Abstract
There are many instances of hollow-structure morphogenesis in the development of tissues. Thus, the fabrication of hollow structures in a simple, high-throughput and homogeneous manner with proper natural biomaterial combination is valuable for developmental studies and tissue engineering, while it is a significant challenge in biofabrication field. We present a novel method for the fabrication of a hollow cell module using a coaxial co-flow capillary microfluidic device. Sacrificial gelatin laden with cells in the inner layer and GelMa in the outer layer are used via a coaxial co-flow capillary microfluidic device to produce homogenous micro-beads. The overall and core sizes of core–shell microbeads were well controlled. When using human vein vascular endothelial cells to demonstrate how cells line the inner surface of core–shell beads, as the core liquifies, a hollow cell ball with asymmetric features is fabricated. After release from the GelMa shell, individual cell balls are obtained and deformed cell balls can self-recover. This platform paves way for complex hollow tissue modeling in vitro, and further modulation of matrix stiffness, curvature and biochemical composition to mimic in vivo microenvironments.
Keywords: core–shell beads, hollow cell ball, microfluidics, hydrogels, soft matter
Graphical Abstract

Introduction
A lot of our understanding of the cell mechanism comes from 2D culture, in which cells lay flattened on a rigid surface. However, in vivo, cells are embedded in the extracellular matrix (ECM) and interact with other cells and display different morphologies and behaviors. 3D culture technologies can tune the configuration and mechanical and biochemical cues to guide cell proliferation, migration, differentiation and morphogenesis [1–3]. Unlike 2D, a 3D environment can exhibit in vivo-like properties. Consequentially, the application of 3D cell culture has increased in fundamental biology studies, drug testing and tissue engineering [4, 5].
Enclosed 3D hollow structures, such as hollow spheroids, are composed of a cellular monolayer covering the inner surface of the biomaterial structure and filled with a fluid, which showed asymmetric properties from inside, and outside properties like growth factor and oxygen vary. Cells cultured on 3D hollow structures are sensitive to cues not present in a 2D culture environment, such as asymmetric biomolecular gradient cues from the inner and outer structures, the curvature of the structure [6, 7], mechanical force and compositional binding of structural biomaterials. 3D hollow structures are often present in the developing or matured tissue, such as vascular, ocular lens, renal, heart, lung and inner ear [8–14]. In addition, stem cells form hollow chambers in blastocysts during embryonic development [15]. The function and morphogenesis of these tissues and organs are closely linked to their structure. Therefore, the technology for 3D hollow structure fabrication should better mimic the above in vivo conformation as it recapitulates the environmental cues.
Fabricating enclosed hollow cell structures in a high-throughput manner is still a big challenge, although hydrogel spheroid construction has been reported using the templated method or microfluidic method [16]. Previously, a hollow lens epithelial cell structure has been constructed; however, it requires multiple steps and achieves a nonuniform spheroid from the templated method [17]. Microfluidic methods for fabricating hydrogel microbeads might be an ideal tool for hollow cell structures. Among them, core–shell hydrogel microbeads are special hydrogel microstructures with the inner hydrogel (core) layer surrounded by another material (shell). The core–shell microbeads have been applied for cell delivery, tissue development and co-culture. For example, islet cells with a higher encapsulation rate were achieved by alginate core–shell microbeads [18]. Encapsulating HepG2 and 3T3 cells in the core of core–shell beads showed increased biological properties such as albumin and urea secretion [19]. Till now, there have been reports for fabricating solid cell spheroids only, as noncell adhesion biomaterials (alginate) are commonly used [19–22]. Natural derived biomaterials contain cell binding modify sequence and are favorable to cell self-assembly and maturation. And the natural materials are commonly challenged to be shaped with integrity and production rate, as they often have low modulus, temperature sensitive and slow gelation. Till now limited biomaterial has been reported to fabricated as the core–shell microbeads, especially for its shell [23, 24]. The core–shell microbeads fabricated by suitable natural biomaterial combination, with a controlled size and high-throughput manner, and they application for hollow cell structure formation is still a significant challenge and not achieve yet.
In this study, we developed a novel coaxial co-flow capillary microfluidic platform to produce hollow cell balls in a single step, with controlled size, high-throughput and in a homogeneous manner. Two types of natural derived biomaterials were applied in this system, a gelatin-based hydrogel and a photo-crosslinkable matrix hydrogel (GelMa) to fabricate as core–shell microbeads. Gelatin and GelMa, the derivative of collagen, possess most of the biological properties of collagens. GelMa has high potential in tissue engineering and biomedical applications due to its good biocompatible properties and versatile properties in processing. GelMa is temperature sensitive and also can form crosslink hydrogel by photopolymerization with photoinitiator (lithium phenyl-2,4,6-trimethylbenzoylphosphinate, LAP). Gelatin and gelatin methacrylate (GelMa) were liquefied and delivered in laminar flow in a capillary microfluidic device, and then the laminar flow was broken up into core–shell beads by oil flow. Specificity, the cell contained in the gelatin flowed in the inner core of the coaxial capillary, GelMa flowed in the shell of the coaxial capillary and oil was introduced as a continuous flow. Microcore–shell droplets were generated at 30–40°C and gelled into microbeads immediately by blue light curing, and crosslinking was enhanced by cooling near the outlet on ice to form core–shell spheroids. Finally, hollow structures were achieved by liquefying the gelatin core at 37°C during cell culture. Human vein vascular endothelial cells (HUVECs) were used to demonstrate the method of fabricating hollow cell balls. Core (cell-laden gelatin) shell (GelMa) beads were fabricated as a guiding template for the in-situ assemblies of cells along the inner surface or self-assembly to form the hollow cell structures.
Materials and Methods
Gelatin and GelMa preparation
For fabricating core–shell beads without cells, 12% gelatin (Sigma, USA) and 15% GelMa (Engineering for life, China) were used. For cell encapsulation, the cell solution in (1.5 × 107 cells/ml in DMEM medium) was mixed with 12% gelatin in a 1:1 ratio to achieve a final concentration of 6%. Gelatin type A (12%) from porcine skin 300 Bloom was dissolved in PBS (Gibco, USA) and heated at 60°C for 1 h. GelMa (0.075 g) was dissolved in 500 μl of 0.3% LAP solution (Engineering for life, China) and heated at 39°C with shaking to obtain 15% GelMa. LAP (0.3%) was prepared by dissolving 0.5 g LAP in 16.67 ml PBS solution and heating at 50°C for 1 h.
Fabrication of the coaxial co-flow capillary microfluidic platform
The capillary microfluidic platform was designed based on the co-flow principle, in which the core, shell and oil flow were assembled in parallel channels. The capillary microfluidic platform consists of a capillary microfluidic device, heating device (300 W/50-60Hz, Beurer, USA), pump system (Kd Scientific, USA) and photo-curing system (405 nm, UVATA, China). It also consists of two 3D printed connectors, two round capillaries (outer diameter: 1 ± 0.03 mm; inner diameter: 0.6 ± 0.03 mm; G-1, Narishige) with pulled ends (inner diameter 0.2 ± 0.03 mm), two rectangular capillaries (outer diameter: 1.55 ± 0.03 mm; inner diameter: 1.02 ± 0.01 mm) and two round capillaries (outer diameter: 1 ± 0.03 mm; inner diameter: 0.6 ± 0.03 mm; G-1, Narishige), as shown in Supplementary Figure S1. To assemble the co-flow microfluidic device, a round capillary with a pulled end was inserted into the end of another pulled capillary and assembled with a rectangular capillary in the connector. The other rectangular capillary was assembled as an outlet channel for bead collection. An infrared lamp was placed approximately 30 cm away from the near side of capillary device to maintain the temperature of the capillary device at 30–40°C. The three pumps were connected to the corresponding capillary inlet. The photo-curing system was placed at the outlet of the capillary microfluidic device closely; therefore, the fabricated core–shell beads could be cured immediately when the outlet capillary was removed. To lower the diffusion effect of the laminar flow, we attempted to shorten the length of laminar flow by inserting one capillary with two pulled sections deeper into another capillary.
Fabrication of the core–shell GelMa/gelatin microcapsules
To fabricate the core–shell beads, gelatin, GelMa and oil with spans were injected into the capillary microfluidic device. Specifically, 12% gelatin and 15% GelMa solution with 0.3% LAP were sucked into a 1 ml syringe, respectively. Corn oil (Sigma, USA) supplemented with 1% span-80 (Sigma, USA) was sucked into a 50 ml syringe. Three nylon tubes were connected to the corresponding inlet channel of the capillary microfluidic device. Gelatin, GelMa and oil were connected to the inner (Inlet 1), middle (Inlet 2) and outer (Inlet 3) channels. In this study, the GelMa concentration was fixed at 15% except in the concentration optimization experiments. The gelatin and GelMa flow were fixed at 8 and 10 μl/min, respectively, unless optimization experiments were performed. The oil flow was maintained at 1 ml/min. Finally, the cured core–shell beads were collected in a Phosphate Buffer Saline (PBS) solution covered with hexadecane (Aladdin, China). Hexadecane would break the oil emulsion and promote beads to sunk down to the bottom of PBS solution.
HUVECs were encapsulated in core–shell beads
HUVECs were cultured at 37°C with 5% CO2 in Modified Eagle Medium (DMEM) (Gibco, USA) supplemented with 10% FBS (Gibco, USA) and 1% penicillin-streptomycin (Gibco, USA). For cell encapsulation, cells were harvested using 0.25% trypsin (Gibco, USA) and resuspended in DMEM for the core solution. Twelve percent gelatin solution mixed with equal volume of 1.5 × 107 cells/ml cell solution was used as the core solution for encapsulation.
Core–shell beads transferred from oil to culture medium
Core–shell beads with oil, hexadecane and PBS were collected from the collecting container. The collected samples were transferred to a 15 ml EP tube and centrifuged at 500 × g for 5 min. Then, the supernatant was discarded and the beads were transferred to a new 15 ml centrifugation tube using a 3 ml pipet. The beads were then washed with PBS thrice. The culture medium was then replaced with PBS. Finally, cells laden with core–shell beads were moved to culture dishes.
Core–shell structure characterization
The size of the core–shell beads with varying flow rates was captured using an optical microscope (Eclipse Ti2-U, Nikon, Japan) and measured using ImageJ software. Confocal laser scanning microscopy was used to visualize the core–shell structure, in which the GelMA solution and gelatin solutions were labeled with fluorescent polystyrene nanoparticles L4655 and L3280 (Sigma, USA), respectively.
Immunofluorescent staining
To detect endothelial marker CD31 and tight junction protein ZO-1, cell-laden core–shell beads were dissolved using GelMa lyase (Engineering for life, China), and the remaining hollow cell balls were fixed with paraformaldehyde (PFA) at 4°C overnight. The cell balls were then washed with PBS thrice, and treated with 0.5% Triton-100 for 15 min. The cells were then washed with PBS for three times, 10 min each time, before blocking with 10% bovine serum albumin for 1 h. The cell balls were then stained with primary antibodies (CD31, ab24590, Abcam, USA) overnight at 4°C and another 6 h in room 37°C, washed twice with PBS and stained with secondary antibodies, TRITC-labeled goat anti-mouse or rabbit IgG (Abbkine, CA, USA) for 1 h followed by DAPI (Solarbio, China) for 15–30 min. Images were obtained using a fluorescence microscope.
To analyze the cytoskeleton, cell-laden core–shell beads were fixed using 4% PFA, washed thrice with PBS and treated with 0.5% Triton-100 for 15 min. After washing with PBS thrice, F-actin was stained with rhodamine-phalloidin (Solarbio, China) for 1 h. The cell beads were washed thrice with PBS, incubated with DAPI (Solarbio, China) for nuclear staining and observed using a confocal laser scanning microscope (C2+/C2si+, Nikon, Japan).
Core–shell beads observed by scanning electron microscope
Samples were prepared as described previously [43], with some modifications. Briefly, core–shell beads or hollow cell balls were washed with PBS (Gibco, USA) before fixing with 4% PFA (Solarbio, China), dehydrating with a series of ethanol solutions (50%, 70%, 80%, 90% and 100%) for 10 min each, and then freezing at −80°C overnight. The samples were then lyophilized using a freeze-drier (LyoQuest-85 PLUS, Telstar, Spain), sputter-coated with gold and observed by SEM (Phenom, China). For better observation of the cells encapsulated in the GelMa shell, the shell was cut carefully using a sharp scalpel.
Results
Overview of the hollow core–shell beads fabrication platform
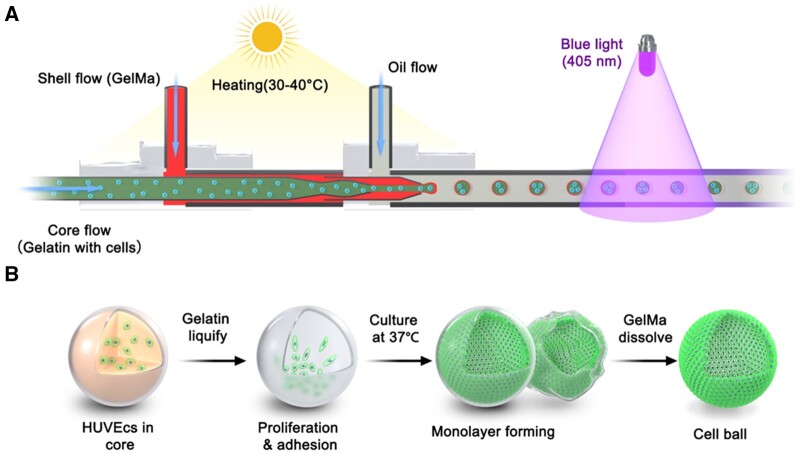
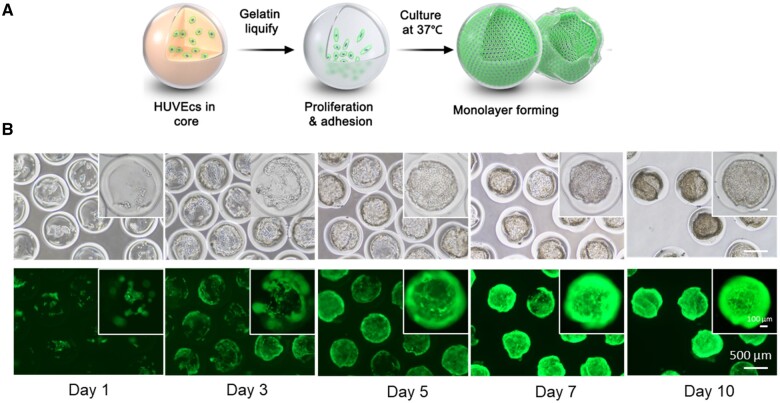
To fabricate the core–shell gelatin/GelMa beads, a combined heating and photo- (405 nm) curing strategy was chosen, as shown in Figure 1. GelMa and gelatin are temperature-responsive sol-gel transition biomaterials. GelMA can form crosslinked networks of hydrogels undergoing photopolymerization with phtotoinitiator (LAP). GelMa and gelatin were melted via infrared lamp heating and delivered into a capillary microfluidic platform and were immediately cross-linked by photo-curing at the outlet of the capillary. Gelatin with cells, GelMa with photoinitiator (LAP), and corn oil were introduced into the inner, middle and outer capillaries, respectively. Next, the gelatin core was melted at 37°C, and the entrapped cells were submerged or attached to the inner surface. Finally, after several days of culture, the cells populated to cover the inner surface. HUVECs labeled with green fluorescent protein (GFP) were chosen to demonstrate the platform.
Figure 1.
Schematic of coaxial co-flow capillary microfluidic platform used to fabricate 3D core–shell beads for hollow ball construction. (A) Gelatin with cells flowed in the inner capillary, GelMa flowed in the middle layer of the capillary. Lamellar flow broke up to form cell-laden core–shell beads by continuous oil flow in the outer layer of the capillary. (B) The inner gelatin cores were melted at 37°C during cell culturing. The cells then adhered to the inner surface of core–shell beads and proliferated to form a hollow cell sheet.
Observation of the fabrication parameters of core–shell beads
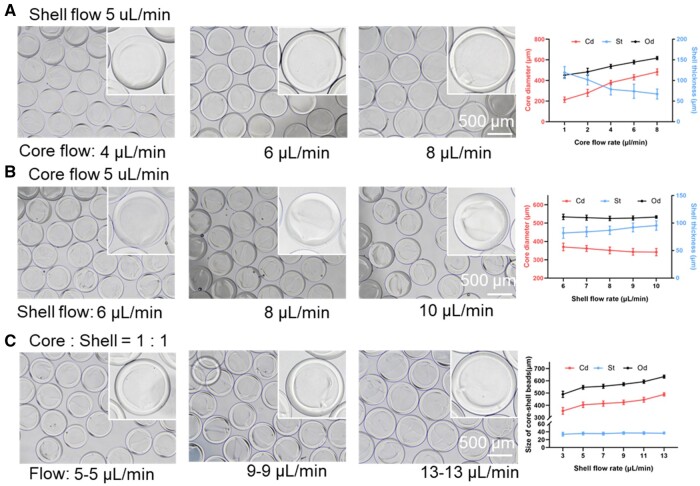
The flow rate and GelMa concentration are the two key parameters affecting the size and integrity of the core–shell beads. To observe their effect on the size of the core–shell beads, we tuned the flow rate of the core, shell and continuous flow. We first fixed the shell flow rate at 5 μl/min and changed the core flow from 1 to 8 μl/min. As shown in Figure 2A, the shell thickness of the core–shell beads decreased from 119.18 ± 14.65 to 67.18 ± 12.24 μm as the core flow increased. Conversely, the core diameter and overall diameter were increased (213.28 ± 26.27–482.86 ± 29.48 and 451.63 ± 25.49–617.22 ± 15.78 μm, respectively). When we fixed the flow rate of the core flow, and then tuned the shell flow from 6 to 10 μl/min, the shell thickness of the core–shell beads increased slightly (Figure 2B). When we fixed the ratio of the core flow and shell flow, with the increasing of the flow, the overall diameter of beads and the core diameter were both increased (489.59 ± 25.34–634.16 ± 12.75 and 354.18 ± 27.45–488.63 ± 13.68 μm, respectively) (Figure 2C).
Figure 2.
The effect of flow rate on the size of core–shell beads. (A) Shell flow = 5 μl/min; core flow = 1–8 μl/min. (B) Core flow = 5 μl/min; shell flow = 6–10 μl/min. (C) The core to shell ratio was fixed at 1:1, and the flow of core–shell was increased from 3 to 13 μl/min. Core–shell beads were immersed in PBS.
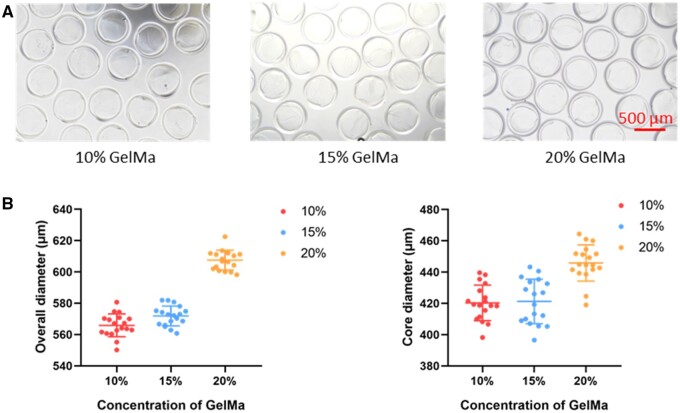
To evaluate the effects of different concentrations of GelMa on the formability and size of the core–shell beads. Three concentrations of 10, 15 and 20% were applied to fabricate the core–shell microbeads. As shown in Figure 3, the overall size of the core–shell microbeads was increased from 565.91 ± 7.12 to 607.54 ± 6.12 μm as the concentration was increased from 10 to 20%. The core size of microbeads had a similar trend, increasing from 420.33 ± 11.04 to 445.85 ± 11.24 μm. These results showed all the concentrations of GelMa (10,15, 20%) were successfully fabricated.
Figure 3.
The fabrication of core–shell beads with different concentrations of GelMa. (A) The bright-field images of the core–shell beads with the concentration of 10%, 15% and 20%. (B) The size of core–shell beads with different concentrations. Core–shell beads were immersed in PBS.
Characterization of the core–shell bead morphology
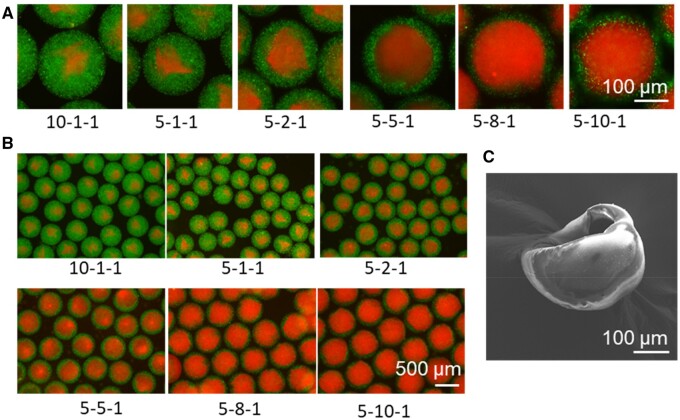
The structural integrity of the core–shell beads is important for further biomedical and drug-related applications. Therefore, we introduced fluorescence beads in the precursor gels to illuminate the core–shell beads for structural observation. Fluorescent polystyrene nanoparticles L3280 and L4655 were co-delivered in the core and shell. The mean diameters of these two nanoparticles were 0.5 and 1 μm, respectively. Moreover, they were trapped and distributed evenly in the hydrogel, owing to their larger size than the pores of GelMa and gelatin. The structure of the core–shell beads was observed under a fluorescence microscope. As shown in Figure 4, the integrity of the core–shell structure was achieved under different flow conditions, and as the core/shell flow rate changed from 1/10–1, the core acquired a rounder morphology.
Figure 4.
The morphology of the core–shell beads. (A) The fluorescent images showed the structure of core–shell beads under different flow rates. (B) The wide-field images at different flow rates. (C) Scanning electron microscopic image of the cut core–shell beads. The shell flow (μl/min), core flow (μl/min) and oil flow (ml/min) were shown under each image. Core (red), shell (green): the gelatin solution and GelMA solution were labeled with fluorescent polystyrene nanoparticles L4655 (red) and L3280 (green) (sigma), respectively.
Cell growth on the inner surface of core–shell beads
To fabricate cell-laden core–shell beads, 6% gelatin laden with HUVECs (∼7.5 × 106 cells/ml) and 15% GelMa were delivered into the inner and middle channels, respectively. After fabrication, core–shell beads were collected and cultured at 37°C to allow gelatin liquefaction and cell growth. HUVECs migrated along the inner surface of the core–shell beads. Figure 5 shows the temporal growth in HUVECs in the inner core–shell beads. Gelatin was liquified after several minutes of culture at 37°C, and the cells migrated easily in this liquid environment. Due to gravity, most of the cells sunk to the bottom during the first 24 h. After culturing, cells began to attach to the GelMa surface and proliferated along the surface. We could observe few dozen cells in the core, and after 3 days, cells covered most of the bottom part of the beads. Between days 3 and 7, the number of cells increased quickly to occupy a large area of the inner surface of the beads. After 10 days of culture, most of the core–shell beads were populated and fully covered with cells assembled over the inner surface. The other core–shell beads can be continuously cultured for full coverage.
Figure 5.
HUVECs Growth within core–shell beads. (A) Schematic illustration of HUVECs growths within core–shell beads. After gelatin liquify, HUVECs proliferation and adhesion to inner surface of GelMa shell and forming cell ball. (B) On Day 1, most of the cells sunk to the bottom of core–shell beads. Cells proliferated to cover the beads in over 10 days. GFP-expressed HUVECs were used for observation in this experiment.
Detached cell structure showed self-standing and deformation recovery properties
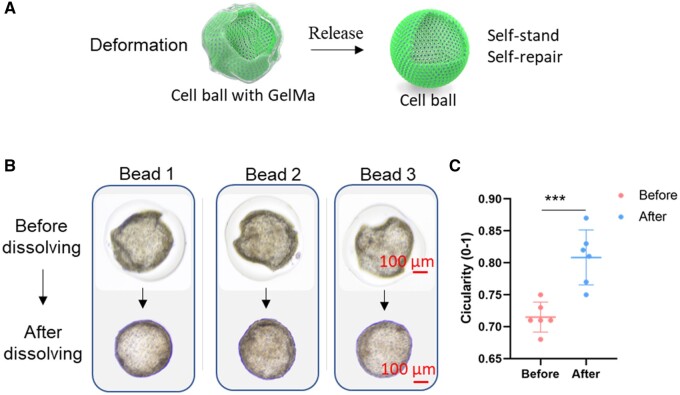
To achieve a pure cell ball, we dissolved the GelMa shell. Interestingly, the cell balls were self-supportive and substantially inflated into a rounder morphology after removal of the GelMa limit (Figure 6A and B). As the cell ball could pump up, it indicated the presence of a hollow chamber inside the cell barrier and that the cells might already have been connected and formed tight junctions. The circularity (0–1) has often been used to describe the morphology of a circle [25]. We also chose those deformed cell balls to demonstrate the deformation recovery process, and the circularity of the cell balls before and after release was calculated. As shown in Figure 6C, the circularity of deformed cell balls increased significantly from 0.715 ± 0.023 to 0.808 ± 0.043, which was in accordance with the inflation into a rounder morphology.
Figure 6.
Deformed cell balls were self-standing and presented deformation recovery after release from the GelMa shell. (A) Schematic section of cell ball releasing from core–shell beads. (B) Three representative deformed beads were chosen for release. (C) Circularity (0-1) of the cell ball in core–shell beads and after releasing. The cell balls were released at Day 12.
Hollow cell structure achievement
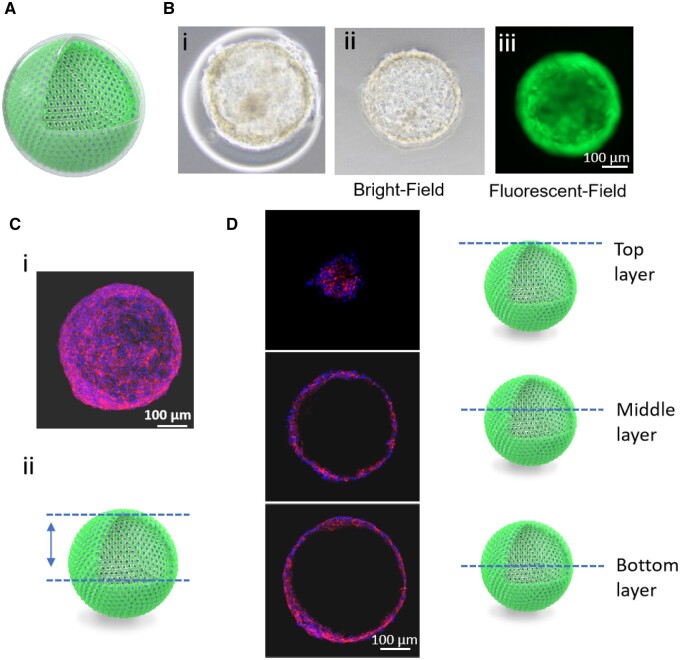
To verify the hollow chamber formation in cell balls, we used confocal microscopy to scan the cell structure stained with phalloidin and 4′, 6-diamidino-2-phenylindole (DAPI). Half structure of the cell balls was scanned and a 3D image as well as a video were reconstructed, as shown in Figure 7C and D, and Supplementary Video S1. The cell balls showed a hollow structure with different layers, confirming the successful fabrication of the hollow cell ball without the support of scaffolds or biomaterials. This was also a further clue on why the cell balls could maintain their structure and expand to a rounder morphology after GelMa dissolution (Figure 6).
Figure 7.
A Hollow cell ball was formed after GelMa dissolution. (A) Section of hollow cell ball. (B) (i) Core–shell beads before GelMa dissolution, (ii) the bright field image and (iii) green fluorescent image of GFP-HUVECs ball after GelMa dissolution. (C) (i) 3D construct of the confocal image of HUVECs ball and (ii) scanned area of the cell ball. (D) On the left are scanned images of a specific layer of the cell ball indicated on the right by schematic images. Cell balls were stained with DAPI (blue) and CD31(red).
To investigate whether the fabricated cell balls exhibited physiological properties, we analyzed the integrity of the 3D endothelial cell balls and the presence of characteristic marker proteins. Immunofluorescence staining was performed to verify the expression of vascular endothelial markers (CD31) in the cell ball. After confocal scanning of the cell ball, the images were reconstructed into a 3D model (Supplementary Figure S3). The presence of CD31 indicated the physiological maintenance ability of the HUVEC phenotype [26].
Discussion
In this study, we developed a novel capillary microfluidic platform to fabricate GelMa core–shell beads for cell ball formation by adhesion-guided self-assembly. To the best of our knowledge, this is the first time that GelMa core–shell beads have been fabricated for cell adhesion on the inner surface. Therefore, we propose a novel strategy for mimicking the structure of hollow tissue.
The major methods for hollow spheroid fabrication rely on self-assembly. For example, Human pancreatic cancer cells of the capan-1 line cultured in suspension and under agitation form hollow spheroids [27]. Magnetic levitation of 3T3 cells that ingested Fe3O4-containing microspheres can also form a millimeter of spheroids with a cavity [28]. These methods are difficult to fabricate high-throughput hollow spheroids with controlled size, cell layers. Recently, a 3D culture technique that combines cell encapsulation in sacrificial microbeads with the formation of spherical chambers in hydrogel matrices has been reported [17]. This method provides the method to fabricate enclosed hollow cell balls in a defined chamber. While this method needs several steps, and microbeads are varied in size. As hollow cell balls were immobilized in a hydrogel, they could not be applied in parallel testing. We provided this high-throughput, homogeneous method for controlled hollow cell ball fabrication, and hollow balls are free for parallel testing. As cell number and geometry can be controlled in this method, we can achieve relatively similar cell balls in a set time. Cell balls can be set as a starting point for growth factors that affect the hollow tissue development test. As cell balls are free in the medium, they also face the challenge of tracking the cell behavior in a defined ball for a long time. Hollow cell balls produced by this method can also be building blocks to the assembly of more complex tissues.
Commonly, alginate was used as the fabricated material in the microfluidic system because of its quick gelation property and fluid state at room temperature. However, alginate lacks the cell adhesion property, and cells encapsulated in the core–shell beads only form solid spheroids. GelMa has good cell adhesion properties because it is derived from gelatin and has an Arg-Gly-Asp (RGD) sequence [29]. It is a temperature-responsive hydrogel, and a microfluidic device requires a liquid state to control the flow of the hydrogel. Therefore, temperature control and quick gelation are key for GelMa microbead fabrication. Moreover, GelMA can be replaced with other photocurable biomaterials to fabricate tissue-specific beads, such as hyaluronic acid methacrylate, chitosan methacrylate and some biomaterials with different integrin-binding sequences [30]. This method is versatile in that we can easily replace core material (gelatin) with other biomaterials, such as collagen, fibrin, fish-gelatin and designed responsive hydrogels [31, 32]. With elaborately choosing a combination of different natural biomaterials, biomimic microenvironments for different tissues might achieved for disease modeling, target screening and tissue engineering.
Fluorinated oils are also often used in droplet microfluidics as they have several advantages in droplet microfluidics. Fluorinated oils are inert, biocompatible and allow gas exchange, making them ideal for biological applications [33, 34]. In this study, we use corn oil as a continuous phase to produce GelMa microbeads. There are already reports using corn oil in droplet microfluidics with no cell cytotoxicity [35, 36]. After microbead fabrication, corn oil could be easily washed away with hexadecane [19]. In the coaxial capillary device, the viscosity of the continuous phase and geometry of capillary tube are important factors for droplet formation [37]. And we found corn oil suitable in our device from GelMa microbeads. Cost is also a consideration factor for oil selection, as we use large amounts of oils, about 1 l/h.
In this study, we demonstrated the model with HUVECs while many other cells with hollow structures in vivo, such as lung alveolar cells, lens epithelial cells and otic-progenitor cells, should be further verified. This platform allows in vitro recapitulation of the tissues which form hollow structures in the early stages of development. This platform can be further developed for precision bead placement by combining a three-axis motion system [38]. Massive parallel of alveolar cell balls can be infected with bacteria and influenza virus to investigate the interaction of pathogen and host. Different antipathogen substances can also applied to screen the effect of drugs. Thus, a high-throughput method for massive hollow tissue platforms can be fabricated for disease modeling and drug screening.
During tissue morphogenesis, cell assembly and hollow structure formation are not clearly understood. Tunable cues affecting the hollow structure formation would be interesting for further work. For example, mechanical force plays an important role in cell differentiation and self-assembly. It provides a model tool for the study of biomechanics in the regulation of cell self-assembly as the mechanical can be tuned by change the concentration of shell materials. HUVECs were chosen for modeling as vascular endothelial cells commonly form hollow structures in vivo. It can also model angiogenesis when embedded in the ECM and stimulated by specific factors. The enclosed endothelial cell ball with tight junctions can serve as a starting point for fabricating the blood barrier in a complex model. For some specific tissues, such as the lens, a hollow structure formation marks the beginning of cell differentiation [39]. Next, lens epithelial cells differentiated into lens fiber cells and filled in the hollow cell ball to form the lens. The hollow lens epithelial cell balls with homogeneous size might be applied for massive screening for exact factors that can induce lens form. Therefore, this platform could be further developed as an efficient differentiation factor screening platform for specific tissue maturation studies.
This core–shell GelMa bead fabrication platform is versatile, although we only verified the hollow tissue fabrication ability. The core–shell structure of GelMa provides variable properties for further bioengineering applications. For single encapsulation, specific tissue cells or tumor cells can be loaded into the core for drug testing or disease modeling. For dual cell encapsulation, a complex hollow-tissue structure can be fabricated. And core–shell beads were suitable for cell encapsulation for in situ cell therapy, as they can protect cells from chemical and mechanical damage [40]. For example, islet cells can be encapsulated in the core and for in situ delivery. Suitable growth factors and drugs loaded in different zones of core–shell beads for controlled release might promote tissue regeneration, such as bone regeneration or wound healing [41, 42].
Conclusion
In conclusion, we demonstrated a novel strategy for fabricating a pure hollow cell structure in a controlled and high-throughput manner which challenges the previous bio-fabrication methods. Core–shell beads were fabricated as a guiding template for cell adhesion along the inner surface, and self-assembly to form a tight hollow cell structure was observed. Going forward, the development of such 3D models for different cell types, using variable differentiation cues, and at different scales can widen the applicability of in vitro tissue and organ modeling.
Supplementary Material
Acknowledgements
This research was supported by Key-Area Research and Development Program of Guangdong Province (2023B0909020003), the National Natural Science Foundation of China (Grant No. 52075285) and the Applied Basic Research Project of Sichuan Province (Grant No. 2021YJ0563).
Conflicts of interest statement. None declared.
Contributor Information
Jianwei Chen, Bio-intelligent Manufacturing and Living Matter Bioprinting Center, Research Institute of Tsinghua University in Shenzhen, Tsinghua University, Shenzhen 518057, People’s Republic of China; Precision Medicine and Healthcare Research Center, Tsinghua-Berkeley Shenzhen Institute (TBSI), Tsinghua University, Shenzhen 518055, People’s Republic of China.
Zeyang Liu, Department of Mechanical and Energy Engineering, Southern University of Science and Technology, Shenzhen, Nanshan District, People’s Republic of China.
Zixian Wang, Precision Medicine and Healthcare Research Center, Tsinghua-Berkeley Shenzhen Institute (TBSI), Tsinghua University, Shenzhen 518055, People’s Republic of China.
Xiuxiu Zhang, Precision Medicine and Healthcare Research Center, Tsinghua-Berkeley Shenzhen Institute (TBSI), Tsinghua University, Shenzhen 518055, People’s Republic of China.
Yi Zhang, Precision Medicine and Healthcare Research Center, Tsinghua-Berkeley Shenzhen Institute (TBSI), Tsinghua University, Shenzhen 518055, People’s Republic of China.
Zhen Zhan, Department of Mechanical and Energy Engineering, Southern University of Science and Technology, Shenzhen, Nanshan District, People’s Republic of China.
Xiaohua Gong, School of Optometry and Vision Science Program, University of California, Berkeley, CA 94720, USA.
Tao Xu, Bio-intelligent Manufacturing and Living Matter Bioprinting Center, Research Institute of Tsinghua University in Shenzhen, Tsinghua University, Shenzhen 518057, People’s Republic of China; Precision Medicine and Healthcare Research Center, Tsinghua-Berkeley Shenzhen Institute (TBSI), Tsinghua University, Shenzhen 518055, People’s Republic of China; Tsinghua Shenzhen International Graduate School, Tsinghua University, Shenzhen 518055, People’s Republic of China.
Supplementary data
Supplementary data are available at Regenerative Biomaterials online.
References
- 1. Baker BM, Chen C.. Deconstructing the third dimension–how 3D culture microenvironments alter cellular cues. J Cell Sci 2012;125:3015–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Griffith LG, Swartz M.. Capturing complex 3D tissue physiology in vitro. Nat Rev Mol Cell Biol 2006;7:211–24. [DOI] [PubMed] [Google Scholar]
- 3. Morizane R, Lam AQ, Freedman BS, Kishi S, Valerius MT, Bonventre J.. Nephron organoids derived from human pluripotent stem cells model kidney development and injury. Nat Biotechnol 2015;33:1193–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Mirbagheri M, Adibnia V, Hughes BR, Waldman SD, Banquy X, Hwang D.. Advanced cell culture platforms: a growing quest for emulating natural tissues. Mater Horiz 2019;6:45–71. [Google Scholar]
- 5. Nunes AS, Barros AS, Costa EC, Moreira AF, Correia IJJB.. Bioengineering, 3D tumor spheroids as in vitro models to mimic in vivo human solid tumors resistance to therapeutic drugs. Biotechnol Bioeng 2019;116:206–26. [DOI] [PubMed] [Google Scholar]
- 6. Callens SJ, Uyttendaele RJ, Fratila-Apachitei LE, Zadpoor AAJB.. Substrate curvature as a cue to guide spatiotemporal cell and tissue organization. Biomaterials 2019;232:119739. [DOI] [PubMed] [Google Scholar]
- 7. Werner M, Kurniawan NA, Bouten CVJM.. Cellular geometry sensing at different length scales and its implications for scaffold design. Materials 2020;13:963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Durruthy-Durruthy R, Gottlieb A, Heller S.. 3D computational reconstruction of tissues with hollow spherical morphologies using single-cell gene expression data. Nat Protoc 2015;10:459–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Alsina B, Whitfield TT.. Sculpting the labyrinth: morphogenesis of the developing inner ear. Semin Cell Dev Biol 2017;65:47–59. [DOI] [PubMed] [Google Scholar]
- 10. Chamberlain CG, McAvoy J.. Induction of lens fibre differentiation by acidic and basic fibroblast growth factor (FGF). Growth Factors 1989;1:125–34. [DOI] [PubMed] [Google Scholar]
- 11. Basil MC, Cardenas-Diaz FL, Kathiriya JJ, Morley MP, Carl J, Brumwell AN, Katzen J, Slovik KJ, Babu A, Zhou S, Kremp MM, McCauley KB, Li S, Planer JD, Hussain SS, Liu X, Windmueller R, Ying Y, Stewart KM, Oyster M, Christie JD, Diamond JM, Engelhardt JF, Cantu E, Rowe SM, Kotton DN, Chapman HA, Morrisey EE.. Human distal airways contain a multipotent secretory cell that can regenerate alveoli. Nature 2022;604:120–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. McAvoy JW, Chamberlain CG, de Iongh RU, Hales AM, Lovicu FJ.. Lens development. Eye (Lond) 1999;13 (Pt 3b):425–37. [DOI] [PubMed] [Google Scholar]
- 13. Koehler KR, Mikosz AM, Molosh AI, Patel D, Hashino E.. Generation of inner ear sensory epithelia from pluripotent stem cells in 3D culture. Nature 2013;500:217–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Hofbauer P, Jahnel S, Papai N, Giesshammer M, Mendjan S.. Cardioids Reveal Self-Organizing Principles of Human Cardiogenesis. Cold Spring Harbor Laboratory, 2020. [DOI] [PubMed] [Google Scholar]
- 15. Wong CC, Loewke KE, Bossert NL, Behr B, Jonge CD, Baer TM, Pera RJNB.. Non-invasive imaging of human embryos before embryonic genome activation predicts development to the blastocyst stage. Nat Biotechnol 2010;28:1115–21. [DOI] [PubMed] [Google Scholar]
- 16. Wang Z, Zhang X, Xue L, Wang G, Li X, Chen J, Xu R, Xu T.. A controllable gelatin-based microcarriers fabrication system for the whole procedures of MSCs amplification and tissue engineering. Regen Biomater 2023;10:rbad068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Wang E, Wang D, Geng A, Seo R, Gong X.. Growth of hollow cell spheroids in microbead templated chambers. Biomaterials 2017;143:57–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Ma M, Chiu A, Sahay G, Doloff JC, Dholakia N, Thakrar R, Cohen J, Vegas A, Chen D, Bratlie KM, Dang T, York RL, Hollister-Lock J, Weir GC, Anderson DG.. Core–shell hydrogel microcapsules for improved islets encapsulation. Adv Healthc Mater 2013;2:667–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Liu Z, Zhang H, Zhan Z, Nan H, Hu CJB. Mild formation of core-shell hydrogel microcapsules for cell encapsulation, 2020. [DOI] [PubMed]
- 20. Agarwal P, Choi JK, Huang H, Zhao S, Dumbleton J, Li J, He X.. P.S. Characterization, a biomimetic core–shell platform for miniaturized 3D cell and tissue engineering. Part Part Syst Charact 2015;32:809–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Alessandri K, Sarangi BR, Gurchenkov VV, Sinha B, Kießling TR, Fetler L, Rico F, Scheuring S, Lamaze C, Simon A, Geraldo S, Vignjevic D, Doméjean H, Rolland L, Funfak A, Bibette J, Bremond N, Nassoy P.. Cellular capsules as a tool for multicellular spheroid production and for investigating the mechanics of tumor progression in vitro. Proc Natl Acad Sci U S A 2013;110:14843–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Wang H, Liu H, Liu H, Su W, Chen W, Qin J. One-step generation of core–shell gelatin methacrylate (GelMA) microgels using a droplet microfluidic system. Adv Mater Technol 2019;4:1–10. [Google Scholar]
- 23. Galogahi FM, Zhu Y, An H, Nguyen NT.. Core-shell microparticles: generation approaches and applications. J Sci Adv Mater Dev 2020;5:417–35. [Google Scholar]
- 24. Huang H, Yin Y, Yong H, He X, Usta OB, Yarmush ML.. Generation and manipulation of hydrogel microcapsules by droplet-based microfluidics for mammalian cell culture. Lab Chip 2017;17:1913–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Cho H, Kim H, Lee K, Lasli S, Ung A, Hoffman T, Nasiri R, Bandaru P, Ahadian S, Dokmeci MR, Lee J, Khademhosseini A.. Liver‐on‐a‐chip: bioengineered multicellular liver microtissues for modeling advanced hepatic fibrosis driven through non‐alcoholic fatty liver disease (small 14/2021). Small 2021;17:1–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Human J, Ayalon O, Sabanai H, Lampugnani M, Dejana E. Spatial and temporal relationships between Cadherins and PECAM-1 in Cell-Cell. Journal of Cell Biology1994;126:247–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Hollande FE. Morphogenesis of “duct-like” structures in three-dimensional cultures of human cancerous pancreatic duct cells (Capan-1). In Vitro Cell Dev Biol Anim 1993;29A:574–84. [DOI] [PubMed] [Google Scholar]
- 28. Lee JH, Hur W.. Scaffold-free formation of a millimeter-scale multicellular spheroid with an internal cavity from magnetically levitated 3T3 cells that ingested iron oxide-containing microspheres. Biotechnol Bioeng 2014;111:1038–47. [DOI] [PubMed] [Google Scholar]
- 29. Van D, Bogdanov B, Rooze ND, Sch Ac Ht EH, Cornelissen M, Berghmans HJB.. Structural and rheological properties of methacrylamide modified gelatin hydrogels. Biomacromolecules 2000;1:31–8. [DOI] [PubMed] [Google Scholar]
- 30. Zhao J, Santino F, Giacomini D, Gentilucci LJB.. Integrin-targeting peptides for the design of functional cell-responsive biomaterials. Biomedicines 2020;8:307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Jiao F, Zhao W, Zhao W, Wang Y, Deng Y, Chang S, Sun J, Lou Q, Wang L, Shan C-X, Xiao Y, Dong L.. Biomass-derived washable composites for accelerating the healing of infected wounds. BMEMat 2023;1:e12055. [Google Scholar]
- 32. Wang S, Wu W-Y, Yeo JCC, Soo XYD, Thitsartarn W, Liu S, Tan BH, Suwardi A, Li Z, Zhu Q, Loh XJ.. Responsive hydrogel dressings for intelligent wound management. BMEMat 2023;1:e12021. [Google Scholar]
- 33. Holtze C, Rowat AC, Agresti JJ, Hutchison JB, Angilè FE, Schmitz CH, Köster S, Duan H, Humphry KJ, Scanga RA, Johnson JS, Pisignano D, Weitz DA.. Biocompatible surfactants for water-in-fluorocarbon emulsions. Lab Chip 2008;8:1632–9. [DOI] [PubMed] [Google Scholar]
- 34. Utech S, Prodanovic R, Mao AS, Ostafe R, Mooney DJ, Weitz DA.. Microfluidic generation of monodisperse, structurally homogeneous alginate microgels for cell encapsulation and 3D cell culture. Adv Healthc Mater 2015;4:1628–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Morimoto Y, Tan WH, Tsuda Y, Takeuchi S.. Monodisperse semi-permeable microcapsules for continuous observation of cells. Lab Chip 2009;9:2217–23. [DOI] [PubMed] [Google Scholar]
- 36. Tan WH, Takeuchi S.. Monodisperse alginate hydrogel microbeads for cell encapsulation. Adv Mater 2007;19:2696–701. [Google Scholar]
- 37. Gruner P, Riechers B, Chacòn Orellana LA, Brosseau Q, Maes F, Beneyton T, Pekin D, Baret J-C.. Stabilisers for water-in-fluorinated-oil dispersions: key properties for microfluidic applications. Curr Opin Colloid Interface Sci 2015;20:183–91. [Google Scholar]
- 38. Jiang S, Zhao H, Zhang W, Wang J, Liu Y, Cao Y, Zheng H, Hu Z, Wang S, Zhu Y. (2020) An automated organoid platform with inter-organoid homogeneity and inter-patient heterogeneity. ScienceDirect2020;1:100161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Mochizuki T, Masai I.. The lens equator: a platform for molecular machinery that regulates the switch from cell proliferation to differentiation in the vertebrate lens. Dev Growth Differ 2014;56:387–401. [DOI] [PubMed] [Google Scholar]
- 40. Wang C, Wang S, Kang DD, Dong Y.. Biomaterials for in situ cell therapy. BMEMat 2023;1:e12039. [Google Scholar]
- 41. Ko A, Liao C.. Hydrogel wound dressings for diabetic foot ulcer treatment: status-quo, challenges, and future perspectives. BMEMat 2023;1:e12037. [Google Scholar]
- 42. Song J, Li L, Fang L, Zhang E, Zhang Y, Zhang Z, Vangari P, Huang Y, Tian F, Zhao Y, Chen W, Xue J.. Advanced strategies of scaffolds design for bone regeneration. BMEMat 2023;1:e12046. [Google Scholar]
- 43. Xuanzhi W, Xingliang D, Xinzhi Z, Cheng M, Xinda LA. 3D bioprinted glioma cell‐laden scaffolds enriching glioma stem cells via epithelial–mesenchymal transition, 2019. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.