Summary
We present a method, named Mx-TOP, for profiling of three epigenetic regulatory layers—chromatin accessibility, general DNA modification, and DNA hydroxymethylation—from a single library. The approach is based on chemo-enzymatic covalent tagging of unmodified CG sites and hydroxymethylated cytosine (5hmC) along with GC sites in chromatin, which are then mapped using tag-selective base-resolution TOP-seq sequencing. Our in-depth validation of the approach revealed its sensitivity and informativity in evaluating chromatin accessibility and DNA modification interactions that drive transcriptional regulation. We employed the technology in a study of chromatin and DNA demethylation dynamics during in vitro neuronal differentiation. The study highlighted the involvement of gene body 5hmC in modulating an extensive decoupling between promoter accessibility and transcription. The importance of 5hmC in chromatin remodeling was further demonstrated by the observed resistance of the developmentally acquired open loci to the global 5hmC erasure in neuronal progenitors.
Keywords: covalent DNA labeling, methyltransferases, synthetic AdoMet cofactors, 5hmC, uCG, TOP-seq, chromatin accessibility, DNA modification, DNA hydroxymethylation
Graphical abstract

Highlights
-
•
Mx-TOP yields one-pot chromatin accessibility, DNA unmethylome, and 5hmCG maps
-
•
Covalent tagging enables tracking of highly and partially accessible chromatin
-
•
Developmental gene expression patterns are best defined by gene body 5hmC changes
-
•
Nascent open chromatin regions show resistance to 5hmC removal at neural progenitors
Skardžiūtė et al. present a chemo-enzymatic tagging-based approach for simultaneous profiling of DNA unmethylome, hydroxymethylome, and chromatin accessibility, named Mx-TOP. The technique proved instrumental for tracking epigenetic transitions during in vitro neural differentiation which demonstrated a prominent 5hmC role in defining gene expression programs.
Introduction
DNA methylation and chromatin structure are important epigenetic factors that control genes by modulating interactions between regulatory proteins and DNA. Analysis of chromatin accessibility in tissues using DNase-seq1 and ATAC-seq2 has identified a variety of cis-regulatory elements that play fundamental roles in development and disease. Methylation of promoters at CG sites is a well-known mechanism for regulation of gene activity. Growing evidence has indicated that DNA methylation outside promoters even better defines gene activation and tissue identity.3,4 Analysis of independent datasets has shown an unanticipated overlap between high levels of DNA methylation and chromatin accessibility at cis-regulatory elements, suggesting functional association of these two epigenetic factors.5,6
The discovery of the TET-dependent DNA demethylation pathway, which removes 5-methylcytosine (5mC) in a stepwise manner,7,8 adds another layer on regulatory mechanisms of a cell. In contrast to 5mC, its oxidized derivative 5hmC acts as a gene activating mark and is a major modifier of the tissue identity.9,10 Therefore, an independent 5hmC analysis in relation to chromatin dynamics will be of critical importance for understanding the complex interactions of different epigenetic layers. Methods that concurrently report on multiple chromatin and DNA features in a single protocol could advance the studies of the complex epigenomic landscape and its functional outcomes.
The whole-genome bisulfite sequencing (WGBS) suffers from the inability to discriminate between 5mC and 5hmC and requires deep sequencing.11,12 We and others have shown that covalent DNA derivatization of specific targets enables robust and sensitive detection of subtle cytosine modification changes in various biological and pathological samples.13,14,15,16,17,18,19,20 Here, we present an adaptation of chemo-enzymatic covalent tagging of cytosine targets for simultaneous profiling of chromatin accessibility, unmodified, and hydroxymethylated DNA by the Mx-TOP approach (multi-omic tethered oligonucleotide-primed sequencing). We demonstrated the utility of distinct combinations of the three covalent tagging modalities using the methyltransferases eM.SssI and M.CviPI and T4 bacteriophage β-glucosyltransferase (BGT) for chromatin analysis in mouse embryonic stem cells (ESC). Using Mx-TOP, we for the first time explored chromatin accessibility dynamics in association with 5mC demethylation during neuronal differentiation of mouse ESC. By combining the chromatin analysis with transcriptome sequencing, we investigated the impact of DNA demethylation on gene regulation during cell fate transitions.
Design
Using chemo-enzymatic labeling approaches, an azide or alkyne group is transferred from a synthetic cofactor analog onto cytosine in unmethylated CGs (uCGs), 5hmC, and GC sites in native nuclei, and then, genomic coordinates of the tagged sites are acquired by single-nucleotide resolution TOP-seq sequencing17,18 (Figure 1A). The uCG site capture using eM.SssI can provide information on both the accessibility and modification status of chromatin, while derivatization of 5hmC sites with an azide-modified glucose13 using BGT informs on DNA demethylation. However, the lack of uCG and 5hmC signals does not give an unequivocal information on the chromatin status. In order to add an independent chromatin accessibility measure that is unaffected by the native DNA modification, we went on to explore M.CviPI-directed covalent labeling of genomic GC sites. In vitro methylation by M.CviPI21 is utilized in nucleosome occupancy and methylome sequencing (NOMe-seq) that analyzes chromatin structure and DNA methylation by WGBS;22 however, the ability of M.CviPI to accept extended side chains from synthetic cofactors has not yet been known. In NOMe-seq, it is intrinsically impossible to distinguish the endogenous CG methylation from in vitro methylated GCs at overlapping GCG sites. As a consequence, a high fraction of CGs has to be discarded from analysis.23 The inherent feature of TOP-seq to define precise cytosine positions could discriminate between the GC and CG counterparts of GCG/CGC targets. Consequently, a greater proportion of the analytical CG sites would expand the genomic coverage of the approach.
Figure 1.
Mx-TOP technology
(A) Outline of the trimodal Mx-TOP procedure. Step 1. Extraction of nuclei. Step 2. One-pot treatment of the nuclei with a cocktail of three enzymes and two synthetic cofactors for selective covalent tagging of GC, uCG, and 5hmC sites. Step 3. DNA fragmentation and addition of terminal adapters (shown in orange) through sonication/ligation or DNA tagmentation with Tn5 transposase. Step 4. Covalent tethering of tag-specific DNA oligonucleotides (ODN) - alkyne-AT-ODN (shown in blue) with azide-labeled 5hmC sites and azide-TT-ODN (shown in red) with alkylated GC and uCG sites. Optional enrichment of the ODN-conjugated DNA by the streptavidin-biotin affinity capture. Step 5. Tethered ODN-primed DNA strand extension. Step 6. Adapter and ODN-directed PCR amplification of the primed DNA products. Step 7. Sequencing and identification of open regions.
(B) Three labeling pipelines of Mx-TOP: scheme a, bimodal tagging of GC and uCG sites using M.CviPI/eM.SssI in the presence of Ado-6-azide; scheme b, bimodal tagging of GC and 5hmC sites with M.CviPI/Ado-6-azide and BGT/UDP-glc-azide, respectively; scheme c, trimodal tagging of GC, uCG, and 5hmC sites using M.CviPI/eM.SssI/BGT in the presence of Ado-6-alkyne and UDP-glc-azide. Below, chemical structures of the covalent tethers linking the tagged cytosine (upper) or 5hmC (lower) and the respective ODN.
(C) Read percentages of the azide-TT-ODN and alkyne-AT-ODN at the stand-alone CG and GC targets, or at the first and second cytosines of the fully pre-hydroxymethylated GhmCGC sites in 3x-TOP analysis of model lambda DNA.
(D) Dependence of the observed 5hmCG/uCG ratio from different 5hmC amounts in a model DNA fragment which was initially pre-hydroxymethylated at two CG sites to 50% (CG#1) or 100% (CG#2) and mixed with unmodified DNA to different ratios. Combined 3x-TOP data of five GC sites are also shown. Linear regression was used to evaluate the relationship between the theoretical and observed 5hmCG/uCG ratio. (p value and R2 are shown). See also Figures S1 and S2.
Altogether, application of the three labeling specificities in different combinations permits to establish three different analytical pipelines of the Mx-TOP technology (Supplemental Item 1): (1) For bimodal (2x-TOP) profiling of CG modification and chromatin accessibility, uCG and GC sites were labeled with an azide group using eM.SssI and M.CviPI, respectively, and a synthetic cofactor analog Ado-6-azide14 (Figures 1A and 1B, scheme a); (2) For 2x-TOP analysis of 5hmC and chromatin accessibility, we combined the BGT and M.CviPI reactivities with their synthetic cofactors UDP-glc-azide13 and Ado-6-azide, respectively (Figures 1A and 1B, scheme b); (3) Trimodal (3x-TOP) mapping of DNA unmethylation, 5hmC levels, and chromatin accessibility was achieved by combining all three enzymes in a single reaction (Figures 1A and 1B, scheme c).
Results
Different configurations of Mx-TOP captures GC in CG sites in open chromatin loci
We optimized conditions for both bimodal and trimodal approaches using model DNA systems (Figures S1 and S2). In our preferred 3x-TOP approach, the unmethylated or hydroxymethylated statuses of CG sites are discriminated by selective covalent tethering of the DNA oligonucleotides (ODN) that carry distinct tag-specific functional groups (Step 4) and also a single A to T replacement in their nucleotide sequences (Figure 1B) to ensure sequence-based assignment to the corresponding CG modification type after sequencing. uCG and GC sites were labeled with a terminal alkyne group using Ado-6-alkyne cofactor24 and subsequently conjugated to the azide-modified ODN (azide-TT-ODN, Figure S2A), while the azide-tagged 5hmC sites were conjugated to the alkyne-modified ODN (alkyne-AT-ODN) (Figure 1B). To minimize the cross-reactivity among the deposited azide and alkyne tags potentially leading to DNA intra or inter-strand crosslinks, the tethering of the ODNs was performed in a stepwise manner using a high excess of alkyne- and then azide-modified ODNs (Figure S2B). Notably, the differences in the chemical linker structures and the minimally distinct sequences of the ODNs used for 5hmCG and uCG tagging (Figure 1B) did not affect the efficiency of the priming reaction (Figure S2C) and enabled discrimination between cytosine modification in complex targets, such as GCG/CGC (Figure 1C). Even though the bulkier 5hmC-ODN conjugates may interfere with other passing-through priming events in heavily labeled DNA templates (Figure S2D), such situations would be rare due to scarcity of 5hmC and would not cause a significant obstacle for data acquisition. We also tested how quantitatively the designed uCG and 5hmC sequence coding system determines the 5hmCG level at a given CG site; the calculated 5hmCG/uCG ratios at two differently pre-hydroxymethylated CG sites in the mixtures of model DNA fragments showed a linear dependence on the 5hmC amount and the expected 5hmC fractions on each strand of the CG sites (Figure 1D). As expected, no changes at alkyne-labeled GCs sites were detected, which confirmed a high specificity of the 3x-TOP tagging configuration in discriminating the three types of genomic targets.
Pilot Mx-TOP analysis of mouse ESC
The performance of Mx-TOP was assessed on mouse ESC. The 2x-TOP and 3x-TOP genomic libraries in two technical and two biological replicates were sequenced using a shallow (∼27 M single-end raw reads for the 2x-TOP and 3x-TOP libraries; Table S1) and medium sequencing (∼128 M reads for the 3x-TOP libraries). Deeper 3x-TOP sequencing increased the number of the captured GCs (idGC) from ∼7.8 to 28.6 M and 5hmCGs from ∼1.3 to ∼2.85 M, while the amounts of uCGs saturated even with shallow sequencing (on average 3.2 M in different libraries), suggesting that higher sequencing depths enable the accessibility measurement of more methylated genomic loci. A good discrimination between the CG and GC labeling in CGC trinucleotides (GCG/CGC sites) was observed, and the 5hmC-specific AT-ODN sequence was confined exclusively to CG sites (Figure S3A). No specific labeling was obtained in other sequence contexts outside CG or GC sites (Figure S3B). Although the read start positions in control samples prepared without the enzymatic labeling step showed weak random amplification (Table S1; Figure S3B), we removed all overlapping GC and CG sites to avoid any false positive target identification.
A genome browser view of the 3x-TOP signal shows a dense coverage of the genome by the captured targets (Figure 2A). To define larger scale contiguous regions of chromatin accessibility and their DNA modification, we employed a cumulative uCG and idGC signal (named uT, or “unmodified targets”), which accounts for the majority of the 3x-TOP signal. In parallel, as a “pure” measure of chromatin openness that is independent from uCG data, we constructed similar regions from the idGC signal alone. Due to their sparsity (an average distance between two closest sites is 348 bp in the medium sequencing 3x-TOP), 5hmCGs were later overlaid on the constructed region sets. For bioinformatic transitions, we transformed the raw site coverage into the weighted idGC- or uT-density estimates (this is not necessary for deeper sequencing samples), which alleviated the variation of the target coverage due to different sequencing depths of the 2x-TOP and 3x-TOP libraries. Then, we applied a seed-and-extend bioinformatic approach (Figure 1A and STAR Methods) to discern open regions. Lastly, we joined closely located (less than 75 bp apart) regions and included 5hmCGs to obtain the final dataset of the peak open chromatin regions (OCRs). We generated approx. 0.75 and 0.66 M OCRs in the shallow sequencing samples, and 1 and 0.9 M OCRs in the medium sequencing samples from the 3x-TOP uT and idGC signals, respectively. The median region length was ∼160 and ∼180 bp for uT and idGC data, respectively (medium sequencing 3x-TOP; Figure S3C), which covered up to 250 Mbp of the genome. Using the two sets of regions, we comprehensively compared the shallow and medium 2x-TOP and 3x-TOP data (Figure S3D). A high overlap (0.96 overlapping fraction) observed between the 2x-TOP libraries demonstrated that the idGC signal is a major contributor to the accessibility measure. There was also a considerable overlap between the shallow and medium 3x-TOP OCR sets, indicating that even low coverage was sufficient to capture the majority of OCRs in ESC. As the idGC and uT signals may influence OCR identification, which is indicated by lower overlap between the datasets, we used both strategies to validate chromatin accessibility in the majority of the downstream 3x-TOP analyses.
Figure 2.
Mx-TOP captures differentially modified open chromatin regions
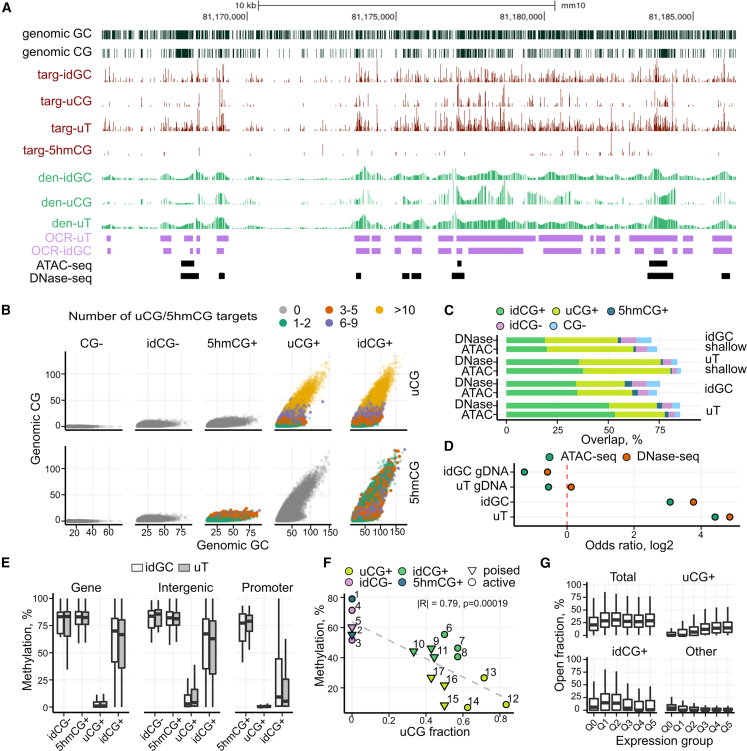
(A) Genome browser view of Mx-TOP data for a region of chr 10 in ESC: separate tracks are shown for the identified targets (idGC, uT, uCG, and 5hmCG), weighted density estimates and OCRs; the tracks of open regions identified by ATAC-seq and DNase-seq are presented. Genomic GC and CG targets are shown in black. Targ, targets; den, density.
(B) Scatter representation of the uCG+, idCG+, 5hmCG+, CG-, and idCG- region types, distributed according to the density of genomic GC and CG targets. Color code represents the amount of identified uCG or 5hmCG targets. Data are shown for a subset of OCRs with 10–49 idGCs (for all groups see Figure S3F).
(C) Overlap of different OCRs with ATAC/DNase-seq regions.
(D) Odds ratio (log2) from Fisher’s exact test for enrichment of Mx-TOP identified sites within ATAC/DNase-seq regions.
(E) Methylation levels (from WGBS) of OCRs calculated from the uT and idGC signals (regions with >10 genomic CGs) in gene bodies, promoters (2 kb upstream TSS) and intergenic areas. The idCG- group was not identified in promoter regions at the used CG threshold.
(F) uCG-fraction compared to bisulfite methylation values in 17 selected OCRs overlapping active and poised enhancers. Linear model was fitted using uCG-fraction and average methylation per region (p value and correlation coefficient are shown).
(G) Relationship of different OCRs that overlap promoters with gene expression. “Other” combines 5hmCG+, CG-, and idCG- regions. See also Figures S3 and S4, Tables S1 and S2.
To define DNA modification of 3x-TOP OCRs, we subdivided them into five categories based on the presence and type of the identified CGs: (1) uCG+; (2) idCG+, containing both uCG and 5hmCG sites; (3) 5hmCG+; (4) CG-, the regions without genomic CGs; and (5) idCG-, the regions without identified uCG/5hmCGs (Figure 2B). The groups differed in the region size and a fraction of the captured targets: the idCG+ and uCG+ groups varied mostly in length (mean length 400 bp) and were relatively uCG-rich, whereas the GC-, idCG-, and 5hmCG+ loci represented CG-poor and potentially methylated genomic areas. The constant idGC fraction value (approx. 0.6, i.e., six-tenths of GC sites are identified) obtained for all region types confirmed the utility of the quite uniformly distributed GCs (on average, one GC at each 8 bp in autosomes) for the region estimation. The distribution of the groups across various genomic elements (Figure S3E) indicated the dominant presence of uCG+ and idCG+ in promoters and enhancers, whereas the other three groups tended to localize within genes and intergenic areas. As both the peak OCRs and the total 3x-TOP data should represent the accessibility, we independently analyzed their enrichment and found them both enriched at active promoters, enhancers, CG islands, and target sites of pluripotency transcription factors NANOG, OCT4, and SOX2 for uCG+ and idCG+, and depleted within silenced chromatin and repetitive elements (Figure S4A).
To validate our accessibility measurements, we compared OCRs with the ATAC-seq and DNase-seq datasets of the same cell type.25,26 Many more regions were identified by 3x-TOP (∼1 M vs. 50 k of ATAC-seq) (see also Figure 2A for a browser view comparison) which include 71%–86% of the ATAC-seq/DNAse-seq regions, specified by our data mostly as the idCG+ and uCG+ loci (Figure 2C). The signal of the 3x-TOP libraries prepared with extracted genomic DNA was underrepresented within the ATAC-seq/DNAse-seq loci, confirming the specificity of the approach for open chromatin (Figure 2D).
DNA methylation levels of OCRs were assessed by comparing their uCG-density or a fraction of uCGs (uCG-fraction) with WGBS methylation data.27 The analysis highlighted a good inverse correlation between the two data types (Pearson r = |0.48|-|0.6|) (Figures S4B and S4C). Notably, uCG+ loci showed the lowest methylation levels, especially at promoters, whereas 5hmCG+ and idCG- loci were highly methylated in most of the genome (Figures 2E and S4D). Although the idCG- group most likely represents intensively methylated regions, it also may result from the missed CGs (Figure S4D) due to some variability in the labeling reaction (Figures S1A and S1B). However, these regions are the shortest and most CG-poor, and represent a minority of OCRs in largely unmethylated promoters and enhancers (Figure S3E). To test the concordance of Mx-TOP and bisulfite methylation values, we analyzed 17 regions in active and poised enhancers by bisulfite sequencing. A good correlation with BS-seq data confirmed the discerned methylation levels of the OCR groups, high methylation of idCG- loci, and the methylation differences of active and poised enhancers (Figure 2F). Furthermore, a good agreement was observed between 3x-TOP and 5hmCG levels in OCRs derived from available TAB-seq data28 (Figure S4E).
Distribution of the protein-coding genes according to their expression levels and the type of OCRs at promoters (2 kb upstream TSS) revealed the positive association of the uCG+ regions with transcription, while idCG+ showed positive correlation for lower expression genes (Figure 2G).
Overall, our in-depth validation showed that Mx-TOP is capable of detecting highly unmodified and accessible open chromatin areas and also those in which DNA modification and chromatin accessibility may not coincide. The analysis confirmed the utility of Mx-TOP for simultaneous analysis of the three important epigenetic factors – DNA methylation, hydroxymethylation, and chromatin accessibility.
Chromatin and DNA modification dynamics along the neuronal differentiation trajectory
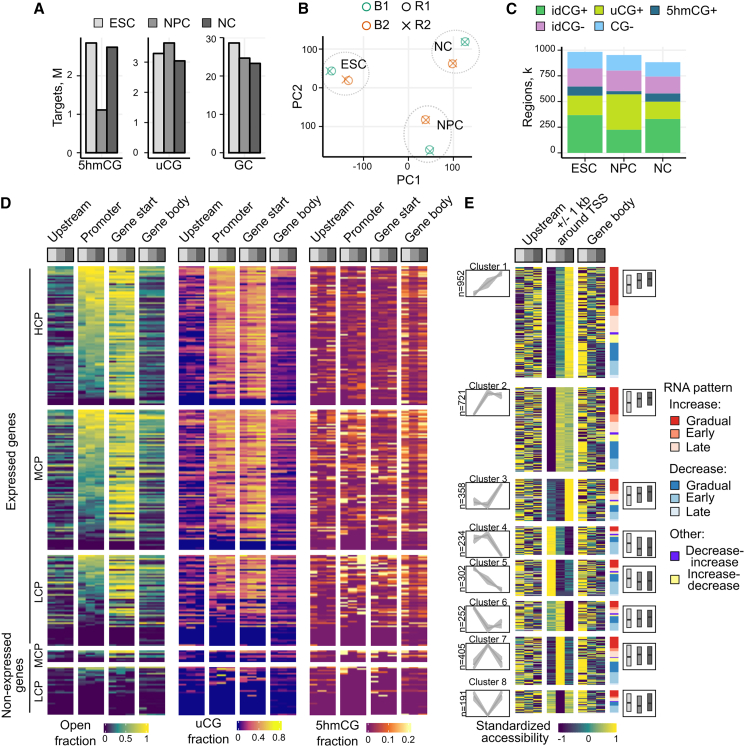
To measure the interplay of the epigenetic factors during development, we performed differentiation of mouse ESC into a population of radial-glial neuronal progenitor cells (day 8 after LIF removal and stimulation with retinoic acid; NPC, S1) and later into terminally differentiated glutamatergic pyramidal neurons (day 13; NC, S2)29 that yielded ∼81% neurons, as characterized by the formation of synaptic connections and expression of neuronal marker proteins (Figures S5A–S5C). In order to relate chromatin dynamics with transcriptional changes, 3x-TOP and RNA sequencing in two technical and two biological replicates was performed for each of the time points (Table S1, medium and shallow Mx-TOP).
The numbers of idGCs and uCGs were similar for all stages (∼3 M uCGs and ∼25 M GCs), whereas the 5hmCG amount dropped during transition from ESC to NPC from ∼2.8 to ∼1.1 M and again increased to ∼2.7 M in NC (Figure 3A). Notably, the uCG and 5hmCG data of 3x-TOP discriminated well among the three cell populations in PCA (Figure 3B). Using our peak OCR calling strategy, we quantified approx. 0.9 M regions in each cell population with slightly decreasing abundance toward NC (Figure 3C), the tendency that was also observed by other in vitro differentiation analyses.30 The idCG+ loci were most abundant in ESC and NC, while uCG+ dominated in NPC most likely due to the 5hmC drop in these cells.
Figure 3.
Mx-TOP captures DNA modification and accessibility changes during mouse ESC differentiation
(A) Numbers of captured GC, uCG, and 5hmCG targets in ESC, NPC, and NC (3x-TOP medium sequencing).
(B) PCA of the cell populations (uCG+5hmCG target data). R1/R2, technical replicates, B1/B2, biological replicates.
(C) Distribution of uCG+, idCG+, 5hmCG+, CG-, and idCG- regions in each of the cell types.
(D) Clusterization of genes according to promoter accessibility (1-kb region upstream TSS) defined as a fraction of bp covered by OCRs (uT signal). Accessibility, uCG, and 5hmCG data are also shown for 1-kb region downstream TSS (GeneStart), upstream distal areas (1–4 kb upstream TSS) and gene bodies. Promoters were divided into HCP, MCP, and LCP.
(E) Heatmap of chromatin accessibility Z scores at promoters, upstream, and gene body areas for DEGs grouped by C-means clustering of promoters (±1 kb around TSS) (shown on the left, gene numbers are presented). Genes within each cluster are ranked by c-means RNR patterns (shown on the right). The boxplots indicate mean expression (FPKM log2) changes across the cell stages. Only genes with cluster membership probability ≥0.25 are plotted. See also Figures S5 and S6, Tables S1 and S3.
To gain an overview of the promoter accessibility and DNA modification changes around genes, we clustered the regions overlapping upstream distal areas (1–4 kb upstream TSS), the 1-kb region upstream (promoter) and downstream TSS (GeneStart), and gene bodies (Figure 3D). As genes with key cellular functions are associated with different promoter CG densities,31 we additionally segregated high-, mid-, or low-CG density promoter groups (HCP, MCP, and LCP, respectively). The highest accessibility was evident 1 kb downstream TSS of the HCP and MCP groups of expressed genes, whereas non-expressed genes, containing mostly LCPs, displayed very limited accessibility, as expected. Notably, all inspected genomic intervals showed the loss of 5hmC in NPC, except for the promoter regions of the HCP and MCP groups, which were generally poor in 5hmCGs.
Next, for investigation of the interdependence of chromatin accessibility and transcription, we applied C-means unsupervised clustering of promoter OCRs (±1 kb around TSS) and also calculated differentially expressed genes (DEGs) among the stages. The promoter analysis identified eight main modes of the accessibility behavior (Table S3): gradual opening (cluster 1), early (cluster 2), or late opening (cluster 3); gradual closing (cluster 4); immediate (cluster 5), or late closing (cluster 6); opening at NPC with subsequent closing (cluster 7) and vice versa – loss of the accessibility in NPC and its re-gain in NC (cluster 8) (Figure 3E). The calculation of DEGs (the 2-fold expression difference in transcript abundance for pairwise comparisons, adjusted p value ≤0.05) revealed 8023 genes in total, whose expression patterns followed similar modes as those of promoter accessibility clusters (Figure S5D). The inspection of several key developmental genes, such as the pluripotency markers SOX2, OCT4, and NANOG, the key 5mC eraser TET1,8 and the neuronal transcription factors PAX6 and SOX11,29,32 revealed the expected concordant behavior between promoter accessibility and transcription (Figure S5E), verifying the sensitivity of 3x-TOP for a gene-level analysis.
While chromatin dynamics at upstream areas (1–4 kb upstream TSS) generally followed the promoter patterns, gene bodies showed decreasing accessibility toward NC. Notably, even though the promoter accessibility and the averaged expression changes in all C-means promoter clusters showed concerted changes (Figure 3E, right boxplots), a variety of transcriptional patterns was observed in each accessibility cluster. The general uncorrelation between promoter accessibility and transcription in neural development was also observed by ATAC-seq analysis.33
Hydroxymethylation of intragenic open chromatin loci defines transcriptional changes in differentiation
We analyzed in more detail the observed transcriptional variability in the gradual opening and gradual closing promoter accessibility clusters (clusters 1 and 4). First, we constructed the distribution profiles of the five OCR types across promoters, gene bodies, and the 3-kb surrounding areas (Figure 4A). Surprisingly, idCG+ loci dominated in all areas, except for promoters in ESC, which were enriched in uCG+ loci. In cluster 1, the idCG+ amounts at promoters increased toward NC, pointing to a growing fraction of promoters experiencing demethylation. The loss of 5hmC in NPC resulted in the drop of idCG+ and the enrichment of uCG+ regions across the gene bodies. The highly diverse chromatin dynamics at promoters mainly translated into two dominant modes of transcriptional behavior – increasing or decreasing (Figure 3E). Therefore, in search of a regulatory signal that drives the chromatin remodeling and gene expression synchronization, we distributed cluster 1 and 4 into the four possible interaction modes and evaluated the chromatin state, uCG and 5hmCG levels (Figure 4B). The analysis revealed the main trend of responsibilities in both clusters when comparing ESC and NC – the two resulting groups of induced genes despite their reciprocal promoter accessibility changes maintained high 5hmCG levels at gene bodies in NC (Figure 4C). Additionally, 5hmC levels increased at their promoter OCRs in NC together with increasing promoter openness toward NC. The 5hmC profiles of the whole acquired 5hmC signal (the total 5hmC signal) confirmed the observed gene body changes, but, in contrast to OCR data, indicated a strong 5hmC drop at promoters, which is consistent with the general absence of 5hmC at these elements.18,34 The decreasing expression was accompanied by the loss of 5hmCGs at gene bodies in NC, while the promoter openness was kept constant or slightly decreased. Importantly, neither accessibility nor uCG levels at OCRs were obviously related to transcriptional changes. The relatively stable promoter openness and unmethylation level of the downregulated genes most likely indicate a time lag necessary to translate chromatin remodeling signals on transcription, for example, to provide access to transcriptional repressors. We suggest that partial demethylation of gene body open loci precedes promoter changes, and the produced 5hmC, which behaves either as a regulator or an indicator of transcriptional activity, together with intragenic enhancers drive transcriptional responses (enhancer histone marks H3K4me1 and H3K27ac are strongly enriched at gene bodies in ESC; OR = 2.7, p value <0.05).
Figure 4.
Relationship between gene body hydroxymethylation and expression of DEGs
(A) Profiles and heatmaps of uCG+, idCG+, 5hmC+, CG-, and idCG- regions across genes and the 3-kb surrounding areas for promoter accessibility clusters 1 and 4.
(B) Profiles of chromatin accessibility (uT signal), uCGs, and 5hmCGs from OCRs and total 3x-TOP signal for the subsets of genes from promoter clusters 1 and 4 that show the concordant and discordant changes in accessibility and transcription.
(C) Boxplot graphs of 5hmCG changes at gene bodies of the gene subgroups from (B) using the signal from OCRs and total 3x-TOP data. Gene numbers are indicated for each group.
(D) GO functional annotation analysis of the concordant and discordant gene subsets of clusters 1 and 4. The group with closing promoters and increasing RNR changes did not show significant enrichments.
Both groups of the upregulated genes demonstrated links to neuronal development and synaptic processes (Figure 4D), whereas the genes with decreasing expression were related to metabolism, mRNA processing, and proliferation, which usually get downregulated during neuronal development.33
Differentiation-affected genes escape complete demethylation in NPC and maintain developmental transcriptional behavior irrespective of their low 5hmC content
We next explored the influence of 5hmC loss in NPC on expression and first calculated the genomic bins (100 bp) with decreased 5hmC (fold change >2; adjusted p value <0.1), which showed a tendency to overlap with genes (OR = 4.7, p value <0.05 for genes; non-significant value for intergenic regions). The analysis disclosed an increasing 5hmC loss toward the 3′ end at both exons and introns, with the least affected first exon (Figure 5A).
Figure 5.
Dynamics of 5hmC during neuronal differentiation
(A) Percentages of exons and introns that lost 5hmCGs in NPC (absolute fold change >1.5) shown by their gene position.
(B) Fold change of the 5hmCG/uCG ratio between ESC and NPC at promoters (1 kb upstream TSS), the 1st exons and introns, and the rest of the metagene.
(C) Patterns of C-means transcriptional clusters of DEGs and their gene body 5hmCG changes. Lines represent average FPKM or 5hmCG fraction values.
(D) Dependence of gene expression (different FPKM groups) on 5hmCG and uCG fractions of gene body OCRs or total 3x-TOP signal for ESC and NPC. The Wilcoxon test, ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001.
(E) Profiles of accessibility and 5hmCGs from all OCRs or total 3x-TOP signal around the exon-intron boundary at expressed and non-expressed genes.
As the uCG amounts suggested a slightly more demethylated status of the NPC genome (Figures 3A and 3D), we next sought to determine the extent of complete gene body demethylation of DEGs, i.e., production of uCGs following 5hmCG loss. The single-C resolution of Mx-TOP enabled the evaluation of the 5hmCG/uCG ratio across gene elements, which, surprisingly, revealed a strong resistance of the whole gene body to full demethylation—only a minuscule fraction of DEGs showed uCG increase (Figure 5B). In contrast, the 1st exons, a fraction of the 1st introns and promoters showed a tendency to gain uCGs. Importantly, the developmental transcriptional patterns of DEGs were not compromised by the decreased genic 5hmC levels in NPC (Figure 5C).
To elucidate how OCR modification influences gene expression in NPC, we calculated 5hmCG or uCG fractions from the peak OCRs or total 3x-TOP signal over genes ranked by expression levels (Figure 5D). The analysis confirmed the positive association of 5hmCG and uCG levels in ESC, but indicated a negative influence of both marks on highly expressed genes in NPC. This again demonstrated that less methylated gene body OCR contributes to gene upregulation (54%–58% of idCG+ regions localize to gene bodies in all cell stages) (see also Figure 2G), whereas the most active genes in NPC have to be kept methylated, consistent with negative effect of low gene body methylation on gene expression.35 Our data confirm the known bidirectional influence of the gene body 5hmC on gene expression: it promotes gene expression in ESC and many other cell types, but can negatively influence transcription in neuronal progenitors.36,37,38
The tight relationship between open chromatin at gene bodies and expression induction is further illustrated by the enrichment of OCRs close to the exon-intron junctions, preferentially at the exonic side (Figure 5E). A wider open area around splice junctions in transcriptionally active genes should contribute to better access for RNA polymerase II and the whole expression/splicing machinery. Importantly, 5hmCGs tend to concentrate close to the transition site from exon to intron, which suggests potential functional implications of 5hmCG accumulation at splice junctions previously observed by us and others.18,34,39 Notably, the total 3x-TOP signal can provide enhanced detection of the 5hmCG accumulation at the exon-intron boundaries.
Despite the global 5hmC loss in NPC, we detected ∼80 genes whose gene body 5hmC levels constantly increased toward NC. Of these, genes with induced transcription were related to neuronal development, and included the transcription factors MEIS1 and NEUROG2 and the known splicing regulators in neuronal cells NOVA1, ROBO2, and DCC.40 A similar positive correlation between expression and intragenic 5hmC in neuronal function-related genes has been also reported in the developing mouse and human brain systems.41,42
Gene body and distal de novo OCRs maintain synchronized chromatin and 5hmC dynamics and escape 5hmC loss in neuronal progenitors
As DEG promoters showed some accessibility in ESC (Figure 4B), we next searched for the so-called de novo loci, whose appearance after the exit of pluripotency would not be affected by the dynamics around pre-existing OCRs (>1-kb distance to the closest region was selected as a threshold). The identified ∼100,000 and ∼30,000 de novo OCRs in NPC and NC, respectively, were shorter (median length 106 bp) than other OCRs and were enriched in the GC-, idCG-, and uCG+ types (Figure 6A). Approximately a half of de novo OCRs were located in genes, while the rest positioned far from TSS (Figure 6B). The regions tended to distribute in the heterochromatin areas of ESC (Figure S6A), which indicates developmental dynamics at previously silenced regions.
Figure 6.
Distinctive dynamics and regulation of de novo OCRs
(A) Amount of de novo OCRs in NPC and NC distributed in the five region types.
(B) Percentages of de novo OCRs localized at different distances to TSS.
(C) Venn diagram demonstrating the distribution of de novo OCRs at gene bodies and at different distances to TSS for S1, S1S2, and S2 de novo gene groups.
(D) Changes of the accessibility, uCG, and 5hmCG signals in OCRs localized in distal intergenic and gene areas for the upregulated and downregulated gene subsets that contain de novo OCRs or without them in NPC and NC in comparison to ESC.
(E) Heatmap representation of the 5hmCG and uCG standardized fractions of distal (10–50 kb upstream TSS) and genic de novo OCRs and their close surroundings (±3 kb) or other OCRs that overlap the gene body. Data are shown for different RNA clusters. See also Figure S6 and Table S4.
We distributed genes which associated with genic and distal (up to 50 kb upstream TSS) de novo loci into three main groups: genes with NPC-specific loci which were not maintained in NC (S1; 1518 genes), genes which gained loci in NPC and additionally in NC (S1S2; 2697 genes), and genes with NC-specific de novo OCRs (S2; 379 genes). Interestingly, only the S1S2 group was assigned with high significance to neuron development terms, whereas S1 and S2 indicated functional links to cell cycle regulation, RNA processing, and covalent chromatin modification (Figure S6B). This suggests that differentiation first affects metabolic and cell cycle processes, while major neuronal changes, though initiated already in NPC, occur during transition from NPC to NC.
Of the three groups, S1S2 demonstrated the highest fraction of genes with both gene-associated and the most distally located regions (Figure 6C). Therefore, we further analyzed the accessibility, 5hmCG, and uCG dynamics for S1S2 genes with genic and distal de novo OCRs in relation to their expression changes. The upregulated and downregulated genes showed a coordinated increase of the 5hmCG signal at gene body and associated distal OCRs in NC as compared to ESC (Figure 6D). In contrast, no such relationships were noticed for the randomly subsampled upregulated genes without de novo OCRs. Promoter evaluation indicated increasing accessibility and uCG levels for both upregulated and downregulated subgroups, again pointing to limited informativity of a “just promoter” analysis.
Considering the observed informativity of the 5hmC signal, we sought to explore whether de novo OCRs experienced 5hmC loss in NPC. Strikingly, for the induced gene clusters, de novo and surrounding OCRs (±3 kb) at both distally located (10–50 kb upstream) and genic positions appeared to be resistant to the 5hmC erasure, in contrast to the rest of their gene body OCRs and all OCRs overlapping downregulated genes (Figure 6E). The uCG amounts started to increase in all OCRs in genic and a subset of distal regions in NPC, suggesting an early involvement of DNA demethylation, but hardly discriminated between the gene groups with de novo and other OCRs. We independently validated the de novo 5hmC changes by the public data of hMeDIP,38 which confirmed the observed relationships (Figure S6C).
The distal intergenic OCRs of S1S2 tended to overlap with genes of known long intergenic non-coding RNAs (OR 0.9–1.1; p value <0.05) and antisense RNAs (OR 0.7–1.9; p value <0.05). Moreover, the genic and most distally located de novo OCRs were enriched in the binding sites of the SOX group transcription factors, mainly SOX3 and SOX10 (see Table S4). The proper SOX3 and SOX10 expression is known to be important for the maintenance of neural lineage potential.43,44 Other enriched TFs, SOX4, SOX6, and SOX21, play various roles in neural differentiation and their functional impairment results in neurodevelopmental disorders and cancer.32,45
Discussion
Studies of complex developmental transitions require approaches that report on multiple epigenetic regulatory layers from a single sample. The common methods for chromatin accessibility or/and DNA methylation analysis ATAC-seq and NOMe-seq limit investigation of chromatin dynamics in relation to DNA demethylation, which is known to accompany developmental reprogramming.46 Our covalent labeling-based molecular tool, Mx-TOP, offers a multimodal base-resolution analysis of three different regulatory layers – DNA methylation through analysis of unmethylome, DNA hydroxymethylation, and chromatin accessibility. The combination of the three DNA-modifying enzymes in 3x-TOP allows identification of larger regulatory regions, and spots of chromatin “breathing”. Due to the high target specificity and single-C resolution, Mx-TOP allows analysis of complex genomic sites, such as GCG, and thus, expands the number of analytic CG sites, as compared to NOMe-seq. We also demonstrated that the total Mx-TOP signal outside the major peak open regions arises from genuine biological data and, therefore, can provide important complementary information.
During the development of Mx-TOP, an ATAC-Me approach has been proposed that combines ATAC-seq with a subsequent WGBS for DNA methylation analysis.47 In contrast to the latter method, which suffers from the known drawbacks of bisulfite treatment and the preferential capture of highly accessible loci, as it is common to ATAC-seq,48 Mx-TOP monitors chromatin remodeling and DNA demethylation dynamics in regions of different chromatin compaction.
Mx-TOP analysis of the mouse neuronal differentiation revealed an extensive loss of 5hmC in NPC, which is then partially re-established in mature neurons. Importantly, the observed 5hmCG loss across gene bodies of differentiation-affected genes was not followed by a related uCG increase, pointing to the existence of a compensatory biological mechanism, which regulates gene activity most likely through restoring 5mC. However, the observed stable uCG-fractions at gene bodies do not reflect the dynamics of other cytosine modifications that may influence transcriptional activity.49
Our investigation revealed a significant discordance between promoter accessibility and expression of developmentally affected genes. This discordance was modulated by hydroxymethylation at gene body OCRs which better defines transcriptional profiles than promoter openness. The enrichment of OCRs and 5hmC at the exon-intron boundaries further points to the association of gene bodies with regulatory processes. Both expressed and non-expressed genes showed increased 5hmC at splice junctions, where it could drive binding of transcription factors or might predetermine alternative splicing, as suggested.50
During neuronal differentiation, abundant de novo chromatin changes in the uCG/5hmCG-poor genic and intergenic areas were induced already at NPC. Strikingly, the areas proximal to de novo OCRs seemed to escape 5hmC loss in NPC, pointing to a distinctive chromatin regulation at these loci. Furthermore, neuronal function-related genes showed a concordance in chromatin activation and 5hmC dynamics at gene and distal areas, while the accessibility and uCG changes were only minimally related. All this supports the value of combined investigation of various epigenetic factors to characterize the mechanisms that link gene transcription and chromosomal architectures to tissue-specific functions.
Limitations of the study
Although Mx-TOP can measure subtle modification differences by inferring relative modification levels of individual CG sites, the accuracy of such semi-quantitative analysis depends on the sequencing depth. In this study, we used shallow and medium sequencing; therefore, OCRs and their modification profiles were mainly derived based on the identification status of the target sites. As discussed previously20 and shown by WGBS analyses, the uCG-fraction quantitatively depends on the region methylation levels. For shallow Mx-TOP libraries, calculation of the regional density profiles of the CG and GC targets before OCR identification should be used to alleviate inter-sample coverage variability. Due to an inherent variability of the enzymatic reactions and highly uneven distribution of CG and GC dinucleotides, some uCG sites might be missed in CG-poor areas, and thus interpreted as methylated. Therefore, despite the ability of a cost-efficient shallow Mx-TOP to capture the majority of uCGs and most open loci, deeper sequencing above 200 M reads is recommended for a more comprehensive analysis.
As all read count-based methods, Mx-TOP is sensitive to copy number variation and is unable to detect epialleles without prior knowledge of their presence.
Significance
The Mx-TOP approach offers new possibilities for simultaneous analysis of chromatin accessibility and cytosine modification and can track DNA demethylation in the accessible sites of various chromatin compaction. The flexibility to select desired tagging combinations of uCG, 5hmC, and GC sites and tag-directed amplification of genomic areas allows tailoring Mx-TOP for studying a variety of epigenetic systems. Application of Mx-TOP along the neural differentiation trajectory uncovered an important role of 5hmC at gene body open loci in modulating expression of differentiation-affected genes. Moreover, Mx-TOP analysis of chromatin dynamics across the entire genome revealed resistance of developmentally induced accessible regions to global 5hmC erasure in neuronal progenitors, suggesting a distinctive chromatin regulation at these loci. Altogether, our validation of Mx-TOP in various biological contexts highlighted its potential to advance the studies of epigenetic transitions and cell fate specifications to a new level.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Biological samples | ||
| Lambda phage DNA (dam–, dcm–) | Thermo Fisher Scientific | Cat#SD0021 |
| Chemicals, peptides, and recombinant proteins | ||
| Proteinase K, recombinant, PCR grade | Thermo Fisher Scientific | Cat#EO0491 |
| CpG Methyltransferase (M.SssI) | Thermo Fisher Scientific | Cat#EM0821 |
| T4 beta-glycosyltransferase (BGT) | Thermo Fisher Scientific | Cat#EO0831 |
| GpC Methyltransferase (M.CviPI) | New England Biolabs | Cat#20227L |
| eM.SssI | Kriukienė et al.14 | N/A |
| DBCO-S-S-PEG3-biotin | BroadPharm | Cat#BP-22453 |
| Dynabeads MyOne Streptavidin C1 | Thermo Fisher Scientific | Cat#65002 |
| Maxima SYBR Green/ROX qPCR Master Mix (2X) | Thermo Fisher Scientific | Cat#K0221 |
| Pfu DNA polymerase (recombinant) | Thermo Fisher Scientific | Cat#EP0502 |
| Phusion U HS polymerase | Thermo Fisher Scientific | Cat#F555S |
| Klenow Fragment, exo- | Thermo Fisher Scientific | Cat#EP0421 |
| T4 DNA Ligase | Thermo Fisher Scientific | Cat#EL0011 |
| dNTP Mix | Thermo Fisher Scientific | Cat#R0241 |
| Nuclease P1 | Sigma-Aldrich | Cat#N8630 |
| Ado-6-azide cofactor | Kriukienė et al.14 Lukinavičius et al.24 | N/A |
| Ado-6-alkyne cofactor | Lukinavičius et al.24 | N/A |
| UDP-glc-azide | Jena Bioscience | Cat#CLK-076 |
| CuBr, 99.999% | Sigma-Aldrich | Cat#254185 |
| THPA | Sigma-Aldrich | Cat#762342 |
| SYBR Green I nucleic acid gel stain | Sigma-Aldrich | Cat#S9430 |
| Azidobutyric acid NHS ester | Lumiprobe | Cat#63720 |
| FastAP Thermosensitive Alkaline Phosphatase | Thermo Fisher Scientific | Cat#EF0654 |
| Platinum SuperFI PCR Master Mix | Thermo Fisher Scientific | Cat#12358010 |
| Critical commercial assays | ||
| DNA Clean & Concentrator kit | Zymo Research | Cat#D4014, Cat#D4034 |
| Oligo Clean and Concentrator kit | Zymo Research | Cat#D4060 |
| Genomic DNA Clean and Concentrator kit | Zymo Research | Cat#D4010 |
| GeneJET Gel Extraction kit | Thermo Fisher Scientific | Cat#K0692 |
| EZ DNA Methylation-Gold Kit | Zymo Research | Cat#D5005 |
| GeneJET NGS Cleanup Kit | Thermo Fisher Scientific | Cat#K0852 |
| MagJET NGS Cleanup and Size Selection Kit | Thermo Fisher Scientific | Cat#K2821 |
| Colibri Library Quantification kit | Thermo Fisher Scientific | Cat#A38524500 |
| Dynabeads mRNA Purification Kit | Thermo Fisher Scientific | Cat#61006 |
| RiboCop rRNA Depletion Kit | Lexogen | Cat#K03724 |
| Ion Total RNA-seq Kit v2 | Thermo Fisher Scientific | Cat#4475936 |
| Fast DNA End Repair Kit | Thermo Fisher Scientific | Cat#K0771 |
| GeneJET PCR Purification Kit | Thermo Fisher Scientific | Cat#K0701 |
| Agilent High Sensitivity DNA Kit | Agilent | Cat#5067-4626 |
| Qubit 1X dsDNA High Sensitivity (HS) Kit | Thermo Fisher Scientific | Cat#Q33230 |
| Deposited data | ||
| Mouse genome sequence build GRCm38 | Ensembl database | http://www.ensembl.org/index.html |
| DNase-seq data | Yue et al.26,26 | GEO: GSE37074 |
| ATAC-seq data | Kim et al.25 | GEO: GSE113912 |
| Histone ChIP-seq data | ENCODE Project Consortium 2012 | https://www.encodeproject.org/ |
| WGBS data | Lu et al.27 | GEO: GSE56986 |
| hMeDIP-seq data | Tan et al.38 | GEO: GSE40810 |
| TAB-seq data | Yu et al.28 | GEO: GSM882244 |
| Mx-TOP and RNA-seq data | This work | GEO: GSE231929 |
| Experimental models: Cell lines | ||
| E14TG2a, mouse embryonic stem cells | American Type Culture Collection (ATCC) | Cat# CRL-1821; RRID: CVCL_9108 |
| Oligonucleotides | ||
| Mx-TOP adapters for Ion Torrent sequencing: A1, 5’-P-GATTGGAAGAGTGGTTCAGCAGGA ATGCTGAG and A2, 5’-ACACTCTTTCCCTAC ATGACACTCTTCCAATCT |
Metabion | N/A |
| 2x-TOP TO (tethered oligonucleotide) for Ion Torrent sequencing: alkyne-TT-biotin-ODN, 5′-TXTTTTGTGTGGTTTGGAGACTGACTACC AGATGTAACA-Biotin (X=C8-alkyne-dU) |
Base-click | N/A |
| 3x-TOP TO for Ion Torrent sequencing: amine-TT-biotin-ODN, 5‘-TXTTTTGTGTGGTT TGGA GACTGACTACCAGATGTAACA-Biotin (X = C2-amine-dT). Preparation of click-reactive azide-TT-biotin-ODN is described in methods Preparation of the click-reactive oligonucleotides section. |
Base-click | N/A |
| 3x-TOP TO for Ion Torrent sequencing: alkyne-AT-biotin-ODN, 5‘-TXTATTGTG TGGTTTGGAGACTGACTACCAGATGT AACA-Biotin (X=C8-alkyne-dU) |
Base-click | N/A |
| TO complementary priming ODN for Ion Torrent sequencing: IT-EP, 5’-TGTTACA TCTGGTAGTCAGTCTCCAAACCACACAA |
Exiqon | N/A |
| Mx-TOP adapters for Illumina sequencing: Ill-P5-Tn-long, 5’-ACACTCTTTCCCTACAC GACGCTCTTCCGATCTTANNNNNNNAGG AGATGTGTATAAGAGACAG and Tn-COM21, 5’-P-CTGTCTCTTATACACATCTCCdd |
Metabion | N/A |
| 3x-TOP TO for Illumina sequencing: amine-TT-ODN, 5‘-TXTTTTGTGTGGTTTG AGATCGGAAGAGCACACGTCTGAACTC CAGTCAC-3' (X = C2-amine-dT). Preparation of click-reactive azide-TT-ODN is described in methods Preparation of the click-reactive oligonucleotides section. |
Base-click | N/A |
| 3x-TOP TO for Illumina sequencing: alkyne-AT-ODN, 5'-TXTATTGTGTGGTTTGAGATCGGAAGAGCACAC GTCTGAACTCCAGTCAC (X = C8-alkyne-dU) |
Base-click | N/A |
| TO complementary priming ODN for Illumina sequencing: S-IL-P7, 5’-GTGACTGGAGTTCA GACGTGTGCTCTTCCGATCTCAAACCACA∗C∗A∗A (∗phosphorothioate (PTO) linkages). |
Exiqon | N/A |
| Primers for region-specific BS-seq bisulfite sequencing, see Table S2 | Metabion | N/A |
| Illumina adapters from TruSeq Nano DNA LT Kit | Illumina | Cat#15041757 |
| NEBNext Multiplex Oligos for Illumina (Index Primers Set 1) | New England Biolabs | Cat#7335L |
| Software and algorithms | ||
| Bismark aligner and methylation caller | Krueger and Andrews63 | www.bioinformatics.bbsrc.ac.uk/projects/bismark |
| BWA | Li and Durbin52 | http://bio-bwa.sourceforge.net |
| Cutadapt | Martin51 | http://cutadapt.readthedocs.io/en/stable/index.html |
| hisat2 | Kim et al.64 | http://daehwankimlab.github.io/hisat2/ |
| stringtie | Pertea et al.58 | https://ccb.jhu.edu/software/stringtie/ |
| R | R Project | https://www.r-project.org |
| ComplexHeatmap | Gu et al.68 | https://jokergoo.github.io/ComplexHeatmap-reference/book/ |
| EnrichedHeatmap | Gu et al.69 | https://github.com/jokergoo/EnrichedHeatmap |
| limma | Ritchie et al.73 | https://bioconductor.org/packages/release/bioc/html/limma.html |
| GenomicRanges | Lawrence et al.54 | https://bioconductor.org/packages/release/bioc/html/GenomicRanges.html |
| DESEQ2 | Love et al.60 | https://bioconductor.org/packages/release/bioc/html/DESeq2.html |
| Mx-TOP data analysis scripts | This work | https://doi.org/10.5281/zenodo.10183722 |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Edita Kriukienė (edita.kriukiene@bti.vu.lt).
Materials availability
This study did not generate new unique reagents.
Data and code availability
-
•
All raw and processed Mx-TOP and RNA-seq data have been submitted to the NCBI Gene Expression Omnibus (GEO; https://www.ncbi.nlm.nih.gov/geo/) under the accession number GSE231929.
-
•
Mx-TOP analysis code has been deposited at Zenodo and is publicly available as of the date of publication. The DOI is listed in the key resources table.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
Experimental model and study participant details
ESCs cultivation
Wt mouse ESC E14TG2a cell line was purchased from ATCC (ATCC Cat# CRL-1821; RRID: CVCL_9108). ESCs were cultured on feeder-free 0.15% gelatin-coated plates in DMEM containing 15% embryonic stem-cell FBS, 50 μg/ml penicillin–streptomycin mix, 2 mM L-glutamine, 1× non-essential amino acids, 1 mM 2-mercaptoethanol (all from Gibco) and 1000 U/mL ESGRO mLIF (Milipore). Cells were maintained at 37°C in a humidified atmosphere containing 5% CO2.
Method details
Preparation of model DNA fragments
For testing the covalent labeling conditions, two model DNA fragments were produced by PCR amplification from mouse gDNA: 171 bp-hmC (171-hmC-dir 5’-CTGGTTXGTCTGAGGAATGAAGGTC and 171-hmc-rev 5’-CTTTGTCACTTCCTGXGAGAGCCC X- 5hmC) and 188 bp-hmC (188-dir 5’-GTGTTGGGGTGACTATTATG and 188-hmc-rev 5‘-GCATCCTGGAGATTGTGGGCAACATCXGG X= 5hmC). Three model DNA fragments were amplified from human gDNA: 155 bp (1H, 155-dir 5’-TGTGTTACTGTGTGGAAAAGACC and 155-rev 5’-CCACTCCTTATAGTTTGGCTG); 201 bp (201-dir 5’- CCTCATGATTTCTGAGTGAAGG and 201-rev 5’-TAGGTTTGGGAGACTTGAGAATG) and 97 bp-hmC (97-dir 5’-GTTCTGGTGAGTAGATGGTTAAAC or 97-hmC-dir 5’-GTTCTGGTGAGTAGATGGTTAAACATTGTAACTAGGAAGTAAXG X- 5hmC and 97-rev 5’- CTTTCAAAGATTCTCATTGTCCACAC). DNA fragments were gel-purified using GeneJET Gel Extraction kit (Thermo Fisher Scientific, TS).
5hmC was introduced at the GCGC site of the 155 bp model 1H DNA fragment with a 5-fold molar excess of M.HhaI and formaldehyde, as described in Ličytė et al.49 The efficiency of hydroxymethylation was approximately 90%, as tested by the R.Hin6I restriction analysis (see below).
Assessment of double and triple labeling in model DNA systems
Double M.CviPI/eM.SssI and M.CviPI/BGT labeling was performed by incubating 50 ng of model DNA fragments with 4 U M.CviPI (New England Biolabs, NEB) and 2-fold molar excess of eM.SssI (TS) over CG targets (for M.CviPI/eM.SssI labeling) or 2.5 U BGT (TS) (for M.CviPI/BGT labeling), and 600 μM Ado-6-azide and 150 μM UDP-glc-azide (Jena Bioscience) cofactors, respectively, in a buffer (10 mM Tris-HCl (pH 7.4), 50 mM NaCl, 0.1 mg/ml BSA, 10 mM DTT). For the triple M.CviPI/eM.SssI/BGT labeling reactions, 50 ng of model DNA fragments were incubated with 4 U M.CviPI, 2-fold molar excess of eM.SssI over CG targets, 2.5 U BGT, 600 μM Ado-6-alkyne and 150 μM UDP-glc-azide in buffer (10 mM Tris-HCl (pH 8.5), 50 mM NaCl, 0.1 mg/ml BSA, 10 mM DTT). Reactions were incubated at 37°C for 1 h, followed by enzyme inactivation at 65°C for 20 min. Then, DNA was treated with 0.2 mg/ml Proteinase K (ProtK, TS) and 0.1% of SDS for 1 h at 50°C, and then for 20 min at 65°C. DNA was purified with a DNA clean and concentrator kit (Zymo research, ZR).
DNA protection assay
10 ng of a model DNA fragment or 80 ng of gDNA was incubated with 5 U of a relevant restriction enzyme (MspI, Hin6I, TaiI, AluI) (TS) in the vendor’s recommended conditions for 1 h and heat inactivated as recommended. Samples were analyzed by qPCR with the specific primers used to prepare model DNA (see preparation of model DNA fragments section) and the amount of undigested DNA was calculated in relation to an uncleaved control sample using a calibration curve.
DNA enrichment
40 ng of the 155 bp (1H) model DNA fragment or 100 ng of fragmented gDNA (peak size ∼500-600 bp) was mixed with 1 mM DBCO-S-S-PEG3-biotin (BroadPharm) in 10 mM Tris-HCl (pH 8.5). Reactions were incubated at 42°C for 2 h and DNA was purified with a DNA clean and concentrator kit (ZR). Labeled DNA was enriched using Dynabeads MyOne Streptavidin C1 (TS) beads as described in Kriukienė et al.14 and amounts of DNA in the bead and supernatant fractions were analyzed by qPCR with the specific primers (described in preparation of model DNA fragments). The amount of DNA bound to magnetic beads was calculated in relation to total DNA mixture.
Quantitative PCR analysis
Quantitative PCR (qPCR) was performed using Maxima SYBR Green/ROX qPCR Master Mix (TS) using the two-step cycling protocol, as recommended by the manufacturer. Reactions were analyzed using the Rotor-Gene Q system (Qiagen).
Nuclei extraction
Buffers, cell suspensions and tubes for nuclei extraction were kept on ice and all centrifugations were done at 4°C. ∼2∗106 cells per tube were centrifuged at 200xg for 5 min, then, cells were resuspended in 500 μl nuclei suspension buffer (10 mM Tris-HCl (pH 7.4), 10 mM NaCl, 0.5 mM spermidine, 1 mM EDTA, 300 mM sucrose) and again centrifuged as described above. Then, cells were resuspended in a 500 μl nuclei suspension buffer with 0.1% Igepal CA-630 (Sigma) and incubated for 7 min on ice, and centrifugation was done at 500xg for 5 min. Nuclei were washed twice with 200 μl nuclei suspension buffer and resuspended in 100 μl of nuclei suspension buffer for double M.CviPI/eM.SssI and M.CviPI/BGT labeling reactions. For triple M.CviPI/eM.SssI/BGT labeling reactions, nuclei were resuspended in the buffer consisting of 10 mM Tris-HCl (pH 8.5), 10 mM NaCl, 0.5 mM spermidine, 1 mM EDTA, 300 mM sucrose.
Double and triple labeling in nuclei suspension
Double M.CviPI/eM.SssI and M.CviPI/BGT labeling of nuclei was performed by mixing 1∗105 nuclei in nuclei suspension buffer at a 1:1 ratio with the labeling reaction mixture consisting of 80 U M.CviPI, 1.88 μM eM.SssI (for M.CviPI/eM.SssI labeling) or 40 U BGT (for M.CviPI/BGT labeling), 1.2 mM Ado-6-azide and 300 μM UDP-glc-azide (for M.CviPI/BGT labeling) cofactors in buffer (20 mM Tris-HCl (pH 7.4), 20 mM NaCl, 0.2 mg/ml BSA, 20 mM DTT). For triple M.CviPI/eM.SssI/BGT labeling, the nuclei suspension mixture consisting of 1∗105 nuclei was mixed at a 1:1 ratio with a labeling reaction mixture consisting of 80 U M.CviPI, 1.88 μM eM.SssI, 40 U BGT, 1.2 mM Ado-6-alkyne and 300 μM UDP-glc-azide cofactors in buffer (20 mM Tris-HCl (pH 8.5), 20 mM NaCl, 0.2 mg/ml BSA, 20 mM DTT). Reactions were incubated at 37°C for 1 h, and then stopped by adding 1 mg/ml ProtK and lysis buffer (400 mM NaCl, 10 mM Tris-HCl (pH 8), 25 mM EDTA, 0.5% SDS), and incubating at 50°C for 2 h. DNA was purified using standard phenol-chloroform extraction.
Analysis of eM.SssI and M.CviPI alkyne-labeling efficiency by high-performance liquid chromatography – mass spectrometry (HPLC-MS)
For eM.SssI alkyne-labeling efficiency analysis, 300 pmol 25-mer oligonucleotide duplex containing a single CG site (5’-taataataaacgtaataataat/ 5’-attattattattacgtttattatta) was treated with 1.2 μM eM.SssI and 600 μM Ado-6-alkyne in buffer of 10 mM Tris-HCl (pH 8.5), 50 mM NaCl, 0.1 mg/ml BSA, 10 mM DTT. For M.CviPI alkyne-labeling efficiency analysis, 300 pmol 25-mer oligonucleotide duplex containing a single GC site (5’-taataataaagctaataataat/ 5’-attattattattagctttattatta) was treated with 80 U M.CviPI and 600 μM Ado-6-alkyne in the same buffer as for eM.SssI. Reactions were incubated for 1 h at 37°C, followed by the enzyme inactivation at 65°C for 20 min. Then, DNA was treated with 0.2 mg/ml Proteinase K (TF) and 0.1% of SDS for 1 h at 50°C, and for 20 min at 65°C. DNA was purified with Oligo clean and concentrator kit (ZR). For nucleoside analysis, 30 pmol of samples were denatured for 10 min at 80°C and digested to nucleosides with Nuclease P1 (Sigma) using ∼0.33 U for 1 μg DNA at 50°C for 4 h in P1 buffer (10 mM NaOAc pH 5.2, 1 mM ZnOAc), then dephosphorylated with FastAP phosphatase (TS) using ∼1 U for 5 μg DNA at 37°C overnight. Reactions were stopped by heating at 75°C for 10 min and centrifuged at 14000xg at 4°C for 30 min. Samples were analyzed on an integrated HPLC/ESI-MS system (Agilent 1290 Infinity/Agilent Q-TOF 6520 mass analyzer, positive ionization mode) equipped with a Supelco Discovery HS C18 column (7.5 cm × 2.1 mm, 3 μm) by elution with a linear gradient of solvents A (0.0075% formic acid in water) and B (0.0075% formic acid in acetonitrile) at a flow of 0.3 ml/min at 30°C as follows: 0-5 min, 0% B; 5-15 min, 0-10% B; 15-20 min, 10-95% B; 20-24 min, 95% B. For oligonucleotide analysis, 100 pmol of samples were first denatured for 10 min at 80°C and analyzed on an integrated HPLC/ESI-MS system (Agilent 1290 Infinity/Agilent Q-TOF 6520 mass analyzer, negative ionization mode) equipped with a Zorbax SB-C18 column (5 cm x 2.1 mm, 1.8 μm, Agilent Technologies) by elution with a linear gradient of solvents A (5 mM ammonium acetate pH 7.0 in water) and B (5 mM ammonium acetate pH 7.0 in methanol) at a flow of 0.2 ml/min at 45°C as follows: 0-2 min, 0% B; 2-22 min, 0-50% B; 22-26 min, 50-95% B; 26-30 min, 95% B. Capillary voltage was set to 2500 V, drying gas temperature 350°C. All results were analyzed with Agilent MassHunter Qualitative Analysis software.
Preparation of the click-reactive oligonucleotides
20 μM amine-containing ODN 5‘-TXTTTTGTGTGGTTTGGAGACTGACTACCAGATGTAACA-Biotin-3' ODN for Ion Proton sequencing or 5‘-TXTTTTGTGTGGTTTGAGATCG GAAGAGCACACGTCTGAACTCCAGTCAC-Biotin-3' ODN (X = C2-Amine-dT, Base-click) for Illumina sequencing was mixed with freshly prepared 120 mM NaHCO3 and 2 mg/ml azidobutyric acid NHS ester (Lumiprobe, dissolved in N,N-dimethylformamide (DMF)) in water. Reactions were incubated for 3 h at room temperature and purified using Oligo clean and concentrator kit (ZR).
Validation of 3x-TOP on model DNA
5hmC was introduced into GCGC sites of 1.5 ug fragmented (peak size ∼200 bp) bacteriophage lambda gDNA, as described in Ličytė et al.49). Incompletely hydroxymethylated GCGC sites were methylated by treating the GCGC-hydroxymethylated lambda DNA with 2-fold molar excess of M.HhaI in its buffer (10 mM Tris-HCl (pH 7.4), 50 mM NaCl, 0.5 mM EDTA) with 300 μM SAM (TS) at 37°C for 2 h, followed by enzyme inactivation at 65°C for 20 min. Then, DNA was treated with ProtK and purified as described in assessment of double and triple labeling in model DNA systems section. Triple M.CviPI/eM.SssI/BGT labeling reactions were performed by incubating 300 ng of hydroxymethylated lambda DNA with 16 U M.CviPI, 2-fold molar excess of eM.SssI over CG targets, 8 U BGT, 600 μM Ado-6-alkyne and 150 μM UDP-glc-azide in the buffer consisting of 10 mM Tris-HCl (pH 8.5), 50 mM NaCl, 0.1 mg/ml BSA, 10 mM DTT at 37°C for 1 h, followed by the enzyme inactivation at 65°C for 20 min. Then, DNA was treated with ProtK and purified as described above. Libraries for Ion Torrent sequencing were prepared as described in Preparation of TOP-seq libraries for Ion Torrent sequencing section with the following changes: 5 ng of the ODN-conjugated DNA was used for the priming reaction without prior enrichment procedure. The priming reaction mixture was added to 100 μl of amplification reaction and PCR amplification was done for 12 cycles.
For validation on a model DNA fragment, 5hmC was introduced at CG sites of the 171 bp model DNA fragment as described above, except that the 5-fold molar excess of M.SssI (TS) over its targets was used in buffer (10 mM Tris-HCl (pH 6.5), 50 mM NaCl, 0.5 mM EDTA). Five different DNA mixtures containing 0%, 10%, 20%, 50%, and 100% 5hmC-modified fragments were produced by mixing the 5hmC-modified and unmodified model DNA fragments. Then, triple M.CviPI/eM.SssI/BGT labeling was performed by incubating 100 ng of DNA samples with 8 U M.CviPI, 2-fold molar excess of eM.SssI over CG targets, 6 U BGT, 600 μM Ado-6-alkyne and 150 μM UDP-glc-azide in buffer (10 mM Tris-HCl (pH 8.5), 50 mM NaCl, 0.1 mg/ml BSA,10 mM DTT) at 37°C for 1 h, followed by enzymes inactivation at 65°C for 20 min. Then, DNA was treated with ProtK and purified as described above. Libraries for Ion Torrent sequencing were prepared as described in preparation of Mx-TOP libraries for Ion Torrent sequencing section with the following changes: 4 ng of the ODN-conjugated DNA was used for the priming reaction without prior enrichment and PCR amplified as above. Samples were purified with DNA clean and concentrator kit (ZR). Libraries were tested on Agilent 2100 Bioanalyzer (Agilent Technologies) and subjected to Ion Proton (TS) sequencing.
Assessment of the 3x-TOP priming reaction
Assessment of DNA strand extension efficiency from ODN-tethered uCG and 5hmCG sites. For 5hmC labeling, 150 ng of hydroxymethylated 155 bp (1H) fragment was incubated with 6 U BGT supplemented with 150 μM UDP-glc-azide in buffer (10 mM Tris-HCl (pH 8.5), 50 mM NaCl, 0.1 mg/ml BSA,10 mM DTT). For alkyne-labeling, 150 ng of 1H was incubated with 2-fold molar excess of eM.SssI over CG targets supplemented with 600 μM Ado-6-alkyne in the same buffer as described above. Reactions were incubated for 1 h at 37°C, followed by enzyme inactivation at 65°C for 20 min. Then, DNA was treated with ProtK and purified as described in Assessment of double and triple labeling in a model DNA system section. The alkyne- and azide-modified samples were processed as described in Step 4 and 5 of the 3x-TOP procedure. Before the priming reaction, the efficiency of click conjugation was assessed by enrichment of 40 ng of the ODN-conjugated DNA on streptavidin-coated magnetic beads and evaluating the amount of the captured DNA by qPCR as described in DNA enrichment and quantitative PCR analysis sections. Then, 3 ng of the alkyne-AT-ODN and 3.96 ng of the azide-TT-ODN modified samples containing equal amounts of the conjugated DNA were used for the priming reactions and the amounts of the formed products from each DNA strand were evaluated by qPCR using 2 μl of the priming reaction mixture and the primers specific to the conjugated ODN and the upper (155-dir) or bottom strand (155-rev) (described in preparation of model DNA fragments section).
Assessment of DNA strand extension at multi-tagged DNA templates. For uCG labeling, 250 ng of 97 bp fragment was incubated with 2-fold molar excess of eM.SssI over CG targets supplemented with 600 μM Ado-6-alkyne in buffer (10 mM Tris-HCl (pH 8.5), 50 mM NaCl, 0.1 mg/ml BSA, 10 mM DTT). For 5hmCG labeling, 250 ng of hydroxymethylated 97 bp-hmC fragment (see preparation of model DNA fragments section) was incubated with 6 U BGT supplemented with 150 μM UDP-glc-azide in the same buffer as described above. For GC labeling, ∼ 180 ng of 97 bp fragment was incubated with 8 U M.CviPI and 600 μM Ado-6-alkyne in the same buffer as described above. Reactions were incubated for 1 h at 37°C, followed by enzymes inactivation at 65°C for 20 min. Then, DNA was treated with ProtK and purified as described in Assessment of double and triple labeling in model DNA fragments section. The alkyne- and/or azide-modified samples were processed as described in Step 4 and 5 of the 3x-TOP procedure. Before the priming reaction, the efficiency of click conjugation was assessed by enrichment of the ODN-conjugated DNA on streptavidin-coated magnetic beads as described above. Then, ∼ 3 ng of the azide-TT-ODN and/or alkyne-AT-ODN modified samples (containing equal amounts of the conjugated DNA) were used for the priming reactions. Amplification of primed DNA was carried out as described in Step 6 of the 3x-TOP procedure with the following changes: 10 μl priming reaction mixture was added to 50 μl of strand specific amplification reaction using primers specific to the conjugated ODN and DNA fragment upper (97-dir) or bottom strand (97-rev) (see preparation of model DNA fragments section). PCR amplification was done for 10 cycles. The amounts of the products from each DNA strand were analyzed by PAGE, and the intensity of each product band was evaluated using Image Lab Software (Bio-Rad).
Assessment of the click reaction
For uCG labeling, 250 ng of 201 bp fragment was incubated with 2-fold molar excess of eM.SssI over CG targets supplemented with 600 μM Ado-6-azide in buffer (10 mM Tris-HCl (pH 8.5), 50 mM NaCl, 0.1 mg/ml BSA, 10 mM DTT). Reaction was incubated for 1 h at 37°C, followed by enzyme inactivation at 65°C for 20 min. Then, DNA was treated with ProtK and purified as described in the Assessment of double and triple labeling in model DNA fragments section. Then, the GC target in the azide-labeled fragment was labeled by incubating with 8 U M.CviPI and 600 μM Ado-6-alkyne in the same buffer as described above. The reaction was performed and DNA purified as described above. The alkyne- and/or azide-modified samples were processed as described in Step 4 of the 3x-TOP procedure. Click reaction products were visualized by PAGE.
ESC differentiation and immunofluorescence
The differentiation protocol of mouse ESCs to neurons was performed as described in.29 Briefly, ESCs were cultured on feeder-free 0.15% gelatin-coated plates for 2 passages in ESC medium containing DMEM supplemented with 15% embryonic stem-cell FBS, 50 μg/ml penicillin–streptomycin mix, 2 mM L-glutamine, 1 × non-essential amino acids, 1 mM 2-mercaptoethanol (all from Gibco) and 1000 U/mL ESGRO mLIF (Milipore). On day 8, the embryoid bodies were dissociated in a single-cell suspension, plated on poly-D-lysine/laminin-coated plates and switched to N2 medium containing 50% DMEM, 50% F12, 2 mM L-glutamine, 50 μg/ml penicillin–streptomycin mix, 1xN2 supplement, 5% KnockOut Serum Replacement (Gibco) and 50 mg/ml BSA (Sigma). N2 medium was changed 2 h after plating and after 1 day switched to N2B27 medium containing 25% DMEM, 25% F12, 50% neurobasal medium, 2 mM L-glutamine, 50 μg/ml penicillin–streptomycin mix, 1xN2 supplement, 1XB27 supplement, 5% KnockOut serum replacement (all from Gibco). N2B27 medium was changed every two days in further cultivation. The neurons were maintained until day 13 for further experiments. Two independent differentiation experiments were performed.
For immunofluorescence, glass coverslips with day 13 cells were washed with PBS, fixed in 4% paraformaldehyde, permeabilized with 0,1% Triton X-100, blocked in 1% goat serum and incubated with primary antibodies for 2 h at room temperature (RT) (rabbit anti-ßTubulin III antibody, Abcam Cat# ab18207; RRID: AB_444319), washed with PBS and incubated with secondary antibodies (Alexa488 goat anti-rabbit IgG (H+L), TS Cat# A-11034 (also A11034); RRID: AB_2576217) for 30 min at RT. Nuclei were stained with DAPI for 15 min at RT, coverslips washed and mounted with “ProLong™ Diamond Antifade Mountant” buffer and kept in +4°C. Images were acquired with a EVOS FL Auto Imaging System microscope (TS). For quantification of neuronal ß-III Tubulin-positive cells, nuclei on 2 fields of view were manually classified into ß-III tubulin positive and negative cells.
Western blot analysis of marker proteins
Samples were collected at days 0, 8, and 13 after LIF withdrawal. Specifically, cell pellets were resuspended in 100 μl of RIPA lysis buffer (150 mM NaCl, 50 mM Tris-Cl pH 8.7, 1% Nonidet P-40, 0.5% sodium deoxycholate, 0.1% SDS, 2 mM dithiothreitol, 1 mM phenylmethylsulfonyl fluoride, and 0.2 mM N-ethylmaleimide), containing a protease inhibitor mixture (Roche Applied Science). Supernatants were separated on a 12% Tris-Glycine PAAG at 40 mA per gel (1.5 mm thickness). Blots were blocked with 5% milk in TBS (20 mM Tris-HCl (pH 7.4), 150 mM NaCl), incubated with primary antibodies (anti-Oct4 Abcam Cat# ab181557; RRID: AB_2687916, anti-Sox2 Abcam Cat# ab92494; RRID: AB_10585428, anti-Nestin Abcam Cat# ab6142; RRID: AB_305313, anti-Pax6 Abcam Cat# ab195045, RRID: AB_2750924, anti-β III Tubulin Abcam Cat# ab52623, RRID: AB_869991, anti-Actin Abcam Cat# ab3280; RRID:AB_303668) overnight at 4°C and then 2 h with secondary antibodies (Dako goat anti-mouse, HPR conjugated Agilent Cat# P0447 (also P044701-2); RRID: AB_2617137 and goat anti-rabbit, HPR conjugated Agilent Cat# P0448 (also P044801-2); RRID: AB_2617138) at RT. Proteins were visualized using TMB (3,3′, 5,5′-tetramethylbenzidine) (TS).
RNA isolation and preparation of RNA-seq libraries
Total RNA was isolated using RNAzol RT (Molecular Research Center) reagent according to the manufacturer’s instructions. mRNA was enriched from 7 μg of total RNA with Dynabeads mRNA Purification Kit (TS). Ribosomal RNA was depleted using RiboCop rRNA Depletion Kit (Lexogen) according to the manufacturer’s recommendations. cDNA libraries for RNA sequencing were prepared with Ion Total RNA-Seq Kit v2 (TS) in two biological replicates following the manufacturer’s protocol. Final libraries were subjected to Ion Proton (TS) sequencing.
Preparation of Mx-TOP libraries for Ion Torrent sequencing
The detailed step-by-step protocol is presented in Supplemental Item 1. Extracted labeled mESC gDNA was sonicated on E220 Evolution focused-ultrasonicator (Covaris) in 10 mM Tris-HCl (pH 8.5) buffer to yield fragments with a peak size of ∼200 bp (Step 3a).
2x-TOP: (Step 3b, adapter introduction) The M.CviPI/eM.SssI or M.CviPI/BGT labeled gDNA extracted from 1∗105 nuclei (300-600 ng) was used for the ligation of adapters as described in Gibas et al.18 and Supplemental Item 1. (Step 4) gDNA was supplemented with 20 μM biotinylated alkyne-TT-ODN (5′-TXTTTTGTGTGGTTTGGAGACTGACTACCAGATGTAACA-Biotin X=C8-alkyne-dU, Base-click) and 8 mM CuBr: 24 mM THPTA mixture (Sigma-Aldrich) in 50% of DMSO, incubated for 20 min at 45°C and subsequently diluted to < 1.5% DMSO before a column purification (GeneJET NGS Cleanup kit, Protocol A (TS)). (Step 4b, optional) The ODN-conjugated biotinylated gDNA was enriched on 0.1 mg Dynabeads MyOne C1 Streptavidin (TS) magnetic beads by incubating in 10 mM Tris-HCl (pH 8.5), 1 M NaCl buffer at room temperature for 3 h on a roller. DNA-bound beads were washed 2x with 15 mM Tris-HCl (pH 7.4), 3 M NaCl, 1.5 mM EDTA, 0.05% Tween 20 buffer; 2x with 5 mM Tris-HCl (pH 7.4), 1 M NaCl, 0.5 mM EDTA, 0.05% Tween 20 buffer; 1x with 100 mM NaCl and finally resuspended in water. DNA was recovered by the incubation for 5 min at 95°C. (Step 5) ODN-conjugated and enriched (if necessary) DNA was subsequently used in a 20 μl-priming reaction in Pfu buffer with 1 U Pfu DNA polymerase (Promega), 0.2 mM dNTP, 0.5 μM complementary priming oligonucleotide (IT-EP; 5‘-TGTTACATCTGGTAGTCAGTCTCCAAACCACACAA-3‘, with custom LNA modifications (Exiqon) and phosphorothioate linkages at the 3’-end). The reaction mixture was incubated at the following cycling conditions: 95°C 2 min; 5 cycles at 95°C 1 min, 65°C 10 min, 72°C 10 min. (Step 6) Amplification of primed DNA was carried out for 11 cycles (for M.CviPI/eM.SssI 2x-TOP) or 10-13 cycles (for M.CviPI/BGT 2x-TOP) using 17 μl of a priming reaction mixture as described in Gibas et al.18 The final DNA libraries were size-selected for ∼300 bp fragments (MagJET NGS Cleanup and Size-selection kit, (TS)), tested on Agilent 2100 Bioanalyzer (Agilent Technologies) and by qPCR (TS), and subjected to Ion Proton (TS) sequencing.
3x-TOP: (Step 3b) 350 ng of gDNA was used for the ligation of adapters as described in Supplemental Item 1. (Step 4) The two different ODNs were attached in two rounds of click conjugation as described for the 2x-TOP library preparation. In the first round, the biotinylated azide-TT-ODN (5‘-TXTTTTGTGTGGTTTGGA GACTGACTACCAGATGTAACA-Biotin-3' X = C2-azide-dT, see Preparation of click reactive oligonucleotides section) was conjugated, and after DNA purification, the biotinylated alkyne-AT-ODN (5‘-TXTATTGTGTGGTTTGGAGACTGACTACCAGATGTAACA-Biotin-3' X=C8-alkyne-dU, Base-click) was attached in the second round. DNA was purified using the GeneJET NGS Cleanup kit. Biotinylated DNA was enriched and used in the priming and amplification reactions as described for the 2x-TOP libraries, except that libraries were amplified for 12 cycles.
Preparation of 3x-TOP-seq libraries for Illumina sequencing
The detailed step-by-step protocol is presented in Supplemental Item 1. (Step 3, an adapter introduction through the Tn5 fragmentation) 250 ng of M.CviPI/eM.SssI/BGT-labeled gDNA was fragmented using Tn5 transposase (Tn5 and oligonucleotide complex was assembled by mixing 3.5 μM Tn5 and 4.38 μM pre-annealed oligonucleotides 5’-ACACTCTTTCCCTACACGACGCTCTTCCGATCTTANNNNNNNAGGAGATGTGTATAAGAGACAG-3’ and 5’-P-CTGTCTCTTATACACATCTCCdd-3’ in 10 μl of buffer 10 mM Tris-HCl (pH 8), 5 mM MgCl2, 0.2% Triton X-100 at room temperature for 1 h. 250 ng of gDNA was mixed with 10 μl Tn5-oligonucletide complex (145 nM Tn5) in buffer (5 mM Tris-HCl (pH 8), 4 mM MgCl2, 5% DMF) and incubated at 55°C for 15 h. Reactions were stopped by adding 20 mM EDTA, 0.2 mg/ml ProtK and 0.1% of SDS and incubating for 1 h at 50°C, and then for 20 min at 65°C. DNA was purified with GeneJET NGS Cleanup kit (TS, protocol A). (Step 4) ODNs were attached to gDNA in two rounds: first, the azide-TT-ODN (5‘-TXTTTTGTGTGGTTTG AGATCGGAAGAGCACACGTCTGAACTCCAGTCAC-3' X = C2-azide-dT, oligonucleotide preparation is described in Preparation of click reactive oligonucleotides section) and after purification using GeneJET NGS Cleanup kit (TS, protocol A), the alkyne-AT-ODN (5'-TXTATTGTGTGGTTTGAGATCGGAAGAGCACACGTCTGAACTCCAGTCAC-3' X = C8-alkyne-dU, Base-click) as described for Torrent sequencing. (Step 5) 10 ng of the ODN-conjugated DNA was subsequently used in a 20 μl-priming reaction in Pfu buffer with 1 U Pfu DNA polymerase, 0.2 mM dNTP, 0.5 μM complementary priming oligonucleotide (S-IL-P7; 5‘-GTGACTGGAGTTCAGACGTGTGCTCTTCCGATCTCAAACCACA∗C∗A∗A-3‘, ∗phosphorothioate (PTO) linkages). Reaction mixtures were incubated at the following cycling conditions: 95°C 2 min; 5 cycles at 95°C 1 min, 66°C 10 min, 72°C 10 min. (Step 6) Amplification of the primed DNA was carried out by adding 17 μl of a priming reaction mixture to a 100 μl-amplification reaction containing 50 μl of 2× Platinum SuperFi PCR Master Mix (TS) and 0.5 μM each of i5/i7 barcoded primers from NEBNext Multiplex Oligos for Illumina (Dual Index Primers Set 1) kit (NEB). Thermocycler conditions were as follows: 94°C for 4 min; 15 cycles at 95°C for 1 min, 69°C for 1 min, 72°C for 1 min. The final DNA libraries were size-selected for ∼350 bp fragments using MagJET NGS Cleanup and Size-selection kit (TS), tested on Agilent 2100 Bioanalyzer (Agilent Technologies) and by qPCR using Colibri Library Quantification kit (TS), and then, subjected to Illumina sequencing (NextSeq2000, GeneCore facility, EMBL, Germany).
Locus-specific 5mC and 5hmC analysis by bisulfite sequencing
E14TG2a gDNA was precleaned with Genomic DNA Clean and Concentrator kit (ZR). 500 ng of DNA in each reaction was BS converted with EZ DNA Methylation-Gold Kit (ZR) using standard protocol and purified DNA was eluted with 10 μl of M-Elution Buffer. Selected regions were then amplified for 25 cycles with Phusion U HS polymerase (TS) using primers specific to each strand (Table S2) and DNA was purified with DNA clean and concentrator kit (ZR). Fragments were PAGE-purified using SYBR Gold staining (Invitrogen), crushed gel was incubated in an elution buffer (0.5 M CH3COONH4, 0.1 mM EDTA, 0.1% SDS) for 2 h at 37°C, shaking at 600 rpm. Samples were filtered through Costar Spin-X (Corning) centrifuge filters and DNA was purified using DNA clean and concentrator kit (ZR). DNA was end-repaired, then 3′-dA mononucleotide extension was added to the end-repaired DNA as described in Gibas et al.18 and barcoded Illumina adapters (from TruSeq Nano DNA Low Throughput Library Prep Kit, Illumina) were ligated. After 4 cycles of amplification with Illumina specific primers and Platinum SuperFi PCR Master Mix (TS), DNA was purified using GeneJET NGS Cleanup Kit (TS). Libraries were tested on Agilent 2100 Bioanalyzer (Agilent Technologies) and subjected to Illumina sequencing (MiniSeq, DNA Sequencing Center, VU Life Sciences Center, Lithuania).
Quantification and statistical analysis
2x-TOP and 3x-TOP data analysis
Raw TOP-seq reads were filtered for the presence of the 5' and 3’ adaptor sequences; reads without the 5’ adaptors were removed from further analysis. The 5’ adaptor (the sequence of the priming oligonucleotide) and the genomic part at close proximity to the 5’ adaptor was then corrected using an in-house PERL script to account for possible starting position shifting. Corrected reads were filtered by length. If not indicated otherwise all read manipulations were performed using cutadapt.51 Cleaned reads were mapped to the mouse genome (GRCm38) or lambda phage (NCBI:J02459.1) reference genome using bwa program52 and filtered using samtools53 (“-q 30 -F 0x800 -F 256” options applied). To generate a coverage per target table, each read was assigned to the nearest CG/GC based on the adaptor sequences and distance to the target using an in-house R script and GenomicRanges package.54 Reads that mapped to GCG/GCGC sequences were assigned to a specific target only by the exact position (no shift in position allowed). For all other genomic contexts, up to +/-2 bp shift was allowed. When it was impossible to identify the exact target, the read mapping position was retained only for the chromatin accessibility analysis. DNA model fragments were analyzed using a similar strategy, but the region length filtering step was omitted from the pipeline and BLAST (2.9.0+)55 was used instead of bwa.
RNA-seq data analysis
RNA-seq read quality was evaluated using FASTQC,56 and then reads were quality trimmed using cutadapt51 (applied options "-q 20 -m 20 --length 150") and later mapped using STAR57 (applied options "--outFilterMismatchNoverReadLmax 0.15 --outFilterMatchNminOverLread 0.5 --outFilterScoreMinOverLread 0.5") and mapped to the Mus musculus genome version GRCm38. Gene expression quantification was performed using stringtie58 using GENECODE59 genome annotation. Differential expression analysis was performed using the DESeq2 package.60 FPKM values were calculated using DESeq2 package.
Bisulfite-seq data analysis
Raw reads were trimmed using TrimGalore!.51 WGBS data was mapped using bwameth (https://github.com/brentp/bwa-meth) program. Mapped data was cleaned using samtools53 and sambamba (https://lomereiter.github.io/sambamba/index.html),61 duplicates were identified using MarkDuplicates program from GATK package.62 Methylation calling was performed using the MethylDackel (https://github.com/dpryan79/MethylDackel) program with standard parameters. For analysis of individual regions (Table S2), BISMARK63 was used for read mapping and methylation calling and duplicates were not removed from the dataset. Methylation per region was calculated as an average methylation level of CG sites. Only CGs with coverage of at least 5 were used. Correlation between bisulfite and 3x-TOP data was calculated using regions with ≥ 5 targets.
hMeDIP raw reads were trimmed using TrimGalore!.51 The data was mapped using hisat264 using “—no-spliced-alignment --no-mixed --no-discordant” options and peaks were called using macs2 peak caller65 using “-nomodel --extsize 350” options. liftOver function from UCSC utilities was used to lift genomic data from mm9 to mm10.
Open chromatin region identification and analysis
To identify OCRs we implemented a seed and extend principle. Seeds were identified as targets in the genome with a density value higher than 75% percentile of S0 stage density values. Seeds within 10 bp were merged to reduce initial seed number. We then applied an iterative extension method to identify the boundaries of OCRs. In brief, each seed region was extended by 10 bp in both directions and a density value was evaluated per each extension individually. In case of the extension value being smaller than the indicated threshold (75% percentile of S0 stage density value), we extended the region two more times in order to overcome possible low coverage "valleys". After the extension step, region coordinates were adjusted to match the positions of identified targets and nearby regions (distance less than 75 bp) were merged. For further analysis only regions of at least 50 bp in length were retained. De novo OCR regions were identified as regions specific to NPC or NC stage and with a minimal 1 kb distance to any other region in any stage.
Accessibility values of genes, promoters (1kb upstream or ±2 kb around TSS as indicated in Results) and upstream regions were calculated as a fraction of base pairs covered by OCRs per a specific feature. Modification (5hmCG or uCG) levels per each of the features were evaluated using a fraction of targets. Overlaps between region datasets were calculated as a fraction of overlapping regions divided by a smaller region set size.
Statistical and biological analysis
All statistical computations were performed using R 4.0.3 (R Core)66 and 4.3.0 (R Core).67 Heatmaps were created using the ComplexHeatmap68 and EnrichedHeatmap69 packages. GO enrichment analysis was performed using the clusterProfiler.70 Target level enrichment was calculated using Fisher exact test. Region-based enrichment was calculated using a bedtools fisher function. Elements clustering was performed using C-means clustering from the R package Mfuzz.71 Work with regions data was performed using the GenomicRanges package.54 Sequence logos were created with qqseqlogo package.72 High-, mid-, low-CG density promoter groups were identified using 500 bp windows (sliding 5 bp at every step) and classified using the observed and expected CG ratio. Promoters with a ratio ≥ 0.8 were assigned to HCP, ≤ 0.45 were assigned to LCP and all other promoters were assigned to MCP group. Genes with differential 5hmCG levels were identified using the limma package.73 Batch effect was removed using removeBatchEffect from the limma package.73 HOMER was used for transcription factor motif analysis at de novo open loci.74
Acknowledgments
We thank Prof. V. Masevičius for Ado-6-alkyne cofactor, A. Rukšėnaitė for Ion Proton sequencing and HPLC-MS/MS, and EMBL GeneCore facility for Illumina sequencing (Germany). We thank M. Rutkauskaitė, Z. Staševskij, and A. Petkus for technical assistance. Graphical abstract was created with BioRender.com (license FZ264L12EI), and Figure 1A with Clip Studio Paint program. The work was supported by the Research Council of Lithuania (project S-MIP-21-1 to E.K.) and the European Research Council (project ERC-AdG-2016/742654 to S.K.).
Author contributions
E.K. conceived the method and coordinated the experimental design, analytical procedures, and genomic analyses. K.S. established the Mx-TOP protocol, performed method validation, and analyzed HPLC-MS/MS data. M.N. prepared Mx-TOP and regional BS-seq libraries. J.L. established the Illumina Mx-TOP protocol and prepared regional BS-seq libraries. I.P. performed and validated differentiation of ESC. K.K. established the data analysis pipelines and carried out all bioinformatic and statistical analyses. P.G. performed initial bioinformatic validation procedures. S.K. contributed to developing DNA labeling schemes. E.K., K.S., K.K., and S.K. wrote the manuscript.
Declaration of interests
S.K. and E.K. are inventors on patents related to TOP-seq analysis: EP2776575B1, US9347093B2, and US9988673B2.
Published: December 27, 2023
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.chembiol.2023.12.003.
Supplemental information
References
- 1.Boyle A.P., Davis S., Shulha H.P., Meltzer P., Margulies E.H., Weng Z., Furey T.S., Crawford G.E. High-resolution mapping and characterization of open chromatin across the genome. Cell. 2008;132:311–322. doi: 10.1016/j.cell.2007.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Buenrostro J.D., Giresi P.G., Zaba L.C., Chang H.Y., Greenleaf W.J. Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position. Nat. Methods. 2013;10:1213–1218. doi: 10.1038/nmeth.2688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Stadler M.B., Murr R., Burger L., Ivanek R., Lienert F., Schöler A., van Nimwegen E., Wirbelauer C., Oakeley E.J., Gaidatzis D., et al. DNA-binding factors shape the mouse methylome at distal regulatory regions. Nature. 2011;480:490–495. doi: 10.1038/nature10716. [DOI] [PubMed] [Google Scholar]
- 4.Bock C., Beerman I., Lien W.H., Smith Z.D., Gu H., Boyle P., Gnirke A., Fuchs E., Rossi D.J., Meissner A. DNA methylation dynamics during in vivo differentiation of blood and skin stem cells. Mol. Cell. 2012;47:633–647. doi: 10.1016/j.molcel.2012.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Donaghey J., Thakurela S., Charlton J., Chen J.S., Smith Z.D., Gu H., Pop R., Clement K., Stamenova E.K., Karnik R., et al. Genetic determinants and epigenetic effects of pioneer-factor occupancy. Nat. Genet. 2018;50:250–258. doi: 10.1038/s41588-017-0034-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mayran A., Khetchoumian K., Hariri F., Pastinen T., Gauthier Y., Balsalobre A., Drouin J. Pioneer factor Pax7 deploys a stable enhancer repertoire for specification of cell fate. Nat. Genet. 2018;50:259–269. doi: 10.1038/s41588-017-0035-2. [DOI] [PubMed] [Google Scholar]
- 7.Kriaucionis S., Heintz N. The nuclear DNA base 5-hydroxymethylcytosine is present in Purkinje neurons and the brain. Science. 2009;324:929–930. doi: 10.1126/science.1169786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tahiliani M., Koh K.P., Shen Y., Pastor W.A., Bandukwala H., Brudno Y., Agarwal S., Iyer L.M., Liu D.R., Aravind L., Rao A. Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science. 2009;324:930–935. doi: 10.1126/science.1170116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wu H., D'Alessio A.C., Ito S., Wang Z., Cui K., Zhao K., Sun Y.E., Zhang Y. Genome-wide analysis of 5-hydroxymethylcytosine distribution reveals its dual function in transcriptional regulation in mouse embryonic stem cells. Genes Dev. 2011;25:679–684. doi: 10.1101/gad.2036011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Szulwach K.E., Li X., Li Y., Song C.X., Han J.W., Kim S., Namburi S., Hermetz K., Kim J.J., Rudd M.K., et al. Integrating 5-hydroxymethylcytosine into the epigenomic landscape of human embryonic stem cells. PLoS Genet. 2011;7 doi: 10.1371/journal.pgen.1002154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Libertini E., Heath S.C., Hamoudi R.A., Gut M., Ziller M.J., Herrero J., Czyz A., Ruotti V., Stunnenberg H.G., Frontini M., et al. Saturation analysis for whole-genome bisulfite sequencing data. Nat. Biotechnol. 2016;34:691–693. doi: 10.1038/nbt.3524. [DOI] [PubMed] [Google Scholar]
- 12.Booth M.J., Branco M.R., Ficz G., Oxley D., Krueger F., Reik W., Balasubramanian S. Quantitative sequencing of 5-methylcytosine and 5-hydroxymethylcytosine at single-base resolution. Science. 2012;336:934–937. doi: 10.1126/science.1220671. [DOI] [PubMed] [Google Scholar]
- 13.Song C.X., Szulwach K.E., Fu Y., Dai Q., Yi C., Li X., Li Y., Chen C.H., Zhang W., Jian X., et al. Selective chemical labeling reveals the genome-wide distribution of 5-hydroxymethylcytosine. Nat. Biotechnol. 2011;29:68–72. doi: 10.1038/nbt.1732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kriukienė E., Labrie V., Khare T., Urbanavičiūtė G., Lapinaitė A., Koncevičius K., Li D., Wang T., Pai S., Ptak C., et al. DNA unmethylome profiling by covalent capture of CpG sites. Nat. Commun. 2013;4:2190. doi: 10.1038/ncomms3190. [DOI] [PubMed] [Google Scholar]
- 15.Li W., Zhang X., Lu X., You L., Song Y., Luo Z., Zhang J., Xu D., Wang Y., Nie J., et al. 5-Hydroxymethylcytosine signatures in circulating cell-free DNA as diagnostic biomarkers for human cancers. Cell Res. 2017;27:1243–1257. doi: 10.1038/cr.2017.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Song C.X., Yin S., Ma L., Wheeler A., Chen Y., Zhang Y., Liu B., Xiong J., Zhang W., Hu J., et al. 5-Hydroxymethylcytosine signatures in cell-free DNA provide information about tumor types and stages. Cell Res. 2017;27:1231–1242. doi: 10.1038/cr.2017.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Staševskij Z., Gibas P., Gordevičius J., Kriukienė E., Klimašauskas S. Tethered oligonucleotide-primed sequencing, TOP-Seq: a high-resolution economical approach for DNA epigenome profiling. Mol. Cell. 2017;65:554–564.e6. doi: 10.1016/j.molcel.2016.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gibas P., Narmontė M., Staševskij Z., Gordevičius J., Klimašauskas S., Kriukienė E. Precise genomic mapping of 5-hydroxymethylcytosine via covalent tether-directed sequencing. PLoS Biol. 2020;18 doi: 10.1371/journal.pbio.3000684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gordevičius J., Narmontė M., Gibas P., Kvederavičiūtė K., Tomkutė V., Paluoja P., Krjutškov K., Salumets A., Kriukienė E. Identification of fetal unmodified and 5-hydroxymethylated CG sites in maternal cell-free DNA for non-invasive prenatal testing. Clin. Epigenet. 2020;12:1–14. doi: 10.1186/s13148-020-00938-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Narmontė M., Gibas P., Daniūnaitė K., Gordevičius J., Kriukienė E. Multiomics analysis of neuroblastoma cells reveals a diversity of malignant transformations. Front. Cell Dev. Biol. 2021;9 doi: 10.3389/fcell.2021.727353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Xu M., Kladde M.P., Van Etten J.L., Simpson R.T. Cloning, characterization and expression of the gene coding for a cytosine-5-DNA methyltransferase recognizing GpC. Nucleic Acids Res. 1998;26:3961–3966. doi: 10.1093/nar/26.17.3961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kelly T.K., Liu Y., Lay F.D., Liang G., Berman B.P., Jones P.A. Genome-wide mapping of nucleosome positioning and DNA methylation within individual DNA molecules. Genome Res. 2012;22:2497–2506. doi: 10.1101/gr.143008.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Clark S.J., Argelaguet R., Kapourani C.A., Stubbs T.M., Lee H.J., Alda-Catalinas C., Krueger F., Sanguinetti G., Kelsey G., Marioni J.C., et al. scNMT-seq enables joint profiling of chromatin accessibility DNA methylation and transcription in single cells. Nat. Commun. 2018;9:781. doi: 10.1038/s41467-018-03149-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lukinavičius G., Tomkuvienė M., Masevičius V., Klimašauskas S. Enhanced chemical stability of AdoMet analogues for improved methyltransferase-directed labeling of DNA. ACS Chem. Biol. 2013;8:1134–1139. doi: 10.1021/cb300669x. [DOI] [PubMed] [Google Scholar]
- 25.Kim K.Y., Tanaka Y., Su J., Cakir B., Xiang Y., Patterson B., Ding J., Jung Y.W., Kim J.H., Hysolli E., et al. Uhrf1 regulates active transcriptional marks at bivalent domains in pluripotent stem cells through Setd1a. Nat. Commun. 2018;9:2583. doi: 10.1038/s41467-018-04818-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yue F., Cheng Y., Breschi A., Vierstra J., Wu W., Ryba T., Sandstrom R., Ma Z., Davis C., Pope B.D., et al. A comparative encyclopedia of DNA elements in the mouse genome. Nature. 2014;515:355–364. doi: 10.1038/nature13992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lu F., Liu Y., Jiang L., Yamaguchi S., Zhang Y. Role of Tet proteins in enhancer activity and telomere elongation. Genes Dev. 2014;28:2103–2119. doi: 10.1101/gad.248005.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yu M., Hon G.C., Szulwach K.E., Song C.X., Zhang L., Kim A., Li X., Dai Q., Shen Y., Park B., et al. Base-resolution analysis of 5-hydroxymethylcytosine in the mammalian genome. Cell. 2012;149:1368–1380. doi: 10.1016/j.cell.2012.04.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bibel M., Richter J., Lacroix E., Barde Y.A. Generation of a defined and uniform population of CNS progenitors and neurons from mouse embryonic stem cells. Nat. Protoc. 2007;2:1034–1043. doi: 10.1038/nprot.2007.147. [DOI] [PubMed] [Google Scholar]
- 30.Meléndez-Ramírez C., Cuevas-Diaz Duran R., Barrios-García T., Giacoman-Lozano M., López-Ornelas A., Herrera-Gamboa J., Estudillo E., Soto-Reyes E., Velasco I., Treviño V. Dynamic landscape of chromatin accessibility and transcriptomic changes during differentiation of human embryonic stem cells into dopaminergic neurons. Sci. Rep. 2021;11 doi: 10.1038/s41598-021-96263-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Saxonov S., Berg P., Brutlag D.L. A genome-wide analysis of CpG dinucleotides in the human genome distinguishes two distinct classes of promoters. Proc. Natl. Acad. Sci. USA. 2006;103:1412–1417. doi: 10.1073/pnas.0510310103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Stevanovic M., Drakulic D., Lazic A., Ninkovic D.S., Schwirtlich M., Mojsin M. SOX transcription factors as important regulators of neuronal and glial differentiation during nervous system development and adult neurogenesis. Front. Mol. Neurosci. 2021;14 doi: 10.3389/fnmol.2021.654031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bunina D., Abazova N., Diaz N., Noh K.M., Krijgsveld J., Zaugg J.B. Genomic rewiring of SOX2 chromatin interaction network during differentiation of ESCs to postmitotic neurons. Cell Syst. 2020;10:480–494.e8. doi: 10.1016/j.cels.2020.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wen L., Li X., Yan L., Tan Y., Li R., Zhao Y., Wang Y., Xie J., Zhang Y., Song C., et al. Whole-genome analysis of 5-hydroxymethylcytosine and 5-methylcytosine at base resolution in the human brain. Genome Biol. 2014;15:R49. doi: 10.1186/gb-2014-15-3-r49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jjingo D., Conley A.B., Yi S.V., Lunyak V.V., Jordan I.K. On the presence and role of human gene-body DNA methylation. Oncotarget. 2012;3:462–474. doi: 10.18632/oncotarget.497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yang J., Bashkenova N., Zang R., Huang X., Wang J. The roles of TET family proteins in development and stem cells. Development. 2020;147:dev183129. doi: 10.1242/dev.183129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shi D.Q., Ali I., Tang J., Yang W.C. New insights into 5hmC DNA modification: generation, distribution and function. Front. Genet. 2017;8:100. doi: 10.3389/fgene.2017.00100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tan L., Xiong L., Xu W., Wu F., Huang N., Xu Y., Kong L., Zheng L., Schwartz L., Shi Y., Shi Y.G. Genome-wide comparison of DNA hydroxymethylation in mouse embryonic stem cells and neural progenitor cells by a new comparative hMeDIP-seq method. Nucleic Acids Res. 2013;41:e84. doi: 10.1093/nar/gkt091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Khare T., Pai S., Koncevičius K., Pal M., Kriukienė E., Liutkevičiūtė Z., Irimia M., Jia P., Ptak C., Xia M., et al. 5-hmC in the brain is abundant in synaptic genes and shows differences at the exon-intron boundary. Nat. Struct. Mol. Biol. 2012;19:1037–1043. doi: 10.1038/nsmb.2372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zheng S. Alternative splicing programming of axon formation. Wiley Interdiscip. Rev. RNA. 2020;11 doi: 10.1002/wrna.1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hahn M.A., Qiu R., Wu X., Li A.X., Zhang H., Wang J., Jui J., Jin S.G., Jiang Y., Pfeifer G.P., Lu Q. Dynamics of 5-hydroxymethylcytosine and chromatin marks in mammalian neurogenesis. Cell Rep. 2013;3:291–300. doi: 10.1016/j.celrep.2013.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jin S.G., Wu X., Li A.X., Pfeifer G.P. Genomic mapping of 5-hydroxymethylcytosine in the human brain. Nucleic Acids Res. 2011;39:5015–5024. doi: 10.1093/nar/gkr120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bergsland M., Ramsköld D., Zaouter C., Klum S., Sandberg R., Muhr J. Sequentially acting Sox transcription factors in neural lineage development. Genes Dev. 2011;25:2453–2464. doi: 10.1101/gad.176008.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kim J., Lo L., Dormand E., Anderson D.J. SOX10 maintains multipotency and inhibits neuronal differentiation of neural crest stem cells. Neuron. 2003;38:17–31. doi: 10.1016/s0896-6273(03)00163-6. [DOI] [PubMed] [Google Scholar]
- 45.Yang J., Smith D.K., Ni H., Wu K., Huang D., Pan S., Sathe A.A., Tang Y., Liu M.L., Xing C., et al. SOX4-mediated repression of specific tRNAs inhibits proliferation of human glioblastoma cells. Proc. Natl. Acad. Sci. USA. 2020;117:5782–5790. doi: 10.1073/pnas.1920200117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Smith Z.D., Chan M.M., Humm K.C., Karnik R., Mekhoubad S., Regev A., Eggan K., Meissner A. DNA methylation dynamics of the human preimplantation embryo. Nature. 2014;511:611–615. doi: 10.1038/nature13581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Barnett K.R., Decato B.E., Scott T.J., Hansen T.J., Chen B., Attalla J., Smith A.D., Hodges E. ATAC-Me captures prolonged DNA methylation of dynamic chromatin accessibility loci during cell fate transitions. Mol. Cell. 2020;77:1350–1364.e6. doi: 10.1016/j.molcel.2020.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nordström K.J.V., Schmidt F., Gasparoni N., Salhab A., Gasparoni G., Kattler K., Müller F., Ebert P., Costa I.G., DEEP consortium, et al. Unique and assay specific features of NOMe-ATAC-and DNase I-seq data. Nucleic Acids Res. 2019;47:10580–10596. doi: 10.1093/nar/gkz799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ličytė J., Gibas P., Skardžiūtė K., Stankevičius V., Rukšėnaitė A., Kriukienė E. A bisulfite-free approach for base-resolution analysis of genomic 5-carboxylcytosine. Cell Rep. 2020;32 doi: 10.1016/j.celrep.2020.108155. [DOI] [PubMed] [Google Scholar]
- 50.Marina R.J., Sturgill D., Bailly M.A., Thenoz M., Varma G., Prigge M.F., Nanan K.K., Shukla S., Haque N., Oberdoerffer S. TET-catalyzed oxidation of intragenic 5-methylcytosine regulates CTCF-dependent alternative splicing. EMBO J. 2016;35:335–355. doi: 10.15252/embj.201593235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet. j. 2011;17:10–12. [Google Scholar]
- 52.Li H., Durbin R. Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Danecek P., Bonfield J.K., Liddle J., Marshall J., Ohan V., Pollard M.O., Whitwham A., Keane T., McCarthy S.A., Davies R.M., Li H. Twelve years of SAMtools and BCFtools. GigaScience. 2021;10:giab008. doi: 10.1093/gigascience/giab008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lawrence M., Huber W., Pagès H., Aboyoun P., Carlson M., Gentleman R., Morgan M.T., Carey V.J. Software for computing and annotating genomic ranges. PLoS Comput. Biol. 2013;9 doi: 10.1371/journal.pcbi.1003118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Altschul S.F., Gish W., Miller W., Myers E.W., Lipman D.J. Basic local alignment search tool. J. Mol. Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 56.Andrews, S. (2010). FastQC: A Quality Control Tool for High Throughput Sequence Data. http://www.bioinformatics.babraham.ac.uk/projects/fastqc/.
- 57.Dobin A., Davis C.A., Schlesinger F., Drenkow J., Zaleski C., Jha S., Batut P., Chaisson M., Gingeras T.R. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Pertea M., Pertea G.M., Antonescu C.M., Chang T.C., Mendell J.T., Salzberg S.L. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat. Biotechnol. 2015;33:290–295. doi: 10.1038/nbt.3122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Frankish A., Diekhans M., Ferreira A.M., Johnson R., Jungreis I., Loveland J., Mudge J.M., Sisu C., Wright J., Armstrong J., et al. GENCODE reference annotation for the human and mouse genomes. Nucleic Acids Res. 2019;47:D766–D773. doi: 10.1093/nar/gky955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Love M.I., Huber W., Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Tarasov A., Vilella A.J., Cuppen E., Nijman I.J., Prins P. Sambamba: fast processing of NGS alignment formats. Bioinformatics. 2015;31:2032–2034. doi: 10.1093/bioinformatics/btv098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Van der Auwera G.A., O'Connor B.D. 1st ed. O'Reilly Media; 2020. Genomics in the Cloud: Using Docker, GATK, and WDL in Terra. [Google Scholar]
- 63.Krueger F., Andrews S.R. Bismark: a flexible aligner and methylation caller for Bisulfite-Seq applications. Bioinformatics. 2011;27:1571–1572. doi: 10.1093/bioinformatics/btr167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kim D., Langmead B., Salzberg S.L. HISAT: a fast spliced aligner with low memory requirements. Nat. Methods. 2015;12:357–360. doi: 10.1038/nmeth.3317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zhang Y., Liu T., Meyer C.A., Eeckhoute J., Johnson D.S., Bernstein B.E., Nusbaum C., Myers R.M., Brown M., Li W., Liu X.S. Model-based analysis of ChIP-Seq (MACS) Genome Biol. 2008;9:R137–R139. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Team R.C. R Foundation for Statistical Computing; 2020. R: A Language and Environment for Statistical Computing. [Google Scholar]
- 67.Team R.C. R Foundation for Statistical Computing; 2023. R: A Language and Environment for Statistical Computing. [Google Scholar]
- 68.Gu Z., Eils R., Schlesner M. Complex heatmaps reveal patterns and correlations in multidimensional genomic data. Bioinformatics. 2016;32:2847–2849. doi: 10.1093/bioinformatics/btw313. [DOI] [PubMed] [Google Scholar]
- 69.Gu Z., Eils R., Schlesner M., Ishaque N. EnrichedHeatmap: an R/Bioconductor package for comprehensive visualization of genomic signal associations. BMC Genom. 2018;19:234–237. doi: 10.1186/s12864-018-4625-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wu T., Hu E., Xu S., Chen M., Guo P., Dai Z., Feng T., Zhou L., Tang W., Zhan L., et al. clusterProfiler 4.0: A universal enrichment tool for interpreting omics data. Innovation. 2021;2:100141. doi: 10.1016/j.xinn.2021.100141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Futschik M.E., Carlisle B. Noise-robust soft clustering of gene expression time-course data. J. Bioinform. Comput. Biol. 2005;3:965–988. doi: 10.1142/s0219720005001375. [DOI] [PubMed] [Google Scholar]
- 72.Wagih O. ggseqlogo: a versatile R package for drawing sequence logos. Bioinformatics. 2017;33:3645–3647. doi: 10.1093/bioinformatics/btx469. [DOI] [PubMed] [Google Scholar]
- 73.Ritchie M.E., Phipson B., Wu D.I., Hu Y., Law C.W., Shi W., Smyth G.K. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43:e47. doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Heinz S., Benner C., Spann N., Bertolino E., Lin Y.C., Laslo P., Cheng J.X., Murre C., Singh H., Glass C.K. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol. Cell. 2010;38:576–589. doi: 10.1016/j.molcel.2010.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
-
•
All raw and processed Mx-TOP and RNA-seq data have been submitted to the NCBI Gene Expression Omnibus (GEO; https://www.ncbi.nlm.nih.gov/geo/) under the accession number GSE231929.
-
•
Mx-TOP analysis code has been deposited at Zenodo and is publicly available as of the date of publication. The DOI is listed in the key resources table.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.