Abstract
Three transcripts from the terminal repeat of the channel catfish virus (CCV; also known as ictalurid herpesvirus 1) genome were mapped by S1 nuclease and primer extension analyses as well as by cDNA sequencing. These transcripts, TR3, TR5/6, and TR6, are encoded by open reading frame (ORF) 3, ORFs 5 and 6, and ORF 6, respectively, and correspond to those previously identified by sequence analysis (A. J. Davison, Virology 186:9–14, 1992). ORF 5 has previously been determined to encode thymidine kinase, but ORF 3 and ORF 6 encode proteins of unknown function. Although all three transcripts accumulate to high levels in cells infected in the presence of cycloheximide, kinetic analysis demonstrates that TR5/6 and TR6 are either early or late transcripts that leak through the cycloheximide block. In addition, two transcripts from the terminal repeat of the CCV genome that were mapped previously and were thought to be immediate-early in character, TR8a/9 and TR9, exhibit kinetics characteristic of early or late transcripts. TR3 is an immediate-early transcript that appears to have a very short half-life. In the 3′ untranslated region of TR3, there are three copies of an AU-rich element which has previously been shown to be involved in destabilization of the oncogene c-fos and granulocyte/macrophage colony-stimulating factor mRNAs. mRNA destabilization may represent another mechanism by which herpesviruses regulate the rapid switch in expression from immediate-early genes to early genes during the transition to the early phase of infection.
Channel catfish virus (CCV), also known as ictalurid herpesvirus 1, may cause fatal disease in 40 to 90% of channel catfish fry, an economically important agricultural species in the southern United States. Discovered by Fijan in 1968 (3), CCV was classified as a herpesvirus based on morphology (23). Analysis of the genome by DNA sequencing has indicated that the CCV genome consists of a unique sequence of 97 kbp bounded by identical, direct terminal repeats of 18.5 kbp, for a total genomic size of 134 kbp. Sequence data indicate that this herpesvirus is very different in genetic content and sequence from all other herpesviruses whose genomes have been sequenced (2).
To understand viral latency and pathogenesis, we set out to identify and characterize the immediate-early (IE) genes of CCV. Previous results demonstrated that IE transcription is restricted to the repeated regions of the genome (19). Four fragments from the terminal repeat regions which apparently encode IE transcripts were cloned into plasmid vectors. Two 3′ coterminal transcripts, ie1 and ie2, encoded by one of the cloned fragments (pPS3927) were identified and mapped. Both ie1 and ie2 contain open reading frame (ORF) 9, and ie2 also contains ORF 8a. ORF 8 and ORF 9 were identified by Davison (2), and ORF 8a is a 5′-truncated version of ORF 8 (2, 19). ie1 is expressed at a high level in cells translationally blocked with cycloheximide, while ie2 is expressed at lower, albeit detectable levels under the same conditions of CCV infection. ie1 appeared to be a classical IE gene that encodes a protein with a basic pI and a zinc-binding motif, both characteristics which might be expected in a transcription factor. However, ie2 encodes a protein which is predicted to be membrane associated, making it an unusual candidate for a transcription factor.
To extend our studies of the IE region of CCV, we have isolated and characterized cDNAs encoded by pPS7707, another fragment derived from the terminal repeats of CCV that encodes putative IE transcripts. These transcripts have been characterized by S1 nuclease and primer extension analyses and by sequencing of their cDNAs. Throughout the course of infection, the levels of expression of these newly mapped transcripts, as well as of the ie1 and ie2 transcripts, have been characterized.
MATERIALS AND METHODS
Cell culture and virus propagation.
Channel catfish ovary (CCO) cells were grown in modified Eagle medium (MEM) supplemented with 10% fetal bovine serum as previously described (19). CCV (Auburn-1 strain, ATCC VR-665) was propagated as previously described (19). Briefly, confluent monolayers of CCO cells were infected with CCV at a multiplicity of infection (MOI) of 0.2. Virus was harvested when the monolayer was completely detached, at which time cells were subjected to three freeze-thaw cycles. Cellular debris was pelleted by slow-speed centrifugation, and virus was concentrated by centrifugation at 65,000 × g for 1 h in a Beckman Ti-60 rotor at 4°C. Pellets were resuspended in 5 to 10 ml of growth medium, titrated, and stored at −80°C (10).
cDNA library screening.
A cDNA library enriched for IE cDNAs was constructed in λ-ZAP (Stratagene, La Jolla, Calif.) as previously described (19). The library was screened by hybridization to the genomic insert of pPS7707 that was labeled with [α-32P]dCTP by random primer extension (Dupont-New England Nuclear, Boston, Mass.). Pure plaques were obtained after four rounds of screening. In vivo excision was used to obtain Bluescript plasmids containing cDNA inserts from pure phage plaques (Stratagene).
Northern (RNA) blots.
Northern blot analysis was performed as previously described (19). Either 1 μg of poly(A)+ RNA or 10 μg of total RNA was fractionated in a 1.2% agarose gel containing 1× MOPS buffer (0.02 M 3-[N-morpholino]propanesulfonic acid [MOPS; pH 7.4], 5.0 mM sodium acetate, 1.0 mM EDTA) and 2.2 M formaldehyde. Capillary blotting was used to transfer RNA to a nylon membrane (Schleicher & Schuell, Keene, N.H.). Membranes were hybridized with [α-32P]dCTP-labeled probes synthesized from cloned viral DNA fragments, or cloned cDNAs, by random primer extension (Dupont-New England Nuclear). Membranes were hybridized at 42°C and washed as previously described (13).
S1 nuclease analysis.
S1 nuclease analysis was performed as previously described (19, 21). 5′-end labeling of probes was performed by using [γ-32P]ATP (3,000 Ci/mmol) and T4 polynucleotide kinase (U.S. Biochemical, Cleveland, Ohio) in accordance with directions provided by the manufacturer. The labeled probe was gel purified and extracted with phenol, phenol-chloroform, and finally chloroform. Labeled DNA (3,600 cpm) was mixed with 1 μg of poly(A)+ RNA isolated from either mock-infected CCO cells or CCO cells that had been infected for 3 h with 5 50% tissue culture infective doses of CCV per cell in the presence of cycloheximide (100 μg/ml). The RNA and probe mixtures were lyophilized and resuspended in PIPES [piperazine-N,N′-bis(2-ethanesulfonic acid); 1,4-piperazinediethanesulfonic acid] hybridization buffer (80% formamide, 40 mM PIPES [pH 6.4], 1 mM EDTA, 0.4 M NaCl). The hybridization mixtures were heated to 85°C for 10 min and then hybridized at 48°C for 12 h. Ice-cold S1 nuclease buffer (0.28 M NaCl, 50 mM sodium acetate [pH 4.5], 4.5 mM zinc acetate, 20 μg of sheared and denatured herring sperm DNA/ml) was added to the hybridization mixtures, along with S1 nuclease at a concentration of 1,000 U/ml. After incubation at room temperature for 45 min, the reaction was stopped by addition of 20% by volume of 5× S1 stop buffer (4.0 M ammonium acetate, 50 mM EDTA [pH 8.0]) and extracted with phenol-chloroform. After ethanol precipitation, protected fragments were electrophoresed on 6% polyacrylamide gels containing 8 M urea and visualized by autoradiography as previously described (19).
Primer extension analysis.
Oligonucleotide primers pr6 (5′-CCCGGAGAGGAGGAAGATGATAGTGAGAGAG-3′, nucleotide [nt] 9540 to 9570), pr3 (5′-GGAGTAACTAGACTCAAGAGTTTCAGAGCTCGCG-3′, nt 4733 to 4766), and pr5 (5′-CATCCTATGTTACCCTCGACACAGAACACTAGCC-3′, nt 8831 to 8864) were synthesized (Life Technologies Inc., Gaithersburg, Md.) and gel purified on a 15% polyacrylamide gel containing 8 M urea (13). Nucleotide numbers are based on the CCV genomic sequence (GenBank accession no. M75136). Primers were labeled with [γ-32P]ATP by using polynucleotide kinase (Life Technologies) as described by the manufacturer. Chromatography on a Sephadex G-50 column was used to eliminate unincorporated nucleotides. Labeled oligonucleotides were then extracted with phenol, phenol-chloroform, and chloroform before precipitation with ethanol and resuspension in H2O.
Primer extension was performed as previously described (13, 19). Briefly, 104 to 105 cpm of primer and 1 μg of poly(A)+ RNA (from either infected or mock-infected cells) were dried under vacuum and resuspended in 30 μl of hybridization buffer (80% formamide, 40 mM PIPES [pH 6.4], 1 mM EDTA, 0.4 M NaCl) and allowed to hybridize at 30°C for 12 h. After hybridization, nucleic acids were precipitated with ethanol and resuspended in reverse transcriptase buffer (50 mM Tris-HCl [pH 8.3], 40 mM KCl, 6 mM MgCl2, 50 mg of actinomycin D/ml, 2 mM dithiothreitol, 1 U of placental RNase inhibitor/ml), and the reaction was initiated by addition of 200 U of Superscript II reverse transcriptase (Life Technologies). The 20-μl reaction was incubated at 42°C for 2 h. DNase-free RNase was then added to a final concentration of 0.25 μg/ml, and incubation continued at 37°C for 30 min. After phenol extraction and ethanol precipitation, the reaction products were analyzed on a 6% polyacrylamide gel containing 8 M urea.
Infection time course.
CCO cells were grown in 100-mm-diameter tissue culture plates at 30°C until approximately 70% confluent. For cycloheximide treatments, cells were washed in phosphate-buffered saline (PBS) containing 100 μg of cycloheximide/ml, which was then replaced with MEM containing 10% (vol/vol) fetal bovine serum and 100 μg of cycloheximide/ml. For infections in the absence of cycloheximide, cells were washed in PBS, which was then replaced with MEM containing 10% (vol/vol) fetal bovine serum. After a 45-min preincubation, medium was aspirated from all of the plates and replaced with MEM containing 10% (vol/vol) fetal bovine serum, 100 μg of cycloheximide/ml, and virus (MOI = 30) for the inhibited infections or with MEM containing 10% (v/v) fetal bovine serum and virus (MOI = 30) for the uninhibited infections. Media and virus, or media, virus, and cycloheximide, were premixed and then aliquoted to each plate to ensure uniform infection. After incubation for 30 min at 30°C, plates were washed two times with 25 ml of either PBS or PBS with cycloheximide (100 μg/ml), as appropriate. MEM containing 10% (vol/vol) fetal bovine serum, either with or without cycloheximide (100 μg/ml), was then added to each plate. Cells were harvested 30 min after virus was washed off with PBS and at 30-min intervals thereafter for 1.5 h and hourly intervals for the next 2 h from the start of the infection. Thus, cells harvested 30 min after the PBS wash represented the 1-h time point, and those harvested 1 h after the PBS wash represented the 1.5-h time point. All infections were performed with 30 50% tissue culture infective doses of CCV per CCO cell. Total cellular RNA was isolated using RNazol (Tel-Test, Friendswood, Tex.) as directed by the manufacturer. Northern blots were prepared and autoradiography was performed as described above. Densitometry was performed with a Tobias model TBX densitometer (Tobias Associates, Ivyland, Pa.).
RESULTS
Identification of cDNAs representing transcripts encoded in pPS7707.
pPS7707 is a construct containing a CCV genomic fragment of 7,707 bp from the terminal repeat region (Fig. 1). Through the use of cDNA probes, the 7,707-bp fragment has been previously shown to encode CCV transcripts expressed in the presence of cycloheximide (19). To obtain cloned cDNAs representing IE transcripts encoded in the 7,707-bp genomic fragment derived from the terminal repeats, a cDNA library enriched for cDNAs representing IE transcripts was screened for clones that hybridize to that cloned CCV genomic fragment. Eighteen clones were plaque purified, and the corresponding Bluescript plasmids were obtained by using in vivo excision. The 5′ and 3′ ends of all inserts were sequenced by using Sequenase version 2.0 (U.S. Biochemical). All clones represented transcripts from either of two groups. The first group represented transcripts containing the coding region for ORF 3 (nt 4886 to 5791). The second group contained the coding regions for ORF 5 (nt 8785 to 9468) and ORF 6 (nt 9535 to 9948). All ORFs noted were those identified by Davison (2). The longest representatives of each group of cDNAs were completely sequenced and shown to represent RNAs which were not spliced. The sizes of all of the cDNA clones, relative to the positions of their ends in the genomic sequence, were also consistent with the absence of RNA splicing.
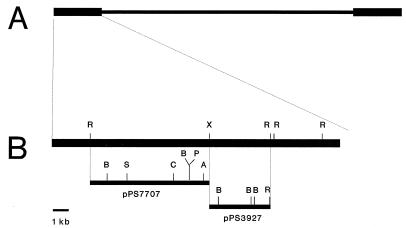
FIG. 1.
Map of the CCV genome showing the positions of viral DNA fragments contained in pPS7707 and pPS3927. (A) Representation of the CCV genome. The thick lines represent the terminal repeats; the thin line represents unique sequence. (B) Map showing positions within the terminal repeat of viral DNA fragments contained in pPS7707 and pPS3927. Abbreviations for restriction sites: A, Asp718; B, BamHI sites are shown only for pPS7707; C, ClaI; P, PstI; R, EcoRI; S, SalI, X, XbaI; B, BstEII sites are shown only for pPS3927.
Identification of transcripts encoded by cDNAs.
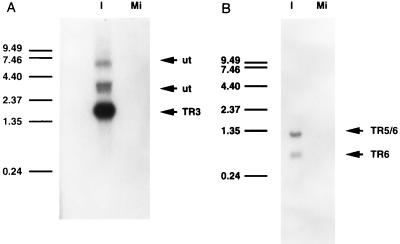
Screening of the cDNA library revealed the presence of two groups of putative IE transcripts. Although the sizes of the cDNAs were known, the sizes of the corresponding transcripts were unknown. To determine the sizes of the transcripts, RNA was isolated from CCO cells infected with CCV in the presence of cycloheximide. This RNA, along with RNA isolated from mock-infected cells, was electrophoresed on formaldehyde-agarose gels and transferred to nylon membranes. Blots from duplicate gels were hybridized separately with radiolabeled cDNAs encoding ORF 3 or ORFs 5 and 6. Hybridization with the ORF 3 cDNA revealed the presence of four transcripts, one of approximately 1.75 kb, designated TR3, two less abundant and unnamed transcripts of approximately 3 kb and a fourth transcript of approximately 7.0 kb (Fig. 2A). The three larger transcripts were not further characterized because they did not appear to be IE transcripts (see below) and no cDNAs representing these transcripts were isolated from the IE transcript-enriched cDNA library. Hybridization with the ORF 5/6 cDNA revealed the presence of two additional transcripts, one of approximately 1.35 kb, designated TR5/6, and another of approximately 620 bases, designated TR6 (Fig. 2B). In each designation, TR indicates that the genomic location is within the terminal repeat the number indicates the ORF encoded. In this case, the two transcripts were of approximately equal abundance. Neither of the cDNAs hybridized to transcripts in the mock-infected samples.
FIG. 2.
Identification of transcripts encoded by CCV cDNAs. Northern blots were prepared with poly(A)+ RNA isolated from CCO cells, either infected with CCV or mock infected. (A) Northern blot probed with a cDNA corresponding to TR3. (B) Northern blot probed with a cDNA corresponding to TR5/6. Lanes: I, RNA isolated from CCO cells infected with CCV in the presence of cycloheximide; Mi, RNA isolated from mock-infected CCO cells. Arrows labeled TR3, TR5/6, TR6, and ut indicate the TR3, TR5/6, and TR6 transcripts and two unnamed transcripts, respectively. Transcript names are based on ORFs encoding the RNAs. Coding capacities of the unnamed transcripts were not determined. The positions and sizes of RNAs in a 0.24- to 9.5-kb ladder are shown on the left.
S1 nuclease and primer extension analyses of transcripts.
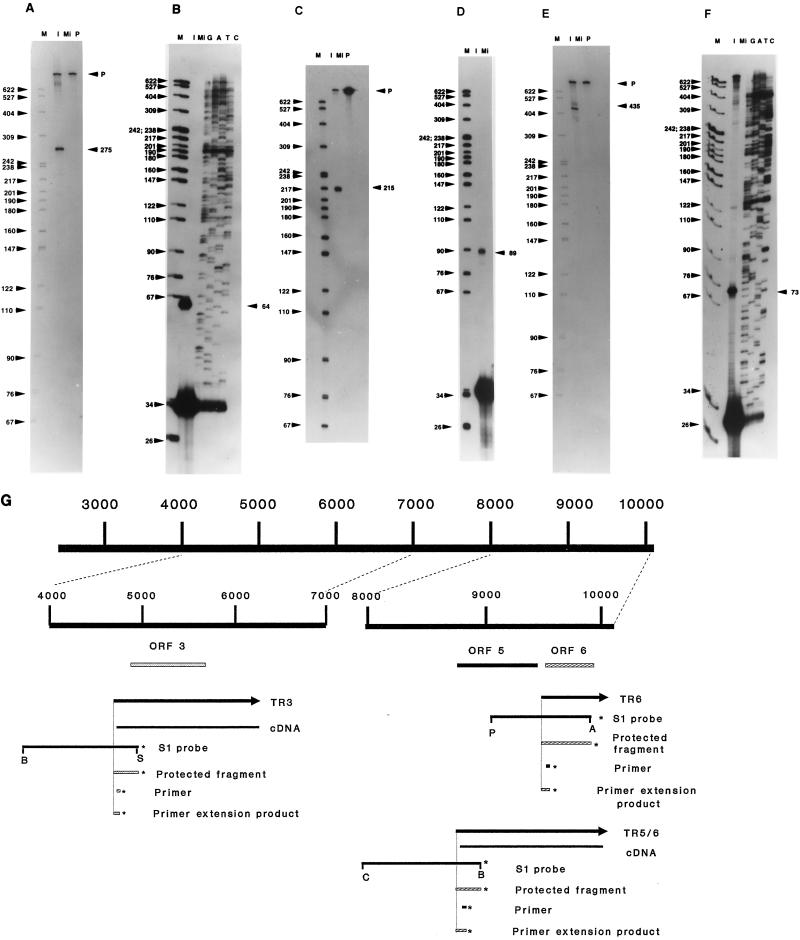
Sequence analysis of the cDNAs demonstrated that the transcripts were unspliced. However, it was necessary to determine the locations of the 5′ ends of the transcripts. The approximate locations of the 5′ ends were determined from the cDNA sequences, and that information was used to design appropriate probes for S1 nuclease analysis. The 5′ end of the longest cDNA representing ORF 3 was located at nt 4722. As the transcript was estimated to be 1.75 kb in length, and the 3′ end of the cDNA was located at nt 6288, the 5′ end was estimated to be located at approximately nt 4550. The S1 probe chosen to map the 5′ end was a SalI-BamHI fragment of 1,281 nt, whose 5′ end is within the ORF 3 cDNA and whose 3′ end extends well beyond the estimated start site of the transcript. This probe was hybridized to RNA isolated at 3 h postinfection (p.i.) from CCO cells infected with CCV in the presence of cycloheximide. S1 analysis revealed a protected fragment of approximately 275 nt, indicating that the 5′ end of the transcript, or a 3′ splice acceptor, is located near nt 4695 (Fig. 3A). Primer extension analysis of the ORF 3 mRNA was done with oligonucleotide pr3, a 34-mer (homologous to nt 4733 to 4766). This primer was hybridized to poly(A)+ RNA isolated at 3 h p.i. from CCO cells infected with CCV in the presence of a cycloheximide block. Extension of the hybridized primer produced a product of 64 nt, indicating that the 5′ end of the transcript is located at nt 4702, confirming the results obtained with S1 nuclease analysis (Fig. 3B).
FIG. 3.
Determination of the 5′ ends of TR3, TR5, and TR6, using S1 nuclease and primer extension analyses. Lanes: M, size marker (MspI digest of pBR322); P, probe; I, poly(A)+ RNA isolated from CCO cells infected in the presence of 100 μg of cycloheximide/ml; Mi, poly(A)+ RNA isolated from mock-infected CCO cells; G, A, T, and C, dideoxynucleotide chain termination sequencing reactions using the primer listed and ddGTP, ddATP, ddTTP, and ddCTP, respectively. In the S1 nuclease analyses, positions of the probes (P) and sizes (in nucleotides) of the protected fragments are indicated by arrowheads on the right. In the primer extension analyses, sizes (in nucleotides) of the extension products are indicated by arrowheads on the right. (A) S1 nuclease analysis of the 5′ end of TR3, using a BamHI-SalI fragment (nt 3681 to 4970); (B) primer extension analysis of the 5′ end of TR3, using primer pr3; (C) S1 analysis of the 5′ end of TR5/6, using a ClaI-BamHI fragment (nt 7970 to 8991); (D) primer extension analysis of the 5′ end of TR5/6, using primer pr5; (E) S1 analysis of the 5′ end of TR6, using a PstI-Asp718 fragment (nt 9062 to 9921); (F) primer extension analysis of the 5′ end of TR6, using primer pr6; (G) map of the insert of pPS7707 indicating the positions of TR3, TR5/6, TR6, ORF 3, ORF 5, ORF 6, and the probes and primers used to map the 5′ ends of these three transcripts. Abbreviations for restriction sites: A, Asp718; B, BamHI; C, ClaI; P, PstI; S, SalI. The left side of the diagram shows an expansion of the region of pPS7707 from nt 4000 to 7000 along with the relationship of that region to the features diagrammed beneath (TR3 transcript, ORF 3, the cDNA representing TR3, etc.). The right side of the diagram shows an expansion of the region of pPS7707 from nt 8000 to 10119 along with the relationship of that region to the features diagrammed beneath (TR5/6 and TR6 transcripts, ORF 5, ORF 6, etc.). The asterisks indicate the ends of the probes, protected fragments, primers, or primer extension products that were labeled. Nucleotide numbers are derived from the published CCV sequence (2).
The 5′ end of the longest cDNA representing ORFs 5 and 6 was located at nt 8795. Considering that the transcript was estimated to be 1.35 kb in length by Northern blot analysis and the 3′ end of the cDNA was at nt 10066, the 5′ end of the transcript should be located approximately at nt 8700. A BamHI fragment was 5′-end labeled with [γ-32P]ATP and cut with ClaI, and the 1,021-bp fragment isolated. This probe was hybridized to RNA isolated at 3 h p.i. from CCO cells infected with CCV in the presence of cycloheximide. Subsequent digestion with S1 nuclease resulted in the protection of a 215-nt fragment, indicating that the 5′ end of the transcript, or a 3′ splice acceptor, is located at nt 8775 (Fig. 3C). Primer extension analysis with pr5 yielded a product of 89 nt, confirming that the 5′ end of the transcript is located at nt 8775 (Fig. 3D).
The smaller transcript of 0.62 kb detected in the Northern blot which hybridized to the ORF 5/6 cDNA was also mapped. Sequencing of cDNAs indicated that all cDNAs from the region terminated at nt 10066. Therefore, it was estimated that the 5′ end of this transcript was near nt 9450. An S1 probe labeled at nt 9921 was generated by 5′-end labeling an Asp718 digest of pPS7707 and isolating the 859 bp Asp718-PstI fragment. This probe was hybridized to RNA isolated at 3 h p.i. from CCO cells infected with CCV in the presence of cycloheximide. Subsequent digestion with S1 nuclease resulted in the protection of a major fragment of 435 nt, indicating that the 5′ end of the transcript, or a 3′ splice acceptor, was located near nt 9486 (Fig. 3E). To confirm that this was the 5′ end of the transcript and not a splice site, primer pr6 was hybridized to RNA isolated at 3 h p.i. from CCO cells infected with CCV in the presence of cycloheximide. Extension of the primer yielded a major product of 73 nt, confirming that the 5′ end of the RNA is located near nt 9497 (Fig. 3F). Minor products of larger size were also visible, probably due to hybridization of the primer to TR5/6. The locations of all probes, primers, and transcripts in relation to the pPS7707 fragment of the CCV genome are shown (Fig. 3G).
Infection time course.
To determine when in the course of infection the various transcripts were expressed, infected cells were harvested at various times p.i. and total RNA was isolated. Samples from both translationally blocked (i.e., with cycloheximide) and nontranslationally blocked cells were analyzed on formaldehyde-agarose gels and transferred to nylon membranes, which were then hybridized to radiolabeled probes specific for each of the transcripts.
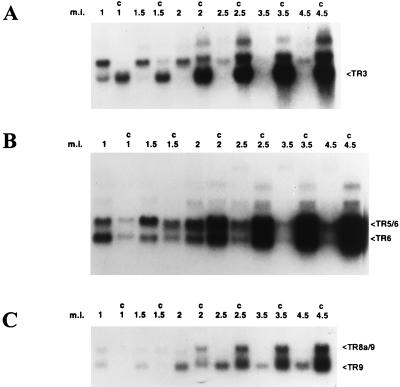
One membrane was probed with a cDNA corresponding to TR3. In cells that were not translationally blocked, the TR3 transcript encoded by ORF 3 was readily detectable at 1 h p.i. (Fig. 4A). By 1.5 h p.i., TR3 abundance was less than at the previous time point, and by 2 h p.i. the transcript was barely detectable. A larger, unnamed transcript of approximately 3.2 kb was more abundant than TR3 at 1 h p.i. The level of this transcript slowly decreased thereafter until 2.5 h p.i., when it was barely detectable. In the presence of cycloheximide, the 3.2-kb transcript appeared to be slightly larger than in the absence of cycloheximide. In the presence of cycloheximide, both transcripts also increased in abundance throughout the infection cycle. A larger transcript, of approximately 7 kb, could be detected only at late times after infection in RNA preparations from translationally blocked cells. In comparing transcript levels in translationally blocked cells with transcript levels in uninhibited cells at 1 and 1.5 h p.i., it is clear that the level of TR3 was enhanced by cycloheximide whereas the level of the 3.2-kb transcript was diminished. As discussed below, this indicates that TR3 is an IE transcript and that the 3.2-kb transcript is either an early or a late transcript.
FIG. 4.
Infection time course of TR3, TR5/6, TR6, TR9, and TR8a/9 transcripts, determined by Northern blot analysis of RNA samples isolated at various times p.i. from CCO cells infected with CCV. Times indicated are measured from the start of infection. Cells were incubated with virus for 30 min, and samples were harvested at various intervals which ranged from 30 min after the PBS wash (the 1-h time point) to 4 h after the end of the infection period (4.5-h time point). Lanes: m.i., total RNA isolated from mock-infected CCO cells; c, total RNA isolated from CCV infected CCO cells harvested at 1, 1.5, 2, 2.5, 3.5, or 4.5 h after the initiation of infection either in the absence or presence (C) of cycloheximide. (A) Blot probed with a labeled cDNA encoding TR3; (B) duplicate blot probed with a labeled cDNA encoding TR5/6; (C) duplicate blot probed with a labeled cloned genomic fragment of 997 bp common to both TR8a/9 and TR9 transcripts. Arrows (right) mark the positions of the TR3, TR5/6, TR6, TR8a/9, and TR9 transcripts.
Another membrane was probed with a cDNA corresponding to TR5/6. In uninhibited infections, the transcripts encoded by ORF 5/6 and ORF 6, TR5/6 and TR6, respectively, were both detectable 1 h p.i. (Fig. 4B). At this time, TR6 was more abundant than TR5/6, a condition that was reversed at the 1.5-h time point. The level of TR5/6 increased until 2 h p.i. and then decreased so that by 3.5 h p.i. it was barely detectable. TR6 was most abundant at 1 h p.i. and then gradually decreased. By 3.5 h p.i., this transcript was barely detectable. In the cycloheximide-blocked infections at 1 h p.i., both TR5/6 and TR6 were expressed at lower levels than in the uninhibited infections. As discussed below, this indicates that TR5/6 and TR6 are either early or late transcripts. Throughout CCV infection in the presence of cycloheximide, the levels of both transcripts gradually increased. As with the other RNAs tested, at later times in infection, both transcripts were more abundant in the presence of cycloheximide than in the absence of cycloheximide.
The two transcripts that were previously described were designated ie1 and ie2, in the belief that both of these transcripts were IE (19). The present work indicates that these transcripts are either early or late transcripts. To use a consistent nomenclature that can accommodate changes in understanding of this virus, the transcripts previously designated ie1 and ie2 are referred to as TR9 and TR8a/9, respectively. Another membrane was probed with the 997-bp EcoRI-BstEII fragment which recognizes the ORF 9 region found in the two previously described transcripts TR9 and TR8a/9 (19). In the absence of cycloheximide, both transcripts were detectable at 1 h p.i. (Fig. 4C). The level of the smaller transcript, TR9, peaked at about 2 h p.i. At 2.5 h p.i., the levels of this transcript were approximately the same as at the previous time point, but they were lower at subsequent time points. The larger transcript, TR8a/9, was readily detectable at 1 h p.i. and steadily decreased in intensity until it was undetectable at 3.5 h p.i. In cells treated with cycloheximide, at 1 h p.i., both transcripts were detectable, but TR8a/9 was more abundant than TR9. In the presence of cycloheximide, both transcripts accumulate throughout the CCV infection time course, and TR9 becomes more abundant than TR8a/9. At the earliest time points, TR8a/9 and TR9 also were less abundant in the presence of cycloheximide than in the absence of cycloheximide. However, at later time points, accumulation of both transcripts was higher in the presence of cycloheximide than in the absence of cycloheximide.
All three blots were stripped and rehybridized with a randomly primed probe for G-actin to control for the quality and quantity of RNA loaded. However, cycloheximide treatment appeared to stabilize actin mRNA throughout the course of infection, thereby causing a small increase in actin RNA levels as the time course progressed. It was also observed that infection with CCV in the absence of cycloheximide caused a decrease in actin mRNA levels as infection progressed (data not shown).
DISCUSSION
Our previous work reported the mapping of two transcripts, TR8a/9 and TR9, that are encoded in the terminal repeat region of CCV (19). The present work describes three additional transcripts from this region: TR3, TR5/6, and TR6. TR3, which extends from nt 4702 to 6288 in the CCV genome, contains ORF 3 (nt 4886 to 5791). The sequence surrounding the initiation codon matches the Kozak consensus sequence at positions −3 and +4, both of which are important for correct function of the initiator codon (7). The ORF encodes a protein with a predicted molecular mass of 32 kDa and a predicted pI of 4.2. Several aspects of the kinetic data for expression indicate that this is an IE transcript. At the 1-h time point, TR3 is far more abundant in cycloheximide-treated cells than in uninhibited infections. By 1.5 h p.i., TR3 is barely detectable in the uninhibited infections but is abundant in the infections subject to cycloheximide block. In addition, in contrast to the other viral RNAs tested, in the absence of cycloheximide, the amount of TR3 mRNA abruptly declines after the first time point at which it is detectable (1 h). These observations suggest that TR3 is an IE transcript. However, an examination of transcript levels, and their response to cycloheximide at early times after infection, clearly demonstrates that TR3 is an IE transcript whereas the other transcripts (TR5/6, TR6, TR8a/9, and TR9) are either early or late transcripts. At 1 h p.i., the presence of cycloheximide enhances the level of the TR3 transcript (Fig. 4A, lanes 1 and c1). That cycloheximide inhibits the levels of TR5/6 and TR6 (Fig. 4B, lanes 1 and c1) and of TR8a/9 and TR9 (Fig. 4C, lanes 1 and c1) can also be readily observed. Taken together, these observations indicate that the TR3 transcript is indeed an IE transcript and that the other transcripts are either early or late transcripts.
TR5/6 is a bicistronic transcript which encodes the enzyme thymidine kinase (TK). Davison predicted, based on sequence analysis, that ORF 5 would encode TK (2). Subsequently, Hanson et al. demonstrated that ORF 5 does encode a functional TK product (5). Kinetic data indicate that TR5/6 is not an IE RNA, as the transcript level at 1 h p.i. is reduced in inhibited infections compared with infections uninhibited by cycloheximide. TR5/6 levels reach maximum at 2 h p.i. and then decline. The peak relatively early in infection suggests that, like TK RNAs in other herpesviruses such as herpes simplex virus type 1 (HSV-1) (11) and bovine herpesvirus 4 (1, 24), CCV TK RNA is an early RNA.
TR6 is a small transcript of unknown function based on sequence analysis. As TR6 peaks earlier than TR5/6 but is inhibited by cycloheximide at very early times, it is also probably an early RNA. The ORF contained in TR6, ORF 6, extends from nt 9535 to 9948. The protein encoded by ORF 6 has a predicted molecular mass of 14.6 kDa and a predicted pI of 4.4. Davison suggested that sequence comparisons using the BlastP and FastA algorithms failed to reveal significant sequence relationships with other proteins (2). However, Davison noted that this protein is very hydrophobic, which may indicate membrane association (2).
Northern blot analysis of RNAs expressed over the course of infection in the presence or absence of cycloheximide produced a number of interesting observations. By the 2-h time point, there was significant leakage through the cycloheximide block of transcripts that were not IE transcripts (e.g., TR9). That these transcripts do not belong to the IE class is evident because their levels are inhibited by cycloheximide at earlier time points (compare TR5/6 and TR6 at 1 h with and without cycloheximide in Fig. 4). Furthermore, in the presence of cycloheximide, all transcripts accumulate to relatively high levels by 3.5 h p.i. In contrast, in the uninhibited infections, the levels of many transcripts are virtually undetectable by that time (e.g., TR3 and TR8a/9 [Fig. 4]). This finding suggests that cycloheximide has a nonspecific stabilizing effect on the transcripts. This lack of specificity was demonstrated with regard to the cellular actin transcripts, which also accumulate to levels slightly higher in the presence of the drug than in the absence of drug.
Our previous investigations indicated that TR8a/9 and TR9 were IE transcripts (19). The present results support a different conclusion. At 1 h p.i., levels of both of these transcripts are inhibited by cycloheximide treatment (Fig. 4), demonstrating that neither transcript is IE. In the previous work, CCO cells were infected with CCV for 4 h in the presence of cycloheximide. The long infection period, combined with leakage through the cycloheximide block and stabilization of the transcripts, resulted in accumulation of high levels of TR9 and somewhat lower levels of TR8a/9. This resulted in the incorrect interpretation that both transcripts belonged to the IE gene class. Thus, caution must be exercised when attempting to define IE transcripts by their level of expression in CCO cells translationally blocked with cycloheximide.
In examining the results of the infection time course, it is evident that different transcripts exhibit dissimilar patterns of regulation. Transcripts reach peak levels at different times and the length of the peak is also variable. TR5/6 peaks at 2 h p.i., while TR6 peaks at approximately 1 h p.i. In fact, at 1 h p.i. the ratio of TR5/6 to TR6 is approximately 2:3, but at 1.5 h p.i. the ratio changes to approximately 2:1 due to an increase in TR5/6 and a decrease in TR6. Although TR6 levels peak early in infection, TR6 transcript levels are inhibited by cycloheximide at these times, and therefore TR6 is not an IE transcript. At 1 h p.i., TR8a/9 and TR9 transcript levels are approximately equal, but by 2 h p.i., TR9 is much more abundant than is TR8a/9. By 2.5 h p.i., the TR9 transcript is at peak levels whereas the TR8a/9 transcript is almost undetectable. As with TR5/6 and TR6, both transcripts are inhibited by cycloheximide early in infection and so are not considered IE transcripts.
TR3 is an IE transcript which is only seen very early in infection. TR3 was readily detectable at 1 h p.i. and by 1.5 h p.i. was almost undetectable. Densitometric analysis reveals that between these two time points, TR3 levels decrease more than threefold. None of the other transcripts exhibits such a dramatic decline in abundance. This result suggests that the TR3 transcript is more unstable than other transcripts. In support of this hypothesis, the 3′ untranslated region (UTR) of TR3 revealed three copies of an AU-rich element (ARE) which has been shown to destabilize mRNAs in other systems. AREs have been found in the 3′ UTRs of mRNAs encoding c-fos and granulocyte/macrophage colony-stimulating factor (16–18). The functional sequence appears to be UUAUUUA(U/A)(U/A), with the efficacy of destabilization determined by the number of elements and the degree of mismatch in the first two and last two nucleotides (9, 25).
The AUUUA core sequences, and the flanking nucleotides, from TR3 were analyzed (Fig. 5). Alignment with the UUAUUUA(U/A)(U/A) consensus demonstrates that TR3 contains the AUUUA core motifs necessary to confer the destabilization phenotype in three different locations within the 3′ UTR. Motif I (nt 6099 to 6107) matches the consensus in seven of nine positions, motif II (nt 6175 to 6183) matches the consensus in eight of nine positions, and motif III (nt 6204 to 6212) matches the consensus in six of nine positions. Thus, the presence of three copies of this element which are closely related to the consensus sequence, when viewed in conjunction with the rapid degradation of the TR3 transcript, suggests that these sequences could be involved in mediating the short half-life of TR3. However, a definitive assignment of such a function to these sequences must await empirical evidence.
FIG. 5.
Alignment of the AU motifs in the 3′ UTR of CCV ORF 3 with the ARE consensus sequence. The three AUUUA motifs and their flanking nucleotides are shown along with the ARE consensus sequence (9). The locations of each motif in the CCV genome are noted (right) with nucleotide numbers derived from the published CCV sequence (2).
Actin mRNA levels progressively decreased throughout the course of CCV infection. Infection is known to destabilize transcripts in at least two other herpesviruses. vhs has been shown to destabilize both cellular and virally encoded transcripts in cells infected with HSV-1 (8, 20). A similar activity is present in HSV-2 (20). The destabilization activity observed during CCV infection differs from that seen with vhs of HSV-1, because the mRNA destabilizing activity of vhs is observed when cells are infected with HSV-1 in the presence of cycloheximide (15). In contrast, when CCO cells are infected with CCV in the presence of cycloheximide, both viral and cellular transcripts appear to be stabilized (Fig. 4 and data not shown).
ACKNOWLEDGMENTS
We thank R. Young-White and E. A. Screws for valued technical support.
This work was supported by NIH, USDA, Morris Animal Foundation, and the Food Animal and Disease Research Program at Auburn University College of Veterinary Medicine.
Footnotes
This work is dedicated to the memory of Doris and Henry Silverstein.
REFERENCES
- 1.Chang, L. Y., and V. L. van Santen. Immediate-early, early, and late RNAs in bovine herpesvirus-4-infected cells. Virology 190:909–920. [DOI] [PubMed]
- 2.Davison A J. Channel catfish virus: a new type of herpesvirus. Virology. 1992;186:9–14. doi: 10.1016/0042-6822(92)90056-u. [DOI] [PubMed] [Google Scholar]
- 3.Fijan N N. Progress report on acute mortality of channel catfish fingerlings caused by a virus. Off Int Epizoot Bull. 1968;69:1167–1168. [PubMed] [Google Scholar]
- 4.Greenberg M E, Hermanowski A L, Ziff E B. Effect of protein synthesis inhibitors on growth factor activation of c-fos, c-myc, and actin gene transcription. Mol Cell Biol. 1986;6:1050–1057. doi: 10.1128/mcb.6.4.1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hanson L A, Kousoulas K G, Thune R L. Channel catfish herpesvirus (CCV) encodes a functional thymidine kinase gene: elucidation of a point mutation that confers resistance to ara-T. Virology. 1994;202:659–664. doi: 10.1006/viro.1994.1387. [DOI] [PubMed] [Google Scholar]
- 6.Jackson R J. Cytoplasmic regulation of mRNA function: the importance of the 3′ untranslated region. Cell. 1993;74:9–14. doi: 10.1016/0092-8674(93)90290-7. [DOI] [PubMed] [Google Scholar]
- 7.Kozak M. An analysis of vertebrate mRNA sequences: intimations of translational control. J Cell Biol. 1991;115:887–903. doi: 10.1083/jcb.115.4.887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kwong A D, Kruper J A, Frenkel N. Herpes simplex virus virion host shutoff function. J Virol. 1988;62:912–921. doi: 10.1128/jvi.62.3.912-921.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lagnado C A, Brown C Y, Goodall G J. AUUUA is not sufficient to promote poly(A) shortening and degradation of an mRNA: the functional sequence within AU-rich elements may be UUAUUUA(U/A)(U/A) Mol Cell Biol. 1994;14:7984–7995. doi: 10.1128/mcb.14.12.7984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lennette E H, Smith N J. Diagnostic procedures for viral, rickettsial, and chlamydial infection. 5th ed. Washington, D.C: American Public Health Association; 1979. pp. 34–35. [Google Scholar]
- 11.Roizman B, Sears A E. Herpes simplex viruses and their replication. In: Fields B N, et al., editors. Fundamental virology. 2nd ed. New York, N.Y: Raven Press Ltd.; 1991. pp. 849–894. [Google Scholar]
- 12.Sachs A B. Messenger RNA degradation in eucaryotes. Cell. 1993;74:413–421. doi: 10.1016/0092-8674(93)80043-e. [DOI] [PubMed] [Google Scholar]
- 13.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Press; 1989. [Google Scholar]
- 14.Savant-Bhonsale S, Cleveland D W. Evidence for instability of mRNAs containing AUUUA motifs mediated through translation-dependent assembly of a >20S degradation complex. Genes Dev. 1992;6:1927–1939. doi: 10.1101/gad.6.10.1927. [DOI] [PubMed] [Google Scholar]
- 15.Schek N, Bachenheimer S L. Degradation of cellular mRNAs induced by a virion-associated factor during herpes simplex virus infection of Vero cells. J Virol. 1985;55:601–610. doi: 10.1128/jvi.55.3.601-610.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shaw G, Kamen R. A conserved AU sequence from the 3′ untranslated region of GM-CSF mRNA mediates selective mRNA degradation. Cell. 1986;46:659–667. doi: 10.1016/0092-8674(86)90341-7. [DOI] [PubMed] [Google Scholar]
- 17.Shyu A-B, Greenberg M E, Belasco J G. The c-fos transcript is targeted for rapid decay by two distinct mRNA degradation pathways. Genes Dev. 1989;3:60–72. doi: 10.1101/gad.3.1.60. [DOI] [PubMed] [Google Scholar]
- 18.Shyu A-B, Belasco J G, Greenberg M E. Two distinct destabilizing elements in the c-fos message trigger deadenylation as a first step in rapid mRNA decay. Genes Dev. 1991;5:221–231. doi: 10.1101/gad.5.2.221. [DOI] [PubMed] [Google Scholar]
- 19.Silverstein P S, Bird R C, van Santen V L, Nusbaum K E. Immediate-early transcription from the channel catfish virus genome: characterization of two immediate-early transcripts. J Virol. 1995;69:3161–3166. doi: 10.1128/jvi.69.5.3161-3166.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Strom T, Frenkel N. Effects of herpes simplex virus on mRNA stability. J Virol. 1987;61:2198–2207. doi: 10.1128/jvi.61.7.2198-2207.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.van Santen V L. Characterization of the bovine herpesvirus 4 major immediate-early transcript. J Virol. 1991;65:5211–5224. doi: 10.1128/jvi.65.10.5211-5224.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.van Santen V L. Characterization of a bovine herpesvirus 4 immediate-early RNA encoding a homolog of the Epstein-Barr virus R transactivator. J Virol. 1993;67:773–784. doi: 10.1128/jvi.67.2.773-784.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wolf K, Darlington R W. Channel catfish virus: a new herpesvirus of ictalurid fish. J Virol. 1971;8:525–533. doi: 10.1128/jvi.8.4.525-533.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang L, van Santen V L. Interaction of bovine herpesvirus 4 (BHV-4) immediate early 2 gene product with BHV-4 thymidine kinase promoter-regulatory region. J Gen Virol. 1995;76:2433–2445. doi: 10.1099/0022-1317-76-10-2433. [DOI] [PubMed] [Google Scholar]
- 25.Zubiaga A M, Belasco J G, Greenberg M E. The nonamer UUAUUUAUU is the key AU-rich sequence motif that mediates mRNA degradation. Mol Cell Biol. 1995;15:2219–2230. doi: 10.1128/mcb.15.4.2219. [DOI] [PMC free article] [PubMed] [Google Scholar]