Abstract
A serologic survey of primates living in a French zoo allowed identification of three cases of infection with simian immunodeficiency virus in sooty mangabeys (Cercocebus atys) (SIVsm). Viral isolates, which were designated SIVsmFr66, SIVsmFr74, and SIVsmFr85, were obtained after short-term culture of mangabey lymphoid cells. Phylogenetic analysis of gag and env sequences amplified directly from mangabey tissues showed that the three SIVsmFr were genetically close and that they constituted a new subtype within the diverse SIVsm–SIVmac–human immunodeficiency virus type 2 (HIV-2) group. We could reconstruct the transmission events that likely occurred in 1986 between the three animals and evaluate the divergence of SIVsmFr sequences since transmission. The estimated rate of mutation fixation was 6 × 10−3 substitutions per site per year, which was as high as the rate found for SIVmac infection in macaques. These data indicated that SIVsmFr replicated at a high rate in mangabeys, despite the nonpathogenic character of infection in this host. The viral load evaluated by competitive PCR reached 20,000 viral DNA copies per 106 lymph node cells. In addition, productively infected cells were readily detected in mangabey lymphoid tissues by in situ hybridization. The amounts of viral RNA in plasma ranged from 105 to 107 copies per ml. The cell-associated and plasma viral loads were as high as those seen in susceptible hosts (humans or macaques) during the asymptomatic stage of HIV or SIVmac infections. Thus, the lack of pathogenicity of SIVsm for its natural host cannot be explained by limited viral replication or by tight containment of viral production.
Simian immunodeficiency viruses (SIV) naturally infect a variety of nonhuman primates of African origin (33, 53). SIV seroprevalence is particularly high in African green monkey (Cercopithecus aethiops) populations and in some troops of sooty mangabeys (Cercocebus atys atys) (15, 38, 43, 47, 55). Although SIV share many structural and biological properties with human immunodeficiency viruses (HIV), they do not seem to induce AIDS in their natural hosts (3, 20, 22, 23, 34, 37, 61). In contrast, SIV from sooty mangabey (SIVsm) induces an immunodeficiency syndrome very similar to AIDS when it is experimentally inoculated in Asian monkey species such as macaques (24, 51, 71). Accidental transmission of SIVsm from mangabeys to macaques housed in primate centers resulted in the emergence of the SIVmac that caused simian AIDS outbreaks during the 1970s (12, 17, 27). The characterization of an SIV strain from African green monkey (SIVagm) that causes disease in pig-tailed macaques (Macaca nemestrina) further suggests that primate lentiviruses occasionally become pathogenic when transmitted to a new species (31). The AIDS viruses HIV HIV-1 and HIV-2 may well be the result of several accidental transmissions of SIV to humans. The evidence for a simian origin of HIV-2 is compelling, since the SIVsm genome is closely related to that of HIV-2 and since the HIV-2 endemic zone in West Africa corresponds to the natural range of SIVsm-carrying sooty mangabeys (26, 34, 47). Several clusters on the HIV-2–SIVsm phylogenetic tree group human and simian viruses together, implying that multiple independent transmission events gave rise to the different HIV-2 subtypes (15, 25). The origins of HIV-1 are less clear, but the genetic proximity of divergent HIV-1 strains with a lentivirus isolated from chimpanzees (SIVcpz) favors the idea that HIV-1 infection also originated as a zoonosis (35, 52, 60).
Both viral and host factors have been shown to control SIV pathogenic potential. A major viral determinant of virulence is the protein Nef, which is essential to disease induction in adult macaques and which contributes to the phenotype of a hyperpathogenic strain of SIVsm named PBj14 (18, 21, 40). Host determinants are less well characterized but clearly play a role, since a SIV molecular clone can induce disease in one species and a chronic but benign infection in another species (31). The antiviral immune response differs in monkeys susceptible or resistant to disease, but the relevance of these differences to the pathogenic process is not yet known (24, 54, 71). The viral load, which is the amount of virus that persists in the organism as a result of viral production minus viral clearance, is thought to be an important determinant of SIV and HIV pathogenicity. A positive correlation between the viral load early in infection and the risk of progression to disease seems to be the rule in SIVmac, SIVagm, and HIV-1 infections (29, 31, 32, 46, 72). To verify whether this rule holds true for every type of SIV infection, we investigated virologic and phylogenetic parameters in naturally infected sooty mangabeys.
The present study reports the characterization of a novel SIVsm subtype isolated from a group of three mangabeys. Since the animals likely belonged to the same infection chain, it was possible to evaluate the number of mutations that had accumulated in viral sequences since transmission. The rate of mutation fixation was found to be high in both gag and env (TM) regions, which suggested that SIVsm replicated rapidly and continuously in vivo. In addition, disease-resistant mangabeys were found to harbor as much virus as hosts susceptible to the pathogenic effects of SIV and HIV infections. These findings indicated that neither the viral load nor the viral replication rate was predictive of SIVsm pathogenicity.
MATERIALS AND METHODS
Animals.
Nonhuman primates of the Ménagerie du Museum National d’Histoire Naturelle in Paris, France, were tested for immunological reactivity with HIV-1 and HIV-2 antigens. Serum samples were collected over a period of 10 years (from 1987 to 1997) and kept frozen at −30°C until they were tested by Western blotting. Three sooty mangabeys (C. atys atys, formerly known as Cercocebus torquatus atys) had antibodies that reacted with HIV-2 antigens. Two of them, M66 and F74, were a male and a female that had been living in the same group for 22 years. These animals were wild caught and of unknown geographical origin. The younger seropositive mangabey, F85, was a female born in captivity in a London zoo. She was introduced into the group of M66 and F74 in November 1985 but had to be isolated after being bitten during a violent fight with other females. F85 was transferred to another zoo in October 1986 to be paired with male M78. Although both animals had been kept together for 10 years, M78 was seronegative at the time of testing. None of the mangabeys studied manifested clinical signs, except for an arthritic condition associated with old age in M66.
Virus isolation.
Blood samples collected in sodium citrate coated tubes were obtained for the three seropositive animals and from M78. Peripheral blood mononuclear cells (PBMC) were separated by gradient centrifugation on Ficoll-Hypaque and were cultivated at a concentration of 106/ml in RPMI medium supplemented with 10% of a supernatant enriched in human interleukin-2 (IL-2; Lymphocult-T-LF; Biotest AG). The mitogen phytohemaglutinin (PHA) was added at a concentration of 5 μg/ml. After 2 days, PHA-stimulated human PBMC were added to the cultures. The cocultures were monitored for SIV production by an antigen-capture assay specific for SIV p27 Gag (Coulter). Peripheral and mesenteric lymph nodes were obtained at necropsy from animals F74, F85, and M78. Lymph nodes were disrupted in RPMI medium by passage through metal grids until a homogeneous suspension was obtained. Half of the lymph node cells (LNC) from F74 were depleted of CD8+ cells by using an anti-CD8 antibody directly coupled to magnetic beads according to the recommendations manufacturer of the (Immunotech, Marseille, France). The efficiency of the depletion was monitored by flow cytometric analysis with the Leu2a antibody coupled to R-phycoerythrin (Becton Dickinson). Less than 2% of the LNC remained CD8+ positive after depletion. The culture conditions for LNC were the same as those used for PBMC.
Western blotting.
Western blot analysis was performed with extracts from CEMx174 cells infected with HIV-2 ROD, SIVsmFr, and SIVmac 251. Briefly, 107 cells were pelleted and resuspended in 100 μl of buffer containing 10 mM Tris-HCl (pH 7.6), 150 mM NaCl, 1 mM EDTA, 0.2 mM phenylmethylsulfonyl fluoride, and 100 U of aprotinin (Iniprol; Choay) before the addition of 100 μl of the same buffer containing 2% (vol/vol) Triton X-100 (lysis buffer). Cell extracts were centrifuged at 12,000 × g for 10 min, and the supernatants were stored at −80°C until use. To prepare protein extracts from viral pellets, the supernatants of infected cultures were first centrifuged at 12,000 × g for 10 min before high-speed centrifugation at 100,000 × g for 15 min in a Beckman TL100 centrifuge. The virus pellet obtained from a culture of 107 cells was solubilized in 200 μl of lysis buffer. Viral proteins were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and transferred onto nitrocellulose membranes as previously described (65). The blots were incubated in 5% nonfat milk in phosphate-buffered saline to block nonspecific binding sites and were then incubated for 2 h at room temperature (r.t.) with each monoclonal antibody (MAb) at a concentration of 5 μg/ml in phosphate-buffered saline containing 10% fetal calf serum. The viral proteins were revealed with anti-mouse immunoglobulin complexed to horseradish peroxidase and chemiluminescence reagents (ECL; Amersham) as recommended by the manufacturer. The enhanced light signal was captured on autoradiographic film (Hyperfilm TM-MP; Amersham). MAbs 125-B and 1H8, specific for HIV-2 ROD surface glycoprotein gp125 and transmembrane glycoprotein (TM) gp36, respectively, were provided by F. Traincard (Hybridolab, Institut Pasteur, Paris, France) (70). To test the reactivities of mangabey sera by Western blotting, a goat anti-human immunoglobulin labeled with horseradish peroxidase (Amersham) was used as the secondary antibody, followed by revelation with ECL reagents.
Generation of riboprobes.
The 35S-labeled RNA probe used for in situ hybridization was derived from the transcription vector pCR2.1 (Invitrogen), into which a 0.9-kb fragment spanning the SIVsmFr74 nef gene was inserted. The antisense riboprobe was generated from the T7 promoter by in vitro transcription of 1 μg of plasmid template with 50 U of T7 RNA polymerase in the presence of 200 μCi of 35S-UTP. After incubation for 1 h at 40°C, the DNA template was digested with 10 U of DNase I for 15 min at 37°C. To enhance the penetration of the probe into tissue sections, 35S-labeled RNA was subjected to mild alkaline hydrolysis in 80 mM NaHCO3–120 mM Na2CO3 at 60°C. The hydrolysis time was optimized to obtain a majority of fragments in the 150- to 200-nucleotide range. After neutralization with 600 mM Na acetate and 167 mM acetic acid, the probe was purified by phenol chloroform extractions and ethanol precipitation. Specific activity ranged between 1 × 108 and 5 × 108 dpm/μg of input DNA.
In situ hybridization.
Hybridization techniques were based on published procedures (68). Lymph nodes were frozen in isopentane cooled in liquid nitrogen. After embedding in OCT compound (Miles, Elkhart, Ind.), the tissues were cryostat sectioned at 4-μm intervals, and the sections were stored at −20°C until use. Hybridization was carried out with freshly cut tissue sections to minimize RNA degradation during storage. Sections were fixed in 4% paraformaldehyde and acetylated in 0.25% acetic anhydride–0.1 M triethanolamine (pH 8) to minimize background. Sections were denatured in 70% formamide at 70°C for 2 min to enhance the accessibility of nucleic acids. The hybridization mix contained the 35S-labeled riboprobe at 50,000 dpm/μl, 50% formamide, 10% (wt/vol) dextran sulfate, 0.3 M NaCl, 20 mM Tris (pH 7.5), 5 mM EDTA, 10 mM NaH2PO4. 1× Denhardt’s solution, 0.5-mg/ml yeast tRNA, and 100 mM dithiothreitol (DTT). The mix was heated at 80°C for 2 min and applied to slides. Coverslips were mounted, and hybridization was carried out at 45°C overnight in a humid chamber. Slides were rinsed successively in 2× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate)–10 mM DTT for 1 h at r.t., in 50% formamide–2×–SSC–10 mM DTT for 30 min at 60°C, in 2× SSC for 30 min at r.t., and in 0.1× SSC for 1 h at r.t. and were then dehydrated in ethanol with 0.3 M ammonium acetate. Slides were coated with LM-1 emulsion (Amersham) diluted 1:1 with 0.6 M ammonium acetate and were autoradiographed for 20 days at 4°C. The long exposure time ensured that weak hybridization signals were detected. After exposure, the slides were treated with Kodak D-19 developer, fixed, and stained with hematoxylin-eosin. Controls included hybridization of lymph node tissue from uninfected monkeys and hybridization with a control probe unrelated to SIV.
Amplification, cloning, and sequencing of SIVsmFr.
Genomic DNA was extracted from mangabey PBMC and LNC by using a DNA extraction kit (Qiagen). Proviral gag and env DNA fragments were amplified by nested PCR with two rounds of the following cycling conditions: 95°C for 2 min followed by 35 cycles of 95°C for 30 s, 55°C for 45 s, and 72°C for 1 min and by a final extension step of 10 min at 72°C. A gag fragment corresponding to positions 1076 to 1587 in the SIVsmmH4 sequence (52) was amplified with the following primers: outer gag pair (1,265 bp) GF1 (5′-TGG GAG ATG GGC GCG AGA AAC TCC GTC-3′) and GR1 (5′-TCC ACA TTT CCA GCA GCC CTG TCT TCT-3′), and inner gag pair (512 bp) GF2A-Cla (5′-ccatcgatGGG AAG AAA GCA GAT GAA TTA GAA-3′) and GAG5-Eco (5′-gggaattCTT CTG ATA GCG CCT GAA ATC CTG GCA CTA C-3′). An env (gp41) fragment corresponding to positions 8329 to 8911 in the SIVsmmH4 sequence was amplified with the following primers: outer env pair (680 bp) TM4mg (5′-GTT GTA TAG CCA CGT CAA GAG GCG-3′) and SE24 (5′-GGG ATA GTG CAG CAA CAG CAA CAG C-3′) and inner env pair (583 bp) TM1-Eco (5′-gggaattCAA GAA TTG TTG CGA CTG ACC G-3′) and TM2mg-Xba (5′-gctctaGAA TAT ATT CTA TTT GCC AAG GC-3′). Primers SE24, GF1, GR1, and GF2A have been described previously (15, 61). The other primers were designed so that they matched with regions conserved in all previously published SIVsm sequences (52). Two of the primers (TM2mg and TM4mg) were modified after analysis of the first env clones so as to better match SIVsmFr sequences. The fragments were cloned into a pBluescript KS vector with the restriction sites included in the primers and were sequenced with an automated sequencer (Applied Biosystems Inc.). The clones were designated by the name of the animal from which they originated, followed by the type of cells from which they were isolated (ln for LNC, pb for PBMC, and cc for coculture), the PCR reaction number, and the bacterial clone number (e.g., F74ln62-7). For each animal, 9 to 11 clones in both gag and env regions were sequenced.
Competitive PCR.
Experimental conditions were optimized so that a single round of PCR with only one pair of primer was sufficient to amplify a SIV-specific band from mangabey DNA. Primers GAG15 (5′-TTA ATA CTG TCT GCG TCA TTT GGT G-3′) and GAG7 (5′-GCC TCC TAC TTG CTG CAC TGG GTA A-3′) were designed to amplify a relatively short gag fragment (191 bp) and to match with the majority of the sequenced SIVsmFr gag clones. Primer GAG7 sequence was 100% conserved in 27 of 30 clones, while GAG15 sequence was 100% conserved in 28 of 30 clones. To enhance specificity, a hot start based on the melting of wax beads (Ampliwax PCR Gem100; Perkin-Elmer) was performed prior to the first PCR cycle. The DNA competitor fragment was engineered by introducing a 36-bp deletion in the 191-bp gag fragment amplified from clone SIVsmFr85 no. 61-6. The 5′ and 3′ fragments that flanked the deletion were amplified separately with primers GAG15 and GAG12-Pst (5′-CTG CAG CTT TTG CTT CCT CTG TAT G-3′) on one side and primers GAG7 and GAG11-Pst (5′-CTG CAG ACA AAA TGC CAG AAA CAA G-3′) on the other side. The resulting amplified products were restricted with enzyme PstI, purified on ChromaSpinTE100 columns (Clontech, Palo Alto, Calif.), and ligated together. The ligation product was reamplified with primers GAG15 and GAG7 and inserted in a pCR2.1 vector with a TA cloning system (Invitrogen). The resulting plasmid served as a matrix to produce the competitor fragment by further PCR with primers GAG15 and GAG7. The amount of competitor DNA was quantified by optical density measurement and by comparison with DNA standards after agarose gel electrophoresis. Known amounts of competitor DNA were diluted in a log-3.16 series in siliconized microcentrifuge tubes starting from 104 to 100.5 copies/μl. Ten microliters of each dilution was then coamplified with 250 ng of the DNA to be tested under the PCR conditions described above. The PCR products resulting from endogenous and competitor SIV DNA were distinguished by their different sizes on 3% agarose gels. The ratios (R) of endogenous to competitor PCR products were determined by densitometric measurement of the DNA bands. R was plotted as a function of the log of competitor concentration, and the endogenous DNA concentration was deduced by linear interpolation for R = 1. Determinations of viral load by competitive PCR were repeated twice for each sample. The interassay variation was found to be less than 0.5 log.
Competitive RT-PCR.
RNA was extracted from plasma with a viral RNA extraction kit (Qiagen). The plasma samples were lysed immediately without prior centrifugation to minimize RNA degradation. cDNA synthesis was primed with the antisense GAG7 primer. The amount of RNA corresponding to 21 μl of plasma was incubated with 50 U of Moloney murine leukemia virus reverse transcriptase (RT; Superscript RNase H-; Gibco BRL) in a final volume of 20 μl. This mixture was incubated for 30 min at 42°C, heated for 5 min at 95°C, and chilled for 5 min at 5°C. Competitive RT-PCR was then performed as described above, with the same competitor as that for DNA PCR. All experiments included amplification from M78 RNA or DNA as a negative control. The assay was calibrated by using known amounts of in vitro-transcribed RNA as the template. The gag clone F74cc55-1 restricted by EcoRI was used as a matrix for transcription with T3 polymerase, which yielded a 0.59-kb transcript in the sense orientation. The conditions used for in vitro transcription were similar to those used to synthesize riboprobes, except that all ribonucleotides were used at a final concentration of 0.4 mM. Unincorporated ribonucleotides were removed by spin column chromatography (Chromaspin-100; Clontech). The resulting RNA was quantified by optical density measurement and by electrophoresis on a denaturing 5% polyacrylamide gel in the presence of RNA size markers (Ambion). Densitometric measurement showed that more than 90% of the synthesized RNA was of the expected size (0.59 kb). The synthesized RNA was diluted in a log-3.16 series in diethylpyrocarbonate-treated water and was used as a template to calibrate the RT-PCR assay.
Phylogenetic analysis.
Amino acid sequences were aligned with the PILEUP program from the Genetics Computer Group (GCG) package (version 8.1). A consensus sequence was deduced from the alignment by using the PRETTY program from the GCG package with a threshold parameter of 1.5. Nucleotidic sequences were aligned for subsequent phylogenetic analysis with the CLUSTAL W (version 1.6) program (30), the final alignment being adjusted by eye. Genetic distances between pairs of DNA sequences were calculated by Kimura’s two-parameter model. Phylogenetic analysis of sequences consisted of minimum evolution estimated by the neighbor-joining method of Saitou and Nei (67) implemented in the CLUSTAL program, without taking gaps into account. The reproducibility of the branching order was estimated by applying a bootstrap procedure to 1,000 replicates of the original data set. We verified that tree topologies obtained by the neighbor-joining method were similar to those obtained by parsimony and maximum likelihood methods. The phylogenetic relationships among closely related SIVsmFr clones were tested directly by maximum likelihood with the fastDNAml program (56) on 100 bootstrap replicates of the data. Tree topology was further evaluated with the PUZZLE (version 3.1) program, which estimates maximum likelihoods for a randomly chosen quartet of sequences and which can be used to rapidly evaluate whether clusters are significant (69). The ratio of synonymous to nonsynonymous mutations (ratio of synonymous substitutions to synonymous sites/ratio of nonsynonymous substitutions to nonsynonymous sites [Ks/Ka]) was computed according to the method described by Li et al. (42) by using the NEWDIVERGE program from the GCG package.
Nucleotide sequence accession numbers.
The nucleotide sequences of the 10 SIVsmFr clones aligned in Fig. 3 have been submitted to GenBank and are available under accession nos. AF041980 to AF0411989.
FIG. 3.
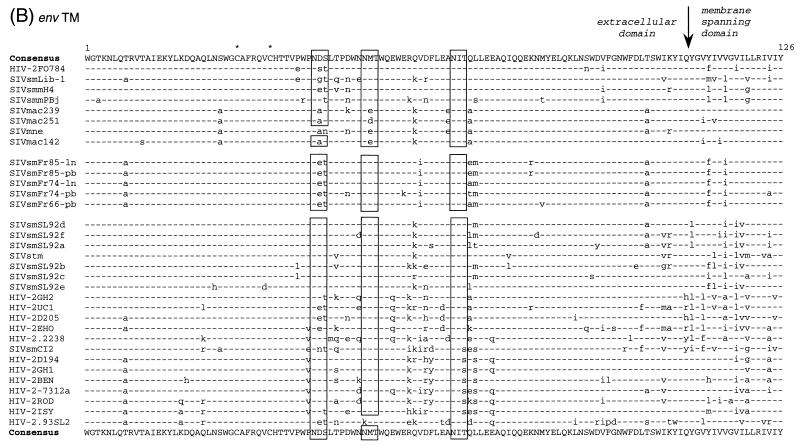
Multiple alignement of amino acid sequences comparing SIVsmFr with other viruses of the HIV-2–SIVsm–SIVmac lineage. Five representative SIVsmFr clones amplified from lymph nodes DNA (ln) or from PBMC DNA (pb) were aligned with previously published sequences (52). (A) Alignment of Gag amino acid sequences. The junction of the matrix p17 with the core protein p27 is indicated by an arrow. A glutamic acid rich sequence unique to SIVsmFr viruses is boxed. (B) Alignment of transmembrane glycoprotein (TM) amino acid sequences. The limits of the extracellular domain with the hydrophobic membrane-spanning domain is indicated by an arrow. Stars, cysteine residues; boxes, conserved glycosylation sites in TM; dashes, sequence identity with the consensus; dots, gaps introduced to optimize the alignment.
RESULTS
Seroepidemiologic screening.
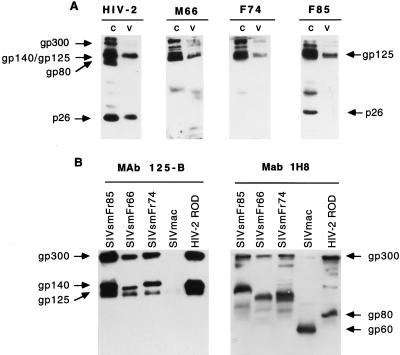
Nonhuman primates from a French zoo were screened for evidence of lentivirus infection. The sera of 18 species of primates were tested by Western blotting for reactivity with HIV-1 and HIV-2 antigens (Table 1). Three of the four sooty mangabeys (C. atys atys) tested had antibodies reacting with HIV-2 proteins. The sera cross-reacted extensively with HIV-2 ROD, since they detected the surface glycoprotein gp125, the dimeric form of the transmembrane glycoprotein (TM) gp80, the dimeric and monomeric forms of the envelope precursor (gp300 and gp140), and, in one case, the major core protein p26 (Fig. 1A). This pattern was indicative of infection with a virus belonging to the HIV-2–SIVsm–SIVmac group. The mangabeys had antibodies directed primarily to envelope antigens, since their serologic reactivities were similar to that obtained with a MAb directed to HIV-2 surface glycoprotein (Fig. 1B; left panel). In contrast, the serologic reactivities to Gag antigens were weak, with only one of three animals positive for antibodies to the p26.
TABLE 1.
Serologic surveya
| Common name (species) | Sex | Origin | Age (yrs) | Serologic status |
|---|---|---|---|---|
| Chimpanzee (Pan troglodytes) | M | W | 25 | − |
| Mandrill (Papio sphinx) | F | C | 8.8 | − |
| F | C | 22.5 | − | |
| M | C | 19 | − | |
| White-cheeked mangabey (Cercocebus albigena) | M | C | 5 days | − |
| F | C | 3.4 | − | |
| M | C | 2.6 | − | |
| M | C | 6.2 | ID | |
| Black mangabey (Cercocebus aterrimus) | M | C | 4.7 | − |
| F | C | 7.6 | − | |
| Crested mangabey (Cercocebus galeritus agilis) | M | C | 3 | − |
| F | W | 3 | − | |
| Golden-bellied mangabey (Cercocebus galeritus chrysogaster) | M | W | 14 | − |
| F | W | 7 | − | |
| F | W | 11 | − | |
| F | W | 17 | − | |
| White-crowned mangabey (Cercocebus atys lunulatus) | M | C | <1 | − |
| M | W | 16 | − | |
| M | C | 0.8 | − | |
| F | C | 15.5 | − | |
| M | C | 14 | − | |
| F | C | 2.8 | − | |
| F | W | 22 | − | |
| F | C | 7.3 | − | |
| F | W | 22.5 | − | |
| Sooty mangabey (Cercocebus atys atys) | M | W | 33 | + |
| F | W | 27 | + | |
| M | C | 18.1 | − | |
| F | C | 15.9 | + | |
| Campbell’s guenon (Cercopithecus campbelli campbelli) | M | W | 8 | − |
| M | W | NA | − | |
| F | C | 4.4 | − | |
| M | C | 3.8 | − | |
| Lowe’s guenon (Cercopithecus campbelli lowei) | M | W | 31 | − |
| F | W | 10 | − | |
| Moustached monkey (Cercopithecus cephus) | F | W | 13 | − |
| Diana monkey (Cercopithecus diana roloway) | F | W | 21 | ID |
| Owl-faced monkey (Cercopithecus hamlyni) | F | W | 11 | − |
| F | W | 11 | − | |
| L’Hoest’s monkey (Cercopithecus l’hoesti) | F | W | 22 | ID |
| F | W | 10 | − | |
| Blue monkey (Cercopithecus mitis) | M | W | 23 | − |
| F | W | 2 | − | |
| De Brazza’s monkey (Cercopithecus neglectus) | F | C | 1.6 | − |
| Greater white-nosed monkey (Cercopithecus nictitans nictitans) | F | C | 3.5 | − |
| F | W | 4 | − | |
| Allen’s swamp monkey (Cercopithecus nigroviridis) | M | C | 5.3 | − |
| F | C | 6.2 | − | |
| F | W | 12 | − |
Sera from 18 species of nonhuman primates were tested for reactivity with HIV-1 and HIV-2 antigens by Western blotting. M, male; F, female; W, wild caught; C, captive borne; −, +, and ID, negative, positive, and indeterminate, respectively, by HIV-2 Western blot; NA, not available.
FIG. 1.
Analysis of mangabey serologic reactivity and of SIVsmFr protein profile by Western blotting. (A) Reactivity of a typical human HIV-2-positive serum (left lane) and of three mangabey serum samples (M66, F74, and F85) with HIV-2 ROD antigens. The sera were tested on proteins extracts from HIV-2-infected cells (c) and from HIV-2 viral pellets (v). The human serum reacted with the glycoprotein precursor (gp 140) and its dimer (gp300), the surface glycoprotein (gp125), the dimer of transmembrane glycoproteins (gp80), and the Gag core p27. The mangabey sera reacted mostly with envelope antigens, while their reactivities to Gag p27 were limited or undetectable. (B) Profile of SIVsmFr proteins detected with anti-HIV-2 MAb directed to the surface glycoprotein (125-B) and to the transmembrane glycoprotein (1H8). Protein extracts from CEMX174 cells infected with either of the three SIVsmFr isolates, SIVmac251, or HIV-2 ROD were tested with MAb 125-B (left panel) or with MAb 1H8 (right panel).
All of the other monkeys and the one chimpanzee included in the survey tested negative. In particular, no seropositivity was detected among the white-crowned mangabeys (Cercocebus atys lunulatus), which are phylogenetically close to the sooty mangabey and whose natural range lies next to that of the sooty mangabeys (28).
Isolation of SIVsm from mangabey tissues.
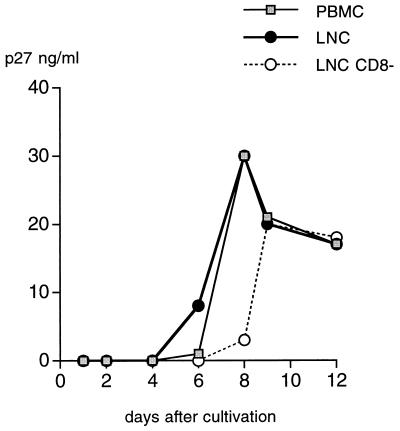
Virus isolation was performed by cocultivating sooty mangabey PBMC with PHA-activated human PBMC. Isolation was also performed from cocultures of LNC obtained from two of the animals (F74 and F85). SIV was readily isolated from the cells of the three seropositive sooty mangabeys. In a typical experiment, shown in Fig. 2, the p27 Gag antigen became detectable as early as day 6 of culture and peaked between days 7 and 11. The depletion of CD8+ lymphocytes, which was performed with F74 LNC prior to cultivation, did not accelerate viral growth (Fig. 2). The new viruses were named SIVsmFr66, SIVsmFr74, and SIVsmFr85, in keeping with the current nomenclature (Fr stands for France).
FIG. 2.
Isolation of SIVsmFr74 from mangabey PBMC and LNC. Virus isolation was performed by coculturing F74 PBMC, LNC, and LNC from which CD8+ cells were depleted (LNC CD8−) with human PBMC in the presence of IL-2 and PHA. Virus production was monitored by an antigen-capture assay specific for SIV p27 Gag.
To analyze viral proteins, SIVsmFr isolates were amplified on the CEMx174 cell line. Proteins were extracted at the peak of infection and analyzed by Western blotting with MAbs directed to HIV-2 envelope. Reactivity with MAb 125-B showed that the surface glycoprotein (SU) from the three SIVsmFr isolates had a size equivalent to that of HIV-2 ROD gp125 (Fig. 1B [left panel]). In contrast, the sizes of the envelope precursor varied, depending on the isolate. SIVsmFr74 and SIVsmFr85 had an envelope precursor with a size similar to that of HIV-2 ROD gp140, while the envelope precursor of SIVsmFr66 was smaller. These findings suggested that SIVsmFr74 and SIVsmFr85 were more closely related to each other than they were to SIVsmFr66. The size difference could be accounted for by different glycosylation patterns or by a premature truncation of the TM of SIVsmFr66. Since the genetic analysis did not reveal the presence of premature stop codons after the transmembrane domain of the TM (see below), differences in the extent of glycosylation were a more likely explanation. Blotting with MAb 1H8 specific for the TM revealed mostly high-molecular-weight species, which corresponded to the envelope precursor gp140, to its dimer gp300, and to oligomeric forms of the TM (Fig. 1B [right panel]). The absence of monomeric forms indicated that the SIVsmFr TM formed particularly stable oligomers, a property which is also characteristic of HIV-2 (65).
Characterization of a novel genetic subtype within the SIVsm group.
To perform genetic analysis, SIVsmFr sequences were amplified by nested PCR from uncultured mangabey tissues. A 538-bp fragment of env coding for part of the extracellular and transmembrane domains of the TM and a 456-bp fragment of gag spanning the p17/p27 junction were amplified, cloned, and sequenced. These regions were chosen so that the new sequences could be aligned over 368 bp with previously published HIV-2 and SIVsm sequences (15, 25). The genetic analysis was done first with one clone representative of each of the three PBMC and two LNC samples. The alignment of deduced protein sequences showed that the three mangabey viruses shared sequence features characteristic of the SIVsm–HIV-2 group such as the positions of cysteines and of glycosylation sites in the TM (Fig. 3B). The premature stop codon that truncates the TM just 3′ to the membrane spanning domain in some SIV clones (12, 19) was not found in SIVsmFr (data not shown). A rapid inspection of the alignments showed that SIVsmFr66, SIVsmFr74, and SIVsmFr85 were genetically close. In particular, they shared a unique glutamic acid rich motif in gag (boxed in Fig. 3A). The three SIVsmFr were nonetheless distinguishable, since sequences obtained from LNC and PBMC from a given animal were more related than sequences obtained from two different animals.
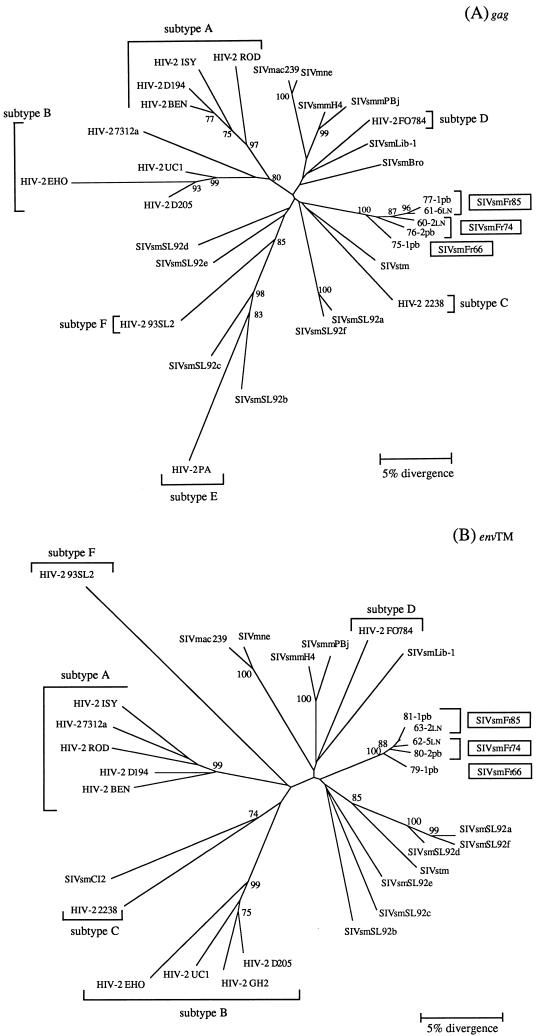
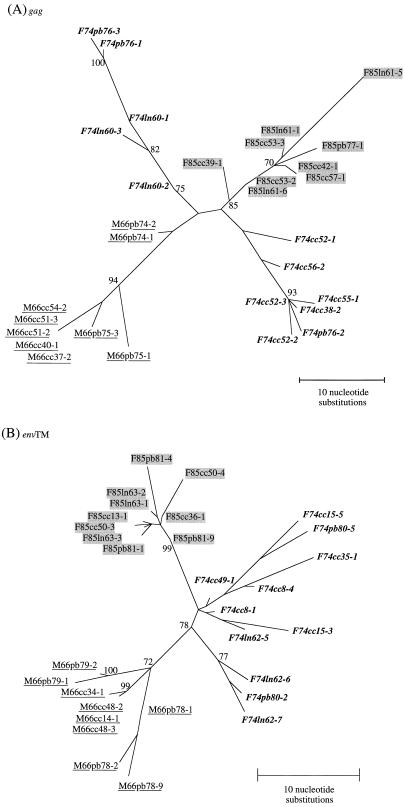
Phylogenetic analysis of gag and env nucleotide sequences was done by the neighbor-joining method (67). The robustness of the phylogenetic tree topology was evaluated by analyzing 1,000 bootstrap replicates of the data. The tree constructed from gag sequences of HIV-2, SIVsm, and SIVmac divided into two main branches, with the HIV-2 subtypes A and B on one side, and the more diverse HIV-2 subtypes C, D, E, and F intermingled with the SIVsm subtypes on the other side. The SIVsmFr sequences formed a new lineage rattached to the second main branch (Fig. 4A). These findings confirmed the extensive genetic diversity of SIVsm isolated to date. The SIVsmFr sequences clearly segregated as a distinct lineage, since their clustering was supported by a 100% bootstrap value in both gag and env trees (Fig. 4A and B). The same clustering was found when trees were constructed by a maximum likelihood method that took into account the nonuniform rates of nucleotide substitutions characteristic of lentivirus sequences (puzzle program with four categories of rate heterogeneity) (69). The estimation of support for the SIVsmFr cluster was 90% by the quartet puzzling method on the gag tree (data not shown). The closest relatives of SIVsmFr were found either in the SIVsmLib-1–SIVsmmPBj group (15% nucleotide divergence in gag) or with SIVsmSL92e from Sierra Leone (13.5% nucleotide divergence in env). Since these associations were not supported by high bootstrap values, the SIVsmFr viruses were best described as forming a new subtype in the diverse HIV-2–SIVsm–SIVmac group.
FIG. 4.
Phylogenetic relationships among the newly characterized SIVsmFr and viruses of the HIV-2–SIVsm–SIVmac lineage. (A) Phylogenetic tree derived from gag sequences. The analysis was performed on a 369-bp fragment that encompassed the Gag p17-p26 junction. (B) Phylogenetic tree derived from env sequences. A 380-bp fragment coding for a portion of the extracellular and membrane-spanning domains of TM was analyzed. Phylogenetic trees were constructed by the neighbor-joining method on 1,000 bootstrap replicates of the data. Values on the nodes are percentages of bootstraps with which the cluster is supported; only values greater than 70% are shown. The new SIVsmFr sequences are boxed; HIV-2 subtypes A to F, as defined in references 14 and 25, are indicated.
The two known clusters of human and simian viruses, namely, HIV-2 FO784–SIVsmLib-1 from Liberia and HIV-2PA–SIVsmSL92b and -c from Sierra Leone, were apparent on the phylogenetic trees. The genetic and geographic proximity of these pairs of viruses has provided the basis for the idea that HIV-2 subtypes emerged as a consequence of multiple independent transmission of SIVsm to humans (14, 15, 25, 26). We identified a possible new HIV-2–SIVsm pair in the env tree, since SIVsmCI2 from Ivory Coast (61) clustered in 74% of the bootstraps with the HIV-2 2238 sequence from Liberia. Although SIVsmCI2 gag sequence was not available to confirm its relatedness to the HIV-2 type C subtype, this observation supports the idea that multiple interspecies transmission events are needed to explain the phylogenetic relationships within the HIV-2–SIVsm–SIVmac group.
Intraindividual genetic variability of SIVsmFr.
We evaluated the intraindividual genetic variability of SIVsmFr by sequencing a total of 60 clones obtained from both mangabey tissues and short-term-cultured isolates. The maximum nucleotide divergence between pairs of SIVsmFr clones was 9% in gag and 6% in env (Table 2). The genetic proximity between the SIVsmFr viruses suggested that viral transmission had occurred among the three seropositive mangabeys. Since these animals were of unrelated geographical origins, their harboring the same SIVsm subtype by chance would have been highly unlikely.
TABLE 2.
Genetic diversity of SIVsmFr sequencesa
| Region sequenced | Intrahost genetic diversity
|
Interhost genetic diversity
|
||||||
|---|---|---|---|---|---|---|---|---|
| Animal | Maximum nucleotide divergence (%) | Animals | Minimum nucleotide divergence (%) | Maximum nucleotide divergence (%) | Clones considered in transmission hypothesis | Nucleotide divergence between clones (%) | Estimated fixation rate of mutations (subst/site/year) | |
| Gag (456 bp) | M66 | 3.39 | M66/F74 | 2.23 | 6.47 | NA | NA | NA |
| F74 | 6.50 | M66/F85 | 2.69 | 8.50 | 74-1/61-5 | 5.77 | 6.01 × 10−3 | |
| F85 | 3.64 | F74/F85 | 2.92 | 9.00 | 60-2/61-5 | 6.26 | 6.52 × 10−3 | |
| TM (538 bp) | M66 | 4.25 | M66/F74 | 2.66 | 5.85 | NA | NA | NA |
| F74 | 4.26 | M66/F85 | 3.45 | 6.09 | 78-1/81-4 | 4.63 | 4.82 × 10−3 | |
| F85 | 2.47 | F74/F85 | 2.08 | 5.05 | 49-1/81-4 | 3.44 | 3.58 × 10−3 | |
Intrahost and interhost genetic divergences were evaluated by comparison of the SIVsmFr sequences, which are presented in Fig. 5. The fixation rate of mutations was computed by dividing the observed divergence by 9.6 years. In each case, a hypothesis was made about which animal was the donor of the virus and which was the recipient. The divergence between the clone of the donor that was closest to the sequences of the recipient and the clone of the recipient that was farthest from the sequences of the donor was computed. NA, not applicable (because time since transmission was not known).
The phylogenetic relationships among SIVsmFr clones were analyzed by the fastDNAml program on 100 bootstrap replicates of the data. The maximum likelihood method implemented in fastDNAml was preferred over a distance method because the data set consisted of closely related sequences. Phylogenetic reconstructions showed that the sequences derived from a given animal clustered together in both gag and env trees (Fig. 5A and B). Thus, the viral quasispecies found in each animal were distinguishable, despite the fact that the range of intraindividual nucleotide divergence overlapped with that of interindividual divergence (Table 2). In addition, the clustering of clones from the same animal suggested that cross-contamination of samples during PCR amplification did not occur.
FIG. 5.
Phylogenetic relationships among SIVsmFr clones. (A) Phylogenetic tree derived from sequence comparison of gag clones. (B) Phylogenetic tree derived from sequence comparison of env (TM) clones. The trees were constructed by the maximum likelihood method implemented in the FastDNAml program, with 100 bootstrap replicates of the data. Values on the nodes are percentages of bootstraps with which the cluster is supported; only values greater than 70% are shown. SIVsmFr66 clones are underlined, SIVsmFr74 clones are in boldface type, and SIVsmFr85 clones are shaded. Abbreviations within the names of the clones indicate whether they were amplified from PBMC (pb), lymph nodes (ln), or short-term cocultures with human PBMC (cc).
Within each viral quasispecies, sequences derived from LNC and PBMC did not cluster separately (see, for example, the cosegregation of clones F74ln62-7 and F74pb80-2 or that of F85ln63-3 and of F85pb81-1 in the env tree). This observation indicated that compartmentalization of the virus between the blood and the lymphoid organs was not stringent.
The clones derived from short-term cultures of SIVsmFr66 and SIVsmFr85 formed a homogeneous population, but those obtained from SIVsmFr74 culture were genetically diverse, with as much as 4.25% nucleotide divergence in env. Similarly, SIVsmFr74 clones from LNC and PBMC exhibited more genetic diversity than those from the other two viruses, with a maximum of 6.5% nucleotide divergence in gag (Table 2). In both gag and env trees, SIVsmFr74 sequences formed two main branches, while SIVsmFr66 and SIVsmFr85 clones clustered in a unique branch. The more diverse viral quasispecies in F74 suggested a more ancient infection in this animal.
The group of F85 clones branched closer to that of F74 clones than to that of M66 clones. In the env tree, the association between F74 and F85 clones was supported by 78% of bootstrap replicates. These data, in addition to the similarity between F74 and F85 envelope precursors, suggested that viral transmission had occurred between F74 and F85 rather than between M66 and F85. Considering the more diverse viral quasispecies in F74, it was likely that SIV was transmitted to the young female F85 from the older female F74. This scenario is consistent with the history of the animals, since F85 had to be separated from the F74-M66 group after she had been severely bitten by other females.
Estimation of the fixation rate of mutations.
F85 had been in contact with the two older mangabeys for a limited time period between November 1985 and October 1986. Tissue samples from F85 were collected in June 1996. We could deduce from this chronology that SIVsmFr transmission between F74 or M66 and F85 had taken place about 10 years ago (minimum duration of infection, 9.6 years; maximum duration of infection, 10.5 years). Given this duration for viral evolution, it is possible to estimate the fixation rate of mutations from the divergence observed between viral sequences of the donor and recipient animals. Considering that SIVsmFr was transmitted from F74 to F85, one can identify among SIVsmFr74 sequences the clone that is the closest to the one that was actually transmitted (F74ln60-2). The rate of mutation fixation between this ancestorlike sequence and the most divergent clone in the recipient (F85ln61-5) is then calculated. Thus, if the 6.2% divergence that separates clone F74ln60-2 from clone F85ln61-5 accumulated over 9.6 years, the estimated fixation rate of mutation would be 6.52 × 10−3 substitutions per site per year (subst/site/yr). The same type of calculation performed with env sequences yields an estimate of 3.58 × 10−3 subst/site/yr (Table 2). Making the hypothesis of viral transmission from M66 to F85 leads to very similar estimates for both gag and env (Table 2). Assuming that the fixation rate in gag (6.52 × 10−3 subst/site/yr) is equivalent to the mutation rate, and considering that the point mutation rate for SIVsm RT is equivalent to that determined for HIV-1 (3.4 × 10−5 subst/base/cycle) (45), the minimum number of consecutive viral replication cycles needed to account for the observed sequence divergence is 190 per year. These calculations indicate that SIVsmFr replicates at a high rate in mangabeys, despite the nonpathogenic character of infection in this host. It should be noted that this rate of virus replication represents a minimal estimate, since the actual mutation rate is higher than the observed fixation rate, especially in relatively conserved regions of the genome such as gag.
Ks/Ka ratio.
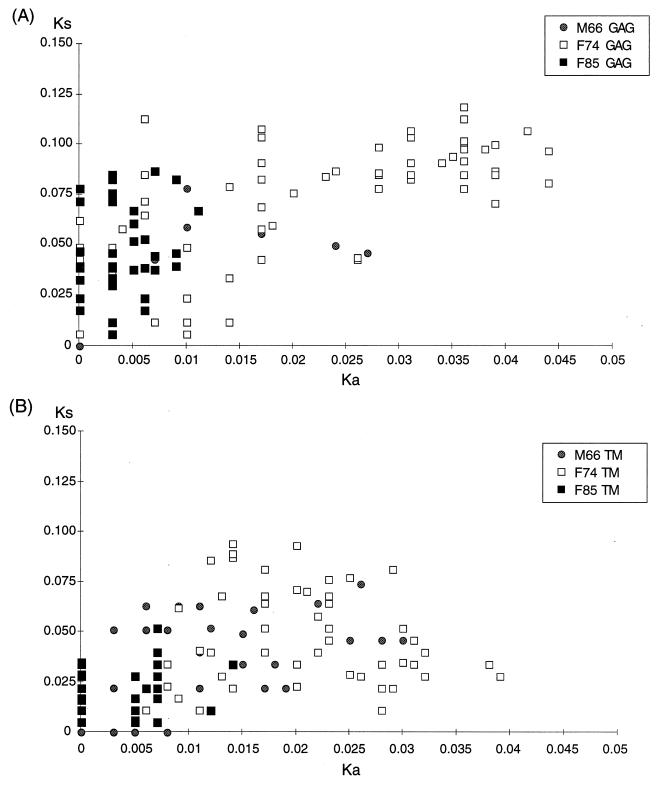
Evaluation of the Ks/Ka ratio gives an indication of the type of selection pressure that contributes to the evolution of viral sequences. A majority of synonymous mutations (Ks/Ka of greater than 1) indicates the predominance of a purifying type of selection, which is associated with a preferential elimination of viruses with variant amino acids. Conversely, a majority of nonsynonymous mutations (Ks/Ka of less than 1) reflects the predominance of a diversifying type of selection, such as the pressure from the immune system which selects for viral escape variants.
The Ks/Ka ratio between pairs of sequences obtained from the same animal was computed according to the method of Li et al. (42). Analysis of SIVsmFr pairs of sequences revealed a high Ks/Ka ratio, with means of 5.47 in gag (standard deviation = 5.08) and of 2.32 in env. (standard deviation = 1.85). Such Ks/Ka ratios were indicative of a predominant purifying selection for viral fitness, which is usually the case in these regions of the lentivirus genome (5, 39). It was interesting to note marked differences between the Ka values in the three infected animals, while the Ks values were similar (Fig. 6). Ka values were particularly low between pairs of SIVsmFr85 sequences, indicating a strong conservation of amino acid sequences for this virus. Since the Ka value was shown to increase over time in the course of HIV infection (44, 75), a similar phenomenon may apply to SIV infection, and the low Ka obtained for SIVsmFr85 may reflect the fact that SIV infection is more recent for F85 than for F74 or M66. Thus, analysis of Ka and Ks parameters supports the hypothesis that SIVsmFr was transmitted to F85 by one of the two older animals, rather than the reverse.
FIG. 6.
Comparison of synonymous and nonsynonymous mutations between pairs of sequences. Ks and Ka values between pairs of clones obtained from the same animal were measured. The range of the Ka parameter was limited for F85, indicating homogeneity of viral amino acid sequences in this animal.
Viral load in SIVsmFr-infected mangabeys.
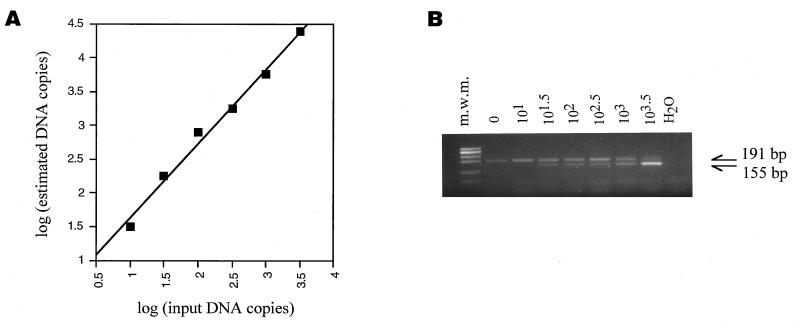
A competitive PCR assay specific for SIVsm sequences was developed to evaluate the viral load in infected mangabey tissues. The assay is based on the coamplification of a known amount of competitor DNA molecules with the endogenous DNA molecules to be quantified. The competitor fragment was engineered by introducing a 36-bp deletion in a SIVsmFr74 gag clone, by PCR-driven mutagenesis (see Materials and Methods). To optimize the efficiency of amplification, the primers were chosen in conserved regions of the previously sequenced gag clones, and the size of the amplified fragment was limited to 191 bp. The competitive PCR assay was calibrated by using known amounts of a plasmid carrying the undeleted gag fragment as samples. The assay gave a linear response over the range of concentrations studied [log (estimated copies) = 1.10 × log (expected copies) + 0.53] (Fig. 7A). The concentration of competitor copies was adjusted in further experiments so that the estimated and expected copy numbers were equal.
FIG. 7.
Quantitation of SIVsmFr by competitive PCR. (A) Calibration of the competitive PCR assay. Test samples containing known amounts of a SIVsmFr74 gag plasmid were assayed by competitive PCR. The log of the DNA copy number estimated in the assay was plotted as a function of the log of input plasmid copy number. Each plot symbol corresponds to the result of a series of amplifications in the presence of different dilutions of competitor DNA fragment. The competitive PCR assay gave a linear response over the range of concentrations studied (y = 1.10 x + 0.53; r = 0.995). (B) Example of a competitive PCR. Lymph node DNA from animal F85 was amplified in the presence of serial dilutions of competitor DNA. The PCR products were separated by electrophoresis on a 3% agarose gel. The endogenous SIV product can be distinguished from the competitor product by its size, which is larger by 36 bp. m.w.m., molecular weight marker.
As shown in Fig. 7B, a single PCR assay with only one set of primers was sufficient to amplify a band visible in a BET-stained agarose gel from SIVsmFr-infected PBMC and LNC DNA. This indicated that the viral load in SIVsmFr-infected animals was relatively high. The quantitation showed that as many as 24,000 viral DNA copies per 106 LNC were detected (Table 3). DNA samples obtained from the seronegative mangabey, M78, remained persistently negative. A serial dilution PCR experiment, which was done by diluting in parallel SIVsmFr85 lymph node DNA and a SIVsmFr gag plasmid in negative mangabey DNA, gave results consistent with those of the competitive PCR assay, with a viral load close to 104 viral copies/106 cells. In infected animals, LNC harbored proportionally more viral DNA copies than PBMC, which was consistent with the lymphoid organs being the main site of SIV replication. The ratios of LNC viral DNA to PBMC viral DNA, which ranged from 2 to 4, were comparable to or lower than those measured for HIV-infected patients, depending on the study considered (49, 58, 59).
TABLE 3.
Viral load in SIVsmFr-infected mangabeysa
| Animal | No. of DNA copies per 106 PBMC | No. of DNA copies per 106 LNC | No. of RNA copies per ml of plasma | LNC DNA copies/PBMC DNA copies |
|---|---|---|---|---|
| M66 | 6,020 | NA | 4,275,600 | NA |
| F74 | 2,450 | 5,140 | 468,800 | 2.10 |
| F85 | 5,380 | 24,000 | 11,529,200 | 4.46 |
The numbers of viral DNA copies in mangabey tissues were evaluated by competitive PCR. Viral RNA in plasma was quantitated by competitive RT-PCR. NA, not available.
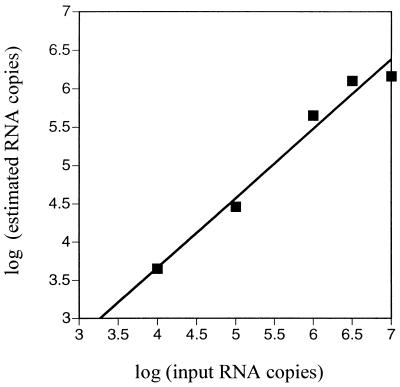
The assay was adapted to amplify viral RNA in plasma by introducing a step for cDNA synthesis prior to the amplification. RT-PCR was calibrated by using known amounts of in vitro-transcribed RNA as samples. The gag plasmid F74cc55-1 was used as a matrix for in vitro transcription. The resulting RNA was quantified by optical density measurement and denaturing PAGE before being tested by RT-PCR (Fig. 8). The estimated RNA copy number was found to be less than the expected copy number [log (estimated copies) = 0.91 × log (expected copies) + 0.03], which indicated that the efficiency of cDNA synthesis was less than 100% (approximate efficiency, 20%). RT-PCR results were corrected in further experiments to account for the cDNA synthesis efficiency. The analysis of mangabey samples showed that the concentrations of viral RNA in plasma ranged from 4 × 105 to 1 × 107 copies per ml (Table 3). Taken together, these data showed that the cell-associated and plasma viral loads detected in SIVsm-infected mangabeys were as high as those seen in susceptible hosts (humans or macaques) during the asymptomatic stage of HIV or SIVmac infection (32, 48, 72).
FIG. 8.
Calibration of the competitive RT-PCR assay. Known amounts of RNA transcribed from a gag plasmid clone (F74cc55-1) were assayed by competitive RT-PCR. The log of the SIV RNA copy number estimated in the assay was plotted as a function of the log of the input RNA copy number. The competitive RT-PCR assay gave a linear response over the range of concentrations studied (y = 0.91 x + 0.03; r = 0.987).
In situ hybridization in lymph nodes.
Histologic examination and in situ hybridization were performed with lymph nodes obtained from F74, F85, and M78. The histology of the lymph nodes from infected mangabeys was normal, with no signs of follicular hyperplasia or of lymphoid depletion. Germinal centers of limited size and normal shape were found scattered at the periphery of the tissue sections (Fig. 9A). Flow cytometry analysis performed with F85 LNC confirmed that there were no obvious signs of CD4 lymphocyte depletion or of abnormal cellular activation in this animal compared to the uninfected control M78. The CD4/CD8 lymphocyte ratios were 1.2 for F85 and 1.5 for M78. In PBMC, the CD4/CD8 ratios were 0.5 and 0.7, respectively. The fraction of lymphocytes expressing the activation marker CD25 was similar in both animals (7% in lymph nodes and less than 2% in PBMC).
FIG. 9.
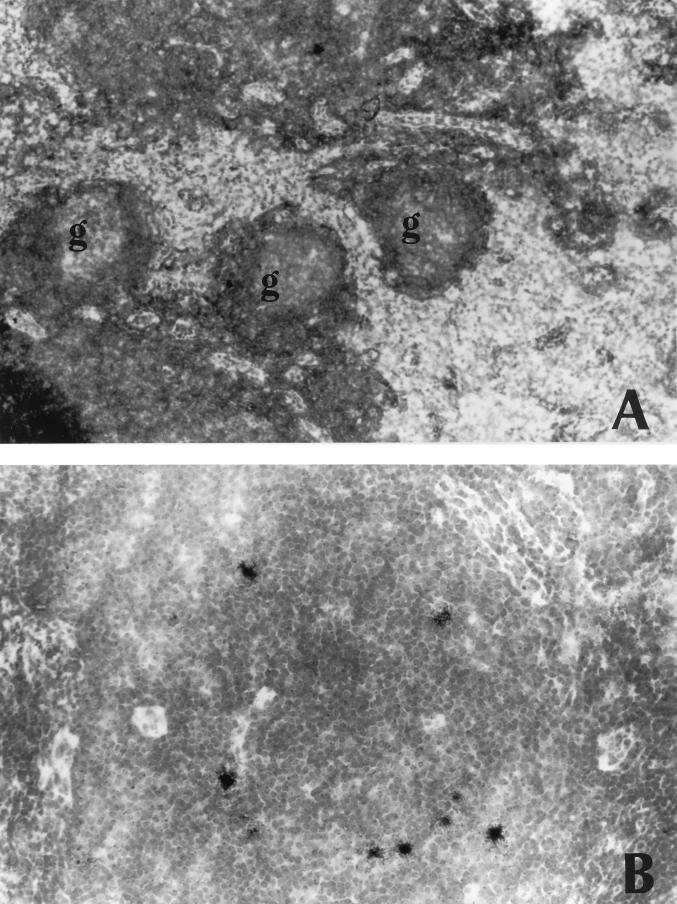
Detection of SIV expression in lymph nodes by in situ hybridization. Frozen lymph node sections were hybridized with a 35S-labeled riboprobe specific for SIVsmFr RNA. (A) Low-magnification view of F85 lymph node. The histology is normal, with follicles of a limited size. No hybridization signal could be detected in the germinal centers (g) (original magnification, ×100); (B) higher-magnification view of F85 lymph node showing numerous infected cells scattered in the paracortical region. The intensity of the hybridization signal indicates that the cells are productively infected (original magnification; ×250).
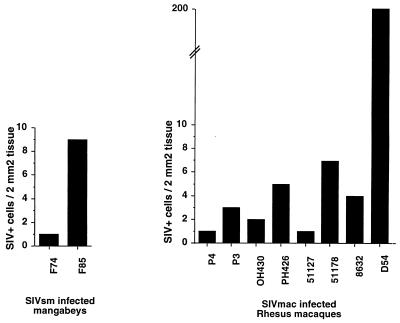
To perform in situ hybridization, the tissue sections were incubated with a 35S-labeled riboprobe derived from a SIVsmFr74 env clone. The riboprobe was synthesized in the antisense orientation so as to detect viral RNA. Numerous infected cells were detected in F85 lymphoid tissue (Fig. 9B), while there were fewer positive cells in the case of F74. Tissue sections from the uninfected mangabey M78 remained negative. The intensity of the hybridization signal in F85 and F74 tissues indicated that productive virus replication took place in the infected cells, which localized mainly in the T-cell areas of the lymph node. We did not detect the diffuse hybridization signal that is usually seen in the germinal centers of hyperplasic HIV- or SIV-infected lymph nodes and which reflects the presence of viral particles trapped at the surface of follicular dendritic cells (Fig. 9A). The viral load was quantitated by counting the number of hybridization spots per surface unit of lymph node section (Fig. 10). A comparison was made with lymph nodes from SIVmac-infected rhesus macaques that were previously studied (11). Macaque lymph nodes were evaluated 2 months after virus inoculation, i.e., early in the asymptomatic stage of the infection. The levels of infection in mangabey lymphoid tissues were in the same range as those found during the asymptomatic stage of SIVmac infection in macaques that were slow or intermediate progressors (1 to 10 positive cells/2 mm2) but were lower than those observed in the lymph nodes of the fastest progressors (as many as 200 positive cells/2 mm2). Thus, the degree of virus replication did not discriminate between the nonpathogenic infection of mangabeys and the slowly progressive infection of macaques.
FIG. 10.
Comparison of the numbers of productively infected cells in mangabey and macaque lymph nodes. The numbers of productively infected cells detected by in situ hybridization in a 2-mm2 area of lymph node section were counted. The mean counts obtained for three sections are indicated. Macaque lymph nodes were analyzed 2 months after inoculation of SIVmac251. Mangabey lymph nodes were obtained from naturally infected animals.
DISCUSSION
Several lines of evidence converge to show that SIVsm replicates actively and to high levels in its natural host: (i) SIVsmFr was rapidly isolated from the PBMC and the LNC of seropositive mangabeys; (ii) the genetic diversity and the estimated rate of mutation fixation were high, which indicated that numerous virus replicative cycles took place in infected animals; (iii) the viral load measured in sooty mangabeys was equivalent to those seen during the asymptomatic stage of pathogenic lentivirus infections in primates; (iv) productively infected cells were detected in lymphoid tissues. Despite its chronically active nature, SIVsm infection does not cause any apparent pathology in sooty mangabeys (22, 23, 47). Indeed, the seropositive animals included in this study had been infected for at least 9 years, the duration of infection being probably greater for the F74 animal. None of the mangabeys exhibited clinical signs, except for an arthritic condition associated with old age in M66. The absence of SIVsm pathogenicity for mangabeys poses a paradox, since there is a clear positive correlation between viral load and pathogenic potenial in other lentivirus infections of primates. Longitudinal virologic studies with HIV-infected patients demonstrated a strong association between viral load in plasma in the asymptomatic stage and the risk of progression to AIDS (48). Levels of 105 to 107 viral RNA copies/ml are associated with a high risk of progression to AIDS in HIV-positive individuals. It is intriguing that similar plasma viral loads are well-tolerated in SIV-infected mangabeys. The same observation holds for the cellular viral load, since levels of 103 to 104 viral DNA copies/106 cells are found in HIV-infected patients (16) as well as in SIV-infected mangabeys (this study and reference 15).
The comparison of SIVsm and SIVmac infections is instructive, since both viruses are of the same phylogenetic origin, and since SIVmac is the result of cross-species transmission of SIVsm to macaques (27, 51). Viral loads similar to those measured in the present study have been described for SIVmac-infected macaques that ultimately progressed to AIDS. Macaques with 105 to 107 viral RNA copies/ml plasma are not among the fastest progressors, which harbor as many as 109 copies/ml, but they do develop disease (32, 72). SIVmac genetic variability has been extensively studied in the hypervariable regions of the envelope. The rate of mutation fixation estimated for V1 and V2 is in the order of 10−2 to 10−3 subst/site/yr (9, 10, 36, 41, 57, 62). It is at least 1 order of magnitude lower in more conserved regions of the genome such as the integrase (10−3 to 10−4 subst/site/yr) (36). Since we analyzed genetic variability in relatively conserved regions of the SIVsm genome (p17 Gag and TM), the rates of 4 × 10−3 to 6 × 10−3 subst/site/yr estimated for these regions are comparable to those found for SIVmac. Thus, SIV replicates at a high rate in mangabeys as well as in macaques, although it induces disease only in the latter species. The idea that SIVsm replicates continuously in its natural host is further supported by the high genetic variability observed in the HIV-2–SIVsm group as a whole. The novel SIVsm subtype identified in this study forms an additional lineage in the already diverse HIV-2–SIVsm phylogenetic tree (14, 15, 25, 26, 61). The facts that SIVsm branches are interspersed with and are as long as HIV-2 branches imply that SIVsm evolves as rapidly as HIV-2.
Comparison of SIVsm and SIVagm models of nonpathogenic lentivirus infections reveals significant differences. The main characteristic of SIVagm infection in African green monkeys is a low viral load, with 20 to 500 provirus per 106 PBMC detected (8, 29). The viral load is thus 1 to 2 orders of magnitude lower in African green monkeys than in mangabeys. In addition, the ratio of LNC viral DNA to PBMC viral DNA is close to 1 in SIVagm infection, suggesting that lymphoid organs do not represent a hidden viral reservoir (8). This ratio ranges from 2 to 4 in SIVsmFr-infected mangabeys, which is closer to that seen in HIV infection and which indicates a preferential infection of lymphoid organs (49, 58, 59). The only reported case of high viral load in SIVagm infection was after experimental inoculation of the SIVagm90 strain in pig-tailed macaques (31). Since the infected macaques developed an AIDS-like syndrome while African green monkeys inoculated with the same strain remained healthy, the data were compatible with a strong association between viral load and pathogenicity. Studies of SIVagm genetic variability revealed that the rate of mutation fixation was high, ranging from 10−2 to 10−3 subst/site/yr in env (4, 19, 50). The conjunction of a low viral load and of a highly variable virus in African green monkeys led to the hypothesis that SIVagm replication was equivalent to that seen in pathogenic lentivirus infections but that the containment of viral replication was more efficient (50). However, the data obtained in the SIVsm-mangabey model indicate that the containment hypothesis is not applicable to every natural SIV infection and that a high viral load is not necessarily associated with disease progression.
The reasons that SIVsm-infected mangabeys do not develop disease remain to be elucidated. It is possible that SIVsm replicates in mangabey cells without inducing major cytopathic effects, although this property has yet to be clearly demonstrated in vitro. In some studies, SIVsm was reported to be less cytopathic for mangabey cells than for macaque cells, while in others the difference was not apparent (21, 74). Another possibility is that the host response to SIV has benign or deleterious effects, depending on the species. The observation that PBMC of SIVsm-infected macaques show an increased susceptibility to apoptosis upon anti-CD3 stimulation, which is not seen with PBMC of SIVsm-infected mangabeys, suggests that immune activation, reflected by apoptosis, is more pronounced in monkeys susceptible to disease (71). This idea is supported by the histology of lymph nodes, which is normal for mangabeys (Fig. 8) and hyperplasic for macaques in the asymptomatic stage of infection (6, 13, 64, 66). A chronic lymphocyte activation, sustained by an abnormal stimulation of the immune response and possibly by a direct action of the viral protein Nef (2, 7, 18), may ultimately impair the renewal capacity of the immune system in susceptible species.
The extent of SIVsmFr genetic variability implies not only that the virus replicates continuously in mangabeys, but also that clearance of virions and of infected cells is relatively efficient. As reasoned for SIVmac infection (62), a clearance mechanism is needed to explain why the rapid rate of SIVsmFr replication does not lead to the exponential accumulation of virions to astronomical numbers. The most likely explanation is that virions and productively infected cells are continuously destroyed by immunological mechanisms. The detection of anti-SIV antibodies and of proliferative responses in SIVsm-infected mangabeys (1, 24) indicates that both the humoral and the cellular arms of the immune response are activated and that natural SIVsm infection does not induce tolerance in the strict sense of the word. Interestingly, however, the intensity of the response seems somewhat lower in mangabeys than that in macaques, as indicated by a generally low anti-SIV antibody titer in the former species (1, 24). The reactivity to Gag antigens is particularly weak, since of the three SIVsmFr-seropositive animals, only one had a serum that detected a p27 Gag band by Western blotting (Fig. 2A). Limited antibody titers may explain why viral particle-antibody complexes are not detected in the germinal centers of lymph nodes in the course of natural SIV infection in mangabeys as well as in African green monkeys (this study and reference 8). Regarding the cellular response, CD8+ lymphocytes from macaques as well as mangabeys and African green monkeys have been found to inhibit SIV replication in vitro through the action of soluble factors (53, 63, 73). However, in our hands, the depletion of CD8+ lymphocytes did not facilitate the recovery of SIVsmFr from mangabey cells (Fig. 2), which suggests that the CD8+ cell-mediated antiviral effect is limited in some cases of SIVsm infection and that it depends on the experimental system. It has long been recognized that SIV is more easily isolated from mangabeys than from macaques in the asymptomatic stage of infection (24, 63), which furthers the idea of a relatively weak CD8+ cell-mediated response in mangabeys. Thus, SIVsm infection seems characterized by a delicate balance between a high viral replication and a moderate immune response, without deleterious effects on the renewal capacity of the immune system or escalation of the viral load.
In conclusion, this study provides evidence that SIVsm replicates actively and establishes a high viral load in its natural host. Since infected mangabeys do not progress to disease, these data indicate that the viral load is not the sole determinant of SIV pathogenic potential.
ACKNOWLEDGMENTS
We thank Alain Blanchard for critical reading of the manuscript and Gérard Masson for help with the analysis of synonymous and nonsynonymous mutations.
This work was supported by grants from the Pasteur Institute and the ANRS (Agence Nationale de la Recherche sur le SIDA).
REFERENCES
- 1.Ahmed-Ansari A, Powell J D, Jensen P E, Yehuda-Cohen T, McClure H M, Anderson D, Fultz P N, Sell K W. Requirement for simian immunodeficiency virus antigen-specific in vitro proliferation of T-cells from infected rhesus macaques and sooty mangabeys. AIDS. 1990;4:399–407. doi: 10.1097/00002030-199005000-00004. [DOI] [PubMed] [Google Scholar]
- 2.Alexander L, Du Z, Rosenzweig M, Jung J U, Desrosiers R C. A role for natural simian immunodeficiency virus and human immunodeficiency virus type 1 Nef alleles in lymphocyte activation. J Virol. 1997;71:6094–6099. doi: 10.1128/jvi.71.8.6094-6099.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Allan J S, Short M, Taylor M E, Su S, Hirsh V M, Johnson P R, Shaw G M, Hahn B H. Species-specific diversity among simian immunodeficiency viruses from African green monkeys. J Virol. 1991;65:2816–2828. doi: 10.1128/jvi.65.6.2816-2828.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baier M, Dittmar M T, Cichutek K, Kurth R. Development of in vivo genetic variability of simian immunodeficiency virus. Proc Natl Acad Sci USA. 1991;88:8126–8130. doi: 10.1073/pnas.88.18.8126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Balfe P, Simmonds P, Ludlam C A, Bishop J O, Brown A J. Concurrent evolution of human immunodeficiency virus type 1 in patients infected from the same source: rate of sequence change and low frequency of inactivating mutations. J Virol. 1990;64:6221–6233. doi: 10.1128/jvi.64.12.6221-6233.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Baskin G B, Martin L N, Murphey-Corb M, Hu F-S, Kuebler D, Davison B. Distribution of SIV in lymph nodes of serially sacrificed rhesus monkeys. AIDS Res Hum Retroviruses. 1995;11:273–285. doi: 10.1089/aid.1995.11.273. [DOI] [PubMed] [Google Scholar]
- 7.Baur A S, Sawai E T, Dazin P, Fantl W J, Cheng-Mayer C, Peterlin B M. HIV-1 Nef leads to inhibition or activation of T cells depending on its intracellular localization. Immunity. 1994;1:373–384. doi: 10.1016/1074-7613(94)90068-x. [DOI] [PubMed] [Google Scholar]
- 8.Beer B, Scherer J, Zur Megede J, Norley S, Baier M, Kurth R. Lack of dichotomy between virus load of peripheral blood and lymph nodes during long-term simian immunodeficiency virus infection in African green monkeys. Virology. 1996;219:367–375. doi: 10.1006/viro.1996.0262. [DOI] [PubMed] [Google Scholar]
- 9.Burns D P, Desrosiers R C. Selection of genetic variants of simian immunodeficiency virus in persistently infected rhesus monkeys. J Virol. 1991;65:1843–1854. doi: 10.1128/jvi.65.4.1843-1854.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Campbell B J, Hirsch V M. Extensive envelope heterogeneity of simian immunodeficiency virus in tissues from infected macaques. J Virol. 1994;68:3129–3137. doi: 10.1128/jvi.68.5.3129-3137.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chakrabarti L, Cumont M C, Montagnier L, Hurtrel B. Variable course of primary simian immunodeficiency virus infection in lymph nodes: relation to disease progression. J Virol. 1994;68:6634–6642. doi: 10.1128/jvi.68.10.6634-6643.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chakrabarti L, Guyader M, Alizon M, Daniel M D, Desrosiers R C, Tiollais P, Sonigo P. Sequence of simian immunodeficiency virus from macaque and its relationship to other human and simian retroviruses. Nature (London) 1987;328:543–547. doi: 10.1038/328543a0. [DOI] [PubMed] [Google Scholar]
- 13.Chakrabarti L, Isola P, Cumont M C, Claessens-Maire M A, Hurtrel M, Montagnier L, Hurtrel B. Early stages of SIV infection in lymph nodes: evidence for high viral load and successive populations of target cells. Am J Pathol. 1994;144:1226–1237. [PMC free article] [PubMed] [Google Scholar]
- 14.Chen Z, Luckay A, Sodora D L, Telfer P, Reed P, Gettie A, Kanu J, Sadek R F, Yee J, Ho D D, Zhang L, Marx P A. Human immunodeficiency virus type 2 (HIV-2) seroprevalence and characterization of a distinct HIV-2 genetic subtype from the natural range of simian immunodeficiency virus infected-sooty mangabeys. J Virol. 1997;71:3953–3960. doi: 10.1128/jvi.71.5.3953-3960.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen Z, Telfier P, Gettie A, Reed P, Zhang L, Ho D D, Marx P A. Genetic characterization of new West African simian immunodeficiency virus SIVsm: geographic clustering of household-derived SIV strains with human immunodeficiency virus type 2 subtypes and genetically diverse viruses from a single feral sooty mangabey troop. J Virol. 1996;70:3617–3627. doi: 10.1128/jvi.70.6.3617-3627.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chun T W, Carruth L, Finzi D, Shen X F, Digiuseppe J A, Taylor H, Hermankova M, Chadwick K, Margolick J, Quinn T C, Kuo Y H, Brookmeyer R, Zeiger M A, Barditchcrovo P, Siliciano R F. Quantification of latent tissue reservoirs and total body viral load in hiv-1 infection. Nature. 1997;387:183–188. doi: 10.1038/387183a0. [DOI] [PubMed] [Google Scholar]
- 17.Daniel M D, Letvin N L, King N W, Kannagi M, Sehgal P K, Hunt R D, Kanki P J, Essex M, Desrosiers R C. Isolation of a T-cell tropic HTLVIII-like retrovirus from macaques. Science. 1985;228:1201–1204. doi: 10.1126/science.3159089. [DOI] [PubMed] [Google Scholar]
- 18.Du Z, Lang S M, Sasseville V G, Lackner A A, Ilynskii P O, Daniel M D, Jung J U, Desrosiers R C. Identification of a nef allele that causes lymphocyte activation and acute disease in macaque monkeys. Cell. 1995;82:665–674. doi: 10.1016/0092-8674(95)90038-1. [DOI] [PubMed] [Google Scholar]
- 19.Fomsgaard A, Johnson P R, London W T, Hirsch V M. Genetic variation of the SIVagm transmembrane glycoprotein in naturally and experimentally infected primates. AIDS. 1993;7:1041–1047. doi: 10.1097/00002030-199308000-00003. [DOI] [PubMed] [Google Scholar]
- 20.Fukasawa M, Miura T, Hasegawa A, Morikawa S, Tsujimoto H, Miki K, Kitamura T, Hayami H. Sequence of simian immunodeficiency virus from African green monkey, a new member of the HIV/SIV group. Nature (London) 1988;333:457–461. doi: 10.1038/333457a0. [DOI] [PubMed] [Google Scholar]
- 21.Fultz P N. Replication of an acutely lethal simian immunodeficiency virus activates and induces proliferation of lymphocytes. J Virol. 1991;65:4902–4909. doi: 10.1128/jvi.65.9.4902-4909.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fultz P N, Gordon T P, Anderson D C, McClure H M. Prevalence of natural infection with simian immunodeficiency virus and simian T cell leukemia virus type 1 in a breeding colony of sooty mangabey monkeys. AIDS. 1990;4:619–625. doi: 10.1097/00002030-199007000-00002. [DOI] [PubMed] [Google Scholar]
- 23.Fultz P N, McClure H M, Anderson D C, Swenson R B, Anand R, Srinivasan A. Isolation of a T-lymphotropic retrovirus from naturally infected sooty mangabey monkeys (Cercocebus atys) Proc Natl Acad Sci USA. 1986;83:5286–5290. doi: 10.1073/pnas.83.14.5286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fultz, P. N., R. B. Stricker, H. M. McClure, D. C. Anderson, W. M. Switzer, and C. Horaist. 1990. Humoral response to SIV/SMM infection in macaque and mangabey monkeys. J. Acquired Immune Defic. Syndr. [PubMed]
- 25.Gao F, Yue L, Robertson D L, Hill S C, Hui H, Biggar R J, Neequaye A E, Whelan T M, Ho D D, Shaw G M, Sharp P M, Hahn B H. Genetic diversity of human immunodeficiency virus type 2: evidence for distinct sequence subtypes with differences in virus biology. J Virol. 1994;68:7433–7447. doi: 10.1128/jvi.68.11.7433-7447.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gao F, Yue L, White A T, Pappas P G, Barchue J, Hanson A P, Greene B M, Sharp P M, Shaw G M, Hahn B H. Human infection by genetically diverse SIVSM-related HIV-2 in West Africa. Nature. 1992;358:495–499. doi: 10.1038/358495a0. [DOI] [PubMed] [Google Scholar]
- 27.Gardner M B. The history of simian AIDS. J Med Primatol. 1996;25:148–157. doi: 10.1111/j.1600-0684.1996.tb00011.x. [DOI] [PubMed] [Google Scholar]
- 28.Grooves C P. Phylogenetics and population systematics of mangabeys (Primates: Cercopithecoidea) Primates. 1978;19:1–34. [Google Scholar]
- 29.Hartung S, Boller K, Cichutek K, Norley S G, Kurth R. Quantitation of a lentivirus in its natural host: simian immunodeficiency virus in African green monkeys. J Virol. 1992;66:2143–2149. doi: 10.1128/jvi.66.4.2143-2149.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Higgins D G, Thompson J D, Gibson T J. Using CLUSTAL for multiple sequence alignments. Methods Enzymol. 1996;266:383–402. doi: 10.1016/s0076-6879(96)66024-8. [DOI] [PubMed] [Google Scholar]
- 31.Hirsch V M, Dapolito G, Johnson P R, Elkins W R, London W T, Montali R J, Goldstein S, Brown C. Induction of AIDS by simian immunodeficiency virus from an African green monkey: species-specific variation in pathogenicity correlates with the extent of in vivo replication. J Virol. 1995;69:955–967. doi: 10.1128/jvi.69.2.955-967.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hirsch V M, Fuerst T R, Sutter G, Carroll M W, Yang L C, Goldstein S, Piatak M, Jr, Elkins W R, Alvord W G, Montefiori D C, Moss B, Lifson J D. Patterns of viral replication correlate with outcome in simian immunodeficiency virus (SIV)-infected macaques: effect of prior immunization with a trivalent SIV vaccine in modified vaccinia virus Ankara. J Virol. 1996;70:3741–3752. doi: 10.1128/jvi.70.6.3741-3752.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hirsch V M, Johnson P R. Pathogenic diversity of simian immunodeficiency viruses. Virus Res. 1994;32:183–203. doi: 10.1016/0168-1702(94)90041-8. [DOI] [PubMed] [Google Scholar]
- 34.Hirsch V M, Olmsted R A, Murphey-Corb M, Purcell R H, Johnson P R. An African primate lentivirus (SIVsm) closely related to HIV-2. Nature (London) 1989;339:389–391. doi: 10.1038/339389a0. [DOI] [PubMed] [Google Scholar]
- 35.Huet T, Cheynier R, Meyerhans A, Roelants G, Wain-Hobson S. Genetic organization of a chimpanzee lentivirus related to HIV-1. Nature. 1990;345:356–359. doi: 10.1038/345356a0. [DOI] [PubMed] [Google Scholar]
- 36.Johnson P R, Hamm T E, Goldstein S, Kitov S, Hirsch V M. The genetic fate of molecularly cloned simian immunodeficiency virus in experimentally infected macaques. Virology. 1991;185:217–228. doi: 10.1016/0042-6822(91)90769-8. [DOI] [PubMed] [Google Scholar]
- 37.Jolly C J, Phillips-Conroy J E, Turner T R, Broussard S, Allan J S. SIVagm incidence over two decades in a natural population of Ethiopian grivet monkeys (Cercopithecus aethiops aethiops) J Med Primatol. 1996;25:78–83. doi: 10.1111/j.1600-0684.1996.tb00198.x. [DOI] [PubMed] [Google Scholar]
- 38.Kanki P J, Kurth R, Becker W, Dreesman G, McLane M F, Essex M. Antibodies to simian T-lymphotropic retrovirus type III in African green monkeys and recognition of STLV-III viral proteins by AIDS and related sera. Lancet. 1985;i:1330–1332. doi: 10.1016/s0140-6736(85)92818-1. [DOI] [PubMed] [Google Scholar]
- 39.Kasper P, Simmonds P, Schneweis K E, Kaiser R, Matz B, Oldenburg J, Brackmann H H, Holmes E C. The genetic diversification of the HIV type 1 gag p17 gene in patients infected from a common source. Aids Res Hum Retroviruses. 1995;11:1197–1201. doi: 10.1089/aid.1995.11.1197. [DOI] [PubMed] [Google Scholar]
- 40.Kestler H W, III, Ringler D J, Mori K, Panicalli D, Sehgal P, Daniel M D, Desrosiers R C. Importance of the nef gene for maintenance of high virus loads and for development of AIDS. Cell. 1991;65:651–662. doi: 10.1016/0092-8674(91)90097-i. [DOI] [PubMed] [Google Scholar]
- 41.Kodama T, Mori K, Kawahara T, Ringler D J, Desrosiers R C. Analysis of simian immunodeficiency virus sequence variation in tissues of rhesus macaques with simian AIDS. J Virol. 1993;67:6522–6534. doi: 10.1128/jvi.67.11.6522-6534.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Li W H, Wu C I, Luo C C. A new method for estimating synonymous and nonsynonymous rates of nucleotide substitution considering the relative likelihood of nucleotide and codon changes. Mol Biol Evol. 1985;2:150–174. doi: 10.1093/oxfordjournals.molbev.a040343. [DOI] [PubMed] [Google Scholar]
- 43.Lowenstine N L, Pedersen N C, Higgins J, Pallis K C, Uyeda A, Marx P, Lerche N W, Munn R J, Gardner M B. Seroepidemiologic survey of captive Old World primates for antibodies to human and simian retroviruses, and isolation of a lentivirus from sooty mangabeys (cercocebus atys) Int J Cancer. 1986;38:563–575. doi: 10.1002/ijc.2910380417. [DOI] [PubMed] [Google Scholar]
- 44.Lukashov V V, Kuiken C L, Goudsmit J. Intrahost human immunodeficiency virus type 1 evolution is related to the length of the immunocompetent period. J Virol. 1995;69:6911–6916. doi: 10.1128/jvi.69.11.6911-6916.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mansky L, Temin H M. Lower in vivo mutation rate of human immunodeficiency virus type 1 than that predicted from the fidelity of purified reverse transcriptase. J Virol. 1995;69:5087–5094. doi: 10.1128/jvi.69.8.5087-5094.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Marthas M L, van Rompay K K, Otsyula M, Miller C J, Canfield D R, Pedersen N C, McChesney M B. Viral factors determine progression to AIDS in simian immunodeficiency virus-infected newborn rhesus macaques. J Virol. 1995;69:4198–4205. doi: 10.1128/jvi.69.7.4198-4205.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Marx P A, Li Y, Lerche N W, Sutjipto S, Gettie A, Yee J A, Brotman B H, Prince A M, Hanson A, Webster R G, Desrosiers R C. Isolation of simian immunodeficiency virus related to human immunodeficiency virus type 2 from a West African pet sooty mangabey. J Virol. 1991;65:4480–4485. doi: 10.1128/jvi.65.8.4480-4485.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mellors J W, Rinaldo C, Jr, Gupta P, White R M, Todd J A, Kingsley L A. Prognosis in HIV-1 infection predicted by the quantity of virus in plasma. Science. 1996;272:1167–1170. doi: 10.1126/science.272.5265.1167. [DOI] [PubMed] [Google Scholar]
- 49.Meylan P R, Bürgisser P, Weyrich-Suter C, Spertini F. Viral load and immunophenotype of cells obtained from lymph nodes by fine needle aspiration as compared with peripheral blood cells in HIV-infected patients. J Acquired Immune Defic Syndr. 1996;13:39–47. doi: 10.1097/00042560-199609000-00007. [DOI] [PubMed] [Google Scholar]
- 50.Müller-Trutwin M C, Corbet S, Dias Tavares M, Herve V M A, Nerrienet E, Georges-Courbot M C, Saurin W, Sonigo P, Barré-Sinoussi F. The evolutionary rate of nonpathogenic simian immunodeficiency virus (SIVagm) is in agreement with a rapid and continuous replication in vivo. Virology. 1996;223:89–102. doi: 10.1006/viro.1996.0458. [DOI] [PubMed] [Google Scholar]
- 51.Murphey-Corb M, Martin L N, Rangan S R S, Baskin G B, Gormus B J, Wolf R H, Andes W A, West M, Montelaro R C. Isolation of an HTLV-III related retrovirus from macaques with simian AIDS and its possible origin in asymptomatic mangabeys. Nature (London) 1986;321:435–437. doi: 10.1038/321435a0. [DOI] [PubMed] [Google Scholar]
- 52.Myers G, Korber B, Foley B, Jeang K T, Mellors J W, Wain-Hobson S. Human retroviruses and AIDS 1996: a compilation and analysis of nucleic acid and amino acid sequences. Los Alamos, N. Mex: Theoretical Biology and Biophysics Group T-10; 1996. [Google Scholar]
- 53.Norley S G. SIVagm infection of its natural African green monkey host. Immunol Lett. 1996;51:53–58. doi: 10.1016/0165-2478(96)02555-2. [DOI] [PubMed] [Google Scholar]
- 54.Norley S G, Kraus G, Ennen J, Bonilla J, König H, Kurth R. Immunological studies of the basis of apathogenicity of simian immunodeficiency virus from African green monkey. Proc Natl Acad Sci USA. 1990;87:9067–9071. doi: 10.1073/pnas.87.22.9067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ohta Y, Masuda T, Tsujimoto H, Ishikawa K, Kodama T, Morikawa S, Nakai M, Honjo S, Hayami M. Isolation of simian immunodeficiency virus from African green monkeys and seroepidemiologic survey of the virus in various non-human primates. Int J Cancer. 1988;41:115–122. doi: 10.1002/ijc.2910410121. [DOI] [PubMed] [Google Scholar]
- 56.Olsen G J, Matsuda H, Hagstrom R, Overbeek R. fastDNAml: a tool for construction of phylogenetic trees of DNA sequences using maximum likelihood. Comput Appl Biosci. 1994;10:41–48. doi: 10.1093/bioinformatics/10.1.41. [DOI] [PubMed] [Google Scholar]
- 57.Overbaugh J, Rudensey L M, Papenhausen M D, Benveniste R E, Morton W R. Variation in simian immunodeficiency virus env is confined to V1 and V4 during progression to simian AIDS. J Virol. 1991;65:7025–7031. doi: 10.1128/jvi.65.12.7025-7031.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Pantaleo G, Graziosi C, Demarest J F, Butini L, Montroni M, Fox C H, Orenstein J M, Kotler D P, Fauci A S. HIV infection is active and progressive in lymphoid tissue during the clinically latent stage of disease. Nature (London) 1993;362:355–358. doi: 10.1038/362355a0. [DOI] [PubMed] [Google Scholar]
- 59.Pantaleo G, Menzo S, Vaccarezza M, Graziosi C, Cohen O J, Demarest J F, Montefiori D, Orenstein J M, Fox C, Schrager L K, et al. Studies in subjects with long-term nonprogressive human immunodeficiency virus infection. N Engl J Med. 1995;332:209–216. doi: 10.1056/NEJM199501263320402. [DOI] [PubMed] [Google Scholar]
- 60.Peeters M, Honoré C, Huet T, Bedjabaga L, Ossari S, Bussi P, Cooper R W, Delaporte E. Isolation and partial characterization of an HIV-related virus occurring naturally in chimpanzees in Gabon. AIDS. 1989;3:625–630. doi: 10.1097/00002030-198910000-00001. [DOI] [PubMed] [Google Scholar]
- 61.Peeters M, Janssens W, Fransen K, Brandful J, Heyndrickx L, Koffi K, Delaporte E, Piot P, Gershy-Damet G M, van der Groen G. Isolation of simian immunodeficiency viruses from two sooty mangabeys in Cote d’Ivoire: virological and genetic characterization and relationship to other HIV type 2 and SIVsm/mac strains. AIDS Res Hum Retroviruses. 1994;10:1289–1294. doi: 10.1089/aid.1994.10.1289. [DOI] [PubMed] [Google Scholar]
- 62.Pelletier E, Saurin W, Cheynier R, Letvin N L, Wain-Hobson S. The tempo and mode of SIV quasispecies development in vivo calls for massive viral replication and clearance. Virology. 1995;208:644–652. doi: 10.1006/viro.1995.1195. [DOI] [PubMed] [Google Scholar]
- 63.Powell J D, Yehuda-Cohen T, Villinger F, McClure H M, Sell K W, Ahmed-Ansari A. Inhibition of SIV/SMM replication in vitro by CD8+ cells from SIV/SMM infected seropositive clinically asymptomatic sooty mangabeys. J Med Primatol. 1990;19:239–249. [PubMed] [Google Scholar]
- 64.Reimann K A, Snyder G B, Chalifoux L V, Waite B C, Miller M D, Yamamoto H, Spertini O, Letvin N L. An activated CD8+ lymphocyte appears in lymph nodes of rhesus monkeys early after infection with simian immunodeficiency virus. J Clin Invest. 1991;88:1113–1120. doi: 10.1172/JCI115410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Rey M A, Laurent A G, McLure J, Krust B, Montagnier M, Hovanessian A G. Transmembrane envelope glycoproteins of human immunodeficiency virus type 2 and simian immunodeficiency virus SIVmac exist as homodimers. J Virol. 1990;64:922–926. doi: 10.1128/jvi.64.2.922-926.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ringler D J, Wyand M S, Walsh D G, Mackey J J, Chalifoux L V, Popovic M, Minassian A A, Sehgal P K, Daniel M D, Desrosiers R C, King N W. Cellular localization of simian immunodeficiency virus in lymphoid tissues. Am J Pathol. 1989;134:373–383. [PMC free article] [PubMed] [Google Scholar]
- 67.Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 68.Singer R H, Lawrence J B, Rashtchian R N. Toward a rapid and sensitive hybridization methodology using isotopic and non isotopic probes. In: Valentino K L, Eberwine J H, Barchas J D, editors. In situ hybridization: application to neurobiology. New York, N.Y: Oxford University Press; 1987. pp. 71–96. [Google Scholar]
- 69.Strimmer K, Von Haeseler A. Quartet puzzling—a quartet maximum-likelihood method for reconstructing tree topologies. Mol Biol Evol. 1996;13:964–969. [Google Scholar]
- 70.Traincard F, Rey-Cuille M A, Huon I, Dartevelle S, Mazie J C, Benichou S. Characterization of monoclonal antibodies to human immunodeficiency virus type 2 envelope glycoproteins. AIDS Res Hum Retroviruses. 1994;10:1659–1667. doi: 10.1089/aid.1994.10.1659. [DOI] [PubMed] [Google Scholar]
- 71.Villinger F, Folks T M, Lauro S, Powell J D, Sundstrom J B, Mayne A, Ansari A A. Immunological and virological studies of natural SIV infection of disease-resistant non-human primates. Immunol Lett. 1996;51:59–68. doi: 10.1016/0165-2478(96)02556-4. [DOI] [PubMed] [Google Scholar]
- 72.Watson A, Ranchalis J, Travis B, McClure J, Sutton W, Johnson P R, Hu S L, Haigwood N L. Plasma viremia in macaques infected with simian immunodeficiency virus: plasma viral load early in infection predicts survival. J Virol. 1997;71:284–290. doi: 10.1128/jvi.71.1.284-290.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Yasutomi Y, Reimann K A, Lord C I, Miller M D, Letvin N L. Simian immunodeficiency virus-specific CD8+ lymphocyte response in acutely infected rhesus monkeys. J Virol. 1993;67:1707–1711. doi: 10.1128/jvi.67.3.1707-1711.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Yehuda-Cohen T, Powell J D, Villinger F, McClure H M, Sell K W, Ansari A A. Comparison of SIV/SMM replication in CD4+ T cell and monocyte/macrophage cultures from Rhesus macaques and sooty mangabeys. J Med Primatol. 1990;19:251–267. [PubMed] [Google Scholar]
- 75.Zhang L, Diaz R S, Ho D D, Mosley J W, Busch M P, Mayer A. Host-specific driving force in human immunodeficiency virus type 1 evolution in vivo. J Virol. 1997;71:2555–2561. doi: 10.1128/jvi.71.3.2555-2561.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]