Abstract
Human T-cell leukemia virus type 1 (HTLV-1) is associated with adult T-cell leukemia/lymphoma, HTLV-1-associated myelopathy/tropical spastic paraparesis, and other diseases. For prevention of the transmission of HTLV-1 and manifestation of these diseases, a small-animal model, especially a mouse model, would be useful. We injected HTLV-1-producing T cells (MT-2) intraperitoneally into neonatal C3H/HeJ mice. While the antibody against HTLV-1 antigens was not detectable in C3H/HeJ mice, HTLV-1 provirus was frequently detected in the spleen, lymph nodes, and thymus by PCR. HTLV-1 provirus was present at the level of 0 to 30 molecules in 105 spleen cells at the age of 15 weeks. In addition, a 59-bp flanking sequence of the HTLV-1 integration site was amplified from the spleen DNA by linker-mediated PCR and was confirmed to be derived from the mouse genome. HTLV-1 provirus was found in the T-cell fraction of the mouse spleen. These results indicate that mice can be infected by HTLV-1 and could serve as an animal model for the study of HTLV-1 infection and its pathogenesis in vivo.
Human T-cell leukemia virus type 1 (HTLV-1) is known to be associated with the pathogenesis of adult T-cell leukemia (ATL) (5, 8, 29, 30, 39, 44), HTLV-1-associated myelopathy/tropical spastic paraparesis (HAM/TSP) (6, 28), HTLV-1-associated arthropathy (26, 31), HTLV-1-associated uveitis (25), and HTLV-1-associated bronchopneumopathy (45). Transmission of HTLV-1 via blood can be prevented by anti-HTLV-1 antibody screening, which was started in Japan in 1987 (13). The main route of HTLV-1 transmission is now from mother to child (7, 37). Therefore, the majority of HTLV-1 carriers are those who were infected with HTLV-1 in childhood. Only a small percentage of carriers of HTLV-1 are patients with HTLV-1-associated diseases (36). In addition, there is a long latency period before manifestation of HTLV-1-associated diseases (14, 38). The pathophysiological status of these carriers is not well understood. These facts make analysis of the pathogenesis of HTLV-1-associated diseases and their prevention difficult. For analysis of the pathophysiology of HTLV-1 carriers, a small-animal model of HTLV-1 infection is useful. Such a model can also be helpful for the development of therapeutics and protective measures against the HTLV-1-associated diseases.
The transmission of HTLV-1 in animals has been reported for rabbits (1, 18, 32, 40) and monkeys (15, 42, 43). Previously, we succeeded in establishing persistent infection with HTLV-1 in rats (34). We and others showed HAM/TSP-like paraparesis in WKA rats (11, 20). Mice have some advantages over other animal models, because more information on genetics and on biological techniques for working with embryos is available for mice (3, 12). In addition, the dose of reagents and the space required should be less than for monkeys and rabbits. In this work, we report on a mouse model of HTLV-1 infection by intraperitoneal injection of HTLV-1-producing human T cells into newborn C3H/HeJ mice.
(A preliminary report of this work has been presented previously [19]).
MATERIALS AND METHODS
Cells.
An HTLV-1-producing human T-cell line, MT-2 (24), was cultured in RPMI 1640 medium supplemented with 10% fetal calf serum.
Animals.
Pregnant C3H/HeJ mice were purchased from Clea, Inc., Tokyo, Japan. Twenty-nine C3H/HeJ offspring born to four dams were each injected intraperitoneally with 2.5 × 106 MT-2 cells within 24 h after birth and again at the age of 1 week. After injection, two offspring were lost by maternal cannibalism and nine offspring were lost by accidents in blood sampling and were not analyzed. Two offspring at the 7th week and 5 offspring at the 11th week were dead at the time of blood sampling, and at the 15th week, 11 offspring were sacrificed after ether anesthesia. Finally, these 18 offspring were analyzed for provirus detection.
PCR.
DNA from peripheral-blood mononuclear cells (PBMC) of the mice was prepared by using a QIAamp Blood Kit (Qiagen, Hilden, Germany). DNA from the thymus, lung, liver, spleen, kidney, and peritoneal lymph nodes was prepared by sodium dodecyl sulfate-proteinase K digestion, followed by phenol extraction. One microgram of DNA was used in each PCR. Detection of the pX sequence of HTLV-1 provirus was performed by PCR and Southern blot analysis as described previously (21, 22, 34). A duplicate PCR was performed with the same DNA (Fig. 1 and 2; Tables 1 and 2). The c-myc sequence of the mouse was amplified in the reaction tube used for multiplex PCR. Visualization of the c-myc band in the gel serves as an internal control to confirm the quality of template DNA and the conditions for successful PCR (21, 34). We also examined the human endogenous retrovirus R (HERV-R) sequence (27) by PCR in order to exclude the possibility of MT-2 cells remaining in MT-2 cell-injected animals (34).
FIG. 1.
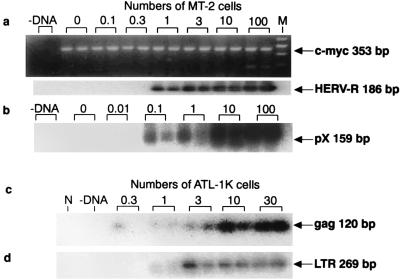
Sensitivity of PCR for detecting HERV-R, pX, gag, and LTR sequences. The PCR templates were prepared so that DNA from 0 to 100 MT-2 (a and b) or ATL-1K (c and d) cells is contained in a tube by dilution with normal mouse spleen DNA, keeping the total DNA content at 1 μg in each reaction. One microgram of DNA is calculated to correspond to 105 cells. (a) A 353-bp c-myc fragment was amplified from mouse c-myc and stained with ethidium bromide. A 186-bp HERV-R fragment was amplified from HERV-R sequence in MT-2 cells and was hybridized with HERV-R probe (34). −DNA, no DNA added in PCR tube; M, DNA size markers, indicating 502, 398, 299, 200, and 101 bp. (b) A 159-bp pX fragment was amplified from MT-2 cell DNA that contained six molecules of pX sequences per cell (17), and this fragment was hybridized with pX probe (34). (c) A 120-bp gag fragment was amplified from ATL-1K cell DNA and was hybridized with gag probe (34). N, normal mouse spleen DNA. (d) A 269-bp LTR fragment was amplified from ATL-1K cell DNA and was hybridized with LTR probe (34).
FIG. 2.
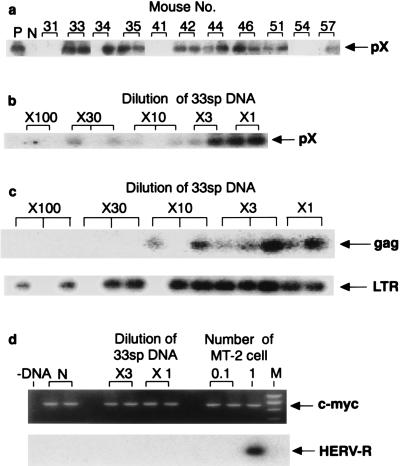
Detection by PCR of HTLV-1 provirus from spleen DNA of MT-2 cell-injected mice. (a) Detection of pX sequence in spleen DNA derived from 11 mice at the age of 15 weeks. Mouse numbers are the same as in Table 2. P, positive control (DNA from 10 MT-2 cells diluted with normal mouse spleen DNA amplified by PCR); N, normal mouse spleen DNA). (b and c) Quantitation of pX, gag, and LTR sequences in the spleen DNA of mouse 33 (33sp DNA). Spleen DNA from mouse 33 was diluted with normal mouse spleen DNA, keeping the total DNA to 1 μg. (d) No human-genome-specific HERV-R was detected in the spleen DNA of mouse 33. Dilution was carried out as described above. −DNA, no DNA added; M, DNA size markers, indicating 502, 398, 299, and 200 bp.
TABLE 1.
Detection of HTLV-1 provirus and anti-HTLV-1 antibody in PBMC
| Mouse no. | No. of positive results/no. of PCRs at the age of:
|
Anti-HTLV-1 antibody resulta | ||||
|---|---|---|---|---|---|---|
| 7 wk
|
11 wk
|
15 wk
|
||||
| gag | pX | gag | pX | pX | ||
| 31 | 0/2 | 0/2 | 0/1 | 0/1 | 1/2 | − |
| 32 | 0/2 | 0/2 | 0/2 | 1/2 | —b | − |
| 33 | 0/2 | 1/2 | 0/2 | 0/2 | 2/2 | − |
| 34 | 0/2 | 0/2 | 0/1 | 0/2 | 0/2 | − |
| 35 | 0/2 | 0/2 | 0/2 | 0/2 | 2/2 | − |
| 41 | 0/2 | NEc | 0/2 | 0/2 | 0/2 | − |
| 42 | 1/2 | 2/2 | 1/2 | 0/2 | 0/2 | − |
| 43 | 0/2 | 0/2 | 0/2 | 2/2 | — | − |
| 44 | 0/2 | 0/2 | 0/2 | 0/2 | 0/2 | − |
| 45 | 0/2 | 1/2 | 2/2 | 2/2 | — | − |
| 46 | 0/2 | 0/2 | 0/2 | 0/2 | 0/2 | − |
| 51 | NE | NE | 0/2 | 0/2 | 0/2 | − |
| 52 | NE | NE | 0/2 | 0/2 | — | − |
| 53 | NE | NE | 1/2 | 2/2 | — | − |
| 54 | NE | NE | 0/2 | 1/2 | 0/2 | − |
| 55 | NE | NE | — | — | — | − |
| 56 | NE | NE | — | — | — | − |
| 57 | NE | NE | NE | NE | 0/2 | − |
| No. of positive animals | 1/11 | 3/10 | 3/15 | 5/15 | 3/11 | 0/18 |
| % Positive animals | 9 | 30 | 20 | 33 | 27 | 0 |
−, anti-HTLV-1 antibody titer of <1:16.
—, no sample available.
NE, not examined.
TABLE 2.
Detection of HTLV-1 pX region in mouse organ DNA
| Mouse no. | Result at the age ofa:
|
|||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 7 wk
|
11 wk
|
15 wk
|
||||||||||||||||||
| TH | LU | LI | SP | K | LN | H | TH | LU | LI | SP | K | LN | H | TH | LU | LI | SP | K | LN | |
| 31 | − | − | − | − | − | + | ||||||||||||||
| 32 | + | − | + | + | − | − | − | |||||||||||||
| 33 | + | + | − | + | + | + | ||||||||||||||
| 34 | + | − | − | + | − | − | ||||||||||||||
| 35 | + | + | − | + | + | + | ||||||||||||||
| 41 | + | − | + | − | + | + | ||||||||||||||
| 42 | + | − | − | + | − | + | ||||||||||||||
| 43 | + | + | − | + | − | − | − | |||||||||||||
| 44 | − | + | − | + | + | + | ||||||||||||||
| 45 | − | + | − | + | − | − | − | |||||||||||||
| 46 | − | + | − | + | − | + | ||||||||||||||
| 51 | − | − | − | + | − | + | ||||||||||||||
| 52 | NE | − | − | + | − | − | NE | |||||||||||||
| 53 | − | NE | − | + | − | + | − | |||||||||||||
| 54 | NE | − | − | − | + | + | ||||||||||||||
| 55 | + | + | − | − | − | + | NE | |||||||||||||
| 56 | − | + | − | − | − | − | − | |||||||||||||
| 57 | + | − | − | + | − | + | ||||||||||||||
| Total positive | 1/2 | 2/2 | 0/2 | 0/2 | 0/2 | 1/2 | 0/1 | 2/4 | 2/4 | 1/5 | 5/5 | 0/5 | 1/5 | 0/4 | 6/10 | 4/11 | 1/11 | 8/11 | 5/11 | 10/11 |
+, one or two positive results in two independent PCRs; −, negative result in two independent PCRs. TH, thymus; LU, lung; LI, liver; SP, spleen; K, kidney; LN, lymph node; H, heart.
Amplification of the integration sites of HTLV-1 by linker-mediated PCR was carried out according to the procedure described by Wattel et al. (41) with slight modifications (reference 4 and unpublished data).
Statistical analysis.
The probability of frequencies for detecting pX and HERV-R sequences was calculated by Poisson distribution, and the parameters were estimated by the maximum likelihood method.
Antibody detection.
The antibodies against HTLV-1 proteins (mainly Gag; in part, Env) in the plasma were assayed with a particle agglutination kit (Serodia HTLV-1; Fujirebio, Tokyo, Japan) (10).
Histological analysis.
The organs of mice were fixed in 4% cacodylate-buffered paraformaldehyde (pH 7.2). Three-micrometer-thick paraffin sections were stained with hematoxylin and eosin and were examined microscopically.
Analysis of mRNA for pX.
RNA was extracted from three mice at the age of 16 weeks with the ISOGEN kit (Nippongene, Osaka, Japan). RNA from MT-2 cells was used as a positive control. The primers used were RPX3 and RPX4, described by Kinoshita et al. (16). One-half microgram of RNA was used for reverse transcription-PCR (RT-PCR) analysis. Reverse transcription was performed with Moloney murine leukemia virus reverse transcriptase. PCR was performed with Ampli Taq Gold (Perkin-Elmer), with 50 cycles of amplification reaction.
Cell sorting by flow cytometry.
For cell sorting, two C3H/HeJ mice injected with MT-2 cells (mouse 2 and mouse 3) were used. They were sacrificed at the age of 12 weeks. Splenocytes from MT-2 cell-injected mice were prepared and doubly stained with phycoerythrin-labeled rat anti-mouse B220 immunoglobulin G2A (IgG2a) and fluorescein isothiocyanate-labeled hamster anti-mouse CD3e IgG (both from PharMingen). In order to exclude dead cells, 1 μg of propidium iodide/ml was added and fractionated with a flow cytometer (FACS VANTAGE; Becton Dickinson). The cells with high expression of B220 or CD3e were sorted as B-cell and T-cell fractions, respectively, and aliquots of 105 cells per tube were collected. The cells were washed with phosphate-buffered saline, and their lysates were analyzed for HTLV-1 pX sequence by PCR and Southern hybridization.
Nucleotide sequence accession number.
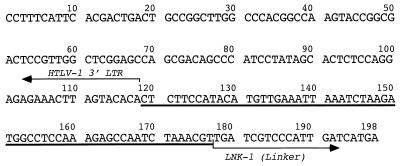
The sequence data shown in Fig. 3 for a part of the 3′ long terminal repeat (LTR) and its flanking mouse genomic sequence are available in the GenBank database under accession no. AF005373.
FIG. 3.
HTLV-1 3′ LTR and flanking cellular sequence identified from the spleen DNA of mouse 51 by linker-mediated PCR and DNA cloning. It is composed of 3′ LTR (nucleotides [nt] 1 to 118; 118 bp), flanking cellular sequence (nt 119 to 177; 58 bp), and linker sequence (nt 178 to 198; 21 bp).
RESULTS
Sensitivity of detection of HERV-R, pX, gag, and LTR sequences by PCR.
We checked the sensitivity of detection of pX and HERV-R sequences by PCR-Southern blot analysis using a serial dilution of MT-2 cell DNA with normal mouse spleen DNA, since we injected MT-2 cells into mice. The HERV-R sequence was detectable down to 1 MT-2 cell equivalent of DNA, which should contain four molecules of LTR of HERV-R in one diploid human cell (27) in the reaction mixture (Fig. 1a). We detected pX sequence from a minimum of 0.1 MT-2 cell equivalent of DNA (Fig. 1b), which should contain 0.6 molecule of pX sequence, since 6 molecules of pX sequence are reported to be present in each MT-2 cell (17). Our results confirmed that one molecule of pX sequence in the reaction tube is detectable in our PCR system. Since gag and 5′ LTR are mostly defective in MT-2 cells (17), we used ATL-1K cells, each of which contains one complete molecule of HTLV-1 provirus (9), as the standard for detection of gag and LTR sequence. As shown in Fig. 1c, gag was detectable to the level of 0.3 to 1 ATL-1K cell equivalent of DNA, indicating that one molecule of gag is detectable. LTR was detectable to 1 ATL-1K cell equivalent of DNA, indicating that one to two molecules of HTLV-1 LTR are detectable (Fig. 1d). Visualization of the amplified mouse c-myc sequence indicates that the PCR conditions were satisfactory (Fig. 1a).
Detection of HTLV-1 provirus sequence in MT-2 cell-injected mice.
We analyzed PBMC and several other organs in the mice to detect HTLV-1 provirus sequence. HTLV-1 pX sequence was detected both in PBMC (Table 1) and in the other organs, including the thymus, lung, liver, spleen, kidney, and lymph nodes, in C3H/HeJ mice (Table 2). gag and pX sequences in PBMC were detected in 9 and 30% of the mice, respectively, at the age of 7 weeks, and in 20 and 33% of the mice, respectively, at the age of 11 weeks. pX sequence was detected in PBMC in 27% of the mice at the age of 15 weeks (Table 1). Failure to detect the provirus in PBMC does not necessarily mean that the provirus is not present in other organs of the same animal (Table 2). The rate of detection of HTLV-1 pX sequence was higher in the spleen (13 of 18 mice [72%]), the lymph nodes (12 of 18 mice [67%]), and the thymus (9 of 18 mice [50%]). Every mouse contained the provirus in at least one organ. The detection rate was lowest in the liver DNA. No HTLV-1 pX sequence was detected in PBMC and the other organs of negative-control mice which had not received MT-2 cells.
Quantitation of HTLV-1 provirus and HERV-R molecules in spleen DNA.
For quantitation of the provirus molecules, we analyzed spleen DNA, since we frequently detected the provirus in the spleen (Table 2; Fig. 2a). We diluted the spleen DNA with normal mouse spleen DNA. pX sequence was detected in 1- to 30-fold dilutions of the positive DNA samples. We further analyzed the amount of the pX sequence in the spleen DNA of mouse 33, since this mouse had the largest amount of provirus in the spleen DNA. We detected the pX sequence in a 30-fold dilution of the spleen DNA of mouse 33 (Fig. 2b). The small, irregular signals in one of the two lanes at a 100-fold dilution are stains that appeared during Southern hybridization. gag sequence was detected in 1- to 10-fold dilutions, and LTR sequence was detected in 1- to 100-fold dilutions, of the spleen DNA of mouse 33 (Fig. 2c). No HERV-R sequence could be detected in the spleen DNA of mouse 33, even at a onefold dilution (Fig. 2d). From the comparison of the calculated and observed probabilities of frequencies for detecting pX and HERV-R sequences in spleen DNA from mouse 33 by Poisson distribution, the parameter was calculated by maximum likelihood. The probability of MT-2 contamination in spleen DNA from mouse 33 was calculated to be 0.0019, which is very low.
These results are consistent with the integration of HTLV-1 provirus in the spleen DNA of mouse 33.
Identification of the flanking sequence of the HTLV-1 integration site.
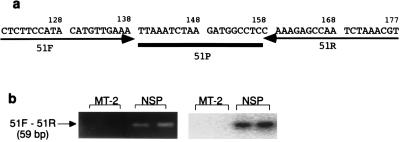
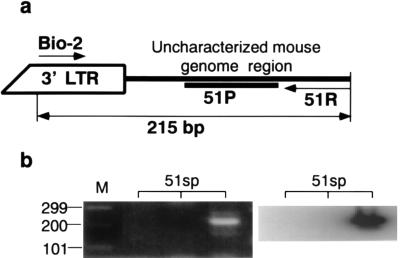
To get more direct evidence of the integration of HTLV-1 in the mouse genome and to show that it is not the contamination of MT-2 cells which might be in the mouse spleen, we isolated the integration site by linker-mediated PCR as described by Wattel et al. (41). We used the spleen DNA of mouse 51, since we had used up most of the spleen DNA of mouse 33. As shown in Fig. 3, we cloned and identified a flanking sequence of the HTLV-1 integration site. The flanking sequence has no significant homology to the available database sequences (see “Nucleotide sequence accession number” under Materials and Methods). To prove that the integration site is of mouse genome origin, we designed a primer set, 51F and 51R, as shown in Fig. 4a, to amplify the 59-bp flanking sequence from the template DNA, using normal mouse spleen DNA or MT-2 cell DNA as the template. The expected 59-bp DNA fragment was amplified from normal mouse spleen DNA but not from MT-2 cell DNA (Fig. 4b). In a parallel set of PCRs, pX fragment was successfully amplified from MT-2 cell DNA but not from mouse spleen DNA, indicating that the quality of MT-2 cell DNA is suitable for use as a template (data not shown). The results show that the flanking sequence is derived from the mouse genome. To further confirm that the cloned sequence was present as the flanking sequence to the HTLV-1 LTR, PCR was performed with a forward primer (Bio-2) complementary to a part of the 3′ LTR, with 51R as the reverse primer, and with spleen DNA from mouse 51 or MT-2 cell DNA as the template (Fig. 5a). If the integrated HTLV-1 provirus sequence as shown in Fig. 3 or 4 originated from the spleen DNA of mouse 51, the PCR would yield a 215-bp DNA fragment from the spleen DNA of mouse 51 but not from the MT-2 cell DNA. The results show that the expected band was observed from one of the three DNA samples from the spleen of mouse 51 (Fig. 5b) and that its DNA sequence was identical to the previously cloned 59-bp flanking sequence (Fig. 4) and to 156 bp of 3′ LTR.
FIG. 4.
Identification of 3′ LTR flanking sequence in spleen DNA from mouse 51 as being of mouse genome origin. (a) Oligonucleotides used for PCR as a primer set (51F and 51R) and probe (51P). (b) Results of PCR for detection of the predicted 59-bp fragment by using 0.16 μg of DNA from MT-2 cells or from normal mouse spleen (NSP) as the template. Ten microliters of each PCR product was electrophoresed in a 2% agarose gel, followed by ethidium bromide staining (left panel). Southern hybridization was carried out with probe 51P (right panel).
FIG. 5.
Direct PCR amplification of the integration site composed of HTLV-1 3′ LTR and flanking cellular sequence in spleen DNA from mouse 51 (51sp). (a) PCR using Bio-2 (a portion of 3′ LTR of HTLV-1 provirus; nt 8899 to 8922) and 51R (flanking sequence of spleen DNA from mouse 51 [Fig. 4, nt 177 to 158]) as primers should generate a 215-bp fragment (Fig. 3). (b) Results of three independent experiments using 1 μg of spleen DNA from mouse 51 in each PCR; the PCR products were separated in an agarose gel. Shown are results from ethidium bromide staining (left panel) and the corresponding Southern hybridization with probe 51P (right panel). M, DNA size markers indicating 299, 200, and 101 bp.
Detection of antibody response.
An antibody titer of 16-fold or more by the particle agglutination method was considered positive. Antibodies against HTLV-1 antigens were not detected in the plasma of any of the 18 C3H/HeJ mice at the ages of 7 to 15 weeks (Table 1).
Histological findings for the HTLV-1 carrier mice.
No abnormal histological finding was observed in the thymus, lung, heart, liver, spleen, kidney, stomach, small intestine, colon, peritoneal lymph nodes, ovary, or uterus for the MT-2 cell-injected mice (data not shown).
RT-PCR analysis of pX mRNA.
One-half microgram of mouse RNA containing RNA from 10 to 100 MT-2 cells was detected by RT-PCR, as revealed by ethidium bromide staining on an agarose gel. However, 1/2 μg of RNA from the spleen of each of three mice examined did not show the corresponding signal on agarose gel electrophoresis.
Analysis of the infected cell types in the spleen.
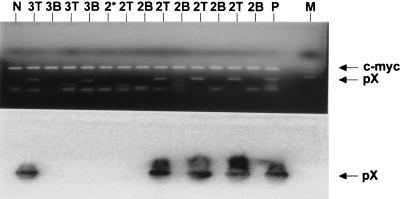
The splenocytes were fractionated into T-cell and B-cell fractions by a cell sorter. Three of four tubes of T-cell fraction from mouse 2 and one of two tubes of T-cell fraction from mouse 3 showed the presence of pX sequence. Four tubes of B-cell fraction and one tube of non-T, non-B fraction from mouse 2, and two tubes of B-cell fraction from mouse 3, did not show the presence of pX sequence (Fig. 6).
FIG. 6.
Detection of HTLV-1 pX sequence in T-cell fraction of splenocytes from MT-2 cell-injected C3H/HeJ mice. Four aliquots of T-cell and B-cell fractions and one aliquot of non-T, non-B fraction from mouse 2 (2T, 2B, and 2★, respectively), as well as four aliquots of T-cell and B-cell fractions from mouse 3 (3T and 3B, respectively) were analyzed. Each lane shows an independent aliquot of T-cell and B-cell fractions. N, negative control (normal rat PBMC lysate); P, positive control (the cell lysate equivalent to 100 ATL-1K cells was diluted with the lysate equivalent to 105 normal rat PBMC). A rat c-myc fragment of 345 bp could also be amplified with the primer set that was used for mouse c-myc. M, DNA size markers indicating 502, 398, 299, and 200 bp.
DISCUSSION
We detected HTLV-1 provirus sequence in DNA of PBMC and several organs in C3H/HeJ mice that received injections with HTLV-1-producing human T cells. The quantitation of HTLV-1 provirus by PCR and statistical analyses of the spleen DNA from the mice strongly suggested that the HTLV-1 provirus was derived from infected mouse cells, not from contamination from MT-2 cells remaining in the mouse spleen. Furthermore, we directly identified a flanking sequence of the proviral integration site from the spleen DNA of a mouse and demonstrated that the flanking sequence was the mouse genomic sequence. These results show that HTLV-1 transmission occurred from MT-2 cells to mouse cells in vivo, demonstrating that mice can be infected by HTLV-1.
The rate of detection of HTLV-1 provirus in the HTLV-1-injected C3H/HeJ mice was higher in the spleen and lymph nodes than in PBMC. These results suggest that the susceptible cells are enriched in the spleen or the lymph nodes or that the infected cells have tropism to the lymphatic tissues. The analysis of the cell types in the spleen in HTLV-1-infected mice showed that the T-cell fraction gave the positive PCR band of HTLV-1 pX sequence. This is consistent with the notion that HTLV-1 is a T-lymphotropic virus. We are now analyzing whether other types of cells are also infected in this mouse system, since we could find the provirus in the lung and other tissues but not in the spleen in some of the mice (Table 2), and previous reports also showed infection of cell types other than T cells in vitro (23).
The level of provirus as high as 1 molecule in 104 spleen cells, as shown in spleen DNA from mouse 33, indicates efficient replication and infection in mouse cells. However, there is no detectable antibody to HTLV-1 antigens in the mice. Three possibilities may be considered. First, the viral antigens might not be persistently expressed. RT-PCR showed that there was negligible mRNA for pX in the spleen cells of the infected mice. This result, together with flanking sequence data showing two proviruses with identical integration sites in DNA from the spleen of mouse 51, was consistent with the possibility that this virus replicates mainly by cell division rather than by viral gene expression leading to viral amplification like human immunodeficiency virus type 1 (HIV-1) (41). Second, MT-2 cells were injected within 24 h after birth. This might make the mouse tolerant to HTLV-1 antigens expressed in MT-2 cells at the time of injection. Third, the host strain, C3H/HeJ, is known to have a low level of response to antigens such as bacterial lipopolysaccharide (2). In another set of experiments, in which BALB/c mice were similarly injected with MT-2 cells, anti-HTLV-1 antibodies were detected in the plasma of four of eight BALB/c mice at the age of 12 weeks (data not shown). However, further study is required to clarify the precise mechanisms of different antibody responses in different mouse strains.
There have been no reports to date of HTLV-1 infection in mice. In in vitro experiments, mouse cell lines have been shown to be resistant to vesicular stomatitis virus pseudotypes bearing HTLV-1 envelope glycoproteins or to HTLV-1-induced syncytium formation (33). Thus, mouse cell lines were suggested to be devoid of HTLV-1 receptor. Our data show HTLV-1 transmission in mice. It is possible that cells in vivo express variable numbers of HTLV-1 receptor or coreceptors, although most mouse cell lines may express less. Our in vivo data are consistent with the recent report that some mouse cell lines are susceptible, from a study using a sophisticated pseudotype virus of HIV(HTLV-1) (35).
The low level of expression of HTLV-1 antigen or mRNA has been well observed in carriers and patients with ATL (16, 23); however, the mechanism by which viral expression in vivo is inhibited is still an enigma. It has been reported that PBMC derived from ATL patients expressed HTLV-1 antigen only when they were cultured in vitro (8, 9). Therefore, the demonstration of HTLV-1 antigen expression in an ex vivo culture is necessary to establish a useful model system for HTLV-1 pathogenesis.
ACKNOWLEDGMENTS
J. Fang and S. Kushida contributed equally to this work.
We thank H. Takahashi for his advice in statistical analysis and H. Nakauchi, Y. Matsuzaki, and Y. Takahama for suggestions. We also thank T. Mogi for histological preparations of mice organs, and Y. Morita and N. Arashi for technical assistance.
This work was supported in part by a Grant-in-Aid for the 2nd Term of the Comprehensive 10-Year Strategy for Cancer Control and Cancer Research from the Ministry of Health and Welfare of Japan and by a special grant for scientific research from the University of Tsukuba.
REFERENCES
- 1.Akagi T, Takeda I, Oka T, Ohtsuki Y, Yano S, Miyoshi I. Experimental infection of rabbits with human T-cell leukemia virus type I. Jpn J Cancer Res. 1985;76:86–94. [PubMed] [Google Scholar]
- 2.Coutinho A. Genetic control of B-cell responses. II. Identification of the spleen B-cell defect in C3H/HeJ mice. Scand J Immunol. 1976;5:129–140. doi: 10.1111/j.1365-3083.1976.tb02999.x. [DOI] [PubMed] [Google Scholar]
- 3.Dietrich W F, Miller J, Steen R, Merchant M A, Damron-Boles D, Husain Z, Dredge R, Daly M J, Ingalls K A, O’Connor T J, Evans C A, DeAngelis M M, Levinson D M, Kruglyak L, Goodman N, Copeland N G, Jenkins N A, Hawkins T L, Stein L, Page D C, Lander E S. A comprehensive genetic map of the mouse genome. Nature. 1996;380:149–152. doi: 10.1038/380149a0. [DOI] [PubMed] [Google Scholar]
- 4.Fang J, Kushida S, Feng R, Tanaka M, Kikukawa H, Kawamura T, Uchida K, Miwa M. Integration of HTLV-1 provirus into mouse transforming growth factor-α gene. Biochem Biophys Res Commun. 1997;233:792–795. doi: 10.1006/bbrc.1997.6557. [DOI] [PubMed] [Google Scholar]
- 5.Franchini G. Molecular mechanism of human T-cell leukemia/lymphotropic virus type I infection. Blood. 1995;86:3619–3639. [PubMed] [Google Scholar]
- 6.Gessain A, Vernant J C, Maurs L, Barin F, Gout O, Calendar A, de Thé G. Antibodies to human T-lymphotrophic virus type-I in patients with tropical spastic paraparesis. Lancet. 1985;ii:407–409. doi: 10.1016/s0140-6736(85)92734-5. [DOI] [PubMed] [Google Scholar]
- 7.Hino S, Yamaguchi K, Katamine S, Sugiyama H, Amagasaki T, Kinoshita K, Yoshida Y, Doi H, Tsuji Y, Miyamoto T. Mother-to-child transmission of human T-cell leukemia virus type-I. Jpn J Cancer Res. 1985;76:474–480. [PubMed] [Google Scholar]
- 8.Hinuma Y, Nagata K, Hanaoka M, Nakai M, Matsumoto M, Kinoshita T, Shirakawa S, Miyoshi I. Adult T-cell leukemia: antigen in an ATL cell line and detection of antibodies to the antigen in human sera. Proc Natl Acad Sci USA. 1981;78:6476–6480. doi: 10.1073/pnas.78.10.6476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hoshino H, Esumi H, Miwa M, Shimoyama M, Minato K, Tobinai K, Hirose M, Watanabe S, Inada N, Kinoshita K, Kamihira S, Ichimaru M, Sugimura T. Establishment and characterization of 10 cell lines derived from patients with adult T-cell leukemia. Proc Natl Acad Sci USA. 1983;80:6061–6065. doi: 10.1073/pnas.80.19.6061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ikeda M, Fujino R, Matsui T, Yoshida T, Komoda H, Imai J. A new agglutination test for serum antibodies to adult T-cell leukemia virus. Gann. 1984;75:845–848. [PubMed] [Google Scholar]
- 11.Ishiguro N, Abe M, Seto K, Sakurai H, Ikeda H, Wakisaka A, Togashi T, Tateno M, Yoshiki T. A rat model of human T lymphocyte virus type I (HTLV-I) infection. 1. Humoral antibody response, provirus integration, and HTLV-I-associated myelopathy/tropical spastic paraparesis-like myelopathy in seronegative HTLV-I carrier rats. J Exp Med. 1992;176:981–989. doi: 10.1084/jem.176.4.981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jordan E, Collins F S. A march of genetic maps. Nature. 1996;380:111–112. doi: 10.1038/380111a0. [DOI] [PubMed] [Google Scholar]
- 13.Kamihira S, Nakashima S, Oyakawa Y, Moriuti Y, Ichimaru M, Okuda H, Kanamura M, Oota T. Transmission of human T cell lymphotropic virus type I by blood transfusion before and after mass screening of sera from seropositive donors. Vox Sang. 1987;52:43–44. doi: 10.1111/j.1423-0410.1987.tb02987.x. [DOI] [PubMed] [Google Scholar]
- 14.Kawano F, Yamaguchi K, Nishimura H, Tsuda H, Takatsuki K. Variation in the clinical courses of adult T-cell leukemia. Cancer. 1985;55:851–856. doi: 10.1002/1097-0142(19850215)55:4<851::aid-cncr2820550424>3.0.co;2-j. [DOI] [PubMed] [Google Scholar]
- 15.Kinoshita K, Yamanouchi K, Ikeda S, Momita S, Amagasaki T, Soda H, Ichimaru M, Moriuchi R, Katamine S, Miyamoto T, Hino S. Oral infection of a common marmoset with human T-cell leukemia virus type-I (HTLV-I) by inoculating fresh human milk of HTLV-I carrier mothers. Jpn J Cancer Res. 1985;76:1147–1153. [PubMed] [Google Scholar]
- 16.Kinoshita T, Shimoyama M, Tobinai K, Ito M, Ito S, Ikeda S, Tajima K, Shimotohno K, Sugimura T. Detection of mRNA for the tax1/rex1 gene of human T-cell leukemia virus type I in fresh peripheral blood mononuclear cells of adult T-cell leukemia patients and viral carriers by using the polymerase chain reaction. Proc Natl Acad Sci USA. 1989;86:5620–5624. doi: 10.1073/pnas.86.14.5620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kobayashi N, Konishi H, Sabe H, Shigesada K, Noma T, Honjo T, Hatanaka M. Genomic structure of HTLV (human T-cell leukemia virus): detection of defective genome and its amplification in MT-2 cells. EMBO J. 1984;3:1339–1343. doi: 10.1002/j.1460-2075.1984.tb01974.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kotani S, Yoshimoto S, Yamato K, Fujishita M, Yamashita M, Ohtsuki Y, Taguchi H, Miyoshi I. Serial transmission of human T-cell leukemia virus type I by blood transfusion in rabbits and its prevention by use of X-irradiated stored blood. Int J Cancer. 1986;37:843–847. doi: 10.1002/ijc.2910370608. [DOI] [PubMed] [Google Scholar]
- 19.Kushida S, Maeda N, Fang J, Uchida K, Miwa M. Abstracts of the 18th Symposium, Leukemia and Lymphoma/Pathogenesis and Treatment/Molecular Aspects. International Association for Comparative Research on Leukemia and Related Diseases, Kyoto, Japan. 1995. Establishment of HTLV-1 carrier mice by injection with HTLV-1-producing T cells, abstr; p. 86. [Google Scholar]
- 20.Kushida S, Mizusawa H, Matsumura M, Tanaka H, Ami Y, Hori M, Yagami K-I, Kameyama T, Tanaka Y, Yoshida A, Nyunoya H, Shimotohno K, Iwasaki Y, Uchida K, Miwa M. High incidence of HAM/TSP-like symptoms in WKA rats after administration of human T-cell leukemia virus type 1-producing cells. J Virol. 1994;68:7221–7226. doi: 10.1128/jvi.68.11.7221-7226.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kushida S, Matsumura M, Tanaka H, Ami Y, Hori M, Kobayashi M, Uchida K, Yagami K, Kameyama T, Yoshizawa T, Mizusawa H, Iwasaki Y, Miwa M. HTLV-1-associated myelopathy/tropical spastic paraparesis-like rats by intravenous injection of HTLV-1-producing rabbit or human T-cell line into adult WKA rats. Jpn J Cancer Res. 1993;84:831–833. doi: 10.1111/j.1349-7006.1993.tb02052.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Matsumura M, Kushida S, Ami Y, Uchida K, Kameyama T, Terano A, Shiraki H, Sato H, Miwa M. A simple and reliable method for the detection and quantitation of the human T-cell leukemia virus type-I provirus in peripheral blood mononuclear cells of seropositive blood donors. Jpn J Clin Oncol. 1992;22:335–341. [PubMed] [Google Scholar]
- 23.Miwa M. Mechanism of oncogenesis of adult T-cell leukemia/lymphoma. Hematol Rev. 1990;3:247–255. [Google Scholar]
- 24.Miyoshi I, Kubonishi I, Yoshimoto S, Akagi T, Ohtsuki Y, Shiraishi Y, Nagata K, Hinuma Y. Type C virus particles in a cord T-cell line derived by co-cultivating normal human cord leukocytes and human leukaemic T cells. Nature. 1981;294:770–771. doi: 10.1038/294770a0. [DOI] [PubMed] [Google Scholar]
- 25.Mochizuki M, Watanabe T, Yamaguchi K, Takatsuki K, Yoshimura K, Shirao M, Nakashima S, Mori S, Araki S, Miyata N. HTLV-I uveitis: a distinct clinical entity caused by HTLV-I. Jpn J Cancer Res. 1992;83:236–239. doi: 10.1111/j.1349-7006.1992.tb00092.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nishioka K, Maruyama I, Sato K, Kitajima I, Nakajima Y, Osame M. Chronic inflammatory arthropathy associated with HTLV-I. Lancet. 1989;i:441. doi: 10.1016/s0140-6736(89)90038-x. [DOI] [PubMed] [Google Scholar]
- 27.O’Connell C D, Cohen M. The long terminal repeat sequences of a novel human endogenous retrovirus. Science. 1984;226:1204–1206. doi: 10.1126/science.6505687. [DOI] [PubMed] [Google Scholar]
- 28.Osame M, Matsumoto M, Usuku K, Izumo S, Ijichi N, Amitani H, Tara M, Igata A. Chronic progressive myelopathy associated with elevated antibodies to HTLV-1 and adult T-cell leukemia cells. Ann Neurol. 1987;21:117–122. doi: 10.1002/ana.410210203. [DOI] [PubMed] [Google Scholar]
- 29.Poiesz B J, Ruscetti F W, Gazdar A F, Bunn P A, Minna J D, Gallo R C. Detection and isolation of type C retrovirus particles from fresh and cultured lymphocytes of a patient with cutaneous T-cell lymphoma. Proc Natl Acad Sci USA. 1980;77:7415–7419. doi: 10.1073/pnas.77.12.7415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Robert-Guroff M, Nakao Y, Notake K, Ito Y, Sliski A, Gallo R C. Natural antibodies to human retrovirus HTLV-I in a cluster of Japanese patients with adult T-cell leukemia. Science. 1982;215:975–978. doi: 10.1126/science.6760397. [DOI] [PubMed] [Google Scholar]
- 31.Sato K, Maruyama I, Maruyama Y, Kitajima I, Nakajima Y, Higaki M, Yamamoto K, Miyasaka N, Osame M, Nishioka K. Arthritis in patients infected with human T lymphotropic virus type I. Arthritis Rheum. 1991;34:714–721. doi: 10.1002/art.1780340612. [DOI] [PubMed] [Google Scholar]
- 32.Seto A, Isono T, Ogawa K. Infection of inbred rabbits with cell-free HTLV-I. Leuk Res. 1991;15:105–110. doi: 10.1016/0145-2126(91)90090-g. [DOI] [PubMed] [Google Scholar]
- 33.Sommerfelt M A, Williams B P, Clapham P R, Solomon E, Goodfellow P N, Weiss R A. Human T cell leukemia viruses use a receptor determined by human chromosome 17. Science. 1988;242:1557–1559. doi: 10.1126/science.3201246. [DOI] [PubMed] [Google Scholar]
- 34.Suga T, Kameyama T, Kinoshita T, Shimotohno K, Matsumura M, Tanaka H, Kushida S, Ami Y, Uchida M, Uchida K, Miwa M. Infection of rats with HTLV-1: a small animal model for HTLV-1 carriers. Int J Cancer. 1991;49:764–769. doi: 10.1002/ijc.2910490522. [DOI] [PubMed] [Google Scholar]
- 35.Sutton R E, Littman D R. Broad host range of human T-cell leukemia virus type 1 demonstrated with an improved pseudotyping system. J Virol. 1996;70:7322–7326. doi: 10.1128/jvi.70.10.7322-7326.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tajima K, et al. The T and B Cell Malignancy Study Group. The 4th nationwide study of adult T cell leukemia/lymphoma (ATL) in Japan: estimates of risk of ATL and its geographical and clinical features. Int J Cancer. 1990;45:237–243. doi: 10.1002/ijc.2910450206. [DOI] [PubMed] [Google Scholar]
- 37.Tajima K, Tominaga S, Suchi T, Kawagoe T, Komoda H, Hinuma Y, Oda T, Fujita K. Epidemiological analysis of the distribution of antibody to adult T-cell leukemia virus-associated antigen: possible horizontal transmission of adult T-cell leukemia virus. Gann. 1982;73:893–901. [PubMed] [Google Scholar]
- 38.Takatsuki, K., K. Yamaguchi, F. Kawano, T. Hattori, H. Nishimura, H. Tsuda, I. Sanada, K. Nakada, and Y. Itai. 1985. Clinical diversity in adult T-cell leukemia-lymphoma. Cancer Res. 45(Suppl.):4644S–4645S. [PubMed]
- 39.Uchiyama T, Yodoi J, Sagawa K, Takatsuki K, Uchino H. Adult T-cell leukemia: clinical and hematologic features of 16 cases. Blood. 1977;50:481–492. [PubMed] [Google Scholar]
- 40.Uemura Y, Kotani S, Yoshimoto S, Fujishita M, Yamashita M, Ohtsuki Y, Taguchi H, Miyoshi I. Mother-to-offspring transmission of human T cell leukemia virus type I in rabbits. Blood. 1987;69:1255–1258. [PubMed] [Google Scholar]
- 41.Wattel E, Vartanian J-P, Pannetier C, Wain-Hobson S. Clonal expansion of human T-cell leukemia virus type I-infected cells in asymptomatic and symptomatic carriers without malignancy. J Virol. 1995;69:2863–2868. doi: 10.1128/jvi.69.5.2863-2868.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yamamoto N, Hayami M, Komuro A, Schneider J, Hunsmann G, Okada M, Hinuma Y. Experimental infection of cynomolgus monkeys with a human retrovirus, adult T-cell leukemia virus. Med Microbiol Immunol. 1984;172:57–64. doi: 10.1007/BF02123570. [DOI] [PubMed] [Google Scholar]
- 43.Yamanouchi K, Kinoshita K, Moriuchi R, Katamine S, Amagasaki T, Ikeda S, Ichimaru M, Miyamoto T, Hino S. Oral transmission of human T-cell leukemia virus type-I into a common marmoset (Callithrix jaccus) as an experimental model for milk-borne transmission. Jpn J Cancer Res. 1985;76:481–487. [PubMed] [Google Scholar]
- 44.Yoshida M, Seiki M, Yamaguchi K, Takatsuki K. Monoclonal integration of human T-cell leukemia provirus in all primary tumors of adult T-cell leukemia suggests causative role of human T-cell leukemia virus in the disease. Proc Natl Acad Sci USA. 1984;81:2534–2537. doi: 10.1073/pnas.81.8.2534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yoshioka R, Yamaguchi K, Yoshinaga T, Takatsuki K. Pulmonary complications in patients with adult T-cell leukemia. Cancer. 1985;55:2491–2494. doi: 10.1002/1097-0142(19850515)55:10<2491::aid-cncr2820551030>3.0.co;2-i. [DOI] [PubMed] [Google Scholar]