Abstract
Polygenic risk scores (PRSs) summarize the genetic predisposition of a complex human trait or disease and may become a valuable tool for advancing precision medicine. However, PRSs that are developed in populations of predominantly European genetic ancestries can increase health disparities due to poor predictive performance in individuals of diverse and complex genetic ancestries. We describe genetic and modifiable risk factors that limit the transferability of PRSs across populations and review the strengths and weaknesses of existing PRS construction methods for diverse ancestries. Developing PRSs that benefit global populations in research and clinical settings provides an opportunity for innovation and is essential for health equity.
Introduction
Genome-wide association studies (GWAS) have discovered thousands of variants associated with complex human traits, illustrating the high polygenicity of many common diseases and emphasizing the potential of leveraging these findings for genetic prediction of health outcomes. Although each individual associated variant accounts for a small proportion of phenotypic variance, aggregating information across multiple variants into a single score creates a compendium of an individual’s genetic predisposition for a given complex trait or disease. Calculated as a sum of alleles weighted by their estimated effect sizes, these cumulative genetic profiles are commonly referred to as polygenic risk scores (PRSs) when derived for a binary disease outcome or polygenic scores when calculated for a general trait.
PRSs have the potential to advance precision medicine by improving disease stratification and prioritization of high-risk individuals for appropriate interventions, enabling more accurate diagnoses and predicting therapeutic outcomes1. Nearly two decades of GWAS have provided a rich foundation of discoveries to facilitate PRS construction2. However, more than 85% of GWAS have been undertaken in individuals of European ancestries, and PRSs derived from these studies can be substantially less predictive for other genetic ancestries, potentially exacerbating health disparities3,4. The growing recognition of this issue has spurred methodological innovations to integrate data from diverse populations, enabling more powerful analyses of existing studies, as well as the establishment of new biobanks, consortia and data collection initiatives of diverse populations5.
Here, we consider the principles and methods of applying PRSs across global populations, complementing existing reviews on PRS methodology and clinical utility1,6-10. Specifically, we consider how genetic and non-genetic factors impact PRS performance, how new methods might improve their transferability across populations and implications for PRS clinical utility. We describe the technical challenges associated with the development of PRS predictive modelling within the context of social and environmental influences, and discuss the evaluation of model performance and clinical utility in populations of diverse ancestral backgrounds. These aspects are especially important given that many individuals have complex genetic ancestries shaped by recent admixture (Fig. 1). In addition to these challenges, we highlight emerging resources and opportunities for the development of new methods to enable more widespread and equitable translation of PRSs.
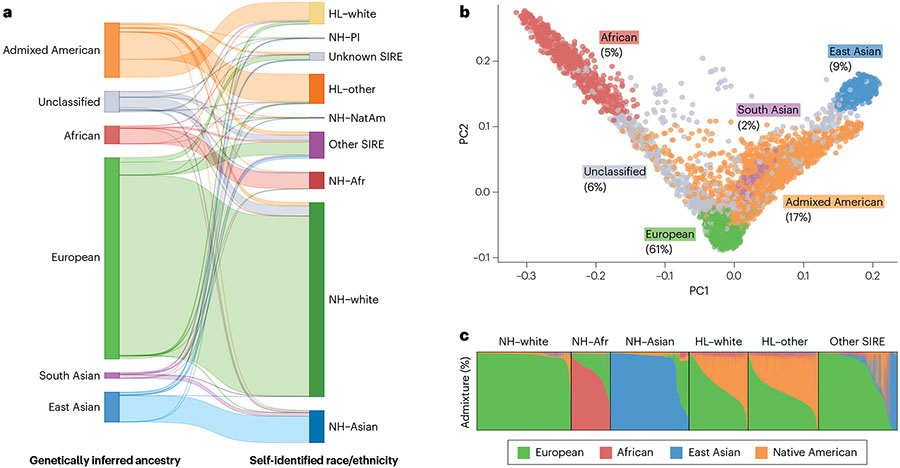
Fig. 1 ∣. Complex genetic ancestries and admixture using data from UCLA-ATLAS.
a, Comparison between genetically inferred ancestry and self-identified race and ethnicity (SIRE): Hispanic/Latino (HL), non-Hispanic/Latino (NH), Pacific Islander (PI), Native American (NatAm) and African/African American (Afr). Genetically inferred ancestry labels are assigned based on proximity to 1000 Genomes reference populations in principal component (PC) space using the k-nearest neighbour algorithm. SIRE is a composite label based on separate entries in the ‘Race’ and ‘Ethnicity’ fields extracted from medical records. b, First two PCs of the genetic data. Each dot represents an individual, with colours corresponding to their assigned genetically inferred ancestry cluster. A non-trivial percentage of individuals could not be categorized into a ‘homogeneous’ or ‘continental’ population. c, Unsupervised clustering of the genetic data. Each column represents the proportion of the global genetic ancestry of an individual with respect to 1000 Genomes reference populations.
Genetic factors influencing PRS performance
PRSs have lower accuracy in cross-population prediction when the target sample is genetically distant from the discovery GWAS sample, regardless of the PRS construction method3,11-16 (Fig. 2a,b). This is akin to agricultural genetics, in which the accuracy of genomic prediction decreases as the genetic distance between the training and target populations increases17-20. Several genetic factors can influence the genetic architecture of a complex trait and may limit the transferability of PRSs across populations, including differences in heritability, causal allele frequencies, allelic effect sizes and linkage disequilibrium (LD) patterns.
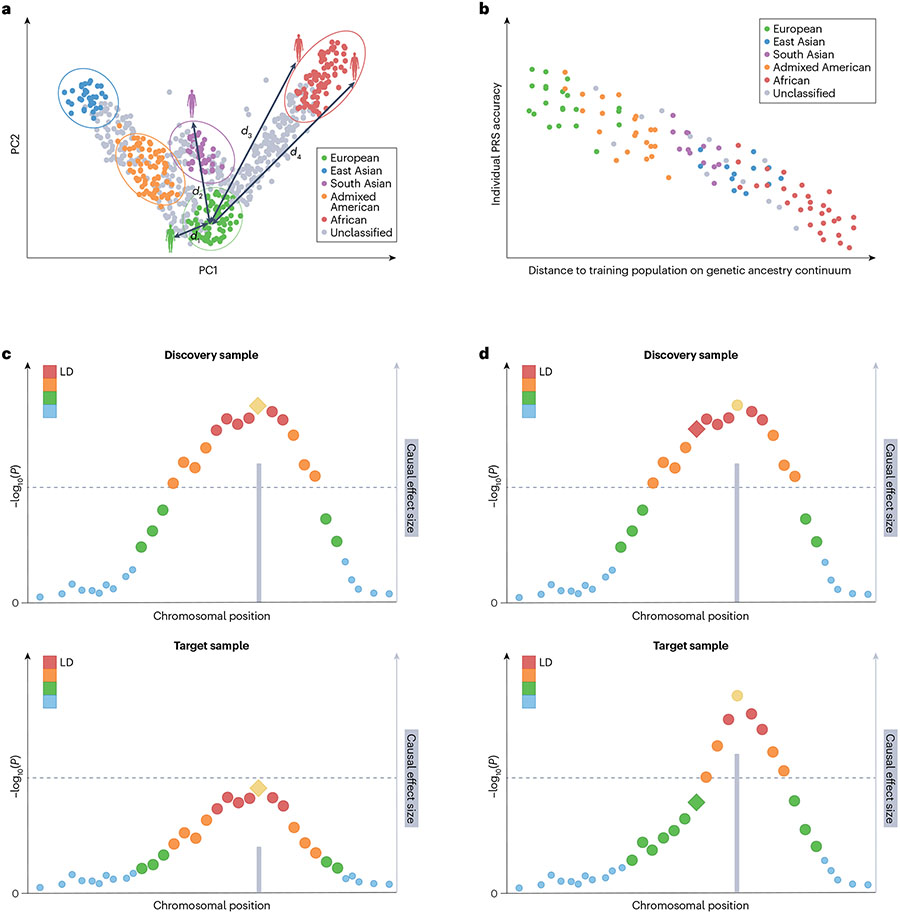
Fig. 2 ∣. Genetic factors that can influence PRS performance.
a, First two principal components (PCs) of the genetic data. Each dot represents an individual. Individuals are assigned discrete population labels by applying arbitrary cut-offs to the genetic ancestry continuum. Different colours represent different population labels. Grey dots represent individuals who are unclassified. A genetic distance (d) can be calculated between each individual and the centre of the discovery genome-wide association study (GWAS) samples in the PC space. b, Prediction accuracy of the polygenic risk score (PRS) shows individual to individual variation and decreases along the genetic ancestry continuum when the genetic distance between the training and target samples increases. c, Differences in causal allelic effect size between the discovery (upper graph) and target (lower graph) samples can influence the accuracy of PRS across populations. d, Differences in linkage disequilibrium (LD) patterns between the discovery (upper graph) and target (lower graph) samples can influence the accuracy of PRS across populations. In panels c and d, each dot represents the marginal association strength of a genetic variant. The lead (most associated) variant in yellow represents the causal variant and the grey bar represents its effect size. Other variants are coloured by descending degrees of LD with the causal variant (ordered red, orange, green and blue dots). Diamond represents the variant (which may be a tagging variant) used in PRS construction. Dashed line represents genome-wide significance.
Heritability, the percentage of variation in a trait attributed to genetics, can differ across populations. Array heritability21, the proportion of phenotypic variation that can be captured by the additive effects of SNPs assayed and imputed from a GWAS array, limits the prediction accuracy of a PRS based on common genetic variation in a given population22-26. As heritability measures the relative contribution of genetic factors to total inter-individual trait variation in a specific population or context, differences in sociocultural factors, environmental exposures and measurement errors between populations that affect total phenotypic variance can alter heritability estimates and influence the accuracy of PRSs (discussed further below). It has been shown that even within a relatively homogeneous population, heritability – and thus the predictive accuracy of PRSs – can differ by major demographic variables such as age, sex and socioeconomic status27,28.
The transferability of PRSs across populations might also be impacted by differences in the frequency of causal variants and their allelic effect sizes (Fig. 2c). These factors can vary due to demographic histories (such as mutations in historical migrant groups followed by genetic drift or population bottlenecks) or gene by environment interactions. Although existing studies have shown extensive genetic overlap across ancestries for a range of complex traits and diseases both at the variant level and the genome-wide level (via cross-population genetic correlation analyses), especially between European and East Asian populations29-32, widespread allelic effect heterogeneity has also been observed33,34. Even assuming identical allelic effects, population differences in allele frequencies can impact the amount of phenotypic variance explained by these variants, and thus PRS performance. As the vast majority of GWAS have been conducted in individuals of European ancestries, most existing PRSs have been constructed from variants that are common in European populations, which may have substantially lower frequencies in other populations. Moreover, causal variants in non-European ancestry populations may not be detectable in European ancestry GWAS due to low allele frequencies in European ancestries, and thus limited statistical power, further reducing the generalizability of PRSs.
The marked differences in LD patterns across populations can also impact PRS transferability. For example, the size of LD blocks is, on average, much smaller in African populations than in European or East Asian populations35,36. As a result, many more variants are needed to capture the same level of genetic variation in African populations than in other populations. Many of the variants used in PRS construction are not causal but are merely in LD with causal variants, and many early genotyping arrays were designed based on European-enriched variants. Thus, differences in LD across populations can affect the transferability of PRSs derived from European GWAS (Fig. 2d). Taken together, the number of causal alleles and their allelic effect size distributions25,26 coupled with their frequencies and local LD patterns can have a complex impact on the performance of PRSs across populations. Furthermore, although most PRSs only capture additive genetic effects tagged by assayed or imputed variants, non-linear genetic effects including allelic heterogeneity, haplotype effects and gene by gene interactions can influence the genetic basis of a complex trait or disease37 and further complicate the transferability of PRSs.
It is important to recognize that the analysis and comparison of genetic architecture of a trait or disease between populations depend on how the continuum of genetic ancestry is operationalized into discrete categories and population labels (Box 1). However, genetic diversity exists even within populations with a relatively high degree of genetic similarity or within geographically constrained regions38. Genetic admixture presents further challenges to the characterization of genetic architecture and the development of PRS construction methods because the genomes of individuals are mosaics of their ancestors, and when individuals have different local genetic ancestries across the genome, the genetic effects can broadly vary from individual to individual.
Box 1. Population descriptors and concepts.
Historically, most genetic association studies have involved assigning participants to discrete clusters to facilitate statistical analyses. Variability in criteria for defining such clusters and inconsistent use of labels for the resulting populations not only creates challenges for analysis and interpretation but might also contribute to harmful misuse of findings from genome-wide association studies (GWAS) and polygenic risk scores (PRSs). Here, we provide a brief overview of common population descriptors and discuss distinct, but related, concepts of ancestry161 that are relevant for genetic association studies.
Race, ethnicity and ancestry
Both race and ethnicity have a history of being used as a misleading shorthand for groups of individuals with shared genetic ancestry. Race is a socially constructed system for classifying humans based on erroneous beliefs about innate biological differences, often proxied by physical features (such as skin colour) and sociocultural characteristics. Ethnicity is a sociopolitical identity that is assumed by or assigned to a group of individuals, typically in a contiguous geographic area, based on shared heritage and cultural similarities, such as language, religion or beliefs. Individuals of the same ethnicity often share genetic or genealogical heritage, but as this system varies globally, in certain regions ethnicity may be primarily a sociocultural identity. Ancestry is a complex and context-dependent term that encompasses both the biological and social components of an individual’s or population’s descent. In the western world, it has an aspect of both sociocultural and continental origin, whereas in the eastern and southern hemispheres there is an aspect of either shared genealogical or genetic heritage, or both, on a smaller regional scale. Race and/or ethnicity should not be conflated with ancestry or used as synonyms for population genetic differences.
Different types of ancestry
Unlike race or ethnicity, which are subjective constructs, genetic ancestry is a fixed characteristic of the genome. Genealogical ancestry describes an individual’s lineage based on family trees of known ancestors that have been traced back over multiple generations. Genealogical ancestry is inferred using oral and written historical records, and, more recently, genetic information. Genetic ancestry refers to segments of an individual’s genome that have been inherited through a subset of realized paths from their ancestors, and unlike genealogical ancestry does not require reconstructing a family pedigree. The complete record of coalescent and recombination events (that is, the convergence of two lineages into a single population and the process of shuffling of alleles on the chromosome to produce novel combinations of alleles, respectively) in the history of an individual’s genetic lineage is called an ancestral recombination graph, which is the fundamental representation of genetic ancestry. Genetic similarity is a quantitative measure of genetic sharing between individuals or populations. Most studies that develop and evaluate PRSs approximate genetic ancestry from measures of genetic similarity, typically with respect to reference ancestral populations. We note that these types of ancestry are rarely distinguished in the literature and the research community frequently uses genetic ancestry as a catch-all term for all dimensions of ancestry. Throughout our review of PRS methods we refer to genetically inferred ancestry, unless otherwise specified. When describing findings from the literature, we use the same terminology, including population labels, as the original publication(s).
Measuring genetic ancestry
Coalescent models provide a statistical framework for reconstructing evolutionary history to determine how alleles sampled from a group of individuals link to a common ancestor. Quantitative models for ancestry inference incorporate patterns of linkage disequilibrium (LD) and allele frequencies, which are also fundamental features for describing the genetic architecture of complex traits. Global (genome-wide) genetic ancestry is the proportion of contributions of different assumed proxy ancestral populations to an individual or a group of individuals’ overall genetic make-up. Global ancestry can be inferred using both model-based162 and data-driven methods163,164. Local ancestry is the genetic ancestry of an individual at a particular location in a chromosomal segment. Local ancestry can be inferred by computational approaches, often using discriminative modelling165 or generative hidden Markov models166 with modifications to improve on efficiency, accuracy and the number of ancestral populations considered167-169.
Admixture
Populations from which an individual or group of individuals have inherited their genome are referred to as ancestral populations. Admixture is the process that brings together individuals from two or more ancestral populations that were previously isolated for a period of evolutionary time, allowing distinct haplotypes to be combined in a gene pool. Although admixture is a pervasive phenomenon, the term ‘admixed’ typically refers to individuals with recent admixture (<100 generations). For much of human history, admixture has occurred through mass migration, colonization or forced displacement. However, in today’s increasingly globalized and interconnected society, novel patterns of recent admixture are emerging and shaping the ancestry of modern human populations (Fig. 1). Genetic ancestry in admixed populations varies between individuals and along haplotypes. The proportion of populations contributing to an individual’s genome can be represented on global and local levels that both attempt to determine the ancestral origin of polymorphisms or chromosomal segments in the admixed individual.
Recent theoretical and empirical studies have attempted to separate the contributions of the different genetic factors discussed above to the transferability of PRSs across diverse populations. These studies support the view that allelic effects of causal variants are similar across ancestries, and the attenuation of PRS accuracies can be primarily attributed to differences in allele frequencies and LD structure, although none of the factors can fully explain the power loss23,32,39,40. In addition, improving PRS accuracy in African-ancestry populations can be especially challenging due to the existence of a much larger number of genetic variants in these groups41. Although these findings are promising to facilitate the dissection of cross-population genetic architecture and inform the development of PRS construction methods, our current understanding of the genetic basis of complex traits and common diseases is still hindered by small sample sizes for under-represented populations. A full accounting of the genetic architecture across the phenotypic spectrum and diverse ancestries, and its impact on the transferability of PRSs, requires continued expansion of non-European genomic resources and a comprehensive catalogue of global genetic and phenotypic variation.
Social and environmental factors influencing PRS performance
Many complex diseases arise from both genetic and environmental risk factors, which may together impact PRS performance across populations. Environmental factors include individual-level exposures (such as cigarette smoking, diet or physical activity), and macro-environmental factors (such as health policies or neighbourhood characteristics including degree of urbanization, green space, available facilities, environmental noise and air pollution, and so on). Risk factors and health outcomes may be shaped by the broader conditions in which populations live, work and age, referred to as social determinants of health (SDOH) (Fig. 3a). SDOH relate to an individual’s place in society, including access to healthcare resources, and capture experiences of social exclusion, such as racism and discrimination. For disease-specific PRS development and evaluation, there is a need to create a conceptual framework that specifies relationships between PRSs, genetic ancestry and specific social and environmental risk factors.
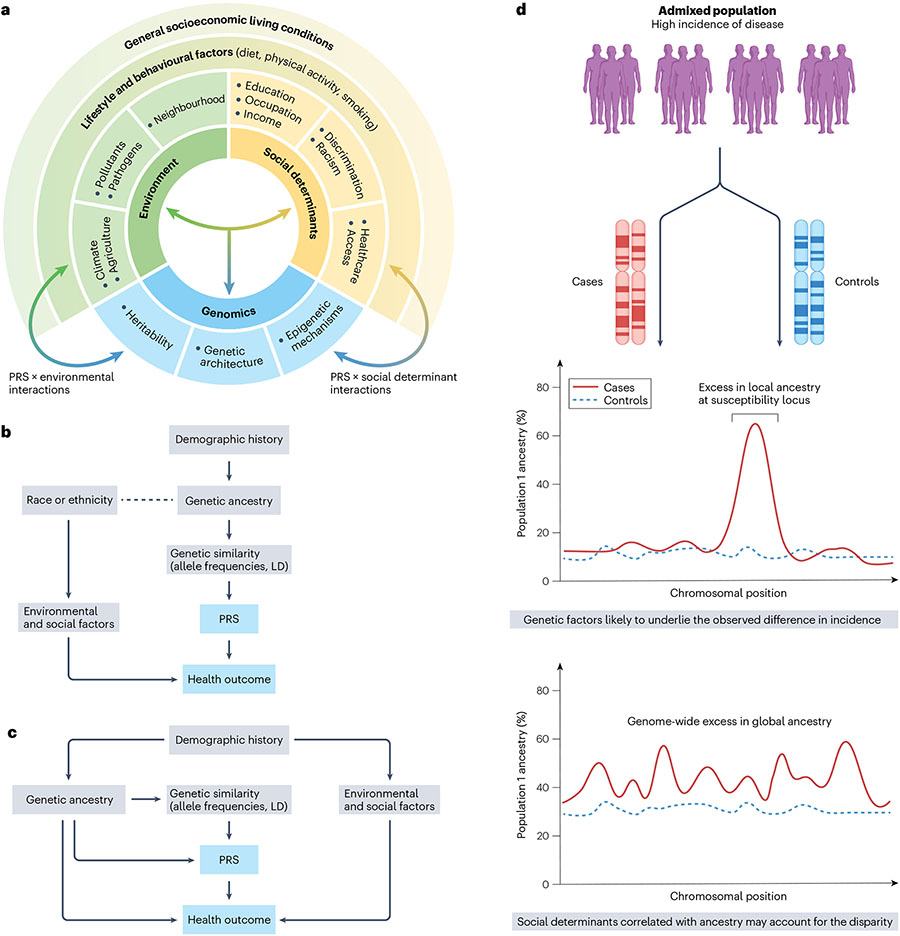
Fig. 3 ∣. Interplay between social, environmental and genetic determinants of health.
a, Complex interrelationship among different risk factors for ill health and poor disease outcomes. These include living and working conditions (such as environmental exposures and social determinants of health (SDOH)) and genomic factors. b, Race and/or ethnicity can confound polygenic risk score (PRS) associations with health outcomes if a correlation exists with genetic ancestry (dotted line). In this case, correction for population structure using methods such as principal component analysis (PCA) that captures similarity in allele frequencies and linkage disequilibrium (LD) structure that arises due to shared demographic histories between populations can mitigate the confounding effect. c, Residual confounding may bias PRS associations when genetic ancestry is correlated with environmental and/or social factors due to shared demographic histories. For instance, when asthma is the health outcome and exposure to air pollution is the non-genetic risk factor, standard methods such as PCA may under-correct for population structure. d, Admixture mapping detects disease-associated loci and patterns of excess local ancestry that help disentangle the contribution of genetic factors to observed disparities in risk.
SDOH encompass a wide array of factors, each of which may act differently in relation to PRSs, as an effect modifier, a confounder or a partial mediator of the observed PRS effects (Fig. 3b,c). To understand and account for the influence of SDOH, these factors must first be accurately measured and operationalized. For instance, if the contribution of important environmental risk factors is lower in the training versus testing populations, the proportion of variance in the trait explained by the PRS may appear lower in the target population. Using structured approaches such as directed acyclic graphs42,43 to clarify assumptions and inform appropriate analytic strategies, for example covariate adjustment for population structure44, may help interpret differences in PRS accuracy. As an example, directed acyclic graphs can identify scenarios in which PRS effects may be partially mediated by genetically inferred ancestry, leading to a different interpretation of adjusted estimates of PRS performance compared with a case in which ancestry acts as a confounder45,46. Furthermore, as PRS performance is context-dependent, identifying and characterizing gene–environment interactions may inform PRS implementation efforts by identifying groups of individuals who may experience a greater or smaller degree of risk stratification benefits from PRS. For example, previous studies have reported a synergistic interaction effect between PRSs and childhood trauma on depression, suggesting that individuals with both high PRSs and exposure to childhood trauma are particularly at risk for developing depression and could form a target group for interventions47. However, efforts to replicate this effect have led to mixed results48, and evidence supporting robust PRSs by environment interactions in general has been limited to date7.
It is important to assess the potential impact of PRS implementation on health disparities and recognize that genetic factors do not capture the full understanding of an individual’s health. The contribution of heritable factors can vary depending on how a disparity is quantified. In the United States, individuals identified as African American or Black, Hispanic or Latino and Native American are disproportionately affected by numerous conditions, such as hypertension49, chronic kidney disease50,51 and certain cancers52-55, compared with those identified as white. Trends in the incidence of such conditions that are not easily attributed to diagnostic biases or differences in risk factor profiles may offer clues to a potential role of genetic factors. Disparities in mortality are largely driven by inequities in healthcare access, although for certain cancers, germ-line genetic factors have been shown to underlie differences in the prevalence of actionable tumour mutations between populations55,56. In this context, race poses a unique confounding challenge because it is correlated with measures of SDOH and genetic ancestry, hence setting the stage for confounding of PRS associations when considering trait predictions.
If disease susceptibility varies across ancestral groups, the degree of admixture and the extent to which SDOH and other modifiable factors correlate with genetic ancestry will affect PRS performance. If differences in disease risk between two ancestral populations arise partly due to differences in the frequencies of risk alleles, we would expect to observe an enrichment of one ancestry in cases compared with controls at specific loci in the genome (Fig. 3d). Admixture mapping can be used to identify genetic loci that contribute to differences in disease risk, while controlling for global ancestral differences as well as confounding due to non-genetic factors that differ between populations, thus providing insights into the genetic causes of health disparities in recently admixed populations. To date, admixture mapping has refined important risk loci for prostate cancer57, breast cancer58, asthma59, multiple sclerosis60 and coronary heart disease61, which may complement PRS development efforts by uncovering ancestry-specific causal variants.
Methods to improve PRS transferability across populations
Recent shifts in GWAS towards increasing the diversity of populations included33,34,62,63, in parallel with rapid advances in methods that leverage these more diverse studies as well as existing Eurocentric GWAS, aim to improve the transferability of PRSs across populations. These methods can be broadly grouped into approaches that combine or jointly model population-specific summary statistics (Table 1).
Table 1 ∣.
Polygenic prediction methods integrating GWAS summary statistics from diverse populations
| Category | Method | Input | Variants for prediction |
Need validation data set |
Tuning parameters | Algorithm | Ref. |
|---|---|---|---|---|---|---|---|
| Combining approaches | Meta-analysis | Population-specific GWAS and LD | Up to all 1000 Genomes common variants | Optional | Depends on the single-population PRS method | Fixed-effect meta-analysis and a single-population PRS method | 62 |
| ShaPRS | Population-specific GWAS and LD | Up to all 1000 Genomes common variants | Optional | Depends on the single-population PRS method | Meta-analysis accounting for heterogeneity between GWAS and a single-population PRS method | 64 | |
| MultiPRS | Population-specific GWAS and LD | Up to all 1000 Genomes common variants, depending on the C + T parameters | Yes | C + T parameters and linear combination weights for population-specific PRS | C + T | 67 | |
| Joint modelling of two populations | XP-BLUP | GWAS in the auxiliary population; individual-level data for the target population | Genotyped SNPs in the target data set | Yes | P-value threshold for the auxiliary GWAS (optional) | Local FDR for variant selection; restricted maximum likelihood (ReML) for model fitting; best linear unbiased prediction | 71 |
| XPASS(+) | GWAS and LD from the auxiliary and target populations | All available variants | No | None | Best linear unbiased prediction and conjugate gradient for solving linear systems | 72 | |
| BridgePRS | GWAS and LD from the auxiliary and target populations; individual-level data for the auxiliary population | All available variants | Yes | Ridge shrinkage parameters and linear combination weights for PRS generated under different prior parameters and loci selection criteria | Best linear unbiased prediction | 73 | |
| TL-Multi | GWAS and LD from the auxiliary and target populations | HapMap3 | Optional | Regularization parameters in LASSO | Coordinate descent for model fitting | 74 | |
| SDPRX | GWAS and LD from the auxiliary and target populations | HapMap3 | Yes | Linear combination weights for population-specific PRS | Markov chain Monte Carlo | 76 | |
| Joint modelling of two or more populations | CT-SLEB | Population-specific GWAS and LD | Up to all 1000 Genomes common variants, depending on the C + T parameters | Yes | C + T parameters and parameters in the super learning model (such as LASSO, ridge regression and neural networks) | Two-dimensional C + T; empirical Bayes (for SNP effect estimation); super learning (for combining PRSs generated under different C + T parameters) | 41 |
| TL-PRS/MTL-PRS | Population-specific GWAS and LD | HapMap3 | Yes | Learning rate and number of iterations in the gradient descent algorithm | Gradient descent for model fitting | 77 | |
| PROSPER | Population-specific GWAS and LD | HapMap3 + MEGA chip array | Yes | Regularization parameters in LASSO, ridge regression and LD matrix estimation; tuning parameters in the ensemble regression | Coordinate descent and super learning (for combining PRSs generated under different tuning parameters) | 78 | |
| ME-Bayes SL | Population-specific GWAS and LD | HapMap3 + MEGA chip array | Yes | Parameters in the super learning model (such as linear regression, ridge regression and elastic net) | LDpred2 (for estimating causal SNP proportions and heritability); ME-Bayes (for SNP effect estimation); super learning (for combining PRSs generated under different ME-Bayes parameters) | 79 | |
| PRS-CSx(-auto) | Population-specific GWAS and LD | HapMap3 | Optional | The global shrinkage parameter and linear combination weights for population-specific PRS; none for the auto algorithm | Markov chain Monte Carlo | 68 | |
| Incorporating information beyond GWAS | XPXP | Population-specific GWAS (for both the target trait and its genetically correlated traits) and LD | All available variants | No | None | Best linear unbiased prediction and conjugate gradient for solving linear systems | 81 |
| X-Wing | Population-specific GWAS and LD | HapMap3 | No | None | Scan statistics (for local genetic correlation estimation); Markov chain Monte Carlo (for PRS model fitting); summary statistics based repeated learning (for combining population-specific PRSs) | 82 | |
| PolyPred-S+/PolyPred-P+ | Population-specific GWAS and LD; functional annotations | All available variants for PolyFun-pred; HapMap3 for SBayesR and PRS-CS | Yes | Linear combination weights for population-specific PRS | PolyFun + SuSiE (for fine-mapping informed PRS) and Markov chain Monte Carlo (for SBayesR and PRS-CS) | 69 |
C + T, clumping and P-value thresholding; FDR, false discovery rate; GWAS, genome-wide association studies; LD, linkage disequilibrium; PRS, polygenic risk score.
One approach combines GWAS from multiple ancestry groups using fixed-effect meta-analysis and constructs PRSs using the meta-GWAS and a single-population PRS method. This approach is widely used in many large-scale multi-ancestry GWAS and can improve the transferability of PRSs relative to population-specific PRSs34,62,63. However, it can require potentially difficult parsing of mixed LD patterns in the meta-GWAS and makes strong assumptions of homogeneous allelic effects across ancestries, which may limit the accuracy of the resulting PRSs. Restricting the meta-analysis to variants that are common across all populations will further limit PRS transferability. More flexible meta-analysis methods, such as ShaPRS64 that incorporates heterogeneity of SNP effects across GWAS coupled with an appropriate LD reference panel for the derived summary statistics, might improve downstream PRS performance. Alternatively, another approach linearly combines PRSs constructed from each population-specific discovery GWAS using clumping and P-value thresholding (C + T)65,66; this method improved risk prediction in recently admixed individuals67. Subsequent work in combining PRSs across populations has replaced C + T with more sophisticated polygenic prediction methods to further improve prediction68,69. However, this method cannot fully exploit cross-population models of genetic architecture that incorporate heritability, genetic correlation and polygenicity to inform PRS construction.
Another approach uses one large-scale GWAS – often conducted in European populations and termed the ‘auxiliary GWAS’ – to improve prediction in a target non-European population where there are smaller GWAS. This method leverages the observation that many causal signals are shared among populations29 and cross-ancestry genetic correlations for human complex traits and diseases are often moderate to high30,31,70. For example, XP-BLUP71 builds PRSs using a two-component linear mixed-effects model, with one component including variants that show associations in the auxiliary GWAS and the other component including all available variants in the target data set to capture the polygenic background. XPASS(+)72 uses a bivariate linear mixed-effects model with a multivariate normal prior to jointly model SNP effect sizes from the auxiliary and non-European GWAS. BridgePRS73 fits a Bayesian ridge regression to the auxiliary GWAS and uses the posterior SNP effect size estimates as the prior when fitting a second Bayesian ridge regression to the GWAS in the target non-European population. TL-Multi74 transfers the SNP weights estimated by Lassosum75 in the auxiliary GWAS to the target population, with an assumption that effect sizes across populations are largely similar. SDPRX76 models the auxiliary and non-European GWAS using a hierarchical non-parametric Bayesian model with a prior that characterizes the joint effect size distribution of each variant in the two populations to be null, population-specific or shared with correlation.
Recent work has also allowed for the modelling of GWAS summary statistics from more than two populations. For example, CT-SLEB41 expanded the C + T algorithm to the multi-ancestry setting for SNP selection, and uses an empirical Bayes algorithm for computationally efficient SNP effect size estimation. TL-PRS77 fine-tunes effect sizes estimated from large-scale training GWAS to the target population using transfer learning and a gradient descent algorithm. PROSPER78 uses a combination of LASSO (L1) and Ridge (L2) penalties to regularize SNP effect sizes, encouraging a sparse genetic architecture within populations and similar genetic effects across populations. ME-Bayes SL79 performs Bayesian hierarchical modelling of SNP effect size distributions under a multivariate spike-and-slab prior and integrates information across different tuning parameter settings and ancestry groups using ensemble learning. Lastly, PRS-CSx68 extended the single-population polygenic prediction method, PRS-CS80, to jointly model GWAS summary statistics from an arbitrary number of populations, using Bayesian regression and a continuous shrinkage prior on SNP effect sizes that is coupled across populations.
Although these methods have different variant selection procedures or make different assumptions about the prior distribution of SNP effect sizes, they all aim to account for different degrees of polygenicity of the underlying SNP effect size distribution, integrate GWAS summary statistics from two or more populations in a principled statistical framework that accounts for allele frequency and LD differences, and leverage cross-ancestry genetic correlation to borrow information from well-powered European GWAS to improve PRS performance in non-European populations.
Other work has incorporated information beyond GWAS for the trait of interest into PRS construction algorithms. For example, XPXP81 extends XPASS to enable joint modelling of multiple genetically correlated traits in both auxiliary and target populations. X-Wing82 expands the modelling framework of PRS-CSx to allow for annotation-dependent priors and uses trans-ancestry local genetic correlation to up-weight variants whose effect sizes are more concordant across populations. PolyPred-S+/PolyPred-P+69 uses functionally informed statistical fine-mapping to identify and prioritize functional variants, whose effects are often more portable than tagging variants due to assumed shared mechanisms of biology83 and minimal impact of differential LD patterns across populations. The fine-mapping informed PRSs can then be combined with population-specific genome-wide PRSs to capture signals at polygenic loci that are difficult to fine-map.
Considerations of PRS methods for diverse populations
Although methodological developments can help improve PRS transferability, several limitations merit consideration. First, many of the methods require a validation data set with individual-level phenotypes and genotypes to tune an algorithm’s hyper-parameters. Although this can maximize the accuracy of PRSs for specific populations, it also increases the risk of overfitting. In addition, there may not exist a sufficiently large independent validation data set in the target non-European population, especially when the hyper-parameter space is large. Fully Bayesian models84, pseudo-validation methods74,75 and repeated learning techniques82,85 that can automatically learn model parameters from summary-level data may facilitate cross-population PRS development.
Second, many methods require categorizing individuals into a genetic ancestry group before a PRS can be optimized and applied (Box 1). However, this poses challenges when implementing the PRS in clinical settings, where admixed individuals can be difficult to assign to a discrete population cluster, and genetically inferred ancestry can differ from self-reported race or ethnicity.
A third consideration is that existing methods need to balance trade-offs between prediction accuracy and computational complexity. For example, the infinitesimal model, which assumes that prior SNP effect sizes are independent and normally distributed, has a closed-form posterior distribution and is highly scalable41,72,73,81. However, this approach shrinks the effects of all variants towards zero at the same constant rate, and is thus less adaptive to varying genetic architectures. More sophisticated Bayesian methods assume that the prior distribution is a mixture of two or more normals76,86,87, which allows for flexible modelling of the genetic architecture but makes model fitting challenging and potentially unstable. Continuous shrinkage priors68,80 can balance modelling flexibility and computational efficiency, but full posterior inference remains infeasible to scale to all common variants across the genome. As a result, most Bayesian methods use HapMap3 variants to construct PRSs. Although they might have provided a good balance between computational cost and genetic variation captured within European populations, HapMap3 variants do not tag genetic variation in non-European populations equally well and can miss population-specific signals. This coupled with unequal imputation accuracy across populations can limit the transferability of PRSs.
Fourth, although it has been shown that the polygenic background can modify the penetrance of monogenic variants88,89, studies and methods that explore the integration of genome-wide rare and common variants for prediction are limited90-93. The prediction accuracy of rare and low-frequency variants across populations has not been systematically assessed, although a recent study suggests that rare coding variants will likely contribute only modestly to population risk stratification94. Whether rare variants provide value for the prediction of complex traits depends on the number of rare causal variants and their effect sizes, which will likely be trait-specific. Although the contribution of rare high-penetrance variants to overall trait variation may be small relative to common-variant PRSs, they can be valuable to identify high-risk individuals.
Finally, the methods described above primarily use GWAS summary statistics from relatively homogeneous populations as input and only model allele frequency and LD differences at the continental level. Much work remains to develop best practices to integrate GWAS from admixed or under-represented populations (such as the PAGE study33) with large genomic diversity and heterogeneous LD patterns. As noted above, mismatch between the LD structure of the GWAS discovery sample and the reference panel is likely an important contributing factor to the loss in PRS prediction accuracy. Moreover, when the target sample is admixed, most methods weight population-specific PRSs globally without modelling local ancestry and individual-level proportions of admixture. Initial attempts to infer local ancestry tracts, estimate local ancestry-specific effect sizes95 and build local ancestry-aware PRSs40,96 have shown increased power for loci discovery and improved prediction accuracy in admixed populations. A recently developed method, GAUDI97, explicitly models local ancestry using a fused LASSO framework that encourages similar effects across ancestries but allows for population-specific effects. These methods, however, have only been evaluated in two-way recently admixed populations, and the predictive performance of PRSs has not been fully benchmarked against other cross-ancestry polygenic prediction methods.
Benchmarking PRS methods may require a reference-standardized set-up98, whereby a common set of variants and individuals are used to build reference panels and assess the accuracy of PRSs. However, the optimal prediction method might depend on a range of factors including the genetic architecture, the diversity and sample size of the discovery GWAS, and the ancestry composition of the target sample99. For example, clumping and fine-mapping based methods may work well for traits that have a handful of large-effect causal variants but not as well when predicting highly polygenic traits. In contrast, genome-wide PRS approaches may capture signals that do not reach statistical significance, but they may also include a large number of non-informative variants when the genetic architecture is sparse and the power of GWAS is limited. Future methodological efforts that focus on integrating multiple data modalities (such as variants across the allele frequency spectrum, functional annotations and cross-ancestry fine-mapping results100,101) from diverse resources and better local and global ancestry modelling in a computationally efficient and robust framework, coupled with increasing size of non-European genomic resources, hold promise to improve the accuracy and generalizability of PRSs.
Evaluation of PRS clinical utility
Demonstrating the clinical utility of PRSs across diverse populations requires careful consideration of suitable performance metrics, which depend on the intended use of the PRS in a specific clinical context. The area under the receiver operating characteristic curve (AUC) for binary health outcomes and the C-index, an analogous measure of concordance for time to event outcomes102, are widely used to report discriminatory abilities of PRSs. The AUC can take values between 0.5 and 1.0, ranging from completely random, clinically useless predictions to perfect classification. In practice, the AUC of a PRS has an upper bound based on disease heritability and is thus always less than 1. For many common complex diseases, such as breast cancer103-106, coronary heart disease107,108 and type 2 diabetes mellitus84,107, the best-performing PRS has achieved AUC values in the range of 0.6–0.7 across different populations, demonstrating moderate discriminatory abilities. However, the AUC and related measures do not account for disease rates in the underlying population, and therefore cannot be translated directly into the predictive value of PRSs at the individual level and provide little insight regarding clinical utility109.
Many studies use rank-based risk classifications to identify high-risk individuals and report, for example, an odds ratio that compares individuals in the top decile of the PRS distribution with those around the median. These measures, however, depend on the reference population for the PRS distribution. Metrics of PRS utility are also affected by operationalization of the PRS. For instance, when a PRS cut-off is used for classifying individuals as high risk, it is important to select an optimal threshold that maximizes discrimination. This requires calibration of the PRS distributions across populations, which can have different mean values and spread (Fig. 4). Existing studies express polygenic risk on the same scale across ancestrally diverse individuals by removing gross cross-population differences in the mean and variance of the PRS distribution that can be captured by genetic principal components84,110-112; however, these methods could remove real risk differences explainable by genetic and non-genetic risk factors that are correlated with population structure, reducing the predictive power of PRSs.
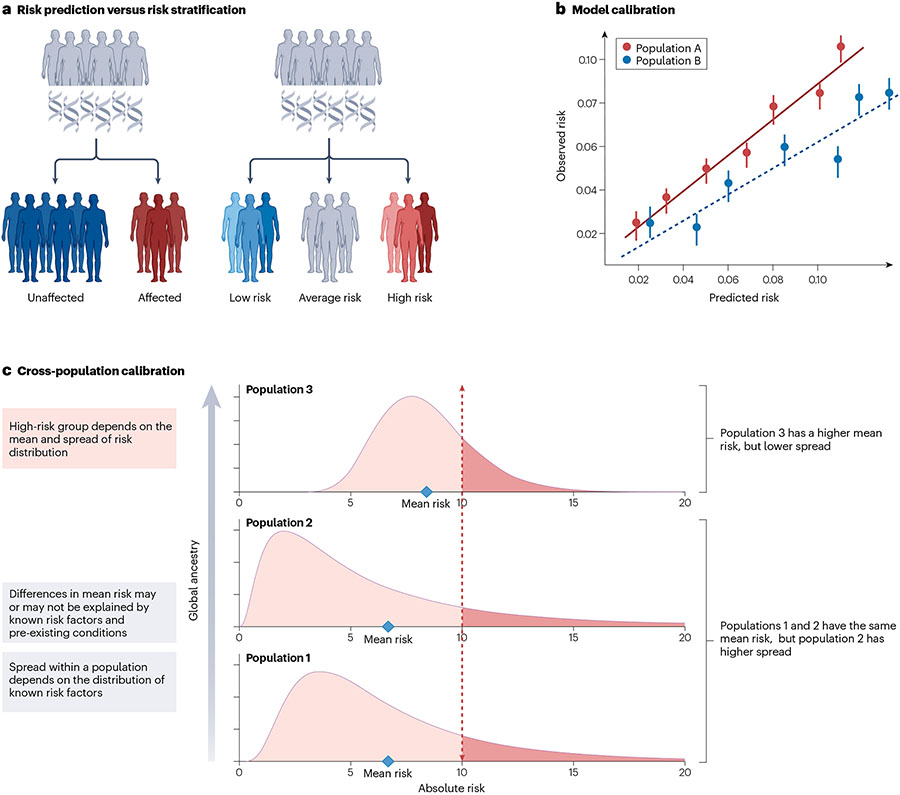
Fig. 4 ∣. Considerations for the assessment of PRS clinical utility.
a, Visual representation of the difference between risk prediction and risk stratification. b, An example of model calibration, that is, the agreement between observed and estimated disease risk. Accurate estimation of absolute risks requires well-calibrated models. For instance, risks are systematically overestimated for Population B compared with Population A. c, Cross-population calibration of the polygenic risk score (PRS) distributions, which can have different mean and spread. Differences in calibration between populations arise due to a combination of the genetic and clinical risk factors. Cross-population calibration is important when selecting a single cut-off to identify individuals as high risk across samples with diverse ancestral and sociocultural backgrounds.
In general, relative risk estimates (such as those based on the odds ratio) do not provide information that can be readily translated to risk thresholds for clinical action, such as diagnostic and treatment decisions. Instead, these typically require estimates on the absolute risk scale. Positive and negative predictive values, which represent the proportions of individuals who test positive (or negative) who will (or will not) have the disease, respectively, give absolute risk estimates and are more clinically relevant metrics. For diseases that have clinical guidelines based on absolute risk thresholds (for example, breast cancer and coronary heart disease), absolute risk models with and without the PRS can be compared to quantify the expected increase in individuals who now meet this threshold. Additionally, we can examine the increase in disease cases detected among the high-risk individuals identified using the PRS108,113. Measures of net reclassification indices114 – which summarize the net number of cases and controls who move into higher and lower risk categories, respectively, according to a new model compared with an existing model – can be used to evaluate the added value of PRSs in relation to existing clinical risk factors115. However, net reclassification indices, similar to the AUC, do not take into account disease rates in the underlying population and do not directly quantify clinical utility. In addition, caution is needed for inappropriate use of such measures in the absence of predefined risk categories116. When specific risk thresholds are not available, one can also carry out decision curve analysis117 to evaluate the net benefit associated with decision-making at different absolute risk thresholds under alternative models118. An important consideration, when evaluating the added value of PRSs in diverse populations, is whether existing risk-threshold guidelines themselves are universally suitable across groups that may have different loss–benefit balances for underlying decisions.
For diseases that do not have established absolute risk models, absolute risk can be estimated by combining overall or relative risk estimates with disease incidence rates observed internally in cohort studies or approximated by external information on population incidence rates119,120. Studies evaluating absolute risk have shown that PRSs for some common diseases, even with their modest discriminatory performance, can now identify a substantial fraction of the population who would be considered as high risk to warrant drug therapy, or invasive and potentially costly interventions not appropriate for the general population108. However, as rates of many diseases as well as SDOH are typically estimated with stratification by self-identified race or ethnicity, and are known to vary widely by racial groups, it is particularly important to incorporate absolute risk considerations when the utility of PRSs is being evaluated across diverse populations. Future research is merited to explore the ability of genetic ancestry to explain known variations in disease incidence rates across population groups121. Furthermore, as the severity, prognosis and financial burdens of diseases can vary widely by socioeconomic status, it will be important to demonstrate the utility of PRSs considering the risk of different types of adverse outcomes.
Another dimension of PRS performance that is not commonly assessed is the uncertainty in relative and absolute risks derived from the PRS and the stability of PRS-based risk classifications across different PRS modelling approaches and choice of LD reference panels. Recent work has demonstrated substantial variability in individual-specific PRS estimates for 13 complex traits122. As an example, for high-density lipoprotein (HDL) PRSs, the 95% credible interval for an individual at the 90th PRS percentile spans widely between the 41st and 99th percentiles. There are multiple sources of uncertainty in PRSs, including error in estimates of SNP weights, and this is likely to be higher for non-European ancestry populations because of the smaller sample sizes used to create PRSs. Quantifying the impact of this uncertainty on clinical decisions will be important for PRS implementation across diverse populations.
The appropriate choice of clinical utility metrics must also be considered in the development stage of PRSs in diverse ancestry settings. Current multi-ancestry methods allow the development of improved population-specific PRSs by borrowing data across diverse multi-ancestry populations. Although these approaches have the advantage of being able to generate the best possible PRS for a given ancestry group using all existing data, in a country such as the United States where there are many diverse groups, and many admixed sub-groups, delivery of optimized PRSs for each distinct group is not practical. Additionally, optimizing PRSs for each ancestry group separately might still exacerbate inequity in their performance across groups because of differences in sample sizes and genetic architecture of traits across groups. Therefore, an alternative goal may be to develop more universal PRSs that can be applied across populations by using suitable loss functions that take into account fairness constraints so that prediction performance is not driven by a majority group. Although there is an emerging body of literature in machine learning theory to incorporate different types of fairness constraints123-126, considerations of such a framework are currently lacking in PRS development. An additional challenge relates to defining what fairness means in the context of PRSs and prioritizing trade-offs, as simultaneously satisfying all constraints may not be feasible127.
Remaining challenges and future directions
As we strive towards creating and improving PRSs across diverse populations, several challenges lie ahead. First, in the shift away from the use of race and ethnicity in biomedical research and clinical practice, genetic ancestry is often put forth as a suitable replacement128. Although this has numerous advantages, the use of discrete population categories, including groups derived from genetic ancestry or genetic similarity, is also problematic129. Global ancestry cut-offs used to create more homogeneous groups are often arbitrary, study-specific and primarily driven by considerations of statistical power. Sensitivity analyses at different ancestry thresholds are typically not performed or reported. The most fundamental consideration for PRS accuracy is the genetic distance between the PRS training population, such as the source GWAS sample, and the target population where the PRS is intended to be applied15. Gauging expected PRS performance using genetic distance does not require forming discrete population clusters, and the development of robust PRSs in the future will benefit from this approach.
Although race, a socially defined construct, is not an acceptable proxy for genetic ancestry or patterns of admixture, it remains extensively used in administrative databases, disease surveillance systems and healthcare records in the United States, and thus has been used in the design and analysis of GWAS. Participants are sampled or enrolled based on race and/or ethnicity rather than genetic ancestry, which can induce systematic differences in ascertainment, phenotyping and measurement of potential confounders or effect modifiers, especially for SDOH. In some GWAS, samples have been genotyped using different arrays depending on participants’ race (self-reported or assigned by healthcare providers), which has implications for imputation quality and downstream analyses130. Ignoring race/ethnicity, especially when it is a study design feature, can bias estimates of PRS accuracy when considering continuous measures of genetic diversity. Differences in study design are a major source of heterogeneity in GWAS meta-analyses, and this heterogeneity can disproportionately affect studies conducted in admixed and non-European ancestry populations. Together with the smaller GWAS sample sizes in these populations, this may further reduce the signal-to-noise ratio in data used to construct PRSs and limit PRS transferability and generalizability.
Another challenge in transitioning to continuous genetic ancestry relates to operationalizing analyses that require additional inputs, such as incidence or mortality rates. This issue applies not only to integration of PRSs with population-level health indicators but also to more basic analyses, such as converting heritability estimates from the observed to the liability scale using lifetime risks131. Although tracking disease morbidity and mortality by race or ethnicity is suboptimal, these metrics are not collected alongside genetic ancestry, and outside the United States, surveillance systems such as disease registries are often agnostic to any metrics of race, ethnicity or ancestry. In developing countries and low-resource settings, surveillance systems may be extremely limited, leading to sparse or non-existent data on disease burden and non-genetic risk factors. Further, as observed variations in disease incidence, outcomes and mortality rates by race, ethnicity and other categorical descriptors (such as immigration status) could be due to life experiences and environmental exposures unrelated to genetics, removing such proxy information prematurely can hinder contextualizing clinical utility of PRSs in the absolute risk scale.
This leads to a broader but related challenge for PRS translation. Once a PRS demonstrating acceptable within-population and cross-population predictive performance has been developed, an appropriate statistical framework for integrating the PRS with other clinical predictors and established risk assessment tools must be established. For conditions with existing risk calculators, the simplest approach is to multiply risk estimates generated by the PRS and the clinical model. However, this assumes that PRSs are independent of other predictors, which may not be valid for all health outcomes, especially if the PRS captures indirect genetic effects partly mediated by clinical or modifiable risk factors. Furthermore, most existing models were not developed and validated in ancestrally diverse populations. For instance, pooled cohort equations, QRISK and Framingham scores for atherosclerotic cardiovascular disease have shown worse performance in African Americans, African Caribbeans and South Asians based on studies in the United States and the United Kingdom132,133. Lung cancer screening criteria from the US Preventive Services Task Force have been shown to significantly underestimate screening eligibility in African Americans compared with white Americans134. This disparity was attenuated, but not eliminated, with the PLCOm2012 model135. Therefore, even if PRS-based risk estimates are well calibrated, the accuracy of the overall risk prediction model may suffer due to poor performance of the non-genetic components.
Simultaneously combining data on all relevant risk factors, including family history136,137 and relevant pre-existing conditions, into a framework that returns a single estimate of absolute risk would accelerate the uptake of PRSs into clinical practice. Currently, few such models exist, and they have been validated only in individuals of European ancestries138. For instance, the BOADICEA model for breast and ovarian cancer has successfully integrated modifiable risk factors with rare, high-penetrance mutations and PRSs139,140. A PRS for coronary heart disease was shown to be largely uncorrelated with pooled cohort equations and QRISK, suggesting that incorporating PRSs may further improve these models if they are transferable across ancestries141. Given the population differences in performance that exist for both genetic and non-genetic predictors, developing reliable risk prediction tools will likely require fitting and validating new ancestry-aware models that incorporate joint effects of PRSs and other risk factors. To achieve this, large cohorts would be needed to measure the calibration of the joint model. The All of US Research Program142 is one such promising cohort. In addition, multiple medical centres have created their own biobanks. Forming a network of these types of cohorts would provide additional avenues to evaluate whether the developed models offer accurate predictions in terms of calibration.
Generating new data in under-represented populations will undoubtedly have the largest impact on precision medicine efforts by providing the information necessary to develop effective, evidence-based tools, including PRSs. For instance, African populations have the greatest genetic diversity, the largest number of population-specific alleles and the smallest LD blocks, providing a wealth of information to enable globally relevant genetic discoveries143,144. However, until systems are in place to support large-scale collection of genetic, clinical and epidemiologic data in Africa and globally144, and until such data collection efforts mature, utilizing existing resources remains important. Furthermore, the success of new data collection efforts will require a commitment to transparency and community engagement in order to build trust with populations who have been historically under-represented and exploited in biomedical research. To improve participation and ensure ethical translation of genetic discoveries, concerns regarding potential for misinterpretation, stigma and discrimination, conflicts of interest, data sovereignty and premature commercialization of PRSs must be understood and addressed145,146.
Data aggregation and pooling efforts are crucial for advancing genetic research in under-represented populations, and must be supported by the development of best practices for phenotyping and data harmonization. Use of external controls may be an effective way of leveraging limited resources, particularly for studies of rare conditions using whole-genome sequencing; however, matching closely on genetic ancestry is critical for avoiding bias and this may be more challenging for admixed populations147. Imputation with more diverse reference panels, such as TOPMed148, will improve the utility of existing genetic data, but this will not fully compensate for the bias in genotyping arrays that were optimized for European LD structure towards alleles that segregate at intermediate frequencies in non-African populations149-152. Until whole-genome sequencing data are more widely available, choice of genotyping arrays and imputation panels will continue to limit PRS transferability and accuracy, especially for highly polymorphic and complex regions, such as HLA152,153.
Understanding the primary drivers of health disparities is critical for contextualizing PRS performance and informing appropriate public health interventions. In addition to comprehensively measuring different dimensions of SDOH, methods are needed to account for complex confounding structures and population stratification that may arise. Detailed information on social constructs, environmental exposures and behavioural factors is often absent from genomic studies, and these variables are less amenable to pooling across studies due to differences in data collection and exposure assessment. Careful consideration of non-genetic risk factors is important both for PRS evaluation and for covariate adjustment in GWAS used to generate summary statistics for PRS development44. Some variables or underlying constructs may only be applicable to specific countries or communities. For instance, although race or ethnicity is an imperfect surrogate for the effects of discrimination and access to health care, for studies conducted in the United States it might be the only available measure of these constructs154. Methods are thus needed for reconciling risk information captured by genetic ancestry and other population descriptors, and PRS evaluations will continue to need to account for disparities that cannot be explained by genetic ancestry alone. Although GWAS have historically prioritized achieving large sample sizes by including the minimal set of covariates available across the largest number of individuals or studies, smaller studies with deep phenotyping and more comprehensive risk factor assessment will be equally important for PRS development.
Conclusions
Although progress has been made towards the improvement of PRS prediction accuracy in non-European populations, substantial efforts are needed to improve PRS transferability, integrate PRSs into routine health care and equitably deliver PRS to global populations. In addition to further narrowing the gap in the prediction and stratification capabilities of PRSs between European and non-European populations, novel statistical and computational methods are needed to construct, validate and optimize PRSs that can be applied to any individual along the continuum of genetic ancestry. Admixture-aware and clinically informative metrics are also needed to assess the accuracy, calibration, uncertainty and stability of PRS prediction in ancestrally diverse samples. These methodological development efforts must be coupled with data generation initiatives to diversify samples in genomic research as well as to collect and harmonize measures for SDOH across studies. Release of ancestry-specific GWAS summary statistics155 in addition to multi-ancestry meta-analysis results is critical to the characterization of the comparative genetic architectures between populations and will facilitate the development of more flexible and accurate PRS construction methods (as shown in Table 1). A comprehensive catalogue of genetic, phenotypic, environmental and behavioural variation in diverse populations will not only inform and facilitate the development of more accurate and generalizable PRSs but also help disentangle the genetic and non-genetic contributions to disease burden and health disparities between populations, and characterize the relationships between PRSs and social and environmental factors.
Finally, future work is critically needed to contextualize PRS performance in real-world healthcare settings, and to develop integrative clinical models that combine PRSs with established risk factors into reliable and unbiased absolute risk assessment tools for patients of diverse ancestral and sociocultural backgrounds. Importantly, all PRS development, evaluation and implementation efforts should adhere to the latest reporting standards156 and promote data sharing and transparency to facilitate reproducibility, replication and benchmarking157.
Encouragingly, the field is rapidly advancing on all fronts, including method development, data generation and clinical implementation. For example, the National Institutes of Heath (NIH)-funded Polygenic Risk Methods in Diverse Populations (PRIMED) Consortium (https://primedconsortium.org) is developing methods and pooling genomic and phenotypic information to improve the use of PRSs for disease prediction in diverse populations. The growth of global biobanks and national health registries5 has substantially expanded sample diversity, accelerated genomic discovery and already informed PRS construction, evaluation and interpretation158. The emerging data sets from medical systems linked to electronic health records and a range of physical measurements and questionnaires on lifestyle, family history, socioeconomic factors and environment, such as the All of Us Research Program142 (https://allofus.nih.gov), the BioMe Biobank, the Mayo Clinic Biobank, the Vanderbilt’s BioVU resource, the Mass General Brigham Biobank and the UCLA Precision Health Biobank, will substantially expand data from groups that are historically under-represented in biomedical research and provide unprecedented opportunities to contextualize PRSs of specific diseases in real-world clinical settings. The NIH-initiated Electronic Medical Records and Genomics (eMERGE) IV study (https://emerge-network.org) has pioneered the return and communication of PRS results along with monogenic risks, family history and clinical risk assessments via a genome-informed risk assessment report to participants and their healthcare providers across ten conditions159,160, and will assess the uptake of care recommendations after the return of results. By combining new PRS construction methods, evaluation metrics, data resources and clinical implementation efforts that focus on diverse populations, the PRS may be well suited to realize its potential to advance precision medicine that benefits global populations.
Acknowledgements
This Review was supported by the National Institutes of Health (NIH) for the Polygenic Risk Methods in Diverse Populations (PRIMED) Consortium, with grant funding for the Coordinating Center (U01HG011697) and the study sites PREVENT (U01HG011710), CAPE (U01HG011715), CARDINAL (U01HG011717), FFAIRR-PRS (U01HG011719), EPIC-PRS (U01HG011720), D-PRISM (U01HG011723) and PRIMED-Cancer (U01CA261339). Additional funding was received from the NIH: R00CA246076 (to L.K.), R01HG010480 and U01CA249866 (to N.C.), R35GM140487 (to D.J.S.), R01CA241410 (to J.S.W.) and R01HG012354 (to T.G.). The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH. The authors thank Y. Ding and H. Zhang for their help with creating the figures in this Review.
Glossary
- Absolute risk
The probability that a person or group of individuals who are free of a certain disease at a given point in time will develop that disease over a certain time period. Absolute risks are typically expressed as proportions from 0 to 100%.
- Admixture
The process by which two or more previously separated populations come into contact, often through migration, generating a descendant population with a mixed mosaic of genetic material.
- Admixture mapping
An approach that consists of inferring local genetic ancestry and testing for association between local ancestry segments derived from different ancestral populations and the phenotype.
- Area under the receiver operating characteristic curve
(AUC). The ability of a model to discriminate between diseased and disease-free individuals is calculated as the AUC, which compares the true positive rate (sensitivity) with the false positive rate (1 – specificity). An AUC of 0.50 indicates that the classification accuracy of a model is equal to chance; an AUC of 1.0 indicates perfect discrimination.
- Clumping
A procedure that iteratively selects the variant with the lowest P-value within a specified window from genome-wide association study (GWAS) results and removes nearby variants that are correlated with the selected variants above a specific linkage disequilibrium (LD) threshold.
- Genetic architecture
The genetic basis of a trait described by the number, frequency and magnitude of effect size of genetic variants contributing to its heritability.
- Genetic correlation
The correlation between the genetic influences on two traits, or the proportion of variance that two traits share due to genetics.
- Haplotype
A cluster of polymorphisms or alleles that typically reside near each other on a chromosome and tend to be inherited together.
- Linkage disequilibrium
(LD). Non-random association of alleles at different genetic loci, often measured as the square of the correlation coefficient between two alleles. LD is, on average, lower in African populations compared with European and Asian populations.
- Meta-analysis
Statistical analysis that combines results from multiple studies.
- Net reclassification indices
Metrics that measure the extent to which a new model improves classification as compared with an old model, calculated as the difference between the proportion of individuals who are correctly reclassified and the proportion of individuals who are incorrectly reclassified.
- P-value thresholding
A procedure that selects the genetic variants whose P-value is below a threshold in a genome-wide association study (GWAS).
- Polygenic risk scores
(PRSs; also known as genetic risk scores). Single values that quantify an individual’s genetic predisposition to a discrete health outcome, calculated as a sum of alleles weighted by effect sizes corresponding to a relative magnitude of association.
- Polygenic scores
Single values that quantify an individual’s genetic predisposition calculated as a sum of trait-associated alleles weighted by their additive, per-allele effect sizes, typically derived from genome-wide association studies (GWAS).
- Population structure
The presence of multiple genetically distinct subpopulations that differ in their allele frequencies and mean phenotypic values. Not accounting for this structure can lead to spurious associations in genome-wide association studies (GWAS) and polygenic risk score (PRS) analyses.
- Relative risk
The probability that a certain health outcome will occur in a person or group of individuals relative to the probability that this event will occur in a reference population. Relative risks are typically expressed as ratios, with 1.0 indicating no difference between the comparison groups.
- Risk stratification
The process of classifying and ordering individuals according to their specific risk estimates.
Polygenic Risk Methods in Diverse Populations (PRIMED) Consortium Methods Working Group
Paul L. Auer20, Nilanjan Chatterjee3, Matthew P. Conomos21, David V. Conti22,23, Yi Ding24, Tian Ge17,18,19, Jibril Hirbo4,5 Linda Kachuri1,2, Eimear E. Kenny9,10,11, Iftikhar J. Kullo8, Iman Martin7, Bogdan Pasaniuc12,13,14, Daniel J. Schaid6, Ying Wang19,25,26, John S. Witte1,2,15,16, Haoyu Zhang27,28 & Yuji Zhang29
20Division of Biostatistics, Institute for Health and Equity, and Cancer Center, Medical College of Wisconsin, Milwaukee, WI, USA. 21Department of Biostatistics, University of Washington, Seattle, WA, USA. 22Center for Genetic Epidemiology, Department of Population and Preventive Health Sciences, Keck School of Medicine, University of Southern California, Los Angeles, CA, USA. 23Norris Comprehensive Cancer Center, Keck School of Medicine, University of Southern California, Los Angeles, CA, USA. 24Bioinformatics Interdepartmental Program, UCLA, Los Angeles, CA, USA. 25Analytic and Translational Genetics Unit, Massachusetts General Hospital, Boston, MA, USA. 26Program in Medical and Population Genetics, Broad Institute of MIT and Harvard, Cambridge, MA, USA. 27Department of Biostatistics, Harvard T.H. Chan School of Public Health, Boston, MA, USA. 28Division of Cancer Epidemiology and Genetics, National Cancer Institute, Bethesda, MD, USA. 29Department of Epidemiology and Public Health, University of Maryland School of Medicine, Baltimore, MD, USA.
Footnotes
Competing interests
The authors declare no competing interests.
Related links
BridgePRS: https://github.com/clivehoggart/BridgePRS
CT-SLEB: https://github.com/andrewhaoyu/CTSLEB
ME-Bayes SL: https://github.com/Jin93/MEBayesSL
PolyPred-S+/PolyPred-P+: https://github.com/omerwe/polyfun
PROSPER: https://github.com/Jingning-Zhang/PROSPER
PRS-CSx(-auto): https://github.com/getian107/PRScsx
SDPRX: https://github.com/eldronzhou/SDPRX
ShaPRS: https://github.com/mkelcb/shaprs
TL-Multi: https://github.com/mxxptian/TLMulti
TL-PRS/MTL-PRS: https://github.com/ZhangchenZhao/TLPRS
X-Wing: https://github.com/qlu-lab/X-Wing
XP-BLUP: https://github.com/tanglab/XP-BLUP
XPASS(+): https://github.com/YangLabHKUST/XPASS
References
- 1.Kullo IJ et al. Polygenic scores in biomedical research. Nat. Rev. Genet 23, 524–532 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Abdellaoui A, Yengo L, Verweij KJH & Visscher PM 15 years of GWAS discovery: realizing the promise. Am. J. Hum. Genet 110, 179–194 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Martin AR et al. Clinical use of current polygenic risk scores may exacerbate health disparities. Nat. Genet 51, 584–591 (2019). This paper demonstrates that PRSs have limited generalizability across populations and emphasizes the importance of diversity to realize the full and equitable potential of PRSs.
- 4. Fatumo S. et al. A roadmap to increase diversity in genomic studies. Nat. Med 28, 243–250 (2022). This paper presents an updated ancestry tabulation for participants in GWAS catalogue and discusses strategies for increasing diversity in genomic studies.
- 5.Zhou W. et al. Global Biobank Meta-analysis Initiative: powering genetic discovery across human disease. Cell Genom. 2, 100192 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang Y, Tsuo K, Kanai M, Neale BM & Martin AR Challenges and opportunities for developing more generalizable polygenic risk scores. Annu. Rev. Biomed. Data Sci 5, 293–320 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lewis CM & Vassos E Polygenic risk scores: from research tools to clinical instruments. Genome Med. 12, 44 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Torkamani A, Wineinger NE & Topol EJ The personal and clinical utility of polygenic risk scores. Nat. Rev. Genet 19, 581–590 (2018). [DOI] [PubMed] [Google Scholar]
- 9.Choi SW, Mak TS-H & O’Reilly PF Tutorial: a guide to performing polygenic risk score analyses. Nat. Protoc 15, 2759–2772 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Polygenic Risk Score Task Force of the International Common Disease Alliance. Responsible use of polygenic risk scores in the clinic: potential benefits, risks and gaps. Nat. Med 27, 1876–1884 (2021). [DOI] [PubMed] [Google Scholar]
- 11.Duncan L. et al. Analysis of polygenic risk score usage and performance in diverse human populations. Nat. Commun 10, 3328 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Martin AR et al. Human demographic history impacts genetic risk prediction across diverse populations. Am. J. Hum. Genet 100, 635–649 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mars N. et al. Genome-wide risk prediction of common diseases across ancestries in one million people. Cell Genom. 2, 100118 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Privé F. et al. Portability of 245 polygenic scores when derived from the UK Biobank and applied to 9 ancestry groups from the same cohort. Am. J. Hum. Genet 109, 373 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Ding Y. et al. Polygenic scoring accuracy varies across the genetic ancestry continuum. Nature 618, 774–781 (2023). This paper shows that the prediction accuracy of PRSs decreases from individual to individual along the continuum of genetic ancestries.
- 16.Cavazos TB & Witte JS Inclusion of variants discovered from diverse populations improves polygenic risk score transferability. HGG Adv. 2, 100017 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wientjes YCJ et al. Empirical and deterministic accuracies of across-population genomic prediction. Genet. Sel. Evol 47, 5 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pszczola M, Strabel T, Mulder HA & Calus MPL Reliability of direct genomic values for animals with different relationships within and to the reference population. J. Dairy. Sci 95, 389–400 (2012). [DOI] [PubMed] [Google Scholar]
- 19.Wientjes YCJ, Veerkamp RF & Calus MPL The effect of linkage disequilibrium and family relationships on the reliability of genomic prediction. Genetics 193, 621–631 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Habier D, Fernando RL & Dekkers JCM The impact of genetic relationship information on genome-assisted breeding values. Genetics 177, 2389–2397 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yang J, Zeng J, Goddard ME, Wray NR & Visscher PM Concepts, estimation and interpretation of SNP-based heritability. Nat. Genet 49, 1304–1310 (2017). [DOI] [PubMed] [Google Scholar]
- 22.Daetwyler HD, Villanueva B & Woolliams JA Accuracy of predicting the genetic risk of disease using a genome-wide approach. PLoS ONE 3, e3395 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Wang Y. et al. Theoretical and empirical quantification of the accuracy of polygenic scores in ancestry divergent populations. Nat. Commun 11, 3865 (2020). This paper theoretically and empirically investigates the impact of various genetic factors on the transferability of PRSs across populations.
- 24.Dudbridge F. Power and predictive accuracy of polygenic risk scores. PLoS Genet. 9, e1003348 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang Y, Qi G, Park J-H & Chatterjee N Estimation of complex effect-size distributions using summary-level statistics from genome-wide association studies across 32 complex traits. Nat. Genet 50, 1318–1326 (2018). [DOI] [PubMed] [Google Scholar]
- 26.Chatterjee N. et al. Projecting the performance of risk prediction based on polygenic analyses of genome-wide association studies. Nat. Genet 45, 400–405, 405e1–405e3 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ge T, Chen C-Y, Neale BM, Sabuncu MR & Smoller JW Phenome-wide heritability analysis of the UK Biobank. PLoS Genet. 13, e1006711 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Mostafavi H. et al. Variable prediction accuracy of polygenic scores within an ancestry group. eLife 9, e48376 (2020). This paper demonstrates that the predictive accuracy of PRSs can depend on sample characteristics such as age, sex and socioeconomic status even within a group that has relatively homogeneous genetic ancestries.
- 29.Shi H. et al. Localizing components of shared transethnic genetic architecture of complex traits from GWAS summary data. Am. J. Hum. Genet 106, 805–817 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shi H. et al. Population-specific causal disease effect sizes in functionally important regions impacted by selection. Nat. Commun 12, 1098 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lam M. et al. Comparative genetic architectures of schizophrenia in East Asian and European populations. Nat. Genet 51, 1670–1678 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hou K. et al. Causal effects on complex traits are similar for common variants across segments of different continental ancestries within admixed individuals. Nat. Genet 55, 549–558 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wojcik GL et al. Genetic analyses of diverse populations improves discovery for complex traits. Nature 570, 514–518 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chen M-H et al. Trans-ethnic and ancestry-specific blood-cell genetics in 746,667 individuals from 5 global populations. Cell 182, 1198–1213.e14 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.International HapMap Consortium. The International HapMap Project. Nature 426, 789–796 (2003). [DOI] [PubMed] [Google Scholar]
- 36.1000 Genomes Project Consortium et al. A global reference for human genetic variation. Nature 526, 68–74 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zuk O, Hechter E, Sunyaev SR & Lander ES The mystery of missing heritability: genetic interactions create phantom heritability. Proc. Natl Acad. Sci. USA 109, 1193–1198 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zaidi AA & Mathieson I Demographic history mediates the effect of stratification on polygenic scores. eLife 9, e61548 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Saitou M, Dahl A, Wang Q & Liu X Allele frequency differences of causal variants have a major impact on low cross-ancestry portability of PRS. Preprint at bioRxiv 10.1101/2022.10.21.22281371 (2022). [DOI] [Google Scholar]
- 40.Bitarello BD & Mathieson I Polygenic scores for height in admixed populations. G3 10, 4027–4036 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhang H. et al. Novel methods for multi-ancestry polygenic prediction and their evaluations in 5.1 million individuals of diverse ancestry. Preprint at bioRxiv 10.1101/2022.03.24.485519 (2022). [DOI] [Google Scholar]
- 42.Digitale JC, Martin JN & Glymour MM Tutorial on directed acyclic graphs. J. Clin. Epidemiol 142, 264–267 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lipsky AM & Greenland S Causal directed acyclic graphs. J. Am. Med. Assoc 327, 1083–1084 (2022). [DOI] [PubMed] [Google Scholar]
- 44.Aschard H, Vilhjálmsson BJ, Joshi AD, Price AL & Kraft P Adjusting for heritable covariates can bias effect estimates in genome-wide association studies. Am. J. Hum. Genet 96, 329–339 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tennant PWG et al. Use of directed acyclic graphs (DAGs) to identify confounders in applied health research: review and recommendations. Int. J. Epidemiol 50, 620–632 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Socrates A. et al. Investigating the effects of genetic risk of schizophrenia on behavioural traits. NPJ Schizophr. 7, 2 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Peyrot WJ et al. Effect of polygenic risk scores on depression in childhood trauma. Br. J. Psychiatry 205, 113–119 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Peyrot WJ et al. Does childhood trauma moderate polygenic risk for depression? A meta-analysis of 5765 subjects from the psychiatric genomics consortium. Biol. Psychiatry 84, 138–147 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Dorans KS, Mills KT, Liu Y & He J Trends in prevalence and control of hypertension according to the 2017 American College of Cardiology/American Heart Association (ACC/AHA) guideline. J. Am. Heart Assoc 7, e008888 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Centers for Disease Control and Prevention. Chronic kidney disease in the United States. CDC https://www.cdc.gov/kidneydisease/publications-resources/ckd-national-facts.html (2021). [Google Scholar]
- 51.Chu CD et al. Trends in chronic kidney disease care in the US by race and ethnicity, 2012–2019. JAMA Netw. Open 4, e2127014 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Siegel RL, Miller KD, Fuchs HE & Jemal A Cancer statistics, 2022. CA Cancer J. Clin 72, 7–33 (2022). [DOI] [PubMed] [Google Scholar]
- 53.Zavala VA et al. Cancer health disparities in racial/ethnic minorities in the United States. Br. J. Cancer 124, 315–332 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Marinac CR, Ghobrial IM, Birmann BM, Soiffer J & Rebbeck TR Dissecting racial disparities in multiple myeloma. Blood Cancer J. 10, 19 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Daly B & Olopade OI A perfect storm: how tumor biology, genomics, and health care delivery patterns collide to create a racial survival disparity in breast cancer and proposed interventions for change. CA Cancer J. Clin 65, 221–238 (2015). [DOI] [PubMed] [Google Scholar]
- 56.Carrot-Zhang J. et al. Genetic ancestry contributes to somatic mutations in lung cancers from admixed Latin American populations. Cancer Discov. 11, 591–598 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Freedman ML et al. Admixture mapping identifies 8q24 as a prostate cancer risk locus in African-American men. Proc. Natl Acad. Sci. USA 103, 14068–14073 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Fejerman L. et al. Admixture mapping identifies a locus on 6q25 associated with breast cancer risk in US Latinas. Hum. Mol. Genet 21, 1907–1917 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Gignoux CR et al. An admixture mapping meta-analysis implicates genetic variation at 18q21 with asthma susceptibility in Latinos. J. Allergy Clin. Immunol 143, 957–969 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Chi C. et al. Admixture mapping reveals evidence of differential multiple sclerosis risk by genetic ancestry. PLoS Genet. 15, e1007808 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Tcheandjieu C. et al. Large-scale genome-wide association study of coronary artery disease in genetically diverse populations. Nat. Med 28, 1679–1692 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Trubetskoy V. et al. Mapping genomic loci implicates genes and synaptic biology in schizophrenia. Nature 604, 502–508 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Mahajan A. et al. Multi-ancestry genetic study of type 2 diabetes highlights the power of diverse populations for discovery and translation. Nat. Genet 54, 560–572 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kelemen M, Vigorito E, Fachal L, Anderson CA & Wallace C ShaPRS: leveraging shared genetic effects across traits or ancestries improves accuracy of polygenic scores. Preprint at bioRxiv 10.1101/2021.12.10.21267272 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.International Schizophrenia Consortium et al. Common polygenic variation contributes to risk of schizophrenia and bipolar disorder. Nature 460, 748–752 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Choi SW & O’Reilly PF PRSice-2: polygenic risk score software for biobank-scale data. Gigascience 8, giz082 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Márquez-Luna C, Loh P-R, South Asian Type 2 Diabetes (SAT2D) Consortium, SIGMA Type 2 Diabetes Consortium, & Price AL Multiethnic polygenic risk scores improve risk prediction in diverse populations. Genet. Epidemiol 41, 811–823 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Ruan Y. et al. Improving polygenic prediction in ancestrally diverse populations. Nat. Genet 54, 573–580 (2022). This paper introduces a Bayesian model that can integrate GWAS summary statistics from multiple populations to improve the predictive performance of PRSs across diverse populations.
- 69. Weissbrod O. et al. Leveraging fine-mapping and multipopulation training data to improve cross-population polygenic risk scores. Nat. Genet 54, 450–458 (2022). This paper leverages functionally informed fine-mapping to improve cross-population polygenic prediction.
- 70.Brown BC, Asian Genetic Epidemiology Network Type 2 Diabetes Consortium, Ye CJ, Price AL & Zaitlen N Transethnic genetic-correlation estimates from summary statistics. Am. J. Hum. Genet 99, 76–88 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Coram MA, Fang H, Candille SI, Assimes TL & Tang H Leveraging multi-ethnic evidence for risk assessment of quantitative traits in minority populations. Am. J. Hum. Genet 101, 638 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Cai M. et al. A unified framework for cross-population trait prediction by leveraging the genetic correlation of polygenic traits. Am. J. Hum. Genet 108, 632–655 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Hoggart C. et al. BridgePRS : a powerful trans-ancestry polygenic risk score method. Preprint at bioRxiv 10.1101/2023.02.17.528938 (2023). [DOI] [Google Scholar]
- 74.Tian P. et al. Multiethnic polygenic risk prediction in diverse populations through transfer learning. Front. Genet 13, 906965 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Mak TSH, Porsch RM, Choi SW, Zhou X & Sham PC Polygenic scores via penalized regression on summary statistics. Genet. Epidemiol 41, 469–480 (2017). [DOI] [PubMed] [Google Scholar]
- 76.Zhou G, Chen T & Zhao H SDPRX: a statistical method for cross-population prediction of complex traits. Am. J. Hum. Genet 110, 13–22 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Zhao Z, Fritsche LG, Smith JA, Mukherjee B & Lee S The construction of cross-population polygenic risk scores using transfer learning. Am. J. Hum. Genet 109, 1998–2008 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Zhang J. et al. An ensemble penalized regression method for multi-ancestry polygenic risk prediction. Preprint at bioRxiv 10.1101/2023.03.15.532652 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Jin J. et al. ME-Bayes SL: enhanced Bayesian polygenic risk prediction leveraging information across multiple ancestry groups. Preprint at bioRxiv 10.1101/2023.04.12.536510 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Ge T, Chen C-Y, Ni Y, Feng Y-CA & Smoller JW Polygenic prediction via Bayesian regression and continuous shrinkage priors. Nat. Commun 10, 1776 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Xiao J. et al. XPXP: improving polygenic prediction by cross-population and cross-phenotype analysis. Bioinformatics 38, 1947–1955 (2022). [DOI] [PubMed] [Google Scholar]
- 82.Miao J. et al. Quantifying portable genetic effects and improving cross-ancestry genetic prediction with GWAS summary statistics. Nat. Commun 14, 832 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Amariuta T. et al. Improving the trans-ancestry portability of polygenic risk scores by prioritizing variants in predicted cell-type-specific regulatory elements. Nat. Genet 52, 1346–1354 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Ge T. et al. Development and validation of a trans-ancestry polygenic risk score for type 2 diabetes in diverse populations. Genome Med. 14, 70 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Zhao Z. et al. PUMAS: fine-tuning polygenic risk scores with GWAS summary statistics. Genome Biol. 22, 257 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Vilhjálmsson BJ et al. Modeling linkage disequilibrium increases accuracy of polygenic risk scores. Am. J. Hum. Genet 97, 576–592 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Lloyd-Jones LR et al. Improved polygenic prediction by Bayesian multiple regression on summary statistics. Nat. Commun 10, 5086 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Fahed AC et al. Polygenic background modifies penetrance of monogenic variants for tier 1 genomic conditions. Nat. Commun 11, 3635 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Oetjens MT, Kelly MA, Sturm AC, Martin CL & Ledbetter DH Quantifying the polygenic contribution to variable expressivity in eleven rare genetic disorders. Nat. Commun 10, 4897 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Dornbos P. et al. A combined polygenic score of 21,293 rare and 22 common variants improves diabetes diagnosis based on hemoglobin A1C levels. Nat. Genet 54, 1609–1614 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Lali R. et al. Calibrated rare variant genetic risk scores for complex disease prediction using large exome sequence repositories. Nat. Commun 12, 5852 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Chen C-Y et al. The impact of rare protein coding genetic variation on adult cognitive function. Nat. Genet 55, 927–938 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Fiziev PP et al. Rare penetrant mutations confer severe risk of common diseases. Science 380, eabo1131 (2023). [DOI] [PubMed] [Google Scholar]
- 94.Weiner DJ et al. Polygenic architecture of rare coding variation across 394,783 exomes. Nature 614, 492–499 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Atkinson EG et al. Tractor uses local ancestry to enable the inclusion of admixed individuals in GWAS and to boost power. Nat. Genet 53, 195–204 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Marnetto D. et al. Ancestry deconvolution and partial polygenic score can improve susceptibility predictions in recently admixed individuals. Nat. Commun 11, 1628 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Sun Q. et al. Improving polygenic risk prediction in admixed populations by explicitly modeling ancestral-specific effects via GAUDI. Preprint at bioRxiv 10.1101/2022.10.06.511219 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Pain O. et al. Evaluation of polygenic prediction methodology within a reference-standardized framework. PLoS Genet. 17, e1009021 (2021). This study establishes a reference-standardized framework for fair comparison of PRS construction methods.
- 99.Wang Y. et al. Polygenic prediction across populations is influenced by ancestry, genetic architecture, and methodology. Preprint at bioRxiv 10.1101/2022.12.29.522270 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Shen J. et al. Fine-mapping and credible set construction using a multi-population joint analysis of marginal summary statistics from genome-wide association studies. Preprint at bioRxiv 10.1101/2022.12.22.521659 (2022). [DOI] [Google Scholar]
- 101.Yuan K. et al. Fine-mapping across diverse ancestries drives the discovery of putative causal variants underlying human complex traits and diseases. Preprint at medRxiv 10.1101/2023.01.07.23284293 (2023). [DOI] [PubMed] [Google Scholar]
- 102.Harrell FE Jr, Califf RM, Pryor DB, Lee KL & Rosati RA Evaluating the yield of medical tests. J. Am. Med. Assoc 247, 2543–2546 (1982). [PubMed] [Google Scholar]
- 103.Mavaddat N. et al. Polygenic risk scores for prediction of breast cancer and breast cancer subtypes. Am. J. Hum. Genet 104, 21–34 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Ho W-K et al. European polygenic risk score for prediction of breast cancer shows similar performance in Asian women. Nat. Commun 11, 3833 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Shieh Y. et al. A polygenic risk score for breast cancer in US Latinas and Latin American women. J. Natl Cancer Inst 112, 590–598 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Du Z. et al. Evaluating polygenic risk scores for breast cancer in women of African ancestry. J. Natl Cancer Inst 113, 1168–1176 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Khera AV et al. Genome-wide polygenic scores for common diseases identify individuals with risk equivalent to monogenic mutations. Nat. Genet 50, 1219–1224 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Dikilitas O. et al. Predictive utility of polygenic risk scores for coronary heart disease in three major racial and ethnic groups. Am. J. Hum. Genet 106, 707–716 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Chatterjee N, Shi J & García-Closas M Developing and evaluating polygenic risk prediction models for stratified disease prevention. Nat. Rev. Genet 17, 392–406 (2016). This paper provides a comprehensive review of concepts and methods relevant for the development and evaluation of risk prediction models that incorporate genetic susceptibility factors.
- 110.Wang M. et al. Validation of a genome-wide polygenic score for coronary artery disease in South Asians. J. Am. Coll. Cardiol 76, 703–714 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Khera AV et al. Whole-genome sequencing to characterize monogenic and polygenic contributions in patients hospitalized with early-onset myocardial infarction. Circulation 139, 1593–1602 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Khan A. et al. Genome-wide polygenic score to predict chronic kidney disease across ancestries. Nat. Med 28, 1412–1420 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Hurson AN et al. Prospective evaluation of a breast-cancer risk model integrating classical risk factors and polygenic risk in 15 cohorts from six countries. Int. J. Epidemiol 50, 1897–1911 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Leening MJG, Vedder MM, Witteman JCM, Pencina MJ & Steyerberg EW Net reclassification improvement: computation, interpretation, and controversies: a literature review and clinician’s guide. Ann. Intern. Med 160, 122–131 (2014). [DOI] [PubMed] [Google Scholar]
- 115. Kachuri L. et al. Pan-cancer analysis demonstrates that integrating polygenic risk scores with modifiable risk factors improves risk prediction. Nat. Commun 11, 6084 (2020). This paper quantifies the added predictive value of PRSs for 16 cancer types when added to models that contain extensive clinical and environmental risk factors.
- 116.Kerr KF et al. Net reclassification indices for evaluating risk prediction instruments: a critical review. Epidemiology 25, 114–121 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Vickers AJ & Elkin EB Decision curve analysis: a novel method for evaluating prediction models. Med. Decis. Mak 26, 565–574 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Pal Choudhury P. et al. Comparative validation of breast cancer risk prediction models and projections for future risk stratification. J. Natl Cancer Inst 112, 278–285 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Pal Choudhury P. et al. iCARE: an R package to build, validate and apply absolute risk models. PLoS ONE 15, e0228198 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Pain O, Gillett AC, Austin JC, Folkersen L & Lewis CM A tool for translating polygenic scores onto the absolute scale using summary statistics. Eur. J. Hum. Genet 30, 339–348 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Naret O. et al. Improving polygenic prediction with genetically inferred ancestry. HGG Adv. 3, 100109 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122. Ding Y. et al. Large uncertainty in individual polygenic risk score estimation impacts PRS-based risk stratification. Nat. Genet 54, 30–39 (2022). This paper estimates the variance of an individual’s PRS and highlights the importance of incorporating uncertainty into the interpretation of individual PRS estimates.
- 123.Chouldechova A & Roth A The frontiers of fairness in machine learning. Preprint at 10.48550/arXiv.1810.08810 (2018). [DOI] [Google Scholar]
- 124.Komiyama J, Takeda A, Honda J & Shimao H in Proc. 35th Int. Conf. Machine Learning Vol. 80 (eds Dy J & Krause A) 2737–2746 (PMLR, 2018). [Google Scholar]
- 125.Agarwal A, Dudik M & Wu ZS in Proc. 36th Int. Conf. Machine Learning Vol. 97 (eds Chaudhuri K & Salakhutdinov R) 120–129 (PMLR, 2019). [Google Scholar]
- 126.Rajkomar A, Hardt M, Howell MD, Corrado G & Chin MH Ensuring fairness in machine learning to advance health equity. Ann. Intern. Med 169, 866–872 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Kleinberg J, Mullainathan S & Raghavan M Inherent trade-offs in the fair determination of risk scores. Preprint at 10.48550/arXiv.1609.05807 (2016). [DOI] [Google Scholar]
- 128.Oni-Orisan A, Mavura Y, Banda Y, Thornton TA & Sebro R Embracing genetic diversity to improve Black health. N. Engl. J. Med 384, 1163–1167 (2021). [DOI] [PubMed] [Google Scholar]
- 129.Lewis ACF et al. Getting genetic ancestry right for science and society. Science 376, 250–252 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Banda Y. et al. Characterizing race/ethnicity and genetic ancestry for 100,000 subjects in the Genetic Epidemiology Research on Adult Health and Aging (GERA) Cohort. Genetics 200, 1285–1295 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Lee SH, Wray NR, Goddard ME & Visscher PM Estimating missing heritability for disease from genome-wide association studies. Am. J. Hum. Genet 88, 294–305 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Tillin T. et al. Ethnicity and prediction of cardiovascular disease: performance of QRISK2 and Framingham scores in a U.K. tri-ethnic prospective cohort study (SABRE-Southall And Brent REvisited). Heart 100, 60–67 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Rodriguez F. et al. Atherosclerotic cardiovascular disease risk prediction in disaggregated Asian and Hispanic subgroups using electronic health records. J. Am. Heart Assoc 8, e011874 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Aldrich MC et al. Evaluation of USPSTF lung cancer screening guidelines among African american adult smokers. JAMA Oncol. 5, 1318–1324 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Pasquinelli MM et al. Risk prediction model versus United States Preventive Services Task Force lung cancer screening eligibility criteria: reducing race disparities. J. Thorac. Oncol 15, 1738–1747 (2020). [DOI] [PubMed] [Google Scholar]
- 136.Mars N. et al. Systematic comparison of family history and polygenic risk across 24 common diseases. Am. J. Hum. Genet 109, 2152–2162 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Hujoel MLA, Loh P-R, Neale BM & Price AL Incorporating family history of disease improves polygenic risk scores in diverse populations. Cell Genom. 2, 100152 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Mars N. et al. Polygenic and clinical risk scores and their impact on age at onset and prediction of cardiometabolic diseases and common cancers. Nat. Med 26, 549–557 (2020). [DOI] [PubMed] [Google Scholar]
- 139.Pal Choudhury P. et al. Comparative validation of the BOADICEA and Tyrer–Cuzick breast cancer risk models incorporating classical risk factors and polygenic risk in a population-based prospective cohort of women of European ancestry. Breast Cancer Res. 23, 22 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Lee A. et al. Comprehensive epithelial tubo-ovarian cancer risk prediction model incorporating genetic and epidemiological risk factors. J. Med. Genet 59, 632–643 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Riveros-Mckay F. et al. Integrated polygenic tool substantially enhances coronary artery disease prediction. Circ. Genom. Precis. Med 14, e003304 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.NIH. The ‘All of Us’ Research Program. N. Engl. J. Med 381, 668–676 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Choudhury A. et al. High-depth African genomes inform human migration and health. Nature 586, 741–748 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Pereira L, Mutesa L, Tindana P & Ramsay M African genetic diversity and adaptation inform a precision medicine agenda. Nat. Rev. Genet 22, 284–306 (2021). [DOI] [PubMed] [Google Scholar]
- 145.Chapman CR Ethical, legal, and social implications of genetic risk prediction for multifactorial disease: a narrative review identifying concerns about interpretation and use of polygenic scores. J. Community Genet 10.1007/s12687-022-00625-9 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Lemke AA et al. Addressing underrepresentation in genomics research through community engagement. Am. J. Hum. Genet 109, 1563–1571 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Wojcik GL et al. Opportunities and challenges for the use of common controls in sequencing studies. Nat. Rev. Genet 23, 665–679 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Taliun D. et al. Sequencing of 53,831 diverse genomes from the NHLBI TOPMed Program. Nature 590, 290–299 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Bien SA et al. Strategies for enriching variant coverage in candidate disease loci on a multiethnic genotyping array. PLoS ONE 11, e0167758 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Kim MS, Patel KP, Teng AK, Berens AJ & Lachance J Genetic disease risks can be misestimated across global populations. Genome Biol. 19, 179 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Martin AR et al. Low-coverage sequencing cost-effectively detects known and novel variation in underrepresented populations. Am. J. Hum. Genet 108, 656–668 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Emde A-K et al. Mid-pass whole genome sequencing enables biomedical genetic studies of diverse populations. BMC Genomics 22, 666 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Kim MS et al. Testing the generalizability of ancestry-specific polygenic risk scores to predict prostate cancer in sub-Saharan Africa. Genome Biol. 23, 194 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Borrell LN et al. Race and genetic ancestry in medicine—a time for reckoning with racism. N. Engl. J. Med 384, 474–480 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Reales G & Wallace C Sharing GWAS summary statistics results in more citations. Commun. Biol 6, 116 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156. Wand H. et al. Improving reporting standards for polygenic scores in risk prediction studies. Nature 591, 211–219 (2021). This paper outlines a framework for systematic reporting of methods and results from PRS studies that is necessary to build a high-quality evidence base for informing PRS translational efforts.
- 157.Lambert SA et al. The polygenic score catalog as an open database for reproducibility and systematic evaluation. Nat. Genet 53, 420–425 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Wang Y. et al. Global Biobank analyses provide lessons for developing polygenic risk scores across diverse cohorts. Cell Genom. 3, 100241 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159. Linder JE et al. Returning integrated genomic risk and clinical recommendations: the eMERGE study. Genet. Med 25, 100006 (2023). This paper describes the ongoing prospective eMERGE study that returns integrated genetic risk assessment including monogenic risks, PRSs and family history to high-risk individuals for 11 conditions.
- 160.Lennon NJ et al. Selection, optimization, and validation of ten chronic disease polygenic risk scores for clinical implementation in diverse populations. Preprint at bioRxiv 10.1101/2023.05.25.23290535 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Mathieson I & Scally A What is ancestry? PLoS Genet. 16, e1008624 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Pritchard JK, Stephens M & Donnelly P Inference of population structure using multilocus genotype data. Genetics 155, 945–959 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Price AL et al. Principal components analysis corrects for stratification in genome-wide association studies. Nat. Genet 38, 904–909 (2006). [DOI] [PubMed] [Google Scholar]
- 164.Patterson N, Price AL & Reich D Population structure and eigenanalysis. PLoS Genet. 2, e190 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Maples BK, Gravel S, Kenny EE & Bustamante CD RFMix: a discriminative modeling approach for rapid and robust local-ancestry inference. Am. J. Hum. Genet 93, 278–288 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Li N & Stephens M Modeling linkage disequilibrium and identifying recombination hotspots using single-nucleotide polymorphism data. Genetics 165, 2213–2233 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Browning SR, Waples RK & Browning BL Fast, accurate local ancestry inference with FLARE. Am. J. Hum. Genet 110, 326–335 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Price AL et al. Sensitive detection of chromosomal segments of distinct ancestry in admixed populations. PLoS Genet. 5, e1000519 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Salter-Townshend M & Myers S Fine-scale inference of ancestry segments without prior knowledge of admixing groups. Genetics 212, 869–889 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]