Abstract
Parrots have remarkable plumage coloration that result in part from a unique ability to produce pigments called psittacofulvins that yield yellow to red feather colors. Little is known about the evolution of psittacofulvin-based pigmentation. Widespread color mutations of captive-bred parrots provide perfect opportunities to study the genetic basis of this trait. An earlier study on blue budgerigars, which do not possess psittacofulvins, reveals the involvement of an uncharacterized polyketide synthase (MuPKS) in yellow psittacofulvin synthesis. The blue phenotype had repeatedly appeared in different parrot species, similar to independent experimental replications allowing the study of convergent evolution and molecular mechanism of psittacofulvin-based pigmentation. Here, we investigated the genetic basis of the blue phenotypes in two species of Agapornis parrots, Fischer's lovebird (A. fischeri) and Yellow-collared lovebird (A. personatus). Using whole-genome data, we identified a single genomic region with size <2 Mb to be strongly associated with the color difference between blue and wild-type (WT) birds in both species. Surprisingly, we discovered that the mutation associated with the blue Agapornis phenotype was identical to the previously described substitution causing the functional change of MuPKS in budgerigars. Together with the evidence of shared blue-associated haplotypes and signatures of a selective sweep in this genomic region in both species, we demonstrated both de novo mutation and interspecific introgression play a role in the evolution of this trait in different Agapornis species. The convergent substitution in the same gene in both lovebirds and budgerigars also indicates a strong evolutionary constraint on psittacofulvin-based coloration.
Keywords: genetic convergence, avian coloration, positive selection, parrot feather pigment, psittacofulvin
Introduction
Plumages of many avian species are under sexual selection and have evolved to become colorful to attract mates (1). Many birds use carotenoids, a pigment solely derived from dietary sources, to color their feathers (2, 3). In contrast, parrots (order Psittaciformes), which have one of the most striking coloration in nature, have evolved the ability to produce a unique pigment—psittacofulvin—to make their feathers yellow, orange, and red (4, 5). Many parrots have a green plumage that is a combination of the yellow psittacofulvin-based pigmentation and blue structural color (4). Plumage coloration can serve as important social or sexual signals in parrots. Therefore, understanding the genetic basis of psittacofulvin-based coloration is crucial for gaining insights into the evolution of parrot ecology and behavior.
As popular pets (6), captive parrots have been under strong artificial selection for new plumage colors. Selective breeding has led to the emergence of the same color phenotypes across different parrot species. Independent evolution of identical phenotypes offers an ideal system to study the evolution and developmental mechanisms of plumage coloration. The most outstanding phenotypic convergence of plumage color in different parrot species is the repeated evolution of the blue phenotype, due to independent losses of psittacofulvin-based coloration that leaves the feather structurally blue. This convergent evolution of parrot plumage color provides an unparalleled replicate set to investigate the genetic basis and functional constraints underlying the evolution of psittacofulvin-based feather coloration.
Little was known about psittacofulvin pigmentation (7), until an uncharacterized polyketide synthase (MuPKS) was recently found to be involved in yellow psittacofulvin synthesis (8). Substitution of an amino acid from arginine (R) to tryptophan (W) at residue 644 (R644W) in MuPKS was shown to cause yellow pigmentation loss, and hence the blue phenotype, in captive budgerigars (Melopsittacus undulatus) (8). However, the underlying genetic mechanisms of the blue phenotype in other species, specifically whether the same gene or even the same causative mutation is involved, remains unknown. Studying other parrot species will unravel the evolutionary constraints of psittacofulvin-based pigmentation and if loci other than MuPKS also play a role in parrot plumage color development and evolution.
Here, we determine the genetic basis of the blue phenotype in two species of parrots in the genus Agapornis, Fischer's lovebird (A. fischeri) and Yellow-collared lovebird (A. personatus) (9) (Fig. 1A). The first reported blue A. personatus was a wild-caught individual in 1928, while blue A. fischeri were reported in captive-bred individuals in the early 1970s (10). To avoid overestimating genetic convergence due to only examining a priori candidate genes (i.e. MuPKS), we first examined the whole genomes of blue vs. wild-type (WT; or non-blue) individuals to identify the candidate genomic region(s) and gene(s) underlying the blue phenotype. Genes with different functional roles and belonging to multiple genetic networks have been identified in melanin- and carotenoid-based coloration (2, 11). By conducting whole-genome analysis of independently evolved phenotypes in different parrot species, we can investigate whether convergent evolution has occurred at both the genetic and phenotypic levels, and if the same substitution and gene were recurrently recruited. Additional individuals were genotyped using targeted Sanger sequencing after the identification of candidate gene(s) using whole-genome data.
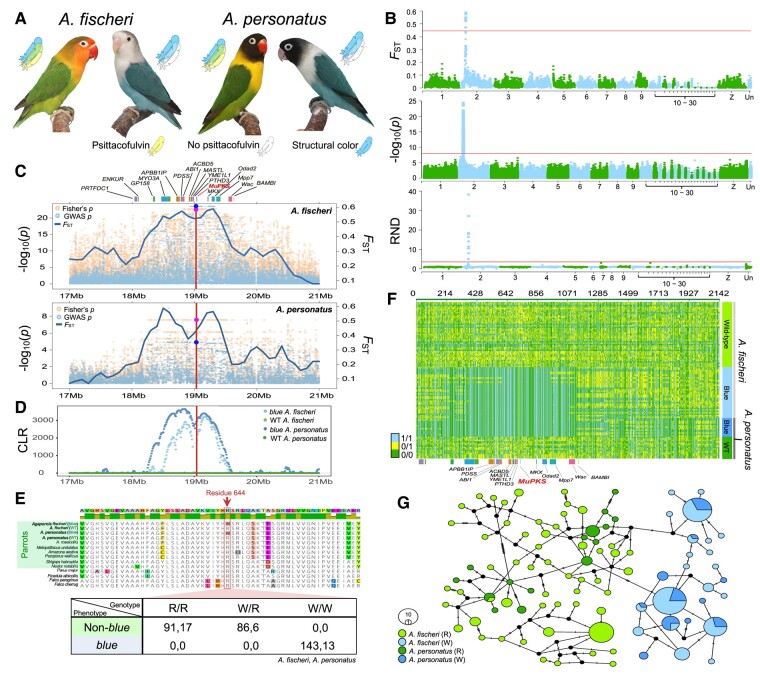
Fig. 1.
A) WT and blue Agapornis fischeri and A. personatus. Photos © Dirk Van den Abeele. B) Genome-wide differentiation (FST) between WT and blue individuals, single nucleotide polymorphism (SNP) associations with the color, and RND values between blue and WT individuals across the genome were shown for A. fischeri. The same peak of FST and SNP association in scaffold 2 was identified for A. personatus (results are not shown). Points indicate overlapping sliding window FST and RND values in 200 kb windows. Red horizontal lines indicate the 99.9th percentile for FST, Bonferroni-corrected P value for genome-wide association studies (GWAS), and 99.9th percentile for RND values, respectively. Chromosome-level scaffold identities for the reference A. roseicollis genome are shown at the bottom. C) FST, SNP associations, and Fisher's exact test P values of the highly differentiated region in A. fischeri (top) and A. personatus (bottom). Annotated genes in this region are depicted by blocks at the top. The red line indicates the position of the R644W substitution. The blue and pink solid dots indicate the association and Fisher's exact test P value of the R644W substitution. D) Signatures of selective sweep in the blue A. fischeri and blue A. personatus but not WT individuals. The red line indicates the position of the R644W substitution. CLR, composite likelihood ratio values. E) Alignment of MuPKS sequences from different avian species showing the region with the R644W substitution. Bottom table shows the nonsynonymous substitution of arginine (R) by tryptophan (W) segregates completely with the color difference in both A. fischeri (left) and A. personatus (right), based on both Sanger sequencing and whole-genome data. F) Genotypes at SNPs between WT and blue A. fischeri and A. personatus in the MuPKS region (scaffold 2, 18−21 Mb), indicating the blue-associated region with the signature of selective sweep is highly similar in both species. Each row represents one individual. Green, light blue, and yellow indicate positions homozygous for the allele that was the same as the reference genome, homozygous for the allele different from the reference, and heterozygous for both alleles, respectively. Annotated genes in this region are depicted by blocks at the bottom. G) Relationship between haplotypes of the divergent region from WT and blue A. fischeri and A. personatus, showing a high level of blue-associated haplotypes (residue 644: W) sharing but little overlaps in the WT haplotypes (residue 644: R) between the two species.
Results and discussion
In both A. personatus and A. fischeri, a single genomic island of differentiation between blue and WT (or non-blue) individuals was identified in the same position of the genome (Fig. 1B). This divergent genomic region of size <2 Mb contains 14 genes, with the MuPKS gene located in the middle (Fig. 1C). SNPs in this region were highly associated with the color difference (Fig. 1C). In A. fischeri, which had a larger sample size than A. personatus, the association was the strongest in a < 1 Mb region that contains MuPKS and six other genes (Fig. 1C). We further investigated the whole MuPKS gene and identified only a single nucleotide and amino acid substitution that completely segregated with color phenotype (Fig. 1E). Surprisingly, it was an identical nucleotide (C1930T) and amino acid substitution (R644W) in the MuPKS gene of both Agapornis species as that which abolished MuPKS activity and caused the blue phenotype in the budgerigar (8). We further genotyped 108 blue and 132 non-blue A. fischeri and eight non-blue A. personatus, using Sanger sequencing and demonstrated a complete segregation of a recessive Mendelian blue allele in this position of MuPKS with the coloration (Fig. 1E). The convergent evolution between the Agapornis species and budgerigars at the genetic level is exceptional given the same mutation was independently recruited out of the 6,318 nucleotides and 2,106 amino acids of the MuPKS gene. Recurrent de novo mutation is one of the mechanisms responsible for convergent evolution (12). The finding of the same substitution recurrently recruited in the same gene in different parrot species, which caused a functional change of the MuPKS and phenotypic difference, suggests that this might be one of very few substitutions able to completely abolish MuPKS function to produce a blue phenotype with a single mutation. Mutation of the arginine at residue 644 likely diminished the ability of the malonyl-CoA:acyl carrier protein (ACP) transacylase (MAT) domain active site to bind malonate substrates (8, 13). That the causative mutation occurred in the same gene also indicates the key role of MuPKS in the development and evolution of pigment-based coloration in parrots. Our finding does not rule out the involvement of other genes in the use of psittacofulvin to color the feather. Indeed, multiple genes are often required for different stages of pigmentation development (2, 11). Future studies of other parrots that have mutations in psittacoculvin-based coloration will be a fruitful area to further explore the constraints of the psittacoculvin production pathway and evolution of plumage coloration in parrots.
We also detected a strong signature of selective sweep in the divergent MuPKS genomic region in both blue A. personatus and blue A. fischeri, but not the WT individuals of these species (Fig. 1D). The strong signature of selective sweep was consistent with a strong artificial selection by pet bird breeders for individuals with a color different from the WT. The intense and recent selective breeding for the blue phenotype had caused a selective sweep at the MuPKS locus underlying the trait, which reduced levels of genetic diversity near the locus. Genotypes of the polymorphic sites in this genomic region showed the blue-associated haplotypes were highly homogeneous in the blue individuals (Fig. 1F). In contrast, the regions flanking the genomic island of differentiation and the region with MuPKS in WT individuals were mostly heterozygous (Fig. 1F). Furthermore, blue birds in both Agapornis species shared the same or highly similar haplotypes (Fig. 1G), indicating a single origin of the blue mutation in the two species and therefore an introgression of the blue allele between them. A. personatus and A. fischeri are closely related, and they, together with A. nigrigenis and A. lilianae but no other species, can hybridize and produce fertile offspring (14). Blue A. fischeri was thought to originate from an introgression of the blue mutation from A. personatus (10). The and relative node depth (RND) statistics indicated the introgression of the genomic region containing MuPKS from blue A. personatus to blue A. fischeri (Fig. 1B; result not shown). Interspecific introgression, in addition to recurrent de novo mutation, is therefore another evolutionary process that can result in repeated involvement of the same gene in phenotypic changes (12).
Our results support the role of both de novo mutation and interspecific introgression in the evolution of the blue phenotype in different Agapornis species. We discovered convergent substitution underlies the independent evolution of blue plumage coloration in Agapornis and budgerigars, whereas introgression between A. personatus and A. fischeri spread the de novo Agapornis mutation between these two lineages. The sharing of the identical substitution and mutated gene with budgerigars indicates a strong constraint on functional change in psittacofulvin production at the molecular level. Future studies of the genetic basis of blue phenotypes in other parrots investigating whether the same substitution was recurrently recruited and whether genes other than MuPKS are involved will provide important insights into the evolutionary constraint at the gene vs. molecular level. An understanding of the genetic mechanism will also shed light on the evolution of the unique psittacofulvin pigmentation in parrots, contributing to our knowledge of the evolution, ecology, and behavior of this diverse group of colorful birds.
Materials and methods
We sequenced the whole-genomes of WT and blue A. fischeri and A. personatus individuals to identify genomic region(s) of differentiation and nucleotide sites that were strongly associated with the plumage color difference. The genes in the region of differentiation were identified and analyzed. Targeted Sanger sequencing was performed on additional 248 individuals to genotype the MuPKS gene. We also identified signatures of a selective sweep in the candidate genomic region and tested for introgression between the two Agapornis species. Details are available in Supporting Information.
Supplementary Material
Acknowledgments
We thank all the breeders for assistance with sample collection. The computations were performed using research computing facilities offered by the Information Technology Services, HKU.
Contributor Information
Fushi Ke, School of Biological Sciences, The University of Hong Kong, Hong Kong, China.
Henriëtte van der Zwan, Focus Area for Human Metabolomics, North-West University, Potchefstroom 2531, South Africa.
Emily Shui Kei Poon, School of Biological Sciences, The University of Hong Kong, Hong Kong, China.
Alison Cloutier, School of Biological Sciences, The University of Hong Kong, Hong Kong, China.
Dirk Van den Abeele, Ornitho-Genetics VZW, 9260 Wichelen, Belgium.
Rencia van der Sluis, Focus Area for Human Metabolomics, North-West University, Potchefstroom 2531, South Africa.
Simon Yung Wa Sin, School of Biological Sciences, The University of Hong Kong, Hong Kong, China.
Supplementary Material
Supplementary material is available at PNAS Nexus online.
Funding
Funding was provided by the Research Grant Council, University Grants Committee (Hong Kong) to S.Y.W.S., and SABDI grant (407/01.SABDI.16/1016) to R.v.d.S.
Author Contributions
S.Y.W.S. designed the research; H.v.d.Z., R.v.d.S., and D.V.d.A. collected the samples; E.S.K.P. and H.v.d.Z. performed the research; F.K. and S.Y.W.S. analyzed the whole-genome data; H.v.d.Z. analyzed the Sanger sequencing data; S.Y.W.S. wrote the paper with input from all authors; A.C. edited the paper; S.Y.W.S. and R.v.d.S. acquired the funding; and S.Y.W.S. supervised the project.
Data Availability
The sequencing data have been archived in NCBI under the BioProject accession number PRJNA1048627.
References
- 1. Hill GE. 2007. Melanins and Carotenoids as feather colorants and signals. In: Reproductive biology and phylogeny of birds, Part B: Sexual selection, behavior, conservation, embryology and genetics. Boca Raton (FL): CRC Press. p. 41–74. [Google Scholar]
- 2. Toews DP, Hofmeister NR, Taylor SA. 2017. The evolution and genetics of carotenoid processing in animals. Trends Genet. 33:171–182. [DOI] [PubMed] [Google Scholar]
- 3. Toomey MB, et al. 2022. A mechanism for red coloration in vertebrates. Curr Biol. 32:4201–4214.e12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Berg ML, Bennett AT. 2010. The evolution of plumage colouration in parrots: a review. Emu Austral Ornithol. 110:10–20. [Google Scholar]
- 5. McGraw KJ. 2006. Mechanics of uncommon colors: pterins, porphyrins, and psittacofulvins. In: Bird coloration: mechanisms and measurements. Cambridge (MA): Harvard University Press. p. 354–3981. [Google Scholar]
- 6. Chan DTC, Poon ESK, Wong ATC, Sin SYW. 2021. Global trade in parrots–influential factors of trade and implications for conservation. Global Ecol Conserv. 30:e01784. [Google Scholar]
- 7. Price-Waldman R, Stoddard MC. 2021. Avian coloration genetics: recent advances and emerging questions. J Hered. 112:395–416. [DOI] [PubMed] [Google Scholar]
- 8. Cooke TF, et al. 2017. Genetic mapping and biochemical basis of yellow feather pigmentation in budgerigars. Cell. 171:427–439.e21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Van der Zwan H, Visser C, Van der Sluis R. 2019. Plumage colour variations in the Agapornis genus: a review. Ostrich. 90:1–10. [Google Scholar]
- 10. Van den Abeele D. 2016. Lovebirds compendium: genus Agapornis: species, breeding, genetics, mutations. Warffum: About Pets. [Google Scholar]
- 11. Roulin A, Ducrest A-L. 2013. Genetics of colouration in birds. Semin Cell Dev Biol. 24:594–608. [DOI] [PubMed] [Google Scholar]
- 12. Christin P-A, Weinreich DM, Besnard G. 2010. Causes and evolutionary significance of genetic convergence. Trends Genet. 26:400–405. [DOI] [PubMed] [Google Scholar]
- 13. Rangan VS, Smith S. 1997. Alteration of the substrate specificity of the malonyl-CoA/acetyl-CoA: acyl carrier protein-S-acyltransferase domain of the multifunctional fatty acid synthase by mutation of a single arginine residue. J Biol Chem. 272:11975–11978. [DOI] [PubMed] [Google Scholar]
- 14. Huynh S, Cloutier A, Sin SYW. 2023. Museomics and phylogenomics of lovebirds (Psittaciformes, Psittaculidae, Agapornis) using low-coverage whole-genome sequencing. Mol Phylogenet Evol. 185:107822. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The sequencing data have been archived in NCBI under the BioProject accession number PRJNA1048627.