Abstract
Functional traits influence the assembly of microbial communities, but identifying these traits in the environment has remained challenging. We studied ectomycorrhizal fungal (EMF) communities inhabiting Populus trichocarpa roots distributed across a precipitation gradient in the Pacific Northwest, USA. We profiled these communities using taxonomic (meta-barcoding) and functional (metagenomic) approaches. We hypothesized that genes involved in fungal drought-stress tolerance and fungal mediated plant water uptake would be most abundant in drier soils. We were unable to detect support for this hypothesis; instead, the abundance of genes involved in melanin synthesis, hydrophobins, aquaporins, trehalose-synthases, and other gene families exhibited no significant shifts across the gradient. Finally, we studied variation in sequence homology for certain genes, finding that fungal communities in dry soils are composed of distinct aquaporin and hydrophobin gene sequences. Altogether, our results suggest that while EMF communities exhibit significant compositional shifts across this gradient, coupled functional turnover, at least as inferred using community metagenomics is limited. Accordingly, the consequences of these distinct EMF communities on plant water uptake remain critically unknown, and future studies targeting the expression of genes involved in drought stress tolerance are required.
Keywords: ectomycorrhizal fungi, metagenomics, drought, gradient, profiling, community assembly
Introduction
Identifying functional traits that mediate the distribution and functioning of microbial communities represents an urgent area of research [1]. This is because microbial trait distributions can promote understanding of the role of microbes in key biogeochemical transformations, such as those required for adaptation to altered precipitation regimes. Historically, the analysis of microbial traits has largely relied on morphological or process-based measurements. The abundance of molecular genes can serve as proxies for microbial functioning and traits, and they have the advantage of being readily studied using whole-community metagenomic profiling [2, 3].
Ectomycorrhizal fungi (EMF) are dominant microbial members of forest ecosystems. Well-known for their role in plant nutrient uptake, EMF may also improve plant drought stress tolerance via specialized water transporters that increase root hydraulic conductivity [4, 5] and by extending the surface area of plant roots. Through their role in plant water uptake, EMF support the evolution of drought tolerance in host plants [6]. However, the extent to which EMF serve to extend plant drought tolerance and increase plant water uptake under field conditions has remained inconclusive [4].
In light of laboratory evidence that EMF influence plant water relations, widespread observations of EMF community turnover across precipitation gradients is notable [7]. Two coupled processes could generate turnover in EMF composition and function across precipitation gradients. First, these patterns could result from variation in inherent EMF physiological tolerance to soil water availability or conditions which are modified by increased water availability. Secondly, plant water demand may influence EMF community composition if plants reward EMF that transfer greater water resources [8]. Such non-mutually exclusive processes would result in coupled compositional and functional trait turnover, whereby traits involved in water acquisition and fungal drought tolerance would exhibit the greatest turnover [9].
Shifts in gene counts per genome, or gene counts measured at the community level, represent tractable molecular approaches to study shifts in microbial trait profiles [2]. Metagenomic measures of microbial genes have been used to identify genes that could serve to determine the suitability of organisms to the local environment [2, 3]. This approach is conceptually similar to analysing plant leaf or root traits along ecological gradients where coupled shifts are considered indicative of trait-based environmental filtering [9].
In the current study, we studied EMF communities inhabiting the roots of Populus trichocarpa distributed across an ecosystem-scale gradient of water availability in the Pacific Northwest, USA (Fig. 1A). P. trichocarpa is a widespread and important pioneer tree species in riparian habitats; we studied this species as a standardized host to remove the potentially confounding effects of host differences on EMF communities. We tested the hypothesis that EMF communities exhibit coupled taxonomic and functional shifts across a precipitation gradient, and that EMF communities inhabiting drier soils are enriched in genes that promote drought stress tolerance and host plant water uptake. We focused on a core set of gene families which previous work identified as most likely involved in fungal drought stress tolerance and water uptake. Due to the relatively small sample size, we wanted to test a priori hypotheses regarding specific gene families rather than focusing on global scale genome analysis. I we focused on core gene families such as fungal hydrophobins, aquaporins, melanins, and trehalose synthases, collectively some of the best characterized genes related to drought tolerance for fungi [10–13]. Fungal aquaporins are thought to play a critical role in transmembrane water transfer to host roots and may beneficially influence root water uptake [5]. Melanin is a component of fungal cell wall and has been shown to improve drought stress tolerance by reducing osmotic stress [14]. Fungal hydrophobins are small amphiphilic molecules that coat hyphae, serving to facilitate growth across air-filled soil pores. Finally, we also studied certain carbohydrate active enzymes (CAZy), which could be involved in cell wall remodeling and transmembrane water transfer [13].
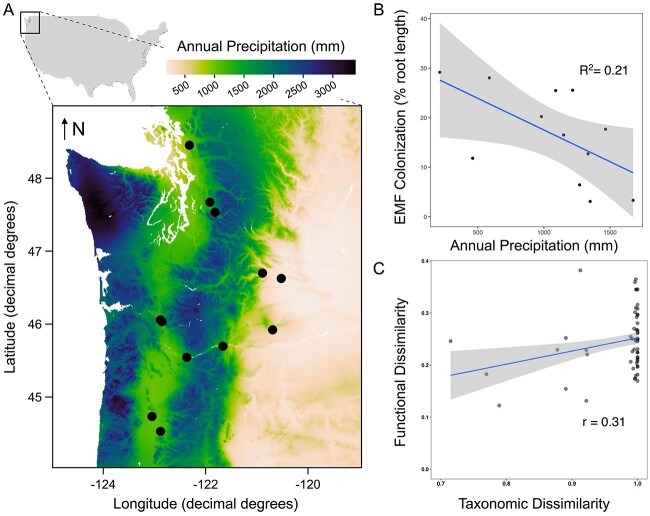
Figure 1.
(A). Sites across the Pacific Northwest (USA), where ectomycorrhizal fungal (EMF) communities associated with Populus trichocarpa were sampled. (B) EMF colonized root length was inversely correlated with mean annual precipitation (MAP; P = .04; R2 = 0.21). (C) Significant taxonomic and functional coupling for EMF communities, as measured using counts of Pfam annotated genes (Bray–Curtis distances). Taxonomic dissimilarity is measured using fungal amplicon sequence variants (ASV). For (B and C), Lines represent linear splines with 95% confidence intervals.
Results and discussion
In July and August of 2017, we conducted a field survey of root samples from 12 forest sites dominated by P. trichocarpa [15]. These sites represent a subset of a continental scale sampling network [16]. The 12 field sites differ markedly in mean annual precipitation (MAP: range = 213–1674 mm yr−1; Fig. 1a), and we used MAP as a coarse proxy for soil water availability. EMF root-tip colonization (% root-length colonized: grid-intersect method [17]) ranged from approximately 4–29%, and colonization was inversely correlated with mean annual precipitation (MAP; P = 0.04; Fig. 1b). We characterized EMF communities in P. trichocarpa using metabarcoding of the ITS1 region of rDNA. Soil properties such as pH, and soil carbon and nitrogen were also measured from the sampled soil cores (Supplementary Methods).
The relative abundance of Basidiomycete and Ascomycete EMF were invariant across the sampled gradient (Supplementary Figure 1). However, at finer taxonomic scales, EMF communities exhibited significant compositional turnover. Community dissimilarity as measured using fungal ASVs, was primarily associated with variation in soil pH (P = 0.01), and marginally with MAP (P = 0.06; PERMANOVA; Supplementary Table 1; Supplementary Figure 2), but non-significantly by other soil chemical parameters such as soil carbon, or nitrogen release. It should be noted that soil pH and MAP were significantly correlated, with drier sites being more alkaline (R2 = 0.25, P < 0.001); this correlation is often observed, in part because increased precipitation reduces the availability of buffering ions (i.e. carbonates; [18]). Overall, EMF genera such as Tuber and Wilcoxina were dominant in drier sites, whereas Scleroderma and Geopora were most abundant in wetter sites (Supplementary Figure 3).
The same DNA pool was used for the construction of metagenomic libraries, sequenced using Illumina NovaSeq. We employed the JGI IMG pipeline to filter and annotate fungal reads to Pfam domains, whilst removing plant and bacterial sequences [19] (Supplementary Methods). Overall, soil chemistry and climatic variables were insignificant predictors of community-scale variation in fungal gene dissimilarity (all Pfam domains: Supplementary Figure 4). We further studied composition-function linkages, finding that fungal communities and functional gene composition were significantly correlated after accounting for geographic distances among samples (Partial Mantel r = 0.31, P = 0.007; Fig. 1c).
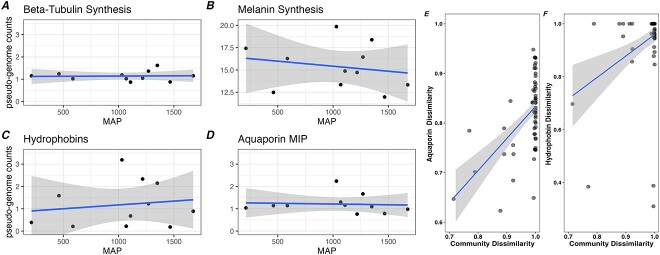
We hypothesized that trees occurring in comparatively drier soils would host EMF enriched in genes related to drought stress and water acquisition. Although individual gene families involved in melanin synthesis exhibited hypothesized trends across the precipitation gradient, these relationships were statistically insignificant (Supplementary Figure 5). We studied the cumulative abundance of genes involved in melanin synthesis, aquaporins, trehalose synthases, hydrophobins, respectively, as well as other genes encoding abundant carbohydrate active enzymes (CAZymes). Overall, we detected no statistically significant shifts in gene abundances for any gene category (Fig. 2; Supplementary Fig. 6). Because soil pH was a strong predictor of EMF community composition, we additionally studied correlations in soil pH and abundance of targeted gene families; we observed broadly similar but overall insignificant relationships (Supplementary Fig. 7).
Figure 2.
(A-D). Normalized abundance of gene family counts across the precipitation gradient: Mean annual precipitation (MAP). In order to account for variation in sequencing depth and the compositional nature of the metagenomic data, we performed a procedure that normalizes gene counts by those encoding the near single-copy gene Asparaginase. This additive log-ratio procedure, allows for the calculation of pseudo-genome counts for key gene families. Lines represent linear splines with 95% confidence intervals. (E-F). Correlations between EMF community dissimilarity (Bray–Curtis), and aquaporin and Hydrophobin sequence dissimilarity based on Pfam homology.
While the above analyses measured gene abundances on a statistically derived genome level, we additionally reasoned that P. trichocarpa occurring in dry soils would host a greater total abundance of EMF genes involved in drought stress tolerance. Estimating the net abundance of EMF genes involved in water relations captures relative plant investment and reliance on the EMF community. To estimate plant investment in EMF across this gradient we weighted metagenomic gene-counts by the percentage of root-length colonized by EMF (Supplementary Fig. 8); this weighting procedure serves as an estimate of the cumulative biomass of EMF on roots [3]. We detected strong negative relationships in the weighted abundance of genes involved in melanin synthesis (P = .03, R2 = 0.36), aquaporins (P = .02, R2 = 0.43), trehalose synthases (P = .03, R2 = 0.35), alpha amylase (PF00128; P = .03, R2 = 0.35), melanin synthases (P = .02, R2 = 0.36), but not fungal hydrophobins (P = .86) (Supplementary Fig. 8). Similar patterns were observed when weighted gene counts were regressed against soil pH (Supplementary Fig. 9). We acknowledge that this weighting-procedure is coarse, and that the effect of MAP for individual target gene families must be compared with genes that serve as a statistical null. In this case, Beta-tubulins, a gene at near-single copy in fungal genomes and plausibly under minimal ecological selective pressure across this soil gradient, exhibited similar relationships as observed for functional genes (P = .02, R2 = 0.40). This therefore limits understanding of the significance of the cumulative abundance of these gene families in plant water relations. Sampling from other host-trees would be useful to help understand the generality of the patterns observed here.
Finally, we investigated potential intra-genetic variation in functional genes by focusing on gene sequence homology. Using a partial mantel test, we identified aquaporin (Mantel r = 0.50, P = .001) and hydrophobin (Mantel r = 0.33, P = .023) sequence dissimilarity (Bray–Curtis) as positively correlated with EMF community dissimilarity (Fig. 2E and F). Consequentially, the functioning of aquaporins or hydrophobins could co-vary with EMF communities across the precipitation gradient irrespective of shifts in gene abundances. Additional targeted gene families either did not have sufficient data for calculation of distance matrices or exhibited insignificant relationships. Further functional analysis is required to understand if fungal aquaporins or hydrophobins vary in their activity across this gradient.
Conclusions
EMF communities associated with P. trichocarpa exhibited large compositional shifts across a natural precipitation gradient. Moreover, P. trichocarpa root-systems were significantly more colonized in drier soils. Despite distinct EMF communities occurring in drier soils, the abundance of genes hypothesized to be involved in drought stress tolerance was relatively invariant. One scenario is that EMF differentially express genes putatively involved in drought-stress tolerance despite possessing a similar core repertoire. In addition, our results demonstrating significant variation in sequence homology for aquaporin and hydrophobin genes could be suggestive of differences in the functioning of these genes across the studied precipitation gradient. In both cases metagenomic profiling would be unable to differentiate amongst these possibilities. Our results therefore suggest that meta-transcriptomic, or metabolomic profiling could be necessary to infer the physiological attributes of ECM drought stress tolerance and plant water uptake [11, 20]. Finally, due to the small number of communities studied here, additional sampling is necessary to infer the role of MAP and soil water availability in structuring these communities. Moreover, we acknowledge that the patterns observed here are challenging to disentangle from soil variables like pH.
Supplementary Material
Contributor Information
Peter T Pellitier, Department of Biology, Stanford University, Stanford CA 94305, United States; Department of Earth System Science, Stanford University, Stanford CA 94305, United States.
Michael Van Nuland, Department of Biology, Stanford University, Stanford CA 94305, United States; Society for the Protection of Underground Networks, SPUN, Dover, DE 19901, United States.
Asaf Salamov, Department of Energy Joint Genome Institute, Lawrence Berkeley National Laboratory, Berkeley, CA 94720 United States.
Igor V Grigoriev, Department of Energy Joint Genome Institute, Lawrence Berkeley National Laboratory, Berkeley, CA 94720 United States; Department of Plant and Microbial Biology, University of California Berkeley, Berkeley, CA 94720, United States.
Kabir G Peay, Department of Biology, Stanford University, Stanford CA 94305, United States; Department of Earth System Science, Stanford University, Stanford CA 94305, United States.
Conflicts of interest
The authors declare no conflicts of interest.
Funding
P.T.P. was supported by NSF PRFB 2010897. K.G.P. and M.V.N. were supported by DOE award DE-SC0016097. The work (proposal: 10.46936/10.25585/60000790) conducted by the U.S. Department of Energy Joint Genome Institute (https://ror.org/04xm1d337), a DOE Office of Science User Facility, is supported by the Office of Science of the U.S. Department of Energy operated under Contract No. DE-AC02-05CH11231.
Data availability
ITS amplicon sequence data are archived in SRA SUB13575173, and bioproject accession PRJNA987748. Metagenomic data is available through JGI, IMG # 10.46936/10.25585/60000790. No restrictions to data utilization.
References
- 1. Malik AA, Martiny JBH, Brodie EL. et al. Defining trait-based microbial strategies with consequences for soil carbon cycling under climate change. The ISME Journal 2020;14:1–9. 10.1038/s41396-019-0510-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Bouma-Gregson K, Olm MR, Probst AJ. et al. Impacts of microbial assemblage and environmental conditions on the distribution of anatoxin-a producing cyanobacteria within a river network. The ISME Journal 2019;13:1618–34. 10.1038/s41396-019-0374-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Pellitier PT, Zak DR. Ectomycorrhizal fungal decay traits along a soil nitrogen gradient. New Phytol 2021;232:2152–64. 10.1111/nph.17734 [DOI] [PubMed] [Google Scholar]
- 4. Lehto T, Zwiazek JJ. Ectomycorrhizas and water relations of trees: a review. Mycorrhiza 2011;21:71–90. 10.1007/s00572-010-0348-9 [DOI] [PubMed] [Google Scholar]
- 5. Xu H, Zwiazek JJ. Fungal Aquaporins in ectomycorrhizal root water transport. Front Plant Sci 2020;11:521729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Cosme M. Mycorrhizas drive the evolution of plant adaptation to drought. Commun Biol 2023;6:1–10. 10.1038/s42003-023-04722-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Erlandson SR, Savage JA, Cavender-Bares JM. et al. Soil moisture and chemistry influence diversity of ectomycorrhizal fungal communities associating with willow along an hydrologic gradient. FEMS Microbiol Ecol 2016;92:fiv148. 10.1093/femsec/fiv148 [DOI] [PubMed] [Google Scholar]
- 8. Bogar L, Peay K, Kornfeld A. et al. Plant-mediated partner discrimination in ectomycorrhizal mutualisms. Mycorrhiza 2019;29:97–111. 10.1007/s00572-018-00879-7 [DOI] [PubMed] [Google Scholar]
- 9. Cornwell WK, Ackerly DD. Community assembly and shifts in plant trait distributions across an environmental gradient in coastal California. Ecol Monogr 2009;79:109–26. 10.1890/07-1134.1 [DOI] [Google Scholar]
- 10. Branco S, Schauster A, Liao H-L. et al. Mechanisms of stress tolerance and their effects on the ecology and evolution of mycorrhizal fungi. New Phytol 2022;235:2158–75. 10.1111/nph.18308 [DOI] [PubMed] [Google Scholar]
- 11. Erlandson SR, Margis R, Ramirez A. et al. Transcriptional acclimation and spatial differentiation characterize drought response by the ectomycorrhizal fungus Suillus pungens. New Phytol 2022;234:1910–3. 10.1111/nph.17816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Koide RT, Fernandez C, Malcolm G. Determining place and process: functional traits of ectomycorrhizal fungi that affect both community structure and ecosystem function. New Phytol 2014;201:433–9. 10.1111/nph.12538 [DOI] [PubMed] [Google Scholar]
- 13. Treseder KK, Lennon JT. Fungal traits that drive ecosystem dynamics on land. Microbiol Mol Biol Rev 2015;79:243–62. 10.1128/MMBR.00001-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Fernandez CW, Koide RT. The function of melanin in the ectomycorrhizal fungus Cenococcum geophilum under water stress. Fungal Ecol 2013;6:479–86. 10.1016/j.funeco.2013.08.004 [DOI] [Google Scholar]
- 15. Barge EG, Leopold DR, Peay KG. et al. Differentiating spatial from environmental effects on foliar fungal communities of Populus trichocarpa. J Biogeogr 2019;46:2001–11. 10.1111/jbi.13641 [DOI] [Google Scholar]
- 16. Van Nuland M, Daws ECS, Bailey JK. et al. Above– and belowground fungal biodiversity of Populus trees on a continental scale. Nat Microbiol 2023;8:2406–19 [DOI] [PubMed] [Google Scholar]
- 17. McGONIGLE TP, Miller MH, Evans DG. et al. A new method which gives an objective measure of colonization of roots by vesicular—arbuscular mycorrhizal fungi. New Phytol 1990;115:495–501. 10.1111/j.1469-8137.1990.tb00476.x [DOI] [PubMed] [Google Scholar]
- 18. Slessarev EW, Lin Y, Bingham NL. et al. Water balance creates a threshold in soil pH at the global scale. Nature 2016;540:567–9. 10.1038/nature20139 [DOI] [PubMed] [Google Scholar]
- 19. Chen I-MA, Chu K, Palaniappan K. et al. IMG/M v.5.0: an integrated data management and comparative analysis system for microbial genomes and microbiomes. Nucleic Acids Res 2019;47:D666–77. 10.1093/nar/gky901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Malik AA, Swenson T, Weihe C. et al. Drought and plant litter chemistry alter microbial gene expression and metabolite production. ISME J 2020;14:2236–47. 10.1038/s41396-020-0683-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
ITS amplicon sequence data are archived in SRA SUB13575173, and bioproject accession PRJNA987748. Metagenomic data is available through JGI, IMG # 10.46936/10.25585/60000790. No restrictions to data utilization.