Abstract
The papillomavirus E2 proteins can function as sequence-specific transactivators or transrepressors of transcription and as cofactors in viral DNA replication. We previously demonstrated that acute expression of the bovine papillomavirus type 1 (BPV1) E2 protein in HeLa and HT-3 cervical carcinoma cell lines greatly reduced cellular proliferation by imposing a specific G1/S phase growth arrest. In this report, we analyzed the effects of a panel of point mutations in the BPV1 E2 protein to identify the functional requirements for acute growth inhibition. Disruption of E2-specific transactivation by mutations within either the transactivation domain or the DNA binding domain severely impaired E2-mediated growth inhibition in HeLa and HT-3 cells, even though these mutants retain various other E2 activities. This result indicates that functional transactivation activity is required for acute E2-mediated growth inhibition. HeLa cells, which contain a wild-type p53 gene, and HT-3 cells, which contain a transactivation-defective p53 gene, exhibited similar responses to the E2 mutants, suggesting that identical functions of the E2 protein were required for growth arrest regardless of p53 status. Replacement of the E2 transactivation domain with that of the herpes simplex virus VP16 generated a chimeric transactivator that efficiently stimulated expression of an E2-responsive reporter plasmid yet was completely defective for growth inhibition, suggesting that an E2-specific transactivation function is required for growth arrest. Surprisingly, the transactivation-defective E2 mutants were also markedly defective in their ability to repress transcription of the native human papillomavirus type 18 (HPV18) E6/E7 oncogenes in HeLa cells and of the HPV18 promoter present in a transfected reporter plasmid. These mutants were also defective in their ability to increase p53 levels. Therefore, efficient repression of the HPV18 promoter in HeLa cells is not merely a consequence of the binding of an E2 protein to appropriately situated binding sites in the promoter.
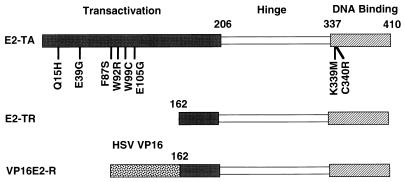
Papillomavirus E2 proteins participate in a variety of essential regulatory functions during the viral life cycle (35, 67). The full-length bovine papillomavirus type 1 (BPV1) E2 protein, E2-TA, is a dimeric sequence-specific DNA binding protein that regulates viral gene transcription (2, 35, 52). In transfection studies, the E2 protein is able to inhibit transcription from the human papillomavirus type 16 (HPV16) or HPV18 E6/E7 promoter, an effect which is thought to occur by the E2 protein binding to the promoter and displacing other necessary transcription factors (4, 5, 16, 19, 20, 25, 28, 33, 42, 45, 54–60). E2-TR, an amino-terminal-truncated form of the E2 protein expressed by BPV1, is expressed from an internal methionine initiation codon and interferes with the activity of E2-TA by competing for DNA binding sites and by forming inactive heterodimers with E2-TA (3, 35). E2-TA also binds to the viral E1 replication protein and is required for efficient viral DNA replication (37, 49, 61, 68, 69). The BPV1 E2 protein has been subjected to detailed mutational analysis which revealed that the conserved amino-terminal domain is required for transactivation, E1 binding, and viral replication activities, while the carboxy-terminal domain is sufficient for sequence-specific DNA binding and dimerization (1, 6, 8, 23, 27, 35, 41, 66) (see Fig. 1).
FIG. 1.
Domain structure of wild-type and mutant E2 proteins. The full-length E2 transactivator (E2-TA), internally initiated transcriptional repressor (E2-TR), and VP16 chimeric transactivator (VP16E2-R) proteins are shown schematically, with the conserved DNA binding domain, the less conserved “hinge” region, and the transactivation domain indicated. The chimeric VP16E2-R protein contains the transactivation domain of the HSV VP16 protein fused to the E2-TR. The numbers refer to the amino acid positions in the full-length E2-TA. Mutations analyzed here are identified by the single-letter amino acid abbreviation of the wild-type amino acid followed by the position of the mutation and then the abbreviation for the substituted amino acid.
The E2 gene also appears to play a role during HPV-induced carcinogenesis. The great majority of human cervical cancers contain integrated HPV DNA and express the viral E6 and E7 oncogenes, which appear to exert their proliferative effects, at least in part, by binding to the tumor suppressor proteins p53 and p105Rb (62, 64). In contrast, the E2 gene is usually disrupted in cervical cancers, suggesting that the loss of the E2 protein is an important step in the development of cervical cancer (13, 39, 48, 62). The ability of E2-TA to inhibit the expression of the HPV18 E6/E7 promoter suggested that disruption of the E2 gene during carcinogenic progression relieved repression of the E6/E7 genes, resulting in increased E6/E7 expression and delivery of an enhanced proliferative signal (46, 60). This model was further supported by the finding that mutations in the HPV16 E2 gene increased the ability of transfected viral DNA to immortalize primary human keratinocytes (44).
To explore the effects of the E2 protein on cell proliferation, the E2 gene was introduced into cell lines derived from cervical cancers, including HeLa cells, which express HPV18 E6 and E7. We used a BPV1/simian virus 40 (SV40) recombinant virus to drive expression of the BPV1 E2 gene in the vast majority of infected cells and demonstrated that expression of the wild-type E2-TA protein in HeLa cells caused an acute and profound decrease in cellular proliferation and a dramatic inhibition of HPV18 E6/E7 expression (31, 32). Growth inhibition was also observed in HT-3 cells, which possess transactivation-defective p53 and were initially classified as HPV DNA negative (14, 15, 40, 47, 71). However, a recent analysis indicates that HT-3 cells do harbor HPV DNA sequences (18). Acute E2-mediated inhibition of cellular proliferation did not appear to be due to toxicity or apoptosis but rather to a block near the G1-to-S-phase cell cycle transition (21, 31). In HeLa cells, the E2 protein activated the dormant p53-dependent growth inhibitory pathway by increasing levels of p53 and the p53-inducible cyclin dependent kinase (cdk) inhibitor p21 (21, 31). Lowered cdk activity resulted in the accumulation of hypophosphorylated p105Rb and decreased E2F expression, thereby preventing the expression of genes required for transit through the G1/S boundary. Because E2 expression led to a dramatic decrease in the E6/E7 mRNA accumulation in HeLa cells, it was plausible that the E2 protein merely reinstated normal growth control by preventing the E6 and E7 proteins from interfering with p53 and pRb function. However, HT-3 cells possess a mutated and defective form of p53 (14, 15, 47), so p53 function was dispensable for growth arrest, at least in this cell line.
In contrast to the G1/S phase arrest seen in HeLa or HT-3 cells infected with the BPV/SV40 recombinant, others have reported that the E2 protein can cause apoptosis (17) or S-phase arrest due to an uncoupling of S phase and mitosis (24). Because these systems appear to express high levels of the E2 protein or to express adenovirus proteins which may affect the cellular phenotype, it appears that the acute cellular response to E2 expression depends on the particular E2 strain, host cell, and expression vector used. The antiproliferative effect of the E2 protein has also been measured in a long-term assay in which cells are cotransfected with E2 expression vectors and a neomycin-selectable marker (21, 60). The E2 gene from BPV1, HPV16, or HPV18 caused a significant reduction in the number of drug-resistant colonies formed by HeLa cells, though no difference in colony numbers was noted for HT-3 cells. Mutants with large deletions in the transactivation domain of E2-TA or with point mutations in the DNA binding domain did not inhibit colony formation, suggesting that the DNA binding and transactivation functions of the E2 protein are required for the reduction of the number of colonies (21). However, such an assay cannot differentiate between specific growth arrest, toxicity, or apoptosis, and it is not readily amenable to biochemical analysis.
To identify the activities of the E2 protein involved in the acute G1-to-S-phase growth inhibition in HeLa and HT-3 cells, we used the BPV1/SV40 recombinant virus system to test the effects of BPV1 E2 mutants containing specific amino acid substitutions in the transactivation and DNA binding domains of the E2 protein. We also tested the effects of these mutants on the expression of HPV18 E6/E7 mRNA and p53 in HeLa cells. Intact transactivation and DNA binding domains were required for efficient growth inhibition in both cell types despite their difference in p53 status. Furthermore, although a heterologous transactivation domain was able to convert the E2-TR protein into a powerful E2 binding site-specific transactivator, it was unable to inhibit cellular proliferation. Unexpectedly, the transactivation function of the E2 protein was also required for efficient repression of HPV18 E6/E7 expression.
MATERIALS AND METHODS
Plasmids and mutagenesis.
The eight E2 point mutations shown in Fig. 1 and the VP16E2-R fusion (which contains amino acids 410 to 490 of the herpes simplex virus [HSV] VP16 transactivation domain linked in frame to BPV1 E2 residues 162 to 410) were transferred into the BPV1/SV40 expression system to allow production of high-titer viral stocks (6, 41, 50). The parental pPava1 BPV1/SV40 plasmid was first modified by inserting a BglII linker (New England Biolabs catalog no. 1001) into the unique BstEII site 5′ of the BPV1 E2 gene to create pPava-5′Bgl. The E5 gene was then disrupted by linearizing pPava-5′Bgl at the unique SpeI site, filling in the 5′ extensions with Klenow DNA polymerase plus all four deoxyribonucleoside triphosphates followed by intramolecular ligation to generate pPava-5′BΔS. The E2 mutants and the chimera were transferred into pPava-5′BΔS by ligating the BamHI/BstXI fragment containing the E2 mutation to the large BglII/BstXI fragment of pPava-5′BΔS. pPava-E2am1 containing an amber stop codon at amino acid 158 has been previously described (50).
For the experiments examining the effect of the transfected E2-TA gene on repression of the E6/E7 promoter, E2amber, E39G, W92R, K399M, and wild-type pPava-5′BΔS were modified to change the E2-TR initiator methionine into threonine (43) by using the QuickChange Site Directed Mutagenesis Kit (Stratagene) with appropriate primers according to the manufacturer’s instructions.
Cells and virus stocks.
HeLa cells were maintained in Dulbecco’s modified Eagle medium (DMEM) supplemented with 10% fetal bovine serum (FBS), and HT-3 cells were maintained in McCoys 5a medium supplemented with 15% FBS. Recombinant BPV1/SV40 stocks were prepared essentially as described previously (31, 50), with the modification that infected-cell pellets were resuspended in Tris-EDTA (pH 8.7) and freeze-thawed three times, and the cell debris was removed by centrifugation. Viral stocks were titered in eight-chamber slides by infecting CMT-4 cells with serial dilutions of virus followed by indirect immunofluorescence of the SV40 VP1 protein (38). This method of titering virus does not depend on E2-mediated transactivation or site-specific DNA binding.
Immunoprecipitation and immunoblotting of BPV1 E2.
HeLa cells (6 × 105) were plated in 100-mm-diameter dishes and allowed to grow overnight. Sufficient virus stock was added to 2 ml of DMEM supplemented with 2% FBS to generate a multiplicity of infection (MOI) of 20. After 2 h of viral absorption, 8 ml of DMEM supplemented with 12% FBS was added, and the cells were incubated at 37°C for an additional 2 days. The cells were washed twice in 4°C phosphate-buffered saline (PBS), scraped from the tissue culture dish in 1 ml of cold PBS, and transferred to a 1.5-ml microcentrifuge tube. After a low-speed spin, the cell pellets were resuspended in 1-ml portions of RIPA buffer (50 mM Tris-Cl [pH 8.0], 150 mM NaCl, 1% Nonidet P-40 [NP-40], 10 mM sodium deoxycholate, 0.1% sodium dodecyl sulfate [SDS], 1 mM phenylmethylsulfonyl fluoride, 5 μg of leupeptin per ml, 5 μg of aprotinin per ml, 1 mM NaF, 0.1 M sodium orthovanadate), disrupted by pipetting, and extracted by rotating 30 min at 4°C. Cellular debris was removed by centrifugation, and the protein concentration was determined by the detergent-resistant bicinchoninic acid (BCA) protein assay (Pierce). Protein (500 μg) from each sample was mixed with 0.2 μg of the BPV1 E2-specific monoclonal antibody B202 (37) purified from mouse ascitic fluid and incubated for 2 h at 4°C. Twenty microliters of a 50% slurry of protein A Sepharose CL-4B (Pharmacia) was then added, and an additional 1-h incubation was carried out. The antibody complexes were then washed twice in RIPA buffer containing protease and phosphatase inhibitors and eluted by boiling for 5 min in 25 μl of 2× Laemmli sample buffer. Proteins were electrophoresed on a 10% polyacrylamide gel containing SDS, transferred to Immobilon P (Millipore) in 200 mM glycine–25 mM Tris, blocked in 5% bovine serum albumin in Tris/S buffer (13 mM Tris-HCl, 170 mM NaCl; buffer pH 7.4) and incubated with a 1/100 dilution of B202 tissue culture supernatant in Tris/S plus 0.01% NP-40 for 2 h. The filter was washed twice for 10 min in Tris/S buffer, once with Tris/S plus 0.1% NP-40, and twice more with Tris/S, followed by a 1-h incubation with a 1:10,000 dilution of a protein A-horseradish peroxidase conjugate (Amersham), subjected to washes as described above, and visualized with the enhanced chemiluminescence (ECL) reagent (Amersham).
Subcellular localization of BPV1 E2.
The localization of DNA binding mutants K339M and C340R was tested in infected COS7 cells. At 20 to 24 h after infection, the cells were fixed in 4% formaldehyde (Buffered Formalde-Fresh; Fisher) for 10 min at room temperature, washed four times with PBS, and made permeable and blocked by incubation with 0.2% Triton X-100 and 0.1 M glycine in PBS for 10 min. The cells were incubated for 1 h at 37°C with a 1:20 dilution of B201 (56) monoclonal antibody supernatant in PBS. Cells were washed with PBS four times, and then a 1:100 dilution of goat anti-mouse fluorescein-conjugated secondary antibody (Jackson Laboratories) was added and incubated for 1 h at 37°C. Cells were washed in PBS four times and mounted with a cover slip using Gel Mount (Biomeda).
To localize the E2-TA transactivation domain mutants, the hemagglutinin (HA) epitope from the plasmid pBSHA (a gift from M. Barbosa, Signal Pharmaceuticals) was fused in frame to the amino termini of the wild-type and mutant E2 coding sequences expressed from pcDNA3 (Invitrogen). HeLa cells were transfected by the method of Chen and Okayama (11), and after 2 days, the cells were fixed with periodate-lysine-paraformaldehyde (36). The cells were made permeable as described above, incubated for 3 h in a 1:30,000 dilution of anti-HA1 12CA5 ascitic fluid in PBS containing 2% control goat serum, washed three times in PBS, and incubated with a 1:300 dilution of a CY3-labeled, goat anti-mouse antibody for 1 h at room temperature. After three washes in PBS, the nuclei were counterstained by treatment with 0.4% Hoechst 33258 in PBS for 5 min and mounted by using a SlowFade Antifade Kit (Molecular Probes).
BPV1 E2 transactivation and transrepression assays.
To measure the transactivation potential of E2 proteins, pPava plasmids expressing wild-type and mutant E2 genes were cotransfected with an E2-responsive reporter construct (p407-1) containing the long control region (LCR) of BPV1 coupled to an enhancerless SV40 early promoter 5′ of the chloramphenicol acetyltransferase (CAT) gene (52). HeLa or HT-3 cells (5 × 105) were plated in 35-mm-diameter dishes and transfected with 1 μg of p407-1 and 0.5 μg of the appropriate BPV1/SV40 E2 expression vector, using 9 or 6 μl of Lipofectamine (Life Technologies), respectively. Between 24 and 48 h posttransfection, cell extracts were prepared by using Reporter Lysis Buffer (Promega) and CAT activity was determined by using the QUAN-T-CAT assay system (Amersham). The results were normalized to a constant amount of protein by the BCA protein assay (Pierce), and a CAT standard curve was used to generate milliunits of CAT activity per milligram of total protein. To facilitate comparisons between experiments and cell lines, the activity of a mutant was divided by that of the wild-type E2 protein to yield the percentage of wild-type activity.
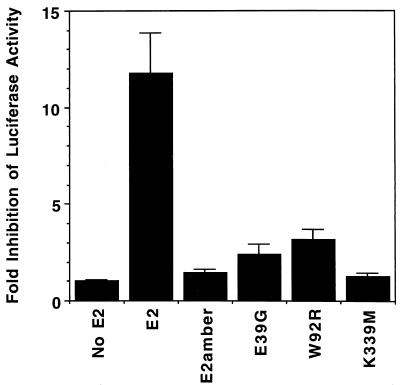
To measure the E2-mediated repression of the HPV18 p105 promoter, a 1,053-bp BamHI fragment of HPV18 (nucleotides 6929 to 119) containing the LCR and p105 promoter was inserted into the BglII site of pGL3-basic vector (Stratagene). In the resulting plasmid, pGL3-HPV18LCR, the HPV sequences were responsible for expression of the luciferase gene. To assay the repression activity due to E2-TA in the absence of E2-TR, the initiator methionine of E2-TR was converted into a threonine in the wild-type and mutant BPV1/SV40 pPava plasmids. Plasmids (0.5 μg) expressing E2, E2amber, the transactivation mutants E39G and W92R, or the DNA binding mutant K339M construct, each containing the E2-TR mutation, were cotransfected with 1 μg of pGL3-HPV18LCR in HeLa cells as described above. Twenty-four hours after transfection, luciferase activity was determined by using the Luciferase Assay System (Promega). Two experiments were performed in duplicate or triplicate, and essentially identical results were obtained. The data were combined and presented as the luciferase activity from pGL3-HPV18LCR cotransfected with a vector unable to produce the E2 protein divided by the activity present after cotransfection with one of the E2 expression vectors to yield fold inhibition of luciferase activity (see Fig. 7).
FIG. 7.
Repression of the HPV18 E6/E7 promoter by wild-type and mutant E2 proteins. A construct containing the HPV18 LCR upstream of a promoterless luciferase gene was cotransfected with vectors expressing wild-type or mutant E2-TA, as indicated, and luciferase activity was determined 24 h posttransfection. All E2 expression vectors contained the mutation at the E2-TR initiation codon (43). The results are the averages of two independent experiments and are expressed as fold inhibition of the HPV18 E6/E7 promoter activity, which is calculated as a ratio of luciferase activity from the HPV18LCR vector cotransfected with a nonspecific plasmid to that from cotransfection with a plasmid expressing E2-TA.
Growth inhibition assays.
Cells (2.5 × 104 HeLa or 3 × 104 HT-3 cells) were plated in each well of a 24-well tissue culture plate and allowed to grow overnight to approximately 5 × 104 cells per well. The cells were infected by the indicated viral stocks at MOIs ranging from 2.5 to 20 in 0.2 ml of the appropriate media supplemented with 2% FBS. Two hours later, 0.8 ml of media containing sufficient FBS was added to produce a final concentration of 10 or 15% FBS, respectively, for HeLa or HT-3 cells. Cell proliferation was determined by measuring [3H]thymidine incorporation into trichloroacetic acid-insoluble material 48 to 60 h postinfection (31). Fold growth inhibition was calculated by dividing the incorporated counts per minute of mock-infected cells by incorporated counts per minute from cells infected with virus. Generally, assays were performed in quadruplicate, and the mutant viruses were assayed in 3 to 11 separate experiments. The data were averaged, and a standard deviation of the mean was calculated for the whole population.
RNA analysis.
Total RNA was prepared from cells 48 h postinfection with Trizol reagent (Life Technologies). Five micrograms of RNA was subjected to formaldehyde-agarose gel electrophoresis, transferred to a nitrocellulose membrane (Biotrace NT from Gelman) or nylon membrane (Nytran from Schleicher & Schuell), and hybridized with [α-32P]dATP random-primed HPV18 E6/E7 probe (22). Amounts of RNA were normalized by probing for γ-actin followed by quantitation on a PhosphorImager (Molecular Dynamics), using volume integration. Fold inhibition of E6/E7 mRNA was calculated by dividing the PhosphorImager signal from mock-infected cells by that from cells infected with the wild-type and mutant viruses. Growth inhibition assays performed in parallel with the RNA extractions confirmed efficient growth inhibition by the wild-type E2 protein.
p53 protein detection.
HeLa cells (2 × 105) were plated in 35-mm-diameter dishes and infected with virus stocks at an MOI of 20. Thirty-two hours postinfection, the cells were washed with PBS and lysed in 1 ml of Trizol reagent. After the standard RNA and DNA fractionation (Life Technologies protocol), protein was quantitatively precipitated by the addition of 3 volumes of acetone, incubation at 4°C for 1 h, and centrifugation. The pellets were washed in 1-ml portions of 75% cold acetone and dissolved in 200-μl portions of 2% SDS. Following the quantitation of protein by the BCA protein assay, 20 μg of total cellular protein from each sample was electrophoresed in an 8% acrylamide–SDS gel and transferred to Immobilon P in a solution consisting of 25 mM Tris, 192 mM glycine, 0.1% SDS, and 20% methanol. The membrane was blocked in TBST (10 mM Tris [pH 7.2], 150 mM NaCl, 0.1% Tween 20) containing 5% nonfat dry milk and sodium azide. A 1-μg/ml solution of a mouse monoclonal anti-p53 antibody (Ab-6; Oncogene Research Products) in TBST with 5% milk and with azide was incubated with the membrane for 1 h, and then the membrane was washed in four changes of TBST over a 30-min period. Following incubation of the membrane with a 1:10,000 dilution of a horseradish peroxidase-labeled goat anti-mouse antibody (Jackson Immunoresearch) in TBST with 5% milk without azide and washes as described above, the p53 protein was visualized by ECL.
RESULTS
Description of E2 mutants.
The domain structure of the full-length BPV1 E2 transactivator (E2-TA) consists of a long amino-terminal transactivation domain, a carboxyl-terminal domain sufficient for DNA binding and dimerization, and an intervening region designated the hinge region (Fig. 1). Both BPV1 and the BPV1/SV40 recombinant also encode E2-TR, a transactivation-defective carboxyl-terminal fragment of the E2 protein. E2-TR has little, if any, growth inhibitory activity under our conditions (32). Figure 1 also shows the locations of the point mutations studied here, consisting of single-amino-acid substitutions within the transactivation and DNA binding domains of the E2 protein. These mutants were previously identified on the basis of their decreased activity as sequence-dependent transcription factors in a genetic screen performed with Saccharomyces cerevisiae (6, 26, 41). A virus expressing an E2 gene with an amber stop codon inserted at position 158 (E2amber) was included in many experiments as a negative control. This mutant encodes the E2-TR protein and a carboxy-truncated E2 protein which is defective for DNA binding, transactivation, papillomavirus DNA replication, and induction of growth inhibition. In addition, a chimeric transactivator that recognizes the E2 DNA binding site was constructed by fusing the heterologous transactivation domain of the HSV VP16 protein to the E2-TR protein (VP16E2-R).
These mutants were transferred to the BPV1/SV40 expression system and characterized. We confirmed by electrophoretic mobility shift assays that both mutations in the DNA binding domain, K339M and C340R (41), disrupted binding to a DNA fragment containing E2 binding sites (data not shown). In the wild-type DNA binding domain, residues 339 and 340 directly contact the E2 binding site (30). In contrast, the mutations in the transactivation domain did not interfere with site-specific DNA binding, as assessed by a two-hybrid assay in yeast (6, 56, 70).
Accumulation of mutant BPV1 E2 proteins within HeLa cells.
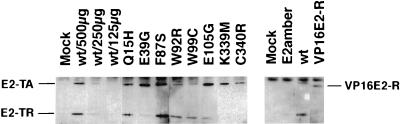
Before examining the biological properties of these mutants, it was necessary to demonstrate that the mutant E2 proteins accumulated in infected cells. HeLa cells were infected with wild-type virus and with each of the mutants at a MOI of 20. To detect the E2 protein, 500 μg of cellular extract prepared 60 h after infection was immunoprecipitated with the anti-BPV1 E2 monoclonal antibody B202, resolved on a denaturing polyacrylamide gel, and immunoblotted with the same antibody. In addition, various amounts of extract from cells expressing the wild-type E2 protein were immunoprecipitated to allow a rough comparison of protein levels in the mutants. The results in Fig. 2 demonstrated that all the full-length mutant E2 proteins stably accumulated in HeLa cells. Several mutants (Q15H, E39G, F87S, E105G, K339M, and C340R) accumulated to levels similar to that of the wild-type protein, whereas W92R and W99C were approximately twofold less abundant. Thus, all of the mutants accumulated to levels at which the wild-type E2 protein caused substantial growth inhibition (see below). In addition, cells infected with each of the viruses also expressed the ∼30-kDa E2-TR protein. These results demonstrated that gross deletions or rearrangements affecting the E2 gene had not occurred during amplification of the virus stocks and established that none of the mutations markedly affected the expression or stability of the mutant E2 proteins.
FIG. 2.
Accumulation of wild-type and mutant E2 proteins in infected HeLa cells. HeLa cells were infected with the indicated viral stocks, and protein was isolated 60 h later. Total protein (500 μg) (except for the wild-type virus [wt] titration) was immunoprecipitated and then immunoblotted with the B202 monoclonal antibody. The migration positions of the full-length E2 protein (E2-TA) and transcriptional repressor (E2-TR) are noted. Various amounts of wild-type virus extract ranging from 125 to 500 μg, as indicated, were assayed in parallel to assess the signal resulting from different levels of the E2 protein. In a separate experiment, viruses expressing the E2amber, VP16E2-R, and wild-type E2 proteins were assayed. The VP16E2-R protein has a lower molecular weight than wild-type E2 and thus migrates more rapidly during electrophoresis.
Nuclear localization of the BPV1 E2 mutants.
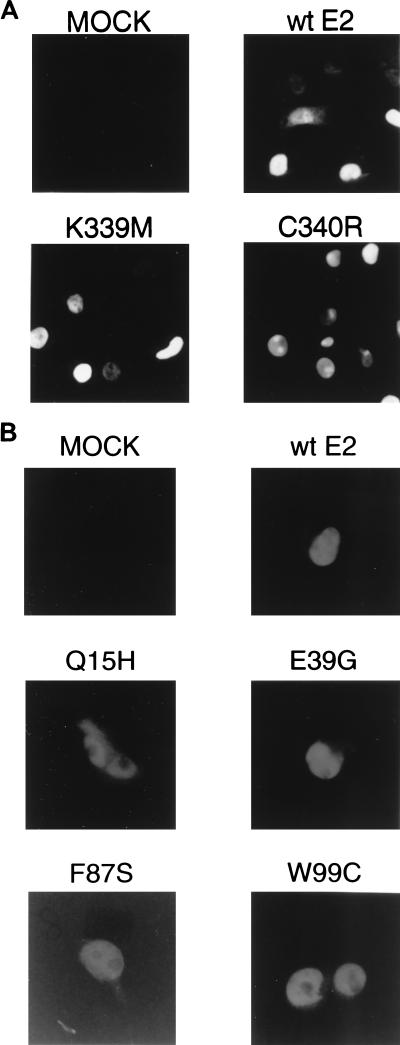
Although none of these mutations are located within signals known to be required for nuclear localization of E2-TA (51), it was possible that the E2 mutant proteins might be unable to enter the nucleus due to disruption of unidentified localization signals or to catastrophic misfolding. Two approaches were used to determine the locations of the E2 proteins. First, COS7 cells were infected with viruses expressing the wild-type E2 protein or DNA binding mutants, which contain mutations in the domain common to E2-TA and E2-TR. After 20 to 24 h, the cells were immunostained with an anti-E2 antibody which detects both the transactivator and transcriptional repressor forms of the E2 protein. As shown in Fig. 3A, the wild-type protein and both of these mutants displayed nuclear localization. A different approach was used to examine the localization of E2 mutants with substitutions in the transactivation domain. Because viruses encoding these mutants also expressed the wild-type E2-TR, which was expected to localize normally, we did not attempt to determine localization by using the anti-E2 antibody. Instead, an HA epitope was inserted at the amino termini of the wild-type and mutant E2-TA proteins, and plasmids encoding these proteins were transfected into HeLa cells. The cells were then immunostained with an anti-HA antibody to determine the subcellular localization of the full-length E2-TA proteins. As shown for representative examples in Fig. 3B, all the mutant and wild-type E2-TA proteins tested in this fashion accumulated within the nucleus, although some mutants, in particular Q15H, displayed some cytoplasmic staining as well.
FIG. 3.
Subcellular localization of wild-type and mutant E2 proteins. (A) COS7 cells mock infected or infected with virus expressing wild-type (wt) E2 or the DNA binding mutant K339M or C340R were fixed and stained for E2 localization with the B201 monoclonal antibody. (B) HeLa cells mock transfected or transfected with vectors expressing HA-tagged wild-type E2-TA or the indicated HA-tagged mutants were fixed and stained for E2 localization with an anti-HA antibody.
Transactivation activity of the E2 mutants.
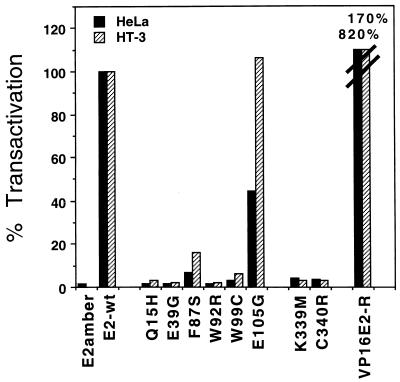
Because the mutants were classified as transactivation defective on the basis of studies in yeast, we determined their transactivation potential in HeLa and HT-3 cells, the same cells used for the growth inhibition assays. The ability of the mutants to transactivate was assayed by cotransfection of the BPV1/SV40 plasmid containing the E2 mutant of interest with a reporter plasmid containing an E2-responsive promoter driving expression of the bacterial CAT gene. Extracts were prepared 48 h later and assayed for CAT activity. This transactivation assay required that the E2 transactivation domain be functional and that the E2 mutant be able to bind to its recognition sequence. As shown in Fig. 4, the wild-type E2 gene efficiently transactivated the reporter plasmid in both cell lines, whereas the amber mutant was defective. Most of the point mutants displayed <5% of the wild-type levels of transactivation in both HeLa and HT-3 cells. Mutant E105G, which was originally classified as leaky for transactivation in yeast, displays substantial activity in both human cell lines. F87S shows slightly more transactivation activity than the markedly defective mutants.
FIG. 4.
Transactivation assay of wild-type and mutant E2 proteins. The indicated E2 mutants were cotransfected with p407-1, a reporter vector responsive to E2-specific transactivation. Twenty-four hours posttransfection, HeLa and HT-3 cell extracts were assayed for CAT activity. The results are presented as percentages of transactivation by the wild-type E2 plasmid (E2-wt). The slashed lines on the two VP16E2-R bars represent values of 820 and 170% in HeLa and HT-3 cells, respectively, that are off the scale.
Growth-inhibitory activity of BPV1 E2 mutants.
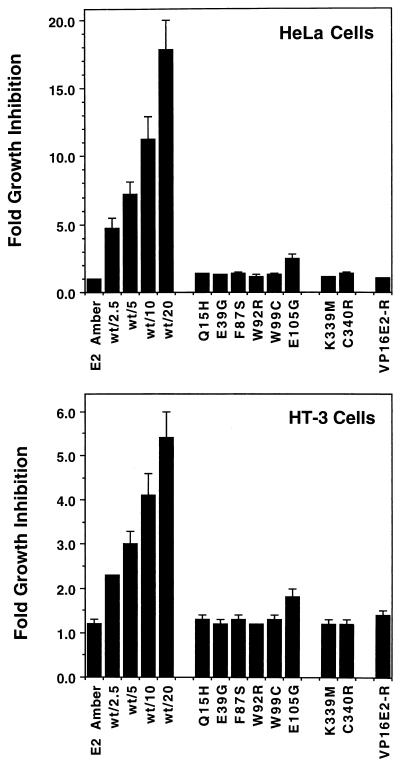
To explore the relationship between the various functions of the E2 protein and its growth-inhibitory activity, we tested the E2 mutants for their ability to inhibit DNA synthesis in HeLa and HT-3 cells. First, we determined whether slight variations in E2 protein levels due to minor differences in viral titer or stability of the mutant proteins could mask an effect of the mutation on growth inhibition. HeLa and HT-3 cells were infected with the BPV1/SV40 recombinant virus containing the wild-type E2 gene at MOIs ranging from 2.5 to 20. The amber mutant was included at an MOI of 20 as a control for nonspecific effects of viral infection. Growth inhibition was assayed by determining incorporation of tritiated thymidine into trichloroacetic acid-precipitable material 48 to 60 h postinfection. In a representative experiment, HeLa cells incorporated 415,000 cpm after mock infection, 335,000 cpm after infection with the E2amber virus, and 11,000 cpm after infection with the wild-type virus at an MOI of 20. In HT-3 cells, incorporation was 82,000, 89,000, and 19,000 cpm, respectively. The results of multiple such experiments are combined and presented in Fig. 5. Although growth inhibition by wild-type E2 clearly decreased with decreasing amounts of virus, even a low MOI of 2.5 was sufficient for readily detectable growth inhibition in both HeLa and HT-3 cells, whereas cells infected at a high MOI with the E2amber mutant showed negligible growth inhibition.
FIG. 5.
Growth inhibition of HeLa and HT-3 cells by the E2 protein. Viral stocks expressing wild-type or mutant E2 were used to infect HeLa or HT-3 cells, and incorporation of tritiated thymidine was assayed 40 to 60 h postinfection. The data are expressed as fold inhibition, which is calculated as the ratio of the thymidine incorporation in mock-infected cells divided by that of infected cells. The left set of bars in each graph show the activity of the E2amber mutant following infection at an MOI of 20 compared to a titration of wild-type E2 following infection at the indicated MOIs. The remainder of the bars in each graph show the fold growth inhibition values of the indicated mutants and the heterologous VP16E2-R transactivator, all following infection at a MOI of 20. The results of multiple experiments have been combined, and the error bars represent 2 standard deviations of the mean.
Extracts from HeLa cells infected at the various MOIs with the wild-type virus were prepared, and E2 protein levels were examined by immunoblotting. The data indicated that the levels of E2 protein roughly correlate with the MOI (data not shown). Thus, growth inhibition was clearly detectable at a concentration of wild-type E2 protein that was lower than the least abundant transactivation-defective mutant protein. Based on these experiments, growth assays of the E2 mutants were carried out after infection at an MOI of 20, conditions under which minor variations in the levels of the mutant E2 protein would evidently not interfere with our ability to measure growth inhibition.
To assess the effects of the mutations on growth inhibition, HeLa or HT-3 cells were infected at an MOI of 20 with viruses containing a wild-type or mutant E2 gene, and DNA synthesis was assayed as described above. The averaged results of these experiments using several preparations of virus for each mutant are presented in Fig. 5. The transactivation-defective E2 mutants were significantly impaired in their ability to inhibit DNA synthesis in both HeLa and HT-3 cells. These mutants were able to suppress DNA synthesis only about 1.5-fold compared to the 20-fold inhibition by the wild-type protein in HeLa cells and 6-fold inhibition in HT-3 cells. The E105G mutant, which retained significant transactivation activity, exhibited an intermediate level of growth suppression that was greater than the other mutants but still less than that induced by wild-type E2 protein even at low MOIs. The pattern of growth suppression by the panel of point mutants was very similar in p53-positive HeLa cells and in p53-negative HT-3 cells. Thus, the ability of the E2 protein to transactivate appears to be an absolute requirement for efficient growth suppression in both cell lines. We note, however, that all of the point mutants retained modest growth inhibitory activity in both cell types, as did the E2 amber mutant in HT-3 cells. This activity was not dependent on site-specific DNA binding since the DNA binding-defective mutants also display this property.
A heterologous transactivator is unable to substitute for the E2 transactivation domain.
The results presented above indicated that transactivation-competent E2 protein was required for efficient growth suppression. To explore the specificity of this requirement for transactivation, we tested whether another strong transactivation domain could substitute for the E2 transactivation domain. We fused the HSV VP16 transactivation domain to the amino terminus of E2-TR, which contains the DNA binding domain of the E2 protein, to construct VP16E2-R and generated virus stocks expressing the fusion protein. As shown in Fig. 2, this chimeric protein accumulated in infected cells. In addition, Fig. 4 shows that in both HeLa and HT-3 cells, the VP16E2-R chimera exhibited substantially higher levels of transactivation of a gene linked to an E2 binding site than did the wild-type E2 protein. Nevertheless, the chimera was markedly defective in its ability to inhibit DNA synthesis, being indistinguishable from the transactivation-defective E2 mutants (Fig. 5). Thus, targeting a strong transactivation domain to E2 binding sites was not sufficient to induce acute growth inhibition, rather some specific feature of the E2 transactivation domain was required. Dowhanick et al. (21) previously reported that a similar chimera did not inhibit colony formation in transfected HeLa cells.
Repression of HPV18 E6/E7 mRNA by the E2 mutants.
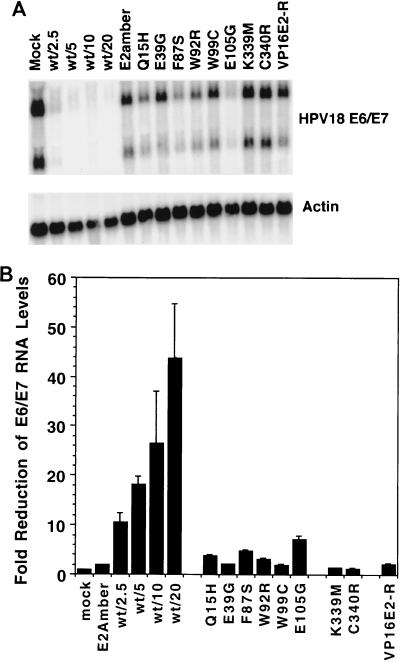
We and others previously showed that expression of the E2 protein in HeLa cells led to the repression of the endogenous HPV18 E6 and E7 oncogene mRNAs (17, 21, 32). Studies in transfected cells suggested that the ability of the E2 protein to bind to E2 binding sites within the HPV18 promoter was essential for repression, presumably by occluding binding sites for necessary cellular transcription factors. To test the effects of the transactivation-defective E2 mutants on expression of the E6 and E7 genes, HeLa cells were infected with virus at an MOI of 20, and after 60 h, total RNA was harvested and analyzed for HPV18 E6/E7 RNA by Northern blotting. An autoradiogram from one experiment is presented in Fig. 6A, and the data from several experiments were quantitated with a PhosphorImager and combined in Fig. 6B. The wild-type E2 protein caused a marked reduction of E6/E7 message levels, ranging from 10-fold at an MOI of 2.5 to ∼45-fold at an MOI of 20. As expected, the DNA binding mutants had no effect on E6/E7 message levels. Surprisingly, though able to bind DNA, the transactivation domain mutants also repressed message levels poorly, reducing expression no more than two- to fivefold compared to the 45-fold repression caused by wild-type E2 at the same MOI. In HeLa cells, the E105G mutant, which retains significant transactivation activity, also repressed E6/E7 to a greater extent than the other mutants. Although the VP16E2-R chimera exhibited strong transactivation activity in HeLa cells, it was unable to repress E6/E7 message to a significant degree. These results demonstrated an unexpected requirement for E2-specific transactivation activity for efficient repression of E6/E7 expression and revealed a strong correlation between the extent of E6/E7 repression and growth inhibition in HeLa cells.
FIG. 6.
Repression of HPV18 E6/E7 message in HeLa cells by BPV1 E2. HeLa cells were infected with BPV1/SV40 expressing wild-type (wt) or mutant E2 proteins, and total RNA was harvested 60 h postinfection. RNA (5 μg) was resolved by formaldehyde agarose gel electrophoresis, transferred to a solid support, and hybridized with a radiolabeled HPV18 E6/E7 probe. After analysis on a PhosphorImager, the probe was removed and the transferred RNA was rehybridized to a radiolabeled actin probe to facilitate comparisons between samples. (A) Autoradiography of the transferred RNA revealed two major E6/E7 messages expressed from integrated copies of the HPV18 genome in HeLa cells. The left set of lanes display the effects of the wild-type E2 protein on E6/E7 message levels following infection at the indicated MOI, while the remainder of the lanes display the indicated E2 mutants following infection at a MOI of 20. The actin-specific signal is shown in the bottom gel. (B) The E6/E7-specific signals were measured by using the PhosphorImager with filters obtained in several independent experiments, and the results are presented in graphical form. The data are expressed as fold inhibition, which is calculated as a ratio of E6/E7 signal from mock-infected cells to that from cells infected with wild-type or mutant E2 viruses.
We also tested the ability of the full-length E2 proteins to repress the HPV18 E6/E7 promoter in a transient-transfection assay. The HPV18 LCR, which contains the HPV18 E6/E7 promoter and the four E2 binding sites that regulate it, was cloned into a promoterless luciferase reporter vector such that expression of the luciferase gene was under the control of the HPV sequence. To prevent expression of E2-TR and focus on E2-TA in these experiments, we mutated the E2-TR initiator methionine to a threonine in the wild-type pPava vector as well as the E2amber, E39G, W92R, and K339M mutants. This mutation does not alter the growth arrest activity of the wild-type BPV1 E2 protein in HeLa or HT-3 cells (32). To measure repression of the HPV18 E6/E7 promoter by wild-type and mutant E2 proteins, we cotransfected HeLa cells with the LCR-luciferase reporter plasmid and various E2 expression vectors. Luciferase activity was measured after 24 h, and the averaged results of two experiments are shown in Fig. 7. The wild-type E2-TA repressed luciferase activity approximately 12-fold, to levels similar to those in cells transfected with the luciferase gene without the HPV LCR. In contrast, the E2amber mutant, the transactivation domain mutants E39G and W92R, and the DNA binding mutant K339M caused only minor decreases in luciferase activity. These results provided direct evidence that transactivation-defective E2-TA proteins are not able to efficiently repress the HPV E6/E7 promoter.
Induction of p53 protein by the E2 mutants in HeLa cells.
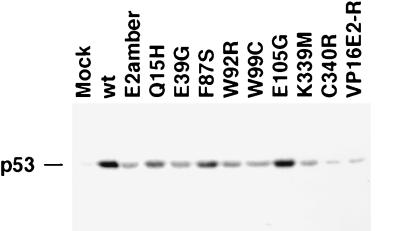
We previously demonstrated that acute E2 expression in HeLa cells led to a dramatic increase in the steady-state level of p53 due to stabilization of the protein (31). To test the ability of the E2 mutants to induce p53, we prepared protein extracts from HeLa cells infected with the various E2 mutants and detected p53 by immunoblotting. As shown in Fig. 8, the wild-type E2 protein caused a substantial increase in the level of p53 compared to that in the mock-infected sample. The transactivation-defective E2 point mutants induced p53 poorly. For the mutants, there was a good correlation between their ability to induce p53 and their diminished ability to repress E6/E7 mRNA expression. The transactivation-competent but growth arrest-negative VP16E2-R mutant was unable to increase the levels of p53. Surprisingly, the leaky E105G mutant led to essentially the same levels of p53 accumulation as did the wild-type E2 protein, despite the intermediate phenotype of this mutant with respect to its ability to arrest cell growth and to repress E6/E7 transcription.
FIG. 8.
Induction of p53 in HeLa cells by BPV1 E2. HeLa cells were infected with BPV1/SV40 recombinant viruses expressing wild-type (wt) or mutant E2 proteins at an MOI of 20. Total protein was isolated 32 h postinfection, and 20 μg was subjected to SDS-polyacrylamide gel electrophoresis and transferred to a polyvinylidene difluoride membrane. A mouse monoclonal antibody directed against human p53 was used to detect the p53 protein by ECL.
DISCUSSION
To investigate the biochemical mechanisms responsible for acute growth inhibition by the BPV1 E2 protein, we analyzed a series of point mutants with single-amino-acid substitutions in either the DNA binding or transactivation domain of this protein. These mutants were chosen with the expectation that such subtle mutations would have minimal effect on the overall conformation, localization, or stability of the E2 protein. In fact, our results demonstrated that all of the mutants tested here retained predominantly nuclear localization and exhibited minimal differences in their expression levels. Furthermore, the transactivation domain mutants retained DNA binding activity (6, 56). In addition, most of the mutants retained the ability to bind the E1 viral replication protein, and several supported viral DNA replication (Table 1). Therefore, it appears that these mutations caused relatively specific defects in E2 function, with ablation of the transcriptional activation function being the one feature common to all the mutants. Thus, any common phenotypic effects of these mutations are likely to be due to the lack of transactivation function rather than to a global disruption of the E2 protein. In contrast, several of the mutants examined by Dowhanick et al. (21) contained large deletions in the E2 protein, which may have affected multiple activities.
TABLE 1.
Summary of E2 mutant phenotypes
| E2 protein | Transactivationa | DNA bindingb | Viral DNA replicationb | Growth inhibitionc |
|---|---|---|---|---|
| Wild-type E2 | ++ | + | ++ | ++ |
| E2amber | − | +d | − | − |
| Q15H | − | + | + | − |
| E39G | − | + | ++ | − |
| F87S | +/− | + | + | − |
| W92R | − | + | − | − |
| W99C | − | + | + | − |
| E105G | +; ++e | + | ++ | +/− |
| K339M | − | − | ND | − |
| C340R | − | − | ND | − |
| VP16E2R | +++ | + | − | − |
Level of transactivation compared to that by wild-type E2. Symbols: −, 0 to 10%; +/−, 10 to 40%; +, 40 to 80%; ++, 80 to 120%; +++, >120%.
Data for DNA binding and viral DNA replication are from references 27, 41, and 56. Symbols for DNA binding: −, no detectable binding; +, readily detectable binding. Symbols for DNA replication: −, <10% of wild-type levels; +, 10 to 50%; ++, >90%. ND, not done.
Fold growth inhibition in infected HeLa cells relative to that in mock-infected cells. Symbols: −, zero to twofold; +/−, two- to fourfold; ++, >10-fold.
Activity of the E2-TR protein which is expressed by this mutan.
E105G scored + in HeLa cells and ++ in HT-3 cells.
Requirements for E2-mediated growth inhibition.
Our results demonstrated that all of the mutants that were severely compromised for transcriptional activation were also severely impaired in their ability to induce growth inhibition. Mutants in this class included Q15H, E39G, F87S, W92R, and W99C with substitutions in the transactivation domain, as well as the DNA binding mutants K339M and C340R. In contrast, the ability of the mutant E2 proteins to bind the viral E1 protein or to support viral DNA replication did not correlate with growth inhibition (Table 1). The defect in growth inhibition displayed by the transactivation domain mutants (which still bind DNA and express the wild-type E2-TR which is itself competent for DNA binding) indicated that binding of an E2 protein at the E2 recognition sites was not sufficient for efficient growth inhibition. The inability of the DNA binding mutants to act as sequence-specific transactivators may underlie their poor growth inhibitory activity, or the DNA binding activity of the E2 protein may play an independent role in growth inhibition. We have been unable to complement the defect in growth inhibition by coinfection with pairs of DNA binding and transactivation domain mutants (data not shown), a result that is consistent with the interpretation that these two classes of mutants share the same fundamental defect.
The E105G mutant was leaky for transactivation in HeLa (and yeast) cells, but it transactivated efficiently in HT-3 cells. This mutant displayed modest growth-inhibitory activity in both human cell types. This phenotype suggests that transactivation is not sufficient for growth inhibition in HT-3 cells. Alternatively, although the transfected mutant efficiently transactivated a cotransfected artificial E2-responsive construct in HT-3 cells, it is possible that this mutant had reduced transactivation or transrepression activity at natural, integrated promoters following delivery of the E2 gene by infection. Thus, a threshold level of E2-mediated transcriptional regulation may not be achieved at these target promoters by E105G.
Dowhanick et al. (21) also concluded that transactivation and DNA binding were required for growth suppression by the BPV1 E2 protein. However, this group did not test the effects of point mutations in the transactivation domain. Furthermore, the mutants were assayed in a colony reduction assay following transfection of HeLa cells, an activity of the E2 protein that may be quite different from the acute G1/S growth inhibition induced in our studies by the E2 gene delivered by infection with the BPV1/SV40 recombinant. In support of this view, HT-3 cell colony formation was not reduced by transfection of the E2 gene, suggesting that the E2 activity measured in this fashion was not equivalent to the acute growth-inhibitory activity measured here. In fact, the observation that transfection of the E2 gene into HeLa cells can lead to apoptosis (17) raises the possibility that a colony reduction assay following transfection may be scoring apoptosis rather than growth arrest.
The BPV1 E2 protein binds to a variety of cellular transcription proteins including TFIIB, TBP, SP1, and AMF-1 (7, 16, 42, 55, 57, 70). Presumably, the assembly of a multiprotein complex consisting of the E2 protein and cellular transcription factors is responsible for its ability to transactivate. The transactivation-defective point mutants studied here displayed varied abilities to bind these cellular partners. For example, Q15H and F87S retained wild-type binding to TFIIB and AMF-1, whereas W99C displayed a specific defect in binding TFIIB (7, 56). Such specific alterations in the protein partners bound by the E2 mutants further emphasized that these mutations did not cause global changes in the structure of the E2 protein. The finding that mutations scattered throughout the first 100 amino acids of the transactivation domain of the E2 protein consistently impaired both transactivation and growth inhibition, even though they differentially affected the ability of the E2 protein to bind various cellular factors, implied that growth inhibition requires an intact transactivation domain rather than the ability of the E2 protein to bind any particular factor. Moreover, the transactivation-competent VP16 chimera was also defective for growth inhibition, indicating that some specific aspect of the E2 transactivation domain was required for growth inhibition.
The requirement for an intact transactivation function is not shared by another viral transactivator, the Zta protein of Epstein-Barr virus, which also inhibits cell growth at the G1/S boundary of the cell cycle (10). The transactivation function of the Zta protein is not required for growth inhibition; rather the zinc finger motif is sufficient (9). This difference indicates that the E2 protein and Zta initiate growth inhibitory cascades by fundamentally different biochemical mechanisms and implies that DNA viruses have evolved multiple distinct mechanisms to impose cell cycle arrest.
Requirements for E2-mediated repression of E6/E7 expression.
Transient-transfection studies showed that repression of the HPV18 E6 and E7 genes requires the presence of E2 binding sites in the p105 promoter, with the E2 binding site proximal to the transcription initiation site having the greatest influence on E2-mediated repression (45, 54, 59). The binding of the E2 protein to this site is thought to interfere with the assembly of a functional transcriptional complex, resulting in repression (20, 58). Consistent with this simple notion, the wild-type E2 protein caused a dramatic inhibition of expression of the endogenous HPV18 E6/E7 genes in HeLa cells, whereas the DNA binding mutants had no effect. However, surprisingly, expression of DNA-binding-competent, transactivation-negative E2 mutants did not cause high-level repression of E6/E7 expression, although these mutants did cause modest repression. We interpret this result to show that E2-mediated assembly of a multiprotein complex within the p105 promoter was responsible for high-level repression and that DNA binding by the E2 protein only or by the E2 protein in association with an incomplete set of cellular partners was not sufficient. Furthermore, the absence of repression by VP16E2-R indicates that such an inhibitory complex cannot be constituted from any assemblage of transcription factors at the promoter but requires some specific feature of the E2 transactivation complex.
E2-TR, which is expressed by all of the mutants including E2amber, did not cause substantial inhibition of E6/E7 repression in our experiments. In contrast, Desaintes et al. (17) reported that transfection of a plasmid expressing the E2-TR protein is sufficient to repress the E6/E7 promoter in HeLa cells. Desaintes et al. may not have been able to distinguish the modest inhibition caused by expression of E2-TR from the more substantial inhibition caused by the transactivation-competent, full-length E2 protein. Alternatively, in their experiments, high levels of expression of the transfected E2 gene from the cytomegalovirus immediate-early promoter may have been sufficient to drive repression by all forms of the E2 protein.
Implications for the mechanism of E2-mediated growth arrest.
The experiments reported here provide several new insights into the mechanism of acute growth inhibition by the BPV1 E2 protein. Our results revealed a strict requirement for an intact transactivation function for acute E2-mediated growth arrest in both HeLa and HT-3 cells. The strikingly similar response of HeLa and HT-3 cells to the various E2 mutants implied that identical functions of the E2 protein were required for growth inhibition in the two cell types, despite their differences in p53 status. Similar functions of the E2 protein appear to be required for the acute growth inhibition studied here and for the reduction in HeLa cell colony formation reported by Dowhanick et al. (21). The requirement for transactivation function in both systems appeared E2 specific because fusion of the VP16 transactivation domain to the E2 DNA binding domain did not restore growth inhibition, even though it generated a strong transactivator protein. The inability of VP16E2-R and the DNA binding mutants to inhibit cell growth ruled out the possibility that growth inhibition resulted merely from transcriptional squelching as a consequence of the expression of a strong transactivator in the cells.
The requirement for E2-mediated transactivation suggests that the E2 protein causes growth inhibition by binding to specific sites in DNA and influencing transcription of growth regulatory genes. Since the mutants described here were defective for both transactivation and transrepression, our results did not establish whether these putative E2 target genes are induced or repressed by the E2 protein. In fact, the phenotype of the transactivation-competent, repression-defective VP16E2-R chimera indicated that growth inhibition correlated better with repression. Furthermore, although the HPV18 E6 and E7 genes are regulated by the E2 protein and their repression is likely to contribute to growth inhibition in HPV-positive cells, cellular genes may also be transcriptional targets of the E2 protein. The extent of acute E2-mediated growth inhibition achieved in HeLa cells, usually greater than 90%, greatly exceeded the growth inhibition attained in these and similar HPV-positive cells by approaches that directly target E6/E7 expression (12, 29, 30a, 34, 53, 63, 65). In addition, the ability of the HPV16 E2 gene to modulate E6/E7 expression does not appear to fully account for the inhibitory effect of HPV16 E2 on viral immortalization (44). Thus, repression of E6/E7 may not be the sole mechanism responsible for the growth-inhibitory effects of papillomavirus E2 proteins. Indeed, the observation that the E105G mutant induces p53 to a level similar to that of wild-type E2 indicates that p53 induction is not sufficient for high-level growth arrest in HeLa cells. The ability of the E2 protein to inhibit the growth of p53-negative HT-3 cells provides further evidence that the E2 protein is able to activate p53-independent growth-inhibitory signals. It is possible that efficient growth inhibition in HeLa cells requires both E6/E7 repression, resulting in restored p53 and pRb activity, and HPV-independent effects of the E2 protein. According to this view, the E2 protein is a less potent inhibitor in HT-3 cells than in HeLa cells, since p53 function cannot be restored in the former cell type. The elucidation of the mechanisms responsible for E2-mediated growth inhibition and the identification of E2-responsive cellular growth regulatory genes will provide further insights into growth control in human cancer cells and may identify new therapeutic targets and suggest new approaches to therapy.
ACKNOWLEDGMENTS
This work was supported by grants from the National Cancer Institute (CA16038 and CA58376) and the American Cancer Society. E.C.G. was supported by an NIH Postdoctoral Training Grant, and L.K.N. was supported in part by postdoctoral fellowships from the American Cancer Society and the Anna Fuller Fund for Cancer Research.
We thank M. Botchan (University of California at Berkeley), T. Nottoli (Yale University), and E. Harlow (Harvard University) for important reagents. We also thank J. Zulkeski for assistance in preparing the manuscript.
REFERENCES
- 1.Abroi A, Kurg R, Ustav M. Transcriptional and replicational activation functions in the bovine papillomavirus type 1 E2 protein are encoded by different structural determinants. J Virol. 1996;70:6169–6179. doi: 10.1128/jvi.70.9.6169-6179.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Androphy E J, Lowy D, Schiller J. Bovine papillomavirus E2 transactivating gene product binds to specific sites in papillomavirus DNA. Nature. 1987;325:70–73. doi: 10.1038/325070a0. [DOI] [PubMed] [Google Scholar]
- 3.Barsoum J, Prakash S S, Han P, Androphy E J. Mechanism of action of the papillomavirus E2 repressor: repression in the absence of DNA binding. J Virol. 1992;66:3941–3945. doi: 10.1128/jvi.66.6.3941-3945.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bernard B A, Bailly C, Lenoir M-C, Darmon M, Thierry F, Yaniv M. The human papillomavirus type 18 (HPV18) E2 gene product is a repressor of the HVP18 regulatory region in human keratinocytes. J Virol. 1989;63:4317–4324. doi: 10.1128/jvi.63.10.4317-4324.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bouvard V, Storey A, Pim D, Banks L. Characterization of the human papillomavirus E2 protein: evidence of trans-activation and trans-repression in cervical keratinocytes. EMBO J. 1994;13:5451–5459. doi: 10.1002/j.1460-2075.1994.tb06880.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Breiding D E, Grossel M J, Androphy E J. Genetic analysis of the bovine papillomavirus E2 transcriptional activation domain. Virology. 1996;221:34–43. doi: 10.1006/viro.1996.0350. [DOI] [PubMed] [Google Scholar]
- 7.Breiding D E, Sverdrup F, Grossel M J, Moscufo N, Boonchai W, Androphy E J. Functional interaction of a novel cellular protein with the papillomavirus E2 transactivation domain. Mol Cell Biol. 1997;17:7208–7219. doi: 10.1128/mcb.17.12.7208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brokaw J L, Blanco M, McBride A A. Amino acids critical for the functions of the bovine papillomavirus type 1 E2 transactivator. J Virol. 1996;70:23–29. doi: 10.1128/jvi.70.1.23-29.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cayrol C, Flemington E. G0/G1 growth arrest mediated by a region encompassing the basic leucine zipper (bZIP) domain of the Epstein-Barr virus transactivator Zta. J Biol Chem. 1996;271:31799–31802. doi: 10.1074/jbc.271.50.31799. [DOI] [PubMed] [Google Scholar]
- 10.Cayrol C, Flemington E K. The Epstein-Barr virus bZIP transcription factor Zta causes G0/G1 cell cycle arrest through induction of cyclin-dependent kinase inhibitors. EMBO J. 1996;15:2748–2759. [PMC free article] [PubMed] [Google Scholar]
- 11.Chen C, Okayama H. High-efficiency transformation of mammalian cells by plasmid DNA. Mol Cell Biol. 1987;7:2745–2752. doi: 10.1128/mcb.7.8.2745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen Z, Kamath P, Zhang S, St. John L, Adler-Storthz K, Shillitoe E J. Effects on tumor cells of ribozymes that cleave the RNA transcripts of human papillomavirus type 18. Cancer Gene Ther. 1996;3:18–23. [PubMed] [Google Scholar]
- 13.Cripe T P, Haugen T H, Turk J P, Tabatabai F, Schmid III P G, Dürst M, Gissmann L, Roman A, Turek L P. Transcriptional regulation of the human papillomavirus-16 E6-E7 promoter by a keratinocyte-dependent enhancer, and by viral E2 trans-activator and repressor gene products: implications for cervical carcinogenesis. EMBO J. 1987;6:3745–3753. doi: 10.1002/j.1460-2075.1987.tb02709.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Crook T, Marston N J, Sara E A, Vousden K H. Transcriptional activation by p53 correlates with suppression of growth but not transformation. Cell. 1994;79:817–827. doi: 10.1016/0092-8674(94)90071-x. [DOI] [PubMed] [Google Scholar]
- 15.Crook T, Wrede D, Vousden K H. p53 point mutation in HPV-negative human cervical carcinoma cell lines. Oncogene. 1991;6:873–875. [PubMed] [Google Scholar]
- 16.Demeret C, Yaniv M, Thierry F. The E2 transcriptional repressor can compensate for Sp1 activation of the human papillomavirus type 18 early promoter. J Virol. 1994;68:7075–7082. doi: 10.1128/jvi.68.11.7075-7082.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Desaintes C, Demeret C, Goyat S, Yaniv M, Thierry F. Expression of the papillomavirus E2 protein in HeLa cells leads to apoptosis. EMBO J. 1997;16:504–514. doi: 10.1093/emboj/16.3.504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.deVilliers, E.-M. Personal communication.
- 19.Dong G, Broker T R, Chow L T. Human papillomavirus type 11 E2 proteins repress the homologous E6 promoter by interfering with the binding of host transcription factors to adjacent elements. J Virol. 1994;68:1115–1127. doi: 10.1128/jvi.68.2.1115-1127.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dostatni N, Lambert P F, Sousa R, Ham J, Howley P M, Yaniv M. The functional BPV-1 E2 trans-activating protein can act as a repressor by preventing formation of the initiation complex. Genes Dev. 1991;5:1657–1671. doi: 10.1101/gad.5.9.1657. [DOI] [PubMed] [Google Scholar]
- 21.Dowhanick J J, McBride A A, Howley P M. Suppression of cellular proliferation by the papillomavirus E2 protein. J Virol. 1995;69:7791–7799. doi: 10.1128/jvi.69.12.7791-7799.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Feinberg A P, Vogelstein B. A technique for radiolabeling DNA restriction endonuclease fragments to high specific activity. Anal Biochem. 1983;132:6–13. doi: 10.1016/0003-2697(83)90418-9. . (Addendum, 137:266–267, 1984.) [DOI] [PubMed] [Google Scholar]
- 23.Ferguson M K, Botchan M R. Genetic analysis of the activation domain of bovine papillomavirus protein E2: its role in transcription and replication. J Virol. 1996;70:4193–4199. doi: 10.1128/jvi.70.7.4193-4199.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Frattini M G, Hurst S D, Lim H B, Swaminathan S, Laimins L A. Abrogation of a mitotic checkpoint by E2 proteins from oncogenic human papillomaviruses correlates with increased turnover of the p53 tumor suppressor protein. EMBO J. 1997;16:318–331. doi: 10.1093/emboj/16.2.318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Garcia-Carranca A, Thierry F, Yaniv M. Interplay of viral and cellular protein along the long control region of human papillomavirus type 18. J Virol. 1988;62:4321–4330. doi: 10.1128/jvi.62.11.4321-4330.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Grossel M J, Barsoum J, Prakash S S, Androphy E J. The BPV-1 E2 DNA-contact helix cysteine is required for transcriptional activation but not replication in mammalian cells. Virology. 1996;217:301–310. doi: 10.1006/viro.1996.0117. [DOI] [PubMed] [Google Scholar]
- 27.Grossel M J, Sverdrup F, Breiding D E, Androphy E J. Transcriptional activation function is not required for stimulation of DNA replication by bovine papillomavirus type 1 E2. J Virol. 1996;70:7264–7269. doi: 10.1128/jvi.70.10.7264-7269.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Guido M C, Zamorano R, Garrido-Guerrero E, Gariglio P, Garcia-Carranca A. Early promoters of genital and cutaneous human papillomaviruses are differentially regulated by the bovine papillomavirus type 1 E2 gene product. J Gen Virol. 1992;73:1395–1400. doi: 10.1099/0022-1317-73-6-1395. [DOI] [PubMed] [Google Scholar]
- 29.Hamada K, Sakaue M, Alemany R, Zhang W W, Horio Y, Roth J A, Mitchell M F. Adenovirus-mediated transfer of HPV16 E6/E7 antisense RNA to human cervical cancer cells. Gynecol Oncol. 1996;63:219–227. doi: 10.1006/gyno.1996.0310. [DOI] [PubMed] [Google Scholar]
- 30.Hegde R S, Grossman S R, Laimins L A, Sigler P B. Crystal structure at 1.7 Å of the bovine papillomavirus-1 E2 DNA-binding domain bound to its DNA target. Nature. 1992;359:505–512. doi: 10.1038/359505a0. [DOI] [PubMed] [Google Scholar]
- 30a.Hu G, Liu W, Hanania E G, Fu S, Wang T, Deisseroth A B. Suppression of tumorigenesis by transcription units expressing the antisense E6 and E7 messenger RNA (mRNA) for the transforming proteins of the human papilloma virus and the sense mRNA for the retinoblastoma gene in cervical carcinoma cells. Cancer Gene Ther. 1995;2:19–32. [PubMed] [Google Scholar]
- 31.Hwang E-S, Naeger L K, DiMaio D. Activation of the endogenous p53 growth inhibitory pathway in HeLa cervical carcinoma cells by expression of the bovine papillomavirus E2 gene. Oncogene. 1996;12:795–803. [PubMed] [Google Scholar]
- 32.Hwang E, Riese D R, Settleman J, Nilson L, Honig J, Flynn S, DiMaio D. Inhibition of cervical carcinoma cell line proliferation by introduction of a bovine papillomavirus regulatory gene. J Virol. 1993;67:3720–3729. doi: 10.1128/jvi.67.7.3720-3729.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jackson M E, Campo M S. Both viral E2 protein and the cellular factor PEBP2 regulate transcription via E2 consensus sites within the bovine papillomavirus type 4 long control region. J Virol. 1995;69:6038–6046. doi: 10.1128/jvi.69.10.6038-6046.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Madrigal M, Janicek M F, Sevin B U, Perras J, Estape R, Penalver M, Averette H E. In vitro antigene therapy targeting HPV-16 E6 and E7 in cervical carcinoma. Gynecol Oncol. 1997;64:18–25. doi: 10.1006/gyno.1996.4515. [DOI] [PubMed] [Google Scholar]
- 35.McBride A A, Romanczuk H, Howley P M. The papillomavirus E2 regulatory proteins. J Biol Chem. 1991;266:18411–18414. [PubMed] [Google Scholar]
- 36.McLean I W, Nakane P K. Periodate-lysine-paraformaldehyde fixative. A new fixation for immunoelectron microscopy. J Histochem Cytochem. 1974;22:1077–1083. doi: 10.1177/22.12.1077. [DOI] [PubMed] [Google Scholar]
- 37.Mohr I J, Clark R, Sun E, Androphy E J, Macpherson P, Botchan M R. Targeting the E1 replication protein to the papillomavirus origin of replication by complex formation with the E2 transactivator. Science. 1990;250:1694–1699. doi: 10.1126/science.2176744. [DOI] [PubMed] [Google Scholar]
- 38.Naeger, L. K., and D. DiMaio. Unpublished data.
- 39.Park J S, Hwang E S, Park S N, Ahn H K, Um S J, Kim C J, Kim S J, Namkoong S E. Physical status and expression of HPV genes in cervical cancers. Gynecol Oncol. 1997;65:121–129. doi: 10.1006/gyno.1996.4596. [DOI] [PubMed] [Google Scholar]
- 40.Pater M M, Pater A. Human papillomavirus type 16 and 18 sequences in carcinoma cell lines of the cervix. Virology. 1985;145:313–318. doi: 10.1016/0042-6822(85)90164-3. [DOI] [PubMed] [Google Scholar]
- 41.Prakash S S, Grossman S R, Pepinsky R B, Laimins L A, Androphy E J. Amino acids necessary for DNA contact and dimerization imply novel motifs in the papillomavirus E2 trans-activator. Genes Dev. 1992;6:105–116. doi: 10.1101/gad.6.1.105. [DOI] [PubMed] [Google Scholar]
- 42.Rank N M, Lambert P F. Bovine papillomavirus type 1 E2 transcriptional regulators directly bind two cellular transcription factors, TFIID and TFIIB. J Virol. 1995;69:6323–6334. doi: 10.1128/jvi.69.10.6323-6334.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Riese D J, Settleman J, Neary K, DiMaio D. Bovine papillomavirus E2 repressor mutant displays a high-copy-number phenotype and enhanced transforming activity. J Virol. 1990;64:944–949. doi: 10.1128/jvi.64.2.944-949.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Romanczuk H, Howley P M. Disruption of either the E1 or the E2 regulatory gene of human papillomavirus type 16 increases viral immortalization capacity. Proc Natl Acad Sci USA. 1992;89:3159–3163. doi: 10.1073/pnas.89.7.3159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Romanczuk H, Thierry F, Howley P M. Mutational analysis of cis elements involved in E2 modulation of human papillomavirus type 16 P97 and type 18 P105 promoters. J Virol. 1990;64:2849–2859. doi: 10.1128/jvi.64.6.2849-2859.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sang B-C, Barbosa M S. Increased E6/E7 transcription in HPV 18-immortalized human keratinocytes results from inactivation of E2 and additional cellular events. Virology. 1992;189:448–455. doi: 10.1016/0042-6822(92)90568-a. [DOI] [PubMed] [Google Scholar]
- 47.Scheffner M, Münger K, Byrne J C, Howley P M. The state of the p53 and retinoblastoma genes in human cervical carcinoma cell lines. Proc Natl Acad Sci USA. 1991;88:5523–5527. doi: 10.1073/pnas.88.13.5523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Schneider-Maunoury S, Croissant O, Orth G. Integration of human papillomavirus type 16 DNA sequences: a possible early event in the progression of genital tumors. J Virol. 1987;61:3295–3298. doi: 10.1128/jvi.61.10.3295-3298.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sedman J, Stenlund A. Co-operative interaction between the initiator E1 and the transcriptional activator E2 is required for replicator specific DNA replication of bovine papillomavirus in vivo and in vitro. EMBO J. 1995;14:6218–6228. doi: 10.1002/j.1460-2075.1995.tb00312.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Settleman J, DiMaio D. Efficient transactivation and morphologic transformation by bovine papillomavirus genes expressed from a bovine papillomavirus/simian virus 40 recombinant virus. Proc Natl Acad Sci USA. 1988;85:9007–9011. doi: 10.1073/pnas.85.23.9007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Skiadopoulos M H, McBride A A. The bovine papillomavirus type 1 E2 transactivator and repressor proteins use different nuclear localization signals. J Virol. 1996;70:1117–1124. doi: 10.1128/jvi.70.2.1117-1124.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Spalholz B A, Yang Y-C, Howley P M. Transactivation of a bovine papilloma virus transcriptional regulatory element by the E2 gene product. Cell. 1985;42:183–191. doi: 10.1016/s0092-8674(85)80114-8. [DOI] [PubMed] [Google Scholar]
- 53.Steele C, Cowsert L M, Shillitoe E J. Effects of human papillomavirus type 18-specific antisense oligonucleotides on the transformed phenotype of human carcinoma cell lines. Cancer Res. 1993;53:2330–2337. [PubMed] [Google Scholar]
- 54.Steger G, Corbach S. Dose-dependent regulation of the early promoter of human papillomavirus type 18 by the viral E2 protein. J Virol. 1997;71:50–58. doi: 10.1128/jvi.71.1.50-58.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Steger G, Ham J, Lefebvre O, Yaniv M. The bovine papillomavirus 1 E2 protein contains two activation domains: one that interacts with TBP and another that functions after TBP binding. EMBO J. 1995;14:329–340. doi: 10.1002/j.1460-2075.1995.tb07007.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sverdrup, F., D. E. Breiding, and E. J. Androphy. Unpublished data.
- 57.Tan S H, Leong L E, Walker P A, Bernard H U. The human papillomavirus type 16 E2 transcription factor binds with low cooperativity to two flanking sites and represses the E6 promoter through displacement of Sp1 and TFIID. J Virol. 1994;68:6411–6420. doi: 10.1128/jvi.68.10.6411-6420.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tan S H, Gloss B, Bernard H U. During negative regulation of the human papillomavirus-16 E6 promoter, the viral E2 protein can displace Sp1 from a proximal promoter element. Nucleic Acids Res. 1992;20:251–266. doi: 10.1093/nar/20.2.251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Thierry F, Howley P M. Functional analysis of E2-mediated repression of the HPV18 P105 promoter. New Biol. 1991;3:90–100. [PubMed] [Google Scholar]
- 60.Thierry F, Yaniv M. The BPV1-E2 trans-acting protein can be either an activator or a repressor of the HPV18 regulatory protein. EMBO J. 1987;6:3391–3397. doi: 10.1002/j.1460-2075.1987.tb02662.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ustav M, Stenlund A. Transient replication of BPV-1 requires two viral polypeptides encoded by the E1 and E2 open reading frames. EMBO J. 1991;10:449–457. doi: 10.1002/j.1460-2075.1991.tb07967.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Villa L L. Human papillomaviruses and cervical cancer. Adv Cancer Res. 1997;71:321–341. doi: 10.1016/s0065-230x(08)60102-5. [DOI] [PubMed] [Google Scholar]
- 63.von Knebel Doeberitz M, Oltersdorf T, Schwarz E, Gissmann L. Correlation of modified human papilloma virus early gene expression with altered growth properties in C4-1 cervical carcinoma cells. Cancer Res. 1988;48:3780–3786. [PubMed] [Google Scholar]
- 64.Vousden K. Interactions of human papillomavirus transforming proteins with the products of tumor suppressor genes. FASEB J. 1993;7:872–879. doi: 10.1096/fasebj.7.10.8393818. [DOI] [PubMed] [Google Scholar]
- 65.Watanabe S, Kanda T, Yoshiike K. Growth dependence of human papillomavirus 16 DNA-positive cervical cancer cell lines and human papillomavirus 16-transformed human and rat cells on the viral oncoproteins. Jpn J Cancer Res. 1993;84:1043–1049. doi: 10.1111/j.1349-7006.1993.tb02799.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Winokur P L, McBride A A. Separation of the transcriptional activation and replication functions of the bovine papillomavirus-1 E2 protein. EMBO J. 1992;11:4111–4118. doi: 10.1002/j.1460-2075.1992.tb05504.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wu X, Xiao W, Brandsma J L. Papilloma formation by cottontail rabbit papillomavirus requires E1 and E2 regulatory genes in addition to E6 and E7 transforming genes. J Virol. 1994;68:6097–6102. doi: 10.1128/jvi.68.9.6097-6102.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Yang L, Li R, Mohr I J, Clark R, Botchan M R. Activation of BPV-1 replication in vitro by the transcription factor E2. Nature. 1991;353:628–632. doi: 10.1038/353628a0. [DOI] [PubMed] [Google Scholar]
- 69.Yang L, Mohr I, Li R, Nottoli T, Sun S, Botchan M. Transcription factor E2 regulates BPV-1 DNA replication in vitro by direct protein-protein interaction. Cold Spring Harbor Symp Quant Biol. 1991;LVI:335–346. doi: 10.1101/sqb.1991.056.01.040. [DOI] [PubMed] [Google Scholar]
- 70.Yao J-M, Breiding D E, Androphy E J. Functional interaction of the papillomavirus E2 transactivation domain with TFIIB. J Virol. 1998;72:1013–1019. doi: 10.1128/jvi.72.2.1013-1019.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Yee C I, Krishnan-Hewlett I, Baker C C, Schlegel R, Howley P M. Presence and expression of human papillomavirus sequences in human cervical carcinoma cell lines. Am J Pathol. 1985;119:361–366. [PMC free article] [PubMed] [Google Scholar]