Abstract
It is well established that glycosylation is essential for assembly of enveloped viruses, but no information is yet available as to the function of carbohydrates on the nonenveloped but glycosylated rotavirus. We show that tunicamycin and, more pronouncedly, a combination of tunicamycin and brefeldin A treatment caused misfolding of the luminal VP7 protein, leading to interdisulfide bond aggregation. While formation of VP7 aggregates could be prevented under reducing conditions, they reoccurred in less than 30 min after a shift to an oxidizing milieu. Furthermore, while glycosylated VP7 interacted during maturation with protein disulfide isomerase, nonglycosylated VP7 did not, suggesting that glycosylation is a prerequisite for protein disulfide isomerase interaction. While native NSP4, which does not possess S-S bonds, was not dependent on N-linked glycosylation or on protein disulfide isomerase assistance for maturation, nonglycosylated NSP4 was surprisingly found to interact with protein disulfide isomerase, further suggesting that protein disulfide isomerase can act both as an enzyme and as a chaperone. In conclusion, our data suggest that the major function of carbohydrates on VP7 is to facilitate correct disulfide bond formation and protein folding.
Several studies have shown that glycosylation and oligosaccharide trimming in the endoplasmic reticulum (ER) are required for proper folding and assembly of secretory proteins (9, 11, 17). Without oligosaccharides, many glycoproteins misfold, aggregate, and are degraded without transport from the ER to the Golgi complex and beyond (11). While the requirements for glycosylation are well documented for cellular and viral proteins that traverse the secretory pathway, it has not been established how critical these modifications are for maturation of either ER resident proteins or for the nonenveloped rotavirus.
Rotavirus is one of the very few viruses that utilizes the ER for assembly and has therefore been an attractive model with which to study ER translocation, retention, and protein folding (1, 18, 20, 23, 28). Of the 12 structural and nonstructural proteins of rotavirus, two have been of particular interest from the viral assembly point of view. The outer capsid VP7 protein is an integral membrane polypeptide with luminal orientation that contains only N-linked high-mannose oligosaccharide residues (7). The ER retention motif of VP7, which does not include KDEL or lysine residues, has been the focus of numerous studies, and more recent data propose that three amino acids in the amino terminus are involved in ER retention (18). We and others have previously shown that calcium and disulfide bonds are required for infectivity and correct folding of VP7 (6, 23, 25, 28, 29) and that VP7, which is normally endo-β-N-acetylglucosaminidase H sensitive, is processed to endo-β-N-acetylglucosaminidase H resistance during brefeldin A (BFA) treatment (20).
Early studies established that inhibition of glycosylation by tunicamycin (TM) resulted in reduced infectivity and accumulation of immature enveloped rotavirus particles in the ER (22, 24). The exact role of the carbohydrates for morphogenesis and infectivity remains to be determined, however, but it has been proposed that one of the functions of the carbohydrates may be to assist in survival of the virus in the gastrointestinal tract, and yet another is that the carbohydrates modulate serotype specificity (13). The nonstructural NSP4 protein is a novel type of trans-ER-resident glycoprotein which functions not only as a receptor for subviral particles in the cytoplasm (1, 19), but also, as has been shown, as an enterotoxin capable of inducing diarrhea (2). Furthermore, it has been shown that depletion of cellular Ca2+ and prevention of NSP4 glycosylation by TM treatment cause accumulation of NSP4 in the virion-associated envelope (22–24).
To our knowledge, there are no reports addressing the function of carbohydrates in folding of ER-resident proteins, including VP7 and NSP4 of rotavirus. In this study, we have treated rotavirus-infected cells with TM and BFA and analyzed the folding of rotavirus glycoproteins. BFA is an antiviral agent that inhibits protein transport out of the ER and also induces recycling of Golgi-specific enzymes back to the ER (20).
We have found that treatment of the luminal VP7 protein with TM plus BFA prevented association with protein disulfide isomerase (PDI), which led to interdisulfide bond aggregates of VP7 and in turn affected proper virus assembly. We also found that the trans-ER NSP4 protein is not dependent on N-linked glycosylation and PDI assistance for correct folding under native conditions, but that PDI interacts with the nonglycosylated form of NSP4. These two modes of action suggest that PDI can act both as an enzyme and as a chaperone. This work suggests for the first time a direct function of carbohydrates for rotavirus. The principal role of the carbohydrate on VP7 is to facilitate correct disulfide bond formation and folding and, ultimately, viral assembly.
MATERIALS AND METHODS
Cells, viruses, and antibodies.
MA-104 cells were grown in Dulbecco’s modified Eagle’s minimal essential medium (MEM) supplemented with 10% fetal calf serum. Rhesus rotavirus (RRV) was obtained from infected MA-104 cells by freezing and thawing. The monoclonal antibodies (MAbs) used in this study include the MAb M60, which recognizes a cross-reactive nonneutralizing epitope on VP7 (26) that is dependent on correct disulfide bond formation (28); 7A12, a neutralizing anti-VP4 MAb which recognizes VP4 and neutralizes RRV (16, 26); and a rabbit polyclonal antibody against PDI, supplied by Kari Kivirikko (Department of Medical Biochemistry, University of Oulu, Oulu, Finland).
Rotavirus infection.
RRV was activated with 10 μg of trypsin per ml for 30 min at 37°C before inoculation of MA-104 cells in serum-free medium. After 1 h of infection, the inoculum was replaced with fresh Eagle’s MEM. The titers of RRV were determined by peroxidase staining as previously described (28).
Metabolic labeling of viral proteins.
To produce metabolically labeled cell lysates, MA-104 cells were infected with trypsin-activated RRV at a multiplicity of infection (MOI) of 10 as described previously (28). At 7 h postinfection (hpi), infected cells were starved for 1 h in methionine- and cysteine-free medium before being labeled with 50 μCi (0.17 μCi/μl) or 200 μCi (0.67 μCi/μl) of [35S]methionine-cysteine (Trans-label; Dupont) for various periods of time. For chase experiments, cells were washed and incubated with medium containing an excess of methionine (10 mM) and 1 mM cycloheximide (Sigma). At the end of each radioactive pulse or after a chase period, cells were incubated with ice-cold phosphate-buffered saline (PBS) containing 40 mM N-ethylmaleimide (NEM) (Sigma) for 2 min to prevent disulfide bond rearrangements. Cells were then lysed in ice-cold sodium dodecyl sulfate (SDS) lysis buffer (10 mM Tris-HCl [pH 7.5], 150 mM NaCl, 1% Triton X-100, 0.5% SDS, 6 μg of leupeptin per ml, 3 μg of antipain per ml, 1 μg of aprotinin per ml, and 0.1 mg of pefabloc per ml) or gentle lysis buffer {150 mM NaCl, 2% 3-[(3-cholamidopropyl)-dimethylammonio]-1-propanesulfonate (CHAPS), 50 mM HEPES} containing protease inhibitors. Cell lysates were clarified of cell debris by centrifugation at 13,000 × g for 2 min in a microcentrifuge before use.
Treatment of cells with dithiotritiol, TM, and BFA.
BFA was purchased from Sigma. A stock solution was prepared as previously described (20). To obtain metabolically labeled proteins in the presence of BFA, 2 μg/ml was added at 1 hpi and was maintained through the chase. BFA was replaced with fresh BFA every 4 h. To inhibit N-linked glycosylation, 2 μg of TM per ml was added to media 3 h before pulse-labeling and maintained through the chase.
To obtain metabolically labeled proteins synthesized under reduced conditions, dithiothreitol (DTT) was added to the media 20 min before a pulse and maintained during the chase period (28).
RIPA.
Immunoprecipitation was performed as previously described (20). Briefly, radiolabeled lysates (50 μl) were incubated with 1 μl of the desired antibody and 450 μl of radioimmunoprecipitation assay (RIPA) buffer (10 mM Tris-HCl [pH 7.5], 150 mM NaCl, 0.6 M KCl, 4 mM EDTA, 1% Triton X-100) or CHAPS buffer (150 mM NaCl, 50 mM HEPES, 0.5% CHAPS) overnight at 4°C. Twenty-five μl of Staphylococcus aureus protein A-Sepharose CL-4B (Pharmacia, Uppsala, Sweden) was subsequently added to the mixture, which was then incubated for 2 h at 4°C or 1 h at room temperature. The protein A-Sepharose-coupled immune complexes were then pelleted at 13,000 × g for 30 s in a microcentrifuge and washed three times with RIPA buffer and twice with 10 mM Tris-HCl (pH 8.0)–150 mM NaCl or CHAPS buffer. The immune complexes were suspended in 30 μl of nonreducing sample buffer (10 mM Tris-HCl [pH 6.8], 0.5% SDS, 10% glycerol) or reducing sample buffer (nonreducing sample buffer including 2% β-mercaptoethanol). Unless otherwise indicated, samples were boiled for 5 min before separation by SDS-polyacrylamide gel electrophoresis (PAGE).
SDS-PAGE.
Polypeptide separation was performed by SDS PAGE with a 4.5% stacking gel and 10% separation gel as previously described (28). Electrophoresis was carried out at a constant voltage of 50 V at room temperature, followed by fixation with 10% glacial acetic acid and 35% methanol for 1 h at room temperature. Autoradiography was performed as previously described (28). Molecular mass standards included myosin (200 kDa), phosphorylase b (97 kDa), bovine serum albumin (69 kDa), ovalbumin (46 kDa), carbonic anhydrase (30 kDa), and lysozyme (14 kDa).
Protein recovery from SDS-PAGE.
To elute a specific protein from the SDS-PAGE, the band of a desired protein was excised from the dried gel and incubated with elution buffer (10 mM Tris-HCl [pH 7], 0.1% SDS) for 2 h at 37°C. The acrylamide fragments were pelleted at 13,000 × g for 10 min in a microcentrifuge. The supernatants were used for further experiments.
RESULTS
Absent or impaired glycosylation of rotavirus proteins leads to formation and accumulation of a novel 78-kDa protein.
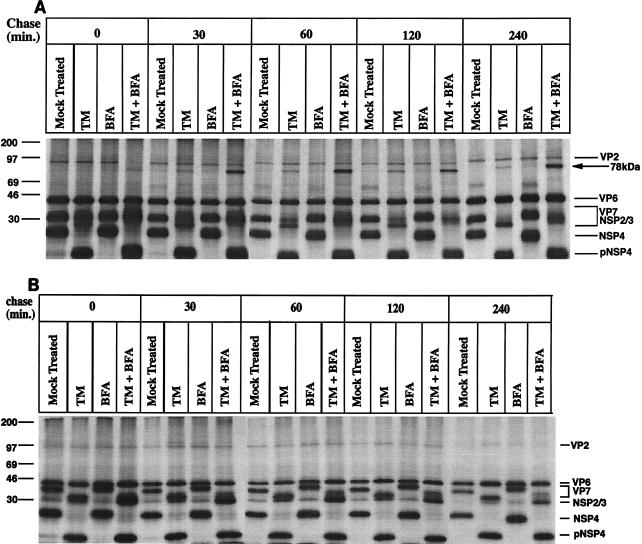
To investigate the role of glycosylation in correct folding of the ER-associated VP7 and NSP4 proteins, RRV-infected cells were treated with BFA (2 μg/ml) at 1 hpi and/or with TM (2 μg/ml) at 5 hpi. After 1 h of methionine starvation, infected cells were metabolically labeled at 8 hpi for 30 min, followed by a chase as described in Methods and Materials. At indicated times, monolayers were briefly incubated with 40 mM NEM to prevent artificial disulfide bond formation and then lysed and analyzed by SDS-PAGE. When the cell lysates were analyzed under nonreducing conditions, a 78-kDa band was observed in TM-treated cells and, more pronouncedly, in TM- and BFA-treated cells (Fig. 1A). A close examination of Fig. 1A also revealed that the intensity of the 78-kDa band increased from the pulse to a 60-min chase but that no significant amount of the 78-kDa protein had further accumulated from that time, which suggests that the formation of the 78-kDa protein was a posttranslational process, predominantly occurring within 60 min after biosynthesis. It was also observed that the 78-kDa protein was not found to be significantly degraded within the first 4 h after biosynthesis.
FIG. 1.
Role of glycosylation and oligosaccharide processing in folding of NSP4 and VP7. Cells infected with RRV (MOI of 10) were mock or BFA (2 μg/ml) treated at 1 hpi. At 5 hpi, TM (2 μg/ml) was added to the medium of the same monolayers (as indicated) and was maintained through the experiment. At 7 hpi, cells were starved for 1 h in methionine-cysteine-free media and then subsequently metabolically labeled (50 μCi) for 30 min. To examine posttranslational processing, labeled proteins were chased in Eagle’s MEM supplemented with 1 mM cycloheximide and 10 mM methionine for the lengths of time indicated over the lanes. At the end of the pulse or chase, monolayers were incubated with ice-cold PBS with 40 mM NEM for 2 min, and cells were then harvested in SDS lysis buffer and mixed with nonreducing sample buffer (A) or reducing sample buffer (B), boiled, and analyzed by SDS-PAGE.
To uncover whether the 78-kDa protein was held together by covalent bonds, the same lysates presented in Fig. 1A were examined under reducing conditions (Fig. 1B). After disulfide bond reduction, the 78-kDa band completely disappeared and a protein band with a molecular mass of 36 to 38 kDa, depending on the treatment (TM, BFA, etc.), appeared (Fig. 1B). This strongly suggests that the 78-kDa protein complex consisted of interdisulfide cross-linked polypeptide(s) with a molecular mass range of 36 to 38 kDa.
Upon carbohydrate manipulation, the luminal and ER-associated VP7 misfolds and accumulates into 78-kDa interdisulfide aggregates.
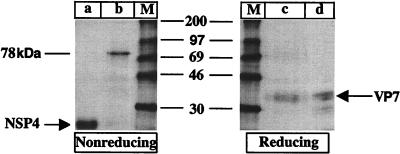
To identify the component of the interdisulfide bond aggregate identified in Fig. 1A, the 78-kDa protein was eluted from the TM-plus-BFA 60-min chase (Fig. 1A) and analyzed under reducing and nonreducing SDS-PAGE conditions (Fig. 2). To monitor the elution efficiency, the NSP4 protein was also eluted and analyzed. Lane a in Fig. 2 shows the elution efficiency of NSP4, and lane b shows the elution efficiency of the 78-kDa protein. Lanes a and b (Fig. 2) not only show that the elution was efficient, but, more importantly, also show that only a single protein was purified. By comparing the migration mobility of VP7 immunoprecipitated with a VP7-specific MAb, (Fig. 2, lane d) with that of the eluted and disulfide bond-reduced 78-kDa protein (Fig. 2, lane c), it can be concluded that the 78-kDa protein consists of multimers of a protein with a molecular mass identical to that of the VP7 glycoprotein.
FIG. 2.
To identify the component of the disulfide aggregate presented in Fig. 1A, the 78-kDa and NSP4 bands from the gel presented in Fig. 1A (TM+BFA, 60-min chase) were excised and incubated with elution buffer (10 mM Tris-HCl [pH 7], 0.1% SDS) for 2 h at 37°C. The eluted 78-kDa protein was analyzed by nonreducing (lane b) and reducing (lane c) SDS-PAGE. The eluted NSP4 protein was analyzed by nonreducing SDS-PAGE (lane a). The lysate from the pulse-chase experiment presented in Fig. 1A (TM+BFA, 60-min chase) was immunoprecipitated with a VP7 MAb (M60) (lane d). M, molecular mass markers. The molecular masses (kilodaltons) of the markers are shown in the middle of the panel.
A reducing milieu prevents aggregation of VP7 with manipulated carbohydrates.
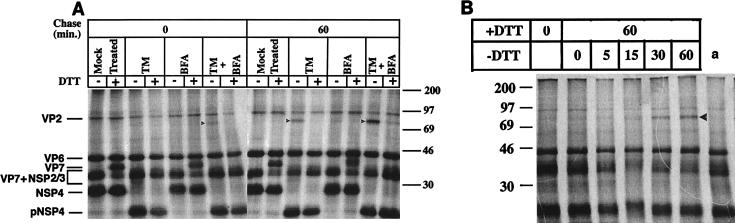
We have previously shown that a brief exposure of rotavirus-infected cells to the reducing agent DTT inhibits formation of correct intradisulfide bonds on VP7 and that prolonged exposure permanently misfolds VP7 (28). To analyze if a reducing milieu also could prevent aggregation of VP7, DTT (2 mM) was added to methionine-cysteine-deficient medium 20 min before a metabolic pulse and maintained during a 60-min chase. At the end of each radioactive pulse or after a chase period, cells were washed twice with MEM and incubated with ice-cold PBS containing 40 mM NEM for 2 min to alkylate free thiol groups. Cells were then lysed in ice-cold SDS lysis buffer. As illustrated in Fig. 3A, aggregation of VP7 was prevented, both in TM and in TM plus BFA-treated cells. The dramatic effect of DTT on VP7 folding is furthermore illustrated in mock-treated cells, in which VP7 migrates significantly faster under oxidizing conditions than during reducing conditions. To examine if VP7 becomes permanently protected from aggregation after a 60-min incubation in a reducing milieu, DTT was washed out and cells were incubated in oxidizing media up to 60 min. As illustrated in Fig. 3B, VP7 had already begun to aggregate after 30 min in an oxidizing milieu, indicating that a reducing milieu prevents aggregation of rotavirus VP7, while aggregation occurs rapidly after a switch to an oxidizing milieu.
FIG. 3.
(A) Effect of a reducing milieu on folding of TM- and/or BFA-treated VP7. Infected cells were TM and/or BFA treated as described in the legend to Fig. 1. At 7 hpi, cells were starved for 1 h in methionine-cysteine-free media. Twenty minutes before a 30-min metabolical 35S pulse, 2 mM DTT was added to the cells for the duration of the experiment. After a 30-min pulse with 50 μCi of 35S Trans-label, cells were chased for 60 min in Eagle’s MEM supplemented with 1 mM cycloheximide and 10 mM methionine. At the end of the pulse or chase, cells were washed twice with MEM and incubated with ice-cold PBS containing 40 mM NEM for 2 min to alkylate free thiol groups. Cells were subsequently harvested in SDS lysis buffer and mixed with nonreducing sample buffer, boiled, and analyzed by SDS-PAGE. The arrowheads indicate disulfide aggregates. The molecular masses (kilodaltons) of the markers are shown on the right side of the panel. (B) Effect of intracellular reoxidation on folding of non-N-linked glycosylated VP7. Infected cells were BFA and TM treated as described in the legend to Fig. 1. At 7 hpi, cells were starved for 1 h in methionine-cysteine-free media. Twenty minutes before a 30-min pulse, 4 mM DTT was added, and it remained in the media through the pulse and chase. After a 30-min pulse with 50 μCi of 35S Trans-label, cells were chased for 60 min in Eagle’s MEM supplemented with 1 mM cycloheximide, 10 mM methionine, and 4 mM DTT. The monolayers were then washed with MEM and incubated with chase medium without DTT for different periods of time as indicated. At the end of the pulse or chase, cells were washed twice with MEM and incubated with ice-cold PBS containing 40 mM NEM for 2 min to alkylate free thiol groups. Cells were subsequently harvested in SDS lysis buffer and mixed with nonreducing sample buffer, boiled, and analyzed by SDS-PAGE. a, TM- and BFA-treated infected cells chased for 120 min in the presence of 4 mM DTT. The molecular masses (kilodaltons) of the markers are shown on the left side of the panel.
Disulfide bond-dependent antigenicity is altered in VP7 with manipulated carbohydrates.
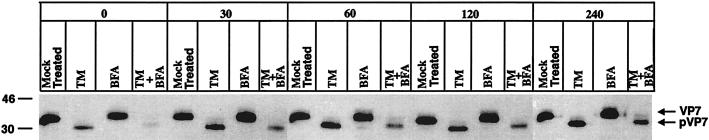
It is generally believed that the main function of N-linked carbohydrates is to provide increased solubility, which in turn facilitates correct folding and antigenicity of proteins. However, the importance of N-linked carbohydrates with respect to folding and antigenicity varies among viruses and strains and the backbone localization of the glycosylation site (11). To examine if carbohydrate manipulation of VP7 affects disulfide bond-dependent antigenicity, cell lysates from the pulse-chase experiment presented in Fig. 1 were immunoprecipitated with M60, a disulfide bond-dependent VP7 MAb (20, 28), and analyzed under nonreducing conditions. As shown in Fig. 4, significantly less VP7 was immunoprecipitated from lysates treated with TM and TM plus BFA than from mock- or BFA-treated cells. Quantification by scanning densitometry showed that 67.5% of total mock-treated VP7 was immunoprecipitated by M60 at a 240-min chase compared to 31.3% of TM-plus-BFA-treated VP7. As expected, M60 did not immunoprecipitate the 78-kDa VP7 aggregate (data not shown).
FIG. 4.
Cell lysates from the pulse-chase experiment presented in Fig. 1 immunoprecipitated with a VP7 MAb (M60) and analyzed by nonreducing SDS-PAGE.
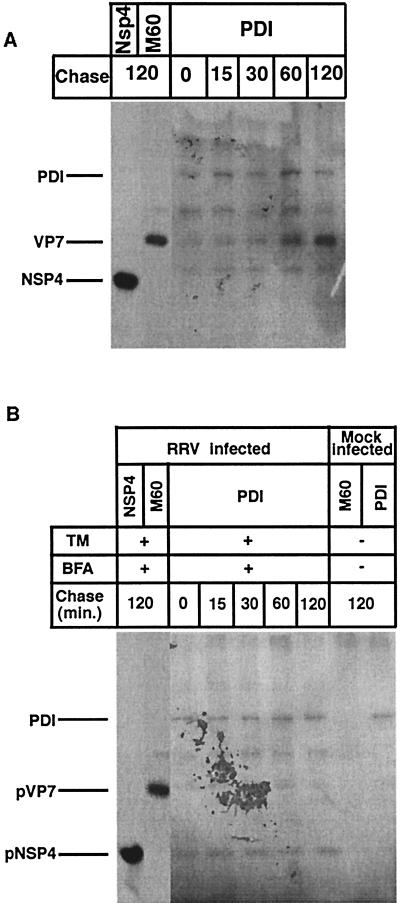
PDI associates with glycosylated VP7 and with nonglycosylated NSP4.
PDI is an ER-associated enzyme that assists in protein folding by catalyzing the formation of disulfide bonds (8). More recent data also suggest that PDI has a chaperone-like activity (4, 27, 30). Because, to our knowledge, no or only limited information is available concerning if and how PDI participates in folding of ER-resident proteins, we decided to examine if the ER-associated VP7 and NSP4 of rotavirus associate with PDI during maturation. To address this question, RRV-infected cells were mock treated or treated with BFA at 1 hpi and with TM at 5 hpi. After 1 h of methionine starvation, cells were metabolically labeled at 8 hpi for 15 min, followed by chase periods. At indicated times, the monolayers were briefly incubated with 40 mM NEM to prevent artificial disulfide bond formation and then lysed in gentle lysis buffer followed by immunoprecipitation. As shown in Fig. 5A, PDI associated with VP7 in a kinetic manner, and at 120 min postchase, a significant amount of PDI was seen associated with VP7. Figure 5A and B also identify PDI at a molecular mass of approximately 60 kDa. In contrast to the glycosylated and disulfide bond-rich VP7 (28), no interaction was observed between NSP4, which lacks disulfide bonds (28), and PDI. A most surprising observation, however, was that manipulation of N-linked glycosylation resulted in PDI association with NSP4 (Fig. 5B). Furthermore, inhibition of N-linked glycosylation abolished the PDI interaction with VP7, suggesting that correct N-linked glycosylation is a prerequisite for PDI interaction with VP7. The NSP4 results support the hypothesis, which suggests that PDI, in addition to promoting correct disulfide bond formation, may react as a chaperone for proteins lacking disulfide bonds (4, 27).
FIG. 5.
Cells infected with RRV (MOI of 10) were mock (A) or BFA (2 μg/ml) treated (B) at 1 hpi. At 5 hpi, TM (2 μg/ml) was added to the medium of the monolayers (as indicated) and was maintained through the experiment. At 7 hpi, cells were starved for 1 h in methionine-cysteine-free media and then metabolically labeled (200 μCi) for 15 min. Labeled proteins were chased with Eagle’s MEM supplemented with 1 mM cycloheximide and 10 mM methionine for the lengths of time indicated over the lanes. At the end of the pulse or chase, monolayers were incubated with ice-cold PBS with 40 mM NEM for 2 min, and cells were harvested in gentle lysis buffer. The cell lysates were immunoprecipitated with MAbs to VP7 and NSP4 and rabbit polyclonal antibodies to PDI.
DISCUSSION
Rotavirus is unique among animal viruses in being a glycosylated, nonenveloped virus that matures in the ER. While the function of carbohydrates is well documented for viral proteins that traverse the secretory pathway (11, 14, 15), no function has yet been recognized for the carbohydrates of rotavirus VP7 or NSP4, nor has it been established how critical glycosylation is for the maturation of ER-resident proteins.
Previous studies have shown not only that inhibition and aberrant glycosylation of rotavirus proteins by TM and BFA reduce infectivity and cause accumulation of immature enveloped particles in the ER (20, 22, 24), but also that BFA may induce O-linked glycosylation of VP7 (20). Furthermore, a clone of rotavirus strain SA-11 designated 28 has been shown to be infectious, yet it has no glycosylated VP7 (22), suggesting that carbohydrates on VP7 are not per se required for cell-receptor interaction or viral entry. We found that carbohydrate manipulation with either TM or TM plus BFA, but not BFA alone, rapidly caused misfolding of VP7, leading to interdisulfide bond aggregation and aberrant antigenicity within 60 min after biosynthesis. This dramatic effect of misfolding is interesting, because VP7, in contrast to most secretory glycoproteins, only possesses a single high-mannose oligosaccharide (9, 11). We suggest, by combining this body of work with the observations described above, that the principal function of N-linked carbohydrates on rotavirus VP7 is to facilitate correct protein folding and viral assembly.
We have previously shown that glycosylated VP7 must be in an oxidized form to fold correctly (28). A reducing milieu not only prevents correct folding, but also causes permanent misfolding upon prolonged exposure to DTT. In this study, we found that aggregation of VP7 with manipulated carbohydrates was prevented by a reducing milieu; however, soon after the return to an oxidizing milieu, aggregation of VP7 occurred, indicating that glycosylation and an oxidizing milieu are crucial for correct posttranslational folding of VP7.
In contrast to the luminal and ER-associated VP7, no misfolding or aggregation of the trans-ER-associated NSP4 was noted. Previous studies, however, have shown that membrane-bound secretory proteins may very well aggregate upon misfolding (17). A possible explanation for the different properties could be that NSP4, in contrast to most secretory proteins, does not possess luminal thiol groups and therefore cannot aggregate into disulfide bond complexes. Another explanation is that few (less than 28 or 67, depending on the model [3, 5]) of the 175 amino acids of NSP4 are luminally oriented.
In recent years, it has become clear that many proteins do not fold spontaneously in vivo, but require the help of other proteins, often called chaperones (10). Among the enzymes and chaperones that assist in protein folding in the ER, the role of PDI for the folding of rotavirus VP7 and NSP4 was examined in this study. Because PDI catalyzes formation of native disulfide bonds and prevents aberrant disulfide bond formation, it may play an important role in the folding of ER-resident proteins.
While it is clear that PDI participates in protein folding (8), there are few reports clearly demonstrating an interaction between PDI and maturation of secretory proteins (21), and there is no information available regarding the role of PDI in the maturation of ER-resident proteins. We found that PDI only interacted with glycosylated VP7 with kinetics correlating with the processing of carbohydrates of membrane- and virus-associated VP7 to Man6GlcNAc2 (12). It has previously been reported that VP7 exists in two forms—one membrane associated and one virus associated—with apparent differences in conformation and localization (12). Because triple-shelled particles are assembled within 15 to 20 min after protein synthesizes and PDI remains associated with VP7 at 120 min postsynthesis, it is reasonable to believe that the final disulfide bond formation on VP7 occurs on the virus particles together with the final oligosaccharide trimming (12), rather than on membrane-associated VP7. This proposal is further supported by work with conformation-dependent MAbs showing that neutralizing and disulfide bond-dependent MAbs only recognize fully oxidized and virus-assembled VP7 (28).
The fact that PDI only interacted with correctly N-linked glycosylated VP7 suggests that carbohydrates and PDI are closely associated with correct folding of the ER-resident VP7. The exact role of the N-linked oligosaccharides on VP7 for PDI interaction is not established. Our suggestion is that no or aberrant glycosylation modifies the folding of VP7 and thereby prevents the interaction between VP7 and PDI. A reasonable role of the N-linked carbohydrate on VP7 could therefore be to prevent VP7 from premature folding and nonspecific aggregation before it is presented to PDI and becomes correctly folded. Judging by the time kinetics, this would be logical, since glycosylation is a cotranslational event, whereas disulfide bond formation is posttranslational. As illustrated in Fig. 5A, PDI immunoprecipitated not only VP7, but also a small amount of a protein with a molecular mass of around 40 kDa. This protein most likely represents VP6 (41 kDa), the major inner capsid protein of rotavirus that is translocated as a constituent of double-shelled particles into the ER lumen and that associates with VP7 within 15 min after protein synthesis (12). That the 40-kDa protein is of cellular origin can be ruled out, because no cellular proteins were metabolically labeled (Fig. 1), nor did the anti-PDI antibody recognize any cellular proteins except PDI (Fig. 5B). The most reasonable explanation is therefore that PDI, under the gentle RIPA conditions used, immunoprecipitated mature virus containing both VP6 and VP7.
Previous studies have shown that neither of the two thiol groups on NSP4 is localized on the luminal part of ER (1, 3), a positioning which excludes them from being oxidized (28). It was therefore expected that PDI would not recognize native NSP4. However, it was surprising to note that TM-BFA-treated NSP4 was recognized by PDI (Fig. 5B). Presently, we have no explanation for this interaction, but it should be mentioned that recent studies propose that PDI, in addition to being an enzyme, has chaperone-like activity and can interact with proteins lacking disulfide bonds (4, 27, 30). As illustrated in Fig. 5B, a small amount of VP6 was immunoprecipitated together with TM-BFA-treated NSP4. The most reasonable explanation for this is that impaired viral assembly induced by TM treatment leads to accumulation of ER-derived envelope particles (22, 24) and, possibly, to entrapment of double-shelled particles (particularly VP6) on NSP4-ER membranes, instead of a release into the ER lumen during ER translocation under normal conditions.
ACKNOWLEDGMENTS
This project received financial support from the Swedish Medical Council (K97-06X-10392-05A) and the European Community (ERBIC18CT960027).
We are grateful to Harry Greenberg for the M60 MAb and Kari Kivirikko for the PDI antibody.
REFERENCES
- 1.Au K-S, Chan W-K, Burns J W, Estes M K. Receptor activity of rotavirus nonstructural glycoprotein NS28. J Virol. 1989;63:4553–4562. doi: 10.1128/jvi.63.11.4553-4562.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ball J M, Tian P, Zeng C Q, Morris A P, Estes M K. Age-dependent diarrhea induced by a rotaviral nonstructural glycoprotein. Science. 1996;272:101–104. doi: 10.1126/science.272.5258.101. . (See comments.) [DOI] [PubMed] [Google Scholar]
- 3.Bergmann C C, Maass D, Poruchynsky M S, Atkinson P H, Bellamy A R. Topology of the non-structural rotavirus receptor glycoprotein NS28 in the rough endoplasmic reticulum. EMBO J. 1989;8:1695–1703. doi: 10.1002/j.1460-2075.1989.tb03561.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cai H, Wang C C, Tsou C L. Chaperone-like activity of protein disulfide isomerase in the refolding of a protein with no disulfide bonds. J Biol Chem. 1994;269:24550–24552. [PubMed] [Google Scholar]
- 5.Chan W K, Au K S, Estes M K. Topography of the simian rotavirus nonstructural glycoprotein (NS28) in the endoplasmic reticulum membrane. Virology. 1988;164:435–442. doi: 10.1016/0042-6822(88)90557-0. [DOI] [PubMed] [Google Scholar]
- 6.Dormitzer P R, Greenberg H B. Calcium chelation induces a conformational change in recombinant herpes simplex virus-1-expressed rotavirus VP7. Virology. 1992;189:828–832. doi: 10.1016/0042-6822(92)90616-w. [DOI] [PubMed] [Google Scholar]
- 7.Estes M K, Cohen J. Rotavirus gene structure and function. Microbiol Rev. 1989;53:410–449. doi: 10.1128/mr.53.4.410-449.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Freedman R B, Hirst T R, Tuite M F. Protein disulphide isomerase: building bridges in protein folding. Trends Biochem Sci. 1994;19:331–336. doi: 10.1016/0968-0004(94)90072-8. [DOI] [PubMed] [Google Scholar]
- 9.Hammond C, Braakman I, Helenius A. Role of N-linked oligosaccharide recognition, glucose trimming, and calnexin in glycoprotein folding and quality control. Proc Natl Acad Sci USA. 1994;91:913–917. doi: 10.1073/pnas.91.3.913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hammond C, Helenius A. Quality control in the secretory pathway. Curr Opin Cell Biol. 1995;7:523–529. doi: 10.1016/0955-0674(95)80009-3. [DOI] [PubMed] [Google Scholar]
- 11.Helenius A. How N-linked oligosaccharides affect glycoprotein folding in the endoplasmic reticulum. Mol Biol Cell. 1994;5:253–265. doi: 10.1091/mbc.5.3.253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kabcenell A K, Poruchynsky M S, Bellamy A R, Greenberg H B, Atkinson P H. Two forms of VP7 are involved in assembly of SA11 rotavirus in endoplasmic reticulum. J Virol. 1988;62:2929–2941. doi: 10.1128/jvi.62.8.2929-2941.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lazdins I, Coulson B S, Kirkwood C, Dyall-Smith M, Masendycz P J, Sonza S, Holmes I H. Rotavirus antigenicity is affected by the genetic context and glycosylation of VP7. Virology. 1995;209:80–89. doi: 10.1006/viro.1995.1232. [DOI] [PubMed] [Google Scholar]
- 14.Machamer C E, Rose J K. Influence of new glycosylation sites on expression of the vesicular stomatitis virus G protein at the plasma membrane. J Biol Chem. 1988;263:5948–5954. [PubMed] [Google Scholar]
- 15.Machamer C E, Rose J K. Vesicular stomatitis virus G proteins with altered glycosylation sites display temperature-sensitive intracellular transport and are subject to aberrant intermolecular disulfide bonding. J Biol Chem. 1988;263:5955–5960. [PubMed] [Google Scholar]
- 16.Mackow E R, Shaw R D, Matsui S M, Vo P T, Dang M N, Greenberg H B. The rhesus rotavirus gene encoding protein VP3: location of amino acids involved in homologous and heterologous rotavirus neutralization and identification of a putative fusion region. Proc Natl Acad Sci USA. 1988;85:645–649. doi: 10.1073/pnas.85.3.645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Marquardt T, Helenius A. Misfolding and aggregation of newly synthesized proteins in the endoplasmic reticulum. Cell Biol. 1992;117:505–513. doi: 10.1083/jcb.117.3.505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mass D R, Atkinson P H. Retention by the endoplasmic reticulum of rotavirus VP7 is controlled by three adjacent amino-terminal residues. J Virol. 1994;68:366–378. doi: 10.1128/jvi.68.1.366-378.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Meyer J C, Bergmann C C, Bellamy A R. Interaction of rotavirus cores with the nonstructural glycoprotein NS28. Virology. 1989;171:98–107. doi: 10.1016/0042-6822(89)90515-1. [DOI] [PubMed] [Google Scholar]
- 20.Mirazimi A, von Bonsdorff C H, Svensson L. Effect of brefeldin A on rotavirus assembly and oligosaccharide processing. Virology. 1996;217:554–563. doi: 10.1006/viro.1996.0150. [DOI] [PubMed] [Google Scholar]
- 21.Persson R, Pettersson R F. Formation and intracellular transport of a heterodimeric viral spike protein complex. J Cell Biol. 1991;112:257–266. doi: 10.1083/jcb.112.2.257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Petrie B L, Estes M K, Graham D Y. Effects of tunicamycin on rotavirus morphogenesis and infectivity. J Virol. 1983;46:270–274. doi: 10.1128/jvi.46.1.270-274.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Poruchynsky M S, Maass D R, Atkinson P H. Calcium depletion blocks the maturation of rotavirus by altering the oligomerization of virus-encoded proteins in the ER. J Cell Biol. 1991;114:651–656. doi: 10.1083/jcb.114.4.651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sabara M, Babiuk L A, Gilchrist J, Misra V. Effect of tunicamycin on rotavirus assembly and infectivity. J Virol. 1982;43:1082–1090. doi: 10.1128/jvi.43.3.1082-1090.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shahrabadi M S, Babiuk L A, Lee P W. Further analysis of the role of calcium in rotavirus morphogenesis. Virology. 1987;158:103–111. doi: 10.1016/0042-6822(87)90242-x. [DOI] [PubMed] [Google Scholar]
- 26.Shaw R D, Vo P T, Offit P A, Coulson B S, Greenberg H B. Antigenic mapping of the surface proteins of rhesus rotavirus. Virology. 1986;155:434–451. doi: 10.1016/0042-6822(86)90205-9. [DOI] [PubMed] [Google Scholar]
- 27.Song J L, Wang C C. Chaperone-like activity of protein disulfide-isomerase in the refolding of rhodanese. Eur J Biochem. 1995;231:312–316. doi: 10.1111/j.1432-1033.1995.tb20702.x. [DOI] [PubMed] [Google Scholar]
- 28.Svensson L, Dormitzer P R, von Bonsdorff C-H, Maunula L, Greenberg H B. Intracellular manipulation of disulfide bond formation in rotavirus proteins during assembly. J Virol. 1994;68:5204–5215. doi: 10.1128/jvi.68.8.5204-5215.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tian P, Hu Y, Schilling W P, Lindsay D A, Eiden J, Estes M K. The nonstructural glycoprotein of rotavirus affects intracellular calcium levels. J Virol. 1994;68:251–257. doi: 10.1128/jvi.68.1.251-257.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang C C, Tsou C L. Protein disulfide isomerase is both an enzyme and a chaperone. FASEB J. 1993;7:1515–1517. doi: 10.1096/fasebj.7.15.7903263. [DOI] [PubMed] [Google Scholar]