Abstract
We have studied the effect of mutations in the human immunodeficiency virus type 1 (HIV-1) nucleocapsid (NC) sequence on tRNA3Lys genomic placement, i.e., the in vivo placement of primer tRNA3Lys on the HIV-1 primer binding site (PBS). HIV-1 produced from COS cells transfected with wild-type or mutant proviral DNA was used in this study. We have found that mutations in the amino acid sequences flanking the first Cys-His box in the NC sequence produce the maximum inhibition of genomic placement. A similar finding was obtained when the NC-facilitated annealing of primer tRNA3Lys to the HIV PBS in vitro was studied. However, since the genomic placement of tRNA3Lys occurs independently of precursor protein processing, the NC mutations studied here have probably exerted their effect through one or both of the precursor proteins, Pr55gag and/or Pr160gag-pol. One mutation in the linker region between the two Cys-His boxes, P31L, prevented packaging of both Pr160gag-pol and tRNA3Lys and prevented the genomic placement of tRNA3Lys. Both packaging and genomic placement were rescued by cotransfection with a plasmid coding for wild-type Pr160gag-pol. For other linker mutations [R7R10K11 S, R32G, and S3(32-34)], packaging of Pr160gag-pol and tRNA3Lys was not affected, but genomic placement was, and placement could not be rescued by cotransfection with plasmids coding for either Pr55gag or Pr160gag-pol. After placement, the initiation of reverse transcription within extracellular virions is characterized by a 2-base DNA extension of the placed tRNA3Lys. This process requires precursor processing, and those NC mutations which showed the most inhibition of initiation were in either of the two NC Cys-His boxes. Destabilization of a U5 stem-A-rich loop immediately upstream of the PBS (through deletion of four consecutive A’s in the loop) did not affect the in vivo genomic placement of tRNA3Lys but resulted in the presence in the extracellular virus of longer cDNA extensions of tRNA3Lys, with a corresponding decrease in the presence of unextended and 2-base-extended tRNA3Lys.
In human immunodeficiency virus type 1 (HIV-1), the synthesis of minus-strand cDNA is initiated from a tRNA3Lys primer. This cellular tRNA is selectively incorporated into the assembling virus independently of both genomic RNA packaging and precursor protein proteolysis (28). tRNA3Lys interacts with a site near the 5′ end of the genomic RNA known as the primer binding site (PBS). The PBS is an 18-nucleotide stretch complementary to the 3′-terminal 18 nucleotides of the primer tRNA. Chemical and enzymatic probing and computer modeling suggest that additional interactions occur between regions upstream of the HIV-1 PBS and the D, TΨC, and anticodon loops of tRNA3Lys (20, 22). In HIV-1, a specific interaction may also occur between the anticodon of tRNA3Lys and A-rich regions located upstream or downstream of the PBS in the HIV-1 genome (20, 22, 26). An interaction between regions upstream of the PBS and the TΨC loop has been proposed to exist in non-HIV retroviruses as well (1, 2).
Both the tRNA primer and the genomic RNA sequences which bind to the tRNA contain double-stranded regions, and the annealing of tRNA3Lys to genomic RNA might be expected to require denaturation of both molecules. In vitro, tRNA3Lys does not anneal to genomic RNA without some help, either in the form of heat or through the presence of the HIV-1 nucleocapsid (NC) protein. Mature (NCp7) or partially processed NC sequences have been shown to interact with, and denature, primer tRNA3Lys in vitro (5, 25) and to facilitate the annealing of tRNA3Lys to in vitro-transcribed genomic RNA sequence (12). Similar observations have been made with the Rous sarcoma virus and Moloney murine leukemia virus systems, where the NC sequence promotes the annealing of tRNATrp and tRNAPro to the PBSs of the Rous sarcoma virus and Moloney murine leukemia virus genomic RNAs, respectively (34). However, the mature NC protein probably does not play a role in the genomic placement of tRNA3Lys in vivo, since the placement of primer tRNA on the genomic RNA within HIV-1 and murine and avian retroviruses occurs independently of precursor protein processing (10, 19, 40). The ability of NCp7 to facilitate tRNA placement in vitro (5, 12, 34) may reflect a similar function for NC sequences present within the Gag or Gag-Pol precursor.
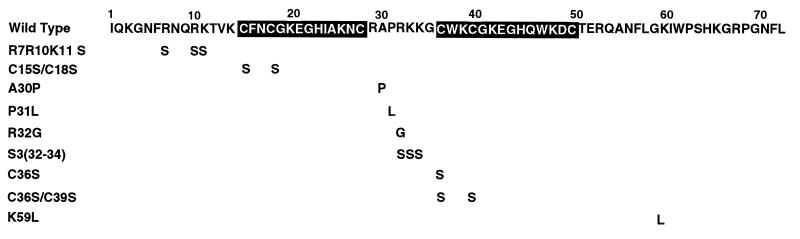
Mature HIV-1 NC (NCp7) contains two subdomains known either as Cys-His boxes (because they contain the CCHC motif, Cys-Xaa2-Cys-Xaa4-His-Xaa4-Cys) or as Zn2+ fingers because of their ability to bind Zn2+. The amino acid sequence of NCp7 is shown in Fig. 1. The positions of the two Cys-His boxes in HIV-1 define other subdomains of NCp7. From the N to the C terminus, these may be termed the N-terminal subdomain, Cys-His box 1, the linker subdomain, Cys-His box 2, and the C-terminal domain (7). In vitro studies have indicated that the NC-facilitated annealing of tRNA3Lys to HIV genomic RNA does not depend on the presence of the two Cys-His boxes but depends on the presence of the amino acids flanking the first Cys-His box (11, 12). Point mutations in the coding sequences for these flanking amino acids resulted in noninfectious viral particles (31). Since the protein responsible for tRNA3Lys placement in vivo is likely to be a precursor such as Pr55gag or Pr160gag-pol, we have examined whether the in vitro effects of NC mutations on tRNA3Lys placement are also seen when the in vivo placement of tRNA3Lys in HIV-1 is examined.
FIG. 1.
Schematic representations of wild-type and mutant NCp7 proteins. The two Cys-His boxes are in black, and their positions define other subdomains of this protein, such as the N and C subdomains and the 7-amino-acid linker subdomain between the two boxes.
After genomic placement of tRNA3Lys, the reverse transcription of minus-strand strong-stop DNA can be resolved into initiation and elongation steps (21). In vitro work, using heat-annealed primer tRNA3Lys, has shown that there is an initial buildup of 3- and 5-base extensions in the initiation phase, followed by a rapid conversion to a more processive cDNA synthesis during the elongation phase. In HIV-1 produced either from COS cells transfected with HIV-1 proviral DNA or from a variety of chronically infected human lymphocytic or monocytic cell lines, the placed primer tRNA3Lys exists in two abundant forms: unextended by reverse transcriptase (RT) or extended by the first two DNA bases incorporated, C and T (19). While proteolysis of the precursor proteins Pr55gag and Pr160gag-pol is not required for genomic placement of tRNA3Lys, it is required for the 2-base initiation of reverse transcription (19), suggesting a requirement for mature viral proteins such as p66/p51 RT and NCp7. In this work we have shown that specific mutations in the NC sequence which minimally affect tRNA3Lys placement can inhibit the initiation of reverse transcription.
In vitro experiments suggest that this early pause in reverse transcription may be due to the presence of a U5 stem/A-rich loop structure immediately upstream of the PBS (4, 21) and that the interaction of the anticodon loop of tRNA3Lys with an A-rich region within this A-rich loop may serve to destabilize the stem-loop structure and allow elongation to proceed. In this study, we have examined the effect of destabilizing the U5 stem/A-rich loop (through deletion of the four A’s) on tRNA3Lys placement and initiation in vivo and have obtained evidence indicating that the intact stem-loop structure is not required for tRNA3Lys placement in vivo but serves to block elongation beyond the 2-base cDNA extension in the extracellular virion.
(This work was performed by Y.H. in partial fulfillment of the requirements for the Ph.D. degree from McGill University, Montreal, Canada.)
MATERIALS AND METHODS
Plasmid construction.
SVC21.BH10 is a simian virus 40-based vector which contains full-length wild-type HIV-1 proviral DNA (27, 41) and was a gift from E. Cohen, University of Montreal. Site-directed mutagenesis was carried out to create the NC mutations (15, 31). All mutations (Fig. 1) were verified by direct sequence analysis of the SpeI-SalI fragment after reconstructing the full-length proviral genome, using Sequenase (United States Biochemical Corp.). The construction and characterization of the Dr2 mutation have been previously described (29). It is a 6-bp nucleotide insertion within the connection domain of RT (at nucleotide position 3715 in the HXB2 HIV-1 strain), resulting in the insertion of two additional amino acids, alanine and glycine, after the amino acid phenylalanine. This mutation inhibits the incorporation of both Pr160gag-pol and tRNA3Lys into the virion during assembly. pSVGAG-RRE-R and pSVFS5TprotD25G have been described previously (37–39) and were donated by D. Rekosh and M. L. Hammarskjold. NC mutants C15S/C18S, C36S, and C36S/C39S were donated by A. Rein and R. Gorelick (15).
Production of wild-type and mutant HIV-1 virus and viral RNA isolation.
Transfection of COS-7 cells with 10 μg of the above-described plasmids by the calcium phosphate method was as previously described (18). Virus was isolated from the cell culture medium at 63 h posttransfection. The supernatant was first centrifuged in a Beckman GS-6R rotor at 3,000 rpm for 30 min, and the virus particles were then pelleted from the resulting supernatant by centrifugation in a Beckman Ti45 rotor at 35,000 rpm for 1 h. The viral pellet was then purified by centrifugation at 26,500 rpm for 1 h through 15% sucrose onto a 65% sucrose cushion, using a Beckman SW41 rotor. Attempts to rescue genomic placement of tRNA3Lys in mutant virions were performed by cotransfecting COS cells with 10 μg of the mutant HIV-1 proviral DNA plasmid and 10 μg of a plasmid coding for either Pr55gag (pSVGAG-RRE-R) or Pr160gag-pol (pSVFS5TprotD25G).
Total viral RNA was extracted from viral pellets by the guanidinium isothiocyanate procedure (9). The pellets were dissolved in 5 mM Tris buffer, pH 7.5.
Annealing of tRNA3Lys to synthetic genomic RNA.
Synthetic genomic RNA (497 bases) used for annealing with purified tRNA3Lys was synthesized from an AccI-linearized DNA plasmid, pHIV-PBS, by using T7 RNA polymerase (Ambion, Austin, Tex.). The synthetic genomic RNA comprises the complete U5 region, the PBS, and a part of the Gag-coding region (HIV-1111B DNA sequence positions 473 to 958). The purification of tRNA3Lys from human placenta was performed as previously described (23). To anneal tRNA3Lys to synthetic HIV-1 genomic RNA, 0.5 pmol of synthetic genomic RNA was incubated with 0.5 pmol of tRNA3Lys in RT buffer (50 mM Tris-HCl [pH 7.5], 60 mM KCl, 3 mM MgCl2, 10 mM dithiothreitol) at 85°C for 2.5 min, at 50°C for 8 min, and then at 37°C for 10 min. The annealed tRNA primer was stored at −70°C for future use.
Quantitation of unspliced viral genomic RNA in total viral RNA by RPA.
A DNA template for the synthesis of radioactive probes, KSIIΨCS, was a gift from A. M. Lever, Cambridge, United Kingdom (24), and represents a ScaI/ClaI fragment (positions 313 to 830 of HXBc2) inserted into EcoRV and ClaI sites in the polylinker of Bluescript KSII (Stratagene). KSIIΨCS was linearized with XbaI, and 32P-labelled antisense riboprobes were synthesized with T3 RNA polymerase with an in vitro transcription kit (Ambion) and purified from 5% polyacrylamide–8 M urea gels prior to use in RNase protection assays (RPA). Quantitative RPA was performed by using an RPAII kit (Ambion) according to the instructions of the manufacturer. Total viral RNA was incubated with 2 × 105 cpm of 32P-labelled probe in 20 μl of hybridization buffer (80% formamide, 100 mM sodium citrate [pH 6.4], 300 mM sodium citrate [pH 6.4], 1 mM EDTA) for 16 h at 42°C. Unhybridized regions of the probe were then degraded by the addition of 0.5 U of RNase A and 20 U of RNase T1 in 200 μl of RNase digestion buffer (Ambion). Protected fragments were precipitated in ethanol, resuspended in RNA loading buffer, separated on 5% polyacrylamide–8 M urea gels, and analyzed by autoradiography or phosphorimaging (Bio-Rad). To quantitate the amount of unspliced genomic RNA, standard RNA was used, which was synthesized from KSIIΨCS after cutting out a BamHI-to-BglII fragment and then synthesizing template RNA representing positions 473 to 828 of HIV-1 (HXBc2).
Analysis of the placement and endogenous initiation of reverse transcription.
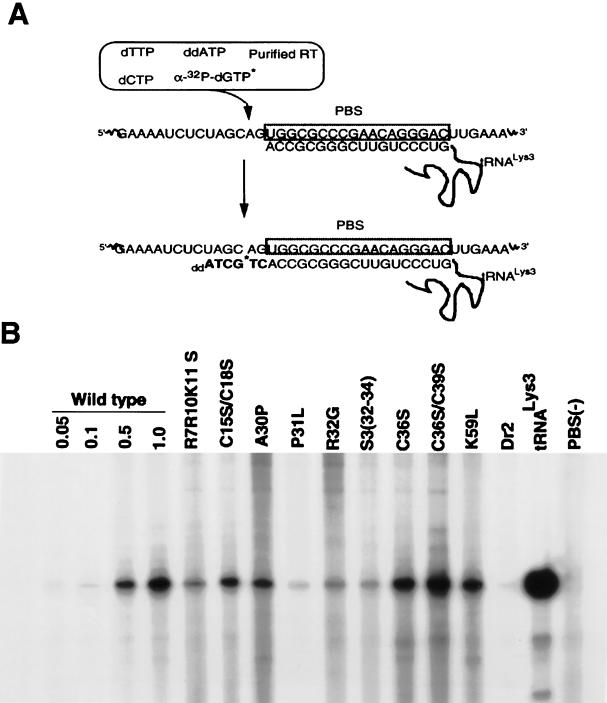
Total viral RNA, containing approximately 0.5 × 108 molecules of viral genomic RNA (as determined by RPA), was incubated for 15 min at 37°C in 20 μl of RT buffer containing 40 ng of purified HIV RT, 10 U of RNasin, and various radioactive α-32P-labelled deoxynucleoside triphosphates (dNTPs). To measure total tRNA3Lys placement (which includes both unextended and 2-base-extended forms of tRNA3Lys), the reaction mixture contained 0.2 mM dCTP, 0.2 mM dTTP, 5 μCi of [α-32P]dGTP (Dupont; 3,000 Ci/mmol, 10 mCi/ml), and 0.05 mM ddATP (see Fig. 3A). To resolve unextended and 2-base-extended tRNA3Lys by one-dimensional (1D) polyacrylamide gel electrophoresis (PAGE), the reaction mixture contained only 5 μCi each of [α-32P]dGTP and [α-32P]dCTP (Dupont; 3,000 Ci/mmol, 10 mCi/ml), as previously described (19). The extended primer was ethanol precipitated, resuspended, and analyzed on 6% polyacrylamide–7 M urea–1× Tris-borate-EDTA.
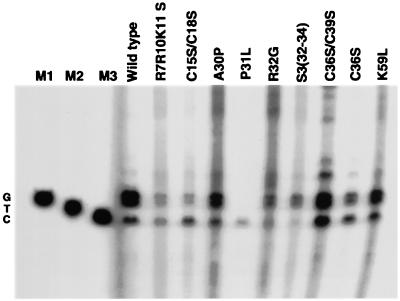
FIG. 3.
tRNA3Lys placement in wild-type and mutant virions. (A) Placement was measured by the ability of tRNA3Lys to be extended 6 bases in an in vitro reverse transcription reaction with HIV-1 RT and total viral RNA as the source of primer-template. In the presence of dCTP, dTTP, [α-32P]dGTP, and ddATP instead of dATP, extension terminated after 6 bases. (B) Resolution by 1D PAGE of 6-base extension products of tRNA3Lys in an in vitro reverse transcription reaction with total RNA from wild-type and mutant viruses as the source of primer-template, as described for panel A. Each viral RNA sample (including wild-type lane 1.0) contained 0.5 × 108 molecules of unspliced genomic RNA (determined by RPA). The first three wild-type lanes contained 0.05, 0.1, and 0.5 times this amount of genomic RNA. After the wild-type lanes, the next nine lanes represent NC mutant virions. Dr2 is an RT mutant virus. The tRNA3Lys lane represents a 6-base extension of purified tRNA3Lys annealed in vitro with synthetic genomic RNA; PBS(−) represents total viral RNA extracted from a mutant virus missing the PBS.
RESULTS
Effect of NC mutations on genomic placement of tRNA3Lys.
Mature NCp7 has been shown to facilitate the in vitro annealing of tRNA3Lys to the PBS in HIV genomic RNA (12), and in this work, we have examined the role of NC sequence in the in vivo genomic placement of tRNA3Lys by studying the effect of NC mutations on this process. The NC mutations tested are shown in Fig. 1. Total viral RNA isolated from wild-type or mutant virions was used as the source of primer tRNA-template in an in vitro reverse transcription reaction used to measure tRNA3Lys placement. This reaction utilized exogenous HIV-1 RT and was carried out in the presence of dTTP, dCTP, [α-32P]dGTP, and ddATP. The first six bases incorporated are, sequentially, CTGCTA (see Fig. 3A), and only a 6-base extension of tRNA3Lys, terminating in ddA, will occur under these reaction conditions. This species was used to indicate the amount of tRNA3Lys genomic placement per given amount of genomic RNA.
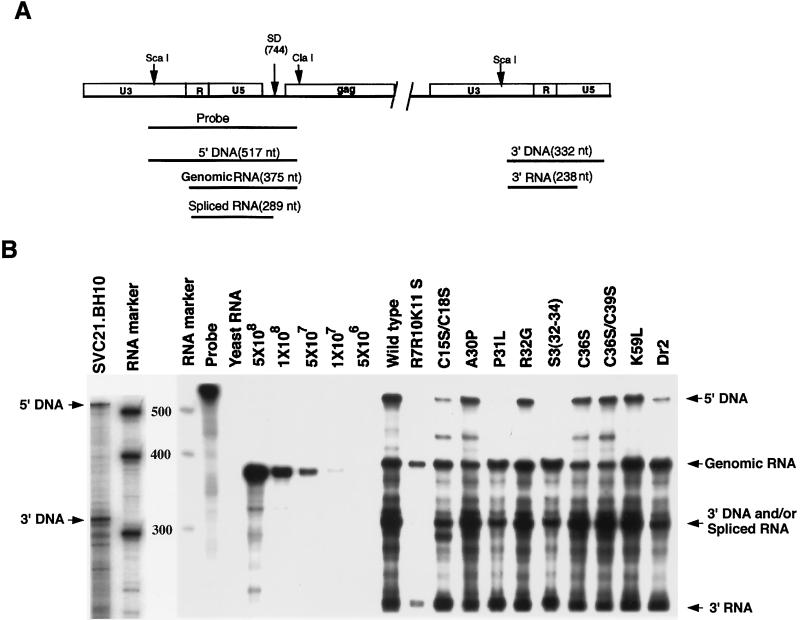
Equal amounts of full-length genomic RNA in the total viral RNA were used in each reverse transcription reaction. The amount of full-length genomic RNA was quantitated by RPA (24). As shown in Fig. 2A, this assay will in theory distinguish between HIV DNA, unspliced genomic RNA, and spliced HIV RNA. Hybridization of DNA and RNA species to the radioactive RNA probe will protect different lengths of the probe against RNase digestion. Figure 2B shows the products of the RNase protection assay, as resolved by 1D PAGE. The first two lanes represent a separate electrophoresis run from the other samples. The position of the digested radioactive probe protected by the 5′ and 3′ termini of HIV-1 DNA is shown in lane 1 (SVC21.BH10). As has been previously reported (24), the data in Fig. 2B indicates that HIV-1 produced from transfected COS cells contains, in addition to unspliced genomic RNA, a variable amount of the transfecting viral DNA. The size of the labelled RNA probe protected by the 3′ terminus of HIV-1 DNA is very similar to the size of the probe protected by spliced viral RNA, and we have been unable to resolve these two species under our electrophoretic conditions. We therefore cannot accurately assess the amount of packaged spliced RNA by using this assay. The presence of the probe fragment representing 5′ DNA varied with the preparation and not with the mutant type. Lanes 7 through 11 in Fig. 2B show a standard curve generated by using known amounts of synthetic genomic RNA, which allowed us to determine the number of copies of unspliced genomic RNA present in the wild type and different mutant viral RNA preparations.
FIG. 2.
RPA used to determine the amount of full-length genomic RNA present in viral preparations. (A) Model showing how unspliced viral RNA is distinguishable from spliced viral RNA and proviral DNA by the RPA. The 32P-labelled ScaI-ClaI RNA probe is complementary to a region of the RNA genome which goes from the 3′ region of U3 to the 5′ region of the gag gene, and the fragment sizes of the probe which are protected from RNase degradation when hybridizing to the different nucleic acids are shown. nt, nucleotides. (B) 1D PAGE separation of radioactive probe fragments protected from RNase digestion by hybridizing the RNA probe with total viral RNA isolated from wild-type and mutant virions. Lane 1, HIV-1 (BH10) proviral DNA in an HpaI-linearized plasmid DNA (SVC21.BH10); lanes 2 and 4, RNA size markers (RNA Century Marker template set; Ambion); lane 3, molecular weights of the RNA size markers; lane 5, undigested RNA probe; lane 6, yeast RNA; lanes 7 to 11, standard curve with synthetic HIV-1 RNA which will protect a probe fragment similar in size (18 bases shorter) to that protected by unspliced genomic RNA (see Materials and Methods). The number of molecules used in each lane is listed.
Figure 3B shows the 6-base extension products of reverse transcription as resolved by 1D PAGE, with total viral RNA as the source of primer-template. Equal amounts of unspliced genomic RNA (0.5 × 108 copies) in total viral RNA isolated from wild-type and mutant HIV-1 were used in each reaction to examine tRNA3Lys placement. A standard curve was also generated (lanes 1 to 4), in which various amounts of total wild-type viral RNA were used. The intensities of each band were quantitated by phosphorimaging, and the intensities relative to those for wild-type virus are listed in Table 1. It can be seen in Table 1 that the maximum inhibition of tRNA3Lys placement occurred with mutations in regions flanking Cys-His box 1, i.e., N terminal to this box (R7R10K11S) or in the linker region between the two Cys-His boxes [P31L, R32G, and S3(32-34)]. Mutations in the two Cys-His boxes showed either moderate inhibition (first Cys-His box) or little or no inhibition (second Cys-His box), and the K59 mutation in the C-terminal region also showed only very weak inhibition of placement. While the R7R10K11 S, R32G, and S3(32-34) mutations strongly inhibited tRNA3Lys genomic placement, they have previously been shown to not inhibit either tRNA3Lys or Pr160gag-pol incorporation into the virus (17). On the other hand, the P31L mutation has been shown to inhibit incorporation of both tRNA3Lys and Pr160gag-pol into the virion (17). Also shown in this experiment (Fig. 3B, lane 14) is the strong inhibitory effect on tRNA3Lys placement of the RT connection domain mutation, Dr2, which, like P31L, inhibits viral incorporation of both tRNA3Lys and Pr160gag-pol (29). Lane 15 (positive control) represents a reaction in which purified tRNA3Lys annealed in vitro to synthetic genomic RNA served as the source of primer-template, while lane 16 (negative control) shows the absence of priming when RNA isolated from a virion lacking the PBS was used.
TABLE 1.
Placement and 2-base extension of tRNA3Lys within wild-type and mutant HIV-1 viruses
| Virus | Genomic placement (% of wild type)a | 2-base extension (% of tRNA3Lys placed)b |
|---|---|---|
| Wild type | 100 | 80 ± 3 |
| Mutants | ||
| R7R10K11 S | 24 ± 5 | 60 ± 2 |
| C15S/C18S | 54 ± 8 | 40 ± 2 |
| A30P | 61 ± 9 | 83 ± 3 |
| P31L | 7 ± 4 | NDc |
| R32G | 19 ± 8 | 98 ± 3 |
| S3(32-34) | 25 ± 9 | 91 ± 2 |
| C36S | 94 ± 10 | 65 ± 2 |
| C36S/C39S | 105 ± 10 | 55 ± 2 |
| K59L | 87 ± 9 | 79 ± 2 |
| Dr2 | 5 ± 2 | ND |
| Cotransfection with pSVGag-RRE | ||
| R7R10K11 S | 20 ± 5 | |
| P31L | 5 ± 2 | |
| R32G | 12 ± 6 | |
| S3(32-34) | 17 ± 5 | |
| Cotransfection with pSVFS5TproD25G | ||
| R7R10K11 S | 22 ± 5 | |
| P31L | 120 ± 12 | |
| R32G | 10 ± 5 | |
| S3(32-34) | 20 ± 6 |
Results are means and standard deviations for single transfection experiments performed three times and cotransfection experiments performed two times.
Percentage of total tRNA3Lys placed = [2-base extended/(unextended + 2-base extended) × 100. Results are means and standard deviations for experiments performed three times.
ND, not detected.
Attempts to rescue genomic placement of tRNA3Lys in virions containing mutant NC.
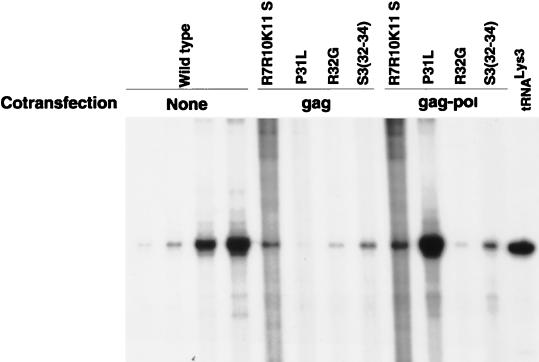
The NC mutations inhibiting tRNA3Lys genomic placement may act through either Pr55gag, Pr160gag-pol, or a complex of both precursors. To define the role of either precursor in the placement process, we have investigated whether either wild-type Pr55gag or wild-type Pr160gag-pol can rescue tRNA3Lys genomic placement in the R7R10K11 S, P31L, R32G, and S3(32-34) mutant virions, using cotransfection experiments. Pr55gag is capable of particle assembly in the absence of other viral proteins (13, 16, 30, 38), while in COS cells, Pr160gag-pol alone does not form particles (24, 32, 38). When these two precursors are expressed in the same cell from different plasmids, the Pr55gag particles package the Pr160gag-pol (17, 32, 38, 39). We have previously used this cotransfection system to show that the defect in tRNALys packaging in the P31L mutant could be rescued by cotransfecting mutant proviral DNA with a plasmid coding for wild-type Pr160gag-pol (17). In this paper, we report the results of experiments designed to test whether tRNA3Lys placement can be rescued by cotransfecting the mutant proviral DNA with a second plasmid coding for either unprocessed wild-type Pr55gag (pSVGAG-RRE) or unprocessed wild-type Pr160gag-pol (pSVFS5TprotD25G). The RT-generated 6-base extension products, resolved by 1D PAGE, are shown in Fig. 4, and the results of quantitation of the bands by phosphorimages analysis are listed in Table 1. The defects in tRNA3Lys placement in the R7R10K11 S, R32G, and S3(32-34) mutants could not be rescued by cotransfection with either wild-type Pr55gag or Pr160gag-pol. On the other hand, wild-type Pr160gag-pol (but not wild-type Pr55gag) did rescue tRNA3Lys placement in the P31L mutation, just as it rescued Pr160gag-pol and tRNALys packaging in this mutant (17).
FIG. 4.
Attempts to rescue genomic placement of tRNA3Lys in mutant NC virions with wild-type Pr55gag or wild-type Pr160gag-pol. COS cells were cotransfected with mutant proviral DNA and with a plasmid coding for either Pr55gag (pSVGAG-RRE-R) or Pr160gag-pol (pSVFS5TprotD25G). Total RNA was isolated from the virions produced, and placement was measured by the ability of the RNA to produce a 6-base extension of tRNA3Lys as described for Fig. 3. Each viral RNA sample (including wild-type lane 4) contained 0.5 × 108 molecules of unspliced genomic RNA (determined by RPA). The first three wild-type lanes contained 0.05, 0.1, and 0.5 times this amount of genomic RNA.
Effect of NC mutations on the initiation of reverse transcription.
In the wild-type virions, tRNA3Lys exists in two major forms: unextended and extended by the first two deoxynucleotides to be incorporated, C and T (19). The placement of tRNA3Lys on the genome occurs independently of proteolysis, but extension of the tRNA requires proteolysis (19), perhaps because of a requirement for mature RT and/or mature NCp7. It is therefore possible that any effect that NC sequences have upon tRNA3Lys extension will be through mature NCp7 and not through a precursor protein. Therefore, in addition to examining the effect of NC mutations on tRNA3Lys genomic placement (see above), we have also examined the effect of these same mutations on tRNA3Lys extension in the mature virion. We have studied the effect of NC mutations on the ability of placed tRNA3Lys to be extended in vivo, using the in vitro reverse transcription extension assay to detect both unextended and 2-base-extended tRNA3Lys. The first 6 bases incorporated during reverse transcription are CTGCTA. In the presence of only two dNTPs, dCTP and dGTP (both radioactive), unextended tRNA3Lys is extended by 1 base (C), and tRNA3Lys, which was extended by 2 bases in the virion, will be able to incorporate the third base, G, and the fourth base, C. These results have previously been reported for wild-type virus (19), and resolution by 1D PAGE of the unextended and extended tRNA3Lys is shown in Fig. 5, representing reactions which used total viral RNA from wild-type and mutant virus particles as the source of primer-template. The ratios of extended tRNA3Lys to extended plus unextended tRNA3Lys were determined from this data, and the results are listed in Table 1. It can be seen from Table 1 that the mutations which exerted the greatest inhibitory effect on the initiation of reverse transcription occurred in the two Cys-His boxes, regions which affect tRNA3Lys placement either moderately (Cys-His box 1) or very little (Cys-His box 2). The R32G and S3(32-34) mutations, which showed the greatest effect on genomic placement of tRNA3Lys, had little effect on tRNA3Lys extension. The P31L mutant virus showed no tRNA3Lys extension, which is not surprising since this mutation prevents the incorporation of Pr160gag-pol (and therefore RT) into the virion (29).
FIG. 5.
Analysis of tRNA3Lys placement and extension by RT in wild-type and mutant viruses. Similar to the case for the experiments represented in Fig. 3 and 4, total viral RNA isolated from wild-type and mutant viruses was used as the source of primer-template in the in vitro reverse transcription reaction. However, only [α-32P]dCTP and [α-32P]dGTP were used. Using Fig. 3A as a guide, unextended tRNA3Lys will be extended 1 base by dCTP, while 2-base-extended tRNA3Lys will be extended 3 and 4 bases by dGTP and dCTP, respectively. Lanes M1 to M3, size markers generated in the in vitro reverse transcription reaction with tRNA3Lys annealed to synthetic genomic RNA as the primer-template. Reaction mixtures generating M1, M2, and M3 each contained [α-32P]dCTP and the following dNTPs: M1 (1-base extension), none; M2 (2-base extension), ddTTP; M3 (3-base extension), dTTP and ddGTP.
Role of the U5 stem/A-rich loop in tRNA3Lys genomic placement and tRNA3Lys extension by reverse transcription.
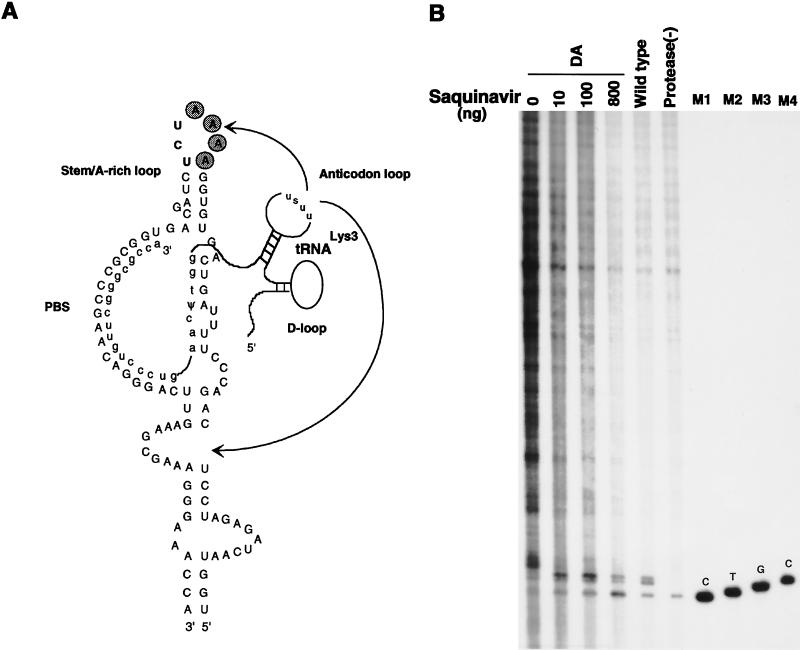
Figure 6A shows the postulated interactions which may occur between primer tRNA3Lys and the HIV-1 genome. This figure represents a modified version of Fig. 1A in reference 3, since we have determined a somewhat different sequence of bases immediately upstream of the PBS, which results in just one base, G, occurring between the 5′-terminal U in the PBS and the beginning of a U5 stem-loop structure. Several regions of potential interaction between the tRNA3Lys and the RNA genome are indicated. HIV-1 contains a run of four A’s in the loop of the U5 stem-loop structure upstream of the PBS, which may interact with the tRNA3Lys anticodon loop (20–22). Figure 6A presents this postulated interaction and also indicates another potential interaction between the TΨC loop of tRNA3Lys and the U5 sequences in the HIV-1 genome, an interaction first postulated to occur in avian retroviruses between primer tRNATrp and the avian retroviral genome (1, 2).
FIG. 6.
Effect of deletion of the four A’s in the A-rich loop on tRNA3Lys genomic placement and extension of tRNA3Lys by RT. (A) Proposed regions of base pairing between tRNA3Lys and the HIV-1 genome. This figure is modified from reference 3. In addition to the interaction between the 3′-terminal 18 nucleotides of tRNA3Lys and the PBS, other proposed interactions include ones between the tRNA3Lys anticodon loop and A-rich regions in the genome both upstream (20–22) and downstream (26) of the PBS (arrows), as well as a proposed interaction of the TΨC loop in the primer tRNA with a U5 region upstream of the PBS, which was initially proposed for avian retroviruses (1, 2). (B) Effect of A-rich loop deletion on tRNA3Lys placement and extension by RT. In vitro reverse transcription reactions were run as described in the Fig. 5 legend. Wild-type and protease(−) lanes represent total RNA isolated from wild-type and protease-negative virions, showing that only unextended tRNA3Lys is detected in protease-negative virions. The four DA lanes represent reactions with total RNA isolated from virus in which the four A’s of the A-rich loop have been deleted. The transfected cells, exposed to DNA for 15 h, were washed with fresh medium and were grown in increasing concentrations of the viral protease inhibitor Saquinovir for an additional 48 h before isolation of the virus. Lanes M1, M2, and M3, size markers generated from tRNA3Lys annealed to synthetic genomic RNA, as described in the legend to Fig. 5. Lane M4, size marker (4-base tRNA3Lys extension) generated by first extending tRNA3Lys 1 base with RT in the presence of [α-32P]dCTP and then adding dTTP, dGTP, and an excess of ddCTP before additional incubation.
Analysis of reverse transcription in vitro, using homologous RT and genomic RNA for various lentiviruses that use tRNA3Lys as a primer, including HIV-1, has shown that there exist both an initiation phase and an elongation phase of reverse transcription (4, 25). The initiation phase is manifest by a limited 1- to 12-base extension of primer tRNA3Lys. The variation in extension depends on the source of viral RNA; i.e., the length of this extension appears to be correlated with the distance of the stem-loop structure upstream of the PBS, which varies in different lentiviruses (4). However, other lentiviruses that use tRNA3Lys as a primer do not contain consecutive A’s in the U5 stem-loop structures associated with the early pausing (4). This suggests that the overall conformation of the stem-loop structure may be more important for its interaction with a tRNA3Lys-RT complex than the specific interaction postulated to occur in HIV-1 between the consecutive A’s in the loop and the USUU anticodon loop of tRNA3Lys. Figure 6A shows that in HIV-1, the first stem base pair, AU, is encountered 2 bases upstream of the PBS, and the second stem base pair is GC. The limited 2-base DNA extension of tRNA3Lys that we detect in mature extracellular HIV-1 (23) reflects the reverse transcription of only the unpaired G and the A in the first stem base pair.
Deletion of the four A’s in the loop of the U5 stem/A-rich loop structure does not appear to affect the in vitro placement of primer tRNA3Lys on the PBS (4, 21) but does result in the elimination of pausing (i.e., short DNA extensions) during in vitro reverse transcription, with larger minus-strand cDNA synthesized (4). In this study, we have examined whether the deletion of the four A’s affects either tRNA3Lys placement or extension in vivo, and the data is shown in Fig. 6B. In vitro primer extension with RT was used to measure unextended and 2-base-extended tRNA3Lys as discussed above (Fig. 5). In Fig. 6B, lane 5 shows the unextended and extended forms of tRNA3Lys when wild-type viral RNA was used as the source of primer-template, while lane 6 shows that tRNA3Lys remained unextended in viral RNA isolated from a protease-negative virus. The first four lanes used total viral RNA isolated from the virus with the A-rich loop deleted as the source of primer-template. We see in the first lane that the unextended and 2-base-extended forms of tRNA3Lys were strongly reduced in favor of the increased synthesis of longer fragments of cDNA. Since tRNA3Lys extension requires the presence of precursor proteolysis, in lanes 2 to 4 we added increasing amounts of the protease inhibitor Saquinovir (Hoffman-La Roche). We have previously documented the inhibitory effect of this drug on precursor proteolysis in HIV-1 produced from transfected COS-1 cells (17, 29). As the Saquinovir concentration is increased, there are increased amounts of 2-base-extended and unextended tRNA3Lys, with the loss of longer cDNA extensions. At the highest concentrations of the inhibitor, the unextended form of tRNA3Lys predominates. These results indicate that genomic placement of tRNA3Lys occurred in the absence of an intact A-rich loop and that disruption of the A-rich loop allowed for greater cDNA extensions from tRNA3Lys. However, this data also shows that during partial inhibition of precursor proteolysis, the predominant pausing of reverse transcription after a tRNA3Lys 2-base extension was still seen even in the absence of the intact A-rich loop.
DISCUSSION
In this work, we have shown that mutations in the HIV-1 NC sequence, as well as in the connection domain of RT (the Dr2 mutant), inhibit the in vivo genomic placement of tRNA3Lys. NC mutations which showed the strongest inhibition of placement are those found in the amino acid regions flanking the first Cys-His box [R7R10K11 S, P31L, R32G, and S3(32-34)]. Both the P31L mutation in NC and the Dr2 mutation in the connection domain of RT strongly inhibited genomic placement of tRNA3Lys and have previously been shown to inhibit the incorporation of both tRNA3Lys and Pr160gag-pol into viral particles (17, 29). Previous results have indicated that Pr160gag-pol packaging is required for tRNA3Lys packaging (28, 29). For P31L (17) and Dr2 (unpublished data), cotransfection with a plasmid coding for wild-type Pr160gag-pol rescues the packaging of both Pr160gag-pol and tRNA3Lys. As shown in this report, this cotransfection also rescues genomic placement of tRNA3Lys in these two mutants. It is, however, not clear from these experiments if the rescue of genomic placement in these mutants is a result of rescuing Pr160gag-pol packaging or tRNA3Lys packaging. However, the direct involvement of a precursor protein (Pr55gag and/or Pr160gag-pol) in genomic placement is likely. Evidence for this consists of the fact that while genomic placement of tRNA3Lys occurs independently of precursor proteolysis, it is nevertheless inhibited by mutations in NC sequences [R7R10K11 S, R32G, and S3(32-34)] (19), and these mutations do not affect Pr160gag-pol or tRNA3Lys packaging.
Unlike the case for the P31L or Dr2 mutation, we were unable to rescue genomic placement in the R7R10K11 S, R32G, and S3(32-34) viruses by cotransfection with wild-type plasmids coding for either Pr55gag, or Pr160gag-pol. The inability of wild-type Pr160gag-pol to rescue genomic placement could reflect a role in this process played by Pr55gag, not Pr160gag-pol, or it could imply that genomic placement of tRNA3Lys is facilitated by a Pr55gag-Pr160gag-pol complex which cannot be formed properly when composed of either two mutant precursors or a wild-type and a mutant precursor. Because much more mutant Pr55gag than mutant Pr160gag-pol is made in the virus, the inability to rescue genomic placement in these mutants with wild-type Pr55gag is a result more difficult to interpret. In addition to the possibility that the improper formation of a wild-type Pr55gag–mutant Pr160gag-pol complex inhibited genomic placement of tRNA3Lys, there may be a technical problem in producing sufficient wild-type Pr55gag to compete with mutant Pr55gag.
The ability of the R7R10K11 S, R32G, and S3(32-34) mutations to allow packaging of Pr55gag, Pr160gag-pol, genomic RNA, and tRNA3Lys, yet to inhibit tRNA3Lys genomic placement, indicates a step in placement which is currently not understood, and one more influenced by the amino acid sequences flanking the first Cys-His box than by amino acid sequences within either Cys-His box or C terminal to the second Cys-His box. While the Cys-His boxes themselves have been implicated as the sequences interacting with the genomic RNA in both avian and mammalian retroviruses (8, 14, 24, 31, 33–36, 42), it is possible that the amino acid sequences flanking the first Cys-His box are more involved in the nucleic-acid-unwinding activity of NC protein, an activity probably required for tRNA3Lys placement. These same flanking amino acid sequences have also been shown to be important for facilitating HIV-1 RNA dimerization in vitro (12).
The effect of mutations in synthetic NCp7 on the in vitro NC-facilitated annealing of tRNA3Lys to HIV-1 RNA (positions 1 to 415) has been reported (12). It was found that the deletion of both Cys-His boxes, or deletion of both boxes and the first 12 N-terminal amino acids and the last 8 C-terminal amino acids, did not affect tRNA3Lys annealing in vitro. While that data and ours point to the importance of amino acids flanking the first Cys-His box in tRNA3Lys genomic placement, some differences in the results exist. In the sequences N terminal to the first Cys-His box, only the presence of V13 and K14 was required for wild-type placement activity in vitro, while we have found that the mutations in the R7R10K11 mutant also strongly affect tRNA3Lys placement in vivo. This could reflect differences in conformation of NC sequences found within the precursor protein or mature NCp7.
While mutations within the Cys-His boxes had relatively little effect on genomic placement of tRNA3Lys, they more strongly inhibit initiation of reverse transcription, i.e., the limited 2-base DNA extension of tRNA3Lys seen in extracellular HIV-1. Since this extension requires precursor proteolysis (19), the NC mutations may act through mature NCp7 and may inhibit the normal formation of a reverse transcription complex which includes RT, NCp7, and tRNA3Lys. The existence of such a complex in vitro has been reported (6). Alternatively, the mutations in the Cys-His box could alter the nature of the placement of tRNA3Lys on the genomic RNA by precursor protein and thereby indirectly affect the ability of this placed tRNA3Lys to be extended by RT.
The data in Fig. 6B indicates that, in vivo, tRNA3Lys is placed on the genome independently of its interaction with the A-rich loop, a conclusion also reached when studying the in vitro annealing of tRNA3Lys with HIV-1 RNA (4, 21). The results in Fig. 6B also indicate that pausing at the 2-base cDNA extension created in vivo was greatly diminished when the four A’s in the A-rich loop were removed, and this was accompanied by larger cDNA extensions which terminated at nonrandom sites before completion of the synthesis of full-length minus-strand cDNA. These results may be explained by the conclusions reached from in vitro reverse transcription studies, which indicate that RT shows lower processivity during the initiation phase of reverse transcription than during the elongation phase (21). It has been suggested that the U5 stem/A-rich loop causes a pause in reverse transcription which produces a more processive enzyme resulting from an alteration in the RT conformation (21). Destabilizing the stem-loop structure may, by diminishing the pausing time, minimize the opportunity for an RT conformation change to a more processive enzyme, resulting in the synthesis of the multiple-sized fragments of strong-stop minus-strand cDNA that we find in the mature extracellular virion. These results indicate that the absence of a transition from the initiation to the elongation phase of reverse transcription in extracellular virions is not the result of insufficient dNTP pools to maintain elongation but could involve a requirement for cellular factors not present in the virus.
ACKNOWLEDGMENTS
This work was supported by grants from the Medical Research Council and Health Canada.
We thank David Rekosh, Mary Lou Hammarskjold, Andrew Lever, Alan Rein, and Robert Gorelick for the gifts of plasmids used in this work, and we thank Sandy Fraiberg for assistance in preparation of the manuscript.
REFERENCES
- 1.Aiyar A, Cobrinik D, Ge Z, Kung H J, Leis J. Interaction between retroviral U5 RNA and the TΨC loop of the tRNATrp primer is required for efficient initiation of reverse transcription. J Virol. 1992;66:2464–2472. doi: 10.1128/jvi.66.4.2464-2472.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aiyar A, Ge Z, Leis J. A specific orientation of RNA secondary structures is required for initiation of reverse transcription. J Virol. 1994;68:611–618. doi: 10.1128/jvi.68.2.611-618.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Arts E J, Ghosh M, Jacques P S, Ehresmann B, LeGrice S F J. Restoration of tRNALys3-primed (−) strand DNA synthesis to an HIV-1 reverse transcriptase mutant with extended tRNAs. J Biol Chem. 1996;271:9054–9061. doi: 10.1074/jbc.271.15.9054. [DOI] [PubMed] [Google Scholar]
- 4.Arts E J, Stetor S R, Li X, Rausch J W, Howard K J, Ehresmann B, North T W, Wohrl B M, Goody R S, Wainberg M A, LeGrice S F J. Initiation of (−) strand DNA synthesis from the tRNALys3 on lentiviral RNAs: implications of specific HIV-1 RNA-tRNALys3 interactions inhibiting primer utilization by retroviral reverse transcriptases. Proc Natl Acad Sci USA. 1996;93:10063–10068. doi: 10.1073/pnas.93.19.10063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Barat C, Lullien V, Schatz O, Keith G, Darlix J L. HIV-1 reverse transcriptase specifically interacts with the anticodon domain of its cognate primer tRNA. EMBO J. 1989;8:3279–3285. doi: 10.1002/j.1460-2075.1989.tb08488.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Barat C, Schatz O, Le Grice S, Darlix J-L. Analysis of the interactions of HIV-1 replication primer tRNALys3 with nucleocapsid protein and reverse transcriptase. J Mol Biol. 1993;231:185–190. doi: 10.1006/jmbi.1993.1273. [DOI] [PubMed] [Google Scholar]
- 7.Berkowitz R, Fisher J, Goff S P. RNA packaging. In: Krausslich H G, editor. Morphogenesis and maturation of retroviruses. Vol. 214. Berlin, Germany: Springer-Verlag; 1996. pp. 177–218. [Google Scholar]
- 8.Bowles N E, Damay P, Spahr P-F. Effect of rearrangements and duplications of the Cys-His motifs of Rous sarcoma virus nucleocapsid protein. J Virol. 1993;67:623–631. doi: 10.1128/jvi.67.2.623-631.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chomczynski P, Sacchi N. RNA isolation from cultured cells. Anal Biochem. 1987;162:156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- 10.Crawford S, Goff S P. A deletion mutation in the 5′ part of the pol gene of Moloney murine leukemia virus blocks proteolytic processing of the gag and pol polyproteins. J Virol. 1985;53:899–907. doi: 10.1128/jvi.53.3.899-907.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dannull J, Surovoy A, Jung G, Moelling K. Specific binding of HIV-1 nucleocapsid protein to PSI RNA in vitro requires N-terminal zinc finger and flanking basic amino acid residues. EMBO J. 1994;13:1525–1533. doi: 10.1002/j.1460-2075.1994.tb06414.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.De Rocquigny H, Gabus C, Vincent A, Fournie-Zaluski M-C, Roques B, Darlix J-L. Viral RNA annealing activities of human immunodeficiency virus type 1 nucleocapsid protein require only peptide domains outside the zinc fingers. Proc Natl Acad Sci USA. 1992;89:6472–6476. doi: 10.1073/pnas.89.14.6472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gheysen D, Jacobs E, de Foresta F, Thiriart C, Francotte M, Thines D, De Wilde M. Assembly and release of HIV-1precursor Pr55gag virus-like particles from recombinant baculovirus-infected insect cells. Cell. 1989;59:103–112. doi: 10.1016/0092-8674(89)90873-8. [DOI] [PubMed] [Google Scholar]
- 14.Gorelick R J, Chabot D J, Rein A, Henderson L E, Arthur L O. The two zinc fingers in the human immunodeficiency virus type 1 nucleocapsid protein are not functionally equivalent. J Virol. 1993;67:4027–4036. doi: 10.1128/jvi.67.7.4027-4036.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gorelick R J, Nigida J S M, Bess J J W, Arthur L O, Henderson L E, Rein A. Noninfectious human immunodeficiency virus type 1 mutants deficient in genomic RNA. J Virol. 1990;64:3207–3211. doi: 10.1128/jvi.64.7.3207-3211.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hong S S, Boulanger P. Assembly-defective point mutants of the human immunodeficiency virus type 1 Gag precursor phenotypically expressed in recombinant baculovirus-infected cells. J Virol. 1993;67:2787–2798. doi: 10.1128/jvi.67.5.2787-2798.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Huang Y, Khorchid A, Wang J, Parniak M A, Darlix J, Wainberg M A, Kleiman L. Effect of mutations in nucleocapsid protein (NCp7) upon Pr160gag-pol and tRNALys incorporation into HIV-1. J Virol. 1997;71:4378–4384. doi: 10.1128/jvi.71.6.4378-4384.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Huang Y, Mak J, Cao Q, Li Z, Wainberg M A, Kleiman L. Incorporation of excess wild type and mutant tRNA3Lys into HIV-1. J Virol. 1994;68:7676–7683. doi: 10.1128/jvi.68.12.7676-7683.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Huang Y, Wang J, Shalom A, Li Z, Khorchid A, Wainberg M A, Kleiman L. Primer tRNA3Lys on the viral genome exists in unextended and two base-extended forms within mature human immunodeficiency virus type 1. J Virol. 1997;71:726–728. doi: 10.1128/jvi.71.1.726-728.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Isel C, Ehresmann C, Keith G, Ehresmann B, Marquet R. Initiation of reverse transcription of HIV-1: secondary structure of the HIV-1 RNA/tRNALys3 (template/primer) complex. J Mol Biol. 1995;247:236–250. doi: 10.1006/jmbi.1994.0136. [DOI] [PubMed] [Google Scholar]
- 21.Isel C, Lanchy J, LeGrice S F J, Ehresmann C, Ehresmann B, Marquet R. Specific initiation and switch to elongation of human immunodeficiency virus type 1 reverse transcription require the post-translational modifications of primer tRNALys3. EMBO J. 1996;15:917–924. [PMC free article] [PubMed] [Google Scholar]
- 22.Isel C, Marquet R, Keith G, Ehresmann C, Ehresmann B. Modified nucleotides of tRNALys3 modulate primer/template loop-loop interaction in the initiation complex of HIV-1 reverse transcription. J Biol Chem. 1993;268:25269–25272. [PubMed] [Google Scholar]
- 23.Jiang M, Mak J, Ladha A, Cohen E, Klein M, Rovinski B, Kleiman L. Identification of tRNAs incorporated into wild-type and mutant human immunodeficiency virus type 1. J Virol. 1993;67:3246–3253. doi: 10.1128/jvi.67.6.3246-3253.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kaye J F, Richardson J H, Lever A M L. cis-acting sequences involved in human immunodeficiency virus type 1 RNA packaging. J Virol. 1995;69:6588–6592. doi: 10.1128/jvi.69.10.6588-6592.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Khan R, Giedroc D P. Recombinant human immunodeficiency virus type 1 nucleocapsid (NCp7) protein unwinds tRNA. J Biol Chem. 1992;267:6689–6695. [PubMed] [Google Scholar]
- 26.Kohlstaedt L A, Steitz T A. Reverse transcriptase of human immunodeficiency virus can use either human tRNALys3 or Escherichia coli tRNAGln2 as a primer in an in vitro primer-utilization assay. Proc Natl Acad Sci USA. 1992;89:9652–9656. doi: 10.1073/pnas.89.20.9652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Levy J A, Cheng M C, Dina D, Luciw P A. AIDS retrovirus (ARV-2) clone replicates in transfected human and animal fibroblasts. Science. 1986;232:998–1001. doi: 10.1126/science.3010461. [DOI] [PubMed] [Google Scholar]
- 28.Mak J, Jiang M, Wainberg M A, Hammarskjold M-L, Rekosh D, Kleiman L. Role of Pr160gag-pol in mediating the selective incorporation of tRNALys into human immunodeficiency virus type 1 particles. J Virol. 1994;68:2065–2072. doi: 10.1128/jvi.68.4.2065-2072.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mak J, Khorchid A, Cao Q, Huang Y, Lowy I, Parniak M A, Prasad V R, Wainberg M A, Kleiman L. Effects of mutations in Pr160gag-pol upon tRNALys3 and Pr160gag-pol incorporation into HIV-1. J Mol Biol. 1997;265:419–431. doi: 10.1006/jmbi.1996.0742. [DOI] [PubMed] [Google Scholar]
- 30.Mergener K, Facke M, Welker R, Brinkmann V, Gelderblom H R, Krausslich H G. Analysis of HIV particle formation using transient expression of subviral constructs in mammalian cells. Virology. 1992;186:25–39. doi: 10.1016/0042-6822(92)90058-w. [DOI] [PubMed] [Google Scholar]
- 31.Ottmann M, Gabus C, Darlix J-L. The central globular domain of the nucleocapsid protein of human immunodeficiency virus type 1 is critical for virion structure and infectivity. J Virol. 1995;69:1778–1784. doi: 10.1128/jvi.69.3.1778-1784.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Park J, Morrow C D. The nonmyristylated Pr160gag-pol polyprotein of human immunodeficiency virus type 1 interacts with Pr55gag and is incorporated into virus-like particles. J Virol. 1992;66:6304–6313. doi: 10.1128/jvi.66.11.6304-6313.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Poon D T K, Wu J, Aldovini A. Charged amino acid residues of human immunodeficiency virus type 1 nucleocapsid p7 protein involved in RNA packaging and infectivity. J Virol. 1996;70:6607–6616. doi: 10.1128/jvi.70.10.6607-6616.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Prats A C, Sarih L, Gabus C, Litvak S, Keith G, Darlix J L. Small finger protein of avian and murine retroviruses has nucleic acid annealing activity and positions the replication primer tRNA onto genomic RNA. EMBO J. 1988;7:1777–1783. doi: 10.1002/j.1460-2075.1988.tb03008.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rein A. Retroviral RNA packaging: a review. Arch Virol Suppl. 1994;9:513–522. doi: 10.1007/978-3-7091-9326-6_49. [DOI] [PubMed] [Google Scholar]
- 36.Rein A, Harvin D P, Mirro J, Ernst S M, Gorelick R J. Evidence that a central domain of nucleocapsid protein is required for RNA packaging in murine leukemia virus. J Virol. 1994;68:6124–6129. doi: 10.1128/jvi.68.9.6124-6129.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Smith A J, Cho M I, Hammarskjöld M L, Rekosh D. Human immunodeficiency virus type 1 Pr55gag and Pr160gag-pol expressed from a simian virus 40 late replacement vector are efficiently processed and assembled into viruslike particles. J Virol. 1990;64:2743–2750. doi: 10.1128/jvi.64.6.2743-2750.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Smith A J, Srivivasakumar N, Hammarskjöld M-L, Rekosh D. Requirements for incorporation of Pr160gag-pol from human immunodeficiency virus type 1 into virus-like particles. J Virol. 1993;67:2266–2275. doi: 10.1128/jvi.67.4.2266-2275.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Srinivasakumar N, Hammarskjöld M-L, Rekosh D. Characterization of deletion mutations in the capsid region of human immunodeficiency virus type 1 that affect particle formation and Gag-Pol precursor incorporation. J Virol. 1995;69:6106–6114. doi: 10.1128/jvi.69.10.6106-6114.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Stewart L, Schatz G, Vogt V M. Properties of avian retrovirus particles defective in viral protease. J Virol. 1990;64:5076–5092. doi: 10.1128/jvi.64.10.5076-5092.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Terwilliger E F, Cohen E A, Lu Y C, Sodroski J G, Haseltine W A. Functional role of human immunodeficiency virus type 1 Vpu. Proc Natl Acad Sci USA. 1989;86:5163–5167. doi: 10.1073/pnas.86.13.5163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhang Y, Barklis E. Nucleocapsid protein effects in the specificity of retrovirus RNA encapsidation. J Virol. 1995;69:5716–5722. doi: 10.1128/jvi.69.9.5716-5722.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]