Abstract
Available evidence indicates that the transcription of the late class of vaccinia virus genes requires the participation of several virus-encoded proteins in addition to the viral RNA polymerase. In this report we describe the identification of a protein present in extracts of uninfected HeLa cells that binds avidly to viral late promoter DNA. The protein bound specifically to several different vaccinia virus late promoters but not an early nor an intermediate promoter. DNase I footprinting localized the protein’s binding site to nucleotides surrounding the transcriptional start site of the I1L promoter. Optimal promoter binding required sequences in the highly conserved TAAAT motif at the transcriptional start site as well as sequences immediately upstream; however, one variation on the motif’s sequence did not affect promoter binding by the protein. Partially purified late promoter binding protein (LPBP) was capable of stimulating the transcription activity of extracts depleted of LPBP on a late promoter-driven template, establishing LPBP as a transcription activator in vitro. These results suggest that a cellular protein is responsible for targeting vaccinia virus late promoters for initiation of transcription.
Vaccinia virus is a member of the poxvirus family, whose members characteristically have a large DNA genome and replicate in the cytoplasmic compartment of the cell. Progression through the virus life cycle is orchestrated through control of the timing of expression of individual gene products functioning in DNA replication and virus assembly. Generally, proteins functioning in DNA replication are synthesized early in the infectious cycle, and those functioning in assembly are made later (reviewed in reference 13). Vaccinia virus gene expression is regulated primarily at the level of transcription initiation. All viral mRNAs are believed to be synthesized by a virus-encoded, multisubunit RNA polymerase which, despite its complexity, lacks the ability to recognize any of the three classes of transcription promoters in the viral genome. The initiation of transcription of the early, intermediate, and late gene classes by the RNA polymerase appears to require separate and nonoverlapping sets of auxiliary proteins. Early gene transcription appears to require a single protein, ETF, which targets early promoters as sites of initiation of mRNA synthesis (1, 11). Intermediate gene transcription requires three proteins: the viral mRNA capping enzyme (21), the viral E4L gene product (15), and another as yet undefined protein that apparently is not virus encoded (16). Five different proteins have been described as being required for high-level transcription from vaccinia virus late promoters in vivo and/or in vitro. These include the vaccinia virus A1L (9), A2L (8, 14), G8R (25), and H5R (10) gene products. A fifth factor, termed VLTF-X (8), has not been assigned to a vaccinia virus gene. Two other proteins, the products of the G2R (5) and A18L (3) genes, have been implicated in elongation and/or termination of late transcripts. The specific role of any of these proteins in the transcription of late genes is not known, nor is it known which, if any, is responsible for targeting late promoters as sites for initiation of transcription. In this report, we describe a cellular protein that has high affinity for late promoters and activates transcription in vitro.
MATERIALS AND METHODS
Promoter DNAs.
All late promoter DNAs were originally generated by PCR on viral genomic DNA with Vent DNA polymerase (New England Biolabs). For the 11-kDa (gene F18R) and I1L promoters. PCR primers were designed to produce DNA fragments that included nucleotides −100 to +46, relative to the first A residue in the TAAAT motif at the start site for transcription. PCR products were ligated into the SmaI site of plasmid pBluescript II KS(+) (Stratagene). The sequences of all plasmid inserts were confirmed by thermal cycle DNA sequencing with a kit (Epicenter Technologies) according to the manufacturer’s instructions. For all promoters except H1L, probes were constructed by excision at flanking restriction sites and 3′ end labeled with the Klenow fragment of DNA polymerase I and an α-32P-labeled deoxynucleoside triphosphate (18) whose choice was determined by the restriction enzyme cleavage site. The H1L promoter probe was constructed by PCR with primers previously labeled at their 5′ ends with polynucleotide kinase and [γ32P]ATP. The H1L probe was designed include nucleotides −80 to +40. The vaccinia virus growth factor gene (C11R) early promoter DNA (nucleotides −35 to +15, relative to the transcriptional start site) and the I3L gene intermediate promoter (nucleotides −35 to +15, relative to the transcriptional start site) were excised from plasmids pSB63 and pSB104, respectively, and similarly radiolabeled at their 3′ ends. All DNA probes were purified by polyacrylamide gel electrophoresis.
DNA affinity matrices were constructed with the 11-kDA and I1L promoter fragments described above. Promoter fragments were released from plasmids by cleavage with restriction endonucleases at flanking sites and were tagged with biotin on their 3′ ends with the Klenow fragment of DNA polymerase I and biotin-16-dUTP. Biotinylated DNA was linked to streptavidin-conjugated paramagnetic beads (Boehringer Mannheim) according to the manufacturer’s instructions.
Cell extracts and protein purification.
HeLa S-3 cells were grown in suspension culture to a density of 5 × 105/ml in volumes up to 12 liters and were harvested for extraction. For virus infections, HeLa cells growing in monolayers in 150-cm2 dishes were infected with vaccinia virus western reserve strain at a ratio of 10 PFU/cell. After virus absorption in 3 ml of medium for 2 h, fresh medium was substituted and the cells were incubated for 16 h, unless otherwise stated, prior to being harvested. All further manipulations were conducted at 4°C. Protein purification was initiated with 2 × 1010 cells. Cell pellets were resuspended in 40 ml of 10 mM Tris (pH 9.0)–0.5 mM phenylmethylsulfonyl fluoride and allowed to swell on ice for 30 min. Cell lysis was completed by Dounce homogenization with a tight-fitting pestle. Nuclei and cell debris were removed by centrifugation at 12,000 × g for 30 min. The supernatant was mixed with 18 ml of Ni2+-nitrilo-agarose (Ni-agarose; Qiagen) and incubated with rocking for 3 h. The resin was washed three times by centrifugation and resuspension in 40 ml of buffer A (150 mM NaCl, 50 mM Tris [pH 8.0], 0.01% Nonidet P-40). A final wash was done with 20 mM imidazole in buffer A. Protein was eluted with two 10-ml washes with 300 mM imidazole in buffer A.
Ni-agarose-purified protein was mixed with an equal volume of buffer B (50 mM Tris [pH 8.0], 0.1 mM EDTA, 1 mM dithiothreitol, 0.01% Nonidet P-40, 10% glycerol) and applied to a 1-ml phosphocellulose column equilibrated with 0.1 M NaCl in buffer B. After the protein was washed with the same solution, promoter binding activity was eluted with 0.3 M NaCl in buffer B. The phosphocellulose eluate was diluted with 2 volumes of buffer B and mixed with 100 μl of a late promoter DNA affinity matrix. The beads were washed with 0.1 M NaCl in buffer B, and late promoter binding activity was eluted with 0.3 M NaCl in buffer B. DNA affinity-purified protein was layered onto an 11-ml 15 to 35% glycerol gradient in buffer B containing 0.2 M NaCl and subjected to velocity sedimentation at 40,000 rpm for 48 h in a Beckman SW41 rotor. Fractions were collected by pumping from the tube bottom. The late promoter binding activity was found to sediment at a rate of about 3.8S relative to sedimentation standards. The protein preparation at this stage is best characterized as partially purified, and we were not able to assign a particular polypeptide(s) as being responsible for promoter binding activity.
DNA binding assays.
Late promoter DNA binding was determined by electrophoretic mobility shift analysis essentially as described previously (4). A typical binding reaction mixture contained 1 to 2 ng of radiolabeled probe DNA. DNA probes were mixed with poly(dI-dC) (Pharmacia), where indicated, prior to the addition of protein. Protein-DNA complexes were resolved by electrophoresis in a nondenaturing polyacrylamide gel and visualized by autoradiography (4).
DNase I footprinting experiments were performed with 32P-labeled promoter DNA segments. The DNA was incubated either alone or with 100 ng of protein in 50 mM Tris (pH 8.0)–1 mM MgCl2 for 30 min at 22°C. DNase I (Worthington Biochemicals) was added to a concentration of 8 ng/ml, and incubation was continued for an additional 2 min, after which the cleavage reaction was terminated by addition of an equal volume of a stop solution (0.2 M NaCl, 30 mM EDTA, 1% sodium dodecyl sulfate, and 100 μg of glycogen/ml). DNA cleavage products were extracted with phenol-chloroform, precipitated with ethanol, and analyzed on a 6% polyacrylamide DNA sequencing gel. Sequence markers were generated by subjecting the DNA fragments to a G+A chemical cleavage reaction (12).
Reporter gene experiments.
The green fluorescent protein (GFP) from Aequorea victoria was used as a reporter for I1L promoter activity. The GFP coding sequences were isolated from plasmid pEGFP-N1 (Clontech) (6) by PCR with Vent polymerase as described above. A NdeI restriction site was engineered into the sequence at the initiation codon, and the fragment was inserted into the SmaI site of plasmid pBluescript II KS(+). Double-stranded synthetic oligonucleotides with the indicated sequences were ligated between the NdeI site and an upstream ClaI site to generate the final promoter-GFP gene constructs. Plasmids were purified on Qiagen columns as recommended by the manufacturer. Ten micrograms of plasmid was transfected by the calcium phosphate precipitation method (18) into 106 HeLa cells that were previously infected with 10 PFU of vaccinia virus/cell. After 17 h, the cells were harvested by scraping and lysed by resuspension in 1 mM Tris (pH 9.0) and Dounce homogenization. Cell debris was removed by sedimentation at 10,000 × g for 20 s, and the supernatant was assayed for fluorescence at 510 nm upon excitation at 475 nm in a Hitachi F-2000 fluorescence spectrophotometer. Fluorescence values were normalized to the mass of the protein as determined with Bio-Rad protein reagent, using bovine serum albumin as a standard.
In vitro transcription reactions.
Cytoplasmic extracts of 108 HeLa cells infected with virus for 16 h were mixed with 0.2 ml of Ni-agarose and placed on a rocking platform for 1 h at 4°C. The beads were subjected to a brief centrifugation, and the supernatant was retained as the “depleted extract.” Transcription reactions were conducted in solutions that were essentially identical to those previously described for early gene transcription (11). The template for transcription was pCFW9, which has the 11-kDa promoter followed by a 400-nucleotide G-less cassette (23). The template was mixed with the indicated amounts of extract, purified promoter binding protein, buffer, and nucleotides, using [α-32P]UTP as the label, and incubated at 30°C for 30 min. At that time, 100 U of RNase T1 (Sigma) was added, and the incubation was resumed for an additional 15 min. The reaction products were extracted with phenol-chloroform, precipitated with ethanol, and resolved by electrophoresis on a denaturing 4% polyacrylamide gel (11). The gel was dried and exposed to X-ray film for autoradiography. Radiolabeled RNA was quantitated with a Packard InstantImager.
RESULTS
Detection of a late promoter DNA binding protein in cell extracts.
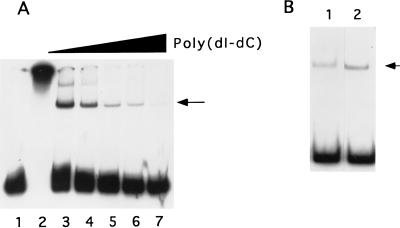
During the course of experiments on histidine-tagged recombinant proteins expressed by vaccinia virus-based expression systems, we detected a protein with vaccinia virus late promoter binding activity that bound Ni-agarose. The vaccinia virus 11-kDa (F18R gene) promoter was used initially for binding experiments because it is a strong late promoter and its sequence requirements for promoter activity in vivo have been characterized in some detail (2). The minimal promoter requires less than 20 nucleotides 5′ to the transcriptional start site, with little obvious requirement for sequences 3′ to the start site. To ensure that all required promoter elements were present, DNA binding probes were designed to encompass nucleotides −100 to +46, relative to the first A in the TAAAT motif at the transcriptional start site. Electrophoretic mobility shift analysis with the 11-kDa promoter DNA demonstrated that protein purified from cytoplasmic extracts by Ni-agarose chromatography had nonspecific DNA binding activity, resulting in aggregation of the probe (Fig. 1A, lane 2); however, a discrete protein-DNA complex was observed when binding reactions were conducted in the presence of low concentrations of the nonspecific competitor poly(dI-dC). The complex was resistant to a 1,000-fold excess of the polymer, suggesting that binding was specific. Subsequent protein fractionation experiments showed that the protein with promoter binding activity was distinct from the expressed recombinant proteins (data not shown) and, indeed, was not dependent on virus infection. Extracts from uninfected HeLa cells contained levels of promoter binding activity similar to those of cells infected with virus (Fig. 1B), indicating that the late promoter binding protein is not virus encoded or virus induced.
FIG. 1.
(A) Detection of late promoter binding activity in Ni-agarose-purified protein. Extracts from vaccinia virus-infected cells were chromatographed on Ni-agarose and assayed for binding to the 32P-labeled 120-nucleotide 11-kDa promoter probe. Lane 1 contains probe alone; lanes 2 to 7 contain probe plus 200 ng of protein. Poly(dI-dC) was included in binding reactions at 0 (lane 2), 100 (lane 3), 200 (lane 4), 500 (lane 5), 1,000 (lane 6), and 2,000 ng (lane 7). Protein-DNA complexes were resolved by electrophoresis in a native gel and visualized by autoradiography. The location of the major protein-probe complex is indicated with an arrow. (B) Ni-agarose-purified protein from uninfected HeLa cells (lane 1) or vaccinia virus-infected cells (lane 2) was tested for binding to the 11-kDa promoter probe. The major protein-probe complex is indicated with an arrow.
To determine whether the late promoter binding activity identified in uninfected cells and that found in vaccinia virus-infected cells were due to the same protein, the protein with late promoter binding activity was purified from both uninfected and virus-infected cells in parallel as described in Materials and Methods. The two proteins behaved identically when chromatographed on Ni-agarose and phosphocellulose, after purification by DNA affinity matrix and glycerol gradient sedimentation and produced protein-promoter DNA complexes with the same mobility in an electrophoretic gel shift experiment (Fig. 1B). It was concluded that the late promoter binding protein is of cellular origin and is not significantly altered by virus infection. All further experiments were conducted with protein purified from uninfected cells.
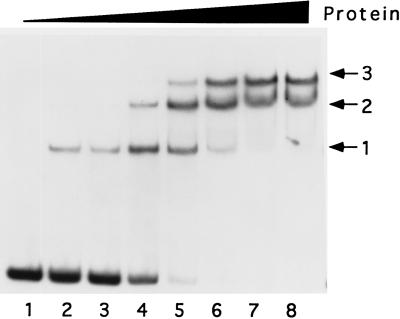
The promoter binding activity of the cellular protein was further characterized by titration of the protein on the 11-kDa promoter (Fig. 2). At low concentrations of protein, a single complex was observed (complex 1). As the protein concentration was increased, two additional slower-migrating complexes (complexes 2 and 3) were observed and appeared to accumulate at the expense of the faster-moving complex. The level of binding of the three complexes together was quantitated, and Scatchard analysis of the data yielded a Kd of 3 nM. This number can be considered to be an averaged value of the affinity of the three combined complexes. It is emphasized that formation of multiple protein-promoter DNA complexes appears to be a feature unique to the 11-kDa promoter. With all other late promoters tested, a single major complex has been observed.
FIG. 2.
Titration of the 11-kDa promoter with Ni-agarose-purified protein. 32P-labeled 120-nucleotide 11-kDa promoter DNA was mixed with 0 (lane 1), 100 (lanes 2 and 3), 200 (lane 4), 300 (lane 5), 400 (lane 6), 500 (lane 7), and 600 ng (lane 8) of protein in the presence of 40 ng of poly(dI-dC). The mobilities of complexes 1, 2, and 3 are indicated. The reaction corresponding to lane 3 included 1 mM MgCl2 in addition to the standard binding buffer.
Specificity of DNA binding.
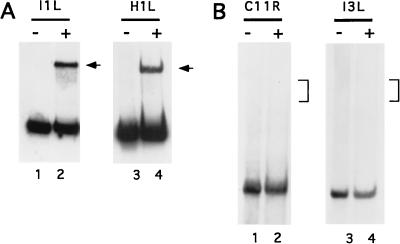
Late promoters other than the 11-kDa promoter were tested for binding to the cellular protein. The I1L (19) and H1L (17) genes have been described as being transcribed from promoters that are uniquely late. The promoters for both of these genes were found to form complexes with the cellular protein, at levels similar to that of the 11-kDa promoter (Fig. 3A). In contrast, the vaccinia virus growth factor (gene C11R) promoter, a representative early promoter (20), and the I3L intermediate promoter, a representative intermediate promoter (22), did not form detectable complexes with the protein (Fig. 3B).
FIG. 3.
Promoter specificity for protein binding. (A) Ni-agarose-purified protein binding to the I1L (lanes 1 and 2) and H1L (lanes 3 and 4) late promoters. Lanes 1 and 3 contain probe alone, and lanes 2 and 4 contain probe plus 300 ng of protein. Major protein-DNA complexes are indicated with arrows. (B) Protein binding to the VGF early (gene C11R) promoter (lanes 1 and 2) and I3L intermediate promoter (lanes 3 and 4). Lanes 1 and 3 contain probe alone, and lanes 2 and 4 contain probe plus 300 ng of protein. The brackets indicate the expected mobilities of protein-DNA complexes with fragments of these sizes.
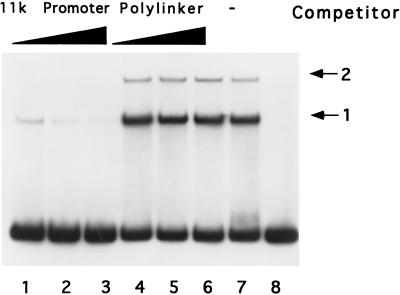
The specificity of DNA binding was also assessed in competition binding experiments in which the effect of excess nonradiolabeled DNA fragments on binding to 32P-labeled 11-kDa promoter was determined. Excess 11-kDa promoter DNA predictably reduced binding to the 11-kDa promoter DNA to a level that was almost undetectable at an 80-fold molar excess (Fig. 4). Challenge of 11-kDa promoter DNA binding with the same excess of an unrelated but similarly sized DNA fragment derived from the polylinker of plasmid pBluescript II KS(+) had little effect. Clearly, this protein has specificity for late promoter DNA relative to other DNAs tested. Hereafter we will refer to the protein as the late promoter binding protein (LPBP) until its identity becomes known.
FIG. 4.
Competition assay for late promoter binding specificity. Binding assays were conducted with 100 ng of Ni-agarose-purified protein and 32P-labeled 120-nucleotide 11-kDa promoter probe in the presence of 40 ng of poly(dI-dC). As a competitor, 0 (lane 7), 20 (lanes 1 and 4), 40 (lanes 2 and 5), or 80 ng (lanes 3 and 6) of nonlabeled 120-nucleotide 11-kDa promoter DNA or plasmid DNA was included in the reaction. Lane 8 is probe alone. The mobilities of protein-DNA complexes 1 and 2 are indicated.
Localization of protein binding in a late promoter.
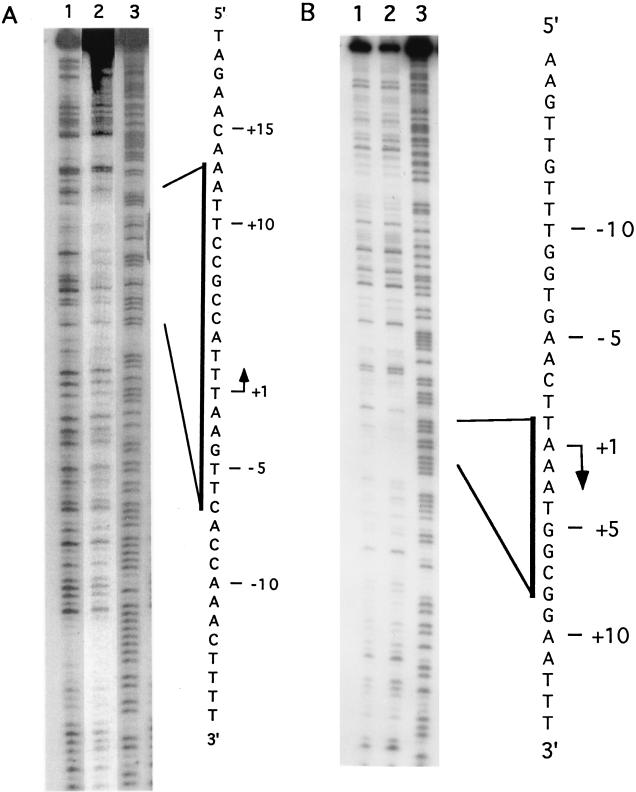
DNase I footprinting was used to localize the site(s) of interaction of the LPBP with late promoter DNAs. Initially the 11-kDa promoter was selected for this purpose, since its ability to support transcription in vivo was previously characterized by deletion analysis (2). When complex 1 (as shown in Fig. 3) was analyzed by DNase I footprinting, no protected sequences were observed, and when complexes 2 and 3 were analyzed, a general protection of the entire probe DNA was observed (data not shown). Because the 11-kDa promoter had the unusual property of forming multiple stable complexes with LPBP, it seemed likely that this promoter has multiple binding sites for LPBP, and the identification of the boundaries for any one specific site might prove difficult. Therefore, the I1L promoter was chosen for further study. Protection from DNase I cleavage by LPBP was detected on the template strand extending from nucleotides −8 to +13, relative to the first A residue in the TAAAT motif at the transcriptional start site (Fig. 5). On the nontemplate strand, weak protection at nucleotides −1 to +8 was observed.
FIG. 5.
DNase I footprinting of protein bound to the I1L promoter. I1L promoter 120-nucleotide DNA uniquely labeled on the template (A) or nontemplate (B) strand was treated with DNase I in the absence (lanes 1) or presence (lanes 2) of glycerol gradient-purified LPBP. Cleavage products were resolved on a 6% polyacrylamide DNA sequencing gel. Lanes 3 contain probe DNA chemically cleaved at purine residues. The nucleotide sequence of each strand of the I1L promoter is shown on the right.
Nucleotide sequences in the I1L promoter required for interactions with LPBP.
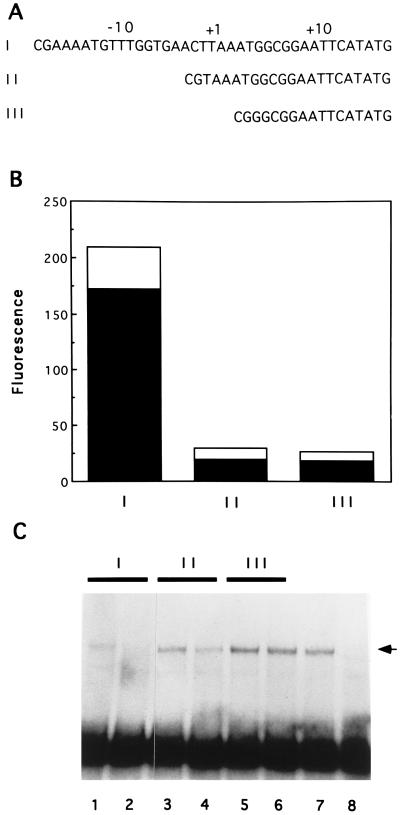
Our understanding of the architecture of vaccinia virus late promoters remains poor. A thorough mutagenesis study of the sequences immediately surrounding the transcriptional start sites of the vaccinia virus 28-kDa promoter and a synthetic promoter identified only the TAAAT motif as important for transcription (7). Runs of T residues 5′ to this motif were implicated in the function of synthetic promoter constructs, but the promoter elements upstream of the TAAAT motif have not been precisely defined. The vaccinia virus 11-kDa promoter sequence requirements for transcriptional activity in vivo have also been characterized. Promoter deletion studies indicated that deletion of upstream nucleotides to −17, relative to the first A in the TAAAT motif, permitted high-level transcription activity; however, further deletion to nucleotide −1 resulted in complete inactivation of the promoter (2). We have performed a similar analysis of the I1L promoter. Nucleotides −16 to +13 of the I1L promoter (referred to here as full length) were linked to the coding sequence for GFP. Transfection of this construct into vaccinia virus-infected cells resulted in significant fluorescence of the reporter protein (Fig. 6B). The DNA synthesis inhibitor, cytosine arabinoside, inhibited the appearance of fluorescence (data not shown), consistent with the promoter being a late class vaccinia virus promoter. Deletion of the I1L promoter to −1 resulted in a decrease in fluorescence to about 10% of that of the full-length promoter. Further deletion of the promoter to +5, removing the TAAAT element, did not result in further reduction of expression of GFP. These results suggest the existence of an important promoter element upstream of the TAAAT motif. These same oligonucleotides were then tested for their affinity for LPBP in a competition assay in which binding of LPBP to the full-length I1L promoter was challenged by an excess of DNA fragments bearing part or all of the I1L promoter. As expected, full-length promoter I (nucleotides −16 to +13) effectively competed with itself for binding to LPBP (Fig. 6C). Essentially all binding activity was lost in the presence of a 100-fold excess of competitor. Promoter II (nucleotides −1 to +13) was able to partially compete for binding to LPBP but was estimated to have an approximately fivefold reduction in affinity relative to that of the full-length promoter. Promoter III (nucleotides +5 to +13), which lacks the TAAAT motif, was completely ineffective in competing with promoter I for LPBP binding. Because deletion to nucleotide −1 partially attenuated binding and loss of the TAAAT motif resulted in nearly total loss of binding by LPBP, these results suggest that LPBP requires sequences both upstream of and within the TAAAT motif in order to interact with the I1L promoter.
FIG. 6.
Effect of promoter deletions on activity of the I1L promoter in vivo and on binding to LPBP. (A) Nucleotide sequences of promoters bearing the vaccinia virus DNA nucleotides −16 to +13 (promoter I), −1 to +13 (promoter II), or +5 to +13 (promoter III). Nucleotides are numbered in reference to the first A residue in the TAAAT motif. The open reading frame encoding GFP begins with the ATG beginning at nucleotide +17. (B) Fluorescence from cell extracts resulting from GFP expression driven by promoter constructs I, II, and III. Solid bars represent fluorescence normalized to the mass of the protein, and open bars indicate the mean standard deviations of the values from three experiments. (C) Competition assays for affinity of LPBP for promoters I, II, and III. 32P-labeled DNA fragments bearing the sequence of promoter I were exposed to glycerol gradient-purified LPBP in the presence of a 50- (lanes 1, 3, and 5) or 100-fold (lanes 2, 4, and 6) molar excess of DNA fragments with the sequence of promoter I (lanes 1 and 2), promoter II (lanes 3 and 4), or promoter III (lanes 5 and 6). Protein-DNA complexes were resolved by electrophoretic mobility shift. The arrow at the right indicates the mobility of the DNA-protein complex. Lane 7 contains LPBP binding in the absence of a competitor, and lane 8 contains probe alone.
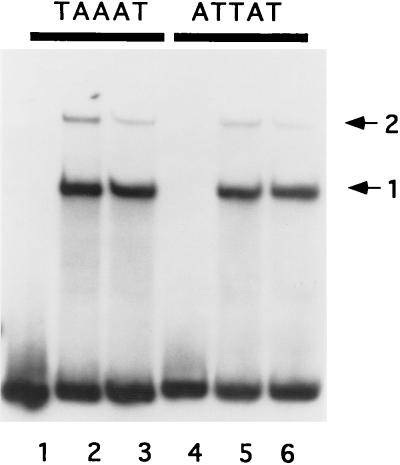
The most highly conserved feature of vaccinia virus late promoters is the TAAAT sequence at the site of initiation of transcription. The effects of replacement of the nucleotides in this motif vary among different late promoters, but the A residues in the motif appear to be absolutely essential for transcription activity in vivo (7). Because the binding site for LPBP was localized to the vicinity of this sequence in the I1L promoter, it was of interest to determine whether the TAAAT sequence is a determinant for binding by LPBP. To test this idea, the abilities of DNA segments with the wild-type 11-kDa promoter sequence and with a mutant variant of the 11-kDa promoter to bind to LPBP were compared. In the mutant variant, the first three nucleotides of the TAAAT motif are replaced with ATT, a change expected to have severe effects on late promoter binding in vivo (7). LPBP bound both the wild-type and mutant variant probes to the same extent (Fig. 7). Because the 11-kDa promoter appears to have multiple LPBP binding sites, a promoter (I1L) with an apparently single binding site was also examined. Identical results were seen when the binding of LPBP to the I1L promoter and to the mutant variant of the 11-kDa promoter were compared (data not shown).
FIG. 7.
An alteration of the TAAAT motif in the 11K promoter does not affect LPBP binding. Ni-agarose-purified LPBP (100 ng) was mixed with double-stranded oligonucleotide probe with the sequence of the 11K promoter (nucleotides −38 to +18) bearing the wild-type TAAAT motif at the transcriptional start site, or the substituted ATTAT motif, in the presence of 40 ng of poly(dI-dC). Protein-DNA complexes were resolved by native polyacrylamide gel electrophoresis. The primary complexes (labeled 1 and 2) are indicated by the arrows.
Transcription factor activity of LPBP.
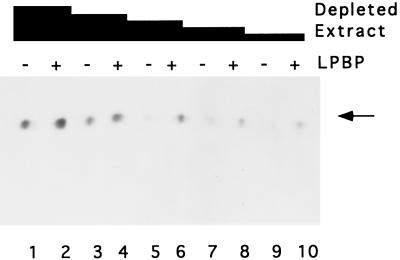
Assigning a functional significance to promoter binding by LPBP required a demonstration of an effect on transcription. Because a system for transcription from vaccinia virus late promoters has yet to be defined, experiments with purified proteins are not possible. As an alternative, we used an approach in which extracts from HeLa cells infected with vaccinia virus were depleted of LPBP by adsorption to Ni-agarose; we then asked whether purified LPBP could restore transcription activity on a G-less template driven by the 11-kDa promoter. High concentrations of the depleted extracts were capable of catalyzing transcription reactions that were only marginally stimulated by addition of LPBP (Fig. 8). As the concentration of extract added to the reaction was reduced, there was a concomitant reduction in the level of transcripts produced. Significantly, purified LPBP was capable of stimulating the transcription activity of reactions having lower concentrations of depleted extracts. The presence of LPBP in these reactions stimulated RNA product formation by an estimated fivefold. The residual transcription activity of the depleted extracts suggests that the extracts were possibly not completely depleted of LPBP by exposure to Ni-agarose, and the dependence of transcription reactions on LPBP was not apparent until the concentration of protein in the reaction was reduced. We cannot determine the extent of depletion of LPBP from the extracts because promoter binding cannot be detected in crude cytoplasmic extracts (data not shown).
FIG. 8.
Transcriptional activation by LPBP. Cytoplasmic extract from virus-infected cells was depleted of LPBP by adsorption to Ni-agarose. Transcription reactions with 195 (lanes 1 and 2), 190 (lanes 3 and 4), 180 (lanes 5 and 6), 170 (lanes 7 and 8), or 160 μg (lanes 9 and 10) of depleted extract (expressed as total protein) were conducted on a G-less cassette DNA template driven by the 11-kDa promoter either in the absence (lanes 1, 3, 5, 7, and 9) or presence of 25 (lane 2), 50 (lane 4), 100 (lane 6), 150 (lane 8), or 200 ng (lane 10) of purified LPBP. The 400-nucleotide RNA products (indicated by the arrow) were resolved by denaturing polyacrylamide gel electrophoresis.
DISCUSSION
During the course of recombinant protein expression studies, we have detected a protein with vaccinia virus late promoter binding activity. This protein, LPBP, exhibits high specificity for late promoters and apparently has little or no affinity for the other two classes of viral promoters. It has a high affinity for late promoters, however, with a Kd in the nanomolar range, a concentration consistent with a function as a promoter DNA binding protein in vivo. LPBP also appears to be a general late promoter binding protein; it has measurable affinities for all late promoters we have tested, although the specific affinities for various late promoters have been observed to vary considerably (data not shown). Whether the affinity of LPBP for late promoters is a determinant of promoter strength, as is believed to be the case for the vaccinia virus early transcription factor, is unknown. The ability of LPBP to stimulate transcription in vitro supports the case for its functioning as a bona fide transcription factor.
Deletion of sequences upstream of the TAAAT motif in the I1L promoter resulted in loss of promoter activity. This is in agreement with a report from Bertholet et al. on their analysis of the 11-kDa promoter (2). Deletion of nucleotides upstream of −1 in the I1L promoter resulted in incomplete impairment of binding of LPBP. Complete impairment of binding was not achieved until the TAAAT motif was also deleted. These results indicate that LPBP recognized the I1L promoter through contacts in and 5′ to the TAAAT motif.
The nearly absolute conservation of the TAAAT motif at the start site for transcription of late genes and the demonstration that this motif is absolutely essential for late promoter activity (7) are compelling arguments for a vital function for this sequence. While it is clear that this site is used to generate the 5′ poly(A) leader on late mRNAs, it is not unreasonable to predict that the TAAAT motif targets a protein to late promoters as a means to initiate the assembly of a preinitiation complex. Because the LPBP binding site was localized to this region in the I1L promoter, the effect of a substitution in the TAAAT motif was tested for its effect on LPBP binding. While we did not test the effect of the substitution on I1L promoter activity, it is expected to result in essentially total loss of activity. The A residues in this motif have been shown in two cases to be absolutely essential for promoter activity in vivo (7). When the first two A residues in the I1L motif were replaced with T residues, no effect on LPBP binding was observed. We have analyzed the effect of only one variation on the TAAAT motif with respect to interaction with LPBP, but it would appear that interaction with this protein is not the sole function of the TAAAT motif. The basis for the importance of this motif in vaccinia virus late transcription remains uncertain, and further characterization of late viral promoter function is clearly needed.
The finding that LPBP is a cellular protein was unexpected. The poxvirus mRNA synthesis machinery is generally regarded as a virtually autonomous system operating through the activities of a number of virus-encoded proteins. It is clear that LPBP is not virus encoded. It is readily detected in uninfected cells. There is precedent, however, for a host protein functioning as a poxvirus transcription factor. The intermediate promoter-specific factor referred to as VITF-2 (15) has been shown by Rosales et al. (16) to be present in uninfected cells.
The functional significance behind the vaccinia virus strategy of adopting a host protein to control the expression of its late genes, many of which encode highly expressed structural proteins, is unclear. Late transcription is activated only after genome replication begins and continues after the genome is highly amplified in the cell. If most late promoters in most of the amplified genomes are active, then the protein that targets late promoters would be expected to be required at high levels in the cell. Perhaps through the utilization of a preexisting protein that is present in abundance, the virus can ensure that accumulation of LPBP is not the rate-limiting step in the activation of late transcription. Conceptually, this is similar to the situation for early gene transcription, in which the early gene transcription machinery is preexistent in the viral core, awaiting only the uncoating process to initiate high-level transcription. Having LPBP present in the cell prior to activation of late transcription would suggest that the initiation of DNA synthesis and the ensuing activation of intermediate promoters is the primary control point mediating the switch from early to late gene transcription.
It is worth noting that several properties exhibited by LPBP are similar to those of the factor VLTF-X described by Wright and Coroneos (24). Both proteins have affinity for Ni-agarose, both behave similarly upon chromatography on phosphocellulose, and both appear to activate transcription in vitro. VLTF-X is reportedly virus induced (24), however, and LPBP is present in uninfected cells. Whether VLTF-X and LPBP are the same protein remains to be determined.
ACKNOWLEDGMENTS
This work was supported by an award from the National Institute of Allergy and Infectious Diseases.
We are grateful to Gunter Kohlhaw and Jonathan LeBowitz for comments on the manuscript and to Li Ni and Henry Weiner for help with the use of their fluorescence spectrophotometer.
REFERENCES
- 1.Baldick C J, Jr, Cassetti M C, Harris N, Moss B. Ordered assembly of a functional preinitiation transcription complex, containing vaccinia virus early transcription factor and RNA polymerase, on an immobilized template. J Virol. 1994;68:6052–6056. doi: 10.1128/jvi.68.9.6052-6056.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bertholet C, Stocco P, Van Meir E, Wittek R. Functional analysis of the 5′ flanking sequence of a vaccinia virus late gene. EMBO J. 1986;5:1951–1957. doi: 10.1002/j.1460-2075.1986.tb04449.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Black E P, Condit R C. Phenotypic characterization of mutants in vaccinia virus gene G2R, a putative transcription elongation factor. J Virol. 1996;70:47–54. doi: 10.1128/jvi.70.1.47-54.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Broyles S S, Yuen L, Shuman S, Moss B. Purification of a factor required for transcription of vaccinia virus early genes. J Biol Chem. 1988;263:10754–10760. [PubMed] [Google Scholar]
- 5.Condit R C, Xiang Y, Lewis J I. Mutation of vaccinia virus gene G2R causes suppression of gene A18R ts mutants: implications for control of transcription. Virology. 1996;220:10–19. doi: 10.1006/viro.1996.0280. [DOI] [PubMed] [Google Scholar]
- 6.Cormack B P, Valdivia R H, Falkow S. FACS-optimized mutants of the green fluorescent protein (GFP) Gene. 1996;173:33–38. doi: 10.1016/0378-1119(95)00685-0. [DOI] [PubMed] [Google Scholar]
- 7.Davison A J, Moss B. The structure of vaccinia virus late promoters. J Mol Biol. 1989;210:771–784. doi: 10.1016/0022-2836(89)90108-3. [DOI] [PubMed] [Google Scholar]
- 8.Hubbs A E, Wright C W. The A2L intermediate gene product is required for in vitro transcription of a vaccinia virus late promoter. J Virol. 1996;70:327–331. doi: 10.1128/jvi.70.1.327-331.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Keck J G, Kovacs G R, Moss B. Overexpression, purification, and late transcription factor activity of the 17-kDa protein encoded by the vaccinia virus A1L gene. J Virol. 1993;67:5740–5748. doi: 10.1128/jvi.67.10.5740-5748.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kovacs G R, Rosales R, Keck J G, Moss B. Modulation of the cascade model for regulation of vaccinia virus gene expression: purification of a prereplicative, late-stage-specific transcription factor. J Virol. 1994;68:3443–3447. doi: 10.1128/jvi.68.5.3443-3447.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li J, Broyles S S. Recruitment of vaccinia virus RNA polymerase to early gene promoters by the viral early transcription factor. J Biol Chem. 1993;268:2273–2280. [PubMed] [Google Scholar]
- 12.Maxam A M, Gilbert W. A new method for sequencing DNA. Proc Natl Acad Sci USA. 1977;74:560–564. doi: 10.1073/pnas.74.2.560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Moss B. Poxviridae: the viruses and their replication. 3rd ed. Philadelphia, Pa: Lippincott-Raven Publishers; 1996. [Google Scholar]
- 14.Passarelli A L, Kovacs G R, Moss B. Transcription of a vaccinia virus late promoter template: requirement for the product of the A2L intermediate-stage gene. J Virol. 1996;70:4444–4450. doi: 10.1128/jvi.70.7.4444-4450.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rosales R, Harris N, Ahn B-Y, Moss B. Purification and identification of a vaccinia virus-encoded intermediate stage promoter-specific transcription factor that has homology to eukaryotic transcription factor SII(TFIIS) and an additional role as a viral RNA polymerase subunit. J Biol Chem. 1994;269:14260–14267. [PubMed] [Google Scholar]
- 16.Rosales R, Sutter G, Moss B. A cellular factor is required for transcription of vaccinia virus intermediate stage genes. Proc Natl Acad Sci USA. 1994;91:3794–3798. doi: 10.1073/pnas.91.9.3794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rosel J L, Earl P L, Weir J P, Moss B. Conserved TAAATG sequence at the transcriptional and translational initiation sites of vaccinia virus late genes deduced by structural and functional analysis of the HindIII H genome fragment. J Virol. 1986;60:436–449. doi: 10.1128/jvi.60.2.436-449.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 19.Schmitt J F C, Stunnenberg H G. Sequence and transcriptional analysis of the vaccinia virus HindIII I fragment. J Virol. 1988;62:1889–1897. doi: 10.1128/jvi.62.6.1889-1897.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Venkatesan S, Baroudy B M, Moss B. Distinctive nucleotide sequences adjacent to multiple initiation and termination sites of an early vaccinia virus gene. Cell. 1981;25:805–813. doi: 10.1016/0092-8674(81)90188-4. [DOI] [PubMed] [Google Scholar]
- 21.Vos J C, Sasker M, Stunnenberg H G. Vaccinia virus capping enzyme is a transcription initiation factor. EMBO J. 1991;10:2552–2558. doi: 10.1002/j.1460-2075.1991.tb07795.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Vos J C, Stunnenberg H G. Derepression of a novel class of vaccinia virus genes upon DNA replication. EMBO J. 1988;7:3487–3492. doi: 10.1002/j.1460-2075.1988.tb03224.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wright C F, Moss B. Identification of factors specific for transcription of the late class of vaccinia virus genes. J Virol. 1989;63:4224–4233. doi: 10.1128/jvi.63.10.4224-4233.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wright C W, Coroneos A M. Purification of the late transcription system of vaccinia virus: identification of a novel transcription factor. J Virol. 1993;67:7264–7270. doi: 10.1128/jvi.67.12.7264-7270.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wright C F, Keck J G, Tsai M M, Moss B. A transcription factor for expression of vaccinia virus late genes is encoded by an intermediate gene. J Virol. 1991;65:3715–3720. doi: 10.1128/jvi.65.7.3715-3720.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]