Abstract
The herpes simplex virus type 1 (HSV-1) capsid is a T=16 icosahedral shell that forms in the nuclei of infected cells. Capsid assembly also occurs in vitro in reaction mixtures created from insect cell extracts containing recombinant baculovirus-expressed HSV-1 capsid proteins. During capsid formation, the major capsid protein, VP5, and the scaffolding protein, pre-VP22a, condense to form structures that are extended into procapsids by addition of the triplex proteins, VP19C and VP23. We investigated whether triplex proteins bind to the major capsid-scaffold protein complexes as separate polypeptides or as preformed triplexes. Assembly products from reactions lacking one triplex protein were immunoprecipitated and examined for the presence of the other. The results showed that neither triplex protein bound unless both were present, suggesting that interaction between VP19C and VP23 is required before either protein can participate in the assembly process. Sucrose density gradient analysis was employed to determine the sedimentation coefficients of VP19C, VP23, and VP19C-VP23 complexes. The results showed that the two proteins formed a complex with a sedimentation coefficient of 7.2S, a value that is consistent with formation of a VP19C-VP232 heterotrimer. Furthermore, VP23 was observed to have a sedimentation coefficient of 4.9S, suggesting that this protein exists as a dimer in solution. Deletion analysis of VP19C revealed two domains that may be required for attachment of the triplex to major capsid-scaffold protein complexes; none of the deletions disrupted interaction of VP19C with VP23. We propose that preformed triplexes (VP19C-VP232 heterotrimers) interact with major capsid-scaffold protein complexes during assembly of the HSV-1 capsid.
Assembly of progeny virions is an essential stage in the life cycle of every virus. For double-stranded DNA viruses such as bacteriophages (4), adenoviruses (7, 9), and herpesviruses (10, 28), capsid subunits initially form a precursor capsid that is packaged with DNA and subsequently matures into an infectious particle. Assembly of the procapsid frequently requires additional proteins, termed scaffolding proteins, that are not present in the mature capsid.
The mature herpes simplex virus type 1 (HSV-1) capsid is an icosahedral shell that is 125 nm in diameter and 15 nm thick (26, 28, 29). Its major structural features are 162 capsomers (150 hexons and 12 pentons) that lie on a T=16 lattice. The capsomers associate at their proximal ends to create a 3-nm-thick floor layer. The capsomer protrusions project radially to a distance of 11 nm from the floor layer, and each capsomer has an axial channel. The major capsid protein, VP5, is the structural subunit of both the hexons and the pentons (22, 23, 38). Hexons are found on the faces and edges of the icosahedron, while one penton is found at each of the 12 capsid vertices. Two minor capsid proteins, VP19C and VP23, make up trigonal nodules called triplexes (320 in all) found just above the capsid floor layer at the local three-fold positions between adjacent capsomers (22). Triplexes may vary somewhat in composition, but on average they are heterotrimers containing one copy of VP19C and two copies of VP23 per triplex. A third minor capsid protein, VP26, is located at the outer tips of the hexons (3, 37, 40).
Assembly of the HSV-1 capsid requires an internal scaffolding protein called pre-VP22a. The major capsid protein interacts with 25 amino acids in the carboxy-terminal domain of pre-VP22a; these residues are cleaved upon release of the scaffold (14, 16, 25, 34). Although the major capsid protein and the scaffolding protein comprise the majority of the protein mass of the capsid as it is assembled, capsid assembly will not occur in the absence of the triplex proteins (6, 33, 35, 39). The triplex proteins interact with major capsid-scaffold protein complexes, forming arc- or dome-like structures called partial capsids (20). The joining of additional subunits allows partial capsids to grow into a spherical procapsid, which undergoes a morphological transition to the mature icosahedral capsid and is packaged with DNA. Three-dimensional reconstructions computed from cryoelectron micrographs of the procapsid show that it is a spherical structure that appears to be open and porous, unlike the mature capsid, which is angular and tightly sealed (20, 36). In the procapsid, the hexons are asymmetric and only loosely formed, as opposed to the symmetric, highly regular hexons in the mature capsid. In addition, the capsid floor layer, which is smooth and continuous in the mature capsid, is rudimentary and incomplete in the procapsid. Triplexes, which are visible at sites between capsomers in the procapsid, appear to be the only substantial connection between adjacent capsomers when observed at a resolution of 26 Å (36).
As described above, analysis of the procapsid structure has suggested an important role for the triplex proteins in capsid assembly. Although the triplex proteins make up a relatively small percentage of the total capsid protein, the triplexes appear to provide essential support for the capsid shell as it is formed. Here we describe use of an in vitro system comprised of insect cell extracts containing recombinant baculovirus-expressed capsid proteins to examine the role of the triplex proteins in capsid assembly. We asked whether the triplex proteins interact with the nascent capsid as separate polypeptides or as preformed structural units. In addition, we employed deletion analysis to identify domains required for triplex formation and for attachment of the triplex proteins to major capsid-scaffold protein complexes. The results shed new light on the essential role of the triplex proteins in HSV capsid assembly.
MATERIALS AND METHODS
Cell lines.
Thomsen et al. (35) described the construction of the four recombinant baculoviruses containing the HSV-1 UL18 (encoding VP23), UL19 (VP5), UL26.5 (pre-VP22a), and UL38 (VP19C) genes employed in this study. The nonessential UL35 gene (VP26) was not included in assembly reactions. Suspension cultures of Spodoptera frugiperda (Sf9) cells were infected with individual recombinant baculoviruses (multiplicity of infection of 5), and at 64 h postinfection the cells were harvested, frozen, and diluted for use in the in vitro system as previously described (20).
In vitro capsid assembly.
Cell-free capsid assembly was performed by a modification of the procedure of Newcomb et al. (21). Assembly-competent reaction mixtures contained aliquots of each of the four cell suspensions described above. Cells were lysed by five repeated cycles of freezing and thawing, and then the cellular debris was removed by centrifugation at 16,000 × g for 5 mins. The supernatant was decanted, and then EDTA (50 mM), dithiothreitol (10 mM), and protease inhibitors (20 mM phenylmethylsulfonyl fluoride, aprotinin [20 μg/ml], 10 μM leupeptin, and trypsin inhibitor [100 μg/ml]) were added. The extracts were incubated at 37°C for 30 min and clarified by centrifugation as described above. Capsids or partial capsid structures were isolated by precipitation with the VP5-specific monoclonal antibody 6F10 as described previously (20).
Construction of VP19C mutants.
Deletions were made in the UL38 gene by using pAc-UL38 (35) as a template for PCRs. Upstream oligonucleotide primers were complementary to the noncoding strand and contained a BamHI restriction site. Downstream oligonucleotide primers were complementary to the coding strand and contained a KpnI restriction site. The UL38 gene was truncated by placing primers at various increments in from the 5′ or 3′ end of the gene. The pAc-373 vector was digested with BamHI and KpnI, gel purified, ligated to PCR products that had been digested with the same enzymes, and transformed as described previously (30). Plasmid DNA from positive clones was purified and cotransfected to generate recombinant baculoviruses (35).
Electron microscopy.
Cell pellets containing capsid assembly products were prepared for electron microscopy by fixation in 2% glutaraldehyde, embedding in Epon 812, and sectioning as previously described (20). All micrographs were recorded on a JEOL 100CX transmission electron microscope operated at 80 keV.
Sucrose density gradient analysis.
Sucrose gradients (5 to 20% sucrose in phosphate-buffered saline) were prepared and equilibrated to 4°C (11). Extracts containing single capsid proteins or combinations of capsid proteins were loaded onto the gradients and immediately subjected to centrifugation at 115,000 × g for 18 h at 4°C. Centrifugation was performed in a Beckman L5-50 Ultracentrifuge by using an SW50.1 rotor at 35,000 rpm, and gradients were fractionated with a Buchler Polystaltic Pump into 40 fractions of approximately 115 μl each (4 drops). Fractions were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and Western blotting to identify capsid proteins. Coomassie blue-stained gels and X-ray films were digitized with a Molecular Dynamics personal densitometer, and Imagequant software was used to quantitate the capsid proteins in each lane.
Sedimentation coefficient and molecular weight determination.
The calculation of sedimentation coefficients was carried out by comparison with a protein standard by the method of Martin and Ames (17). Residual bovine serum albumin (BSA) from each of the cell cultures served as an internal standard for each gradient. The position of BSA in the gradient was determined by SDS-PAGE and Western blotting. Sedimentation coefficients for capsid proteins were determined by a ratio of distance traveled by the capsid protein to distance traveled by BSA. Molecular weights were approximated from the sedimentation coefficients as described previously (17).
SDS-PAGE and Western immunoblotting.
Gels were prepared with a 4% stacking gel and a 12.5% resolving gel as described previously (22). Gels were either stained with Coomassie blue or electrophoretically transferred to a nitrocellulose membrane for Western immunoblotting (30). Immunoblots were stained either with polyclonal antisera specific for VP19C and/or VP23 or with a pool of monoclonal antibodies (3N-5, 3C-16, 2-11, and 1N-11) specific for BSA. HSV-1 B capsids, used as a standard for gel electrophoresis and immunoblotting, were prepared as previously described from BHK-21 cells infected with the 17MP strain of HSV-1 (23).
RESULTS
In vitro assembly reactions were performed to determine whether the triplex proteins can associate with the nascent capsid independently of each other or whether the capsid assembly requires the presence of preformed triplexes. Previous studies have shown that VP5 associates with pre-VP22a in the absence of triplex proteins to form distinct structures (16). The structures are spherical, but they vary slightly in size, averaging 60 nm in diameter. These major capsid-scaffold protein complexes can be precipitated with the VP5-specific monoclonal antibody 6F10 (13). Therefore, even in the absence of capsid formation, it is possible to determine whether triplex proteins have associated with the major capsid-scaffold protein complex by examining the immunoprecipitate. Figure 1 shows SDS-PAGE and Western immunoblot analysis of assembly reaction mixtures consisting of cell extracts containing HSV-1 capsid proteins.
FIG. 1.
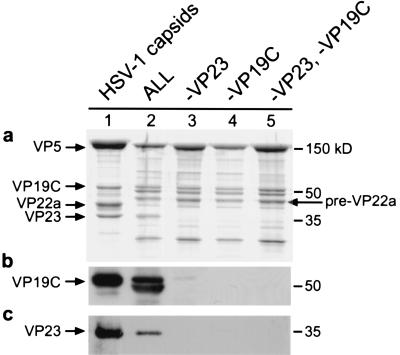
Protein compositions of products formed in the in vitro assembly system. Cell extracts containing recombinant VP5, pre-VP22a, VP23, and VP19C (ALL) were combined and assembly products were precipitated with the VP5-specific monoclonal antibody 6F10. The precipitate was analyzed by SDS-PAGE, and the gel was stained with Coomassie blue (a). In some reactions individual capsid proteins were omitted. Lane 1 contains HSV-1 capsids isolated from infected BHK cells as a protein standard, and lane 2 contains a reaction mixture with all four recombinant proteins. Control HSV-1 capsids from infected cells contain processed scaffolding protein (VP22a), while in vitro assembly reactions contain uncleaved scaffolding protein (pre-VP22a). Lane 3 contains ALL minus VP23, lane 4 contains ALL minus VP19C, and lane 5 contains ALL minus VP23 and VP19C. Two identical gels were blotted and probed with polyclonal antiserum specific for VP19C (b) or VP23 (c). One additional band is visible just below VP19C (panel c, lane 2); this band likely represents a breakdown product resulting from protease activity in the insect cell extract.
Control reaction mixtures contained all four baculovirus-expressed capsid proteins (VP5, pre-VP22a, VP19C, and VP23) and yielded capsids, as viewed by electron microscopy (data not shown). SDS-PAGE analysis revealed that all four capsid proteins were present in the precipitate (Fig. 1, lane 2). Western blot analysis was used to show specifically whether triplex proteins were present in each precipitate (Fig. 1b and c). VP19C, which has a molecular weight of 50,000, migrates to approximately the same position as the antibody heavy chain on the SDS gel, and thus the presence of VP19C could be detected only by immunoblotting (Fig. 1b, lane 2). A band appearing just below the antibody heavy-chain band (Fig. 1a, lanes 2 to 5) does not correspond to VP19C, as shown by the immunoblotting results (Fig. 1b), and probably represents a contaminating cellular protein. In in vitro assembly reactions, VP19C was observed to be very sensitive to proteolysis by enzymes in the cell extract, and even in the presence of protease inhibitors a breakdown product, slightly smaller than the full-length protein, is visible (Fig. 1b, lane 2). In the reaction in which VP23 was omitted, only trace amounts of VP19C were detected in the precipitate (Fig. 1b, lane 3). When VP19C was omitted from the reaction mixture, VP23 was not detected in the pellet (Fig. 1c, lane 4). Omission of both triplex proteins (lane 5) yielded a result that appeared to be identical to that observed when one protein or the other was omitted, suggesting that the triplex proteins are unable to bind to the major capsid-scaffold protein complex independently of each other.
Formation of triplexes in the absence of other capsid proteins.
The possibility that triplex proteins would associate in the absence of other capsid proteins was tested by sucrose density gradient analysis. Insect cells expressing recombinant HSV proteins were lysed and clarified to obtain cell extracts containing VP19C and VP23. The cell extracts were combined and layered on top of 5 to 20% sucrose gradients prepared in phosphate-buffered saline. Cell extracts containing each protein individually were also layered onto separate gradients, and residual BSA from each cell culture served as an internal standard to ensure that differences in protein migration were not due to differences in gradient composition. Gradients were centrifuged at 115,000 × g for 18 h at 4°C. Forty fractions of 115 μl each (4 drops) were collected for each gradient. The fractions were analyzed by SDS-PAGE and Western blotting, and densitometry was used to determine the relative protein intensity for each fraction. Optical density values were normalized for each gradient and plotted versus distance from the meniscus in the gradient, as shown in Fig. 2.
FIG. 2.
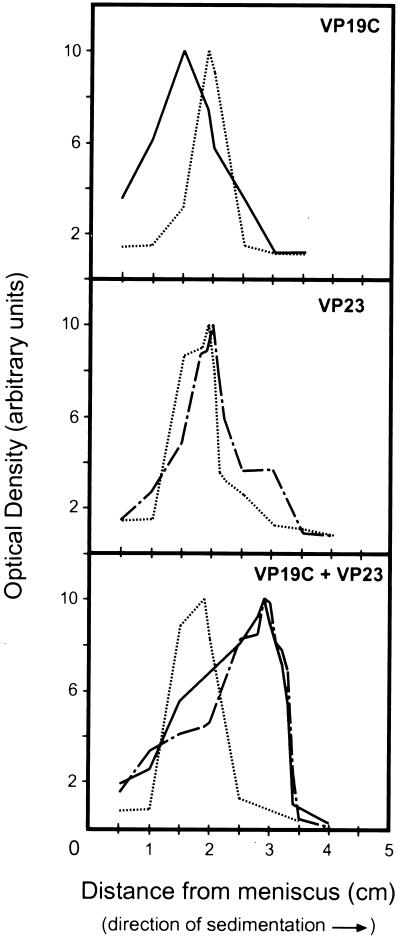
Sedimentation analysis of triplex proteins and protein complexes. Cell extracts containing VP19C, VP23, or VP19C and VP23 were placed on top of 5 to 20% sucrose gradients and centrifuged for 18 h at 115,000 × g. Each gradient was fractionated, and the fractions were analyzed by SDS-PAGE and Western blotting. Protein intensity was determined with Imagequant; the values were normalized, and the relative intensity values were plotted against distance from the meniscus. BSA was included in each gradient as a standard. ——, VP19C; — . — . , VP23; … …, BSA.
In each gradient the BSA standard peaked in fraction 24, a fraction corresponding to a migration of 1.9 cm from the gradient meniscus. VP19C peaked in fraction 27 at 1.5 cm from the meniscus, while VP23 was found in fraction 23 (2.0 cm), sedimenting slightly faster than the BSA. A major shift in migration pattern was seen when the two triplex proteins were combined on the gradient. Both VP19C and VP23 peaked in fraction 15, at a distance of 2.9 cm from the meniscus. The proteins migrated together on the gradient, with the more rapid sedimentation indicating that a larger multiprotein complex had been formed.
The approximate size of the VP19C-VP23 complex observed on the sucrose gradient was determined by comparison with the BSA standard in each gradient. The sedimentation coefficient for each protein or protein complex was calculated (Table 1). The sedimentation coefficient is indirectly proportional to the molecular weight of a protein, because factors such as shape and arrangement of the molecule affect the friction coefficient and thus the sedimentation. However, the approximate molecular weight for each species identified on the sucrose gradients was also calculated (Table 1). For VP19C, such calculations yield a molecular weight of 48,000, a value that is within 5% of the actual molecular weight of 50,260 predicted from the DNA sequence (19). For VP23, both the sedimentation coefficient and the estimated molecular weight of 72,000 were surprising in view of the molecular weight of 34,268 predicted from the DNA sequence (19). However, given the observation that triplexes are heterotrimers composed of one VP19C monomer and one VP23 dimer, we suggest that VP23 may exist as a dimer in solution.
TABLE 1.
Physical properties of triplexes and triplex proteins
| Protein | Distance from meniscus (cm) | S | Experimentally determined mol wt | Actual mol wt from DNA sequence |
|---|---|---|---|---|
| BSAa | 1.9 | 4.7 | 68,000 | 68,000 |
| VP19C | 1.5 | 3.7 | 48,000 | 50,260 |
| VP23 | 2.0 | 4.9 | 72,000 | 34,268 |
| VP19C-VP23 | 2.9 | 7.2 | 129,000 | 118,796b |
BSA was used as a standard to calculate values for other proteins by using the Martin and Ames equation (17).
Molecular weight was calculated for a VP19C-VP232 heterotrimer.
The sedimentation coefficient for the VP19C-VP23 multiprotein complex was computed to be 7.2S, with an estimated molecular weight of 129,000. The theoretical molecular weight for a heterotrimeric complex composed of one VP19C monomer and one VP23 dimer is 118,796. These results support the notion that VP19C and VP23 form triplexes in the absence of other capsid proteins.
Deletion analysis of VP19C.
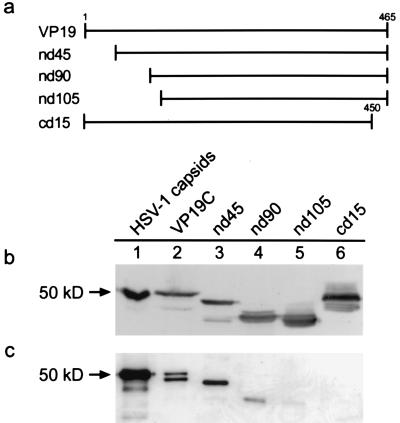
Subsequent experiments were designed to identify protein-protein interactions required for capsid assembly. The VP19C protein was truncated from either the N terminus or the C terminus by using PCR to generate shortened versions of the UL38 coding sequence. Amino acids were removed in increments of 45, 90, and 105 from the N terminus, as shown by the schematic illustration in Fig. 3a. The C terminus was shortened by 15 amino acids. Truncated proteins were expressed by using the recombinant baculovirus system, and the expression of each deletion mutant was verified by Western blotting with polyclonal antiserum specific for VP19C (Fig. 3b). Extracts containing these proteins were then substituted for extracts containing full-length VP19C in the assembly reactions described previously. The reaction mixtures contained VP5, pre-VP22a, VP23, and one of the VP19C deletion mutants. Products of assembly reactions were precipitated with the VP5-specific monoclonal antibody 6F10, and the immunoprecipitate was analyzed by Western blotting to detect the presence of VP19C mutants (Fig. 3c). Deletion of the N-terminal 45 amino acids did not affect the ability of the VP19C protein to associate with the major capsid-scaffold protein complex (Fig. 3b, lane 3). The nd90 protein was present in the precipitate; however, the amount of this protein relative to the full-length and nd45 proteins was significantly decreased (Fig. 3b, lane 4). The nd105 protein did not precipitate with the VP5-VP22a complexes, indicating that a loss of 105 amino acids from the N terminus was too great to support interaction with the assembly complex. In addition, the cd15 protein failed to precipitate (Fig. 3b, lane 6), suggesting that deletion of just 15 amino acids from the C terminus prevents interaction of VP19C with the major capsid-scaffold protein complex.
FIG. 3.
Schematic diagram illustrating the deletions made in the VP19C protein (a) and Western blot detection of VP19C deletion mutants from insect cell extracts (b) or immunoprecipitated assembly complexes (c). VP19C deletion mutants were constructed by cloning truncated genes into baculovirus vectors as described in the text. Proteins that contained deletions at the N terminus or C terminus were expressed in Sf9 cells. Whole-cell lysates were analyzed by SDS-PAGE followed by Western blotting with polyclonal antiserum specific for VP19C (b). Mutants were tested in assembly reactions that included VP5, pre-VP22a, VP23, and either full-length VP19C or one of the deletion mutants. Assembly products were precipitated with monoclonal antibody 6F10 and analyzed by Western blotting as described above (c).
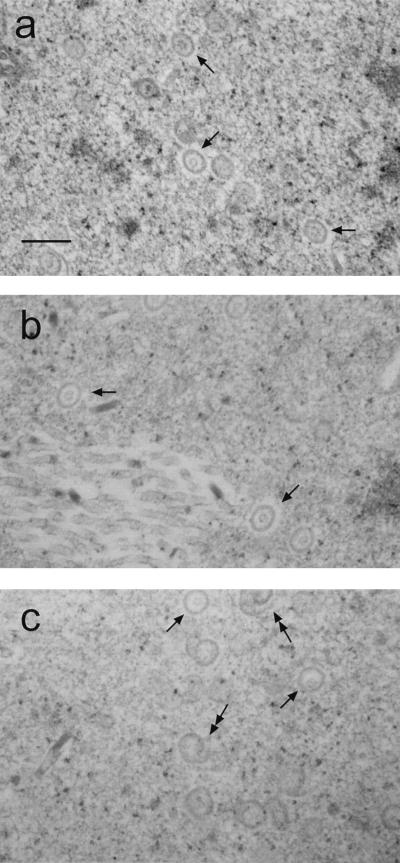
Capsids that incorporated the VP19C deletion mutants were examined by electron microscopy. Insect cells were multiply infected with recombinant baculoviruses expressing each of the capsid proteins (VP5, pre-VP22a, and VP23) and one of the VP19C deletion mutants. At 60 h postinfection the cells were pelleted and thin sectioned to observe capsids or aberrant assembly products. Electron micrographs of cells containing wild-type VP19C (Fig. 4a) showed numerous capsids. The capsids were round and had a measured diameter of approximately 100 nm. An inner core, presumed to be scaffolding protein, was visible in each capsid. Capsids of approximately the same size and shape were also seen in cells containing nd45 and nd90 (Fig. 4b and c), although the number of capsids per field appeared to decrease slightly as the size of the deletion increased. In reactions containing nd90, a number of aberrant structures resembling spirals were also seen, suggesting that assembly with this mutant is much less efficient than with the full-length protein. Aberrant capsids are believed to result from assembly attempts in which the appropriate shell curvature is not maintained. No capsids or aberrant structures were seen in sections of cells containing nd105 or cd15. This was expected based on the observation that these proteins were not detected in immunoprecipitated assembly complexes. Together these results demonstrate that mutants nd105 and cd15 lack the domains required for capsid assembly.
FIG. 4.
Electron micrographs of capsids formed by coinfection of Sf9 cells with recombinant baculoviruses expressing VP5, pre-VP22a, VP23, and either wild-type VP19C or one of the deletion mutants. Cell pellets were fixed and thin sectioned. Capsids (arrows) were observed only in coinfections containing wild-type VP19 (a), nd45 (b), or nd90 (c). Aberrant structures (double arrows) were also seen in infections containing nd90 (c). Bar, 250 nm.
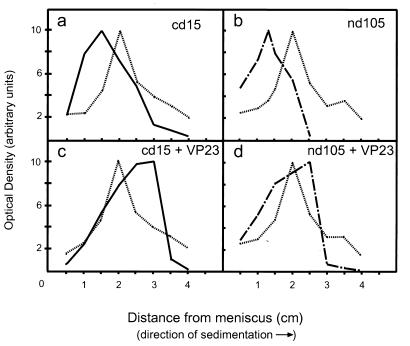
Deletions in the VP19C protein that inhibited capsid assembly could be due to the absence of sequences required for triplex formation or to the lack of sequences involved in attachment of the triplex to the major capsid-scaffold protein complex. To examine whether mutants that failed to support capsid assembly were capable of interacting with VP23, extracts containing either the nd105 or cd15 protein were combined with VP23 and layered onto sucrose gradients, as previously described. The gradients were fractionated and examined by SDS-PAGE and Western blot analysis. Densitometry was performed to determine the concentration of capsid protein in each fraction. The optical density for each protein was plotted against sedimentation distance (distance [centimeters] from the meniscus) to observe migration through the gradient, as shown in Fig. 5. For cell extracts containing only cd15, the protein was observed to sediment slowly, being found 1.5 cm from the meniscus (Fig. 5a). However, when cell extracts contained cd15 combined with VP23, the sedimentation pattern shifted. cd15 and VP23 sedimented together, peaking 3.0 cm from the meniscus, suggesting that a larger protein complex had formed (Fig. 5c). A similar result was observed with the nd105 mutant. For cell extracts containing nd105, the truncated protein was observed to peak 1.4 cm from the meniscus (Fig. 5b). When cell extracts contained nd105 combined with VP23, both proteins were observed to peak 2.5 cm from the meniscus (Fig. 5d). The shifts in sedimentation observed when either mutant was combined with VP23 indicate that both truncated proteins still contained the domains necessary for interaction with VP23. Molecular weight computations confirmed that the observed sedimentation pattern for these protein complexes was consistent with a heterotrimeric triplex containing one copy of the truncated VP19C and one VP23 dimer (Table 2). The predicted molecular weight for a cd15-VP232 heterotrimer was 117,176, and the value for the protein complex derived from gradient analysis was 126,000. The nd105-VP232 complex, predicted to have a molecular weight of 107,456, had a calculated molecular weight of 96,000. Because both proteins were observed to form complexes with VP23, this would suggest that capsids are unable to form in reaction mixtures containing either mutant because the triplexes lack an element required for attachment to the major capsid-scaffold protein complex. Properties of each of the VP19C deletion mutants are summarized in Table 3, with abilities to support capsid assembly and triplex formation indicated. While a substantial portion of the N terminus can be deleted before effects on capsid assembly are observed, a deletion of only 15 amino acids from the C terminus prevents any capsid assembly. The results suggest that two regions of VP19C, amino acids 90 to 105 and 450 to 465 (the C terminus), are required for capsid assembly, although it has not been determined whether they participate in direct interactions or influence other regions that are involved. All of the truncated forms of VP19C examined in this study were able to interact with VP23, demonstrating that the region of VP19C that interacts with VP23 is likely to be located between residues 105 and 450. Finally, the C terminus of VP19C appears to specifically influence attachment of the triplex to the major capsid-scaffold protein complexes during capsid assembly.
FIG. 5.
Sedimentation analysis of VP19C deletion mutants and complexes containing mutant proteins. Cell extracts containing mutant cd15 (a), nd105 (b), cd15 and VP23 (c), or nd105 and VP23 (d) were placed on top of 5 to 20% sucrose gradients and centrifuged for 18 h at 115,000 × g. Each gradient was fractionated and analyzed as described previously, with protein intensity plotted versus distance from the meniscus to show the location of protein peaks. BSA was included in each gradient as a standard. ——, cd15; — . — . , nd105; … …, BSA.
TABLE 2.
Physical properties of truncated triplex proteins and protein complexes
| Protein | Distance from meniscus (cm) | S | Experimentally determined mol wt | Actual mol wt from DNA sequence |
|---|---|---|---|---|
| BSAa | 2.0 | 4.7 | 68,000 | 68,000 |
| cd15 | 1.5 | 3.5 | 44,000 | 48,640 |
| nd105 | 1.4 | 3.3 | 40,000 | 38,920 |
| cd15-VP23 | 3.0 | 7.1 | 126,000 | 117,176b |
| nd105-VP23 | 2.5 | 5.9 | 96,000 | 107,456b |
BSA was used as a standard to determine other values as described in the text.
Molecular weight was calculated for a heterotrimer (VP19C mutant-VP232).
TABLE 3.
VP19C deletion mutant summary
| Protein | Capsid assemblya | Triplex formationb |
|---|---|---|
| Wild-type VP19C | + | + |
| nd45 | + | + |
| nd90 | + | + |
| nd105 | − | + |
| cd15 | − | + |
Capsid assembly was determined by immunoblot analysis of immunoprecipitated in vitro assembly reactions and by electron microscopy of in vivo reactions.
Triplex formation was measured by sedimentation on sucrose gradients as described in the text.
DISCUSSION
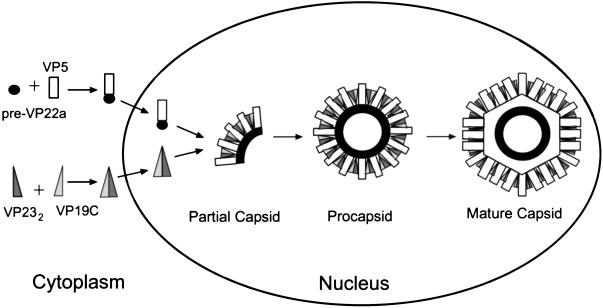
HSV-1 capsid assembly is a complex process in which many proteins are organized into a highly structured shell that houses and protects the viral genome. We have employed a cell-free system to demonstrate that protein interactions required for capsid assembly occur in a specific sequence. Our studies show that assembly products from reaction mixtures containing the major capsid protein, scaffold protein, and one of the two triplex proteins consist only of major capsid and scaffold protein. The triplex proteins, VP19C and VP23, are unable to associate with major capsid-scaffold protein complexes independently of one another, suggesting that VP19C and VP23 must interact with each other prior to binding to major capsid-scaffold protein subunits for capsid assembly. This indicates that the protein interactions are ordered such that the major capsid and scaffolding proteins first interact to form small subunits, as shown in Fig. 6. Heterotrimeric triplexes probably bind to these subunits, connecting them to form arc-shaped or dome-shaped partial capsids. The addition of more subunits results in formation of first a partial capsid and later a spherical procapsid, which is subsequently transformed into the mature icosahedral shell (20). While this proposed pathway is derived from observations in an in vitro system, it seems unlikely that the in vivo pathway would be unrelated.
FIG. 6.
Proposed sequence of events in the HSV-1 capsid assembly pathway, based on intermediates observed in the in vitro assembly system. The major capsid protein and the scaffold protein interact in the cytoplasm, forming VP5–pre-VP22a complexes that are localized to the nucleus (16). VP19C and VP23 interact to form triplexes, which then move to the nucleus. Assembly occurs by interaction of the two types of protein complexes, forming arc-like partial capsids that grow into a procapsid shell. Finally, the spherical procapsid matures by transformation into the icosahedral capsid. In vivo, this transformation would be expected to be accompanied by packaging of viral DNA.
In addition, immunofluorescence studies have shown that in transfected BHK cells expressing HSV capsid proteins, VP5 is transported to the nucleus through interaction with the scaffolding protein pre-VP22a (24). Another study demonstrated that VP23 is transported to the nucleus by VP19C (27), suggesting that such pairwise interactions may be required for all of the assembly components to translocate to the nucleus, where capsid assembly occurs. Studies using the two-hybrid system failed to detect direct interactions between either triplex protein and VP5 or pre-VP22a (5), further supporting the model that the triplex as a unit, rather than individual polypeptides, binds to VP5 and pre-VP22a.
Physical properties of the triplexes.
Although interaction between the triplex proteins, VP19C and VP23, has previously been demonstrated by immunofluorescence (27) and in the two-hybrid system (5), sucrose density gradient analysis was useful for the physical characterization of the triplex. Gradient analysis showed that both proteins had an increased sedimentation coefficient when incubated together compared to the sedimentation coefficient for each protein when incubated alone. Therefore, VP19C and VP23 must come together to form a larger, more rapidly sedimenting protein complex. While immunoprecipitation might have been an easier method for detecting an interaction between the two proteins and determining stoichiometry, precipitating antibodies are not available for either VP23 or VP19C. However, molecular weight estimations based on the sedimentation coefficient are consistent with a heterotrimer composed of one VP19C monomer and one VP23 dimer (the expected molecular weight was 118,796; the observed molecular weight was 129,000). The protein peaks observed were not consistent with a complex composed of one VP19C monomer and one VP23 monomer, which would be predicted to have a molecular weight of 84,528, nor were the derived values consistent with a complex composed of two homodimers (expected molecular weight of 169,056). A complex containing one VP19C dimer and one VP23 monomer would have a molecular weight of 134,788, a value that is close to the calculated molecular weight of 129,000 in this experiment. However, considering the copy number for each protein, as determined by scanning transmission electron microscopy (375 for VP19C and 572 for VP23), this arrangement is unlikely to account for the majority of the triplexes (22). In addition, gradients containing only VP23 indicated that this protein exists as a dimer in solution, which supports the model of the triplex as a heterotrimer. Finally, recent studies of human cytomegalovirus triplex proteins mCP (minor capsid protein) and mC-BP (mCP-binding protein) have shown that these proteins form an mCP-BP-mCP2 heterotrimer and that mCP (VP23 homolog) exists as a dimer when expressed alone (2). These results are consistent with our observations for HSV-1 triplex proteins.
Preliminary evidence suggests that the complexes formed by incubation of cell extracts containing VP19C and VP23 may be competent for capsid assembly. Further studies are required to determine the kinetics of each reaction; however, these studies are complicated by the additional cellular proteins present in the in vitro capsid assembly system. Efforts are currently being directed at purification of each protein for equilibrium studies.
Protein interactions.
Deletion of 15 amino acids from the C terminus of VP19C resulted in a protein that was inactive in capsid assembly. Gradient analysis, however, showed that the truncated protein was still capable of interacting with VP23. Based on this result, it seems likely that the sequence at the C terminus of VP19C (451-LEGVVWRPGGWRACA-465) may be involved in specific interactions required for attachment of the triplex to the major capsid-scaffold protein complex. On the basis of the 1979 report by Zweig et al. (42) of a disulfide linkage between VP19C and VP5, it was hypothesized that the C-terminal cysteine residue might participate in a disulfide bond that links the triplex to the major capsid-scaffold protein complex. However, a single amino acid substitution changing the cysteine to an alanine (C464A) did not inhibit capsid assembly (31), suggesting that another element of the sequence may be important.
At the N terminus of VP19C, 90 amino acids can be removed without interfering with capsid assembly, suggesting that this domain is not involved in essential interactions. However, deletion of 105 amino acids from the N terminus did prevent capsid assembly, possibly indicating that residues 90 to 105 either participate in protein interactions or affect other residues that do. While there are no cysteine residues in the N-terminal domain of the protein, residues in this region may mediate hydrophobic interactions between capsid proteins. Electron micrographs showed a number of aberrant structures in cells expressing the nd90 mutant of VP19C. These structures appear as spirals and probably result from partial capsids that failed to maintain the proper curvature for formation of a closed icosahedral shell. Aberrant capsids may be due to the loss of elements required for hydrophobic interactions that force the subunits into a T=16 lattice.
Deletion analysis of VP23.
In this study deletion analysis was performed only on the VP19C triplex protein. A similar analysis performed on VP23 demonstrated that the loss of 10 residues from the C terminus or 77 amino acids from the N terminus of VP23 inhibited capsid assembly in insect cells infected with recombinant baculoviruses expressing each of the capsid proteins (12). In addition, sites of interaction with VP19C were mapped by using the two-hybrid system. These studies showed that deletion of the N-terminal 77 residues of VP23 prevents interaction with VP19C. Similarly, the loss of 71 residues from the C terminus also inhibited the interaction between the two proteins, although deletion of just 10 C-terminal residues did not. Because the two-hybrid system detects single protein interactions, it is not clear whether deletions that disrupted interaction with VP19C were due to the loss of sequences that mediate binding to VP19C or the loss of sequences that mediate the dimerization of VP23, which may be essential in order for interaction with VP19C to occur. Self-association of VP23 has not been observed in the two-hybrid system (5, 12), indicating that there are limitations to this method, particularly for the analysis of structural proteins.
Triplex structure in the capsid.
In high-resolution studies of the capsid shell, Zhou et al. (41) observed that triplex structure was influenced by the local environment. Six distinct types of triplex based on the type of surrounding capsomers were described. Of particular interest are the triplexes located at the threefold axes of symmetry of each triangular face, between three central hexons. Due to symmetry imposed during reconstruction methods, the structures of these triplexes are in question. While it is interesting to speculate that a homotrimer, rather than a heterotrimer, would likely occupy these sites, no evidence for structures with that stoichiometry was observed during gradient analysis in the experiments reported here.
As higher-resolution images of the HSV-1 capsid and procapsid continue to emerge, it has become clear that the triplexes may perform a role which is different from that of the soc protein of bacteriophage T4, to which the triplexes had been considered analogous. The soc protein is dispensable for phage head formation and binds only to expanded, mature phage heads (1, 15, 32). Given their essential role in maintaining procapsid structure, the HSV-1 triplex proteins seem more comparable to the external scaffolding proteins of bacteriophages such as P4 or φX174. These proteins form cage-like structures around the outside of the phage prohead to maintain its shape and size as it is assembled (8, 17). It has been proposed that the external scaffolding protein Sid controls the incorporation of hexamers into the growing P4 shell (17). We suggest the triplexes may play a similar role in clamping VP5 subunits into the appropriate configuration for maintenance of the icosahedron. One difference between HSV-1 triplex proteins and the external scaffolding proteins of P4 and φX174 is that Sid and gpD are not present in the mature phage heads. However, the triplex proteins of HSV-1, while present in the mature capsid, do not appear to be as important in the mature capsid as they are in the procapsid (36, 40, 41). Treatment of the HSV-1 capsid with 2.0 M guanidine hydrochloride has been observed to result in the loss of pentons and surrounding triplexes without compromising capsid structure (23).
The in vitro capsid assembly system has been extensively employed for the identification of intermediates in the HSV-1 capsid assembly pathway. As the pathway has now been delineated in some detail, the next challenge will be to use this information for the development of therapeutics that prevent assembly of replicating virions. In particular, blocking the transition from procapsid to capsid seems like a feasible target for antiviral agents.
ACKNOWLEDGMENTS
We thank G. Cohen and R. Eisenberg for the VP19C and VP23 antisera, D. Benjamin for the monoclonal antibodies to BSA, and Pam Bruce-Staskal, Patricia Franklin, and Amy Resler for technical assistance.
This work was supported by grants from the National Institutes of Health (AI41644) and the National Science Foundation (MCB-941770).
REFERENCES
- 1.Aebi U, van Driel R, Bijlenga R K, ten Heggeler B, van den Broek R, Steven A C, Smith P R. Capsid fine structure of T-even bacteriophages. Binding and localization of two dispensable capsid proteins into the P23* surface lattice. J Mol Biol. 1977;110:687–698. doi: 10.1016/s0022-2836(77)80084-3. [DOI] [PubMed] [Google Scholar]
- 2.Baxter M K, Gibson W. 22nd International Herpesvirus Workshop, La Jolla, Calif. 1997. The putative cytomegalovirus triplex proteins, minor capsid protein (mCP) and mCP-binding protein (mC-BP) form a heterotrimeric complex that localizes to the cell nucleus in the absence of other viral proteins, abstr. 168. [Google Scholar]
- 3.Booy F P, Trus B L, Newcomb W W, Brown J C, Conway J F, Steven A C. Finding a needle in a haystack: detection of a small protein (the 12-kDa VP26) in a large complex (the 200-MDa capsid of the herpes simplex virus) Proc Natl Acad Sci USA. 1994;91:5652–5656. doi: 10.1073/pnas.91.12.5652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Casjens S, Hendrix R. Control mechanisms in dsDNA bacteriophage assembly. In: Calender R, editor. The bacteriophages. Vol. 1. New York, N.Y: Plenum Publishing Corp.; 1988. pp. 15–91. [Google Scholar]
- 5.Desai P, Person S. Molecular interactions between the HSV-1 capsid proteins as measured by the yeast two-hybrid system. Virology. 1996;220:516–521. doi: 10.1006/viro.1996.0341. [DOI] [PubMed] [Google Scholar]
- 6.Desai P, DeLuca N A, Glorioso J C, Person S. Mutations in herpes simplex virus type 1 genes encoding VP5 and VP23 abrogate capsid formation and cleavage of replicated DNA. J Virol. 1993;67:1357–1364. doi: 10.1128/jvi.67.3.1357-1364.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.D’Halluin J C, Martin G R, Torpier G, Boulanger P A. Adenovirus type 2 assembly analyzed by reversible cross-linking of labile intermediates. J Virol. 1978;26:357–363. doi: 10.1128/jvi.26.2.357-363.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dokland T, McKenna R, Ilag L L, Bowman B R, Incardona N L, Fane B A, Rossmann M G. Structure of a viral procapsid with molecular scaffolding. Nature. 1997;389:308–313. doi: 10.1038/38537. [DOI] [PubMed] [Google Scholar]
- 9.Edvardsson B, Everit E, Jornvall H, Prage L, Philipson L. Intermediates in adenovirus assembly. J Virol. 1976;19:533–547. doi: 10.1128/jvi.19.2.533-547.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gibson W, Roizman B. Proteins specified by herpes simplex virus. VIII. Characterization and composition of multiple capsid forms of subtypes 1 and 2. J Virol. 1972;10:1044–1052. doi: 10.1128/jvi.10.5.1044-1052.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hazlett T L, Dennis E A. Lipid-induced aggregation of phospholipase A2: sucrose density gradient ultracentrifugation and crosslinking studies. Biochim Biophys Acta. 1988;961:22–29. doi: 10.1016/0005-2760(88)90126-9. [DOI] [PubMed] [Google Scholar]
- 12.Homa, F. L. Unpublished data.
- 13.Homa, F. L., and W. W. Newcomb. Unpublished observations.
- 14.Hong Z, Beaudet-Miller M, Durkin J, Zhang R, Kwong A D. Identification of a minimal hydrophobic domain in the herpes simplex virus type 1 scaffolding protein which is required for interaction with the major capsid protein. J Virol. 1996;70:533–540. doi: 10.1128/jvi.70.1.533-540.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ishii T, Yanagida M. The two dispensable structural proteins (soc and hoc) of the T4 phage capsid: their properties, isolation and characterization of deletion mutants and their binding with defective heads in vitro. J Mol Biol. 1977;109:487–514. doi: 10.1016/s0022-2836(77)80088-0. [DOI] [PubMed] [Google Scholar]
- 16.Kennard J, Rixon F J, McDougall I M, Tatman J D, Preston V G. The 25 amino acid residues at the carboxy-terminus of the herpes simplex virus type 1 UL26.5 protein are required for the formation of the capsid shell around the scaffold. J Gen Virol. 1995;76:1611–1621. doi: 10.1099/0022-1317-76-7-1611. [DOI] [PubMed] [Google Scholar]
- 17.Martin R G, Ames B N. A method for determining the sedimentation behavior of enzymes: application to protein mixtures. J Biol Chem. 1961;236:1372–1379. [PubMed] [Google Scholar]
- 18.Marvik O J, Dokland T, Nokling R H, Jacobsen E, Larsen T, Lindqvist B H. The capsid size determining protein sid forms an external scaffold on phage P4 procapsids. J Mol Biol. 1995;251:59–75. doi: 10.1006/jmbi.1995.0416. [DOI] [PubMed] [Google Scholar]
- 19.McGeoch D J, Dalrymple M A, Davison A J, Dolan A, Frame M C, McNab D, Perry L J, Scott J E, Taylor P. The complete DNA sequence of the unique long region in the genome of the herpes simplex virus type 1. J Gen Virol. 1988;69:1531–1574. doi: 10.1099/0022-1317-69-7-1531. [DOI] [PubMed] [Google Scholar]
- 20.Newcomb W W, Homa F L, Thomsen D R, Booy F P, Trus B L, Steven A C, Spencer J V, Brown J C. Assembly of the herpes simplex virus capsid: identification of intermediates in cell-free capsid formation. J Mol Biol. 1996;233:432–446. doi: 10.1006/jmbi.1996.0587. [DOI] [PubMed] [Google Scholar]
- 21.Newcomb W W, Homa F L, Thomsen D R, Ye Z, Brown J C. Cell-free assembly of the herpes simplex virus capsid. J Virol. 1994;68:6059–6063. doi: 10.1128/jvi.68.9.6059-6063.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Newcomb W W, Trus B L, Booy F P, Steven A C, Wall J S, Brown J C. Structure of the herpes simplex virus capsid: molecular composition of the pentons and the triplexes. J Mol Biol. 1993;232:499–511. doi: 10.1006/jmbi.1993.1406. [DOI] [PubMed] [Google Scholar]
- 23.Newcomb W W, Brown J C. Structure of the herpes simplex virus capsid: effects of extraction with guanidine hydrochloride and partial reconstitution of extracted capsids. J Virol. 1991;65:613–620. doi: 10.1128/jvi.65.2.613-620.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nicholson P, Addison C, Cross A M, Kennard J, Preston V G, Rixon F J. Localization of the herpes simplex virus type 1 major capsid protein VP5 to the cell nucleus requires the abundant scaffolding protein VP22a. J Gen Virol. 1994;75:1091–1099. doi: 10.1099/0022-1317-75-5-1091. [DOI] [PubMed] [Google Scholar]
- 25.Oien N L, Thomsen D R, Wathen M W, Newcomb W W, Brown J C, Homa F L. Assembly of herpes simplex virus capsids using the human cytomegalovirus scaffold protein: critical role of the C terminus. J Virol. 1997;71:1281–1291. doi: 10.1128/jvi.71.2.1281-1291.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Perdue M L, Cohen J C, Kemp M C, Randall C C, O’Callaghan D J. Characterization of three species of nucleocapsids of equine herpes virus type 1. Virology. 1975;64:187–205. doi: 10.1016/0042-6822(75)90091-4. [DOI] [PubMed] [Google Scholar]
- 27.Rixon F J, Addison C, MacGregor A, Macnab S J, Nicholson P, Preston V G, Tatman J. Multiple interactions control the intracellular localization of the herpes simplex virus type 1 capsid proteins. J Gen Virol. 1996;77:2251–2260. doi: 10.1099/0022-1317-77-9-2251. [DOI] [PubMed] [Google Scholar]
- 28.Roizman B. Herpesviridae: a brief introduction. In: Fields B N, Knipe D M, Howley P M, Chanock R M, Melnick J L, Monath T P, Strauss S E, Roizman B, editors. Virology. 3rd ed. Philadelphia, Pa: Lippincott-Raven; 1996. pp. 2231–2295. [Google Scholar]
- 29.Schrag J D, Prasad B V, Rixon F J, Chiu W. Three-dimensional structure of the HSV1 nucleocapsid. Cell. 1989;56:651–660. doi: 10.1016/0092-8674(89)90587-4. [DOI] [PubMed] [Google Scholar]
- 30.Spencer J V, Booy F P, Trus B L, Steven A C, Newcomb W W, Brown J C. Structure of the herpes simplex virus capsid: peptide A862-H880 is displayed on the rim of the capsomer protrusions. Virology. 1997;228:229–235. doi: 10.1006/viro.1996.8392. [DOI] [PubMed] [Google Scholar]
- 31.Spencer, J. V., D. R. Thomsen, and F. L. Homa. Unpublished observations.
- 32.Steven A C, Greenstone H L, Booy F P, Black L W, Ross P D. Conformational changes of a viral capsid protein. Thermodynamic rational for proteolytic regulation of bacteriophage T4 capsid expansion, cooperativity, and superstabilization by soc binding. J Mol Biol. 1992;228:870–884. doi: 10.1016/0022-2836(92)90871-g. [DOI] [PubMed] [Google Scholar]
- 33.Tatman J D, Preston V G, Nicholson P, Elliot R M, Rixon F J. Assembly of herpes simplex virus type 1 capsids using a panel of recombinant baculoviruses. J Gen Virol. 1994;75:1101–1113. doi: 10.1099/0022-1317-75-5-1101. [DOI] [PubMed] [Google Scholar]
- 34.Thomsen D R, Newcomb W W, Brown J C, Homa F L. Assembly of the herpes simplex virus capsid: requirement for the carboxyl-terminal twenty-five amino acids of the proteins encoded by the UL26 and UL26.5 genes. J Virol. 1995;69:3690–3703. doi: 10.1128/jvi.69.6.3690-3703.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Thomsen D R, Roof L L, Homa F L. Assembly of herpes simplex virus (HSV) intermediate capsids in insect cells infected with recombinant baculoviruses expressing HSV capsid proteins. J Virol. 1994;68:2442–2457. doi: 10.1128/jvi.68.4.2442-2457.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Trus B L, Booy F P, Newcomb W W, Brown J C, Homa F L, Thomsen D R, Steven A C. The herpes simplex virus procapsid: structure, conformational changes upon maturation, and roles of the triplex proteins VP19c and VP23 in assembly. J Mol Biol. 1996;263:447–462. doi: 10.1016/s0022-2836(96)80018-0. [DOI] [PubMed] [Google Scholar]
- 37.Trus B L, Homa F L, Booy F P, Newcomb W W, Thomsen D R, Cheng N, Brown J C, Steven A C. Herpes simplex virus capsids assembled in insect cells infected with recombinant baculoviruses: structural authenticity and localization of VP26. J Virol. 1995;69:7362–7366. doi: 10.1128/jvi.69.11.7362-7366.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Trus B L, Newcomb W W, Booy F P, Brown J C, Steven A C. Distinct monoclonal antibodies separately label the hexons or the pentons of herpes simplex virus capsid. Proc Natl Acad Sci USA. 1992;89:11508–11512. doi: 10.1073/pnas.89.23.11508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Weller S K, Carmichael E P, Aschman D P, Goldstein D J, Schaeffer P A. Genetic and phenotypic characterization of mutants in four essential genes that map to the left half of HSV-1 UL DNA. Virology. 1987;161:191–210. doi: 10.1016/0042-6822(87)90186-3. [DOI] [PubMed] [Google Scholar]
- 40.Zhou Z H, He J, Jakana J, Tatman J D, Rixon F J, Chiu W. Assembly of VP26 in herpes simplex virus-1 inferred from structures of wild-type and recombinant capsids. Nat Struct Biol. 1995;2:1026–1030. doi: 10.1038/nsb1195-1026. [DOI] [PubMed] [Google Scholar]
- 41.Zhou Z H, Prasad B V, Jakana J, Rixon F J, Chiu W. Protein subunit structures in the herpes simplex virus A-capsid determined from 400 kV spot-scan electron cryomicroscopy. J Mol Biol. 1994;242:456–469. doi: 10.1006/jmbi.1994.1594. [DOI] [PubMed] [Google Scholar]
- 42.Zweig M, Heilman C J, Jr, Hampar B. Identification of disulfide-linked protein complexes in the nucleocapsids of herpes simplex type 2. Virology. 1979;94:442–450. doi: 10.1016/0042-6822(79)90474-4. [DOI] [PubMed] [Google Scholar]