated columns) and in the presence of MG132 (50 μM) or lactacystin (40 μM) for 1 h at 37°C. Cells were then washed to remove unbound virus and further incubated at 37°C in medium containing the indicated inhibitors. At each time point, cells were treated with Pronase to eliminate virus adsorbed at the cell surface and lysed. Postnuclear supernatants were separated in cytosol and pellet fractions. The pellet fraction corresponds to cellular membranes and vesicles. p24Gag contents (in picograms) were measured in the cytosolic fraction. Time zero corresponds to the beginning of infection. Data are representative of three independent experiments.
We determined whether the proteasome affects the stability of incoming virions after entry in target cells. For this, we used an assay based on the measurement of cytosolic p24Gag contents, which reflects productive entry events (28). P4 cells were incubated for 1 h with HIV-1 in the absence or in the presence of MG132 (50 μM) or lactacystin (40 μM). At different periods of time following exposure, cytosolic p24Gag contents were measured (Fig. 4B). In order to remove virus particles adsorbed at the cell surface, cells were treated with Pronase before lysis. Lysates were then ultracentrifuged to purify cytosolic extracts (see Materials and Methods). In control cells, the amount of cytosolic p24Gag did not significantly change during the 8-h period following virus exposure. MG132 or lactacystin treatment induced an accumulation of cytosolic p24Gag (Fig. 4B), indicating that the proteasome affects the fate of incoming viral proteins.
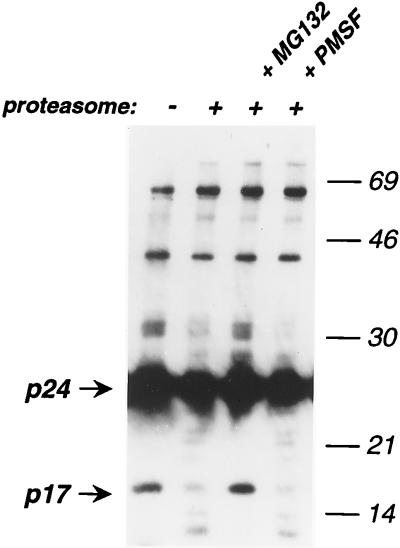
In vitro experiments were performed with catalytic 20S proteasome purified from rat liver (11). HIV-1 virions were pelleted by ultracentrifugation, resuspended in a buffer containing 0.4% Nonidet P-40 to remove viral membranes, and incubated with proteasome for 30 min at 37°C, in the absence or in the presence of MG132 (20 μM) or of the serine protease inhibitor phenylmethylsulfonyl fluoride (50 μM). Viral proteins were then revealed by immunoblotting (Fig. 5). In the absence of proteasome, mature p24Gag (CA) and p17Gag (MA) proteins, as well as Gag precursors (p66Gag) and intermediate cleavage products (p41Gag and p32Gag) were visualized. Incubation with proteasome decreased the amounts of p17Gag, p24Gag, and p32Gag proteins, coincident with the appearance of bands of smaller size, likely corresponding to degradation products. MG132, and not phenylmethylsulfonyl fluoride restored the normal viral protein profile. Interestingly, p66Gag and p41Gag precursors were not degraded by the proteasome. This inaccessibility to the proteolytic activity of the proteasome may result from either the localization of these proteins inside the virion, their oligomeric status, or both. In conclusion, this experiment shows that the proteolytic activity of the 20S proteasome has the ability to degrade components of HIV-1 particles in vitro.
FIG. 5.
In vitro degradation of HIV-1 Gag proteins by purified 20S proteasome. HIV-1 virions were pelleted by ultracentrifugation and resuspended in a buffer containing 0.4% Nonidet P-40 to remove the viral membrane. HIV-1 virions (10 ng of p24Gag) were then incubated with purified 20S proteasome (0.4 μg) for 30 min at 37°C, with or without MG132 (20 μM) or the serine protease inhibitor phenylmethylsulfonyl fluoride (PMSF) (50 μM). Viral proteins were then revealed in a Western blot assay with an anti-HIV-1 serum. Molecular mass markers are indicated (in kilodaltons) on the right. Data are representative of three independent experiments.
The proteasome has an essential antiviral function in vivo. Viral proteins synthetized in infected cells are partially degraded by the proteasome (19, 21, 30). Peptides generated during protein breakdown are bound to major histocompatibility complex class I molecules and delivered to the cell surface. Presentation of these peptides initiates the antiviral immune defenses. Our data indicate that the proteasome has an additional antiviral function at the cellular level, by affecting the fate of incoming viral proteins in naive cells.
Proteosomal degradation may require previous ATP-dependent ubiquitination of target proteins. The presence of ubiquitinated material in HIV particles (1) suggests that at least a fraction of incoming virions are potential targets for ubiquitin-dependent proteasomal degradation. Alternatively, viral proteins may serve as a target for ubiquitin-independent degradation by the 20S proteasome complex, as previously shown for misfolded or normal cellular proteins (8, 12, 24, 36). Although our experiments were performed in the presence of low detergent concentrations, which may affect viral protein conformation, the in vitro degradation of viral proteins by the 20S proteasome is consistent with this hypothesis.
Proteasome inhibitors were efficient when added during the first hours of infection and induced an accumulation of viral DNA and incoming cytosolic p24Gag proteins. On the other hand, MG132 did not affect Tat-induced transactivation of the LTR. Therefore, the proteasome acts on the early steps of the HIV replication cycle. During this still obscure phase of the viral cycle, virion cores are disassembled and reorganized in order to accomplish reverse transcription. Certain cell factors exert antiviral functions at this level. Alpha interferon is a potent inhibitor of HIV replication, acting at multiple steps of the virus cycle, including the initiation of reverse transcription (34, 35). Fv1, which is homologous to human endogenous provirus-like Gag products, inhibits murine leukemia virus replication at a stage after entry and before integration. Interestingly, Fv1 requires extremely low levels of expression to exert strong resistance to viral infection (3, 10, 18). This suggests that at the early phase of infection, the functional organization of incoming viral components is highly susceptible to surrounding host factors. The proteasome likely destabilizes this fragile organization, thus altering proviral DNA synthesis. This activity of the proteasome represents a previously undescribed early intracellular defense against viral infection.
ACKNOWLEDGMENTS
We thank P. Benaroch for critical reading of the manuscript and F. Baleux, N. Landau, and A. Miyanohara for the kind gift of reagents.
This work was supported by grants from the Agence Nationale de Recherche sur le SIDA (ANRS) and the Pasteur Institute.
REFERENCES
- 1.Arthur L O, Bess J W, Sowder II R C, Benveniste R E, Mann D L, Chermann J C, Henderson L E. Cellular proteins bound to immunodeficiency viruses: implications for pathogenesis and vaccines. Science. 1992;258:1935–1938. doi: 10.1126/science.1470916. [DOI] [PubMed] [Google Scholar]
- 2.Arts E J, Wainberg M A. Human immunodeficiency virus type 1 reverse transcriptase and early events in reverse transcription. Adv Virus Res. 1996;46:97–163. doi: 10.1016/s0065-3527(08)60071-8. [DOI] [PubMed] [Google Scholar]
- 3.Best S, Le Tissier P, Towers G, Stoye J P. Positional cloning of the mouse retrovirus restriction gene Fv1. Nature. 1996;382:826–829. doi: 10.1038/382826a0. [DOI] [PubMed] [Google Scholar]
- 4.Brown P O, Bowerman B, Varmus H E, Bishop J M. Correct integration of retroviral DNA in vitro. Cell. 1987;49:347–356. doi: 10.1016/0092-8674(87)90287-x. [DOI] [PubMed] [Google Scholar]
- 5.Bukrinsky M I, Shavora N, McDonald T L, Pushkarskaya T, Tarpley W G, Stevenson M. Association of integrase, matrix, and reverse transcriptase antigens of human immunodeficiency virus type 1 with viral nucleic acids following acute infection. Proc Natl Acad Sci USA. 1993;90:6125–6129. doi: 10.1073/pnas.90.13.6125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Burns J C, Friedmann T, Driever W, Burrascano M, Yee J-K. Vesicular stomatitis virus G glycoprotein pseudotyped retroviral vectors: concentration to very high titer and efficient gene transfer into mammalian and non mammalian cells. Proc Natl Acad Sci USA. 1993;90:8033–8037. doi: 10.1073/pnas.90.17.8033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Charneau P, Mirabeau G, Roux P, Paulous S, Buc H, Clavel F. HIV-1 reverse transcription: a termination step at the center of the genome. J Mol Biol. 1994;241:651–662. doi: 10.1006/jmbi.1994.1542. [DOI] [PubMed] [Google Scholar]
- 8.Ciechanover A, Schwartz A L. The ubiquitin-mediated proteolytic pathway: mechanisms of recognition of the proteolytic substrate and involvement in the degradation of native cellular proteins. FASEB J. 1994;8:182–191. doi: 10.1096/fasebj.8.2.8119489. [DOI] [PubMed] [Google Scholar]
- 9.Clapham P R. HIV and chemokines: ligands sharing cell-surface receptors. Trends Cell Biol. 1997;7:264–268. doi: 10.1016/S0962-8924(97)01075-1. [DOI] [PubMed] [Google Scholar]
- 10.Coffin J. Retrovirus restriction revealed. Nature. 1996;382:762–763. doi: 10.1038/382762a0. [DOI] [PubMed] [Google Scholar]
- 11.Conconi M, Szweda L I, Levine R L, Stadtman E R, Friguet B. Age-related decline of rat liver multicatalytic proteinase activity and protection from oxidative inactivation by heat-shock protein 90. Arch Biochem Biophys. 1996;331:232–240. doi: 10.1006/abbi.1996.0303. [DOI] [PubMed] [Google Scholar]
- 12.Coux O, Tanaka K, Goldberg A L. Structure and functions of the 20S and 26S proteasomes. Annu Rev Biochem. 1996;65:801–847. doi: 10.1146/annurev.bi.65.070196.004101. [DOI] [PubMed] [Google Scholar]
- 13.Drexler H C. Activation of the cell death program by inhibition of proteasome function. Proc Natl Acad Sci USA. 1997;94:855–860. doi: 10.1073/pnas.94.3.855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.D’Souza M P, Harden V A. Chemokines and HIV-1 second receptors. Nat Med. 1996;2:1293–1300. doi: 10.1038/nm1296-1293. [DOI] [PubMed] [Google Scholar]
- 15.Farnet C, Haseltine W A. Determination of viral proteins present in the human immunodeficiency virus type 1 preintegration complex. J Virol. 1991;65:1910–1915. doi: 10.1128/jvi.65.4.1910-1915.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Farnet C M, Bushman F D. HIV-1 cDNA integration—requirement of HMG I(Y) protein for function of preintegration complexes in vitro. Cell. 1997;88:483–492. doi: 10.1016/s0092-8674(00)81888-7. [DOI] [PubMed] [Google Scholar]
- 17.Fenteany G, Standaert R F, Lane W S, Choi S, Corey E J, Schreiber S L. Inhibition of proteasome activities and subunit-specific amino-terminal threonine modification by lactacystin. Science. 1995;268:726–731. doi: 10.1126/science.7732382. [DOI] [PubMed] [Google Scholar]
- 18.Goff S P. Operating under a Gag order: a block against incoming virus by the Fv1 gene. Cell. 1996;86:691–693. doi: 10.1016/s0092-8674(00)80141-5. [DOI] [PubMed] [Google Scholar]
- 19.Goldberg A L, Rock K L. Proteolysis, proteasomes and antigen presentation. Nature. 1992;357:375–379. doi: 10.1038/357375a0. [DOI] [PubMed] [Google Scholar]
- 20.Grimm L M, Goldberg A L, Poirier G G, Schwartz L M, Osborne B A. Proteasomes play an essential role in thymocyte apoptosis. EMBO J. 1996;15:3835–3844. [PMC free article] [PubMed] [Google Scholar]
- 21.Groettrup M, Soza A, Kuckelkorn U, Kloetzel P M. Peptide antigen production by the proteasome: complexity provides efficiency. Immunol Today. 1996;17:429–435. doi: 10.1016/0167-5699(96)10051-7. [DOI] [PubMed] [Google Scholar]
- 22.Groll M, Ditzel L, Lowe J, Stock D, Bochtler M, Bartunik H D, Huber R. Structure of 20S proteasome from yeast at 2.4 Å resolution. Nature. 1997;386:463–471. doi: 10.1038/386463a0. [DOI] [PubMed] [Google Scholar]
- 23.He J, Choe S, Walker R, Di Marzio P, Morgan D O, Landau N R. Human immunodeficiency virus type 1 viral protein R (Vpr) arrests cells in the G2 phase of the cell cycle by inhibiting p34cdc2 activity. J Virol. 1995;69:6705–6711. doi: 10.1128/jvi.69.11.6705-6711.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hochstrasser M. Ubiquitin, proteasomes, and the regulation of intracellular protein degradation. Curr Opin Cell Biol. 1995;7:215–223. doi: 10.1016/0955-0674(95)80031-x. [DOI] [PubMed] [Google Scholar]
- 25.Imajoh-Ohmi S, Kawaguchi T, Sugiyama S, Tanaka K, Omura S, Kikuchi H. Lactacystin, a specific inhibitor of the proteasome, induces apoptosis in human monoblast U937 cells. Biochem Biophys Res Commun. 1995;217:1070–1077. doi: 10.1006/bbrc.1995.2878. [DOI] [PubMed] [Google Scholar]
- 26.Karageorgos L, Li P, Burrel C. Characterization of HIV replication complexes early after cell-to-cell transmission. AIDS Res Hum Retroviruses. 1993;9:817–823. doi: 10.1089/aid.1993.9.817. [DOI] [PubMed] [Google Scholar]
- 27.Mansky L M. Accessory replication proteins and the accuracy of reverse transcription: implications for retroviral genetic diversity. Trends Genet. 1997;13:134–136. doi: 10.1016/s0168-9525(97)01062-7. [DOI] [PubMed] [Google Scholar]
- 28.Maréchal V, Clavel F, Heard J M, Schwartz O. Cytosolic Gag p24 as an index of productive human immunodeficiency virus type 1 entry. J Virol. 1998;72:2208–2212. doi: 10.1128/jvi.72.3.2208-2212.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Matlin K S, Reggio H, Helenius A, Simons K. Pathway of vesicular stomatitis virus entry leading to infection. J Mol Biol. 1982;156:609–631. doi: 10.1016/0022-2836(82)90269-8. [DOI] [PubMed] [Google Scholar]
- 30.Michalek M T, Grant E P, Gramm C, Goldberg A L, Rock K L. A role for the ubiquitin-dependent proteolytic pathway in MHC class-I-restricted antigen presentation. Nature. 1993;363:552–554. doi: 10.1038/363552a0. [DOI] [PubMed] [Google Scholar]
- 31.Miyanohara A, Yee J K, Bouic K, Laporte P, Friedmann T. Efficient in vivo transduction of the neonatal mouse liver with pseudotyped retroviral vectors. Gene Ther. 1995;2:138–142. [PubMed] [Google Scholar]
- 32.Oberlin E, Amara A, Bachelerie F, Bessia C, Virelizier J L, Arenzana-Seisdedos F, Schwartz O, Heard J M, Clark-Lewis I, Legler D F, Loetscher M, Baggiolini M, Moser B. The CXC chemokine, stromal cell derived factor 1 (SDF-1), is the ligand for LESTR/fusin and prevents infection by lymphocyte-tropic HIV-1 syncytium-inducing strains. Nature. 1996;382:833–835. doi: 10.1038/382833a0. [DOI] [PubMed] [Google Scholar]
- 33.Palombella V J, Rando O J, Goldberg A L, Maniatis T. The ubiquitin-proteasome pathway is required for processing the NF-kB1 precursor protein and the activation of NF-kB. Cell. 1994;78:773–785. doi: 10.1016/s0092-8674(94)90482-0. [DOI] [PubMed] [Google Scholar]
- 34.Pitha P M. Multiple effects of interferon on the replication of human immunodeficiency virus type 1. Antivir Res. 1994;24:205–219. doi: 10.1016/0166-3542(94)90068-x. [DOI] [PubMed] [Google Scholar]
- 35.Poli G, Biswas P, Fauci A S. Interferons in the pathogenesis and treatment of human immunodeficiency virus infection. Antivir Res. 1994;24:221–233. doi: 10.1016/0166-3542(94)90069-8. [DOI] [PubMed] [Google Scholar]
- 36.Rock K L, Gramm C, Rothstein L, Clark K, Stein R, Dick L, Hwang D, Goldberg A L. Inhibitors of the proteasome block the degradation of most cell proteins and the generation of peptides presented on MHC I molecules. Cell. 1994;78:761–771. doi: 10.1016/s0092-8674(94)90462-6. [DOI] [PubMed] [Google Scholar]
- 37.Roff M, Thompson J, Rodriguez M S, Jacque J M, Baleux F, Arenzana-Seisdedos F, Hay R T. Role of IkB alpha ubiquitination in signal-induced activation of NF-kB in vivo. J Biol Chem. 1996;271:7844–7850. doi: 10.1074/jbc.271.13.7844. [DOI] [PubMed] [Google Scholar]
- 38.Schwartz O, Maréchal V, Danos O, Heard J-M. Human immunodeficiency virus type 1 Nef increases the efficiency of reverse transcription in the infected cell. J Virol. 1995;69:4053–4059. doi: 10.1128/jvi.69.7.4053-4059.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schwartz O, Virelizier J L, Montagnier L, Hazan U. A microtransfection method using luciferase gene for the study of human immunodeficiency virus long terminal repeat activity. Gene. 1990;88:197–205. doi: 10.1016/0378-1119(90)90032-m. [DOI] [PubMed] [Google Scholar]
- 40.Weiss R A. Cellular receptors and viral glycoproteins involved in retrovirus entry. In: Levy J A, editor. The retroviridae. Vol. 2. New York, N.Y: Plenum Press; 1993. pp. 1–108. [Google Scholar]
- 41.Whitcomb J M, Hughes S H. Retroviral reverse transcription and integration: progress and problems. Annu Rev Cell Biol. 1992;8:275–306. doi: 10.1146/annurev.cb.08.110192.001423. [DOI] [PubMed] [Google Scholar]