Abstract
From mutational analysis of the 3′-terminal hairpin of turnip yellow mosaic virus (TYMV) RNA and use of nonstructured C-rich RNA templates, we conclude that the main determinant in the tRNA-like structure of TYMV RNA for initiation of minus-strand synthesis by the viral RNA-dependent RNA polymerase (RdRp) is the non-base-paired 3′ ACC(A) end. Base pairing of this 3′ end reduces the transcription efficiency drastically, and deletion of only the 3′-terminal A residue results in a fivefold drop in efficiency. The two C residues of the 3′ ACCA end are required for efficient transcription, as shown by substitution mutations. However, the 5′ A residue is not specifically involved in initiation of transcription, as shown by substitution mutations. Furthermore, the hairpin stem and loop upstream of the 3′ ACCA end also do not interact with the RdRp in a base-specific way. However, for efficient transcription, the hairpin stem should be at least five bp in length, while the calculated ΔG value should be less than −10.5 kcal/mol. Unexpectedly, the use of nonstructured C-rich RNA templates showed that the RdRp can start internally on an NCCN or NUCN sequence. Therefore, a possible function of the tRNA-like structure of TYMV RNA may be to prevent internal initiation of minus-strand synthesis.
The first step in the replication of positive-stranded RNA viruses is the production of the minus strand, starting at the 3′ end of the genomic RNA. Therefore, to initiate replication, the replicase complex has to recognize and attach to the 3′ end. In many cases a defined RNA structure is involved in initiation of replication, such as a tRNA-like structure in brome mosaic virus (BMV) RNA (6), a 3′-terminal stem-loop in turnip crinkle virus satellite C RNA (26), or a complicated pseudoknot structure in the genomic RNAs of enteroviruses (15, 24). However, the Qβ replicase is able to transcribe a synthetic RNA with CCC at the 3′ terminus (19), although for efficient RNA synthesis, specific internal regions of the RNA are required (3).
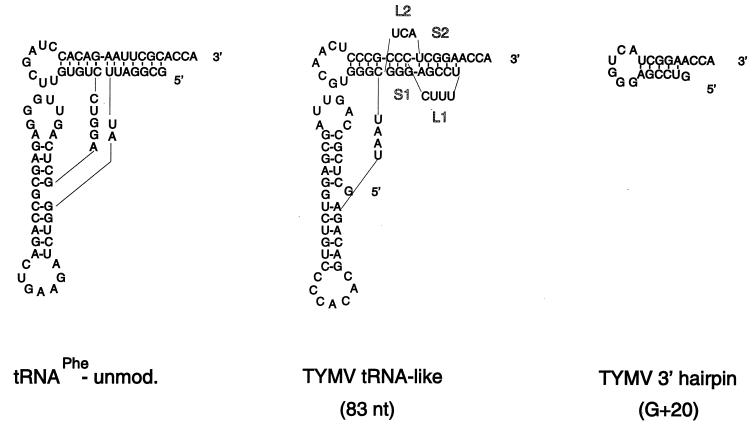
In case of turnip yellow mosaic virus (TYMV), the 3′ end of the genomic RNA is folded into a tRNA-like structure (Fig. 1) (23). As this tRNA-like structure can interact with various tRNA-specific enzymes (10, 13, 21), different functions are expected to be embedded in this part of the genome. Therefore, to be able to investigate RNA replication instead of multiplication of the virus, an in vitro system was developed (5). By using an RNA-dependent RNA polymerase (RdRp) preparation that was shown to be of viral origin and specific for TYMV RNA, it was found earlier that the tRNA-like structure itself could be transcribed very efficiently by the enzyme (5). However, further investigations by mutational analysis showed that the pseudoknot structure at the 3′ end of the tRNA-like structure (Fig. 1) possesses sufficient information for efficient transcription by the RdRp, indicating that the complete tRNA-like structure is not required for initiation of minus-strand synthesis (4). This is in agreement with the inhibitory effect of a fragment consisting of the 3′-terminal 38 nucleotides (nt) of TYMV RNA on the transcription of the viral RNA (9). However, only a twofold drop in efficiency was obtained when stem S1 of the pseudoknot (Fig. 1) was disrupted or when the 3′ hairpin itself (G+20 [Fig. 1]) was used as the template. This indicates that the complete pseudoknot structure is important but, as also concluded for the complete tRNA-like structure, not necessary to start RNA replication (4). Therefore, if base-specific interactions between the RdRp and the 3′ end of TYMV RNA do occur, they can take place only in the 3′-terminal hairpin region.
FIG. 1.
Secondary structures of the 83-nt and G+20 fragments and unmodified tRNAPhe of yeast. The 83-nt fragment consists of the 3′-terminal 83 nt of TYMV RNA and contains the tRNA-like structure (4). The stem regions, S1 and S2, and loop regions, L1 and L2, of the pseudoknot are indicated. The G+20 fragment consists of the 3′-terminal 20 nt of TYMV RNA, and a G residue is added at the 5′ end for T7 transcription (4).
In the present study, the 3′-terminal hairpin (G+20 [Fig. 1]) was used to further characterize the minimal template requirements for initiation of minus-strand synthesis in vitro. These results have led to a better understanding of the presence of a pseudoknotted structure at the 3′ end. The results obtained with a number of different templates, including RNA fragments unrelated to the 3′ end of TYMV RNA, led to the conclusion that the only determinant of the minus-strand promoter located in the tRNA-like structure is the free 3′ ACC(A) end.
MATERIALS AND METHODS
RdRp preparation and RdRp assay.
The RdRp was isolated from Chinese cabbage leaves 10 days after inoculation with TYMV and was purified up to and through the glycerol gradient centrifugation step as described previously (5). Twenty microliters of the glycerol gradient fraction containing the highest RdRp activity was treated with micrococcal nuclease, and in vitro transcription was performed in 100 μl containing 40 mM Tris-HCl (pH 9.0), 8.0 mM MgCl2, 2.5 mM dithiothreitol, 0.8 mM ATP, GTP, and CTP, 10 μCi of [α-32P]UTP (800 Ci/mmol; ICN), 2% (vol/vol) ethanol, 125 ng of actinomycin D, 1 U of RNAguard (Pharmacia) per μl, and an equimolar amount of the various RNA fragments, as described previously (4). The samples were incubated at 37°C for 1 h, and the reaction products were analyzed by gel electrophoresis on an 8 M urea–polyacrylamide gel under denaturing conditions after preheating them for 1 min at 90°C in formamide loading buffer. After electrophoresis, the gel was stained with o-toluidine blue to determine the positions of the template RNAs and to be sure that no degradation took place during the assay. The incorporation of [32P]UMP was determined by Cerenkov counting of the radioactivity of the reaction product in the gel. The relative transcription efficiency was obtained by comparing the [32P]UMP incorporation, corrected for the number of UMP residues in the RdRp transcript, with that obtained for the reference template.
Preparation of G+20 mutants, tetra mutants, G+21, and GG+21.
Oligonucleotides corresponding to the desired RNA fragment were ligated downstream of a T7 promoter in the vector pUC19 as described for the 83-nt, 44-nt, G+30, 28-nt, and G+20 fragments (4, 5). The 3′ CCA end was obtained by digesting the vector with the restriction enzyme MvaI (MBI Fermentas) before T7 transcription. The RNA was purified by electrophoresis on an 8 M–15% polyacrylamide gel as described before (4, 5) or by high-performance liquid chromatography with a DEAE ion-exchange column (Nucleogen DEAE 500-7 IWC) under native conditions (20 mM phosphate, pH 6.0). In the latter case, the RNA was eluted with a 0 to 1.25 M KCl gradient. The fractions containing the RNA were dialyzed against water for about 1 day and freeze-dried to reduce the volume. As a result of T7 transcription, the RNA is sometimes extended at the 3′ end by addition of one or two extra nucleotides. Only fractions containing RNA of the right size, as determined on an 8 M urea–20% polyacrylamide gel, were used.
Preparation of 3′ CCA end (-CC, -C, -CG, and -GC) deletion mutants.
The construction of the various cDNAs was performed as described above. However, to obtain a 3′-terminal C residue, the HindIII and PstI sites of the pUC19 polylinker were used for the ligation of the oligonucleotides and the cDNA was digested with PstI before T7 transcription. To obtain a 3′-terminal G residue, the HindIII and SacI sites of the polylinker were used, and the cDNA was digested with SacI before T7 transcription. Because of the defined secondary structure and high relative transcription efficiency of tetra-G (see Fig. 4A and Table 1), the mutations in the 3′ CCA end were performed with this template in order to increase the reliability of the transcription efficiencies that were expected to be low.
FIG. 4.
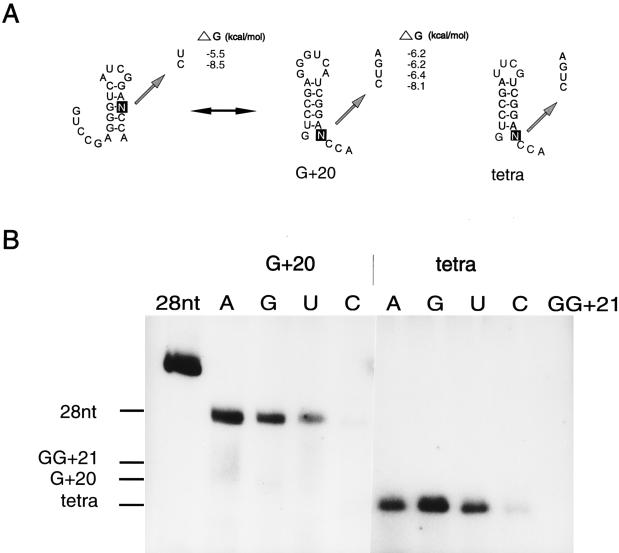
Mutational analysis of the 5′ A residue of the 3′ ACCA end of the G+20 and tetra fragments. (A) Secondary structures of the G+20 and tetra fragments. In the tetra fragments, the viral loop sequence of G+20 is replaced by a stable UUCG tetraloop. The arrow indicates the position of the substitution. For G+20-U and -C, an alternative structure can be formed. The ΔG values were determined by using the program MFold (30). (B) Autoradiograph of the 32P-labeled products with the various templates. The 28-nt and GG+21 fragments (Fig. 2) are used as references for the calculation of the relative efficiencies (Table 1). The positions of the templates, determined by staining, are indicated at the left. As was detected for all templates so far (3, 4), the products of the various G+20 templates migrate more slowly in the 8 M urea–20% polyacrylamide gel than the template itself. It was assumed that the double-stranded product is too stable to be denatured under the conditions used. Remarkably, the products obtained with the tetra mutants migrate at the same positions as their templates. The high stability of the hairpin structure and the small size of the fragment may explain why these double-stranded products can be denatured under the same conditions. This indicates that template and transcript are not covalently linked, which is in agreement with the de novo initiation of transcription.
TABLE 1.
Transcription efficiencies of 83-nt, unmodified tRNAPhe, and 3′ hairpin mutantsa
| Template | Relative efficiency (%)b |
|---|---|
| Wild type 28 nt | 100 |
| 83 nt | 90 |
| tRNAPhe (unmodified) | 50 |
| G+20-A | 46 |
| G+20-G | 45 |
| G+20-U | 31 |
| G+20-C | 8 |
| Tetra-A | 58 |
| Tetra-G | 108 |
| Tetra-U | 61 |
| Tetra-C | 33 |
| G+21 | 10 |
| GG+21 | 2 |
| -CCA | 100 |
| -CC | 18 |
| -C | 6 |
| -CG | 6 |
| -GC | 2 |
| G+20(+)-A | 51 |
| G+20(+)-G | 249 |
| Tetra-G(+) | 50 |
| Tetra-G(−) | 62 |
| Tetra-G-inv | 94 |
The incorporation of [32P]UMP was determined by Cerenkov counting of the radioactivity in reaction products in the gel and was corrected for the various numbers of UMP residues present in the negative-sense reaction product.
Compared to that of the 28-nt fragment. Results are averages for two experiments, except for the G+20 and tetra templates, which are averages for four experiments. Errors are about 5%.
Preparation of SSa, SSa(+), and SSc.
The oligonucleotides used to make the SSa and SSc constructs correspond to the single-stranded regions, nt 6093 to 6138 and 5675 to 5706, respectively, upstream of the tRNA-like structure of TYMV RNA (11). To construct the cDNA of SSa, the HindIII and PstI pUC19 polylinker restriction sites were used, and to construct the cDNA of SSc, the HindIII and EcoRI sites were used in the same way as described above. To synthesize SSa and SSc, the cDNAs were digested with PstI and MvaI, respectively, before transcription with T7 RNA polymerase. As the PstI site in the polylinker is located upstream of the EcoRI site, the latter site was used to obtain SSa(+), resulting in 18 extra nucleotides at the 3′ side of SSa(+), derived from the polylinker of pUC19.
Preparation of PMPK5.
The nucleotide sequences of the oligonucleotides used to construct the cDNA of PMPK5 are derived from the pseudoknot involved in the −1 frameshift event in the gag-pro overlap region of simian retrovirus type 1 (28). The construction of the cDNA was performed as described above with the HindIII and PstI restriction sites of the polylinker of pUC19. The cDNA was digested with PstI before T7 transcription.
Preparation of hp-16S.
The synthesis of hp-16S, corresponding to the 3′-terminal hairpin of Escherichia coli 16S rRNA (nt 1504 to 1534), was described previously (8).
Synthesis of unmodified tRNAPhe.
Unmodified tRNAPhe was obtained by T7 transcription of PT7YP-0, a kind gift from O. Uhlenbeck.
[γ-32P]GTP incorporation.
The RdRp assay was performed as described above except that 10 μCi of [γ-32P]GTP (800 Ci/mmol; ICN) was used instead of [α-32P]UTP (800 Ci/mmol; ICN) and 0.8 mM UTP was used instead of GTP.
Nuclease S1 digestion.
After phenol extraction and precipitation of the reaction products, the pellet was dissolved in 4 μl containing 50 mM Na-acetate (pH 4.6), 200 mM NaCl, 2 mM ZnSO4, and 50 U of nuclease S1 (Pharmacia). An incubation of 30 min at 37°C was performed as described previously (5). The products were separated by electrophoresis on a 20% polyacrylamide gel under native conditions.
RESULTS
Minus-strand synthesis starts with GTP.
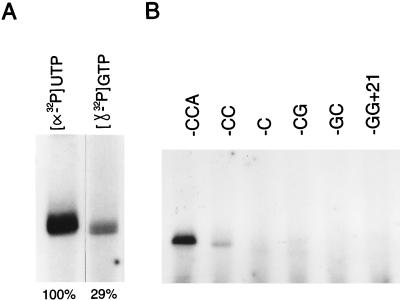
For BMV it has been demonstrated that initiation of minus-strand synthesis takes place at the 3′-proximal C residue of the tRNA-like structure, starting with the incorporation of a GTP (12, 14). For the initiation of tobacco mosaic virus RNA replication, it also was recently shown that initiation starts with incorporation of a GTP (20). Although the results obtained for TYMV so far are in agreement with these findings (2, 17, 18), the exact initiation site still has to be defined. As most templates used in these studies were constructed to have a 3′ CCA end, it is important to know whether the nonviral A residue is transcribed. Therefore, we used [γ-32P]GTP instead of [α-32P]UTP, with a similar specific activity, in the reaction mixture together with a 28-nt fragment (Fig. 2) containing the 3′ pseudoknot of TYMV RNA and previously shown to be transcribed as efficiently as the complete tRNA-like structure (4). Figure 3A shows that with [γ-32P]GTP a labeled product is obtained that migrates at the same position on an 8 M urea–polyacrylamide gel under denaturing conditions as the product obtained with [α-32P]UTP, proving that minus-strand synthesis can start de novo with a GTP. Quantification of both products by Cerenkov counting shows a relative incorporation of 29% in the case of [γ-32P]GTP. This is in agreement with an incorporation of four [32P]UMP residues instead of five. Together, this means that the most 3′-terminal A residue is not transcribed by the RdRp.
FIG. 2.
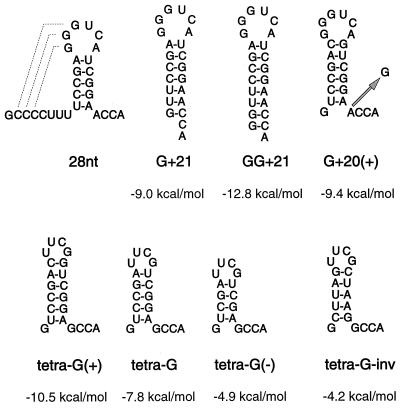
Secondary structures of the 28-nt, G+21, GG+21, and G+20(+) fragments and of the tetra-G mutants. The numbers in the designations indicate the number of viral nucleotides from the 3′ end of TYMV RNA. G+ and GG+ mean that one and two G residues are added at the 5′ end, respectively, for transcription with T7 RNA polymerase. In the tetra-G fragments the viral loop sequence is replaced by a stable UUCG tetraloop, and the A residue directly upstream of the 3′ CCA end is replaced by a G residue. (+) indicates that a nonviral CG base pair is added at the upper part of the stem, (−) indicates that a viral CG base pair is deleted, and inv means that the GC base pairs are replaced by AU and vice versa. The arrow indicates the position of a substitution. The ΔG values were determined by using the program MFold (30). Pseudoknot formation is indicated by dashed lines.
FIG. 3.
Initiation of minus-strand synthesis. (A) Autoradiograph of the 32P-labeled products obtained with [α-32P]UTP and [γ-32P]GTP, using the 28-nt fragment as the template for transcription with the RdRp of TYMV. The relative transcription efficiencies, determined by Cerenkov counting of the radioactivity in the products in the gel and taking the [α-32P]UMP incorporation as 100%, are indicated. (B) Autoradiograph of the [32P]UMP-labeled products obtained with the various 3′ deletion mutants.
The product obtained with [γ-32P]GTP seems to be a doublet. Whether this means that the RdRp starts on both C residues or whether this is the result of an added nucleotide at the 3′ end of the product is unclear.
Unmodified tRNAPhe is used as a template.
We showed recently that tRNAPhe isolated from yeast could not be used as a template by the viral RdRp, although after a long exposure, small products of about 10 nt were detected on the autoradiograph of the gel (4). As some nucleotides in tRNAPhe are modified, we decided to test an RNA fragment corresponding to unmodified yeast tRNAPhe (Fig. 1), obtained by T7 transcription. Table 1 shows that this RNA fragment is used as a template and, surprisingly, has a transcription efficiency which is similar to that previously found for the 3′-terminal hairpin (G+20 [Fig. 1]) (4).
An intact 3′ CCA end is required.
Comparing the secondary structure and nucleotide sequence of tRNAPhe to those of G+20, the most conspicuous resemblance is the 3′ ACCA end (Fig. 1). To test the importance of this free 3′ end, we constructed the fragments G+21 and GG+21, thereby extending the hairpin stem with one and two extra GC base pairs, respectively (Fig. 2). A remarkable reduction in transcription efficiency, to 10 and 2%, respectively, compared to that of the 28-nt fragment was obtained (Table 1). Deletion of the 3′-terminal A residue results in a decrease in efficiency to about 18%, indicating that this nonviral residue does contribute to an efficient start of minus-strand synthesis (Fig. 3B and Table 1). As expected, deletion of the 3′ CA end results in a more drastic drop in efficiency, to about 6%. Replacement of one of the two 3′ C residues by a G also leads to a further decrease in the efficiency: 6% for a 3′ CG end and 2% for a 3′ GC end (Fig. 3B and Table 1). These results show that the 3′ CCA end is indeed part of the promoter sequence for minus-strand synthesis.
Mutational analysis of the A residue upstream of the 3′ CCA end.
The importance of the A residue upstream of the two C residues was studied by mutating this residue in the G+20 fragment (Fig. 4A). Figure 4B and Table 1 show that replacement of A by a G residue (G+20-G) does not have any influence on the transcription efficiency, indicating that the A residue is not specifically recognized by the RdRp. Replacement by U (G+20-U) or C (G+20-C) reduces the transcription efficiency to 31 and 8%, respectively (Fig. 4B; Table 1). However, this could be due to the formation of alternative structures in which the two C residues of the 3′ ACCA end become base paired (Fig. 4A). Especially in the case of G+20-C, most fragments are expected to be in the more stable alternative secondary structure as can be inferred from the calculated ΔG values (Fig. 4A). To avoid formation of these alternative structures, a second set of G+20 mutants was constructed in which the GGGUCA loop was replaced by the more stable UUCG tetraloop (Fig. 4A, tetra). The loop substitution alone (tetra-A) has a slight positive effect on the transcription efficiency (Fig. 4B; Table 1), excluding base-specific interactions of the 3′ hairpin loop with the RdRp. The transcription efficiency of tetra-U is now identical to that of tetra-A (Fig. 4B; Table 1), suggesting that the formation of an alternative conformation was indeed responsible for the reduced transcription efficiency of G+20-U. However, the transcription efficiency of tetra-C is still low in comparison with that of tetra-A (Fig. 4B; Table 1). This can be explained by the formation of a GC base pair between the 5′ unpaired G residue and the introduced C residue, thereby stabilizing the lower part of the hairpin stem. In the case of tetra-U and even tetra-A, the 5′ unpaired G residue can also form base pairs, but these interactions are weaker than that of the GC base pair of tetra-C, explaining the higher transcription efficiencies of these mutants. Surprisingly, the transcription efficiency of tetra-G shows a twofold increase compared to that of tetra-A (Fig. 4B; Table 1) and is identical to that found for the constructs containing a pseudoknot structure (4). As no interaction is expected between the 5′ unpaired G residue and the introduced G residue, this may explain the high relative efficiency. However, this effect was not detected in the G+20-G construct (Fig. 4B; Table 1). Possibly, the combination of stabilization of the upper part of the hairpin stem by a tetraloop and avoidance of stabilization of the lower part of the hairpin stem, a situation comparable to that of the pseudoknot structure, makes this fragment such a good template.
The length and stability of the 3′ hairpin are important.
To test the idea described above, new fragments were constructed in which a CG base pair was added to the hairpin stems of G+20 and the G+20-G mutant; these constructs are designated G+20(+) and G+20(+)-G, respectively (Fig. 2). Table 1 shows that the transcription efficiency of G+20(+) is similar to that of G+20, indicating again that stabilization of the upper part of the stem region is not sufficient to increase the transcription efficiency. However, substitution of G for A upstream of the 3′ CCA end again resulted in an increase in efficiency, this time to about fivefold the value of G+20(+) (Table 1).
In order to see whether we could further increase the transcription efficiency, the loop in G+20(+)-G was replaced by the more stable tetraloop UUCG [tetra-G(+) (Fig. 2)]. However, instead of increasing, the efficiency shows a drop to about 50% (Table 1). The high stability of the structure, as indicated by the ΔG value, may be the reason for this result. Deletion of one GC base pair in tetra-G [tetra-G(−) (Fig. 2)] also results in a reduction of efficiency to about 62% (Table 1), which points to the importance of the length of the stem region.
The RdRp has no base-specific interaction with the stem region.
Because of the different base pair compositions in the corresponding stem regions of G+20 and tRNAPhe (Fig. 1), no base-specific interactions between the RdRp and the 3′-terminal hairpin stem are expected to take place. However, the nucleotides at a few positions are identical. Therefore, a new fragment was constructed, in which all AU base pairs were replaced by GC and vice versa (tetra-G-inv [Fig. 2]). The transcription efficiency obtained (94% [Table 1]), indicates that indeed no base specificity is involved in the interaction between the RdRp and the 3′-terminal hairpin stem.
RNA secondary structure is not required for initiation.
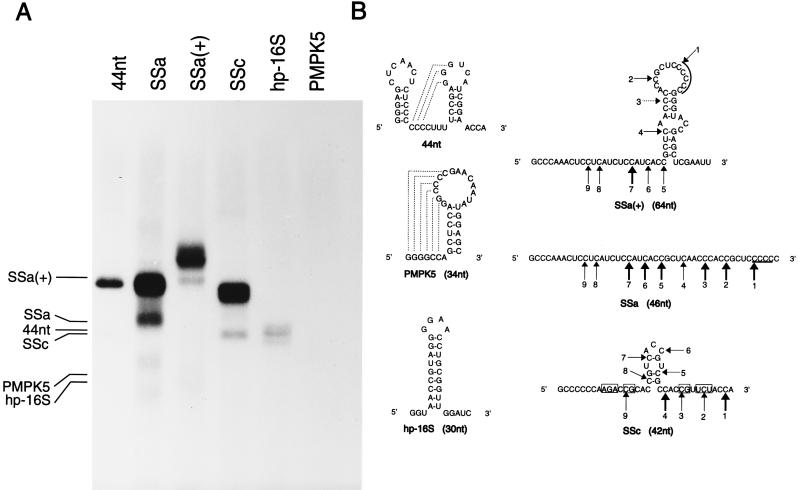
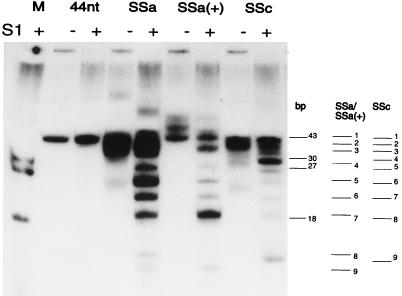
To investigate the relevance of a structured RNA in more detail, fragments which are not related to the 3′ terminus of TYMV RNA were tested as templates. A 44-nt fragment containing the pseudoknot structure and previously shown to have a template efficiency similar to that of the tRNA-like structure (4) was used as a reference (Fig. 5B). Figure 5A and Table 2 show that PMPK5, a pseudoknot derived from simian retrovirus type 1 (Fig. 5B) (28), is not used as template. Fragment hp-16S, a hairpin derived from the 3′ terminus of E. coli 16S rRNA and having a free 3′ GAUC-end (Fig. 5B) (8), has a relative transcription efficiency of only 14% (Fig. 5A; Table 2). Very unexpectedly, two largely nonstructured fragments designated SSa and SSc, both derived from the coat protein cistron of TYMV RNA upstream of the tRNA-like structure (Fig. 5B) (11), appear to be very good templates for the RdRp, having relative efficiencies of 293 and 264%, respectively (Fig. 5A and Table 2). The 3′ terminus of SSc has an ACCA end and therefore was expected to be a template for the viral RdRp. However, the 3′ terminus of SSa has a CCCC end and was therefore not expected to be efficiently transcribed, as removal of the terminal A residue resulted in a drastic decrease in efficiency (see above). Even more surprising was the high relative efficiency (102% [Fig. 5A and Table 2]) obtained with an SSa fragment in which the 3′ end was extended by 18 nt, having a 3′ AAUU end [SSa(+) (Fig. 5B)]. To understand these high efficiencies, the products were treated with nuclease S1 to remove all single-stranded parts and were loaded on a 20% polyacrylamide gel under native conditions. This gel system was used to avoid the problems we have with denaturation of the double-stranded RNA product in a denaturing gel system (see the legend to Fig. 4). As expected and as shown in Fig. 6, no changes in the migration of the product of the 44-nt fragment were detected, indicating that a completely double-stranded product of 43 bp is obtained, taking into account that the 3′ A residue is not transcribed (see above) and removed by nuclease S1. Similar results are obtained with the products of the 28-nt and tetra-A fragments and a G+30 fragment (4), used as references and loaded as a mixture (Fig. 6, lane M). However, a completely different result was obtained with the products of SSa, SSa(+), and SSc (Fig. 6). The high diversity of products can be explained only by the existence of partially double-stranded reaction products which are the result of premature termination or internal initiation on a specific sequence. Recently it was reported for BMV that after the formation of 3 to 13 phosphodiester bonds, the interaction between RdRp and the RNA is very stable (27). Therefore, premature termination after formation of more than 13 phosphodiester bonds, as is the case for most of the products obtained from SSa and SSc, is unlikely. The complete transcription of the 44-nt, G+30, 28-nt, and tetra-A fragments is in agreement with this. Moreover, the striking resemblance between the band patterns of the products obtained from SSa and SSa(+) actually proves that the partially double-stranded products are the result of internal initiation of transcription.
FIG. 5.
Transcription of the 44-nt fragment and of the RNA fragments not related to the 3′ end of TYMV RNA. (A) Autoradiograph of the 32P-labeled products with the 44-nt, PMPK5, hp-16S, SSa, SSa(+), and SSc fragments as templates. The positions of the templates are indicated at the left. (B) Secondary structures of the various fragments. The structures were determined by using the program Mfold (30). The number of nucleotides is indicated in parentheses. Pseudoknot formation is represented by dashed lines. Numbered arrows indicate the positions of internal initiation of transcription as deduced from the migration positions of the size markers (see Fig. 6 and Table 3). A thick arrow means that a relative transcription efficiency of more than 66% is obtained. A dotted arrow is used when no product could be detected with SSa(+) as the template, in comparison with the products obtained with SSa as the template. A line on top of the arrowhead indicates that the initiation site was hard to determine due to the poor resolution of the gel in this region. Nucleotides that may be involved in base pairing, although not found with Mfold, are indicated by boxes.
TABLE 2.
Transcription efficiencies of RNAs unrelated to the 3′ end of TYMV RNAa
| Fragment | Relative efficiency (%)b |
|---|---|
| Wild type 44 nt | 100 |
| SSa | 293 |
| SSa(+) | 102 |
| SSc | 264 |
| hp-16S | 14 |
| PMPK5 | 2 |
The incorporation of [32P]UMP was determined by Cerenkov counting of the radioactivity in the reaction products in the gel and was corrected for the various numbers of UMP residues present in the negative-sense reaction product, assuming that a full-length transcript is synthesized.
Compared to that of the 44-nt fragment. Results are averages for two experiments. Errors are about 5%.
FIG. 6.
Internal initiation of transcription. An autoradiograph of 32P-labeled products with the 44-nt, PMPK5, hp-16S, SSa, SSa(+), and SSc fragments as templates, before (lanes −) and after (lanes +) treatment with nuclease S1, is shown. A 20% nondenaturing polyacrylamide gel was used. A mixture of products, using the G+30 (4), 28-nt, and tetra-A fragments as templates and after treatment with nuclease S1, is used as size markers (lane M) together with the S1-treated product of the 44-nt fragment. The positions of the products of the various templates are copied and indicated on the right. The numbers of base pairs in the markers, corrected for the nontranscribed 3′-terminal A residue, are indicated. The numbers correspond to those in Table 3.
The migration positions of the reference products were used to determine the internal initiation sites in SSa, SSa(+), and SSc. Figure 5B shows that the polymerase recognizes NCCN, and in some cases NUCN, to start transcription. Some internal initiation sites even result in a relative efficiency similar to that of the 44-nt fragment (Table 3). In this respect it is noteworthy that a clear difference in transcription efficiency is obtained with some of the comparable internal initiation sites of SSa and SSa(+) (Fig. 6; Table 3). In Fig. 5B it is shown that because of the 3′ extension in SSa(+), base pairing can take place with the SSa part of the fragment, thereby closing most of the NCCN and NUCN internal initiation sites for which a reduced relative efficiency is determined. In Fig. 5B, the secondary structure of SSc is also presented. Again, a lower transcription efficiency is obtained for those initiation sites that are involved in base pairing (Table 3).
TABLE 3.
Sizes of the double-stranded products of SSa, SSa(+), and SSc
| Template | Migration distance (cm) | bp | Relative efficiency (%)a |
|---|---|---|---|
| 44 nt | 3.70 | 43 | 100 |
| G+30 | 4.60 | 30 | |
| 28 nt | 5.00 | 27 | |
| Tetra-A | 7.00 | 18 | |
| SSa/SSa(+) | |||
| 1 | 3.70 | 42–46d | NDe/42 |
| 2 | 4.00 | 37 | ND/34 |
| 3b | 4.30 | 33 | 67d/0 |
| 4 | 4.85 | 29 | 54/4 |
| 5 | 5.45 | 25 | 109/8 |
| 6 | 6.10 | 22 | 73/16 |
| 7 | 6.90 | 19 | 70/81 |
| 8 | 8.50 | 13c | 16/1 |
| 9 | 9.00 | 11c | 19/9 |
| SSc | |||
| 1 | 3.70 | 41 | 67 |
| 2 | 4.00 | 37 | 34d |
| 3 | 4.30 | 33 | 30d |
| 4 | 4.60 | 30 | 73 |
| 5 | 5.00 | 27 | 15 |
| 6 | 5.50 | 24 | 8 |
| 7 | 6.15 | 22 | 6 |
| 8 | 7.00 | 18 | 9 |
| 9 | 8.70 | 13c | 35 |
Compared to that of the 44-nt fragment.
Not detected for SSa(+).
Hard to determine because of the influence of the nucleotide composition on the migration of the product.
Hard to determine because of poor resolution.
ND, not determined because of poor resolution.
DISCUSSION
In this article we present the minimal template requirements for initiation of minus-strand synthesis by the RdRp of TYMV in vitro. Previously it was shown that the 3′-terminal 28 nt of TYMV RNA, including the pseudoknot structure, contains the minus-strand promoter (4). However, disruption of the 5′ stem (S1 in Fig. 1) of the pseudoknot resulted in only a twofold decrease of transcription efficiency, indicating that the main determinants for initiation of minus-strand synthesis are located in the 3′ hairpin region. By mutational analysis of the 3′-terminal hairpin of TYMV RNA and by using two largely nonstructured C-rich RNA templates, it was found that the main determinant in the tRNA-like structure of TYMV RNA for initiation of minus-strand synthesis by the viral RdRp is the 3′ ACCA end.
By using [γ-32P]GTP we were able to prove that initiation of minus-strand synthesis starts de novo with incorporation of a GTP opposite a C residue upstream of the 3′-terminal A residue. This is in agreement with what was found previously for BMV (12, 14) and tobacco mosaic virus (20) and supports the idea that the RdRps of these viruses may be evolutionarily related, based on the existence of a tRNA-like structure at the 3′ ends of their genomes (7, 10). However, the 3′-terminal A residue is required for efficient transcription of TYMV RNA, as deletion of this residue alone results in a fivefold drop in efficiency. This is remarkable given that this A residue is not present in the viral RNA (16). Therefore, adenylation of the tRNA-like structure is probably required for replication in vivo. The requirement for an intact 3′ CCA end to initiate replication has also been reported for BMV (1, 22).
By mutational analysis it was shown that the two C residues of the 3′ ACCA end are required for efficient transcription but that the 5′ A residue of this 3′ end is not. Furthermore, no base-specific interactions between the RdRp and the stem-and-loop region of the 3′ hairpin structure were found.
Stabilization of the lower part of the hairpin stem by base pairing the nucleotides of the 3′ ACCA end completely blocks transcription. As determined by the calculated ΔG values, the addition of two extra base pairs (G+21 [Fig. 2]) does not result in a structure that is too stable for efficient transcription, as will be discussed below. This suggests that the base pair formation itself and not the increase in stability prohibits efficient transcription, which is again in agreement with what was found for BMV (1, 22). Even the interaction of the non-base-paired 5′ G residue of G+20, added for the sake of T7 transcription, with the 3′ ACCA end reduces the transcription efficiency. However, in mutants in which the upper part of the hairpin stem was stabilized by replacement of the hairpin loop by a stable tetraloop, this drop in efficiency could be overcome by substitution of a G residue for the 5′ A residue of the 3′ ACCA end. The relative efficiency of this mutant (tetra-G [Fig. 4A]) appears to be similar to that obtained for the fragment containing the pseudoknot structure (28 nt [Fig. 2]) (4).
Stabilization of the upper part of the hairpin stem by addition of a CG base pair, instead of the tetraloop substitution, results in an increase in efficiency. However, the combination of the CG base pair addition and the tetraloop substitution [tetra-G(+) (Fig. 2)] results again in a reduction in transcription efficiency, indicating that further stabilization of the upper part of the hairpin stem is prohibitive. On the other hand, a combination of a CG base pair deletion and the tetraloop substitution [tetra-G(−) (Fig. 2)] also reduces the efficiency, suggesting that the length of the stem region is important. We conclude that efficient transcription of about 100% or more is obtained with a stem region of 5 bp or more and a calculated ΔG value of less than −10.5 kcal/mol. The highest transcription efficiency (249%) was obtained with the G+20(+)-G fragment (Fig. 2), in which (i) the 5′ A residue of the 3′ ACCA end of the 3′ hairpin (G+20) was replaced by a G residue and (ii) an extra CG base pair was added at the upper part of the stem region, resulting in a hairpin consisting of 6 bp and having a calculated ΔG value of −9.4 kcal/mol.
The question arises as to why this fragment, in which stabilization of the lower part of the hairpin stem is prevented and the upper part of the hairpin stem is stabilized, is such a good template. Structural analysis of the 3′ pseudoknot of TYMV RNA by probing and nuclear magnetic resonance studies shows that the UA base pair at the bottom of stem S2 (Fig. 1) is frayed (29). On the other side of stem S2, because of the stacking on stem S1, S2 becomes part of a longer, quasicontinuous double helix. Altogether, G+20(+)-G seems to mimic and even optimize the situation normally created by the formation of the pseudoknot structure. The twofold drop in efficiency obtained when stem S1 of the pseudoknot is disrupted (4) can now be explained by the disruption of the longer quasicontinuous helix and the possibility of forming a stabilizing UA base pair at the lower part of stem S2. The fact that a pseudoknot and not something like a G+20-G(+) structure is present at the 3′ end of TYMV RNA probably has to do with different functions embedded in the tRNA-like structure, like adenylation (13) and aminoacylation (21), needed in vivo to regulate multiplication of the virus.
Unexpectedly, the RdRp is able to start internally on NCCN and NUCN sites of two largely nonstructured RNA fragments. Some initiation sites even result in a transcription efficiency which is similar to that obtained with the pseudoknot-containing fragments. This indicates that a secondary structure per se is not required for initiation of minus-strand synthesis. As the genomic RNA of TYMV contains many single-stranded C-rich stretches (11), the replicase complex has to bind to the correct initiation site to prevent production of incomplete genomic RNAs. Possibly, tRNA-specific enzymes play a role in this process (7, 10). As the 83-nt fragment did not give products derived from internal initiation, the tRNA-like structure is sufficiently structured to prevent internal initiation in this part of the genomic RNA.
As mentioned before, in vitro initiation of minus-strand synthesis of positive-stranded RNA viruses often requires a defined secondary structure at the 3′ end of the RNA (6, 26). In this paper we show that this is not the case for TYMV. Interestingly, the minimal template requirements for initiation of minus-strand synthesis of TYMV resemble those found for the Qβ replicase (19). Apparently, for some positive-stranded RNA viruses, the absolute requirements for initiation of minus-strand synthesis in vitro are less stringent than is initially suggested by the presence of a structured 3′ terminus.
The minimal template requirements for initiation of minus-strand synthesis reported here were obtained by using a partially purified viral RdRp. Although the RdRp preparation was shown to be specific for TYMV RNA, some specificity might have been lost during the purification, as was discussed previously (4).
During the preparation of this paper, a paper was published that confirms (i) the de novo initiation of minus-strand synthesis and incorporation of [γ-32P]GTP, (ii) the importance of the 3′-terminal A residue, and (iii) the template capability of unmodified tRNAs (25).
ACKNOWLEDGMENTS
We thank Gied Jaspars for valuable discussions and reading the manuscript, Olke Uhlenbeck for the gift of PT7YP-0, Marinette van de Graaf for preparing the GG+21 fragment, Paul Michiels for providing the PMPK5 fragment, and Leo Formenoy for synthesis of the hp-16S fragment.
This work was performed under the auspices of the BIOMAC Research School of Leiden and Delft Universities.
REFERENCES
- 1.Ahlquist P, Bujarski J J, Kaesberg P, Hall T C. Localization of the replicase recognition site within brome mosaic virus RNA by hybrid-arrested RNA synthesis. Plant Mol Biol. 1984;3:37–44. doi: 10.1007/BF00023414. [DOI] [PubMed] [Google Scholar]
- 2.Boyer J C, Drugeon G, Séron K, Morch-Devignes M D, Agnès F, Haenni A L. In vitro transcripts of turnip yellow mosaic virus encompassing a long 3′ extension or produced from full-length cDNA clone harbouring a 2 kb-long PCR-amplified segment are infectious. Res Virol. 1993;144:339–348. doi: 10.1016/s0923-2516(06)80049-x. [DOI] [PubMed] [Google Scholar]
- 3.Brown D, Gold L. RNA replication by Qβ replicase: a working model. Proc Natl Acad Sci USA. 1996;93:11558–11562. doi: 10.1073/pnas.93.21.11558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Deiman B A L M, Kortlever R M, Pleij C W A. The role of the pseudoknot at the 3′ end of turnip yellow mosaic virus RNA in minus-strand synthesis by the viral RNA-dependent RNA polymerase. J Virol. 1997;71:5990–5996. doi: 10.1128/jvi.71.8.5990-5996.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Deiman B A L M, Séron K, Jaspars E M J, Pleij C W A. Efficient transcription of the tRNA-like structure of turnip yellow mosaic virus by a template-dependent and specific viral RNA polymerase obtained by a new procedure. J Virol Methods. 1997;64:181–195. doi: 10.1016/s0166-0934(96)02166-0. [DOI] [PubMed] [Google Scholar]
- 6.Dreher T W, Hall T C. Mutational analysis of the sequence and structural requirements in brome mosaic virus RNA for minus strand promoter activity. J Mol Biol. 1988;201:31–40. doi: 10.1016/0022-2836(88)90436-6. [DOI] [PubMed] [Google Scholar]
- 7.Dreher T W, Tsai C H, Skuzeski J M. Aminoacylation identity switch of turnip yellow mosaic virus RNA from valine to methionine results in an infectious virus. Proc Natl Acad Sci USA. 1996;93:12212–12216. doi: 10.1073/pnas.93.22.12212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Formenoy L J, Hellendoorn K, van Knippenberg P H, Pleij C W A. Evidence for an identical loop stability in the 3′-terminal hairpin of small subunit ribosomal RNAs, p. 79–90. Ph.D. thesis. Leiden, The Netherlands: Leiden University; 1992. [Google Scholar]
- 9.Gargouri-Bouzid R, David C, Haenni A L. The 3′ promoter region involved in RNA synthesis directed by the turnip yellow mosaic virus genome in vitro. FEBS Lett. 1991;294:56–58. doi: 10.1016/0014-5793(91)81342-6. [DOI] [PubMed] [Google Scholar]
- 10.Giegé R. Interplay of tRNA-like structures from plant viral RNAs with partners of the translation and replication machineries. Proc Natl Acad Sci USA. 1996;93:12078–12081. doi: 10.1073/pnas.93.22.12078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hellendoorn K, Mat A W, Gultyaev A P, Pleij C W A. Secondary structure model of the coat protein gene of turnip yellow mosaic virus RNA: long, C-rich, single stranded regions. Virology. 1996;224:43–54. doi: 10.1006/viro.1996.0505. [DOI] [PubMed] [Google Scholar]
- 12.Kao C C, Sun J-H. Initiation of minus-strand RNA synthesis by the brome mosaic virus RNA-dependent RNA polymerase: use of oligoribonucleotide primers. J Virol. 1996;70:6826–6830. doi: 10.1128/jvi.70.10.6826-6830.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Litvak S, Tarrago-Litvak L, Chapville F. TYMV RNA as a substrate of transfer RNA nucleotidyl-transferase: incorporation of cytidine 5′ monophosphate and determination of short nucleotides sequence at the 3′ end of the RNA. J Virol. 1973;11:238–242. doi: 10.1128/jvi.11.2.238-242.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Miller W A, Bujarski J J, Dreher T W, Hall T C. Minus-strand initiation by brome mosaic virus replicase within the 3′ tRNA-like structure of native and modified RNA templates. J Mol Biol. 1986;187:537–546. doi: 10.1016/0022-2836(86)90332-3. [DOI] [PubMed] [Google Scholar]
- 15.Mirmomeni M H, Hughes P J, Stanway G. An RNA tertiary structure in the 3′ untranslated region of enteroviruses is necessary for efficient replication. J Virol. 1997;71:2363–2370. doi: 10.1128/jvi.71.3.2363-2370.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Morch M D, Boyer J C, Haenni A L. Overlapping open reading frames revealed by complete nucleotide sequencing of turnip yellow mosaic virus genomic RNA. Nucleic Acids Res. 1988;16:6157–6173. doi: 10.1093/nar/16.13.6157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Morch M D, Joshi R L, Denial T M, Haenni A L. A new ‘sense’ RNA approach to block viral replication in vitro. Nucleic Acids Res. 1987;15:4123–4130. doi: 10.1093/nar/15.10.4123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mouchès C, Bové C, Barreau C, Bové J M. TYMV-RNA replicase. In vitro transcription and translation of viral genomes. Coll INSERM. 1975;47:109–120. [Google Scholar]
- 19.Munishkin A V, Voronin L A, Ugarov V I, Bondareva L A, Chetverina H V, Chetverina A B. Efficient templates of Q beta replicase are formed by recombination from heterologous sequences. J Mol Biol. 1991;221:463–472. doi: 10.1016/0022-2836(91)80067-5. [DOI] [PubMed] [Google Scholar]
- 20.Osman T A M, Buck H W. Complete replication in vitro of tobacco mosaic virus RNA by a template-dependent, membrane-bound RNA polymerase. J Virol. 1996;70:6227–6234. doi: 10.1128/jvi.70.9.6227-6234.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pinck M, Yot P, Chapeville F, Duranton H. Enzymatic binding of valine to the 3′ end of TYMV-RNA. Nature (London) 1970;226:954–956. doi: 10.1038/226954a0. [DOI] [PubMed] [Google Scholar]
- 22.Pogue G P, Marsh L E, Connell J P, Hall T C. Requirements for ICR-like sequences in the replication of brome mosaic virus genomic RNA. Virology. 1992;188:742–753. doi: 10.1016/0042-6822(92)90529-x. [DOI] [PubMed] [Google Scholar]
- 23.Rietveld K, van Poelgeest R, Pleij C W A, van Boom J H, Bosch L. The tRNA-like structure at the 3′ terminus of turnip yellow mosaic virus RNA. Differences and similarities with canonical tRNA. Nucleic Acids Res. 1982;10:1929–1946. doi: 10.1093/nar/10.6.1929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rohll J B, Moon D H, Evans D J, Almond J W. The 3′ untranslated region of picornavirus RNA: features required for efficient genome replication. J Virol. 1995;69:7835–7844. doi: 10.1128/jvi.69.12.7835-7844.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Singh R N, Dreher T W. Turnip yellow mosaic virus RNA-dependent RNA polymerase: initiation of minus strand synthesis in vitro. Virology. 1997;233:430–439. doi: 10.1006/viro.1997.8621. [DOI] [PubMed] [Google Scholar]
- 26.Song C, Simon A E. Requirements of a 3′-terminal stem-loop in in vitro transcription by an RNA-dependent RNA polymerase. J Mol Biol. 1995;254:6–14. doi: 10.1006/jmbi.1995.0594. [DOI] [PubMed] [Google Scholar]
- 27.Sun J-H, Kao C C. RNA synthesis by the brome mosaic virus RNA-dependent RNA polymerase: transition from initiation to elongation. Virology. 1997;233:63–73. doi: 10.1006/viro.1997.8583. [DOI] [PubMed] [Google Scholar]
- 28.ten Dam E B, Brierley I, Inglis S C, Pleij C W A. Identification and analysis of the pseudoknot-containing gag-pro ribosomal frameshift signal of simian retrovirus-1. Nucleic Acids Res. 1994;22:2304–2310. doi: 10.1093/nar/22.12.2304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.van Belkum A, Wiersema P J, Joordens J, Pleij C W A, Hilbers C W, Bosch L. Biochemical and biophysical analysis of pseudoknot-containing RNA fragments: melting studies and NMR spectroscopy. Eur J Biochem. 1989;183:591–601. doi: 10.1111/j.1432-1033.1989.tb21088.x. [DOI] [PubMed] [Google Scholar]
- 30.Zuker M. On finding all suboptimal foldings of an RNA molecule. Science. 1989;244:48–52. doi: 10.1126/science.2468181. [DOI] [PubMed] [Google Scholar]