Abstract
The hepatitis C virus (HCV) genome encodes two envelope glycoproteins (E1 and E2) which interact noncovalently to form a heterodimer (E1-E2). During the folding and assembly of HCV glycoproteins, a large portion of these proteins are trapped in aggregates, reducing the efficiency of native E1-E2 complex assembly. To better understand this phenomenon and to try to increase the efficiency of HCV glycoprotein folding, endoplasmic reticulum chaperones potentially interacting with these proteins were studied. Calnexin, calreticulin, and BiP were shown to interact with E1 and E2, whereas no interaction was detected between GRP94 and HCV glycoproteins. The association of HCV glycoproteins with calnexin and calreticulin was faster than with BiP, and the kinetics of interaction with calnexin and calreticulin were very similar. However, calreticulin and BiP interacted preferentially with aggregates whereas calnexin preferentially associated with monomeric forms of HCV glycoproteins or noncovalent complexes. Tunicamycin treatment inhibited the binding of HCV glycoproteins to calnexin and calreticulin, indicating the importance of N-linked oligosaccharides for these interactions. The effect of the co-overexpression of each chaperone on the folding of HCV glycoproteins was also analyzed. However, the levels of native E1-E2 complexes were not increased. Together, our data suggest that calnexin plays a role in the productive folding of HCV glycoproteins whereas calreticulin and BiP are probably involved in a nonproductive pathway of folding.
Hepatitis C virus (HCV) is a positive-strand RNA virus which belongs to the Flaviviridae family (16). Its genome contains a long open reading frame of 9,030 to 9,099 nucleotides that is translated into a single polyprotein of 3,010 to 3,033 amino acids (39). Cleavages of this polyprotein are co- and posttranslational and generate at least 10 polypeptides including 2 glycoproteins, E1 and E2 (54). Since the molecular cloning of HCV (4), characterization of its genomic organization and expression has progressed rapidly. However, despite this progress, data on the HCV life cycle remain scarce. This is due to the poor replication of HCV in cell culture.
HCV glycoproteins E1 and E2 interact to form complexes (10, 20, 34, 53). Characterization of HCV glycoprotein complex formation indicates that a majority of these proteins are misfolded aggregates (6, 10). Since analysis of HCV glycoprotein assembly in viral (10) and nonviral (11) expression systems showed similar results, this tendency toward aggregation does not seem to be due to abnormally high-level production driven by the viral expression systems used. This suggests that their tendency toward aggregation could be an intrinsic property of HCV glycoproteins. Recently, we produced a monoclonal antibody (MAb) which recognizes properly folded E2 and precipitates native HCV glycoprotein complexes but not misfolded aggregates (6). We have shown that properly folded E1 and E2 interact to form a heterodimer stabilized by noncovalent interactions. Formation of stable E1-E2 complexes is slow (t1/2 ≈ 2 h) because of the slow folding of these proteins. Indeed, formation of intramolecular disulfide bonds is slow for E1 (t1/2 > 1 h) (12), and unidentified events following the acquisition of intramolecular disulfide bonds limit the folding of E2 (6, 12, 21 42). In addition, E1 expressed in the absence of E2 does not fold properly, suggesting that E2 plays a chaperone-like role in the folding of E1 (42).
The lack of an efficient system for cell culture replication has so far hampered our understanding of HCV particle assembly. However, the absence of complex glycans, the localization of expressed HCV glycoproteins in the endoplasmic reticulum (ER) (10, 53), and the absence of these proteins on the cell surface (10, 61) suggest that initial virion morphogenesis may occur by budding into intracellular vesicles. More recently, we have confirmed that mature E1-E2 heterodimers do not leave the ER (6), and an ER retention signal has been identified in the C-terminal 29 amino acids of E2 (5).
Folding into a three-dimensional structure in the cell presents a polypeptide chain with some obstacles. First, the nascent polypeptide chain is gradually exposed as it emerges from the ribosome during translation or from the membrane during translocation. Second, many polypeptides are assembled into oligomeric complexes in vivo. Finally, they must fold in a concentrated protein solution that presents many opportunities for inappropriate associations. To overcome these hurdles, the cell expresses molecular chaperones and enzymes responsible for preventing misassociations and facilitating proper folding under intracellular conditions. The ER is a specialized compartment devoted to the maturation of membrane and secretory proteins (23). Along with folding enzymes, such as protein disulfide isomerases and prolyl cis-trans isomerases (19), the ER contains chaperones including immunoglobulin heavy-chain binding protein (BiP or GRP78) (24), GRP94 (28), calnexin (1), and calreticulin (47, 51, 63).
BiP is a soluble member of the heat shock protein 70 (HSP70) family of chaperones (46), which has been shown to associate transiently with folding intermediates of many viral membrane proteins (13, 18, 27, 36). GRP94 is a member of the heat shock protein 90 (HSP90) family of chaperones (28). Based on its association with unassembled oligomeric protein substrates, such as immunoglobulin chains, major histocompatibility complex class II molecules, and a mutant form of the herpes simplex virus type 1 glycoprotein B, it has been proposed that GRP94 acts as a molecular chaperone (40, 41, 48, 58). Calnexin and calreticulin bind selectively and transiently to newly synthesized glycoproteins (22, 50, 51, 63). Their preference for glycoproteins is based on a lectin-like affinity for monoglucosylated N-linked oligosaccharides (Glc1Man9GlcNAc2) (22, 26, 51, 64). The binding of substrate glycoproteins to and release from calnexin and calreticulin depend on trimming and reglucosylation of the N-linked glycans (22, 26). The monoglucosylated oligosaccharides are generated by trimming the two outermost glucose residues from the core oligosaccharides (31). Glucosidase I removes the first of the three glucoses, and glucosidase II removes the second and eventually the third. Monoglucosylated glycans can also be generated by UDP-glucose:glycoprotein glucosyltransferase, a luminal enzyme which adds a glucose residue to glucose-free, high-mannose chains of incompletely folded glycoproteins (59, 60). In a recent model (22, 51), the three enzymes, together with calnexin and calreticulin, provide an ER-specific folding and retention machinery. Calnexin and calreticulin are referred to as molecular chaperones, but there is little direct data that they actually influence the folding of glycoprotein substrates. A recent report suggests that calnexin, at least, could act exclusively as a lectin (66).
In this study, we investigated the interactions of ER chaperones with HCV glycoproteins. We showed that BiP, calnexin, and calreticulin interact with E1 and E2, whereas no interaction could be identified between GRP94 and HCV glycoproteins. The kinetics of association of HCV glycoproteins with calnexin, calreticulin, and BiP were determined. The effect of glycosylation inhibitors such as tunicamycin and castanospermine (CST) on chaperone binding and folding was also studied. In addition, vaccinia virus recombinants expressing calnexin, calreticulin, or BiP were constructed and used to coexpress them with E1 and E2 and to analyze the effects of the overexpression of each chaperone on the folding of HCV glycoproteins.
MATERIALS AND METHODS
Cell culture.
The HepG2, CV-1, BHK-21, and 143B (thymidine kinase-deficient) cell lines were obtained from the American Type Culture Collection, Rockville, Md. Cell monolayers were grown in alpha Eagle’s minimal essential medium (alpha MEM; BHK-21) or Dulbecco’s modified MEM (HepG2, CV-1, and 143B), both supplemented with 10% fetal bovine serum (FBS).
Plasmid constructs.
Plasmids expressing ER chaperones were constructed by standard methods (57). Sequences encoding calnexin (52), calreticulin (14), and BiP (32) were amplified by PCR to position a 5′ NcoI (calnexin and calreticulin) or AflIII (BiP) site and a 3′ XhoI site. The PCR products were digested with NcoI and XhoI or with AflIII and XhoI and cloned into pTM1 (44) to obtain plasmids pTM1/CNX (calnexin), pTM1/CRT (calreticulin), and pTM1/BiP. BiP expressed from pTM1/BiP contains two additional amino acids (Met and Leu) at the N terminus of its signal sequence. The regions of these plasmids amplified by PCR were verified by sequencing.
Generation and growth of viruses.
Vaccinia virus recombinants were generated by homologous recombination essentially as described previously (29) and plaque purified twice on thymidine kinase-deficient 143B cells under bromodeoxyuridine selection (50 μg/ml). Stocks of vTF7-3 (a vaccinia virus recombinant expressing the T7 DNA-dependent RNA polymerase) (17), the wild-type vaccinia virus Copenhagen strain and its thermosensitive ts7 derivative (9), and vaccinia virus recombinants were grown and subjected to titer determination on CV-1 monolayers.
Vaccinia virus-HCV recombinants vHCV1–1488 (expressing C-E1-E2-p7-NS2-NS3181), vHCV170–809 (expressing E1-E2-p7), vHCV371–809 (expressing E2-p7), and vHCV1–383 (expressing C-E1) (15, 20, 42) and a vaccinia virus recombinant generated by recombination with pTM1 lacking a foreign sequence (vpTM1 [38]) have been described previously.
Antibodies.
Anti-HCV E1 (A4) and E2 (A11 and H2) MAbs have been described previously (6, 10) and were produced in vitro by using a MiniPerm apparatus (Heraeus) as recommended by the manufacturer. Anti-BiP antibodies were purchased from Stress Gen (MAb 10C3; SPA-827) and Affinity Bioreagents (PA1-014) or kindly provided by M.-G. Gething (University of Texas, Dallas, Tex.). Anti-calreticulin antibodies were supplied by Affinity Bioreagents (PA3-900) or kindly provided by M. Michalak (University of Alberta, Edmonton, Canada). Anti-GRP94 rat MAb (SPA-850) and anti-calnexin antibodies (SPA-860) were supplied by Stress Gen.
Metabolic labeling and immunoprecipitation.
Subconfluent monolayers in 35-mm dishes were infected with the appropriate recombinant at a multiplicity of infection of 5 to 10 PFU per cell. After 1 h at room temperature, medium containing 5% FBS was added. Between 4 and 4.5 h postinfection, monolayers were washed once with prewarmed medium lacking methionine and cysteine and incubated in the same medium for an additional 30 min. Infected cells were then pulse-labeled for 5 or 15 min with 100 μCi of 35S-protein labeling mix (Dupont, NEN) per ml. The cells were washed twice with prewarmed medium containing a 10-fold excess of both methionine and cysteine and then subjected to a chase for various times. The cells were then lysed with 0.5% Triton X-100 in 20 mM Tris-Cl (pH 7.5)–150 mM NaCl–2 mM EDTA (TBS). Iodoacetamide (20 mM) was included in the lysis buffer for experiments in which disulfide bond formation was being investigated. Cell lysates were clarified by centrifugation in an Eppendorf centrifuge for 5 min at 4°C. In steady-state labeling, the cells were labeled, at 4 h postinfection, with 50 μCi of 35S-protein labeling mix per ml in medium containing 1/40 the normal concentration of methionine and 2% FBS.
Immunoprecipitations were carried out as described previously (12). A 6-μl portion of rabbit anti-mouse immunoglobulin G IgG (Dako) was incubated with protein A-Sepharose (Pharmacia-LKB) for 1 h at 4°C in TBS containing 0.2% Triton X-100 (TBS-T). This step was omitted when polyclonal antibodies were used. The beads were then incubated with 10 μl of MAb or polyclonal antibody, followed by the antigen (each step was performed for 1 h at 4°C). Between each step, the beads were washed once with TBS-T. After the last step, they were washed three times with TBS-T and once with TBS. The precipitates were then boiled for 5 min in sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) sample buffer (under nonreducing conditions, β-mercaptoethanol was omitted) and run on a 10 or 12% polyacrylamide gel (33). After electrophoresis, the gels were treated with sodium salicylate (3), dried, and exposed at −70°C to preflashed Hyperfilm-MP (Amersham). 14C-methylated protein molecular mass markers were purchased from Amersham. For quantitative experiments, the gels were analyzed with a PhosphorImager (Molecular Dynamics). 3-[(Cholamidopropyl)-dimethylammonio]-1-propanesulfonate (CHAPS) and digitonin (1%; Boehringer Mannheim Biochemicals) were used instead of Triton X-100 to solubilize the cells in some experiments. For detection of interactions between BiP and HCV glycoproteins, lysis buffer lacking EDTA was supplemented with apyrase (20 U/ml; Sigma) to deplete the medium of ATP. In some experiments, the cells were treated with tunicamycin (Boehringer Mannheim) or CST (Boehringer Mannheim). Tunicamycin (5 μg/ml) or CST (1 mM) was present during the methionine and cysteine deprivation, the pulse, and the chase.
Western blotting.
Analysis of proteins bound to nitrocellulose membranes (Hybond-ECL; Amersham) was performed by using enhanced chemiluminescence detection (Amersham) as recommended by the manufacturer. Briefly, after separation by SDS-PAGE, proteins were transferred to nitrocellulose membranes by using a Trans-Blot apparatus (Bio-Rad) and revealed with specific antibodies (dilution, 1/1,000) followed by goat anti-mouse or swine anti-rabbit immunoglobulin conjugated to horseradish peroxidase (dilution, 1/1,000; Dako).
RESULTS
Identification of ER chaperones interacting with HCV glycoproteins.
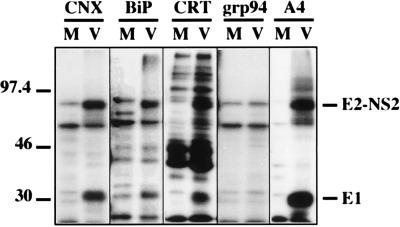
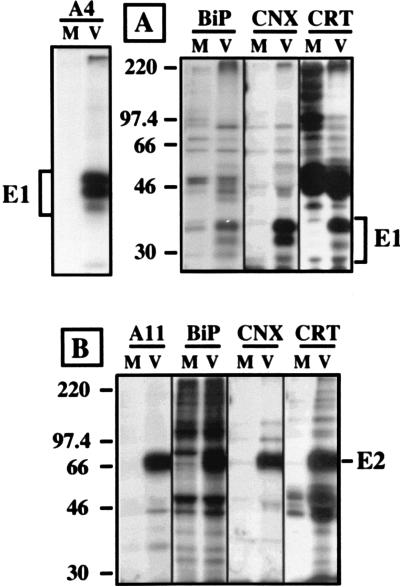
Recently, we reported that calnexin transiently associates with HCV glycoproteins (12). To analyze the potential role of other ER chaperones in early steps of HCV glycoprotein folding, cells infected with a vaccinia virus recombinant expressing a truncated form of HCV polyprotein ending after the serine protease domain of NS3 were pulse-labeled for 15 min, lysed, and immunoprecipitated with anti-BiP, anti-calnexin, anti-calreticulin, or anti-GRP94 antibodies. In addition to other unidentified proteins also present in the control, two HCV proteins, E1 and a precursor of E2 (E2-NS2), coprecipitated with BiP, calnexin, and calreticulin but not with GRP94 (Fig. 1). The absence of coprecipitation of HCV glycoproteins with GRP94 was confirmed by immunoprecipitation with anti-E1 or anti-E2 MAbs followed by Western blotting with a GRP94-specific antibody (data not shown). The stability of potential interactions between HCV glycoproteins and GRP94 could be influenced by the detergent used, and, in addition to Triton X-100, other mild detergents (CHAPS and digitonin) were tested to lyse the cells. However, none of these detergents allowed us to detect interactions between GRP94 and HCV glycoproteins (data not shown), suggesting that GRP94 is not involved in the folding of HCV glycoproteins or that the GRP94-E1 and GRP94-E2 complexes are labile in our experimental conditions.
FIG. 1.
Coprecipitation of HCV glycoproteins with ER chaperones. BHK-21 cells were coinfected with vTF7-3 and vHCV1-1488 (lanes V) or vTF7-3 and vpTM1 (M). At 4.5 h postinfection, infected cells were labeled for 15 min with [35S]methionine and lysed with Triton X-100. Cell lysates were used for immunoprecipitation with anti-E1 (control A4), anti-calnexin (CNX), anti-calreticulin (CRT), anti-BiP, or anti-GRP94 antibodies. Immunoprecipitates were analyzed by SDS-PAGE (10% polyacrylamide). The sizes (in kilodaltons) of molecular mass markers are indicated on the left.
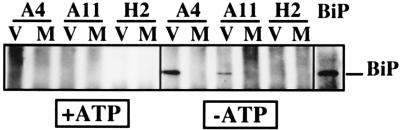
Interactions between HCV glycoproteins and BiP were also studied by immunoprecipitation with anti-HCV glycoprotein MAbs followed by Western blotting with an anti-BiP antibody. As shown in Fig. 2, interactions between BiP and HCV glycoproteins were confirmed when HCV glycoproteins were precipitated with conformation-insensitive MAbs A4 (anti-E1) and A11 (anti-E2) but not with our conformation-sensitive MAb H2. This was expected since MAb H2 recognizes properly folded E2 and precipitates native HCV glycoprotein complexes but not misfolded aggregates (6). Cell lysis was also performed in the presence of excess ATP (Fig. 2). In this case, coprecipitation of BiP with HCV glycoproteins was dramatically reduced. This latter result is in agreement with previous data showing that the addition of ATP to cell extracts causes the disruption of BiP-substrate complexes (45).
FIG. 2.
Western blotting analysis of the interaction between BiP and HCV glycoproteins. BHK-21 cells were coinfected with vTF7-3 and vHCV1-1488 (V) or vTF7-3 and pTM1 (M). At 5 h postinfection, infected cells were lysed with Triton X-100 in buffer containing or lacking 1 mM ATP. Cell lysates were used for immunoprecipitation with anti-E1 (MAb A4), anti-E2 (MAb A11 and H2) or anti-BiP antibodies. Immunoprecipitates were separated by SDS-PAGE (10% polyacrylamide) and revealed by Western blotting with an anti-BiP MAb.
Interactions between HCV glycoproteins and calreticulin were studied as above by immunoprecipitation with anti-HCV glycoprotein MAbs followed by Western blotting with an anti-calreticulin antibody. However, since this chaperone migrates close to the immunoglobulin heavy chain, the two bands could not be resolved (data not shown).
Kinetics of HCV glycoprotein association with calnexin, calreticulin, or BiP.
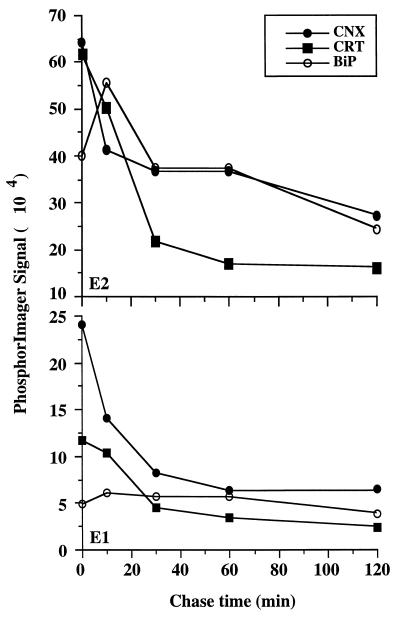
To determine the rate of HCV glycoprotein interactions with calnexin, calreticulin, or BiP, pulse-chase experiments were performed, cell lysates were immunoprecipitated with anti-calnexin, anti-calreticulin, or anti-BiP antibodies, and the coprecipitated HCV glycoproteins were quantified (Fig. 3). The maximum amounts of E1 and E2 that coprecipitated with anti-calnexin or anti-calreticulin antibodies were reproducibly observed during the pulse-labeling, suggesting that HCV glycoproteins associate rapidly with these chaperones. The intensity of labeled HCV glycoproteins associated with calnexin and calreticulin decreased rapidly during the first 30 min of the chase and then decreased slowly until the end of the chase (Fig. 3). Although the kinetics of association between HCV glycoproteins and calnexin or calreticulin were very similar, the maximum amounts of HCV glycoproteins that coprecipitated after 2 h of chase were smaller for calreticulin. Indeed, only about 20% (E1) and 30% (E2) of the maximum amounts of HCV glycoproteins that coprecipitated with anti-calreticulin antibody were still detected after 2 h of the chase, compared to about 30% (E1) or 50% (E2) of the amounts that coprecipitated with anti-calnexin antibody at the same time. The maximum amount of E2 that coprecipitated with anti-BiP antibody was reproducibly observed after a 10-min chase (Fig. 3), suggesting that E2 associates more slowly with BiP than with calreticulin or calnexin. The intensity of labeled E2 associated with BiP decreased very slowly until the end of the chase. The amount of E1 associated with BiP was small and remained rather constant until the end of the chase. The maximum amounts of E2 that coprecipitated with the chaperones reached about 75% of the total labeled protein for calnexin and calreticulin and 70% for BiP. The maximum amounts of E1 that coprecipitated with calnexin, calreticulin, and BiP were about 40, 20, and 10%, respectively.
FIG. 3.
Kinetics of HCV glycoprotein association with calnexin, calreticulin, and BiP. BHK-21 cells coinfected with vHCV170-809 and vTF7-3 were pulse-labeled for 5 min with [35S]methionine and chased for the indicated times. Cell lysates were used for immunoprecipitation with anti-BiP, anti-calnexin, or anti-calreticulin antibodies. Proteins were separated by SDS-PAGE, and quantifications of HCV glycoproteins coprecipitated with chaperones were performed with a PhosphorImager.
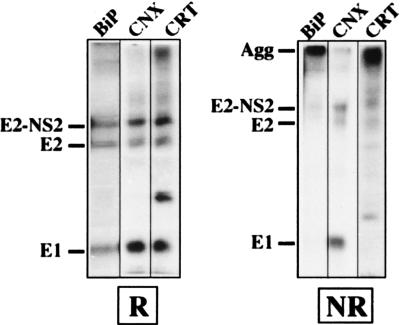
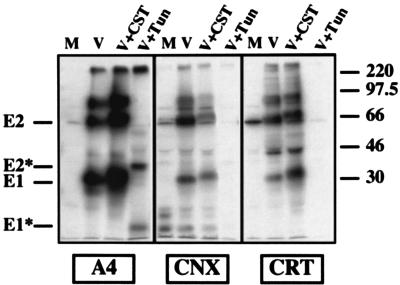
Since a large portion of HCV glycoproteins is involved in nonproductive interactions leading to the formation of large aggregates, we analyzed whether some of these ER chaperones would preferentially bind to such aggregates. However, to reduce the background due to the coprecipitation of other labeled proteins, proteins associated with ER chaperones were released by a mild treatment as previously described (12) and HCV proteins were reprecipitated with anti-E1 or anti-E2 antibodies. Under nonreducing conditions, HCV glycoproteins associated with calnexin were revealed as monomeric bands corresponding to E1 and E2 whereas those associated with calreticulin and BiP antibodies were aggregates (Fig. 4 and data not shown). Similar results were observed at different times of the chase (data not shown). This suggests that proteins recognized by these three chaperones are not in the same state of folding.
FIG. 4.
Analysis of HCV glycoproteins associated with BiP, calnexin (CNX), and calreticulin (CRT) under nonreducing conditions. BHK-21 cells coinfected with vTF7-3 and vHCV1-1488 were pulse-labeled for 15 min with [35S]methionine, chased for 10 min, and lysed with Triton X-100. Cell lysates were used for immunoprecipitation with anti-chaperone antibodies, and proteins associated with these chaperones were released by heating for 5 min at 37°C in 0.5% Nonidet P-40 and reprecipitated with MAb A4 (anti-E1). Immunoprecipitates were analyzed by SDS-PAGE (10% polyacrylamide) under reducing (R) or nonreducing (NR) conditions. Agg, aggregates.
Both E1 and E2 interact with ER chaperones.
Since the HCV glycoproteins E1 and E2 interact to form E1-E2 complexes (10), coprecipitation of E1 or E2 with anti-chaperone antibodies could reflect an interaction with either or both glycoproteins (or an unlabeled cell component). The ability of each glycoprotein to interact with calnexin, calreticulin, or BiP was analyzed by immunoprecipitation of cells infected by vaccinia virus recombinants expressing E1 or E2 alone. As shown in Fig. 5, E1 expressed in the absence of E2 coprecipitated with anti-calnexin, anti-calreticulin, or anti-BiP antibodies. Similarly, E2 expressed in the absence of E1 coprecipitated with the same anti-chaperone antibodies. These results suggest that each of the HCV glycoproteins can interact with these three chaperones. However, when E1 and E2 were coexpressed, BiP and calreticulin interacted with disulfide-bond complexes composed of E1 plus E2 (Fig. 4, NR). Monomeric forms of E1 or E2 can interact with calnexin early after their synthesis, whereas E1-E2 complexes are observed later during the chase (12). E1-E2 complexes associated with calnexin are detected after 10 or 15 min of the chase (Fig. 4; data not shown), whereas complexes recognized by MAb H2 start to be detected after 45 or 60 min of the chase (reference 6 and data not shown), indicating that noncovalent E1-E2 complexes can be formed before the proteins acquire their final state of folding.
FIG. 5.
Interaction of calnexin (CNX), BiP, or calreticulin (CRT) with HCV glycoprotein E1 (A) or E2 (B) expressed alone. BHK-21 cells were coinfected with vTF7-3 and vHCV1-383 (V, panel A) or vHCV371-809 (V, panel B) or vpTM1 (M). At 4 h postinfection, the cells were labeled for 15 min with [35S]methionine and lysed with Triton X-100. Cell lysates were used for immunoprecipitation with anti-chaperone (CNX, BiP, and CRT), anti-E1 (MAb A4), or anti-E2 (MAb A11) antibodies. The immunoprecipitates were analyzed by SDS-PAGE. The sizes (in kilodaltons) of molecular mass markers are indicated on the left.
Folding of HCV glycoproteins and association with ER chaperones in the presence of tunicamycin and CST.
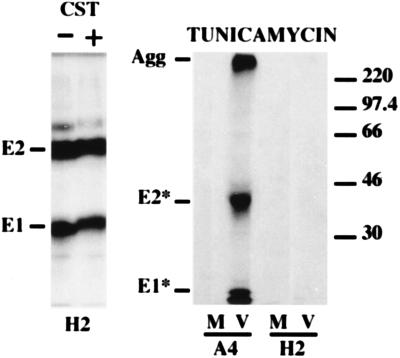
Although protein-protein interactions cannot be excluded (30, 35, 62), recent studies have confirmed that calnexin and calreticulin bind monoglucosylated N-linked oligosaccharides (55, 64). Tunicamycin blocks core glycosylation of nascent glycoprotein precursors. Therefore, we studied the effect of tunicamycin on the interactions of HCV glycoproteins with calnexin and calreticulin. In the presence of tunicamycin, E1 and E2 had an increased mobility due to the absence of N glycosylation, as previously described (15), and coprecipitation of HCV glycoproteins with calnexin or calreticulin was blocked (Fig. 6). When analyzed under nonreducing conditions, HCV glycoproteins from tunicamycin-treated cells formed large aggregates (data not shown). In addition, when our conformation-sensitive MAb H2 was used, HCV glycoproteins from tunicamycin-treated cells were no longer detected by immunoprecipitation (Fig. 7). These results suggest that in the absence of their N-linked oligosaccharides, HCV glycoproteins are unable to fold properly.
FIG. 6.
Effect of CST and tunicamycin (Tun) on the interaction of HCV glycoproteins with calnexin and calreticulin. BHK-21 cells were coinfected with vTF7-3 and vHCV170-809 (V) or vpTM1 (M). Infected cells were incubated in the presence or absence of 1 mM CST or 5 μg of tunicamycin per ml, labeled for 30 min with [35S]methionine in the presence of the same concentrations of drugs, and lysed with Triton X-100. Cell lysates were used for immunoprecipitation with anti-E1 (MAb A4), anti-calnexin (CNX), or anti-calreticulin (CRT) antibodies. Immunoprecipitates were analyzed by SDS-PAGE (10% polyacrylamide). Deglycosylated proteins are indicated by asterisks. The sizes (in kilodaltons) of molecular mass markers are indicated on the right.
FIG. 7.
Formation of native E1-E2 complexes after castanospermine (CST) or tunicamycin treatment, analyzed with the conformation-sensitive MAb H2. BHK-21 cells were coinfected with vTF7-3 and vHCV170-809 (V) or vTF7-3 and vpTM1 (M). Infected cells were incubated in the presence or absence of 1 mM CST or 5 μg of tunicamycin per ml, labeled for 2 h with [35S]methionine in the presence of the same concentrations of drugs, and lysed with Triton X-100. Cell lysates were used for immunoprecipitation with a conformation-sensitive (H2) or -insensitive (A4) MAb and analyzed by SDS-PAGE (10% polyacrylamide). Agg, aggregates. The sizes (in kilodaltons) of molecular mass markers are indicated on the right.
The effect of CST on the interactions of HCV glycoproteins with calnexin and calreticulin was also studied. CST is an α-glucosidase inhibitor that prevents glucose trimming. As such, it inhibits glycoprotein binding to calnexin (22, 26). The mobility of HCV glycoproteins was slightly reduced in CST-treated cells (Fig. 6 and 7), indicating that the trimming of N-linked sugars was inhibited. However, in contrast to what is usually observed for other viral glycoproteins (18, 22, 49, 65), the amount of HCV glycoproteins coprecipitated with calnexin and calreticulin was reproducibly reduced for calnexin and increased for calreticulin (Fig. 6) (see Discussion). For quantitative data, only E1 was analyzed because of the presence of background close to the E2 band. The amounts of E1 coprecipitated with calnexin and calreticulin were approximately 66 and 232%, respectively, of those coprecipitated in the absence of the drug. In addition, the amount of E1 precipitated by MAb A4 was 175% of that precipitated in the absence of the drug, suggesting that the epitope was better exposed in the absence of trimming of the oligosaccharides. In the presence of CST, the formation of HCV glycoprotein aggregates was slightly increased (data not shown). In addition, the amounts of E1 and E2 precipitated by our conformation-sensitive MAb H2 (Fig. 7) were about 80% of those precipitated in the absence of the drug, indicating that the absence of oligosaccharide trimming slightly reduces the formation of native E1-E2 complexes.
Co-overexpression of HCV glycoproteins with ER chaperones does not improve the assembly of native E1-E2 complexes.
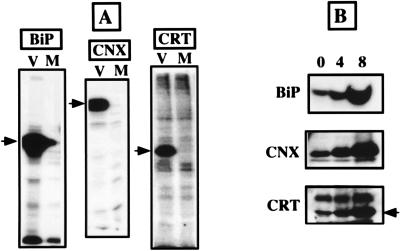
To further investigate the role of ER chaperones in HCV glycoprotein folding, vaccinia virus recombinants expressing calnexin, calreticulin, or BiP were constructed to coexpress these chaperones with HCV glycoproteins. Since the efficiency of folding of HCV glycoproteins is low, we suspected that the coexpression of these glycoproteins with ER chaperones could improve their folding. The expression of calnexin, calreticulin, or BiP by vaccinia virus recombinants was analyzed by immunoprecipitation with specific antibodies. As shown in Fig. 8A, proteins of the expected molecular size were detected when cells were coinfected with vTF7-3 and the recombinants expressing ER chaperones whereas no specific band was detected in control cells infected with vTF7-3 plus vpTM1. Because vaccinia virus infection stops host cell protein synthesis (43), metabolically labeled calreticulin, calnexin, or BiP was not detected in control cells infected with vTF7-3 plus vpTM1. The expression of vaccinia virus-chaperone recombinants was confirmed by Western blotting (Fig. 8B). After 8 h of infection, the intensity of the bands corresponding to the chaperones was approximately seven times higher for BiP and four times higher for calnexin and calreticulin.
FIG. 8.
Expression of the ER chaperones calnexin (CNX), BiP, or calreticulin (CRT) by vaccinia virus recombinants. (A) Analysis of chaperone expression by immunoprecipitation. BHK-21 cells were coinfected with vTF7-3 and the appropriate vaccinia virus-chaperone recombinant (V) or with vTF7-3 and vpTM1 (M). At 4.5 h postinfection, infected cells were labeled for 2 h with [35S]methionine and lysed with Triton X-100. Cell lysates were immunoprecipitated with anti-chaperone antibodies (CNX, BiP, or CRT) and analyzed by SDS-PAGE. Arrows correspond to the migration level of each chaperone. (B) Analysis of chaperone expression by Western blotting. BHK-21 cells were coinfected with vTF7-3 and the appropriate vaccinia virus-chaperone recombinant. At 0, 4, or 8 h postinfection, infected cells were lysed with Triton X-100. Cell lysates were separated by SDS-PAGE and revealed by Western blotting with specific anti-chaperone antibodies.
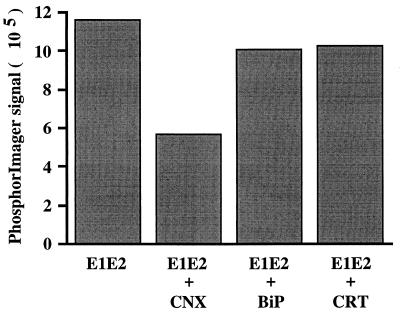
The formation of HCV glycoprotein complexes in cells co-overexpressing either ER chaperone was analyzed by immunoprecipitation with MAb H2. As shown in Fig. 9, the amounts of E1 coprecipitated with MAb H2 did not increase when calnexin, calreticulin, or BiP was co-overexpressed with HCV glycoproteins; when calnexin was overexpressed, a reduction in the assembly of native E1-E2 complexes was observed in a reproducible fashion. The effects of chaperone overexpression on the assembly of native E1-E2 complexes were also analyzed in pulse-chase experiments, and the results were very similar (data not shown). In addition, simultaneous co-overexpression of calnexin, calreticulin, and BiP with HCV glycoproteins did not improve the folding of these glycoproteins (data not shown).
FIG. 9.
Effect of the co-overexpression of calnexin (CNX), BiP, or calreticulin (CRT) with HCV glycoproteins on the formation of native E1-E2 complexes. BHK-21 cells were coinfected with vTF7-3, vHCV170-809, and the appropriate vaccinia virus-chaperone recombinant or vpTM1 (control). At 4.5 h postinfection, infected cells were labeled for 2 h with [35S]methionine and lysed with Triton X-100. Cell lysates were used for immunoprecipitation with the conformation-sensitive MAb H2. Immunoprecipitates were analyzed by SDS-PAGE, and the intensity of coprecipitated E1 was quantified with a PhosphorImager.
DISCUSSION
Molecular chaperones are proteins that associate specifically with incompletely folded or unassembled proteins and increase the efficiency with which they acquire their correct three-dimensional structure. In this report, we showed that the ER chaperones calnexin, calreticulin, and BiP interact with HCV glycoproteins. Calreticulin and BiP interacted preferentially with aggregates, whereas calnexin was shown to associate mainly with noncovalent complexes. This suggests that calnexin plays a role in the productive folding of HCV glycoproteins whereas calreticulin and BiP are probably involved in a nonproductive pathway of HCV glycoprotein folding.
HCV glycoproteins interact with calnexin and calreticulin. Interactions with calnexin and/or calreticulin have been shown for several viral (18, 22, 49, 51, 65) and nonviral (47, 50–52, 63) glycoproteins. Their preference for interaction with glycoproteins is based on a lectin-like affinity for monoglucosylated N-linked oligosaccharides (22, 26, 51, 64). Binding of substrate glycoproteins to and release from calnexin and calreticulin depend on trimming and reglucosylation of the N-linked glycans (22, 26). The absence of interaction between HCV glycoproteins and calreticulin or calnexin after tunicamycin treatment is consistent with the view that these chaperones act as lectins. However, treatment with CST did not abolish the interaction between HCV glycoproteins and calnexin or calreticulin. Indeed, the binding of HCV glycoproteins to calnexin was reduced whereas their binding to calreticulin was increased. This suggests that binding of HCV glycoproteins to and release from calnexin or calreticulin could be independent of trimming and reglucosylation of the N-linked glycans. In some cases, calnexin associates with nonglycosylated proteins (2). However, this is not the case for HCV glycoproteins. Alternatively, the coprecipitation of HCV glycoproteins with these chaperones, especially calreticulin, observed after CST treatment could be indirect. Indeed, a substantial fraction of HCV glycoproteins remain associated with calnexin and calreticulin for a long time and treatment with CST increases the formation of HCV glycoprotein aggregates. It is therefore very likely that in the presence of CST, neosynthesized glycoproteins form aggregates with preformed complexes composed of HCV glycoproteins trapped with calreticulin. This hypothesis is reinforced by the observation that CST can prevent the disassembly of preformed complexes between calnexin or calreticulin and a glycoprotein (25).
Calreticulin interacts preferentially with aggregates of HCV glycoproteins, whereas calnexin associates preferentially with noncovalent E1-E2 complexes. This suggests that these chaperones recognize HCV proteins which are in different states of folding. Our previous studies suggest that HCV glycoproteins can follow two different pathways: a productive pathway leading to the formation of noncovalent native E1-E2 complexes or a nonproductive pathway leading to the formation of large aggregates (6, 10, 12). Since the kinetics of association of HCV glycoproteins with calnexin and calreticulin were parallel, it is likely that instead of interacting sequentially with these glycoproteins, calreticulin is involved in one of these two pathways (the nonproductive pathway) and calnexin is involved in the other (the productive pathway). During their folding, proteins composing the native complex have probably been in contact with calnexin. Indeed, this chaperone was shown to interact with noncovalent complexes, and it has been shown previously to associate with the oxidized form of HCV glycoproteins (12). This suggests that calnexin plays an active role in HCV glycoprotein folding. On the other hand, the aggregates observed in the dead-end pathway have probably preferentially interacted with calreticulin. However, these aggregates are stable in cells expressing HCV glycoproteins (6) whereas the complexes formed between HCV glycoproteins and calreticulin are transient. Dissociation of such aggregates from calreticulin could be due to partial folding of the proteins involved in the aggregates. Indeed, it has been shown that a subdomain of E2 can be folded in such aggregates (21) and that the kinetics of formation of this subdomain are parallel to those of dissociation from calreticulin.
HCV glycoproteins interact with BiP. In pulse-chase experiments, the maximum amount of HCV glycoproteins interacting with calnexin or calreticulin was detected during the pulse whereas the binding of HCV glycoproteins (mainly E2) to BiP rose substantially during the first 10 min of the chase. This suggests that HCV glycoproteins could interact sequentially with calnexin (or calreticulin) and BiP. However, analysis under nonreducing conditions indicated that HCV glycoproteins in association with BiP formed aggregates whereas calnexin interacted essentially with noncovalent E1-E2 complexes. Since BiP associates with HCV glycoprotein aggregates, it is more likely that this chaperone plays a role in trapping some aggregates released from calreticulin. Together with calreticulin, BiP could therefore play a role in the nonproductive assembly pathway of HCV glycoproteins.
Co-overexpression of HCV glycoproteins with ER chaperones does not improve the formation of native E1-E2 complexes. Although molecular chaperones are abundant in the ER (37, 56), a majority of these molecules could already be involved in protein-protein interactions, leaving only a fraction of them free to interact with newly synthesized proteins. Since a large portion of HCV glycoproteins is involved in nonproductive interactions, it is possible that, due to the slow folding of HCV glycoproteins, the ER chaperones cannot fully play their role at physiological concentrations. However, overexpression of calnexin, calreticulin, or BiP did not lead to any improvement in the assembly of native HCV glycoprotein complexes, suggesting that some element might be missing in our system. Modulating the expression of ER chaperones has been tried previously. A reduction in the level of the chaperone BiP increases the secretion of proteins associated with BiP (7), whereas overexpression of this chaperone can lead to a reduction of secretion of several proteins (8). Since several chaperones can be involved in assisted folding of proteins in the ER, it is likely that a proper balance of chaperone activities is required for optimal folding. It is also likely that another chaperone(s) and/or foldase(s), which has not been identified in this work, is necessary to assist in HCV glycoprotein folding.
No interaction between HCV glycoproteins and GRP94 has been detected. It is possible that the association of GRP94 and folding intermediates is weak and that cross-linking is required to detect such an interaction, as it is the case during the folding of the immunoglobulin light chain (41). Alternatively, GRP94 may play no role in the folding and assembly of HCV glycoproteins.
During their folding, HCV glycoproteins interact to form intermediate complexes. Our data suggest that calnexin plays an active role in the folding of HCV glycoproteins. Immediately after their synthesis, monomeric forms of E1 and E2 interact with calnexin. After 10 or 15 min, E1-E2 complexes are detected in association with calnexin, indicating that E1-E2 complexes can be formed before the proteins are extensively folded. Recently, it has been shown that coexpression of E2 is necessary for proper folding of E1 (42), and it is very likely that E2 plays this “chaperone-like” role by interacting directly with E1. This chaperone-like role of E2 could explain the early formation of E1-E2 complexes, which are probably intermediates in the folding of HCV glycoproteins.
Identifying different steps of protein folding in the context of their natural environment is important for our understanding of the mechanisms developed by the cell to help proteins fold properly in subcellular compartments. In addition, a good knowledge of these mechanisms will be helpful to modulate the folding of proteins of interest. In the case of HCV glycoproteins, some folding steps can now be described, but if we want to improve the productive folding of these proteins, additional work is necessary to identify other crucial steps.
ACKNOWLEDGMENTS
We are grateful to M.-J. Gething (University of Texas, Dallas, Tex.), M. Michalak (University of Alberta, Edmonton, Alberta, Canada) and M. Brenner (Harvard Medical School, Boston, Mass.) for the gift of the plasmids containing the sequences of the ER chaperones. We thank Françoise Jacob-Dubuisson for critical reading of the manuscript and André Pillez for excellent technical assistance.
This work was supported by an ATIPE grant from the CNRS, grant 1039 from the ARC, and grant 5FS10 from the INSERM/AFS.
REFERENCES
- 1.Bergeron J J M, Brenner M B, Thomas D Y, Williams D B. Calnexin: a membrane-bound chaperone of the endoplasmic reticulum. Trends Biochem Sci. 1994;19:124–129. doi: 10.1016/0968-0004(94)90205-4. [DOI] [PubMed] [Google Scholar]
- 2.Cannon K S, Hebert D N, Helenius A. Glycan-dependent and -independent association of vesicular stomatitis virus G protein with calnexin. J Biol Chem. 1996;271:14280–14284. doi: 10.1074/jbc.271.24.14280. [DOI] [PubMed] [Google Scholar]
- 3.Chamberlain J P. Fluorographic detection of radioactivity in polyacrylamide gels with the water-soluble fluor, sodium salicylate. Anal Biochem. 1979;98:132–135. doi: 10.1016/0003-2697(79)90716-4. [DOI] [PubMed] [Google Scholar]
- 4.Choo Q-L, Kuo G, Weiner A J, Overby L R, Bradley D W, Houghton M. Isolation of a cDNA clone derived from a blood-borne non-A, non-B viral hepatitis genome. Science. 1989;244:359–362. doi: 10.1126/science.2523562. [DOI] [PubMed] [Google Scholar]
- 5.Cocquerel L, Meunier J-C, Pillez A, Wychowski C, Dubuisson J. A retention signal necessary and sufficient for endoplasmic reticulum localization maps to the transmembrane domain of hepatitis C virus glycoprotein E2. J Virol. 1998;72:2183–2191. doi: 10.1128/jvi.72.3.2183-2191.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Deleersnyder V, Pillez A, Wychowski C, Blight K, Xu J, Hahn Y S, Rice C M, Dubuisson J. Formation of native hepatitis C virus glycoprotein complexes. J Virol. 1997;71:697–704. doi: 10.1128/jvi.71.1.697-704.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dorner A J, Krane M G, Kaufman R J. Reduction of endogenous GRP78 levels improves secretion of a heterologous protein in CHO cells. Mol Cell Biol. 1988;8:4063–4070. doi: 10.1128/mcb.8.10.4063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dorner A J, Wasley L C, Kaufman R J. Overexpression of GRP78 mitigates stress induction of glucose regulated protein and blocks secretion of selective proteins in Chinese hamster ovary cells. EMBO J. 1992;11:1563–1572. doi: 10.1002/j.1460-2075.1992.tb05201.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Drillien R, Spehner D, Kirn A. Complementation and genetic linkage between vaccinia virus temperature-sensitive mutants. Virology. 1982;119:372–381. doi: 10.1016/0042-6822(82)90096-4. [DOI] [PubMed] [Google Scholar]
- 10.Dubuisson J, Hsu H H, Cheung R C, Greenberg H B, Russell D G, Rice C M. Formation and intracellular localization of hepatitis C virus envelope glycoprotein complexes expressed by recombinant vaccinia and Sindbis viruses. J Virol. 1994;68:6147–6160. doi: 10.1128/jvi.68.10.6147-6160.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dubuisson, J., and D. Moradpour. Unpublished results.
- 12.Dubuisson J, Rice C M. Hepatitis C virus glycoprotein folding: disulfide bond formation and association with calnexin. J Virol. 1996;70:778–786. doi: 10.1128/jvi.70.2.778-786.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Earl P L, Moss B, Doms R W. Folding, interaction with GRP78-BiP, assembly, and transport of the human immunodeficiency virus type 1 envelope protein. J Virol. 1991;65:2047–2055. doi: 10.1128/jvi.65.4.2047-2055.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fliegel L, Kimberly B, MacLennan D H, Reithmeier R A F, Michalak M. Molecular cloning of the high affinity calcium-binding protein (calreticulin) of skeletal muscle sarcoplasmic reticulum. J Biol Chem. 1989;264:21522–21528. [PubMed] [Google Scholar]
- 15.Fournillier-Jacob A, Cahour A, Escriou N, Girard M, Wychowski C. Processing of the E1 glycoprotein of hepatitis C virus expressed in mammalian cells. J Gen Virol. 1996;77:1055–1064. doi: 10.1099/0022-1317-77-5-1055. [DOI] [PubMed] [Google Scholar]
- 16.Francki, R. I. B., C. M. Fauquet, D. L. Knudson, and F. Brown. 1991. Classification and nomenclature of viruses. Fifth report of the International Committee on Taxonomy of Viruses. Arch. Virol. 2 (Suppl):223.
- 17.Fuerst T R, Niles E G, Studier F W, Moss B. Eukaryotic transient-expression system based on recombinant vaccinia virus that synthesizes bacteriophage T7 RNA polymerase. Proc Natl Acad Sci USA. 1986;83:8122–8126. doi: 10.1073/pnas.83.21.8122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gaudin Y. Folding of rabies virus glycoprotein: epitope acquisition and interaction with endoplasmic reticulum chaperones. J Virol. 1997;71:3742–3750. doi: 10.1128/jvi.71.5.3742-3750.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gething M-J, Sambrook J. Protein folding in the cell. Nature. 1992;355:33–45. doi: 10.1038/355033a0. [DOI] [PubMed] [Google Scholar]
- 20.Grakoui A, Wychowski C, Lin C, Feinstone S M, Rice C M. Expression and identification of hepatitis C virus polyprotein cleavage products. J Virol. 1993;67:1385–1395. doi: 10.1128/jvi.67.3.1385-1395.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Habersetzer, F., A. Fournillier, J. Dubuisson, D. Rosa, S. Abrigniani, C. Wychowski, I. Nakano, C. Trépo, C. Desgranges, and G. Inchauspé. Unpublished data. [DOI] [PubMed]
- 22.Hammond C, Braakman I, Helenius A. Role of N-linked oligosaccharides, glucose trimming and calnexin during glycoprotein folding in the endoplasmic reticulum. Proc Natl Acad Sci USA. 1994;91:913–917. doi: 10.1073/pnas.91.3.913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hammond C, Helenius A. Quality control in the secretory pathway. Curr Opin Cell Biol. 1995;7:523–529. doi: 10.1016/0955-0674(95)80009-3. [DOI] [PubMed] [Google Scholar]
- 24.Hartl F-U, Hlodan R, Langer T. Molecular chaperones in protein folding: the art of avoiding sticky situations. Trends Biochem Sci. 1994;19:20–25. doi: 10.1016/0968-0004(94)90169-4. [DOI] [PubMed] [Google Scholar]
- 25.Hebert D N, Foellmer B, Helenius A. Calnexin and calreticulin promote folding, delay oligomerization and suppress degradation of influenza hemagglutinin in microsomes. EMBO J. 1996;15:2961–2968. [PMC free article] [PubMed] [Google Scholar]
- 26.Hebert D N, Foellmer B, Helenius A. Glucose trimming and reglucosylation determine glycoprotein association with calnexin in the endoplasmic reticulum. Cell. 1995;81:425–433. doi: 10.1016/0092-8674(95)90395-x. [DOI] [PubMed] [Google Scholar]
- 27.Hurtley S M, Bole D G, Hoover-Litty H, Helenius A, Copeland C S. Interaction of misfolded influenza virus hemagglutinin with binding protein (BiP) J Cell Biol. 1989;108:2117–2126. doi: 10.1083/jcb.108.6.2117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jakob U, Buchner J. Assisting spontaneity: the role of Hsp90 and small Hsps as molecular chaperones. Trends Biochem Sci. 1994;19:205–211. doi: 10.1016/0968-0004(94)90023-x. [DOI] [PubMed] [Google Scholar]
- 29.Kieny M-P, Lathe R, Drillien R, Spehner D, Skory S, Schmitt D, Wiktor T, Koprowski H, Lecocq J-P. Expression of rabies virus glycoprotein from a recombinant vaccinia virus. Nature. 1984;312:163–166. doi: 10.1038/312163a0. [DOI] [PubMed] [Google Scholar]
- 30.Kim P S, Arvan P. Calnexin and BiP act as sequential molecular chaperones during thyroglobulin folding in the endoplasmic reticulum. J Cell Biol. 1995;128:29–38. doi: 10.1083/jcb.128.1.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kornfeld R, Kornfeld S. Assembly of asparagine-linked oligosaccharides. Annu Rev Biochem. 1985;54:631–664. doi: 10.1146/annurev.bi.54.070185.003215. [DOI] [PubMed] [Google Scholar]
- 32.Kozutsumi Y, Normington K, Press E, Slaughter C, Sambrook J, Gething M-J. Identification of immunoglobulin heavy chain binding protein as glucose-related protein 78 on the basis of amino acid sequence, immunological cross-reactivity, and functional activity. J Cell Sci Suppl. 1989;11:115–137. doi: 10.1242/jcs.1989.supplement_11.10. [DOI] [PubMed] [Google Scholar]
- 33.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 34.Lanford R E, Notvall L, Chavez D, White R, Frenzel G, Simonsen C, Kim J. Analysis of hepatitis C virus capsid, E1, and E2/NS1 proteins expressed in insect cells. Virology. 1993;197:225–235. doi: 10.1006/viro.1993.1583. [DOI] [PubMed] [Google Scholar]
- 35.Loo T W, Clarke D M. Prolonged association of temperature-sensitive mutants of human P-glycoprotein with calnexin during biogenesis. J Biol Chem. 1994;269:28683–28689. [PubMed] [Google Scholar]
- 36.Machamer C E, Doms R W, Bole D G, Helenius A, Rose J K. Heavy chain binding protein recognizes incompletely disulfide-bonded forms of vesicular stomatitis virus G proteins. J Biol Chem. 1990;265:6879–6883. [PubMed] [Google Scholar]
- 37.Marquardt M, Hebert D, Helenius A. Post-translational folding of influenza hemagglutinin in isolated endoplasmic reticulum-derived microsomes. J Biol Chem. 1993;268:19618–19625. [PubMed] [Google Scholar]
- 38.Martin A, Escriou N, Chao S F, Lemon S M, Girard M, Wychowski C. Identification and site-directed mutagenesis of the primary (2A/2B) cleavage site of the hepatitis A virus polyprotein: functional impact on infectivity of HAV RNA transcripts. Virology. 1995;213:213–222. doi: 10.1006/viro.1995.1561. [DOI] [PubMed] [Google Scholar]
- 39.Matsuura Y, Miyamura T. The molecular biology of hepatitis C virus. Semin Virol. 1993;4:297–304. [Google Scholar]
- 40.Melnick J, Aviel S, Argon Y. The endoplasmic reticulum stress protein GRP94, in addition to BiP, associates with assembled immunoglobulin chains. J Biol Chem. 1992;267:21303–21306. [PubMed] [Google Scholar]
- 41.Melnick J, Dul J L, Argon Y. Sequential interaction of the chaperones BiP and GRP94 with immunoglobulin chains in the endoplasmic reticulum. Nature. 1994;370:373–375. doi: 10.1038/370373a0. [DOI] [PubMed] [Google Scholar]
- 42.Michalak J-P, Wychowski C, Choukhi A, Meunier J-C, Ung S, Rice C M, Dubuisson J. Characterization of truncated forms of hepatitis C virus glycoproteins. J Gen Virol. 1997;78:2299–2306. doi: 10.1099/0022-1317-78-9-2299. [DOI] [PubMed] [Google Scholar]
- 43.Moss B. Poxviridae: the viruses and their replication. In: Fields B N, Knipe D M, Howley P M, editors. Fields virology. 3rd ed. Philadelphia, Pa: Lippincott-Raven; 1996. pp. 2637–2671. [Google Scholar]
- 44.Moss B, Elroy-Stein O, Mizukami T, Alexander W A, Fuerst T R. New mammalian expression vectors. Nature. 1990;348:91–92. doi: 10.1038/348091a0. [DOI] [PubMed] [Google Scholar]
- 45.Munro S, Pelham H R B. An hsp-70 like protein in the ER: identity with the 78 Kd glucose-regulated protein and immunoglobulin heavy chain binding protein. Cell. 1986;46:291–300. doi: 10.1016/0092-8674(86)90746-4. [DOI] [PubMed] [Google Scholar]
- 46.Munro S, Pelham H R B. An HSP-like protein in the ER: identity with the 78 Kd glucose regulated protein and immunoglobulin heavy chain binding protein. Cell. 1986;46:291–300. doi: 10.1016/0092-8674(86)90746-4. [DOI] [PubMed] [Google Scholar]
- 47.Nauseef W M, McCormick S J, Clark R A. Calreticulin functions as a molecular chaperone in the biosynthesis of myeloperoxidase. J Biol Chem. 1995;270:4741–4747. doi: 10.1074/jbc.270.9.4741. [DOI] [PubMed] [Google Scholar]
- 48.Navarro D, Qadri I, Pereira L. A mutation in the ectodomain of herpes simplex virus 1 glycoprotein B causes defective processing and retention in the endoplasmic reticulum. Virology. 1991;184:253–264. doi: 10.1016/0042-6822(91)90842-y. [DOI] [PubMed] [Google Scholar]
- 49.Otteken A, Moss B. Calreticulin interacts with newly synthesized human immunodeficiency virus type 1 envelope glycoprotein, suggesting a chaperone function similar to that of calnexin. J Biol Chem. 1996;271:97–103. doi: 10.1074/jbc.271.1.97. [DOI] [PubMed] [Google Scholar]
- 50.Ou W-J, Cameron P H, Thomas D Y, Bergeron J J M. Association of folding intermediates of glycoproteins with calnexin during protein maturation. Nature. 1993;364:771–776. doi: 10.1038/364771a0. [DOI] [PubMed] [Google Scholar]
- 51.Peterson J R, Ora A, Nguyen Van P, Helenius A. Transient, lectin-like association of calreticulin with folding intermediates of cellular and viral glycoprotein. Mol Biol. 1995;6:1173–1184. doi: 10.1091/mbc.6.9.1173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rajagopalan S, Xu Y, Brenner M B. Retention of unassembled components of integral membrane proteins by calnexin. Science. 1994;263:387–390. doi: 10.1126/science.8278814. [DOI] [PubMed] [Google Scholar]
- 53.Ralston R, Thudium K, Berger K, Kuo C, Gervase B, Hall J, Selby M, Kuo G, Houghton M, Choo Q-L. Characterization of hepatitis C virus envelope glycoprotein complexes expressed by recombinant vaccinia viruses. J Virol. 1993;67:6753–6761. doi: 10.1128/jvi.67.11.6753-6761.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rice C M. Flaviviridae: viruses and their replication. In: Fields B N, Knipe D M, Howley P M, editors. Fields virology. 3rd ed. Philadelphia, Pa: Lippincott-Raven; 1996. pp. 931–959. [Google Scholar]
- 55.Rodan A R, Simons J F, Trombetta E S, Helenius A. N-linked oligosaccharides are necessary and sufficient for association of glycosylated forms of bovine RNase with calnexin and calreticulin. EMBO J. 1996;15:6921–6930. [PMC free article] [PubMed] [Google Scholar]
- 56.Rowling P, McLaughlin S H, Pollock G S, Freedman R B. A single purification procedure for the major resident proteins of the ER lumen: endoplasmin, BiP, calreticulin and protein disulfide isomerase. Protein Expression Purif. 1994;5:331–336. doi: 10.1006/prep.1994.1049. [DOI] [PubMed] [Google Scholar]
- 57.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 58.Schaiff W T, Hruska K A, McCourt D W, Green M, Schwartz B D. HLA-DR associates with specific stress proteins and is retained in the endoplasmic reticulum in invariant chain negative cells. J Exp Med. 1992;176:657–666. doi: 10.1084/jem.176.3.657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sousa M, Parodi A J. The molecular basis for the recognition of misfolded glycoproteins by the UDP-Glc:glycoprotein glucosyltransferase. EMBO J. 1995;14:4196–4203. doi: 10.1002/j.1460-2075.1995.tb00093.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sousa M C, Ferrero-Garcia M A, Parodi A J. Recognition of the oligosaccharide and protein moieties of glycoproteins by the UDP-Glc:glycoprotein glucosyltranferase. Biochemistry. 1992;31:97–105. doi: 10.1021/bi00116a015. [DOI] [PubMed] [Google Scholar]
- 61.Spaete R R, Alexander D, Rugroden M E, Choo Q-L, Berger K, Crawford K, Kuo C, Leng S, Lee C, Ralston R, Thudium K, Tung J W, Kuo G, Houghton M. Characterization of the hepatitis E2/NS1 gene product expressed in mammalian cells. Virology. 1992;188:819–830. doi: 10.1016/0042-6822(92)90537-y. [DOI] [PubMed] [Google Scholar]
- 62.Van Leeuwen J E M, Kears K P. Calnexin associates exclusively with individual CD3d and T cell antigen receptor (TCR) a protein containing incompletely trimmed glycans that are not assembled into multisubunit TCR complexes. J Biol Chem. 1996;271:9660–9665. doi: 10.1074/jbc.271.16.9660. [DOI] [PubMed] [Google Scholar]
- 63.Wada I, Imai S, Kai M, Sakane F, Kanoh H. Chaperone function of calreticulin when expressed in the endoplasmic reticulum as the membrane-anchored and soluble forms. J Biol Chem. 1995;270:20298–20304. doi: 10.1074/jbc.270.35.20298. [DOI] [PubMed] [Google Scholar]
- 64.Ware F E, Vassilakos A, Peterson P A, Jackson M R, Lehrman M A, Williams D B. The molecular chaperone calnexin binds Glc1Man9GlcNAc2 oligosaccharide as an initial step in recognizing unfolded glycoproteins. J Biol Chem. 1995;270:4697–4704. doi: 10.1074/jbc.270.9.4697. [DOI] [PubMed] [Google Scholar]
- 65.Yamashita Y, Shimokata K, Mizuno S, Daikoku T, Tsurumi T, Nishiyama Y. Calnexin acts as a molecular chaperone during the folding of glycoprotein B of human cytomegalovirus. J Virol. 1996;70:2237–2246. doi: 10.1128/jvi.70.4.2237-2246.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zapun A, Petrescu S M, Rudd P M, Dwek R A, Thomas D Y, Bergeron J J M. Conformation-independent binding of monoglucosylated ribonuclease B to calnexin. Cell. 1997;88:29–38. doi: 10.1016/s0092-8674(00)81855-3. [DOI] [PubMed] [Google Scholar]