Summary
Incomplete immunity in recovered hosts is predicted to favor more virulent pathogens upon re-infection in the population.1 The microbiota colonizing animals can generate a similarly long-lasting, partial immune response, allowing for infection but dampened disease severity.2 We tracked the evolutionary trajectories of a widespread pathogen (Pseudomonas aeruginosa), experimentally passaged through populations of nematodes immune-primed by a natural microbiota member (P. berkeleyensis). This bacterium can induce genes regulated by a mitogen-activated protein kinase (MAPK) signaling pathway effective at conferring protection against pathogen-induced death despite infection.3 Across host populations, this incomplete immunity selected for pathogens more than twice as likely to kill as those evolved in non-primed (i.e., naive) or immune-compromised (mutants with a knockout of the MAPK ortholog) control populations. Despite the higher virulence, pathogen molecular evolution in immune-primed hosts was slow and constrained. In comparison, evolving pathogens in immune-compromised hosts were characterized by substantial genomic differentiation and attenuated virulence. These findings directly attribute the incomplete host immunity induced from microbiota as a significant force shaping the virulence and evolutionary dynamics of novel infectious diseases.
Keywords: pathogen evolution, immune priming, experimental evolution, Pseudomonas aeruginosa, C. elegans
Graphical abstract

Highlights
-
•
Partial immunity from microbiota selects for increased pathogen virulence in hosts
-
•
Pathogen virulence and fitness were decoupled during evolution
-
•
Evolution within immunocompromised hosts leads to pathogen diversification
Organisms generate immune responses during interactions with their microbiota. Hoang et al. found that this immunity selects for higher pathogen virulence but limits molecular evolution.
Results and discussion
When an animal clears an infection, immune memory—a phenomenon that occurs in invertebrates and vertebrates—can protect against future infection.4 Incomplete immunity occurs when a pathogen can re-infect, although the outcome is likely to result in reduced disease severity and death.5 The commensal microbes colonizing hosts (i.e., microbiota) can also generate a protective and long-lasting host immune response, even if the microbes themselves are cleared.6,7,8 Heightened expression of defense genes in the host can be primed through detection of microbe-associated molecular patterns found in both pathogens and microbiota.9 This is a common mechanism in nature by which host microbiota can help against infectious disease.2,10,11 Although direct interactions between commensal microbes and pathogens can select for lower virulence,12,13 immune-mediated mechanisms may have the opposite effect if pathogen colonization can still occur.7,14,15 Incomplete immunity can reduce the costs of virulence to pathogens, an outcome that suggests the leakiness of infection-induced immune protection might favor more virulent pathogens.1 It is unclear whether incomplete immunity from host-microbiota interactions can similarly drive the evolution of pathogens that cause higher host mortality.
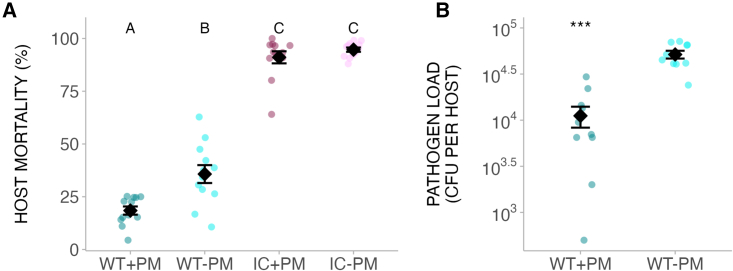
To directly test whether host microbiota can shape pathogen virulence via immune responses, we experimentally evolved a widespread, disease-causing animal pathogen (Pseudomonas aeruginosa) upon introduction to a natural host-commensal interaction. Caenorhabditis elegans nematodes can be infected by the bacterium P. aeruginosa, which harms them by accumulating in the host intestine and destroying tissue over time.16 Nematodes are found naturally with Pseudomonas spp.17 and are frequently associated with a commensal species, Pseudomonas berkeleyensis.3,18 The pathogen isolate used here (PA14), however, was from burn wounds in humans19 and thus novel to C. elegans. Hosts exposed to P. berkeleyensis and subsequently shifted to the pathogen lose their commensal upon pathogen colonization. However, initial exposure to P. berkeleyensis is sufficient to induce genes regulated by mitogen-activated protein kinase (MAPK)—an ancient innate immune pathway found in plants and animals.3,20 Expression of these genes enhances nematode host survival during P. aeruginosa infection (Figure 1A).3 By comparison, immune-compromised mutants were killed readily by P. aeruginosa, with no protective effect elicited by P. berkeleyensis colonization (Figure 1A). The immunity conferred by P. berkeleyensis for wild-type (WT) hosts was incomplete. The pathogen can form a stable infection in protected hosts but had a lower load (Figure 1B). P. berkeleyensis is mildly pathogenic in the absence of threat, similar to other protective microbes (Figure S1A).21,22 Consistent with earlier work on vaccines23 and vertebrate-infectious disease interactions,1 nematode immunity here reduced the costs of virulence by protecting hosts from the disease-induced mortality that would likely limit onward pathogen transmission.24 Reduced pathogen load in immune-primed hosts also exerts strong selection on pathogens that have better abilities to infect and colonize hosts. We thus tested whether incomplete immunity caused by the microbiota favors more virulent pathogens.
Figure 1.
Host microbiota provides incomplete immune protection
(A) Host survival (mean ± SE) upon pathogen infection, with or without exposure to microbiota member ( = 806.48, p < 0.001. Each treatment had six replicates, with ∼100–200 nematodes per replicate).
(B) Pathogen load (mean ± SE) in each host (Student’s t = 7.02, p < 0.001. Each treatment had six replicates, with 10 nematodes per replicate.). WT, wild-type host; IC, immunocompromised host; PM, protective microbiota (P. berkeleyensis). Different letters indicate significant differences. ∗∗∗p < 0.001.
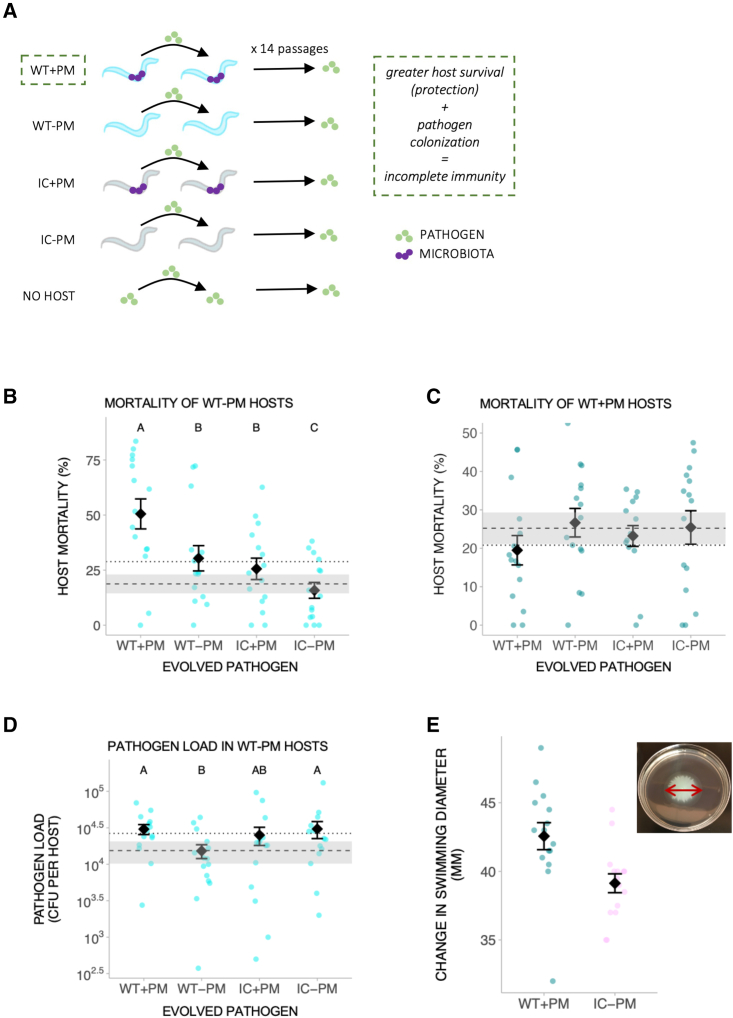
We experimentally passaged pathogen populations independently in nematode populations either previously colonized by P. berkeleyensis or in naive (non-primed) populations (Figure 2A). The pathogen was also passaged in a nematode mutant (pmk-1) not capable of mounting the primed immune response (Figure 2A). These treatments were conducted alongside a no-host control for lab adaptation. We carried out phenotypic assays of host mortality upon infection (metric for pathogen virulence) and load (metric for pathogen fitness) across pathogen generations and treatments. We then used shotgun sequencing of pools of 40 colonies to measure evolutionary changes in the genomic composition of P. aeruginosa populations.
Figure 2.
Incomplete immunity from microbiota selects for more virulent pathogens
(A) Experimental evolution design. WT, wild-type host; IC, immunocompromised host; PM, protective microbiota (P. berkeleyensis, purple dots); green dots, pathogen (P. aeruginosa). Incomplete immunity occurs when hosts exhibit increased survival upon pathogen exposure due to immune priming, but the pathogen is still able to colonize hosts (WT + PM treatment). Greater survival of WT + PM hosts decreases the cost of virulence, while increased defenses in WT + PM hosts may exclude lower virulence strains, establishing the conditions under which high virulence is favored.1
(B) Mortality of wild-type hosts without microbiota (y axis) when infected with pathogen evolved under conditions indicated on x axis ( = 55.39, p < 0.001. Each population had three technical replicates, with ∼100–200 nematodes per replicate).
(C) Mortality of wild-type hosts with prior exposure to protective microbiota (y axis), infected with pathogen evolved under conditions indicated on x axis (microbiota, = 2.36, p = 0.12; host, = 0.066, p = 0.80; interaction, = 2.35, p = 0.13. Each population had three technical replicates, with ∼100–200 nematodes per replicate.).
(D) Load (y axis) of pathogen evolved under conditions indicated on x axis in wild-type hosts without microbiota (5.99, p = 0.014. Each population had three technical replicates, with 10 nematodes per replicate.).
(B–D) Shaded dashed line indicates mean ± SE for hosts infected by no-host control pathogen. Dotted line indicates mean for hosts infected by ancestral pathogen.
(E) Swimming motility of most virulent and least virulent pathogens. Inset: example of bacterial diameter measured for swimming motility assessment.
All error bars are mean ± SE. Different letters indicate significant differences.
See also Figures S1 and S3 and Tables S1 and S2.
Microbiota-induced incomplete immunity favored more virulent pathogens compared with naive hosts (Figure 2B). These findings support theoretical models on incomplete immunity generated from prior pathogen exposure and vaccines.1,23 That microbiota in an invertebrate host can affect pathogens similarly to antibody-generating vaccines, and cross-immunity in vertebrates from previous pathogen exposure, points to a more general role of incomplete immunity in virulence evolution, regardless of the specific priming mechanism. Hosts with only genome-encoded defense maintained the ancestral virulence level, similar to immunocompromised hosts harboring microbiota. Here, weak immune responses may have allowed the microbiota to persist longer in the host. Resource competition between microbiota and pathogen is predicted to select for increased virulence,12,25 which may have favored moderate virulence despite weaker immune protection. Although more virulent, pathogens did not evolve to overcome the protective effects of microbiota exposure (Figure 2C). Immune priming can still offer harm reduction (e.g., WT + protective microbiota [PM] pathogens infecting WT + PM hosts) from increasingly virulent pathogens (e.g., WT + PM pathogens infecting WT − PM hosts) able to colonize (Figure S1B).
Hosts exposed to P. berkeleyensis selected for reduced virulence in naive immune-compromised hosts, but there were no significant host or interaction effects (Figure S1D). This result points to a trade-off in virulence for pathogens evolving in primed hosts. These pathogens had the highest virulence in naive immune-competent hosts relative to other evolved pathogens, but lower virulence in naive immune-compromised hosts. Evolved pathogens did not significantly differ in effects on immune-compromised hosts harboring microbiota (Figure S1E). Collectively, our phenotypic findings demonstrate that the immediate benefits of increased survival and pathogen tolerance conferred by the microbiota can ultimately lead to extremely negative impacts on the host.26
Pathogen virulence and load evolved along different trajectories. The levels of host mortality caused during infection and bacterial accumulation per host were not correlated across treatments (Figure 2D). This result corroborates previous research showing that virulence in novel pathogens can evolve along independent trajectories in experimental replicates and in wild populations.27,28 We hypothesized that density-independent virulence factors, such as toxin production or motility, may be contributors to the higher virulence emerging in pathogens from immune-primed hosts. To identify potential targets of selection on virulence mechanisms, we pool-sequenced evolved pathogen populations (STAR Methods) and quantified the mutations arising over time. Each population had 400–500 mutations, with most partially increasing to <50% of the population (Figure S2A). Further pairwise comparisons between treatments revealed allele frequency differences in genes involved in diverse biological pathways (Figure S2B), and treatment replicates had few unique mutations in common (Figures S2C and S2D). These results suggest that virulence under selection in our experiment has a polygenic basis, as found in other pathogens with broad host ranges.29,30,31
We compared the population genomic composition between treatments with the largest difference in evolved virulence (i.e., immune-primed versus naive, immune-compromised hosts; Figure 2B). We found an intergenic mutation between two genes involved in bacterial flagella function (flgE/flgF). Alterations in regulatory regions are less likely to disrupt function.32,33 Mutation frequency across replicates was positively correlated with infected host mortality (Figures S3A and S3B). Because flagella are virulence factors34,35 and are necessary for motility, we compared the swimming ability of evolved populations (STAR Methods; Figure 2, inset). Pathogen motility significantly differed between these extreme treatments (Figure 2E), although differences across all treatments were marginally insignificant (Figure S3C). Only a small proportion (<30%) of each pathogen population had the flgE/flgF mutation (Figure S3A), suggesting that it is not the sole contributor of virulence. Increased virulence may have emerged from the effects of interactions between this mutation and other loci across the genome.36 A subpopulation of cells with this mutation may alternatively be interacting with cells harboring other mutations.37 By contrast, disruption in metabolism may be playing a role in the reduced virulence34 evolved in immune-compromised hosts. A mutation prominent across treatments and negatively correlated with host mortality (Figures S3D and S3E) was in the fmt (methionyl-tRNA formyltransferase) gene responsible for translation initiation.38 Although P. aeruginosa utilized different genetic pathways to adapt to immune-primed and immune-compromised hosts, both groups converged on similar fitness levels.39,40
The strength of the host immune response induced by microbiota can shape genomic evolution in novel pathogens. Pathogen replication in the presence of weak selection—such as exhibited in immune-compromised hosts41—can make it easier for mutations to accumulate, resulting in extensive genomic diversification. Such rapid changes in genome evolution have been shown in bacterial pathogens responsible for zoonotic diseases42,43 as well as in viral pathogens.44 The initial lower pathogen load in immune-primed hosts (Figure 1B) may also dampen the number of new mutations that can be acquired in these populations.45
We constructed phylogenies based on point mutations to assess the relationship between individual pathogen colonies and the ancestor (Figure 3). Most mutations identified in each individual colony had fixed in the pooled samples (Figures S4A–S4E). Pathogens evolving in immune-compromised hosts diverged substantially from the ancestor (5.57 ± 0.80 mutations per individual colony; Figure 3). These colonies also shared similar distances from the ancestor as those evolving in vitro (Figures 3 and S4F), in addition to converging on similar virulence levels (Figure 2B). The acoA (Acetoin dehydrogenase E1 component alpha-subunit) gene has more mutations and higher proportions of nonsynonymous and small indels in pathogens evolved in naive immune-compromised hosts and without a host compared with those evolved in immune-primed hosts (Figures S4G–S4K). Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis revealed that this gene is involved in microbial metabolism in diverse environments, metabolic pathways, and biosynthesis of secondary metabolites.46 Similar to fmt, mutations in acoA may play some role in the reduced virulence exhibited by these pathogens. These results indicate that mutations acquired from weak selection can reduce virulence and increase genetic diversity. Similar outcomes have been found for pathogens infecting hosts with defects in their immune system,42,47,48 where less virulent pathogens may be able to better compete against more virulent ones.49 By contrast, pathogens evolving in immune-primed hosts had maintained only moderate genetic distance from the ancestor (3.21 ± 0.46 mutations per individual colony; Figure 3), suggesting that the phenotypes we observed were due to interactions of large effect mutations. Despite selecting strongly for high virulence, incomplete immunity ultimately limited pathogen evolution at the molecular level.
Figure 3.
Incomplete immunity from microbiota dampens pathogen molecular evolution
Maximum parsimony phylogeny of colonies sampled from evolved pathogen populations. We sampled more colonies from the two treatments with the most contrast in virulence level: pathogens evolved in immune-primed hosts and naive immune-compromised hosts. Inset: genetic distance from the ancestor (mean ± SE) for colonies isolated from immune-primed hosts and naive immune-compromised hosts. Values were square-root-transformed to meet the condition for normal distribution. WT, wild-type host; IC, immunocompromised host; PM, protective microbiota. ∗∗p < 0.01.
See also Figure S4.
Host immune responses altered the degree of divergence between pathogen populations. Compromised host defenses (i.e., weaker selection) may cause greater pathogen genetic divergence between populations compared to hosts with stronger defenses.50,51,52,53 Strong immune responses can otherwise increase the predictability of microbial adaptation to hosts.41 We calculated pairwise FST for each SNP between replicate populations within each treatment to determine how host defense impacted pathogen population divergence. Pathogens evolving in immune-primed hosts had fewer significant FST loci compared with those evolving in hosts protected only by genome-encoded defense (Figure 4A). Although the absence of microbiota contributed to an increase in significant FST loci across treatments, this effect is likely driven by the differences between the two WT host treatments. There is no host effect, potentially due to other selective forces not tested in our study (e.g., resource competition between microbiota and pathogen in immune-compromised hosts). All treatments exhibited differentiation in genes involved in the bacterial secretion system and two-component systems. These results indicate that incomplete immunity generated by host microbiota limited the genetic differentiation across replicate populations compared with that in non-primed treatments.
Figure 4.
Host defenses induced by microbiota alter pathogen evolutionary paths
Count of loci (A) between replicate populations at passage 14 (treatment, = 10.29, p = 0.016; microbiota, = 4.15, p = 0.042; host, = 0.66, p = 0.42) and (B) between time points (ancestor vs. P7, F3,16 = 5.34, p = 0.010; P7 vs. P14, = 10.09, p = 0.018; ancestor vs. P14, F3,16 = 0.77, p = 0.53) within each treatment, with significant genetic differentiation (FST). Dashed line indicates theoretical expectation. WT, wild-type host; IC, immunocompromised host; PM, protective microbiota; P7, passage 7; P14, passage 14. All error bars are mean ± SE. ∗p < 0.05.
See also Figure S2.
We also evaluated temporal shifts in the genetic composition of the whole population by calculating FST between the ancestral pathogen and evolved populations at the midpoint (i.e., passage 7 [P7]) and endpoint of the experiment. At the midpoint, pathogens evolving in naive hosts had more significant FST loci compared with those evolving in immune-primed hosts (Figure 4B, “ancestor vs. P7”). The absence of microbiota increased the number of significant loci that differed between the ancestor and P7 (p = 0.011), particularly in immune-compromised hosts (p = 0.006). Fewer differences were detected between P7 and passage 14 (P14). Pathogen populations evolved in hosts with only genome-encoded defense differed more across time than those evolved in immune-primed hosts (p = 0.034) and in naive immune-compromised hosts (p = 0.045). By the end of the experiment, treatments no longer varied in terms of the number of significant FST loci (Figure 4B, “ancestor vs. P14”). Earlier in evolutionary time, the absence of commensal microbiota generated more genetic differences between the ancestor and evolved pathogens, but eventually all populations exhibited similar rates of change. Taken together, the results suggest that the dynamics shaping pathogen evolution at the very beginning of emergence can become different after a period of adaptation.43,45
Host microbiota can play a significant role in protecting hosts across the tree of life from harmful infection.11,54,55 Over evolutionary time, however, we found that the incomplete immunity induced by host microbiota can act similarly to evolutionary forecasts of leaky vaccines23,56 and previous infection1 in favoring highly virulent pathogens. Conversely, immune-compromised hosts may serve as environments where pathogens can accumulate mutations, leading to genome degradation and host restriction.42 Host microbiota-immune interactions might therefore be a major source of selection shaping the ongoing evolution of emerging infectious diseases. Usage of probiotic microbes is becoming more prevalent across agricultural and wild systems,57,58 including in species at risk of extinction due to rapid pathogen spread.59,60 For long-lived hosts, application of probiotic microbes is a powerful tool to combat infectious diseases.61,62 Identifying the mechanisms by which these microbes protect seems crucial to predicting their longer-term sustainability in the field. We have found that the efficacy of these microbial therapeutics may be preserved despite pathogen evolution. However, proper precautions should be taken before potentially facilitating the spread of more virulent pathogen variants, balancing future risks with the immediate benefits to host individuals.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Chemicals, peptides, and recombinant proteins | ||
| Lysogeny broth (LB) medium | Prepared by the media kitchen, Department of Biochemistry University of Oxford | N/A |
| M9 medium | Prepared by the media kitchen, Department of Biochemistry University of Oxford | N/A |
| Nematode growth medium (NGM) agar | Prepared by the media kitchen, Department of Biochemistry University of Oxford | N/A |
| Triton X-100 | Sigma-Aldrich | CAS Number: 9036-19-5 |
| Critical commercial assays | ||
| DNeasy Blood and Tissue Kit | Qiagen | Catalog no. 69504 |
| Deposited data | ||
| Whole genome resequencing data of ancestral and evolved pathogen populations | National Center for Biotechnology Information (NCBI) | BioProject: PRJNA998467 |
| Phenotypic and processed genomic data | Mendeley Data | Mendeley Data: https://doi.org/10.17632/xz9t9gjtw6.1 |
| Experimental models: Organisms/strains | ||
| Organism: Caenorhabditis elegans strain N2 Bristol | Caenorhabditis Genetics Center | N2 |
| Organism: Caenorhabditis elegans strain pmk-1 (M03F8.4(op497), pmk-1(km25)) | Jonathan Hodgkin (University of Oxford) | Km25 |
| Organism: Escherichia coli OP50 | Caenorhabditis Genetics Center | OP50 |
| Organism: Pseudomonas aeruginosa PA14-GFP | Kevin Foster (University of Oxford) | PA14 |
| Organism: Pseudomonas berkeleyensis MSPm1 | Michael Shapira (University of California at Berkeley) | MSPm1 |
| Software and algorithms | ||
| R version 4.2.0 | https://www.r-project.org | N/A |
| Breseq | https://barricklab.org/twiki/pub/Lab/ToolsBacterialGenomeResequencing/documentation/ | N/A |
| Popoolation2 | https://sourceforge.net/p/popoolation2/wiki/Home/ | N/A |
| PHYLIP | https://phylipweb.github.io/phylip/ | N/A |
Resource availability
Lead contact
Further information and requests for resources and materials should be directed to and will be fulfilled by the lead contact, Kim Hoang (kim.hoang@emory.edu).
Materials availability
Evolved populations are available on request from Kim Hoang.
Data and code availability
-
•
Raw sequences were deposited in the NCBI Sequence Read Archive under BioProject:PRJNA998467. Phenotypic data have been published in Mendeley Data:https://doi.org/10.17632/xz9t9gjtw6.1.
-
•
R code used for the analyses in the paper is available on request.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
Experimental model and subject details
N2 and pmk-1 C. elegans nematodes were initiated from stocks stored at -80°C and maintained on nematode growth medium (NGM) plates with E. coli OP50 at 20˚C. Pseudomonas berkeleyensis MSPm1, P. aeruginosa PA14-GFP, and E. coli OP50 were initiated from stocks stored at -80˚C and cultured on lysogeny broth (LB) agar plates overnight at 30˚C. Pseudomonas berkeleyensis and E. coli were revived from frozen stock for each passage of experimental evolution and each assay. Stock nematode populations were regularly resurrected from -80˚C throughout experimental evolution and for assays.
Method details
Survival and CFU assays with ancestral P. aeruginosa
Host survival
To prepare the bacteria, we grew one random individual colony of P. berkeleyensis, E. coli, or P. aeruginosa in LB in a shaking incubator at 30°C overnight. We then seeded the bacteria on 9cm NGM plates and incubated them at 30°C for one day. Eggs from N2 and pmk-1 nematodes were collected, surface-sterilized, and age-synchronized following a standard sodium hypochlorite protocol.63 After hatching, about 200 L1 larvae were spotted onto either lawns of P. berkeleyensis or E. coli on NGM. These nematodes were incubated at 20°C for two days. L4/young adults were then transferred to a lawn of P. aeruginosa on NGM and kept at 20°C. After three days, the number of live nematodes were determined by prodding nematodes with a platinum pick to determine signs of movement.
Pathogen CFU
Following the steps above to infect nematodes for three days, we followed a modified protocol from Vega and Gore64 to determine the pathogen load in infected nematodes. Briefly, ten nematodes per population were picked into and washed twice with cold M9 buffer containing 0.01% Triton X-100 (M9-T), then chilled on ice for ∼30 minutes to stop peristalsis. We then added enough cold bleach such that the final concentration is 0.3% in the nematode/M9 mixture. After briefly mixing, the mixture was kept on ice for 10 minutes, then cold M9-T added to stop the bleaching process. Nematodes were washed once more with cold M9-T and supernatant plated to check for efficiency of bleaching. Under a dissecting scope, we pipetted 10 individuals into another tube containing zirconium beads in about 100ul M9-T. Samples were shaken in a bead beater for 2 minutes at 27 1/s in a TissueLyser. After brief centrifugation, serially diluted homogenates were spread onto 9cm LB agar plates and incubated at 30°C. The number of colony forming units were quantified after two days.
Host fecundity
We followed the steps as above to rear N2 or pmk-1 nematodes on either P. berkeleyensis or E. coli. We reared L1s on 9cm NGM plates either seeded with P. berkeleyensis or E. coli until L4/young adulthood (∼2 days at 20°C), then picked individual nematodes onto 6cm NGM plates spotted with the respective bacteria to produce offspring, which were then incubated at 20°C. We counted the number of larvae under a microscope three days later, on the same day we measured host mortality for nematodes infected with P. aeruginosa.
Experimental evolution
We passaged P. aeruginosa PA14-GFP under five treatments (Figure 2A): four host treatments and one no host treatment. To start, one individual colony of PA14-GFP was grown overnight in LB broth and spread onto nematode growth medium,65 with subsequent incubation at 30°C for one day. About 1000 nematodes were transferred from their respective rearing plates (described below) onto the P. aeruginosa plates and incubated at 20°C. Nematodes were washed off each plate after one day, rinsed three times with M9 buffer. Ten percent of the M9/nematode mixture were crushed using a BeadBeater, and homogenates were plated onto LB plates. After overnight incubation, we picked 100 colonies into broth to start the next passage. Each treatment consisted of five replicate rearing and P. aeruginosa plates across 14 passages.
Nematodes were kept evolutionarily static (i.e., not evolving) throughout the experiment. N2 and pmk-1 populations were reared as described in the survival and CFU assays with ancestral P. aeruginosa section. L4/young adults were transferred to P. aeruginosa plates as described above. For each passage, eggs were collected from stock nematode populations that were regularly resurrected from -80°C to limit accumulation of de novo mutations in host lineages throughout the experiment.
Mortality and CFU assays with evolved P. aeruginosa
Mortality and CFU assays for evolved populations follow similar protocols as those for ancestral P. aeruginosa (survival and CFU assays with ancestral P. aeruginosa section). Assays were performed in triplicates. For Figures 2B and 2D, we infected N2 nematodes that had been reared on OP50. For Figures 2C and S1B, we infected N2 nematodes reared on P. berkeleyensis. For Figures S1C and S1D, we infected pmk-1 nematodes reared on E. coli or P. berkeleyensis, respectively, and quantified mortality after two days instead of three days due to high mortality of these hosts.
Swimming motility
To measure motility of ancestral and evolved P. aeruginosa, we followed the protocol from Ha et al.66 to inoculate swimming motility plates. We incubated plates at 30°C for one day as this was the temperature NGM plates were incubated before nematodes were put on the pathogen. We used the diameter of bacterial growth on this day as the initial diameter. We then incubated plates at 20°C for three days following the infection timeline for the host mortality assay, then measured the final diameter. The initial diameter was subtracted from the final diameter to obtain the change in swimming diameter.
DNA extraction and sequencing
For pooled samples, we grew 40 individual colonies for each replicate population separately overnight in LB broth, then standardized the OD600 of each individual colony before pooling them into one tube to perform DNA extraction. For single colony samples, we grew individual colonies separately in LB broth overnight, then performed DNA extraction. We extracted genomic DNA using DNeasy Blood and Tissue Kit (Qiagen) following the manufacturer’s instructions. Sample libraries were prepared using the Illumina DNA Prep kit and sequenced on an Illumina NextSeq 2000. Sequence quality was assessed using FastQC (https://www.bioinformatics.babraham.ac.uk/projects/fastqc/) and sequences were trimmed using fastP.67 We sequenced all five replicate populations of each host treatment and three random populations from the no host treatment. For single colony samples, we sequenced three random individual colonies from each population of the WT+PM and IC-PM treatments, and one random individual colony from each population of the other treatments.
The ancestral PA14-GFP individual colony was sequenced using Oxford Nanopore Technologies (ONT) in addition to Illumina for hybrid assembly. Quality control and adapter trimming was performed with bcl2fastq68 and porechop69 using default parameters for Illumina and ONT sequencing, respectively. Hybrid assembly with Illumina and ONT reads was performed with Unicycler,70 and the resulting assembly was annotated using the Bakta annotation pipeline.71 Coverage of mapped reads was calculated using Samtools.72 Each pooled sample had at least 200X coverage. Each single colony sample had at least 60X coverage.
Quantification and statistical analysis
All statistical analyses for phenotypic data and processed genomic data were carried out in R version 4.2.0.73 Normality of data were assessed using histograms, quantile–quantile plots, and Shapiro-Wilk tests. The significance threshold was defined as P < 0.05. Error bars in figures represent standard errors. The sample size for each assay is indicated in figure legends.
Analysis of ancestral pathogen data
Data for mortality of hosts infected with ancestral pathogen were analyzed using a generalized linear mixed mode with a binomial distribution followed by Tukey multiple-comparison tests to determine pairwise differences. Ancestral pathogen CFU data were analysed using a t-test, and host fecundity data were analyzed using an ANOVA.
Analysis of evolved pathogen data
Mortality of pmk-1 reared on P. berkeleyensis data were analyzed using a linear mixed model. Pathogen CFU data in N2 reared on P. berkeleyensis were square-root transformed to meet assumptions of normality and analyzed using a linear mixed model. Mortality for all other hosts and remaining CFU data were analyzed using generalized linear mixed models (with a binomial distribution or Poisson distribution, respectively) followed by Tukey multiple-comparison tests to determine pairwise differences. Motility data were analyzed using a linear mixed model.
Analysis of pooled samples
We called variants with the ancestor as the reference using the Breseq pipeline polymorphism mode with default parameters.74 We tested whether the frequencies of flgE/flgF and fmt mutations were correlated with mortality using Spearman’s rank correlation.
To determine the allele frequency differences between treatments, we pooled together the reads across all replicate populations for each treatment. We then used the Popoolation2 pipeline75 to calculate the exact allele frequency differences and estimated significance using Fisher's Exact Test. For significant loci found in coding regions, we used the Database for Annotation, Visualization and Integrated Discovery (DAVID)76 tool to map each gene to the KEGG pathways.77
To calculate the per SNP FST within each treatment, we used the Popoolation2 pipeline on all pairwise combinations of replicate populations within a treatment. To calculate the per SNP FST across time points, we used the Popoolation2 pipeline to compare each population when at passage seven with the ancestor, between passages fourteen and seven, and between passage fourteen and the ancestor. For both analyses we estimated significance using Fisher's Exact Test. We then counted the number of significant loci after Bonferroni corrections. We compared the loci count between populations within each treatment using a chi-square test of goodness-of-fit, and across time points using linear models or generalized linear models with Poisson distribution followed by Tukey multiple-comparison tests to determine pairwise differences.
Analysis of individual colony samples
We called variants with the ancestor as the reference using the breseq pipeline with default parameters. We used the output from the breseq gdtools COMPARE command to construct phylogenies with PHYLIP dnapars.78 We then used the cophenetic.phylo function of ape79 to calculate pairwise distances between the ancestor and each individual colony. Data were analysed using linear mixed models.
Acknowledgments
We thank Dana Hawley and Levi Morran for feedback on the manuscript as well as members of the King, Read, and Morran labs for insightful discussions. We are grateful to Steve Diggle for advice on P. aeruginosa, Julia Kreiner for advice on population genomics, and Jelly Vanderwoude for advice on hypermutators. We also thank SeqCenter (seqcenter.com) for generation of the high-throughput sequencing data and assembly of the ancestral P. aeruginosa reference genome. K.L.H. was supported by funding from an NSF Postdoctoral Research Fellowship in Biology (1907076) and a Research Publication Grant in Engineering, Medicine, and Science from the American Association of University Women. K.C.K. was funded by a European Research Council Starting Grant (COEVOPRO 802242) as well as an NSERC Canada Excellence Research Chair.
Author contributions
K.L.H. and K.C.K. conceived and designed the study. K.L.H. collected the data and conducted the data analysis, with guidance from T.D.R. and K.C.K. K.L.H. and K.C.K. drafted the article, with critical revisions provided by all authors.
Declaration of interests
The authors declare no competing interests.
Published: March 1, 2024
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.cub.2024.02.015.
Contributor Information
Kim L. Hoang, Email: kim.hoang@emory.edu.
Kayla C. King, Email: kayla.king@ubc.ca.
Supplemental information
References
- 1.Fleming-Davies A.E., Williams P.D., Dhondt A.A., Dobson A.P., Hochachka W.M., Leon A.E., Ley D.H., Osnas E.E., Hawley D.M. Incomplete host immunity favors the evolution of virulence in an emergent pathogen. Science. 2018;359:1030–1033. doi: 10.1126/science.aao2140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hoang K.L., King K.C. Symbiont-mediated immune priming in animals through an evolutionary lens. Microbiology. 2022;168:1–11. doi: 10.1099/mic.0.001181. [DOI] [PubMed] [Google Scholar]
- 3.Montalvo-Katz S., Huang H., Appel M.D., Berg M., Shapira M. Association with soil bacteria enhances p38-dependent infection resistance in Caenorhabditis elegans. Infect. Immun. 2013;81:514–520. doi: 10.1128/IAI.00653-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Milutinović B., Kurtz J. Immune memory in invertebrates. Semin. Immunol. 2016;28:328–342. doi: 10.1016/j.smim.2016.05.004. [DOI] [PubMed] [Google Scholar]
- 5.Leon A.E., Hawley D.M. Host responses to pathogen priming in a natural songbird host. Ecohealth. 2017;14:793–804. doi: 10.1007/s10393-017-1261-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kim Y., Mylonakis E. Caenorhabditis elegans immune conditioning with the probiotic bacterium Lactobacillus acidophilus strain ncfm enhances gram-positive immune responses. Infect. Immun. 2012;80:2500–2508. doi: 10.1128/IAI.06350-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kwong W.K., Mancenido A.L., Moran N.A. Immune system stimulation by the native gut microbiota of honey bees. R. Soc. Open Sci. 2017;4 doi: 10.1098/rsos.170003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Clarke T.B., Davis K.M., Lysenko E.S., Zhou A.Y., Yu Y., Weiser J.N. Recognition of peptidoglycan from the microbiota by Nod1 enhances systemic innate immunity. Nat. Med. 2010;16:228–231. doi: 10.1038/nm.2087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Selosse M.A., Bessis A., Pozo M.J. Microbial priming of plant and animal immunity: symbionts as developmental signals. Trends Microbiol. 2014;22:607–613. doi: 10.1016/j.tim.2014.07.003. [DOI] [PubMed] [Google Scholar]
- 10.Gabrieli P., Caccia S., Varotto-Boccazzi I., Arnoldi I., Barbieri G., Comandatore F., Epis S. Mosquito trilogy: microbiota, immunity and pathogens, and their implications for the control of disease transmission. Front. Microbiol. 2021;12 doi: 10.3389/fmicb.2021.630438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Khosravi A., Mazmanian S.K. Disruption of the gut microbiome as a risk factor for microbial infections. Curr. Opin. Microbiol. 2013;16:221–227. doi: 10.1016/j.mib.2013.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ford S.A., Kao D., Williams D., King K.C. Microbe-mediated host defence drives the evolution of reduced pathogen virulence. Nat. Commun. 2016;7 doi: 10.1038/ncomms13430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nelson P., May G. Defensive symbiosis and the evolution of virulence. Am. Nat. 2020;196:333–343. doi: 10.1086/709962. [DOI] [PubMed] [Google Scholar]
- 14.Horak R.D., Leonard S.P., Moran N.A. Symbionts shape host innate immunity in honeybees. Proc. Biol. Sci. 2020;287 doi: 10.1098/rspb.2020.1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Corby-Harris V., Snyder L., Meador C.A.D., Naldo R., Mott B., Anderson K.E. Parasaccharibacter apium, gen. Nov., sp. Nov., improves Honey Bee (Hymenoptera: Apidae) resistance to Nosema. J. Econ. Entomol. 2016;109:537–543. doi: 10.1093/jee/tow012. [DOI] [PubMed] [Google Scholar]
- 16.Irazoqui J.E., Troemel E.R., Feinbaum R.L., Luhachack L.G., Cezairliyan B.O., Ausubel F.M. Distinct pathogenesis and host responses during infection of C. elegans by P. aeruginosa and S. aureus. PLoS Pathog. 2010;6 doi: 10.1371/journal.ppat.1000982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dirksen P., Marsh S.A., Braker I., Heitland N., Wagner S., Nakad R., Mader S., Petersen C., Kowallik V., Rosenstiel P., et al. The native microbiome of the nematode Caenorhabditis elegans: gateway to a new host-microbiome model. BMC Biol. 2016;14 doi: 10.1186/s12915-016-0258-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dirksen P., Assié A., Zimmermann J., Zhang F., Tietje A.M., Marsh S.A., Félix M.A., Shapira M., Kaleta C., Schulenburg H., et al. CeMbio - The Caenorhabditis elegans microbiome resource. G3 (Bethesda) 2020;10:3025–3039. doi: 10.1534/g3.120.401309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Grace A., Sahu R., Owen D.R., Dennis V.A. Pseudomonas aeruginosa reference strains PAO1 and PA14: a genomic, phenotypic, and therapeutic review. Front. Microbiol. 2022;13 doi: 10.3389/fmicb.2022.1023523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Engelmann I., Pujol N. Innate immunity in C. elegans. Adv. Exp. Med. Biol. 2010;708:105–121. doi: 10.1007/978-1-4419-8059-5_6. [DOI] [PubMed] [Google Scholar]
- 21.Vorburger C., Ganesanandamoorthy P., Kwiatkowski M. Comparing constitutive and induced costs of symbiont-conferred resistance to parasitoids in aphids. Ecol. Evol. 2013;3:706–713. doi: 10.1002/ece3.491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Łukasik P., Guo H., Van Asch M., Ferrari J., Godfray H.C.J. Protection against a fungal pathogen conferred by the aphid facultative endosymbionts Rickettsia and Spiroplasma is expressed in multiple host genotypes and species and is not influenced by co-infection with another symbiont. J. Evol. Biol. 2013;26:2654–2661. doi: 10.1111/jeb.12260. [DOI] [PubMed] [Google Scholar]
- 23.Read A.F., Baigent S.J., Powers C., Kgosana L.B., Blackwell L., Smith L.P., Kennedy D.A., Walkden-Brown S.W., Nair V.K. Imperfect vaccination can enhance the transmission of highly virulent pathogens. PLoS Biol. 2015;13 doi: 10.1371/journal.pbio.1002198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pike V.L., Stevens E.J., Griffin A.S., King K.C. Within- and between-host dynamics of producer and non-producer pathogens. Parasitology. 2023;150:805–812. doi: 10.1017/S0031182023000586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vorburger C., Perlman S.J. The role of defensive symbionts in host–parasite coevolution. Biol. Rev. Camb. Philos. Soc. 2018;93:1747–1764. doi: 10.1111/brv.12417. [DOI] [PubMed] [Google Scholar]
- 26.Smith C.A., Ashby B. Tolerance-conferring defensive symbionts and the evolution of parasite virulence. Evol. Lett. 2023;7:262–272. doi: 10.1093/evlett/qrad015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tardy L., Giraudeau M., Hill G.E., McGraw K.J., Bonneaud C. Contrasting evolution of virulence and replication rate in an emerging bacterial pathogen. Proc. Natl. Acad. Sci. USA. 2019;116:16927–16932. doi: 10.1073/pnas.1901556116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ekroth A.K.E., Gerth M., Stevens E.J., Ford S.A., King K.C. Host genotype and genetic diversity shape the evolution of a novel bacterial infection. ISME J. 2021;15:2146–2157. doi: 10.1038/s41396-021-00911-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chen H., Bright R.A., Subbarao K., Smith C., Cox N.J., Katz J.M., Matsuoka Y. Polygenic virulence factors involved in pathogenesis of 1997 Hong Kong H5N1 influenza viruses in mice. Virus Res. 2007;128:159–163. doi: 10.1016/j.virusres.2007.04.017. [DOI] [PubMed] [Google Scholar]
- 30.Le Clec’h W., Chevalier F.D., McDew-White M., Menon V., Arya G.A., Anderson T.J.C. Genetic architecture of transmission stage production and virulence in schistosome parasites. Virulence. 2021;12:1508–1526. doi: 10.1080/21505594.2021.1932183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Caseys C., Shi G., Soltis N., Gwinner R., Corwin J., Atwell S., Kliebenstein D.J. Quantitative interactions: the disease outcome of Botrytis cinerea across the plant kingdom. G3 (Bethesda) 2021;11 doi: 10.1093/g3journal/jkab175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sharp C., Foster K.R., Genna V., Vidossich P., Somarowthu S., Pyle A.M., De Vivo M., Marcia M. Author Correction: Visualizing group II intron dynamics between the first and second steps of splicing. Nat. Commun. 2022;13 doi: 10.1038/s41467-021-27699-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rossez Y., Wolfson E.B., Holmes A., Gally D.L., Holden N.J. Bacterial flagella: twist and stick, or dodge across the kingdoms. PLoS Pathog. 2015;11 doi: 10.1371/journal.ppat.1004483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Feinbaum R.L., Urbach J.M., Liberati N.T., Djonovic S., Adonizio A., Carvunis A.R., Ausubel F.M. Genome-wide identification of Pseudomonas aeruginosa virulence-related genes using a Caenorhabditis elegans infection model. PLoS Pathog. 2012;8 doi: 10.1371/journal.ppat.1002813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Duan Q., Zhou M., Zhu L., Zhu G. Flagella and bacterial pathogenicity. J. Basic Microbiol. 2013;53:1–8. doi: 10.1002/jobm.201100335. [DOI] [PubMed] [Google Scholar]
- 36.Marko V.A., Kilmury S.L.N., MacNeil L.T., Burrows L.L. Pseudomonas aeruginosa type IV minor pilins and PilY1 regulate virulence by modulating FimS-AlgR activity. PLoS Pathog. 2018;14 doi: 10.1371/journal.ppat.1007074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ruiz-Bedoya T., Wang P.W., Desveaux D., Guttman D.S. Cooperative virulence via the collective action of secreted pathogen effectors. Nat. Microbiol. 2023;8:640–650. doi: 10.1038/s41564-023-01328-8. [DOI] [PubMed] [Google Scholar]
- 38.Guillon J.M., Mechulam Y., Schmitter J.M., Blanquet S., Fayat G. Disruption of the gene for Met-tRNA(f)/(Met) formyltransferase severely impairs growth of Escherichia coli. J. Bacteriol. 1992;174:4294–4301. doi: 10.1128/jb.174.13.4294-4301.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bailey S.F., Rodrigue N., Kassen R. The effect of selection environment on the probability of parallel evolution. Mol. Biol. Evol. 2015;32:1436–1448. doi: 10.1093/molbev/msv033. [DOI] [PubMed] [Google Scholar]
- 40.Frickel J., Feulner P.G.D., Karakoc E., Becks L. Population size changes and selection drive patterns of parallel evolution in a host-virus system. Nat. Commun. 2018;9 doi: 10.1038/s41467-018-03990-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Barroso-Batista J., Demengeot J., Gordo I. Adaptive immunity increases the pace and predictability of evolutionary change in commensal gut bacteria. Nat. Commun. 2015;6 doi: 10.1038/ncomms9945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Klemm E.J., Gkrania-Klotsas E., Hadfield J., Forbester J.L., Harris S.R., Hale C., Heath J.N., Wileman T., Clare S., Kane L., et al. Emergence of host-adapted Salmonella Enteritidis through rapid evolution in an immunocompromised host. Nat. Microbiol. 2016;1:1–6. doi: 10.1038/nmicrobiol.2015.23. [DOI] [PubMed] [Google Scholar]
- 43.Launay A., Wu C.J., Dulanto Chiang A., Youn J.H., Khil P.P., Dekker J.P. In vivo evolution of an emerging zoonotic bacterial pathogen in an immunocompromised human host. Nat. Commun. 2021;12 doi: 10.1038/s41467-021-24668-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kemp S.A., Collier D.A., Datir R.P., Ferreira I.A.T.M., Gayed S., Jahun A., Hosmillo M., Rees-Spear C., Mlcochova P., Lumb I.U., et al. SARS-CoV-2 evolution during treatment of chronic infection. Nature. 2021;592:277–282. doi: 10.1038/s41586-021-03291-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Day T., Kennedy D.A., Read A.F., Gandon S. Pathogen evolution during vaccination campaigns. PLoS Biol. 2022;20 doi: 10.1371/journal.pbio.3001804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Winsor G.L., Griffiths E.J., Lo R., Dhillon B.K., Shay J.A., Brinkman F.S. Enhanced annotations and features for comparing thousands of Pseudomonas genomes in the Pseudomonas genome database. Nucleic Acids Res. 2016;44:D646–D653. doi: 10.1093/nar/gkv1227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mongelli V., Lequime S., Kousathanas A., Gausson V., Blanc H., Nigg J., Quintana-Murci L., Elena S.F., Saleh M.C. Innate immune pathways act synergistically to constrain RNA virus evolution in Drosophila melanogaster. Nat. Ecol. Evol. 2022;6:565–578. doi: 10.1038/s41559-022-01697-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jansen G., Crummenerl L.L., Gilbert F., Mohr T., Pfefferkorn R., Thänert R., Rosenstiel P., Schulenburg H. Evolutionary transition from pathogenicity to commensalism: Global regulator mutations mediate fitness gains through virulence attenuation. Mol. Biol. Evol. 2015;32:2883–2896. doi: 10.1093/molbev/msv160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Råberg L., De Roode J.C., Bell A.S., Stamou P., Gray D., Read A.F. The role of immune-mediated apparent competition in genetically diverse malaria infections. Am. Nat. 2006;168:41–53. doi: 10.1086/505160. [DOI] [PubMed] [Google Scholar]
- 50.Cruickshank T., Wade M.J. Microevolutionary support for a developmental hourglass: gene expression patterns shape sequence variation and divergence in Drosophila. Evol. Dev. 2008;10:583–590. doi: 10.1111/j.1525-142X.2008.00273.x. [DOI] [PubMed] [Google Scholar]
- 51.Runemark A., Brydegaard M., Svensson E.I. Does relaxed predation drive phenotypic divergence among insular populations? J. Evol. Biol. 2014;27:1676–1690. doi: 10.1111/jeb.12421. [DOI] [PubMed] [Google Scholar]
- 52.MacPherson A., Nuismer S.L. The probability of parallel genetic evolution from standing genetic variation. J. Evol. Biol. 2017;30:326–337. doi: 10.1111/jeb.13006. [DOI] [PubMed] [Google Scholar]
- 53.Scribner M.R., Santos-Lopez A., Marshall C.W., Deitrick C., Coopera V.S., Hogan D.A. Parallel evolution of tobramycin resistance across species and environments. mBio. 2020;11 doi: 10.1128/mBio.00932-20. e00932–e00920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.King K.C. Defensive symbionts. Curr. Biol. 2019;29:R78–R80. doi: 10.1016/j.cub.2018.11.028. [DOI] [PubMed] [Google Scholar]
- 55.Kaltenpoth M., Engl T. Defensive microbial symbionts in Hymenoptera. Funct. Ecol. 2014;28:315–327. [Google Scholar]
- 56.Barclay V.C., Sim D., Chan B.H.K., Nell L.A., Rabaa M.A., Bell A.S., Anders R.F., Read A.F. The evolutionary consequences of blood-stage vaccination on the rodent malaria Plasmodium chabaudi. PLoS Biol. 2012;10 doi: 10.1371/journal.pbio.1001368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Duar R.M., Lin X.B., Zheng J., Martino M.E., Grenier T., Pérez-Muñoz M.E., Leulier F., Gänzle M., Walter J. Lifestyles in transition: evolution and natural history of the genus Lactobacillus. FEMS Microbiol. Rev. 2017;41:S27–S48. doi: 10.1093/femsre/fux030. [DOI] [PubMed] [Google Scholar]
- 58.McKenzie V.J., Kueneman J.G., Harris R.N. Probiotics as a tool for disease mitigation in wildlife: insights from food production and medicine. Ann. N. Y. Acad. Sci. 2018;1429:18–30. doi: 10.1111/nyas.13617. [DOI] [PubMed] [Google Scholar]
- 59.Hoyt J.R., Langwig K.E., White J.P., Kaarakka H.M., Redell J.A., Parise K.L., Frick W.F., Foster J.T., Kilpatrick A.M. Field trial of a probiotic bacteria to protect bats from white-nose syndrome. Sci. Rep. 2019;9 doi: 10.1038/s41598-019-45453-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bletz M.C., Loudon A.H., Becker M.H., Bell S.C., Woodhams D.C., Minbiole K.P.C., Harris R.N. Mitigating amphibian chytridiomycosis with bioaugmentation: characteristics of effective probiotics and strategies for their selection and use. Ecol. Lett. 2013;16:807–820. doi: 10.1111/ele.12099. [DOI] [PubMed] [Google Scholar]
- 61.Cross M.L. Microbes versus microbes: immune signals generated by probiotic lactobacilli and their role in protection against microbial pathogens. FEMS Immunol. Med. Microbiol. 2002;34:245–253. doi: 10.1111/j.1574-695X.2002.tb00632.x. [DOI] [PubMed] [Google Scholar]
- 62.Ouwehand A.C., Forssten S., Hibberd A.A., Lyra A., Stahl B. Probiotic approach to prevent antibiotic resistance. Ann. Med. 2016;48:246–255. doi: 10.3109/07853890.2016.1161232. [DOI] [PubMed] [Google Scholar]
- 63.Stiernagle T. Maintenance of C. elegans. WormBook. 2006;2006:1–11. doi: 10.1895/wormbook.1.101.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Vega N.M., Gore J. Stochastic assembly produces heterogeneous communities in the Caenorhabditis elegans intestine. PLoS Biol. 2017;15 doi: 10.1371/journal.pbio.2000633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Cold Spring Harbor . Cold Spring Harb. Protoc.; 2008. Nematode growth medium (NGM) [DOI] [Google Scholar]
- 66.Ha D., Kuchma S.L., Toole G.A.O. Plate-based assay for swimming motility in Pseudomonas aeruginosa. Methods Mol. Biol. 2014;1149:59–65. doi: 10.1007/978-1-4939-0473-0_7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Chen S., Zhou Y., Chen Y., Gu J. Fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics. 2018;34:i884–i890. doi: 10.1093/bioinformatics/bty560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Illumina . 2013. bcl2fastq: a proprietary Illumina software for the conversion of Bcl files to basecalls. [Google Scholar]
- 69.Wick R. 2018. Porechop: an open source software for the QC and adapter trimming of ONT technologies.https://github.com/rrwick/Porechop [Google Scholar]
- 70.Wick R.R., Judd L.M., Gorrie C.L., Holt K.E. Unicycler: Resolving bacterial genome assemblies from short and long sequencing reads. PLoS Comput. Biol. 2017;13:1–22. doi: 10.1371/journal.pcbi.1005595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Schwengers O., Jelonek L., Dieckmann M.A., Beyvers S., Blom J., Goesmann A. Bakta: Rapid and standardized annotation of bacterial genomes via alignment-free sequence identification. Microb. Genomics. 2021;7 doi: 10.1099/mgen.0.000685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N., Marth G., Abecasis G., Durbin R. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.R Core Team . 2021. R: A Language and Environment for Statistical Computing.https://www.r-project.org/ [Google Scholar]
- 74.Deatherage D.E., Barrick J.E. Identification of mutations in laboratory-evolved microbes from next-generation sequencing data using breseq. Methods Protoc. 2014;1151:165–188. doi: 10.1007/978-1-4939-0554-6_12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kofler R., Pandey R.V., Schlötterer C. PoPoolation2: Identifying differentiation between populations using sequencing of pooled DNA samples (Pool-Seq) Bioinformatics. 2011;27:3435–3436. doi: 10.1093/bioinformatics/btr589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Sherman B.T., Hao M., Qiu J., Jiao X., Baseler M.W., Lane H.C., Imamichi T., Chang W. DAVID: a web server for functional enrichment analysis and functional annotation of gene lists (2021 update) Nucleic Acids Res. 2022;50:W216–W221. doi: 10.1093/nar/gkac194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kanehisa M., Goto S. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res. 2000;28:27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Felsenstein J. 1993. Phylogeny Inference Package (PHYLIP). Version 3.5. University of Washington, Seattle. [Google Scholar]
- 79.Paradis E., Schliep K. Ape 5.0: An environment for modern phylogenetics and evolutionary analyses in R. Bioinformatics. 2019;35:526–528. doi: 10.1093/bioinformatics/bty633. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
-
•
Raw sequences were deposited in the NCBI Sequence Read Archive under BioProject:PRJNA998467. Phenotypic data have been published in Mendeley Data:https://doi.org/10.17632/xz9t9gjtw6.1.
-
•
R code used for the analyses in the paper is available on request.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.