Abstract
Background
Brain metastases (BMs) are common in small cell lung cancer (SCLC), and the efficacy of immune checkpoint inhibitors (ICIs) in these patients is uncertain. In this study we aimed to develop and validate a radiomics nomogram based on magnetic resonance imaging (MRI) for intracranial efficacy prediction of ICIs in patients with BMs from SCLC.
Methods
The training and validation cohorts consisted of 101 patients from two centers. The interclass correlation coefficient (ICC), logistic univariate regression analysis, and random forest were applied to select the radiomic features, generating the radiomics score (Rad‐score) through the formula. Using multivariable logistic regression analysis, a nomogram was created by the combined model. The discrimination, calibration, and clinical utility were used to assess the performance of the nomogram. Kaplan–Meier curves were plotted based on the nomogram scores.
Results
Ten radiomic features were selected for calculating the Rad‐score as they could differentiate the intracranial efficacy in the training (area under the curve [AUC], 0.759) and the validation cohort (AUC, 0.667). A nomogram was created by combining Rad‐score, treatment lines, and neutrophil‐to‐lymphocyte ratio (NLR). The training cohort obtained an AUC of 0.878 for the combined model, verified in the validation cohort (AUC = 0.875). Kaplan–Meier analyses showed the nomogram was associated with progression‐free survival (PFS) (p = 0.0152) and intracranial progression‐free survival (iPFS) (p = 0.0052) but not overall survival (OS) (p = 0.4894).
Conclusion
A radiomics nomogram model for predicting the intracranial efficacy of ICIs in SCLC patients with BMs can provide suggestions for exploring individual‐based treatments for patients.
Keywords: immune checkpoint inhibitors, nomogram, prediction, Radiomics, small cell lung cancer
We developed and validated a radiomics nomogram prediction model for predicting the intracranial efficacy of immune checkpoint inhibitors (ICIs) in small cell lung cancer (SCLC) patients with brain metastases (BMs). This study can help tailor their treatment schedules, improve clinical decision‐making, and offer extensive potential applications.
INTRODUCTION
Lung cancer ranks second among common cancers to be diagnosed and is still the leading reason of cancer death. 1 Approximately 15% of all lung cancer cases worldwide are small cell lung cancer (SCLC) and are characterized by an inferior prognosis. The 5‐year overall survival (OS) rate for SCLC is just 6.7%, primarily due to its propensity for early metastasis during tumor development. 2 Furthermore, it is worth noting that around 10% of patients diagnosed with SCLC have brain metastases (BMs) at the time of their initial diagnosis, and an additional 40% to 60% will eventually develop BMs as the disease progresses. Despite extensive research, the prognosis remains unfavorable. 3
The combination of immune checkpoint inhibitors (ICIs) and platinum‐based chemotherapy has been widely used in extensive‐stage small cell lung cancer (ES‐SCLC) as the first‐line treatment. 4 IMpower 133, a phase 3 trial, demonstrated that the combination of atezolizumab and carboplatin plus etoposide significantly prolonged both progression‐free survival (PFS) and OS in treatment‐naive ES‐SCLC, but for patients with BMs, no significant distinction between the two groups was observed in OS or PFS. 5 , 6 In another phase 3 trial known as CASPIAN research, the updated results showed that compared with the platinum‐etoposide (EP) group, the durvalumab plus EP group continued to show OS benefits (hazard ratio [HR], 0.75; 95% confidence interval [CI]: 0.62–0.91; p = 0.0032). However, the median PFS was similar for all groups. For patients with metastases in the brain, the two groups had no statistical difference in OS (HR, 0.79; 95% CI: 0.44–1.41). 7 , 8 The efficacy of ICIs for SCLC patients is still uncertain, especially for patients with BMs. 9
Programmed death‐ligand 1 (PD‐L1) has been identified as the most reliable indicator of immunotherapy response. 10 Tumor mutational burden (TMB), which reflects the potential for producing new tumor antigens in tumors, may also be a valuable biomarker for cancer immunotherapy benefits in recent years. 11 However, the predictive capacity of these indicators has not been observed in SCLC. Nevertheless, a considerable proportion of patients may not have enough high‐quality tissue samples to conduct biomarker analysis. The above issue arises due to the rare adoption of surgical resection as a therapeutic measure in SCLC, resulting in a dependence on diagnostic biopsy specimens, which are frequently tiny and necrotic. Invasive sampling is not available in BMs, therefore, the study using tissue samples for analysis is limited, emphasizing the necessity of developing a noninvasive predictor for cancer immunotherapy.
Magnetic resonance imaging (MRI), as a powerful noninvasive diagnostic imaging tool for BMs, holds the potential possibility to predict the effectiveness of ICIs. 12 Presently, the best way to diagnose BMs is gadolinium‐enhanced MRI, which is more sensitive than enhanced computed tomography (CT). As a secondary tumor, BMs have the characteristics of latent onset, rapid progression, and poor prognosis, so their accurate diagnosis, precise location, and early treatment effectiveness evaluation are essential for prognosis. 13 However, the subjective assessments of radiologists are susceptible to interobserver heterogeneity and typically exhibit limited robustness and precision. Radiomics, an emerging technology, has the potential to address this issue using quantitative analysis of medical images. Radiomics is a process that transforms encrypted medical images into interpretable data through the extraction of high‐throughput imaging features. This data is then correlated with specific clinical outcomes and analyzed to guide clinical decision‐making. 14
Radiomics has been applied to identify the pathological types of metastatic tumors as well as the prediction of therapeutic efficacy and prognosis in recent years. 15 , 16 , 17 , 18 , 19 However, there is a lack of related exploration in predicting the intracranial efficacy of SCLC patients treated with ICIs and diagnosed with BMs. Consequently, we conducted this study to identify the related radiomics features and clinical factors and construct a radiomics nomogram that could predict the intracranial efficacy of these patients.
METHODS
Patients and study design
The retrospective analysis obtained ethical approval and exempted the informed consent requirement. We identified 101 SCLC patients with BMs who had been treated with ICIs from June 2019 to June 2022, which included a training cohort (n = 70) from Shandong First Medical University Affiliated Provincial Tumor Hospital and an independent external validation cohort (n = 31) from Qilu Hospital of Shandong University. The Supporting Information 1: Figure S1 presents the recruitment pathway for patients in this study. The following were the inclusion criteria: (1) patients with a confirmed diagnosis of SCLC through biopsy, (2) patients who were diagnosed with BMs and had received therapy with ICIs and (3) patients who received an enhanced MRI of the brain performed within 1 month before ICI treatment. The following were exclusion criteria: (a) patients who received immunotherapy less than twice; (b) patients for whom imaging data or medical records necessary for evaluating intracranial efficacy were not available; (c) patients who did not have a qualified MRI before ICIs treatment and (d) patients with BMs having a diameter <5 mm. The independent validation cohort was selected based on the identical criteria used for the training cohort.
The intracranial efficacy was assessed using the Response Assessment in Neuro‐Oncology Brain Metastases (RANO‐BM) criteria, 20 with two neuroradiologists independently classifying the response as good or poor. BMs with any of the following were defined as having a good response: (1) partial response, (2) complete response, and (3) maintaining a stable disease state for a duration of either 3 months or more than 3 months. BMs with any of the following were defined as having a poor response: (1) progressive disease and (2) maintaining a stable disease state for less than 3 months. Baseline demographic and clinical data, such as Eastern Cooperative Oncology Group (ECOG) score, age, gender, smoking history, stage, number of BMs, and blood indicators, were derived from medical reports, and dates of baseline MRI were also recorded. In our study, treatment lines represent which line of treatment ICIs were applied in SCLC, and most ICIs were combined with chemotherapy or angiogenic inhibitors treatments. OS is defined as the time from the date of baseline MRI until death due to any cause. PFS or iPFS is defined as the time from the date of baseline MRI to the progression of systemic or intracranial tumors or death from any cause (whichever occurs first). PFS, iPFS, and OS were determined by calculating the time between the date of the baseline MRI and the last follow‐up if there was no evidence of progression or mortality at the last follow‐up.
The radiomics nomogram workflow applied in this study is presented in Figure 1. The study design was divided into four sections: region of interest (ROI) segmentation, feature extraction and selection, and model construction.
FIGURE 1.
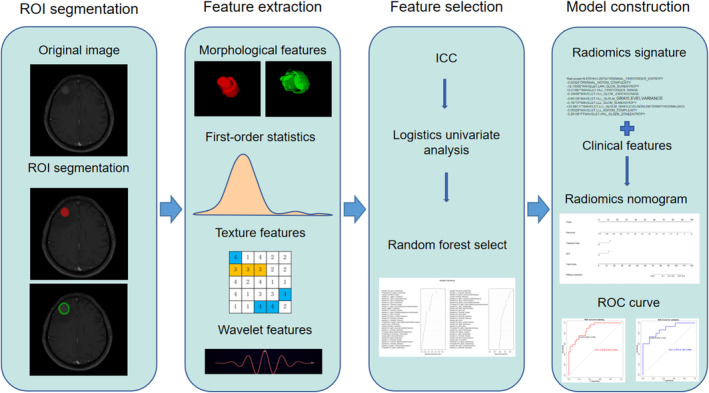
Radiomics nomogram workflow. The radiomics nomogram process included four parts: region of interest (ROI) segmentation, feature extraction, feature selection, and model construction. ROI was delineated on both the tumor and the peritumoral areas on contrast‐enhanced T1 weighted images. Radiomics features were extracted, including nontextural, textural, and wavelet features. We performed interclass correlation coefficient (ICC), logistics univariate analysis, and random forest for feature selection. The radiomics signature was combined with clinical features to build the radiomics nomogram based on multivariable logistic analysis. Further performance evaluation was explored, including ROC curves, calibration curves, decision curves, and prognostic value analysis.
MRI acquisition
All included patients were imaged on the 3.0 T scanner system (Magnetom Verio, Siemens) with an eight‐channel phased‐array surface coil. An intravenous dose of gadodiamide (0.1 mmol/kg, Omniscan, GE Healthcare) was administered before the MRI using a power injector (rate of 2.5 mL/s). Then, 20 mL saline was injected to flush the remaining agents (rate of 2.5 mL/s). The scanning range is covered from the calva to the lower neck in the supine position.
Tumor and peritumoral region segmentation
In order to guarantee the precision of ROI delineation, we conducted a manual ROI by a radiologist with 5 years of work experience (reader 1) on both tumor and peritumoral areas from contrast‐enhanced T1 weighted images (CE‐T1WI), drawing on the opening software ITK‐SNAP (http://www.radiantviewer.com). The tumor area was delineated on the slice with the maximum tumor area but not peritumoral edema, and the peritumor was defined as the 5 mm area around the tumor. A senior radiologist with 12 years (reader 2) of experience verified each ROI.
Feature extraction
All slices were resampled to a standard voxel size of 1 × 1 × 1 mm3. A total of 851 imaging features were finally extracted from CE‐T1WI on tumor and peritumoral areas, respectively. Consequently, a total of 1702 radiomic features were extracted for each lesion. The feature set was divided into four groups: (1) shape and size, (2) first‐order statistics, (3) texture and (4) wavelet features. Radiomics features extracted in this experiment are shown in the Supporting Information file 1: Table S1. All feature extractions were implemented using Python (https://www.python.org/downloads/). Texture features were achieved based on five textural matrixes: (1) The neighborhood gray‐tone difference matrix, (2) the gray level size zone matrix, (3) the gray level dependence matrix, (4) the gray level run‐length matrix, and (5) the gray level co‐occurrence matrix. The images with the wavelet transformation were used to extract wavelet features. The wavelet transform process for CE‐T1WI is given in the Additional file 1: S1.
Feature selection and radiomics signature model building
Interobserver reproducibility of radiomic features was analyzed with 30 randomly chosen patients by readers 1 and 2. The ICC was used to assess the consistency of feature extraction between observers. Only stable features with an ICC >0.75 were qualified. If a patient delineates multiple BMs, the radiomic features of multiple BMs are integrated based on a volume‐weighted formula (Additional file 1: S2). Based on the training cohort, logistic univariate regression analysis and random forest method are used for further feature selection. The Rad‐score for each patient was generated using a logit model, incorporating selected features weighted according to their corresponding coefficients. The AUC obtained from the receiver operating characteristic (ROC) analysis was applied to measure the discriminative ability of the Rad‐score in the training and validation cohorts. The statistical software packages R (http://www.R-project.org; The R Foundation) and EmpowerStats (http://www.empowerstats.com, X&Y Solutions, Inc., Boston, MA) were utilized to carry out the Rad‐score.
Clinical model building
A univariate logistic regression analysis was applied to identify useful clinical candidate factors in the training cohort. Then, the clinical candidate predictors were used to further select and develop a clinical predictive model for intracranial efficacy through multivariable logistic regression analysis. The AUC derived from the ROC analysis was used to measure the discriminative ability of the clinical model in both training and validation cohorts.
Combined model building and developing a radiomics nomogram
The multivariable logistic regression analysis based on the clinical predictors and Rad‐score was applied to develop a combined predictive model for intracranial efficacy in the training cohort. We constructed the radiomics nomogram using the combined model from the training cohort in order to provide the oncologist with a quantitative clinical tool to estimate the individual probability of intracranial effectiveness. The Hosmer–Lemeshow test verified the goodness‐of‐fit of the combined model.
The AUC derived from the ROC analysis was used to measure the discriminative ability of the radiomics nomogram. The radiomics nomogram's calibration was evaluated by plotting the calibration curve. The performance of the nomogram was tested in the independent validation cohort. Finally, the AUC values and calibration curves derived from the training and validation cohorts were plotted.
Demographic and clinical characteristic analysis
The disparities in demographic and clinical factors among all patient cohorts were assessed by Pearson's chi‐square or Fisher's exact tests.
Clinical utility of the radiomics nomogram
The clinical utility of the radiomics nomogram was assessed by analyzing the net benefits under various threshold probabilities in both the training and validation cohorts through decision curve analyses.
Prognostic value analysis
Kaplan–Meier curves were plotted based on the radiomics nomogram scores in order to stratify the OS, PFS, and iPFS in patients of SCLC with BMs treated with ICIs. The median of the nomogram scores were used to divide all patients into high‐ and low‐score groups. The log‐rank test was applied to assess if the survival curves of the two patient groups were significantly different.
Subgroup analyses
A total of 49 patients received treatment with chemotherapy combined with PD‐L1 inhibitors scheme. A total of 52 patients received treatment with chemotherapy combined with programmed death‐1 (PD‐1) inhibitors and 41 patients did not receive brain radiation therapy. We further analyzed the predictive ability of the established combined model in the above subgroups.
Statistical analysis
All statistical analyses were carried out using R software (version 4.2.2; http://www.Rproject.org) and SPSS software (version 26; IBM Corporation). The R packages used in this study are reported in the Supporting Information file 1: S3. A two‐sided p‐value <0.05 was identified for statistical significance.
RESULTS
Clinical characteristics of patients
The baseline characteristics of all patients included are shown in Table 1. We selected 101 patients according to the inclusion criteria: 79 males and 22 females; median age, 58 years; age range, 27 to 77 years. There were 76 patients with the number of BMs ≤3 and 25 patients with the number of BMs >3. A total of 80 patients were found to have extracranial metastasis, and 21 patients did not. Among all patients, 51 received ICIs as first‐line treatment, and 49 received concurrent brain radiotherapy. The training cohorts comprised 70 individuals, while the validation cohorts comprised 31. Additionally, there were no statistically significant differences between the training and validation cohorts concerning demographic or clinical features.
TABLE 1.
Characteristics of patients in the training and validation cohorts.
| Training cohort (n = 70) | Validation cohort (n = 31) | P (inter) | |||||
|---|---|---|---|---|---|---|---|
| Good response (n = 41) | Poor response (n = 29) | P (intra) | Good response (n = 16) | Poor response (n = 15) | P (intra) | ||
| Age, year | |||||||
| ≤60 | 23 (32.9) | 24 (34.3) | 0.019* | 14 (45.2) | 7 (22.6) | 0.023* | 0.953 |
| >60 | 18 (25.7) | 5 (7.1) | 2 (6.5) | 8 (25.8) | |||
| Gender | |||||||
| Male | 30 (42.9) | 24 (34.3) | 0.347 | 14 (45.2) | 11 (35.5) | 0.394 | 0.694 |
| Female | 11 (15.7) | 5 (7.1) | 2 (6.5) | 4 (12.9) | |||
| ECOG score | |||||||
| 0 | 22 (31.4) | 16 (22.9) | 0.900 | 6 (19.4) | 9 (29.0) | 0.289 | 0.584 |
| 1 + 2 + 3 | 19 (27.1) | 13 (18.6) | 10 (32.3) | 6 (19.4) | |||
| Smoking history | |||||||
| Yes | 21 (30.0) | 22 (31.4) | 0.037* | 12 (38.7) | 6 (19.4) | 0.073 | 0.750 |
| No | 20 (28.6) | 7 (10.0) | 4 (12.9) | 9 (29.0) | |||
| T stage | |||||||
| 0 + 1 + 2 | 18 (25.7) | 12 (17.1) | 0.834 | 7 (22.6) | 7 (22.6) | 1.000 | 0.829 |
| 3 + 4 | 23 (32.9) | 17 (24.3) | 9 (29.0) | 8 (25.8) | |||
| N stage | |||||||
| 0 + 1 + 2 | 24 (34.3) | 15 (21.4) | 0.572 | 8 (25.8) | 8 (25.8) | 1.000 | 0.703 |
| 3 | 17 (24.3) | 14 (20.0) | 8 (25.8) | 7 (22.6) | |||
| Number of BM | |||||||
| ≤3 | 32 (45.7) | 19 (27.1) | 0.245 | 14 (45.2) | 11 (35.5) | 0.394 | 0.403 |
| >3 | 9 (12.9) | 10 (14.3) | 2 (6.5) | 4 (12.9) | |||
| Extracranial metastasis | |||||||
| Yes | 31 (44.3) | 26 (37.1) | 0.137 | 13 (41.9) | 10 (32.3) | 0.433 | 0.409 |
| No | 10 (14.3) | 3 (4.3) | 3 (9.7) | 5 (16.1) | |||
| LDH | |||||||
| ≤248 U/L | 28 (40.0) | 18 (25.7) | 0.589 | 10 (32.3) | 10 (32.3) | 1.000 | 0.907 |
| >248 U/L | 13 (32.5) | 11 (15.7) | 6 (19.4) | 5 (16.1) | |||
| Types of immunotherapies | |||||||
| PD‐1 | 21 (30.0) | 15 (21.4) | 0.967 | 10 (32.2) | 6 (19.4) | 0.289 | 0.986 |
| PD‐L1 | 20 (28.6) | 14 (20.0) | 6 (19.4) | 9 (29.0) | |||
| NLR | |||||||
| ≤4.0 | 27 (38.6) | 9 (12.9) | 0.004* | 12 (38.7) | 3 (9.7) | 0.004* | 0.778 |
| >4.0 | 14 (20.0) | 20 (28.6) | 4 (12.9) | 12 (38.7) | |||
| Treatment lines | |||||||
| <2 | 26 (37.1) | 9 (12.9) | 0.008* | 11 (35.5) | 5 (16.1) | 0.076 | 0.881 |
| ≥2 | 15 (21.4) | 20 (28.6) | 5 (16.1) | 10 (32.3) | |||
| Concurrent brain radiotherapy | |||||||
| Yes | 25 (35.7) | 9 (12.9) | 0.014* | 11 (35.5) | 4 (12.9) | 0.032* | 0.986 |
| No | 16 (22.9) | 20 (28.6) | 5 (16.1) | 11 (35.5) | |||
Note: P (Intra) the result of univariable analyses between good and bad groups, P (Inter) significant difference between training and validation cohorts. Data are numbers of patients, with percentages in parentheses.
Abbreviations: BM, brain metastasis; ECOG, Eastern Cooperative Oncology Group; LDH, lactate dehydrogenase; NLR, neutrophil‐to‐lymphocyte ratio; T, tumor.
p < 0.05.
In the training cohort, 41 patients showed a good response of intracranial efficacy, while 29 patients had a poor response. In the validation cohort, 16 patients showed a good response, while 15 patients had a poor response. No significant difference was shown for the intracranial efficacy status distribution in the training and independent validation cohorts (p = 0.515).
Feature selection and radiomics signature model building
A total of 659 tumor features and 296 peritumoral features with an ICC >0.75 were selected for further analysis. Subsequently, a logistic univariate regression analysis was conducted to identify a total of 119 radiomic features of the tumor and peritumoral regions (p < 0.05). Finally, the Rad‐score was calculated using the top 10 features selected based on the random forest. The results of random forest are shown in the Supporting Information file 1: Figure S2. Features extracted from tumor regions performed statistically better than those from the peritumoral regions. A total of 10 potential indicators were selected from 1702 features in the training cohort, including nine tumoral and one peritumoral features. After using a logit model of selected features that were weighted by their respective coefficients, the Rad‐score calculation formula obtained was presented as follows (prefix PT represents the peritumoral feature):
Rad‐score = 9.67819 + 3.28734*ORIGINAL_FIRSTORDER_ENTROPY
−0.00325*ORIGINAL_NGTDM_COMPLEXITY
−12.13092*WAVELET.LHH_GLCM_SUMENTROPY
+0.01997*WAVELET.HLL_FIRSTORDER_RANGE
−0.34446*WAVELET.HLL_GLCM_JOINTAVERAGE
−0.80138*WAVELET.HLL_GLRLM_GRAYLEVELVARIANCE
−0.16710*WAVELET.LLL_GLCM_SUMENTROPY
+23.89111*WAVELET.LLL_GLRLM_GRAYLEVELNONUNIFORMITYNORMALIZED
−0.00529*WAVELET.LLL_NGTDM_COMPLEXITY
−0.29136*PTWAVELET.HHL_GLSZM_ZONEENTROPY
A detailed explanation of each selected feature is shown in Supporting Information file 1: Table S2. Rad‐scores for every patient in each cohort are shown (Figure 2). The AUCs that resulted from the use of Rad‐score for prediction were 0.759 (95% CI: 0.648–0.871) and 0.667 (95% CI: 0.470–0.863) in the training and validation cohorts, respectively.
FIGURE 2.
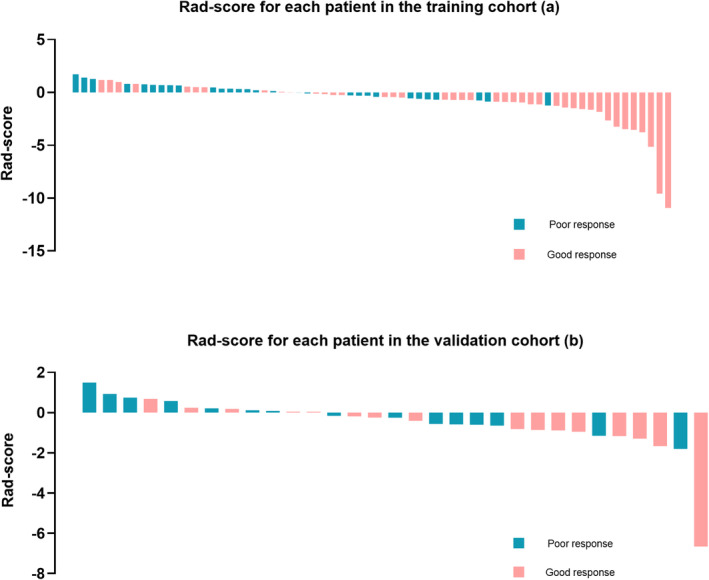
Rad‐score for every patient in each cohort. (a) Rad‐score for every patient in the training cohort. (b) Rad‐score for every patient in the validation cohort. The intracranial efficacy response is marked with different colors.
Clinical model building
The univariate and multivariate logistic regression analysis results of related predictive indicators based on the training cohort are shown in Supporting Information file 1: Table S3. The clinical candidate indicators of age, smoking history, treatment lines, concurrent brain radiotherapy, and NLR were found to have associations with intracranial efficacy in univariate logistic regression analysis. The younger the patient, the poorer the intracranial efficacy of immunotherapy, which is inconsistent with the conclusions of previous studies. 7 , 8 The relationship between smoking history and intracranial efficacy is unstable and opposite in the two cohorts and may be limited by the sample size. Therefore, we excluded age and smoking history from the multivariate analysis. The remaining clinical candidate indicators were further selected through multivariable logistic regression analysis. Finally, the treatment lines and NLR were used to construct a clinical prediction model. The AUCs of the clinical model were 0.754 (95% CI: 0.639–0.869) for the training cohort and 0.825 (95% CI: 0.674–0.976) for the validation cohort.
Combined model building and developing a radiomics nomogram
A multivariable logistic regression analysis was applied to develop a combined predictive model of the Rad‐score, treatment lines, and NLR (Table 2). The independent predictors mentioned above were developed into a combined model and presented as a nomogram (Figure 3a). The p‐value of the Hosmer–Lemeshow test was 0.345, showing that the combined model had a good fit.
TABLE 2.
Risk factors for intracranial efficacy of the combined model.
| Intercept and variable | Model | ||
|---|---|---|---|
| β | Odds ratio (95% CI) | p‐value | |
| Intercept | −1.9939 | 0.0016 | |
| Rad‐score | 1.3285 | 3.775 (1.599 to 8.912) | 0.0024 |
| Treatment lines | 2.1383 | 8.485 (2.092 to 34.409) | 0.0028 |
| NLR | 1.8795 | 6.550 (1.750 to 24.523) | 0.0053 |
Abbreviations: CI, confidence interval; NLR, neutrophil‐to‐lymphocyte ratio.
FIGURE 3.
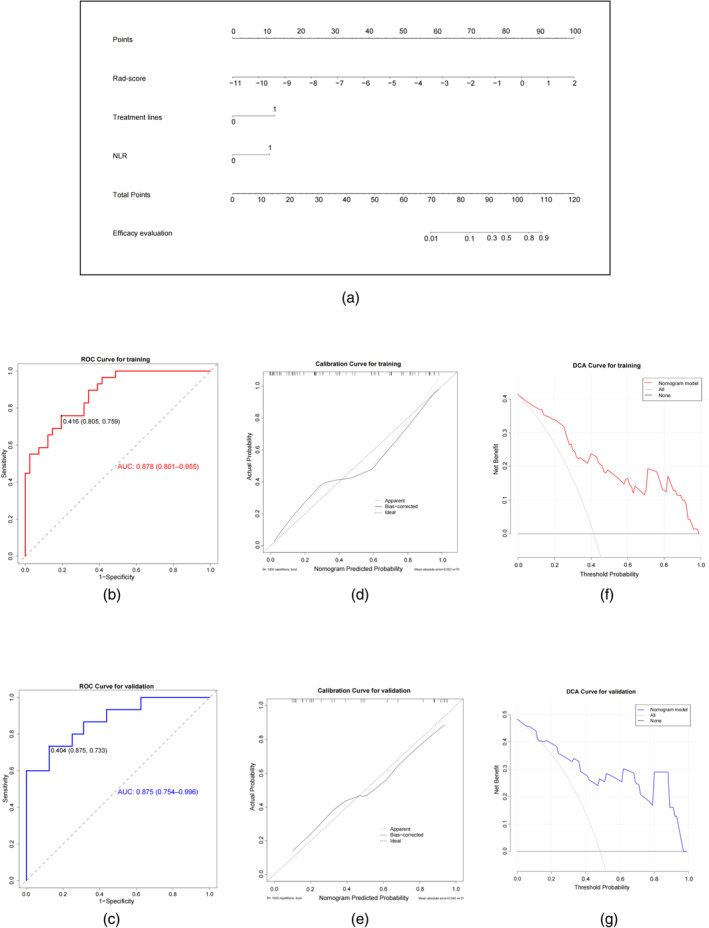
Radiomics nomogram and related model performance evaluation. (a) The radiomics nomogram was developed in the primary cohort, with Rad‐score, treatment lines, and neutrophil‐to‐lymphocyte ratio (NLR) incorporated. The receiver operating characteristic (ROC) curves of the radiomics nomogram in the training cohort (b) and validation cohort (c). The calibration curves of the radiomics nomogram demonstrated satisfactory agreement between prediction and observation in both the training (d) and test cohorts (e). Decision curve analysis for the radiomics nomogram in the training cohort (f) and validation cohort (g). The y‐axis measures the net benefit, and the x‐axis represents the threshold probability. The black line at the bottom represents the hypothesis that no patients had a good intracranial response. The gray line represents the hypothesis that all patients had a good intracranial response. The red and blue lines represent the net benefit of the radiomics nomogram at different threshold probabilities.
The radiomics nomogram based on the combined model achieved AUC values of 0.878 (95% CI: 0.801–0.955) and 0.875 (95% CI: 0.754–0.996) in training (Figure 3b) and validation (Figure 3c) cohorts, respectively. The AUCs of models based on clinical predictors and/or the Rad‐score are shown in the Supporting Information file 1: Table S4. In both the training cohort (Figure 3d) and the independent validation cohort (Figure 3e), the radiomics nomogram for intracranial efficacy showed good agreement between prediction and observation through the calibration curves.
Clinical utility of the radiomics nomogram
Evaluating the clinical usefulness of the radiomics nomogram can assess whether it improves patient outcomes. The decision curve in the training cohort (Figure 3f) shows that when the threshold probability of a patient is between 0% and 99%, using the radiomics nomogram in the current study to predict the intracranial efficacy response, different degrees of net benefit can be obtained. For example, where the personal threshold probability of a patient is 40%, using the radiomics nomogram for treatment determination yields a net benefit of 0.238. This advantage surpasses that of both the treat‐all strategy and the treat‐none strategy. The validation cohort had a similar net improvement when evaluated using the radiomics nomogram (Figure 3g).
Example of how to use nomogram
An SCLC patient is 59 years with BMs. Before immunochemotherapy as first‐line treatment, the Rad‐score obtained through radiomics calculations was −8.5. The NLR value of this patient was <4, so the clinical indicator points are all zero. The nomogram shows that the total points are 20, and the probability of obtaining poor efficacy could be <1%. The lower the total points on the nomogram, the more likely it is to achieve good treatment response. After treatment, the patient's treatment efficacy achieved a complete response.
Prognostic value of the radiomics nomogram
The radiomics nomogram is capable of predicting the prognostic outcomes of all patients. The PFS of patients with a low‐score was longer than that of patients with a high‐score (HR = 0.604, 95% CI: 0.393–0.929, p = 0.0152, Figure 4a). Patients with a low‐score showed a significantly longer iPFS versus patients with a high‐score (HR = 0.541, 95% CI: 0.340–0.862, p = 0.0052, Figure 4b). However, the OS of both patient groups did not exhibit a statistically significant difference (HR = 1.267, 95% CI: 0.647–2.482, p = 0.4894, Figure 4c). In our study, the median PFS (6.50 vs. 4.23 months) of the low and high‐score groups was similar to that of the atezolizumab and placebo groups of IMpower 133 (5.20 vs. 4.30 months), but the median OS was not reached.
FIGURE 4.
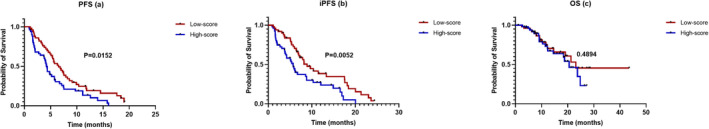
Kaplan–Meier survival analysis of the radiomics nomogram. All patients were divided into low‐score (red line) and high‐score (blue line) groups based on the median of the radiomics nomogram scores. The Kaplan–Meier analysis of (a) progression‐free survival (PFS) and (b) iPFS showed statistically significant differences in the two patient groups but not (c) overall survival (OS).
Subgroup analyses
In the subgroup of 49 patients treated with chemotherapy combined with PD‐L1 inhibitors, the combined model achieved AUC values of 0.892 (95% CI: 0.805–0.978). In the subgroup of 52 patients treated with chemotherapy combined with PD‐1 inhibitors, the combined model achieved AUC values of 0.856 (95% CI: 0.752–0.960). In the subgroup of 41 patients who did not receive brain radiation therapy, the combined model achieved AUC values of 0.905 (95% CI: 0.818–0.992).
DISCUSSION
In this study, a radiomics signature‐based nomogram was established and verified to predict the intracranial efficacy of ICIs in patients with BMs from SCLC. The predictors of this nomogram included the Rad‐score, NLR, and treatment lines. The radiomic signatures were obtained from the pretreatment CE‐T1WI of the MRI in order to assess the morphology, signal intensity, and spatial diversity of the tumoral and peritumoral areas. Finally, we verified the prognostic value of the nomogram for patients.
In the past 10 years, there has been a remarkable transformation in lung cancer treatment. The high mutational burden of SCLC has resulted in the development of ICIs, which can be used alone or in combination with chemotherapy. 21 Despite significant benefits, resistance to ICIs frequently occurs in lung cancer, manifesting as either a lack of response or disease deterioration after an initial positive response. Therefore, the efficacy of the ICIs varies considerably among individual patients. Nevertheless, the task of accurately predicting the individuals who would exhibit a reaction to ICIs has presented significant challenges. There is a higher mortality rate particularly in BM patients. At present, the clinical efficacy of SCLC patients with BMs remains uncertain, and only a limited number of clinical trials have included subgroup analyses focused on BMs. Invasive examinations are also usually unsuitable for patients with BMs, which increases the difficulty of predicting efficacy. Consequently, a pressing requirement exists to identify an accurate and repeatable decision‐making method in clinical applications for selecting the beneficiaries of immunotherapy for SCLC patients with BMs. At present, multiple previous studies indicate that radiomics has the potential to work as a predictive tool for assessing the prognosis and response to treatment in patients with SCLC. The study conducted by Jain et al. 22 showed that radiomic features obtained from CT images of the lung tumor, both inside and around the tumor, were predictors of OS and the effectiveness of chemotherapy in patients with SCLC. Chen et al. 23 established and validated a PFS predictive model based on random forests that performed better than the predictions made by single clinical features in SCLC patients. Furthermore, a number of radiomics model predictions have been established to predict tumor differential diagnosis in SCLC patients. 24 , 25 , 26 , 27 , 28 , 29 These previous studies have demonstrated that the radiomics method is practicable for predicting the intracranial efficacy of ICIs in SCLC.
We chose CE‐T1WI to extract radiomic features. As indicated by multiple previous studies related to the prognosis of brain tumors, there is a significant correlation between prognosis and the radiomic features in the contrast‐enhanced region of MRI. 30 Among all 10 selected features in our study, the tumoral region contributes more than the peritumoral region, with nine features from the tumor and one from the peritumor. Furthermore, more features with an ICC >0.75 selected from the tumor region were more reliable than peritumoral, possibly due to more heterogeneous information within the tumor region. However, according to many previous studies, heterogeneity in peritumoral areas is also an important feature that should be quantified for treatment outcome prediction in BMs. 31 , 32 Consequently, it is reasonable to extract radiomic features from the tumoral and peritumoral regions of the CE‐T1WI, which can include most of the details of the heterogeneous areas of BMs and better predict the efficacy of patients with BMs than the features from a single region. Then, evaluating the combination of radiomics features and clinical factor models or using any one separately, the results showed that the combined model had stronger predictive power, as shown by many previous studies that focused on radiomics nomograms. Jaberipour et al. also reported a 16% relative improvement in AUC for local failure prediction in BMs undergoing hypofractionated stereotactic radiotherapy when radiomic and clinical characteristics were combined, compared to the clinical characteristics alone. 33 In our study, two clinical indicators were significantly associated with the intracranial efficacy of SCLC, namely the treatment lines and NLR. Yang et al. 34 report ES‐SCLC clinical practice should use first‐line ICIs combined with chemotherapy. If the patient has not received ICIs as first‐line treatment, consideration should be given to incorporating ICIs into the second‐line treatment. This study showed that with chemotherapy alone as the first‐line treatment followed by ICIs as a second‐line treatment, PFS was extended but did not reach a significant difference. Two other prospective clinical trials have shown that the efficacy of immunotherapy in the second‐line treatment of SCLC is unclear. CheckMate‐331 35 is a phase III clinical trial primarily targeting SCLC patients who have relapsed after first‐line platinum chemotherapy, making a comparative trial between nivolumab and standard chemotherapy regimens. The main study endpoint OS remained a negative result between the two groups (7.5 vs. 8.4 months, HR = 0.86), while PFS was even worse numerically in the nivolumab group (1.4 vs. 3.8 months, HR = 1.41). The IFCT‐1603 36 study is a phase II clinical trial evaluating the efficacy and safety of atezolizumab compared to chemotherapy after the progression of first‐line EP regimen treatment. The results showed that the 6‐week objective response rate of the atezolizumab group and chemotherapy group were 2.3% and 10%, respectively, with PFS of 4.3 and 1.4 months (HR = 2.26, 95% CI: 1.3–3.93, p = 0.004), but there was no difference in OS between the two groups (9.5 vs. 8.7 months; HR = 0.84, 95% CI: 0.45–1.58, p = 0.6). The above studies are consistent with our research results, showing that applying ICIs in the first‐line treatment of ES‐SCLC can achieve better intracranial efficacy, and both iPFS and PFS are prolonged. However, the efficacy of using ICIs in second‐line treatment of ES‐SCLC is unsatisfactory. Another clinical predictor, NLR, has been confirmed by multiple previous studies 37 , 38 to be associated with the prognosis of ES‐SCLC, with higher NLR typically indicating poorer prognosis. NLR can reflect the systemic inflammatory status. Inflammatory response has a tumor‐promoting effect, which helps the proliferation and survival of tumor cells, promotes angiogenesis and metastasis. 39
The model established in this study was further evaluated for long‐term survival assessment through Kaplan–Meier analysis. The Kaplan–Meier curves of patients in the low and high‐score groups showed statistically significant differences in terms of PFS and iPFS, indicating that the model has the potential to divide pretreatment BM patients into different long‐term prognostic groups. However, it failed to stratify OS, potentially because of the limited sample size. We further analyzed the predictive ability of the established combined model in the different subgroups. The model achieved good performance in the subgroups of chemotherapy combined with PD‐1 or PD‐L1, respectively. Therefore, our model performance is stable, and when PD‐L1 inhibitors are not available, PD‐1 inhibitors can be considered as a substitute. In recent years, serplulimab combined with chemotherapy has become a breakthrough in the first‐line treatment of ES‐SCLC. Another important issue related to the radiomics nomogram is its role in guiding clinical decision‐making. According to the decision curve, if the radiomics nomogram shows that a certain SCLC patient with BMs will respond well to treatment, adopting appropriate intervention strategies will promote the patient's prognosis.
There were still several limitations in this study. First, it was conducted retrospectively and was subject to its inherent limitations, including the presence of unavoidable selection bias. Second, manual segmentation in this research may have introduced potential bias in selecting tumor slices and delineating manual ROIs. Therefore, a reproducibility analysis was conducted in order to verify the repeatability of the delineated ROI, as mentioned in the part on feature selection. Third, the radiomics nomogram was built using a training cohort and verified in the independent external validation cohort. Although the multi‐institutional data guarantees the radiomics nomogram's clinical generalizability and utility, morbidity reduces the sample size, making it lower than other type of tumor radiomics research samples but comparable to SCLC studies.
In conclusion, we developed and validated a radiomics nomogram prediction model for predicting the intracranial efficacy of ICIs in SCLC patients with BMs. This study can help tailor patient treatment schedules, improve clinical decision‐making, and offer extensive potential applications. A larger cohort of patients is still necessary for subsequent studies.
AUTHOR CONTRIBUTIONS
Xiaonan Shi, Peiliang Wang, and Feifei Teng conceived and designed the project. Xiaonan Shi, Yikun Li, and Junhao Xu acquired the data. Xiaonan Shi and Feifei Teng segmented the tumor manually. Xiaonan Shi, Yikun Li, Junhao Xu, and Peiliang Wang analyzed and interpreted the data. Xiaonan Shi, Peiliang Wang, Tianwen Yin, and Feifei Teng inspected the results. All authors read and approved the final manuscript.
FUNDING INFORMATION
This study was supported by grants from the Academic Promotion Program of Shandong First Medical University (2019ZL002), Research Unit of Radiation Oncology, Chinese Academy of Medical Sciences (2019RU071), the foundation of National Natural Science Foundation of China (81627901, 81972863 and 82030082), the foundation of Natural Science Foundation of Shandong (ZR201911040452).
CONFLICT OF INTEREST STATEMENT
The authors declare that this study was conducted without potential conflicts of interest.
Supporting information
Data S1. Supplementary information.
ACKNOWLEDGMENTS
Sincere thanks to all patients and authors involved in this research.
Shi X, Wang P, Li Y, Xu J, Yin T, Teng F. Using MRI radiomics to predict the efficacy of immunotherapy for brain metastasis in patients with small cell lung cancer. Thorac Cancer. 2024;15(9):738–748. 10.1111/1759-7714.15259
Xiaonan Shi and Peiliang Wang are co‐first authors.
REFERENCES
- 1. Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2021;71(3):209–249. [DOI] [PubMed] [Google Scholar]
- 2. Vikis HG. Mouse models of chemically‐induced lung carcinogenesis. Front Biosci. 2013;E5(3):939–946. [DOI] [PubMed] [Google Scholar]
- 3. Gaebe K, Li AY, Park A, Parmar A, Lok BH, Sahgal A, et al. Stereotactic radiosurgery versus whole brain radiotherapy in patients with intracranial metastatic disease and small‐cell lung cancer: a systematic review and meta‐analysis. Lancet Oncol. 2022;23(7):931–939. [DOI] [PubMed] [Google Scholar]
- 4. Zugazagoitia J, Paz‐Ares L. Extensive‐stage small‐cell lung cancer: first‐line and second‐line treatment options. J Clin Oncol. 2022;40(6):671–680. [DOI] [PubMed] [Google Scholar]
- 5. Horn L, Mansfield AS, Szczęsna A, Havel L, Krzakowski M, Hochmair MJ, et al. First‐line Atezolizumab plus chemotherapy in extensive‐stage small‐cell lung cancer. N Engl J Med. 2018;379(23):2220–2229. [DOI] [PubMed] [Google Scholar]
- 6. Liu SV, Reck M, Mansfield AS, Mok T, Scherpereel A, Reinmuth N, et al. Updated overall survival and PD‐L1 subgroup analysis of patients with extensive‐stage small‐cell lung cancer treated with Atezolizumab, carboplatin, and etoposide (IMpower133). J Clin Oncol. 2021;39(6):619–630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Paz‐Ares L, Dvorkin M, Chen Y, Reinmuth N, Hotta K, Trukhin D, et al. Durvalumab plus platinum‐etoposide versus platinum‐etoposide in first‐line treatment of extensive‐stage small‐cell lung cancer (CASPIAN): a randomised, controlled, open‐label, phase 3 trial. Lancet. 2019;394(10212):1929–1939. [DOI] [PubMed] [Google Scholar]
- 8. Goldman JW, Dvorkin M, Chen Y, Reinmuth N, Hotta K, Trukhin D, et al. Durvalumab, with or without tremelimumab, plus platinum‐etoposide versus platinum‐etoposide alone in first‐line treatment of extensive‐stage small‐cell lung cancer (CASPIAN): updated results from a randomised, controlled, open‐label, phase 3 trial. Lancet Oncol. 2021;22(1):51–65. [DOI] [PubMed] [Google Scholar]
- 9. Berghoff AS, Ricken G, Wilhelm D, Rajky O, Widhalm G, Dieckmann K, et al. Tumor infiltrating lymphocytes and PD‐L1 expression in brain metastases of small cell lung cancer (SCLC). J Neurooncol. 2016;130(1):19–29. [DOI] [PubMed] [Google Scholar]
- 10. Iams WT, Porter J, Horn L. Immunotherapeutic approaches for small‐cell lung cancer. Nat Rev Clin Oncol. 2020;17(5):300–312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Kim ES, Velcheti V, Mekhail T, Yun C, Shagan SM, Hu S, et al. Blood‐based tumor mutational burden as a biomarker for atezolizumab in non‐small cell lung cancer: the phase 2 B‐F1RST trial. Nat Med. 2022;28(5):939–945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Derks SHAE, van der Veldt AAM, Smits M. Brain metastases: the role of clinical imaging. Br J Radiol. 2022;95(1130):20210944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Seute T, Leffers P, ten Velde GPM, Twijnstra A. Detection of brain metastases from small cell lung cancer: consequences of changing imaging techniques (CT versus MRI). Cancer. 2008;112(8):1827–1834. [DOI] [PubMed] [Google Scholar]
- 14. Gillies RJ, Kinahan PE, Hricak H. Radiomics: images are more than pictures, they are data. Radiology. 2016;278(2):563–577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Huang Y‐Q, Liang C‐H, He L, Tian J, Liang C‐S, Chen X, et al. Development and validation of a Radiomics nomogram for preoperative prediction of lymph node metastasis in colorectal cancer. J Clin Oncol. 2016;34(18):2157–2164. [DOI] [PubMed] [Google Scholar]
- 16. Liang W, Zhang L, Jiang G, Wang Q, Liu L, Liu D, et al. Development and validation of a nomogram for predicting survival in patients with resected non‐small‐cell lung cancer. J Clin Oncol. 2015;33(8):861–869. [DOI] [PubMed] [Google Scholar]
- 17. Kong C, Zhao Z, Chen W, Lv X, Shu G, Ye M, et al. Prediction of tumor response via a pretreatment MRI radiomics‐based nomogram in HCC treated with TACE. Eur Radiol. 2021;31(10):7500–7511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Zhao L, Gong J, Xi Y, Xu M, Li C, Kang X, et al. MRI‐based radiomics nomogram may predict the response to induction chemotherapy and survival in locally advanced nasopharyngeal carcinoma. Eur Radiol. 2020;30(1):537–546. [DOI] [PubMed] [Google Scholar]
- 19. Li M, Zhang J, Dan Y, Yao Y, Dai W, Cai G, et al. A clinical‐radiomics nomogram for the preoperative prediction of lymph node metastasis in colorectal cancer. J Transl Med. 2020;18(1):46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Lin NU, Lee EQ, Aoyama H, Barani IJ, Barboriak DP, Baumert BG, et al. Response assessment criteria for brain metastases: proposal from the RANO group. Lancet Oncol. 2015;16(6):e270–e278. [DOI] [PubMed] [Google Scholar]
- 21. Esposito G, Palumbo G, Carillio G, Manzo A, Montanino A, Sforza V, et al. Immunotherapy in Small Cell Lung Cancer. Cancers (Basel). 2020;12(9):2522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Jain P, Khorrami M, Gupta A, Rajiah P, Bera K, Viswanathan VS, et al. Novel non‐invasive Radiomic signature on CT scans predicts response to platinum‐based chemotherapy and is prognostic of overall survival in small cell lung cancer. Front Oncol. 2021;11:744724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Chen N, Li R, Jiang M, Guo Y, Chen J, Sun D, et al. Progression‐free survival prediction in small cell lung cancer based on Radiomics analysis of contrast‐enhanced CT. Front Med (Lausanne). 2022;9:833283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Li H, Gao L, Ma H, Arefan D, He J, Wang J, et al. Radiomics‐based features for prediction of histological subtypes in central lung cancer. Front Oncol. 2021;11:658887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Liu S, Liu S, Zhang C, Yu H, Liu X, Hu Y, et al. Exploratory study of a CT Radiomics model for the classification of small cell lung cancer and non‐small‐cell lung cancer. Front Oncol. 2020;10:1268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Dang S, Guo Y, Han D, Ma G, Yu N, Yang Q, et al. MRI‐based radiomics analysis in differentiating solid non‐small‐cell from small‐cell lung carcinoma: a pilot study. Clin Radiol. 2022;77(10):e749–e757. [DOI] [PubMed] [Google Scholar]
- 27. Shah RP, Selby HM, Mukherjee P, Verma S, Xie P, Xu Q, et al. Machine learning Radiomics model for early identification of small‐cell lung cancer on computed tomography scans. JCO Clin Cancer Inform. 2021;5:746–757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Huang J, He W, Xu H, Yang S, Dai J, Guo W, et al. Evaluating histological subtypes classification of primary lung cancers on unenhanced computed tomography based on random Forest model. J Healthc Eng. 2023;2023:8964676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Chen BT, Chen Z, Ye N, Mambetsariev I, Fricke J, Daniel E, et al. Differentiating peripherally‐located small cell lung cancer from non‐small cell lung cancer using a CT Radiomic approach. Front Oncol. 2020;10:593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Chaddad A, Tanougast C. Extracted magnetic resonance texture features discriminate between phenotypes and are associated with overall survival in glioblastoma multiforme patients. Med Biol Eng Comput. 2016;54(11):1707–1718. [DOI] [PubMed] [Google Scholar]
- 31. Karami E, Soliman H, Ruschin M, Sahgal A, Myrehaug S, Tseng C‐L, et al. Quantitative MRI biomarkers of stereotactic radiotherapy outcome in brain metastasis. Sci Rep. 2019;9(1):19830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Mouraviev A, Detsky J, Sahgal A, Ruschin M, Lee YK, Karam I, et al. Use of radiomics for the prediction of local control of brain metastases after stereotactic radiosurgery. Neuro Oncol. 2020;22(6):797–805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Jaberipour M, Soliman H, Sahgal A, Sadeghi‐Naini A. A priori prediction of local failure in brain metastasis after hypo‐fractionated stereotactic radiotherapy using quantitative MRI and machine learning. Sci Rep. 2021;11(1):21620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Yang Y, Ai X, Xu H, Yang G, Yang L, Hao X, et al. Treatment patterns and outcomes of immunotherapy in extensive‐stage small‐cell lung cancer based on real‐world practice. Thorac Cancer. 2022;13(23):3295–3303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Spigel DR, Vicente D, Ciuleanu TE, Gettinger S, Peters S, Horn L, et al. Second‐line nivolumab in relapsed small‐cell lung cancer: CheckMate 331☆. Ann Oncol. 2021;32(5):631–641. [DOI] [PubMed] [Google Scholar]
- 36. Pujol J‐L, Greillier L, Audigier‐Valette C, Moro‐Sibilot D, Uwer L, Hureaux J, et al. A randomized non‐comparative phase II study of anti‐programmed cell death‐ligand 1 Atezolizumab or chemotherapy as second‐line therapy in patients with small cell lung cancer: results from the IFCT‐1603 trial. J Thorac Oncol. 2019;14(5):903–913. [DOI] [PubMed] [Google Scholar]
- 37. Suzuki R, Lin SH, Wei X, Allen PK, Welsh JW, Byers LA, et al. Prognostic significance of pretreatment total lymphocyte count and neutrophil‐to‐lymphocyte ratio in extensive‐stage small‐cell lung cancer. Radiother Oncol. 2018;126(3):499–505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Kang MH, Go SI, Song HN, Lee A, Kim SH, Kang JH, et al. The prognostic impact of the neutrophil‐to‐lymphocyte ratio in patients with small‐cell lung cancer. Br J Cancer. 2014;111(3):452–460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Mantovani A, Allavena P, Sica A, Balkwill F. Cancer‐related inflammation. Nature. 2008;454(7203):436–444. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data S1. Supplementary information.


