Abstract
Methane (CH4) and nitrous oxide (N2O) are major greenhouse gases that are predominantly generated by microbial activities in anoxic environments. N2O inhibition of methanogenesis has been reported, but comprehensive efforts to obtain kinetic information are lacking. Using the model methanogen Methanosarcina barkeri strain Fusaro and digester sludge-derived methanogenic enrichment cultures, we conducted growth yield and kinetic measurements and showed that micromolar concentrations of N2O suppress the growth of methanogens and CH4 production from major methanogenic substrate classes. Acetoclastic methanogenesis, estimated to account for two-thirds of the annual 1 billion metric tons of biogenic CH4, was most sensitive to N2O, with inhibitory constants (KI) in the range of 18–25 μM, followed by hydrogenotrophic (KI, 60–90 μM) and methylotrophic (KI, 110–130 μM) methanogenesis. Dissolved N2O concentrations exceeding these KI values are not uncommon in managed (i.e. fertilized soils and wastewater treatment plants) and unmanaged ecosystems. Future greenhouse gas emissions remain uncertain, particularly from critical zone environments (e.g. thawing permafrost) with large amounts of stored nitrogenous and carbonaceous materials that are experiencing unprecedented warming. Incorporating relevant feedback effects, such as the significant N2O inhibition on methanogenesis, can refine climate models and improve predictive capabilities.
Keywords: nitrous oxide, methane, greenhouse gas emissions, inhibition, feedback loop, climate change
Introduction
Carbon dioxide (CO2) receives primary attention as a driver for climate change, but methane (CH4) and nitrous oxide (N2O) account for ~20% and 7%, respectively, of the net radiative forcing in the atmosphere [1, 2]. The central objective of the Paris Agreement, which is to hold the global average temperature increase to “well below 2°C above preindustrial levels” [3, 4], cannot be met without controlling CH4 and N2O emissions. Global emissions of both CH4 and N2O are ultimately controlled by microbial processes [2, 5]; however, human activities have massively disturbed the natural balance of microbial production and consumption of these greenhouse gases [6]. The current estimated annual net atmospheric emission increases of ~51 Tg of CH4 [7, 8] and 2.2 Tg of N2O [9] are both predicted to accelerate [1, 10]. The key microbial guilds and biogeochemical processes responsible for CH4 and N2O production and consumption are known, and their responses to climate change have been a matter of intense research [11, 12]. One area of considerable uncertainty pertains to positive and negative feedback loops affecting greenhouse gas emissions and climate [13]. An infamous scenario for a positive feedback loop in response to a warming climate is permafrost thawing, which stimulates the microbial activity and the release of CO2, CH4, and N2O from newly bioavailable carbon and nitrogen pools. The massive release of greenhouse gases will further accelerate radiative forcing and in turn cause more thawing. This example illustrates why a comprehensive understanding of both positive and negative feedback loops is essential. Quantitative information is needed to meaningfully incorporate feedback effects into refined climate models.
Methanogenic archaea (methanogens) drive CH4 production by utilizing acetate, H2/CO2, and methylated compounds as substrates, generating around 1 billion metric tons of CH4 annually [14, 15]. Approximately, two-thirds of biogenic CH4 is produced from acetoclastic methanogenesis (i.e. the conversion of acetate to CH4 and CO2), with the remaining one-third attributed to hydrogenotrophic CO2 reduction (hydrogenotrophic pathway) and methylated compound utilization (methylotrophic pathways) [16–20]. While the three major methanogenesis pathways share a core set of enzymes, several mechanistically distinct enzyme systems with corrinoid prosthetic groups (i.e. vitamin B12 derivatives) are involved in methyl group transfer reactions, energy conservation, and CH4 production (Fig. 1) [18, 19, 21].
Figure 1.
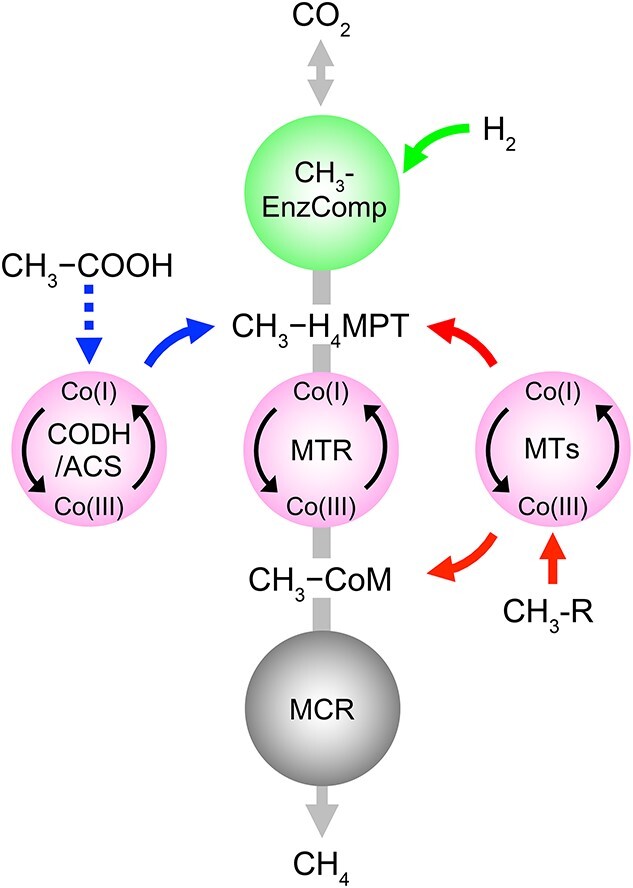
Illustration of the major methanogenic pathways that together account for most of the biogenically produced CH4 in nature; acetoclastic (left), hydrogenotrophic (top), and methylotrophic (right) conversions channel into the methanogenic pathway; the central circles indicate steps catalyzed by corrinoid-dependent enzyme systems potentially susceptible to N2O inhibition; abbreviations: CH3-EnzComp, CH3-formation enzyme complexes; CH3-H4MPT, methyl-tetrahydromethanopterin; MTR, N5-methyltetrahydromethanopterin:CoM methyltransferase; MTs, substrate-specific methyltransferases; CH3-R, methylated compounds (e.g. methanol).
In hydrogenotrophic methanogenesis, CO2 is sequentially reduced to a methyl group carried by tetrahydromethanopterin (H4MPT) with H2 as electron donor. The corrinoid-dependent N5-methyltetrahydromethanopterin:CoM methyltransferase (MTR) complex then transfers the methyl group onto another C1-carrier, coenzyme M, forming CH3–S–CoM, a step associated with energy conservation, followed by CH4 production catalyzed by methyl coenzyme M reductase (MCR) [22]. Acetoclastic methanogens produce CH4 by activating acetate to acetyl-CoA, which is then cleaved by the corrinoid-dependent enzyme system carbon monoxide dehydrogenase/acetyl-CoA synthase (CODH/ACS) to yield an enzyme-bound methyl group and a carbonyl group. The carbonyl moiety is oxidized to CO2 and the methyl group is transferred to H4MPT [23]. The MTR complex catalyzes a methyl group transfer to form CH3–S–CoM, a step associated with energy conservation, before MCR mediates the reduction of the methyl group to CH4 [16, 24]. Methylotrophic methanogens either directly generate CH3–S–CoM from methylated compounds (e.g. MeOH) utilizing substrate-specific, corrinoid-dependent MTRs, or cleave the methyl group from methoxylated compounds (e.g. 2-methoxybenzoate) and form CH3–CoM via H4MPT and the MTR complex [25], followed by CH4 production via MCR. All three methanogenesis pathways utilize corrinoid-dependent enzyme systems for methyl group transfers and have strict requirements for super-reduced Co(I) to generate CH4 and conserve energy [26, 27]. These features render methanogenesis sensitive to oxidative stress (e.g. fluctuating redox conditions and oxygen intrusion) [19, 28].
N2O is an even stronger oxidant than oxygen (Eo’(N2O(g)/N2(g) = +1.355 V > Eo’(O2(g)/H2O(l) = +0.818 V), reacts with Co(I), and has been shown to interfere with Co(I) cobamide-dependent enzyme systems [29, 30]. Demonstrated metabolic consequences include ceased corrinoid-dependent methionine biosynthesis and impaired organohalide respiration [31–33]. Based on this information, elevated N2O in anoxic ecosystems would be expected to inhibit other corrinoid-dependent processes such as methanogenesis; however, available studies investigating the impacts of N2O on methanogenesis have led to inconsistent N2O inhibition patterns for axenic methanogen cultures and CH4 producing microbial communities [34–36]. Laboratory incubations showed that Methanobacterium bryantii strain Bab1 grown with H2/CO2 ceased CH4 production in the presence of 95 μM N2O, whereas Methanosarcina barkeri strain MS maintained some methanogenic activity at 10-fold higher N2O concentrations under the same growth conditions [35]. Different sensitivities to N2O inhibition were also reported for mixed methanogenic cultures maintained with different substrates. For example, inhibition of CH4 production in a mixed community bioreactor occurred only at N2O concentrations exceeding 700 μM [36], whereas 20–28 μM N2O completely inhibited methanogenic activity in salt marsh sediment and Amazon peatland enrichment cultures [34, 37]. These variable sensitivities to N2O suggest that inhibition of methanogenesis by N2O is organism- and possibly substrate-specific; however, the available data are scarce and do not allow a robust, quantitative assessment of N2O inhibition on methanogenesis from relevant methanogenic substrates [34–36].
N2O fluxes in soil–water systems have risen sharply due to the intensified use of synthetic N fertilizer in agriculture [6, 9, 38]. As a result, elevated N2O concentrations occur more frequently in ecosystems with CH4 production such as rice paddy soils [39], wastewater treatment plants [40], sediments [41], and groundwater aquifers [42]. Also, permafrost thawing accelerates N turnover, releasing large amounts of N2O [38, 43]. More detailed knowledge about the interactions between N2O concentrations and methanogenesis is needed to advance the predictive capabilities of climate change impacts on future greenhouse gas emission scenarios. To address the existing knowledge gaps, we assessed the inhibitory effect of N2O on CH4 production from major methanogenic substrates in growth experiments with axenic and mixed methanogenic cultures and in whole-cell suspension assays. Using cultures performing acetoclastic, methylotrophic, and/or hydrogenotrophic methanogenesis, we determined kinetic parameters that quantitatively describe N2O inhibition on methanogenesis.
Materials and methods
Methanogenic cultures and growth inhibition experiments
The methanogenic archaeon M. barkeri strain Fusaro metabolizes acetate, H2/CO2, and MeOH while employing different, substrate-specific methanogenic pathways. To determine if M. barkeri exhibits varied sensitivities to N2O, cultures were pregrown with MeOH, H2/CO2, or acetate for at least three consecutive transfers before the impact of N2O on CH4 production was examined. Also, we analyzed three methanogenic mixed cultures derived from anaerobic digester sludge, which is known to harbor a broad diversity of methanogenic archaea. Three different enrichment cultures from the same source material were obtained with MeOH, H2/CO2, or acetate as growth substrate. The mixed cultures were transferred at least six times on the respective substrate and were used to examine the impact of N2O on CH4 production from different methanogenic substrates (see Supplemental Information for additional information on the mixed methanogenic cultures). Experiments were performed in triplicate 60-ml glass serum bottles with 30 ml N2/CO2 (80/20, v/v) headspace and 30 ml bicarbonate-buffered (50 mM) mineral salt medium (pH 7.2) reduced with 0.2 mM sulfide and 0.2 mM L-cysteine [44]. Acetoclastic, methylotrophic, and hydrogenotrophic cultures received 20 mM acetate, 30 mM MeOH, and 1.24 mmol H2, respectively. To avoid overpressure in bottles with H2 as electron donor, the headspace of culture bottles was replaced with 30 ml of filter-sterilized H2. For M. barkeri cultures, 0.1–1.0 ml of N2O gas (undiluted or 10-fold diluted in N2) was directly added to the incubation vessels to achieve final aqueous N2O concentrations of 100 and 200 μM in cultures with MeOH, 50 and 100 μM in cultures with H2, and 20 and 50 μM in cultures with acetate. For the methanogenic mixed cultures grown with 30 mM MeOH, 30 ml H2 (1.24 mmol), and 20 mM acetate, 0.5–1.6 ml of 10-fold diluted N2O gas (in N2) was introduced to achieve final aqueous phase N2O concentrations of 10 and 30 μM. More detailed information about N2O additions and concentration calculations is provided in the Supplemental Methods. All cultures were incubated without agitation at 37°C in the dark with the stoppers facing up, and replicates without N2O and without inoculum served as positive and negative controls, respectively. CH4 and N2O were analyzed throughout the growth experiments by injecting 100 μl headspace samples into an Agilent 3000A Micro gas chromatograph equipped with thermal conductivity detectors and a molecular sieve column and a PLOT Q column for CH4 and N2O measurements, respectively.
Whole-cell suspension assays to determine N2O inhibition constants for CH4 production
The M. barkeri and the methanogenic mixed cultures were first grown in 1.6 l medium and harvested via centrifugation when about two-thirds of the initial substrate (i.e. MeOH, H2/CO2, or acetate) had been converted to CH4, as calculated based on CH4 production according to Equations (1)–(3) (see below). The cell pellets collected from 1.6 l of medium were washed and suspended in 1.6 ml of reduced mineral salt medium in sealed 2-ml glass vials, resulting in a 1000-fold concentration of the biomass. A 0.2 ml aliquot of the concentrated cell suspension was sacrificed to measure total protein with the Bradford assay [45].
Cell suspension assays were performed at room temperature in 20-ml glass vials flushed with N2/CO2 (80/20, v/v) and were sealed with Teflon-lined butyl rubber stoppers held in place with aluminum crimps. The assay vials received a total of 0.9 ml reduced mineral salt medium, 0.1 ml of cell suspension, and increasing concentrations of substrates (i.e. MeOH, H2, or acetate, with N2O as indicated in Tables S1–S7). For assay vials receiving H2 as electron donor, the headspace was replaced with increasing volumes of premixed H2/CO2 (4/1, v/v) to achieve H2 concentrations ranging from 1.2 to 333 μM (Table S3). Small volumes (89–178 μl) of undiluted or 10-fold diluted (in N2) N2O were directly injected into the 20-ml assay vials, what resulted in small pressure changes with negligible impact on the distribution of N2O between the aqueous phase and the headspace. All cell suspensions were freshly prepared following identical procedures to ensure consistency between independent experiments. Vials that received 0.1 ml of sterile mineral salt medium or 0.1 ml of heat-killed (i.e. autoclaved) cell suspension served as negative controls.
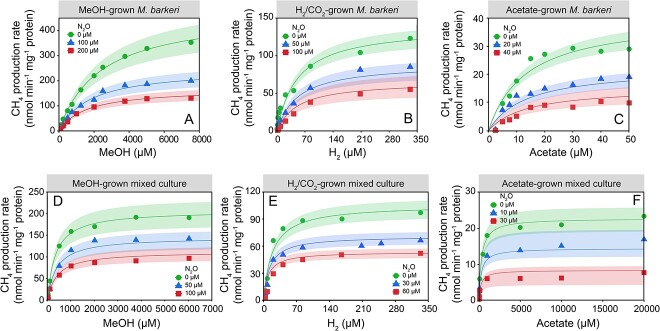
Following 10 min of equilibration, 0.1 ml of cell suspension was added to initiate the assays. Headspace samples (100 μl) were withdrawn with an air-tight syringe with a lock every 30 min over a 3-h incubation period and, for the kinetic experiments, CH4 was analyzed using an Agilent 7890 GC series gas chromatograph equipped with a flame ionization detector and a DB-624 capillary column (60 m length × 0.32 mm diameter, 1.8 μm film thickness). For each treatment at a fixed initial substrate concentration [S], an initial CH4 production rate v, normalized to the amount of protein per vial in the unit of nmol CH4 min−1 mg protein−1, was determined. The determined rate data were fit into Michaelis–Menten competitive, noncompetitive, and uncompetitive inhibition models (Table S8) to determine the maximum CH4 production rate Vmax, the half-velocity constant Km, and the inhibitory constant KI of N2O on CH4 production from the different substrates. The best-fit inhibition model was chosen based on the highest coefficient of determination (R2) and the lowest standard deviation of the residuals (Table S9). Each datum point on the Michaelis–Menten plots (Figure 5) represents a CH4 production rate generated from at least four time points for one substrate concentration [S].
Figure 5.
Kinetics of CH4 production from MeOH, H2, and acetate in whole-cell suspension assays of M. Barkeri and the methanogenic mixed cultures in the presence of increasing concentrations of N2O; the upper panels show the Michaelis–Menten plots of CH4 production rates versus the respective substrate concentrations in cell suspensions of M. Barkeri without and in the presence of increasing N2O concentrations in basal salt medium amended with MeOH (A), H2 (B), or acetate (C); the bottom panels show Michaelis–Menten plots of CH4 production rates versus the respective substrate concentrations in concentrated whole-cell suspensions of the methanogenic mixed cultures without N2O and in the presence of increasing N2O levels in basal salt medium amended with MeOH (D), H2 (E), or acetate (F); the shaded ribbons represent the standard distances (95% confidence interval) between the measured values and the nonlinear regression lines.
Genomic DNA extraction and 16S ribosomal RNA gene amplicon sequencing
DNA extraction and PCR assays followed established procedures [46] and details are provided in the Supplemental Information. The purified DNA samples were processed and barcoded with primers 341F/785R targeting the V3/V4 region of the prokaryotic 16S rRNA gene [47] following established procedures [48]. The resulting sequence data were analyzed using the QIIME 2 v2021.4 environment [49]. The precise programs and settings are described in the Supplemental Information, and the QIIME 2 pipeline script and custom R file employed to parse results are available at https://github.com/rwmurdoch/methanogens_and_N2O. The raw amplicon library reads were deposited in the Sequence Read Archive (SRA) under accessions SRR19782291 to SRR19782296.
Quantitative real-time PCR and growth yield calculations
Quantitative real-time PCR (qPCR) to enumerate archaeal 16S rRNA genes in M. barkeri and methanogenic mixed cultures followed established protocols using primer set Mtgen835F/918R and probe FAM-Mtgen831 (Table S10) [50]. Samples for enumeration of cell numbers were collected at the beginning and at the end of the prolonged growth experiments. Average growth yields of methanogens were calculated from the changes in 16S rRNA gene copy numbers in triplicate culture vessels divided by the total amounts of CH4 produced over the same time period (Table S11). Reported growth yield (i.e. cells produced per μmol of CH4 formed) used conversion factors of 3 and 2.5 16S rRNA gene copies per methanogen cell for M. barkeri [51] and the enrichment cultures [52], respectively. For comparison with theoretical [53] and reported values from the literature (Table 2), growth yields were also calculated as μg of dry biomass per μmol of CH4 formed making the following assumptions: an average methanogen cell has a volume of 2.5 μm3 [54] with a density equal to water [55]. The dry cell biomass is 30% of the wet cell biomass [56], and 90% of the dry biomass represents organic material [55].
Table 2.
Comparison of growth yields of methanogens utilizing different substrates with values collected from the peer-reviewed literature and calculated values based on thermodynamics and bioenergetic principles.
| Growth yield (μg organic matter per μmol CH4 formed) | |||||
|---|---|---|---|---|---|
| Cultures | Substrates | N2O (μM) | Measureda | Predicted from thermodynamicsb | Range (references) |
| M. barkeri | MeOH | 0 | 3.57 ± 0.32 | 5.2 | 3.3–6.4 ([71, 72]) |
| MeOH | 100 | 1.30 ± 0.28 | |||
| MeOH | 200 | 0.62 ± 0.03 | |||
| H2 | 0 | 0.53 ± 0.14 | 3.6 | 5.4–8.6 ([73]) | |
| H2 | 50 | 0.15 ± 0.02 | |||
| H2 | 100 | 0.08 ± 0.02 | |||
| Acetate | 0 | 1.45 ± 0.46 | 2.1 | 1.4–5.7 ([74]) | |
| Acetate | 20 | 1.02 ± 0.16 | |||
| Acetate | 50 | 0.70 ± 0.02 | |||
| Mixed methanogenic cultures | MeOH | 0 | 4.30 ± 0.56 | 5.2 | 3.3–6.4 ([72]) |
| MeOH | 10 | 0.14 ± 0.05 | |||
| MeOH | 30 | NA | |||
| H2 | 0 | 0.43 ± 0.05 | 3.6 | 1.3–7.2 ([14]) | |
| H2 | 10 | 0.16 ± 0.01 | |||
| H2 | 30 | NA | |||
| Acetate | 0 | 0.65 ± 0.05 | 2.1 | 1.4–5.7 ([23, 64, 75]) | |
| Acetate | 10 | 0.31 ± 0.02 | |||
| Acetate | 30 | NA | |||
Cell numbers were determined with qPCR. Because M. barkeri has three copies of the 16S rRNA gene, the qPCR results were divided by a factor of three to obtain cell numbers. Calculation of methanogen cell numbers in the mixed cultures assumed an average 16S rRNA gene content of 2.5.
Theoretical values calculated in this study based on thermodynamics and published information [53].
Results
N2O has distinct impact on CH4 production from different methanogenic substrates
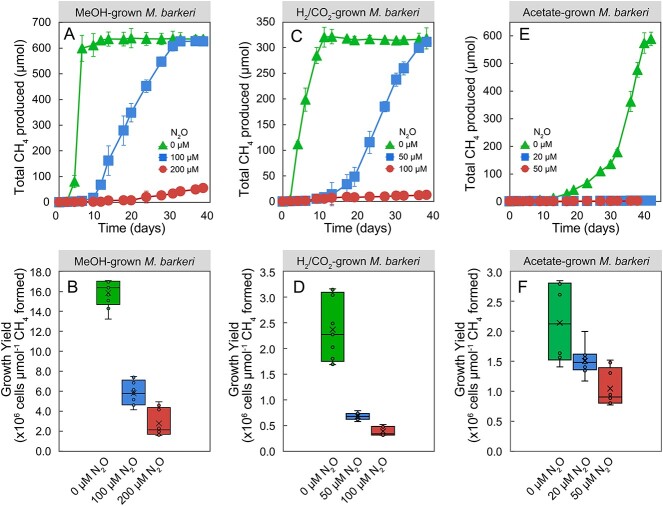
In the absence of N2O, MeOH-grown M. barkeri cultures consumed 895 ± 10 μmol of MeOH within a 6-day incubation period and produced 635 ± 34 μmol of CH4 (Fig. 2A). This stoichiometry closely matched the expected CH4 production based on Equation (1):
Figure 2.
Effect of N2O on CH4 production and growth yields in axenic M. Barkeri cultures; the upper panels show time courses of CH4 production in cultures that received MeOH (A), H2 (C), or acetate (E); the bottom panels display growth yields after 38-day incubation for M. Barkeri growing with MeOH (B), H2 (D), or acetate (F); error bars represent the standard deviation of replicate samples and are not shown when smaller than the symbol size; n = 3 for (A), (C), and (E); n = 9 (including three technical replicates for triplicate biological samples) for (B), (D), and (F).
 |
(1) |
Growth yields of 5.3 × 106 ± 0.3 × 106 cells per μmol CH4 were measured for cultures without N2O (Fig. 2B). By contrast, cultures that received 100 or 200 μM N2O produced negligible amounts of CH4 during the initial 6-day incubation period without any apparent growth after 6 days. Following an 11-day lag phase, cultures with 100 μM N2O started consuming MeOH, and 626 ± 20 μmol of CH4 were produced following a 38-day incubation period, indicating that 100 μM N2O delayed, but did not prevent, CH4 production from MeOH by M. barkeri (Fig. 2A). Although complete conversion of MeOH to CH4 according to Equation (1) was achieved in the presence of 100 μM N2O over a prolonged 38-day incubation period, the growth yield decreased by 63.8% ± 7.8% compared to cultures without N2O, (i.e. 1.9 × 106 ± 0.4 x 106 vs. 5.3 × 106 ± 0.3 × 106 cells were produced per μmol of CH4 formed) (Fig. 2B). In the presence of 200 μM N2O, only 55 ± 11 μmol of CH4 were produced over a 38-day incubation period, and the growth yield decreased by over 80% to 0.9 × 106 ± 0.4 × 106 cells per μmol CH4, indicating a pronounced inhibitory effect of N2O on CH4 production and growth of M. barkeri.
More pronounced N2O inhibition on CH4 production and growth was observed in M. barkeri cultures that received H2 as electron donor (i.e. hydrogenotrophic methanogenesis). In the absence of N2O, M. barkeri cultures, that received H2 as electron donor, produced 318 ± 21 μmol of CH4 from 1.24 mmol of H2 over an 11-day incubation period, consistent with Equation (2):
 |
(2) |
In the absence of N2O, a growth yield of 0.8 × 106 ± 0.2 × 106 cells per μmol of CH4 formed was measured (Fig. 2C and D). In the presence of 50 or 100 μM N2O, CH4 production by M. barkeri cultures commenced after prolonged lag phases ranging from 13 to 17 days. At the end of the 38-day incubation period, M. barkeri cultures with 50 μM N2O and produced 311 ± 7 μmol of CH4. The M. barkeri cultures that had received 100 μM N2O only generated 17 ± 3.2 μmol of CH4, resulting in a 95% decrease in total CH4 production compared to cultures without N2O. The average growth yield in cultures with 50 or 100 μM N2O decreased by ~70% and 85% to 2.3 × 105 ± 0.3 × 105 and 1.2 × 105 ± 0.3 × 104 cells per μmol CH4 formed, respectively, compared to cultures without N2O.
The most pronounced N2O inhibition was observed in acetate-fed M. barkeri cultures, and 20 μM N2O severely diminished CH4 production (Fig. 2E). In the absence of N2O, M. barkeri produced 588 ± 24 μmol of CH4 from 605 ± 17 μmol of acetate over a 38-day incubation period, consistent with Equation (3):
 |
(3) |
In the presence of 20 or 50 μM N2O, M. barkeri cultures only produced 21.3 ± 4.1 and 20.4 ± 3.8 μmol of CH4, respectively, a decline of over 96% in total CH4 production compared to cultures without N2O over the 38-day incubation period. Acetate-grown M. barkeri cultures that had received 20 or 50 μM N2O generated 1.5 ± 0.2 × 106 and 1.0 ± 0.3 × 106 cells per μmol of CH4 produced, decreases of 30.4% ± 3.3% and 52.5% ± 12.6%, respectively, compared to the average yield of 2.1 × 106 ± 0.7 × 106 cells per μmol CH4 in cultures without N2O (Fig. 2F).
In all culture vessels with observed inhibition of methanogenic activity, N2O concentrations remained constant throughout the experiment, consistent with the absence of a “nos” operon on the genome of M. barkeri. Taken together, these results demonstrate that micromolar levels of N2O inhibit methanogenesis, reduce growth yields of this model methanogen, and further reveal that the inhibition is most pronounced for acetoclastic methanogenesis.
N2O adversely affects CH4 production and methanogen growth yields in mixed methanogenic enrichment cultures
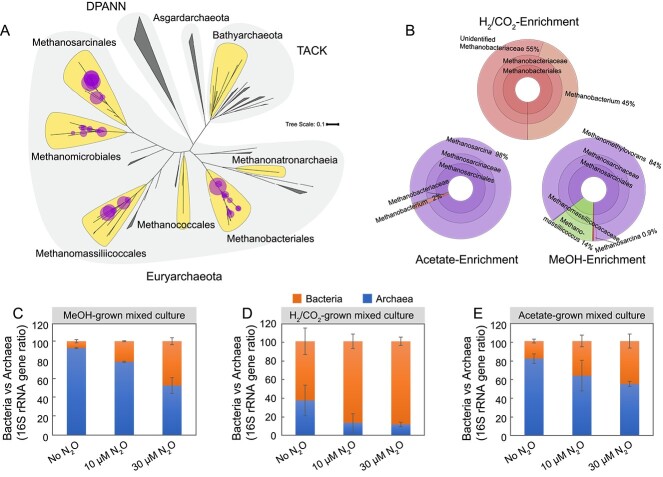
To examine the impact of N2O on mixed methanogen communities, growth and kinetic assays were performed with enrichment cultures derived from digester sludge. Microbial community analysis revealed the presence of diverse methanogen groups known to utilize acetate, H2/CO2, and MeOH as substrates, with distinct methanogen taxa prevalent under the different enrichment conditions (Tables S12 and S13). In cultures that received H2 as electron donor, sequences representing the genus Methanobacterium and other unidentified members of the family Methanobacteriaceae dominated, whereas in acetate-fed cultures, Methanosarcina was the most abundant archaeal taxon. Methanomethylovorans was the most abundant archaeal genus in the MeOH-fed cultures, but sequences representing the genus Methanomassiliicoccus were also prevalent. 16S rRNA gene amplicons representing six families and four of the known eight orders of methanogens were represented in the examined mixed cultures derived from digester sludge (Fig. 3A and B; Table S12).
Figure 3.
The composition and relative abundances of total sequences representing methanogenic archaea in mixed cultures enriched with acetate, H2/CO2, or MeOH; (A) phylogenetic placements of archaeal 16S rRNA gene amplicons detected in the enrichment cultures; the highlighted lobes indicate major clades of archaea known or suspected to produce CH4; the circles indicate best phylogenetic placements of archaeal taxa identified across all enrichment conditions; the size of the circle is proportional to the number of actual sequence variants (ASVs) detected; large shaded areas indicate archaeal superphyla, including Diapherotrites, Parvarchaeota, Aenigmarchaeota, Nanohaloarchaeota, and Nanoarchaeota (DPANN) and Thaumarchaeota, Aigarchaeota, Crenarchaeota, and Korarchaeota (TACK); see Supplemental Information for details on tree construction and fragment placement methodology; (B) relative abundances of total sequences representing methanogenic archaea in mixed cultures; panels (C) MeOH, (D) H2/CO2, and (E) acetate depict qPCR data showing the proportional changes of total bacterial and total archaeal (methanogen) 16S rRNA genes in the mixed cultures without N2O and in the presence of 10 and 30 μM N2O; error bars represent the standard deviation of replicate samples (n = 9, three technical replicates of triplicate biological samples).
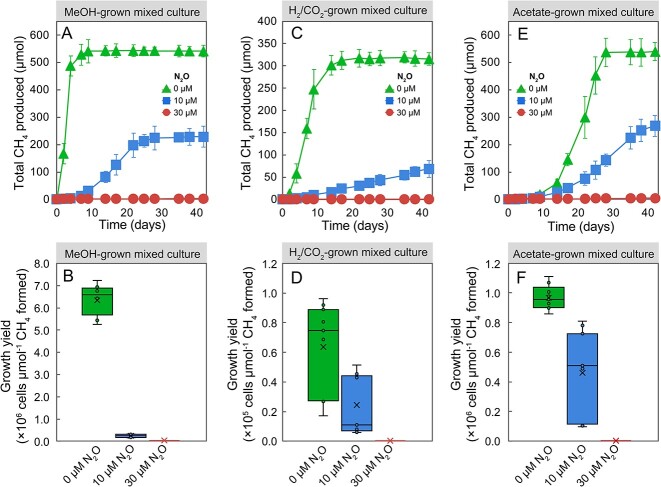
Without N2O addition, the mixed cultures produced 542.6 ± 23.3, 316.5 ± 25.4, and 589.2 ± 33.7 μmol of CH4 from 0.9 mmol of MeOH, 1.24 mmol of H2, or 0.6 mmol of acetate, consistent with Equations (1)–(3). When cultures were amended with 10 or 30 μM N2O, CH4 production in MeOH-, H2-, and acetate-fed methanogenic mixed cultures was substantially or completely inhibited (Fig. 4A, C, and E). Even over an extended 42-day incubation period, the presence 10 μM N2O still repressed the total CH4 production in MeOH-, H2-, or acetate-grown mixed cultures by ~60%, 80% and 50%, respectively, compared to incubations without N2O. With 30 μM N2O, only negligible amounts of CH4 were detected in all incubations over a 42-day incubation period. Some N2O loss was observed in the mixed culture vessels that received H2/CO2 or MeOH as substrates, with no more than 20% of the initial amount of N2O consumed. Taken together, these results demonstrate that N2O exerts a stronger inhibitory effect on methanogenesis in the mixed cultures harboring diverse methanogen populations than in axenic M. barkeri incubations.
Figure 4.
Effects of N2O on CH4 production and growth yields in methanogenic mixed cultures enriched with MeOH, H2, or acetate; the upper panels depict CH4 production from MeOH (A), H2 (C), and acetate (E); the bottom panels demonstrate methanogen growth yield differences in cultures amended with MeOH (B), H2 (D), or acetate (F); error bars represent standard deviation and are not shown when smaller than the symbol size (n = 3 for upper panels; n = 9 for bottom panels [three technical replicates of triplicate biological samples]).
Enumeration of total methanogens and total bacteria in the mixed cultures using qPCR revealed that N2O diminished methanogen growth yields (Fig. 4B, D, and F). The qPCR analysis further revealed significantly decreased ratios of methanogen-to-bacterial 16S rRNA genes in all N2O-treated cultures (Fig. 3C–E), illustrating that N2O impacted the methanogen populations much more strongly than the bacterial populations. In the absence of N2O, the average growth yields of methanogens in the enrichment cultures with MeOH, H2, or acetate were 0.6 × 107 ± 0.1 × 107, 0.6 × 105 ± 0.3 × 105, and 1.0 × 106 ± 0.1 × 106 cells per μmol of CH4 formed, respectively. In the presence of 10 μM N2O, the growth yields of methanogens declined by ~90%, 60%, and 50% in enrichment cultures that received MeOH, H2, and acetate, respectively (Fig. 4B, D, and F). Only negligible CH4 production and methanogen growth were measured in all mixed cultures with 30 μM N2O (Fig. 4B, D, and F). Taken together, the results of the mixed culture studies corroborate that N2O concentrations in the low micromolar range exhibit pronounced inhibitory effects on hydrogenotrophic, methylotrophic, and acetoclastic methanogenesis in microbial communities.
Kinetic studies confirm potent N2O inhibition of methane formation rates
Whole-cell suspension assays using M. barkeri and the methanogenic mixed cultures were performed to quantitatively assess the inhibitory effects of N2O on CH4 production from each methanogenic substrate (Table S1). The Michaelis–Menten single-substrate single-inhibitor model (R2 > 0.95) best explained the trends of CH4 production rates versus increasing substrate concentrations (Fig. 5). Among all assays with M. barkeri and the mixed cultures, maximum CH4 production rates (i.e. Vmax values) of acetate-fed cultures were most strongly affected by increasing N2O concentrations, followed by the H2- and MeOH-fed cultures (Figs 2 and 3).
In the absence of N2O, the Vmax values for CH4 production in MeOH-, H2-, or acetate-amended M. barkeri cell suspension assays were 440.4 ± 10.2, 138.1 ± 6.9, and 40.8 ± 4.7 nmol CH4 min−1 mg protein−1, respectively (Fig. 5A, Table 1). The addition of N2O decreased the Vmax of CH4 production in MeOH-, H2-, and acetate-fed M. barkeri cell suspension assays to different extents. Rate data determined in M. barkeri suspensions assays with MeOH fit the Michaelis–Menten inhibition model best. The Vmax values declined by ~45% and 57% to 244.1 ± 38.4 and 188.8 ± 32.5 nmol CH4 min−1 mg protein−1, respectively, in the presence of 100 and 200 μM N2O. The determined inhibitory constant, KI, of N2O on methylotrophic CH4 production was 130.9 ± 4.7 μM in M. barkeri cell suspensions (Table 1), indicating that N2O concentrations around 130 μM reduced the maximum CH4 production rate (Vmax) by 50%. More pronounced N2O inhibition was observed in M. barkeri whole-cell suspensions assays using H2 as the electron donor for CO2 reduction (Fig. 5B). The addition of 50 and 100 μM N2O reduced the Vmax values in H2-fed M. barkeri cell suspension assays by ~40% and 57% to 82.8 ± 29.5 and 59.9 ± 5.9 nmol of CH4 min−1 mg protein−1, respectively. The model simulation determined a KI value of 90.6 ± 10.8 μM N2O (Fig. 5B, Table 1), indicating a stronger inhibition of N2O on CH4 production in H2- versus MeOH-amended M. barkeri cell suspension assays. The M. barkeri assays with acetate as substrate showed the most pronounced inhibition by N2O (Fig. 5C, Table 1). In assays amended with 20 and 40 μM N2O, the Vmax values decreased by 21% and 44% to 32.1 ± 4.2 and 22.8 ± 2.5 nmol of CH4 min−1 mg protein−1, respectively. From the best-fit inhibition model, a KI value of 24.8 ± 2.6 μM was determined for N2O inhibition of acetoclastic methanogenesis in M. barkeri cell suspensions. Collectively, the cell suspension assays corroborate strong inhibitory effects of N2O on methanogenesis, with the kinetics of acetoclastic methanogenesis being most impacted by N2O.
Table 1.
Kinetic (Vmax, Km) and inhibition (KI) parameters for CH4 production from MeOH, H2, and acetate determined in concentrated whole-cell suspension assays of M. barkeri and methanogenic mixed cultures in response to increasing N2O concentrations.a
| Culture | Substrate | N2O (μM) | V max (nmol CH4 min−1 mg protein−1) | Km (μM) | KI (μM) |
|---|---|---|---|---|---|
| M. barkeri | MeOH | 0 | 440.4 ± 10.2 | 2.1 (±0.1) × 103 | 130.9 ± 4.7 |
| 100 | 244.1 ± 38.4 | ||||
| 200 | 188.8 ± 32.5 | ||||
| M. barkeri | H2 | 0 | 138.1 ± 6.9 | 51.1 ± 7.2 | 90.6 ± 10.8 |
| 50 | 82.8 ± 29.5 | ||||
| 100 | 59.9 ± 5.9 | ||||
| M. barkeri | Acetate | 0 | 40.8 ± 4.7 | 13.4 (±2.7) × 103 | 24.8 ± 3.1 |
| 20 | 32.1 ± 4.2 | ||||
| 50 | 22.8 ± 2.5 | ||||
| Mixed culture | MeOH | 0 | 209.0 ± 4.4 | 359.3 ± 30.8 | 109.9 ± 6.8 |
| 50 | 130.8 ± 22.6 | ||||
| 100 | 51.4 ± 44.7 | ||||
| Mixed culture | H2 | 0 | 105.8 ± 3.7 | 19.0 ± 2.3 | 62.1 ± 6.4 |
| 30 | 71.6 ± 10.4 | ||||
| 60 | 36.6 ± 13.6 | ||||
| Mixed culture | Acetate | 0 | 22.8 ± 0.7 | 302.2 ± 46.2 | 17.7 ± 1.8 |
| 10 | 14.1 ± 1.8 | ||||
| 30 | 5.7 ± 3.6 |
aData listed show results from the best fit Michaelis-Menten single-substrate-single-inhibitor models. Error values represent 95% confidence intervals.
Similar kinetic responses were observed for whole-cell suspension assays conducted with the methanogenic mixed cultures pregrown with the respective substrates (Fig. 5D–F). In the absence of N2O, the determined Vmax values for mixed culture cell suspension assays that received MeOH, H2, or acetate were 209.0 ± 4.4, 105.8 ± 3.7, and 22.8 ± 0.7 nmol CH4 min−1 mg protein−1, respectively (Table 1). Notably, the methanogenic mixed cultures were more sensitive to N2O than axenic M. barkeri cultures irrespective of the type of methanogenic substrate provided. In cell suspension assays that received MeOH, the presence of 50 and 100 μM N2O reduced the Vmax values by 37.4 ± 10.8% and 75.4 ± 21.4%, respectively, compared to assays without N2O. The inhibition model determined a KI value of 109.9 ± 6.8 μM for N2O inhibition on CH4 production in cell suspension assays amended with MeOH (Fig. 5D, Table 1). In H2-amended cell suspension assays, the presence of 30 and 60 μM N2O decreased the Vmax values by 32.3 ± 9.8 and 65.4 ± 12.9%, respectively, compared to assays without N2O (Fig. 5E, Table 1). The best-fit inhibition model determined a KI value for N2O inhibition of 62.1 ± 6.4 μM for CH4 production in assays that received H2 as electron donor. Consistent with the observations made with M. barkeri, the most pronounced N2O inhibition on CH4 production rates was observed in mixed culture cell suspension assays that received acetate as substrate (Fig. 5F). In the presence of 10 and 30 μM N2O, the Vmax values in suspension assays with acetate decreased by 38 ± 7.8% and 74.9 ± 16.0%, respectively, compared to assays without N2O. Using the best-fit Michaelis–Menten inhibition model, a KI value of 17.7 ± 1.8 μM was determined for N2O inhibition of CH4 production in cell suspension assays with acetate (Table 1).
Taken together, the experimental data demonstrate that dissolved N2O concentrations in the low μM range (i.e. 20–100 μM) repress CH4 production and reduce growth yields of M. barkeri in axenic culture and of different methanogen guilds in methanogenic mixed cultures. Evaluation of the kinetic data determined in cell suspension assays revealed distinct N2O inhibition patterns for CH4 production from acetate, H2, and MeOH, with the rate of acetoclastic CH4 production being most sensitive to N2O inhibition. The KI values for N2O inhibition of CH4 production from key methanogenic substrates ranged between 18 and 130 μM, suggesting N2O can impact CH4 production and emissions in diverse ecosystems, including critical zone environments (e.g. thawing permafrost).
Discussion
Microorganisms drive global C and N cycling and ultimately control CH4 and N2O production, consumption, and thus emissions to the atmosphere [10, 11, 13]. Predictive climate models must consider the responses of microbial CH4 and N2O production under environmental change scenarios [10, 57]. Quantitative assessment of feedbacks that affect CH4 production are crucial for refining greenhouse gas emission models. This study investigated the feedback effects between two important greenhouse gases, specifically the inhibitory effect of N2O on archaeal CH4 production, to provide quantitative data that link environmental N2O concentrations with methanogenesis. The findings demonstrate that environmentally relevant, micromolar levels of N2O suppress CH4 production and the growth of methanogens. The determined KI values reveal a concentration-dependent, progressively negative feedback by N2O on archaeal CH4 formation rates and the total amounts of CH4 produced and show that the strength of the inhibition is most pronounced for acetoclastic methanogenesis.
Assuming the current day partial pressure of 335 ppb N2O in the atmosphere, the theoretical concentration of N2O in air-equilibrated water should be around 7 nM; however, substantially elevated levels of dissolved N2O have been observed in groundwater and watersheds [33, 42]. In areas impacted by agricultural activities (e.g. fertilizer application), dissolved groundwater N2O concentrations can exceed 100 μM [42]. Even in some remote, natural aquatic systems, such as ice-covered Antarctic lakes, N2O concentrations of up to 86 μM have been reported [58, 59]. N2O is generated during N cycling and major formation processes include microbial denitrification, ammonia oxidation, and abiotic chemodenitrification [44]. Based on the physiology of the microorganisms (e.g. ammonia oxidizers are strict aerobes) and the thermodynamics of the processes (e.g. nitrate and nitrite reduction are associated with a greater change in Gibbs free energy than methanogenesis), one might argue that N2O formation and methanogenesis are physically separate processes, thus limiting the exposure and inhibitory effects of N2O on methanogens. Such redox stratification does occur; however, most environmental matrices, such as soils, are highly heterogenous and characterized by dynamic spatial and temporal gradients resulting in patchy distribution of redox processes. N2O is water-soluble and, depending on hydrology, can reach other redox zones [60]. Consequently, impacts of elevated N2O on various biogeochemical processes, including those associated with greenhouse gas emissions, are likely. Laboratory studies have reported inhibitory effects of nitrogen oxides (NOx), including N2O, on methanogenesis [34–37, 61, 62]; however, the available data are scarce and no uniform pattern has emerged that would support a quantitative relationship between N2O and microbial CH4 production. Consequently, this negative feedback of N2O on CH4 production has not been considered in greenhouse gas emission models.
The evaluation of N2O inhibition on CH4 production from methanogenic substrates (i.e. MeOH, acetate, and H2/CO2) with both the model methanogen M. barkeri and digester sludge-derived mixed methanogenic cultures quantitatively links N2O concentrations with methanogen activity and growth. The growth experiments illustrate that micromolar N2O concentrations affect CH4 production, and the whole-cell suspension assays and kinetic model simulations provide a plausible explanation for the inconsistent literature reports about N2O effects on methanogenesis. Specifically, acetoclastic methanogenesis was most sensitive to N2O (KI values of 18–25 μM), followed by hydrogenotrophic (KI 60–90 μM) and methylotrophic methanogenesis (KI 110–130 μM), indicating that the type of methanogenic substrate utilized affects the sensitivity of methanogens to N2O. N2O inhibition was significantly more pronounced in methanogenic enrichment cultures than in axenic M. barkeri cultures, and 30 μM N2O prevented CH4 production and methanogen growth in the mixed culture experiments regardless of the type of methanogenic substrate utilized. Previous studies support that mixed methanogenic communities are more sensitive to N2O than commonly studied model methanogen isolates [34, 35, 62, 63]. These observations suggest that the axenic methanogen cultures used to elucidate the biochemistry and genetics of methanogenesis may not serve as general models for other features of methanogen biology (e.g. N2O inhibition). The reasons for the reduced sensitivity of axenic versus mixed methanogen cultures to N2O may be a result of long-term adaptation of the isolates to laboratory cultivation, or not-yet-characterized microbe–microbe interaction networks render mixed cultures more susceptible to N2O inhibition. The enrichment cultures used to determine the KI values for N2O inhibition harbored diverse methanogen groups (Fig. 3A), and kinetic studies (e.g. determination of KI values for N2O inhibition) with representative isolates of the various lineages are warranted to capture the breadth of methanogen responses to N2O.
Taken together, the low μM range KI values of N2O that impact methanogenesis suggest major consequences of rising N2O concentrations for C cycling. Of note, the vast majority of the annual ~1 billion metric tons of biogenic CH4 is generated from acetate- and H2-driven CO2 reduction [16–20, 64], the two processes with the lowest observed KI values for N2O inhibition. It is therefore reasonable to predict that elevated environmental N2O will impact CH4 production and methanogen growth in ecosystems with high bioavailable C and N loads, such as wetlands, sediments, and permafrost soils.
Key enzyme systems involved in CH4 production and energy conservation require cobamide prosthetic groups [65] (Fig. 1). The super-reduced Co(I) form of cobamides is susceptible to oxidants such as N2O, a plausible mechanism for the observed inhibition of methanogenesis [29, 34, 63]. The experimental efforts demonstrated that acetoclastic methanogenesis was most sensitive to N2O, with KI values in the range of 18–25 μM, indicating that N2O concentrations in the range of 20 μM would reduce the Vmax of CH4 production from acetate by 50%. The KI values for N2O inhibition determined for hydrogenotrophic and methylotrophic methanogenesis ranged between 60–90 and 110–130 μM, respectively. The reasons for the apparently substrate-specific KI values for N2O inhibition likely reflect differences in the pathways leading to CH4 production from acetate, H2/CO2 and MeOH (Fig. 1). The acetoclastic pathway involves two steps catalyzed by corrinoid-dependent enzyme systems, CODH/ACS and the MTR corrinoid enzyme complex [16, 65]. (Fig. 1), both of which are targets for N2O inhibition. Distinct cobamide-dependent enzyme systems catalyze the formation of CH3–CoM in the hydrogenotrophic and the methylotrophic pathways [22, 66]. Both the acetoclastic and hydrogenotrophic pathways depend on the MTR corrinoid enzyme complex to generate CH3–CoM and to conserve energy [22, 27]. By contrast, methylotrophic methanogens can directly generate CH3–CoM without the energy-conserving MTR corrinoid enzyme complex from methylated compounds (e.g. MeOH) via a substrate-specific, corrinoid-dependent methyltransferase complex (i.e. MtaA, MtaB, and MtaC in the case of MeOH) [67]. The experimental efforts consistently demonstrated that CH4 production from acetate and H2/CO2 exhibited 7- and 3-fold higher sensitivities, respectively, to N2O than CH4 production from MeOH. These findings suggest that N2O inhibition of methanogenesis is related to the oxidation of the super-reduced Co(I) cobamide, which is essential for the corrinoid enzyme complexes involved in the different methanogenesis pathways. While the differential susceptibilities of pathway-specific corrinoid-dependent enzymatic steps can explain the distinct inhibitory effects of N2O on CH4 production via the acetoclastic, hydrogenotrophic, and methylotrophic pathways, detailed enzymatic studies would be needed to assess the responses of individual enzyme systems to N2O. Kinetic data for the enzymatic regeneration of the super-reduced Co(I) state are lacking, and the rates of reduction may differ between enzymes and pathways, which could contribute to the observed physiological response of decreased CH4 production. Recovery from N2O inhibition was not a focus, but the experimental data indicate partial recovery of methane formation in N2O-treated axenic and mixed cultures; however, the methanogen growth yields remained lower in N2O-treated cultures compared to controls without N2O even over the extended incubation period. Further studies are needed to generate mechanistic insights into modes of recovery from N2O inhibition.
The presence of N2O significantly decreased growth yields or completely abolished growth in the examined methanogenic cultures (Table 2). The type of methanogenic substrate utilized determines the fraction of electrons available from electron donor oxidation directed toward cell synthesis and thus governs the growth yields of methanogens [53, 68]. N2O affects corrinoid-dependent enzymes involved in electron transfer (e.g. the MTR enzyme complex), and it is not surprising that N2O interferes with energy conservation in methanogens. Consistently, enumeration of methanogen 16S rRNA genes at the termination of all growth experiments illustrated that N2O not only negatively impacted CH4 production but also methanogen growth yields. An alternate explanation for the reduced methanogen growth yields in the mixed cultures exposed to N2O could be competition for electron donor (e.g. H2); however, the sequencing and the qPCR data do not support this hypothesis, and N2O inhibition explains the decline of methanogens and the changes of methanogen-to-bacteria ratios. The measured methanogen growth yield data in the absence of N2O were on par with reported experimental data and closely matched the theoretical values (i.e. yields calculated based on thermodynamics) (Table 2). One exception were the growth yields measured in H2/CO2-fed M. barkeri cultures, which were ~10-fold lower than data reported in the literature. The most pronounced growth suppression was observed in the mixed methanogenic cultures, where 30 μM N2O was sufficient to prevent the growth of methanogenic archaea. Collectively, the data show that micromolar concentrations of N2O decrease or abolish CH4 production, reduce methanogen growth yields, and exhibit progressively negative feedback on microbial CH4 production.
Microbial processes are strongly influenced by environmental factors and their responses to climate change vary both spatially and temporally [11, 69]. To improve the predictive power of climate models and potentially justify the application of biotechnological approaches for managing greenhouse gas emissions, interactions and feedbacks between relevant biotic/abiotic processes must be understood and quantitatively captured [70]. Attempts have been made to include microbial data (i.e. biomass, enzyme, and growth kinetics) to improve climate models [70], but the incorporation of multifactorial, multidirectional, and often nonlinear biotic/abiotic feedbacks underlying the global CH4 and N2O budgets is challenging and requires robust quantitative data. The determined KI values for N2O inhibition of methanogenesis reveal a relevant negative feedback effect on CH4 emissions, and the new quantitative information generates opportunities to refine CH4 emission models.
Supplementary Material
Contributor Information
Yongchao Yin, Center for Environmental Biotechnology, University of Tennessee, Knoxville, TN 37996, United States; Department of Microbiology, University of Tennessee, Knoxville, TN 37996, United States; Biosciences Division, Oak Ridge National Laboratory, Oak Ridge, TN 37831, United States.
Fadime Kara-Murdoch, Center for Environmental Biotechnology, University of Tennessee, Knoxville, TN 37996, United States; Biosciences Division, Oak Ridge National Laboratory, Oak Ridge, TN 37831, United States.
Robert W Murdoch, Center for Environmental Biotechnology, University of Tennessee, Knoxville, TN 37996, United States.
Jun Yan, Center for Environmental Biotechnology, University of Tennessee, Knoxville, TN 37996, United States; Department of Microbiology, University of Tennessee, Knoxville, TN 37996, United States; Key Laboratory of Pollution Control and Environmental Engineering, Institute of Applied Ecology, Chinese Academy of Sciences, Shenyang, Liaoning 110016, China.
Gao Chen, Center for Environmental Biotechnology, University of Tennessee, Knoxville, TN 37996, United States; Department of Civil and Environmental Engineering, University of Tennessee, Knoxville, TN 37996, United States.
Yongchao Xie, Center for Environmental Biotechnology, University of Tennessee, Knoxville, TN 37996, United States; Department of Civil and Environmental Engineering, University of Tennessee, Knoxville, TN 37996, United States.
Yanchen Sun, Center for Environmental Biotechnology, University of Tennessee, Knoxville, TN 37996, United States; Department of Civil and Environmental Engineering, University of Tennessee, Knoxville, TN 37996, United States.
Frank E Löffler, Center for Environmental Biotechnology, University of Tennessee, Knoxville, TN 37996, United States; Department of Microbiology, University of Tennessee, Knoxville, TN 37996, United States; Biosciences Division, Oak Ridge National Laboratory, Oak Ridge, TN 37831, United States; Department of Civil and Environmental Engineering, University of Tennessee, Knoxville, TN 37996, United States; Department of Biosystems Engineering and Soil Science, University of Tennessee, Knoxville, TN 37996, United States.
Conflicts of interest
The authors declare no competing interests.
Funding
This work was supported through the Dimensions of Biodiversity Program of the US National Science Foundation (award 1831599 to F.E.L.). Y.Y., Y.X., and Y.S. acknowledge the support from the China Scholarship Council.
Data availability
All data generated or analyzed during this study are included in this published article and its supplementary information files. The raw amplicon library reads are available in the SRA repository.
References
- 1. Intergovernmental Panel on Climate Change (IPCC) . Stocker TF (ed.), Climate change 2013: the physical science basis. Working Group I contribution to the fifth assessment report of the Intergovernmental Panel on Climate Change. New York: Cambridge University Press, 2013. 10.1017/CBO9781107415324 [DOI] [Google Scholar]
- 2. Intergovernmental Panel on Climate Change (IPCC) . Global Warming of 1.5°C: IPCC Special Report on Impacts of Global Warming of 1.5°C above Pre-Industrial Levels in Context of Strengthening Response to Climate Change, Sustainable Development, and Efforts to Eradicate Poverty. Cambridge: Cambridge University Press, 2018. https://www.ipcc.ch/sr15/ [Google Scholar]
- 3. United Nations Framework Convention on Climate Change (UNFCCC) . Adoption of the Paris Agreement. Report No. FCCC/CP/2015/L.9/Rev.1. Geneva: United Nations, 2015. https://unfccc.int/resource/docs/2015/cop21/eng/l09r01.pdf [Google Scholar]
- 4. Intergovernmental Panel on Climate Change (IPCC) . Shukla PR (ed.), Climate Change and Land: An IPCC Special Report on Climate Change, Desertification, Land Degradation, Sustainable Land Management, Food Security, and Greenhouse Gas Fluxes in Terrestrial Ecosystems, Geneva, Switzerland, 2019). https://www.ipcc.ch/srccl/download/.
- 5. Luan J, Wu J, Liu S et al. Soil nitrogen determines greenhouse gas emissions from northern peatlands under concurrent warming and vegetation shifting. Commun Biol 2019;2:132. 10.1038/s42003-019-0370-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Parn J, Verhoeven JTA, Butterbach-Bahl K et al. Nitrogen-rich organic soils under warm well-drained conditions are global nitrous oxide emission hotspots. Nat Commun 2018;9:1135. 10.1038/s41467-018-03540-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Reay DS, Smith P, Christensen TR et al. Methane and global environmental change. Annu Rev Environ Resour 2018;43:165–92. 10.1146/annurev-environ-102017-030154 [DOI] [Google Scholar]
- 8. Jackson RB, Saunois M, Bousquet P et al. Increasing anthropogenic methane emissions arise equally from agricultural and fossil fuel sources. Environ Res Lett 2020;15:071002. 10.1088/1748-9326/ab9ed2 [DOI] [Google Scholar]
- 9. Tian H, Xu R, Canadell JG et al. A comprehensive quantification of global nitrous oxide sources and sinks. Nature 2020;586:248–56. 10.1038/s41586-020-2780-0 [DOI] [PubMed] [Google Scholar]
- 10. Cavicchioli R, Ripple WJ, Timmis KN et al. Scientists' warning to humanity: microorganisms and climate change. Nat Rev Microbiol. 2019;17:569–86. 10.1038/s41579-019-0222-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Singh BK, Bardgett RD, Smith P et al. Microorganisms and climate change: terrestrial feedbacks and mitigation options. Nat Rev Microbiol. 2010;8:779–90. 10.1038/nrmicro2439 [DOI] [PubMed] [Google Scholar]
- 12. Stein LY. The long-term relationship between microbial metabolism and greenhouse gases. Trends Microbiol 2020;28:500–11. 10.1016/j.tim.2020.01.006 [DOI] [PubMed] [Google Scholar]
- 13. Jansson JK, Hofmockel KS. Soil microbiomes and climate change. Nat Rev Microbiol 2020;18(1):35–46. 10.1038/s41579-019-0265-7 [DOI] [PubMed] [Google Scholar]
- 14. Thauer RK, Kaster AK, Seedorf H et al. Methanogenic archaea: ecologically relevant differences in energy conservation. Nat Rev Microbiol. 2008;6:579–91. 10.1038/nrmicro1931 [DOI] [PubMed] [Google Scholar]
- 15. Evans PN, Boyd JA, Leu AO et al. An evolving view of methane metabolism in the archaea. Nat Rev Microbiol 2019;17:219–32. 10.1038/s41579-018-0136-7 [DOI] [PubMed] [Google Scholar]
- 16. Ferry JG. Acetate-based methane production. In: Wall JD, Harwood CS, Demain A (eds.), Bioenergy. Washington, DC. ASM Press: Wiley Online Library, 2008. [Google Scholar]
- 17. Lessner DJ. Methanogenesis Biochemistry. In eLS, John Wiley & Sons Ltd, Hoboken, NJ, USA, 2009.
- 18. Ferry JG. Fundamentals of methanogenic pathways that are key to the biomethanation of complex biomass. Curr Opin Struct Biol 2011;22:351–7. 10.1016/j.copbio.2011.04.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Lyu Z, Shao N, Akinyemi T et al. Methanogenesis. Curr Biol 2018;28:R727–32. 10.1016/j.cub.2018.05.021 [DOI] [PubMed] [Google Scholar]
- 20. Carr SA, Schubotz F, Dunbar RB et al. Acetoclastic Methanosaeta are dominant methanogens in organic-rich Antarctic marine sediments. ISME J 2018;12:330–42. 10.1038/ismej.2017.150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Goetzl S, Jeoung JH, Hennig SE et al. Structural basis for electron and methyl-group transfer in a methyltransferase system operating in the reductive acetyl-CoA pathway. J Mol Biol 2011;411:96–109. 10.1016/j.jmb.2011.05.025 [DOI] [PubMed] [Google Scholar]
- 22. Welander PV, Metcalf WW. Loss of the mtr operon in Methanosarcina blocks growth on methanol, but not methanogenesis, and reveals an unknown methanogenic pathway. Proc Natl Acad Sci USA 2005;102:10664–9. 10.1073/pnas.0502623102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Prakash D, Chauhan SS, Ferry JG. Life on the thermodynamic edge: respiratory growth of an acetotrophic methanogen. Sci Adv 2019;5:eaaw9059. 10.1126/sciadv.aaw9059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Borrel G, Adam PS, Gribaldo S. Methanogenesis and the wood-Ljungdahl pathway: an ancient, versatile, and fragile association. Genome Biol Evol 2016;8:1706–11. 10.1093/gbe/evw114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kurth JM, Nobu MK, Tamaki H et al. Methanogenic archaea use a bacteria-like methyltransferase system to demethoxylate aromatic compounds. ISME J 2021;15:3549–65. 10.1038/s41396-021-01025-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Thauer RK, Kaster AK, Goenrich M et al. Hydrogenases from methanogenic archaea, nickel, a novel cofactor, and H2 storage. Annu Rev Biochem 2010;79:507–36. 10.1146/annurev.biochem.030508.152103 [DOI] [PubMed] [Google Scholar]
- 27. Thauer RK. The Wolfe cycle comes full circle. Proc Natl Acad Sci USA 2012;109:15084–5. 10.1073/pnas.1213193109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Richter N, Zepeck F, Kroutil W. Cobalamin-dependent enzymatic O-, N-, and S-demethylation. Trends Biotechnol 2015;33:371–3. 10.1016/j.tibtech.2015.03.011 [DOI] [PubMed] [Google Scholar]
- 29. Banks RGS, Henderson RJ, Pratt JM. Reactions of gases in solution. Part III. Some reactions of nitrous oxide with transition-metal complexes. J Chem Soc Sec A 1968;0:2886–9. 10.1039/j19680002886 [DOI] [Google Scholar]
- 30. Blackburn R, Kyaw M, Swallow AJ. Reaction of cob(I)alamin with nitrous oxide and cob(III)alamin. J Chem Soc, Faraday Trans I 1977;73:250–5. 10.1039/f19777300250 [DOI] [Google Scholar]
- 31. Drummond JT, Matthews RG. Nitrous oxide inactivation of cobalamin-dependent methionine synthase from Escherichia coli: characterization of the damage to the enzyme and prosthetic group. Biochemistry 1994;33:3742–50. 10.1021/bi00178a034 [DOI] [PubMed] [Google Scholar]
- 32. Drummond JT, Matthews RG. Nitrous oxide degradation by cobalamin-dependent methionine synthase: characterization of the reactants and products in the inactivation reaction. Biochemistry 1994;33:3732–41. 10.1021/bi00178a033 [DOI] [PubMed] [Google Scholar]
- 33. Yin Y, Yan J, Chen G et al. Nitrous oxide is a potent inhibitor of bacterial reductive dechlorination. Environ Sci Technol 2018;53:692–701. 10.1021/acs.est.8b05871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Balderston WL, Payne WJ. Inhibition of methanogenesis in salt marsh sediments and whole-cell suspensions of methanogenic bacteria by nitrogen oxides. Appl Environ Microbiol 1976;32:264–9. 10.1128/aem.32.2.264-269.1976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Klüber HD, Conrad R. Inhibitory effects of nitrate, nitrite, NO and N2O on methanogenesis by Methanosarcina barkeri and Methanobacterium bryantii. FEMS Microbiol Ecol 1998;25:331–9. 10.1016/S0168-6496(97)00102-5 [DOI] [Google Scholar]
- 36. Tugtas AE, Pavlostathis SG. Inhibitory effects of nitrogen oxides on a mixed methanogenic culture. Biotechnol Bioeng 2007;96:444–55. 10.1002/bit.21105 [DOI] [PubMed] [Google Scholar]
- 37. Buessecker S, Zamora Z, Sarno AF et al. Microbial communities and interactions of nitrogen oxides with methanogenesis in diverse peatlands of the Amazon basin. Front Microbiol 2021;12:p. 659079. 10.3389/fmicb.2021.659079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Elberling B, Christiansen HH, Hansen BU. High nitrous oxide production from thawing permafrost. Nat Geosci 2010;3:332–5. 10.1038/ngeo803 [DOI] [Google Scholar]
- 39. Carlson KM, Gerber JS, Mueller ND et al. Greenhouse gas emissions intensity of global croplands. Nat Clim Chang 2016;7:63–8. 10.1038/nclimate3158 [DOI] [Google Scholar]
- 40. Campos JL, Valenzuela-Heredia D, Pedrouso A et al. Greenhouse gases emissions from wastewater treatment plants: minimization, treatment, and prevention. J Chem 2016;2016:1–12. 10.1155/2016/3796352 [DOI] [Google Scholar]
- 41. Davidson TA, Audet J, Svenning JC et al. Eutrophication effects on greenhouse gas fluxes from shallow-lake mesocosms override those of climate warming. Glob Change Biol 2015;21:4449–63. 10.1111/gcb.13062 [DOI] [PubMed] [Google Scholar]
- 42. Jurado A, Borges AV, Brouyere S. Dynamics and emissions of N2O in groundwater: a review. Sci Total Environ 2017;584-585:207–18. 10.1016/j.scitotenv.2017.01.127 [DOI] [PubMed] [Google Scholar]
- 43. Harris E, Yu L, Wang YP et al. Warming and redistribution of nitrogen inputs drive an increase in terrestrial nitrous oxide emission factor. Nat Commun 2022;13:4310. 10.1038/s41467-022-32001-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Onley JR, Ahsan S, Sanford RA et al. Denitrification by Anaeromyxobacter dehalogenans, a common soil bacterium lacking the nitrite reductase genes nirS and nirK. Appl Environ Microbiol 2018;84:e01985–17. 10.1128/AEM.01985-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 1976;72:248–54. 10.1016/0003-2697(76)90527-3 [DOI] [PubMed] [Google Scholar]
- 46. Ritalahti KM, Amos BK, Sung Y et al. Quantitative PCR targeting 16S rRNA and reductive dehalogenase genes simultaneously monitors multiple Dehalococcoides strains. Appl Environ Microbiol 2006;72:2765–74. 10.1128/AEM.72.4.2765-2774.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Klindworth A, Pruesse E, Schweer T et al. Evaluation of general 16S ribosomal RNA gene PCR primers for classical and next-generation sequencing-based diversity studies. Nucleic Acids Res 2013;41(1):e1–1. 10.1093/nar/gks808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Caporaso JG, Lauber CL, Walters WA et al. Global patterns of 16S rRNA diversity at a depth of millions of sequences per sample. Proc Natl Acad Sci USA 2011;108:4516–22. 10.1073/pnas.1000080107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Bolyen E, Rideout JR, Dillon MR et al. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat Biotechnol 2019;37:852–7. 10.1038/s41587-019-0209-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Kleindienst S, Chourey K, Chen G et al. Proteogenomics reveals novel reductive dehalogenases and methyltransferases expressed during anaerobic dichloromethane metabolism. Appl Environ Microbiol 2019;85:e02768–18. 10.1128/AEM.02768-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Maeder DL, Anderson I, Brettin TS et al. The Methanosarcina barkeri genome: comparative analysis with Methanosarcina acetivorans and Methanosarcina mazei reveals extensive rearrangement within methanosarcinal genomes. J Bacteriol 2006;188:7922–31. 10.1128/JB.00810-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Sun D-L, Jiang X, Wu QL et al. Intragenomic heterogeneity of 16S rRNA genes causes overestimation of prokaryotic diversity. Appl Environ Microbiol 2013;79:5962–9. 10.1128/AEM.01282-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Rittmann BE, McCarty PL. Environmental Biotechnology: Principles and Applications. NewYork: McGraw-Hill, 2001 [Google Scholar]
- 54. Blohs M, Moissl-Eichinger C, Mahnert A, et al. Archaea – an introduction. In: Schmidt TM (ed.), Encyclopedia of Microbiology (Fourth Edition). Oxford: Academic Press; 2019, p. 243–52. 10.1016/B978-0-12-809633-8.20884-4 [DOI] [Google Scholar]
- 55. Duhamel M, Edwards EA. Growth and yields of dechlorinators, acetogens, and methanogens during reductive dechlorination of chlorinated ethenes and dihaloelimination of 1,2-dichloroethane. Environ Sci Technol 2007;41:2303–10. 10.1021/es062010r [DOI] [PubMed] [Google Scholar]
- 56. Bratbak G, Dundas I. Bacterial dry matter content and biomass estimations. Appl Environ Microbiol 1984;48:755–7. 10.1128/aem.48.4.755-757.1984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Guo X, Gao Q, Yuan M et al. Gene-informed decomposition model predicts lower soil carbon loss due to persistent microbial adaptation to warming. Nat Commun 2020;11:4897. 10.1038/s41467-020-18706-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Priscu JC, Christner BC, Dore JE et al. Supersaturated N2O in a perennially ice-covered Antarctic lake: molecular and stable isotopic evidence for a biogeochemical relict. Limnol Oceanogr 2008;53:2439–50. 10.4319/lo.2008.53.6.2439 [DOI] [Google Scholar]
- 59. Murray AE, Kenig F, Fritsen CH et al. Microbial life at −13 °C in the brine of an ice-sealed Antarctic lake. Proc Natl Acad Sci USA 2012;109:20626–31. 10.1073/pnas.1208607109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Priscu JC. The biogeochemistry of nitrous oxide in permanently ice-covered lakes of the McMurdo dry valleys. Antarctica Glob Chang Biol 1997;3:301–15. 10.1046/j.1365-2486.1997.00147.x [DOI] [Google Scholar]
- 61. Clarens M, Bernet N, Delgenès J-P et al. Effects of nitrogen oxides and denitrification by Pseudomonas stutzeri on acetotrophic methanogenesis by Methanosarcina mazei. FEMS Microbiol Ecol 1998;25:271–6. 10.1111/j.1574-6941.1998.tb00479.x [DOI] [Google Scholar]
- 62. Klüber HD, Conrad R. Effects of nitrate, nitrite, NO and N2O on methanogenesis and other redox processes in anoxic rice field soil. FEMS Microbiol Ecol 1998;25:301–19. 10.1016/S0168-6496(98)00011-7 [DOI] [Google Scholar]
- 63. Fischer R, Thauer RK. Methanogenesis from acetate in cell extracts of Methanosarcina barkeri: isotope exchange between CO2 and the carbonyl group of acetyl-CoA, and the role of H2. Arch Microbiol 1990;153:156–62. 10.1007/BF00247814 [DOI] [Google Scholar]
- 64. Welte C, Deppenmeier U. Bioenergetics and anaerobic respiratory chains of aceticlastic methanogens. Biochim Biophys Acta 2014;1837:1130–47. 10.1016/j.bbabio.2013.12.002 [DOI] [PubMed] [Google Scholar]
- 65. Banerjee R, Ragsdale SW. The many faces of vitamin B12: catalysis by cobalamin-dependent enzymes. Annu Rev Biochem 2003;72:209–47. 10.1146/annurev.biochem.72.121801.161828 [DOI] [PubMed] [Google Scholar]
- 66. Mand TD, Metcalf WW. Energy conservation and hydrogenase function in methanogenic archaea, in particular the genus Methanosarcina. Microbiol Mol Bio Rev 2019;83:e00020–19. 10.1128/MMBR.00020-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Welander PV, Metcalf WW. Mutagenesis of the C1 oxidation pathway in Methanosarcina barkeri: new insights into the Mtr/Mer bypass pathway. J Bacteriol 2008;190:1928–36. 10.1128/JB.01424-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Karadagli F, Rittmann BE. Kinetic characterization of Methanobacterium bryantii M.o.H. Environ Sci Technol 2005;39:4900–5. 10.1021/es047993b [DOI] [PubMed] [Google Scholar]
- 69. Arneth A, Harrison SP, Zaehle S et al. Terrestrial biogeochemical feedbacks in the climate system. Nat Geosci 2010;3:525–32. 10.1038/ngeo905 [DOI] [Google Scholar]
- 70. McCalley CK, Woodcroft BJ, Hodgkins SB et al. Methane dynamics regulated by microbial community response to permafrost thaw. Nature 2014;514:478–81. 10.1038/nature13798 [DOI] [PubMed] [Google Scholar]
- 71. Hippe H, Caspari D, Fiebig K et al. Utilization of trimethylamine and other N-methyl compounds for growth and methane formation by Methanosarcina barkeri. Proc Natl Acad Sci USA 1979;76(1):494–8. 10.1073/pnas.76.1.494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Müller V, Blaut M, Gottschalk G. Utilization of methanol plus hydrogen by Methanosarcina barkeri for methanogenesis and growth. Appl Environ Microbiol 1986;52:269–74. 10.1128/aem.52.2.269-274.1986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Londry KL, Dawson KG, Grover HD et al. Stable carbon isotope fractionation between substrates and products of Methanosarcina barkeri. Organic Geochem 2008;39:608–21. 10.1016/j.orggeochem.2008.03.002 [DOI] [Google Scholar]
- 74. Peinemann S, Müller V, Blaut M et al. Bioenergetics of methanogenesis from acetate by Methanosarcina barkeri. J Bacteriol 1988;170:1369–72. 10.1128/jb.170.3.1369-1372.1988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Feist AM, Scholten JC, Palsson BO et al. Modeling methanogenesis with a genome-scale metabolic reconstruction of Methanosarcina barkeri. Mol Syst Biol 2006;2:0004. 10.1038/msb4100046 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data generated or analyzed during this study are included in this published article and its supplementary information files. The raw amplicon library reads are available in the SRA repository.