Abstract
Recombinant forms of human immunodeficiency virus type 1 (HIV-1) have been shown to be of major importance in the global AIDS pandemic. Viral RNA dimer formation mediated by the dimerization initiation sequence (DIS) is believed to be essential for viral genomic RNA packaging and therefore for RNA recombination. Here, we demonstrate that HIV-1 recombination and replication are not restricted by variant DIS loop sequences. Three DIS loop forms found among HIV-1 isolates, DIS (CG), DIS (TA), and DIS (TG), when introduced into deletion mutants of HIV-1 recombined efficiently, and the progeny virions replicated with comparable kinetics. A fourth DIS loop form, containing an artificial AAAAAA sequence disrupting the putative DIS loop-loop interactions [DIS (A6)], supported efficient recombination with DIS loop variants; however, DIS (A6) progeny virions exhibited a modest replication disadvantage in mixed cultures. Our studies indicate that the nonhomologous DIS sequences found in different HIV-1 subtypes are not a primary obstacle to intersubtype recombination.
Homologous recombination is an important process in the retrovirus life cycle (15, 23–25, 27, 32, 56). Retrovirus genomes are packaged into virus particles as RNA dimers (3, 4, 8, 14, 41). During replication, the viral reverse transcriptase (RT) can switch templates, creating a recombinant genome from the two RNA molecules (13, 55, 56). Recombination may function in the rapid generation of viral diversity and in the recovery from genomic damage caused by an error-prone RT (24).
Evidence for the direct involvement of recombination in the generation of diversity has emerged from analysis of human immunodeficiency virus type 1 (HIV-1) isolates from multiple geographic locales (48). At least eight genetic subtypes of HIV-1 have been identified to date (42). Some HIV-1 strains of major importance in the AIDS pandemic apparently arose by recombination between two of the subtypes, including the subtype E virus that initiated a major new focus of infection in Southeast Asia (10, 19). However, only some pairs of HIV-1 subtypes have been observed to recombine (39). Factors that influence the frequency of recombination between subtypes are important to define, as they may influence which variants arise and spread in a future pandemic.
Formation of RNA dimers within the virion is essential for the high level of recombination exhibited by retroviruses (24–26, 53, 60, 62). Electron microscopy studies first identified an RNA dimerization linkage structure (DLS) near the 5′ end of the genome (4, 41). The DLS is a complex structure containing elements required for the efficient packaging of viral genomic RNA into virions and dimerization of the RNA genome. While sequences responsible for encapsidation have been identified and characterized for many retroviruses (31, 36, 51, 59), attempts to identify sequence requirements for RNA dimerization were unsuccessful until the development of in vitro dimerization systems (2, 18, 37, 54). Dimerization of purified subgenomic RNA encompassing the 5′ untranslated region and gag gene can be induced in vitro (2, 5, 7, 11, 16, 17, 30, 37, 44, 49, 52, 54, 61). The viral nucleocapsid protein can greatly facilitate RNA dimer formation, probably by activating base pair rearrangements (16, 17, 57). However, under appropriate conditions, spontaneous RNA dimerization can occur in the absence of viral and cellular proteins (2, 37, 40, 54, 61).
Sequences responsible for in vitro HIV-1 RNA dimerization were identified outside of the previously defined DLS (38, 52). The newly identified dimerization initiation site (DIS), located upstream of the major splice donor site in the gag leader region, adopts a stem-loop structure (21). Dimerization is thought to proceed through the interaction of self-complementary DIS loop palindromic sequences. Hybridization of the DIS loops does not propagate to the DIS stem to form an extended duplex (45) as previously proposed (30, 40, 52). Instead, a “loop-loop kissing” complex is maintained, and the dimer is stabilized by other, more distant interactions (38, 44). Some of the main evidence supporting the DIS palindrome interaction in dimerization comes from mutagenesis studies, which show that disruption of base pairing in the loop abolishes RNA dimerization in vitro (44). Several different DIS loop sequences were shown to be functional in the formation of homodimers, while RNA molecules with several combinations of heterologous DIS were unable to dimerize.
Here we report that at least three different DIS loop palindromic sequences occur in HIV-1. An examination of 75 viral isolates showed that the DIS sequence of subtypes B and D was different from that of subtypes A, C, E, F, and G and group O. By construction of deletion mutants of an infectious molecular clone and mutagenesis of the DIS, we evaluated the effect of DIS variants on virus replication and determined whether viruses bearing the nonhomologous DIS found in the different HIV subtypes can recombine. Alterations in the DIS loop palindrome had little effect on replication, and DIS incompatibility did not restrict recombination. We conclude that the different DIS sequences found in HIV-1 subtypes are not a major impediment to intersubtype recombination.
MATERIALS AND METHODS
Restriction analysis of DIS loop sequences in multiple HIV-1 subtypes.
HIV-1 isolates represented a broad geographic distribution and were selected based on their previously sequenced gag and/or envelope (env) genes. They were from subtypes A, B, C, D, F, G, and H, as follows: subtype A, VI32, VI57, VI59, VI310, VI415, K7, K29, K88, K89, K98, K112, CI4, CI20, CI51, CI59, LBV23-10, and DJ258; subtype B, BK132, BZ167, BZ190, PH136, and PH153; subtype C, ZAM18, ZAM20, SM145, VI313, and UG268; subtype D, VI203, K31, G109, SE365, UG270, and UG274; subtype F, BZ126, BZ162, BZ163, VI69, and VI174; and subtype G, LBV2-7 (33, 34). DNA from primary or low-passage-number virus cultures on donor peripheral blood mononuclear cells was used as the template for PCR amplification of a 163-bp segment in the gag leader region. Primers 5′-CTCTCCTTCTAGCCTCCGCTAGTC and 5′-AAATCTCTAGCAGTGGCGCCC (complementary to nucleotides [nt] 784 to 761 and 622 to 642 of HIVMN, respectively) were used with Taq polymerase under standard reaction conditions; the thermocycle routine was 30 cycles of 95°C denaturation for 30 s, 55°C annealing for 30 s, and 72°C extension for 30 s. Products were digested with restriction endonuclease ApaLI, BssHII, or FspI (New England Biolabs, Beverly, Mass.) as directed by the vendor and evaluated on 3% NuSieve–1% agarose gels (FMC BioProducts, Rockland, Maine) after staining with ethidium bromide.
Construction of HIV-1 mutants.
The infectious molecular clone NL4-3 (1) was used to construct DIS, polymerase gene (pol), and env mutants employed in this study. To generate DIS mutants, a 958-bp SacI/SphI fragment was subcloned into pUC31. The unique BssHII site, corresponding to the GCGCGC palindromic loop sequence of the DIS hairpin (nt 257 to 262), was converted to GTGCAC, GTGCGC, or AAAAAA by double-stranded site-directed mutagenesis using a Chameleon kit (Stratagene, Inc., La Jolla, Calif.) and the following complementary primers containing substitution mutations (underlined): 5′CGGCTTGCTGAAGTGCACACGGCAAGAGGC, 5′GCCTCTTGCCGTGTGCACTTCAGCAAGCCC, 5′CGGCTTGCTGAAGTGCGCACGGCAAGAGGC, 5′GCCTCTTGCCGTGCGCACTTCAGCAAGCCG, 5′CGGCTTGCTGAAAAAAAAACGGCAAGAGGC, and 5′GCCTCTTGCCGTTTTTTTTTCAGCAAGCCG.
DIS mutants were initially screened by restriction enzyme digestion with BssHII (cleaves GCGCGC), ApaLI (cleaves GTGCAC), and FspI (cleavage site overlaps by 5 nt with the sequence GTGCGC). The sequence of the entire insert was verified by DNA sequence analysis prior to subcloning into wild-type NL4-3 or pol or env deletion mutants of NL4-3. pol deletion mutants were generated by TthIII and AgeI cleavage of a 4.3-kb SphI/SalI fragment of NL4-3 subcloned in pUC31. The cut plasmid DNA was treated with Klenow polymerase in the presence of deoxynucleoside triphosphates, and the DNA was transformed into Escherichia coli after ligation of the blunt ends. env deletion mutants were constructed by BglII cleavage and religation of the 3.1-kb EcoRI/XhoI fragment of NL4-3 subcloned into pUC31. The 404-bp pol and 580-bp env deletions were verified by restriction enzyme digestion and DNA sequence analysis prior to subcloning into plasmid NL4-3 DIS variants. In all, 12 constructions were generated, with the four DIS variants in NL4-3 Δpol, NL4-3 Δenv, and the wild type.
Cell culture conditions.
SupT1 cells were cultured at 37°C with 5% CO2 in RPMI 1640 medium containing 10% fetal calf serum, 2 mM glutamine, 100 mM pyruvate, penicillin (50 U/ml), and streptomycin (50 mg/ml). Human 293 cells were grown at 37°C and 5% CO2 in Dulbecco modified Eagle medium containing 10% fetal calf serum, 2 mM glutamine, 100 mM pyruvate, penicillin (50 U/ml), and streptomycin (50 mg/ml). MAGI cells were obtained from the NIH AIDS Research and Reference Reagent Program and cultured as described previously (29).
Preparation of viral stocks and infection procedure.
Infectious virus stocks of NL4-3 DIS variants were generated by transient transfection of 293 cells with proviral DNA by the Ca2(PO4)2 DNA precipitation method (47). Heterozygous virions containing combinations of NL4-3 Δpol and NL4-3 Δenv and the corresponding DIS mutants were generated by cotransfection of the parental proviruses. Virus stocks were treated with DNase I (Boehringer Mannheim) for 4 h prior to harvesting to eliminate plasmid DNA clones of proviral genome contaminating the preparation. Virus preparations were analyzed for p24gag content by enzyme-linked immunosorbent assay (Coulter), and titers were determined by a single-cycle infection assay (29). SupT1 cells were exposed to 100 or 1,000 infectious doses of virus at a multiplicity of infection (MOI) ranging from 0.0001 to 0.001. After 4 h at 37°C, the cells were washed three times with phosphate-buffered saline and maintained in RPMI complete medium at a concentration of 0.3 × 106 to 0.5 × 106/ml. Cells were fed every 3 days and split if necessary. At designated time points, cleared supernatant was analyzed for p24gag content and cells were harvested for proviral DNA analysis. In some experiments, proviral DNA was purified from transfected cells at the time of harvest of the virus stock to evaluate the level of DNA recombination occurring. DNA from Hirt supernatants (22) was analyzed by PCR amplification and Southern blot hybridization to detect parental and recombinant proviral forms as described below.
DIS loop sequence analysis in infectious recombinant provirus.
The DIS loop sequence was analyzed as the recombinant virus spread in culture. At designated time points, the DIS stem-loop and flanking sequences were amplified from infected cell lysates by using two primers: upstream (sense) primer DIS1 (5′- AAATCTCTAGCAGTGGCGCCCGAACAG) and downstream (antisense) primer DIS2 (5′-CTCTCCTTCTAGCCTCCGCTAGTC). A 165-bp PCR fragment was generated after 30 cycles of standard PCR amplification. A 10-μl sample of the PCR mixture was subjected to digestion with BssHII, ApaLI, or FspI to identify the DIS phenotype. The PCR-amplified products were also ligated into the TA cloning vector pCRII (Invitrogen) and sequenced with the universal T7 and SP6 primers and a Taq dye primer cycle sequencing kit (Applied Biosystems) on an Applied Biosystems 373A DNA sequencer.
Analysis of recombinant provirus formation.
The rate of homologous recombination was determined during a single cycle of replication. SupT1 cells were exposed to 1,000 MAGI infectious doses of heterozygous virus at an MOI of 0.001. Immediately after infection, an aliquot of cells was harvested and saved for contaminating proviral DNA analysis. After 4 h at 37°C, the cells were washed three times with phosphate-buffered saline and maintained for 20 to 24 h in RPMI complete medium at a concentration of 0.3 × 106 to 0.5 × 106/ml. Recombinant HIV proviral DNA sequences were detected by using a modified version of the long PCR technique employed for subcloning HIV-1 provirus as previously described (50). Briefly, pellets containing 106 cells were lysed in a solution containing 10 mM Tris-HCl (pH 7.5), 2.5 mM MgCl2, 0.45% Triton X-100, 0.45% Tween 20, and 0.12 mg of proteinase K per ml. Wild-type and mutant proviral DNA sequences were amplified from crude lysates by using two primers which flank the deletions in the pol and env genes: Rec1 (sense; 5′-CACCAGGGATTAGATATCAGTACAATGTGCTTCCAC) and Rec2 (antisense; 5′-CACCACTCTTCTCTTTGCCTTGGTGGGTGCTACTCC) (see fig. 3A). The PCR mixture contained 250 nM primers Rec1 and Rec2, 350 μM deoxynucleoside triphosphate, 1.75 mM MgCl2, 50 mM Tris-HCl (pH 9.2), 16 mM (NH4)2SO4, and 3.5 U of polymerase (Boehringer Mannheim). PCRs consisted of 10 cycles of 94°C denaturing for 10 s, 55°C annealing for 30 s, and 68°C extension for 4 min and 24 cycles of 94°C denaturing for 10 s, 55°C annealing for 30 s, and 68°C extension for 4 min, plus an additional 20 s added at each extension cycle. A final extension at 72°C for 10 min was used to complete the reaction. PCR products were separated by agarose gel electrophoresis, transferred onto a Gene Screen, and immobilized, and proviral sequences were identified by hybridization with 5′-end-labeled oligonucleotides. The Std* oligonucleotide (sense; 5′-GCAGGGGAAAGAATAGTAGACATAATAGCAACAGAC) detected all forms of parental and recombinant provirus, while the Pol* (sense; 5′-TCCATCCTGATAAATGGACAGTACAGCCTATAGTGC) and ENV* (antisense; 5′-TTGTAACAAATGCTCTCCCTGGTCCCCTCTGGAT AC) oligonucleotides distinguished env- and pol-containing provirus, respectively. The relative amounts of parental and recombinant provirus were quantitated with a Molecular Dynamics PhosphorImager. To ensure that the amplification and hybridization were in the linear range, reactions with diluted lysates were performed. The ratio of recombinant provirus (full-length and double-deleted provirus) to all proviral forms (NL4-3 Δpol, NL4-3 Δenv, full-length, and double-deleted provirus) gave the frequency of recombination.
RNA secondary-structure analysis.
RNA structure predictions were performed with the MFold program, version 7.2 (63).
RESULTS
DIS loop sequences vary among HIV-1 isolates.
Secondary-structure interactions in the gag leader region of HIV-1 RNA produce a hairpin loop containing a six-base palindrome where the two strands of RNA hybridize to form a dimer. The six-base palindromic sequence of the DIS loop was found to be variable among sequenced HIV-1 isolates, with three forms identified to date (Fig. 1). Variation occurred at the second and fifth positions of the palindrome, which were TA, TG, or CG. Each variant corresponded to or largely overlapped the recognition sequence of an available restriction endonuclease (Fig. 1), which permitted a rapid survey of additional isolates. Genomic segments spanning the DIS and predicted to contain a single cleavage site for either ApaLI, BssHII, or FspI were PCR amplified and digested. The DIS loops of 39 isolates representing HIV-1 subtypes A, B, C, D, F, and G were determined, and when these sequences were combined with available sequences and with our unpublished sequences, a survey of 75 isolates, with at least three from each of subtypes A through G and two from group O, was completed (Table 1).
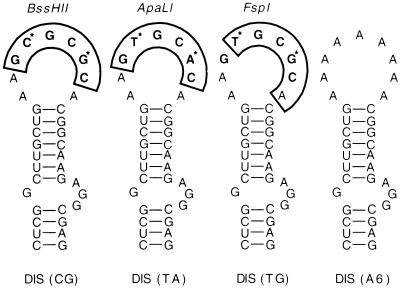
FIG. 1.
Predicted DIS RNA stem-loop structures of NL4-3 and NL4-3 DIS variants. The DIS mutants were generated by site-directed mutagenesis; for details, see Materials and Methods. The MFold program predicts the hairpin structures of all of the DIS variants; thus, alteration of the loop sequence minimally impacts stem structure. The palindromic motifs in the loop are in boldface, and restriction enzymes with corresponding cleavage sites (boxed) are shown above the hairpin loop. Variations at the second and fifth positions of the palindrome occurring in HIV-1 subtypes are marked (*). Phylogenetic association of DIS RNA elements of different HIV-1 strains is given in Table 1.
TABLE 1.
Subtype-specific variation in the HIV-1 DIS
| HIV-1 subtype or group | No. of isolates examined
|
No. with indicated DIS
|
|||
|---|---|---|---|---|---|
| By sequencing | By restriction analysisb | GTGCAC, DIS (TA) | GCGCGC, DIS (CG) | GTGCGC, DIS (TG) | |
| Subtype | |||||
| A | 2 | 18 | 20 | 0 | 0 |
| B | 17 | 5 | 0 | 22 | 0 |
| C | 2 | 5 | 7 | 0 | 0 |
| D | 3 | 5 | 0 | 7 | 1 |
| E | 3 | 0 | 3 | 0 | 0 |
| F | 0 | 5 | 5 | 0 | 0 |
| G | 3 | 1 | 3 | 0 | 1 |
| Othera | 4 | 0 | 4 | 0 | 0 |
| Group O | 2 | 0 | 2 | 0 | 0 |
A/D and A/C intersubtype recombinants.
Restriction endonucleases ApaLI, BssHII, and FspI were used to distinguish DIS sequences as described in Materials and Methods.
The DIS variants were largely, if not entirely, subtype specific. Subtypes B and D harbored the DIS (CG) form, while subtype A, C, E, F, and G and group O isolates had DIS (TA). Recombinants between subtypes A and C or A and D also had DIS (TA). An apparently rare form, DIS (TG), was found in one subtype D and one subtype G isolate.
Based on the in vitro RNA dimerization data, formation of dimers from RNA genomes of different HIV-1 subtypes, a process thought essential in the generation of intersubtype recombinant HIV-1, would be expected to proceed more efficiently when the two subtypes have the same DIS loop sequence; i.e., subtype B would be expected to dimerize more readily with subtype D than with other subtypes, and so forth. The DIS (TG) form would not be expected to support homologous RNA dimer formation with similar efficiency, as it is not a palindrome. However, the HIV-1 DIS sequence analysis suggested that either recombination between HIV-1 genomes with incompatible DIS sequences can occur or DIS sequences such as DIS (TG) facilitate recombination by dimerizing with either DIS (CG) or DIS (TA). Thus, we constructed an experimental system to directly evaluate the effect of DIS sequences on the replication rate and recombination frequency of an HIV-1 isolate in vitro. We used the three naturally occurring DIS variants and a DIS loop with six A residues that should be unable to support RNA dimer formation (Fig. 1).
Multiple DIS variants support HIV-1 replication in vitro.
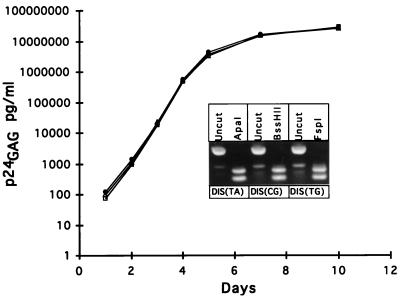
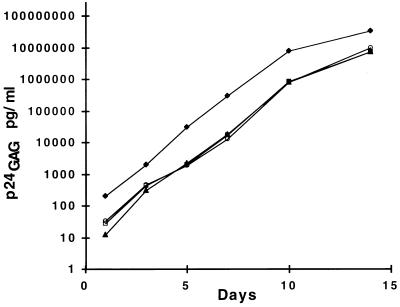
Viral stocks of the infectious molecular clone NL4-3 bearing four different DIS sequences were prepared by transfection of 293 cells and were titrated on MAGI cells. SupT1 cells were infected at an MOI of 0.001 with equivalent infectious units, and virus spread was monitored by measurement of virus-associated p24gag antigen in the cultures. Figure 2 is a representation of two independent experiments. The replication kinetics were virtually identical for viruses bearing the DIS (TA), DIS (CG), and DIS (TG) sequences. Restriction endonuclease digestion of a DNA fragment containing the DIS and PCR amplified by using DNA from the viral cultures harvested at day 10 was used to determine whether there was a measurable accumulation of mutations at the DIS loop; none were observed (Fig. 2, inset). Thus, the absence of a palindromic sequence [as in the DIS (TG) variant] had no appreciable effect on the replication kinetics of NL4-3 in SupT1 cells. No significant selection pressure for reversion of the DIS (TG) sequences was observed over a 10-day interval in culture.
FIG. 2.
Replication kinetics of NL4-3 containing variant DIS. SupT1 cells were infected with NL4-3 DIS (TA) (squares), NL4-3 DIS (CG) (circles), and NL4-3 DIS (TG) (triangles) at an MOI of 0.001, and virus-associated p24gag antigen production was determined at designated time points. The inset shows provirus DIS PCR products at day 12 in culture cleaved with restriction endonucleases capable of distinguishing DIS palindromic sequence. DIS PCR product restriction enzyme digestion results in the production of 89- and 76-bp DNA fragments.
Efficient homologous recombination during a single replication cycle.
The effect of DIS nonhomology on recombination was then investigated. Deletion mutants of NL4-3 were constructed to permit an evaluation of recombination after a single round of replication in cell culture. Figure 3A shows a diagram of their structures and the locations of the PCR primers and probes used to evaluate the products of recombination. One mutant (NL4-3 Δpol) contained a 404-bp deletion in the pol gene, and another (NL4-3 Δenv) had 580 bp deleted from the env gene. When copackaged into virions, a single homologous recombination event within the 3.8-kb segment between the deletions should generate either wild-type or double-deleted genomes. PCR primers positioned outside the two deletions were used to amplify a genomic segment for analysis. Probes located within the pol and env deletions, and a third probe located between the deletions, were used to evaluate and quantitate molecular forms after one replication cycle. Both of the deletion mutants and the wild-type NL4-3 contained the DIS (CG) sequence typical of HIV-1 subtype B.
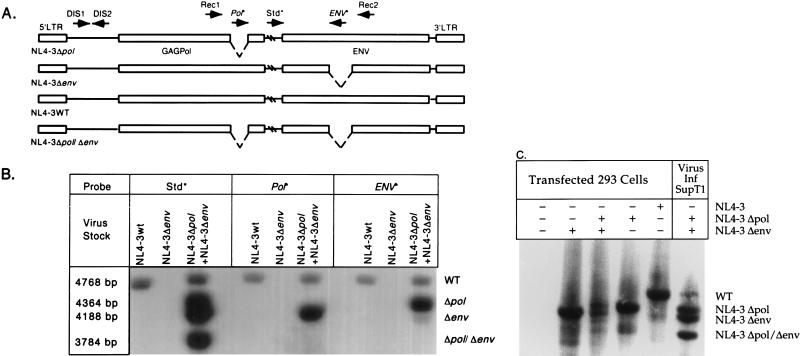
FIG. 3.
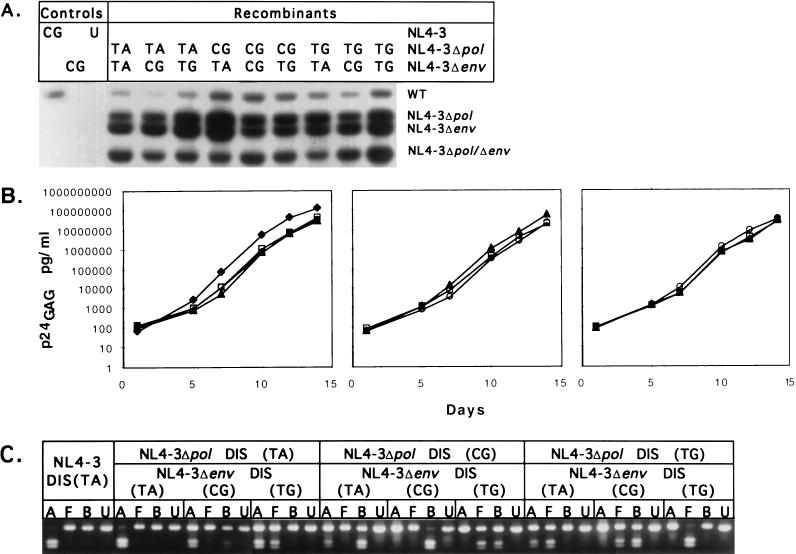
(A) Recombination between heterologous RNA during replication results in four proviral forms, the two parental forms, NL4-3Δpol and NL4-3Δenv, and two recombinant forms, wild-type NL4-3 (NL4-3WT) and NL4-3Δpol/Δenv. Arrows denote oligonucleotides used for PCR amplification (Rec1, Rec2, DIS1, and DIS2) and hybridization (Std*, Pol*, and ENV*) and the direction of sequence complementarity. Oligonucleotides marked with asterisks are probes for detecting PCR products. Hatch marks denote unrepresented sequences in HIV-1. Long terminal repeat (LTR) and structural genes are shown. (B) Provirus resulting from a single-cycle infection of SupT1 cells. Wild-type NL4-3 DIS (CG) and virus generated by cotransfection of NL4-3Δpol DIS (CG) and NL4-3Δenv DIS (CG) were used to infect SupT1 cells. Lysates of infected cells were amplified with primers Rec1 and Rec2, and proviral sequences were identified by hybridization with Std*, Pol*, and ENV* probes. Molecular weights of the PCR products confirm expected recombination products. (C) DNA recombination during virus production. Hirt supernatants from 293 cells transfected with no DNA, pNL4-3Δenv DIS (CG), pNL4-3Δenv DIS (CG) and NL4-3Δpol DIS (CG), NL4-3Δpol DIS (CG), or NL4-3 DIS (CG) were amplified with primers Rec1 and Rec2, and proviral sequences were identified by hybridization with the Std* probe. Lysate of SupT1 cells infected (Inf) with virus generated by cotransfection of NL4-3Δpol DIS (CG) and NL4-3Δenv DIS (CG) was amplified as a control. WT, wild type.
To establish that recombination occurs during replication of NL4-3 in cell culture, viral stocks were prepared by complementing polymerase and envelope function by cotransfection of 293 cells with NL4-3Δpol and NL4-3Δenv. Viral stocks contained RNA dimers of three types: Δpol/Δpol, Δenv/Δenv, and Δpol/Δenv. Upon infection of SupT1 cells at a low MOI with this stock, ensuring that a cell becomes infected with a single virus particle, only the virions containing RNA dimers of the form Δpol/Δenv have the potential to initiate a spreading infection if recombination occurs between the deletions to regenerate a wild-type genome. As negative and positive controls, virus stocks were also prepared by transfection of NL4-3Δenv alone, which should be noninfectious, and by transfection with NL4-3 wild-type DNA, respectively. As an additional control, we determined the maximum extent of DNA recombination occurring during production of the viral stock, as this would confound the analysis of RNA recombination in subsequent steps. We analyzed the DNA from cells cotransfected with the NL4-3 Δpol and NL4-3 Δenv plasmids at the time of harvest of the viral stock. Figure 3C shows that cotransfection of the two plasmid proviral constructs produced, within the sensitivity of detection by Southern blot hybridization, only the parental, and no detectable recombinant, DNA forms as the source of RNA in the viral particles.
Figure 3B shows the analysis of proviral DNA established after a single cycle of replication (24-h time point) after low-MOI (0.001) infection in SupT1 cells by PCR amplification of the 4.65-kb segment encompassing the pol and env deletions. As expected, the wild-type NL4-3 viral stock was infectious and produced proviral DNA that hybridized to all of the probes, while the virus stock produced from NL4-3Δenv was not able to establish proviral DNA. Cotransfection of NL4-3Δenv and NL4-3Δpol produced a virus stock that established four proviral DNA forms in target cells hybridizing to the Std* probe. By their size and pattern of hybridization to the Pol* and ENV* probes, we identify these as wild type (4,768 bp), Δpol (4,364 bp), Δenv (4,188 bp), and Δpol/Δenv (3,784 bp). Packaging of the mutated viral RNA did not appear to be affected by the deletions, as equivalent levels of parental PCR products were observed. Single-cycle replication in SupT1 cells was shown to occur within 24 h by strong stop and gag PCR (data not shown).
These results indicate that recombination between two deletion mutants of NL4-3 bearing homologous DIS readily occurs after infection of SupT1 cells. The frequency of recombination in the 3.8-kb genomic segment was estimated by quantitation of the proviral DNA forms after hybridization with the Std* probe (Fig. 3B, third lane). In two experiments, the recombinant forms that could be scored (wild type and double deleted), which could arise only from one of the three types of viral particles produced by complementation (see above), accounted for approximately one-third of the proviral DNA forms obtained. We conclude that homologous recombination occurred frequently within this 3.8-kb region of the HIV-1 genome.
Effect of DIS nonhomology on recombination.
Pairs of NL4-3 deletion mutants with homologous or heterologous combinations of DIS (TA), DIS (CG), or DIS (TG) were then evaluated for the ability to generate recombinant forms. Figure 4A shows an analysis of proviral DNA forms arising from viral stocks generated from cotransfection of deletion mutants of NL4-3 with homologous or heterologous DIS. Recombinant forms were detected with all combinations of DIS tested. Furthermore, both homologous and heterologous combinations of DIS produced a spreading infection with similar replication kinetics (Fig. 4B). Using PCR amplification and restriction analysis, we determined the DIS variants present in the cultures at day 14 (Fig. 4C). Progeny of recombination in RNA dimers with homologous DIS retained their DIS sequence within the limits of detection of the assay, while those arising from heterologous DIS exhibited a mixture of the input sequences, reflected by partial digestion with each of two restriction endonucleases, as expected (Fig. 4C). Similar results were obtained at days 7 and 10 of culture (data not shown).
FIG. 4.
(A) Recombinant forms of provirus observed in single-cycle infection of SupT1 cells. SupT1 cells were infected with various viral stocks at an MOI of 0.001. Provirus formation was analyzed by PCR, and recombinant viral PCR products were identified by hybridization to Std* probe 24 h after infection (Fig. 3B). PCR products derived from wild-type (WT), parental, and double-deleted mutant provirus are indicated. Provirus bands hybridizing with the probe were quantitated with a PhosphorImager, and the efficiency of recombination was determined by comparing the amount of wild-type and double-deleted provirus formed to the total amount of all provirus structures observed. (B) Replication kinetics of HIV-1 resulting from recombination of NL4-3 DIS variants. SupT1 cells were infected at an MOI of 0.0001 with equivalent MAGI cell infectious units of NL4-3 DIS (TA) and replication-competent recombinant virus resulting from recombination between NL4-3Δpol and NL4-3Δenv DIS variants. Virus-associated p24gag antigen production was measured at the designated time points. Left, kinetics of replication of recombinant virus resulting from recombination between NL4-3Δpol DIS (TA) and NL4-3Δenv DIS (TA) (squares), NL4-3Δenv DIS (CG) (triangles), and NL4-3Δenv DIS (TG) (open circles). Replication kinetics of wild-type NL4-3 DIS (TA) is also shown (diamonds). Center, kinetics of replication of recombinant virus resulting from recombination between NL4-3Δpol DIS (CG) and NL4-3Δenv DIS (TA) (squares), NL4-3Δenv DIS (CG) (triangles), and NL4-3Δenv DIS (TG) (circles). Right, kinetics of replication of recombinant virus resulting from recombination between NL4-3Δpol DIS (TG) and NL4-3Δenv DIS (TA) (squares), NL4-3Δenv DIS (CG) (triangles), and NL4-3Δenv DIS (TG) (circles). (C) DIS element identification in virus recombinants during culture. The DIS loop sequence of replication-competent recombinant provirus was analyzed in cell lysates at day 14 in culture by DIS PCR followed by restriction enzyme digestion of the PCR product (Fig. 3) as follows: B, BssHII; A, ApaLI; F, FspI; U, uncut. Complete restriction enzyme digestion of the DIS PCR product is observed in cultures containing recombinant provirus resulting from recombination of homologous DIS. Partial digestion of DIS PCR product is observed in recombinant provirus resulting from recombination of heterologous DIS. Identical results observed at earlier time points (days 7 and 10) are not shown.
These results indicate that even with only partial sequence homology of DIS, sufficient copackaging of RNA dimers into viral particles occurred to establish a prompt, spreading infection of recombinant viruses in culture. To determine whether homologous DIS resulted in a higher yield of recombinant proviral forms early in infection, we quantitated the PCR products (Fig. 4A) after hybridization with the Std* probe. Table 2 shows that in two experiments, the yields of recombinant forms were similar with homologous and heterologous DIS. We find little evidence that DIS nonhomology significantly inhibits RNA dimerization in the in vitro culture system used.
TABLE 2.
Recombination frequencies between DIS variants
| Expt | % Recombination between NL4-3Δpol/NL4-3Δenva with indicated combination of DIS variants
|
||||||||
|---|---|---|---|---|---|---|---|---|---|
| TA/TA | TA/CG | TA/TG | CG/TA | CG/CG | CG/TG | TG/TA | TG/CG | TG/TG | |
| 1 | 42 | 28 | 42 | 32 | 40 | 41 | 40 | 42 | 42 |
| 2 | 35 | 26 | 24 | 24 | 35 | 31 | 24 | 34 | 36 |
Ratio of recombinant provirus to recombinant and parental provirus multiplied by 100. The amount of recombination product was determined by quantitating hybridization of PCR product to the Std* probe as shown in Fig. 4.
Replication dynamics of recombinant virus populations.
The DIS (A6) variant, when introduced into the infectious molecular clone NL4-3, supported virus replication in SupT1 cells, suggesting that virus replication can occur even in the complete absence of a palindromic DIS (data not shown). We also investigated whether NL4-3 deletion mutants with heterologous combinations of DIS (A6), DIS (TA), DIS (CG), and DIS (TG) could generate recombinant forms capable of replicating in culture. Figure 5 shows that all combinations of DIS generate recombinant virus with comparable replication kinetics; however, the DIS (A6) form exhibited a replication disadvantage. Table 3 shows the relative proportions of DIS (A6) forms over time in the cultures. When naturally occurring DIS forms were present, the proportion of DIS (A6), initially 35% or greater, gradually declined to undetectable levels over a 14-day interval. These data provide evidence that although replication competent, the DIS (A6) form is not as robust as the naturally occurring sequences when virus replication is occurring in a continuous T-cell line.
FIG. 5.
Replication kinetics of NL4-3 DIS (A6). SupT1 cells were infected at an MOI of 0.001 with NL4-3 DIS (A6) (diamonds) and recombinant virus resulting from recombination between NL4-3Δenv DIS (A6) and NL4-3Δpol DIS (TA) (squares), NL4-3Δpol DIS (CG) (triangles), and NL4-3Δpol DIS (TG) (circles), and virus-associated p24gag antigen production was determined at designated time points. Dynamics of recombinant virus populations in culture are shown in Table 3.
TABLE 3.
Replication dynamics of recombinant virus populations
| Days in culture | % DIS (A6) provirusa with indicated DIS combination
|
|||
|---|---|---|---|---|
| DIS (A6) | DIS (TA)/(A6) | DIS (CG)/(A6) | DIS (TG)/(A6) | |
| 5 | 100 | 45 | 70 | 70 |
| 7 | ND | 15 | 30 | 55 |
| 14 | 100 | 2 | 0 | 0 |
Ratio of DIS (A6) clones to total number of clones analyzed multiplied by 100. DIS sequences were identified by restriction endonuclease digestion and DNA sequence analysis of at least 20 clones. ND, not done.
DISCUSSION
The formation of a stable duplex at the DIS is thought to be essential for the efficient formation of the RNA dimers that are packaged into HIV-1 virions. Here we report that two forms of the DIS occur among HIV-1 isolates examined to date: DIS (CG) in HIV-1 subtypes B and D and DIS (TA) in other subtypes and in group O. Unexpectedly, a naturally occurring but apparently rare DIS that is not palindromic and should form a less stable duplex as well as an artificial AAAAAA sequence that should be unable to form a duplex both supported virus replication in vitro when introduced into the infectious molecular clone NL4-3. Furthermore, the recombination frequency between deletion mutants of NL4-3 was not significantly affected by DIS loop nonhomology. We conclude that the difference in DIS found in HIV-1 subtypes B and D is not a primary impediment to RNA dimer formation, and recombination, with other subtypes and the A/D and B/F recombinants, already identified in the global epidemic (37, 45), may be an example of recombination between viruses with different DIS.
Previous studies of RNA dimer formation, in which a cell-free system with highly truncated RNA genomes was used, demonstrated that mutations predicted to destabilize the DIS loop interaction resulted in a markedly delayed dimerization rate and in a lower yield of RNA dimer (44). Contrasting results were found when we used an infectious molecular clone of HIV-1 and measured virus replication in a T-cell line in vitro. The function of the DIS loop palindrome in the virion appears to be distinct from the dimerization properties of DIS sequences in a cell-free system employing purified RNA. Although we did not measure RNA dimer formation directly, the replication kinetics observed with congenic virus stocks containing four different DIS, predicted to form hybrids ranging from stable to completely unstable, were comparable. When the naturally occurring DIS loop sequences were replaced with six A residues, virus replication kinetics were affected, and in mixed cultures, recombinant viruses bearing wild-type DIS gradually increased in proportion to those with six A residues (Table 3), indicating a modest replication disadvantage.
Other studies have shown that deletions or insertions in the DIS loop reduced encapsidation and virus titer (6). The length of the DIS loop may be of greater importance than its sequence when all of the viral components are present in an in vitro system with replicating, infectious viruses. Compensating factors apparently permitted virus replication in the absence of stable, sequence-based interaction of the DIS. Upstream sequences in the gag leader region (20, 28, 58) and others in the 5′ portion of the gag gene (9, 35, 46) that have been shown to influence packaging efficiency may have played a role. RNA dimers may be stabilized by interaction with the nucleocapsid protein. Consistent with infectivity defects in DIS substitution mutants in previous reports (12, 43), DIS (A6) mutant virus produced in nonlymphoid cells (293 cells) is fourfold less infectious than the wild type when normalized to p24 antigen (data not shown).
In the study described here, we evaluated the HIV-1 recombination frequency after a single replication cycle using congenic deletion mutants of NL4-3. While this approach is sensitive to viral input and to the relative efficiency of PCR amplification of recombinant and nonrecombinant forms, we were nonetheless able to quantify proviral DNA forms arising from carefully titered viral stocks with homologous and heterologous DIS and estimate the frequency of recombination. Similar levels of recombination were found with all combinations of DIS (TA), DIS (TG), and DIS (CG) tested. The recombination frequency over the region examined, 3.8 kb in length, approached 10−4/bp. Furthermore, recombinant virus exhibited similar replication kinetics, verifying that recombination between DIS pairs occurred with comparable efficiency and at high levels. The absence of detectable DNA recombination during virus production establishes that recombination occurred during virus replication.
While this approach estimates recombination rates, it is important to compare our results with previous reports using different approaches. In previous studies using a spleen necrosis virus retroviral vector system (24) and dominant selectable markers, the recombination rate was more than twofold lower than that observed here. Differences in the frequency of strand switching by the RTs from different retroviruses, the use of isogenic clones with identical sequences in the region undergoing recombination, and differences in PCR amplification efficiency between the longer, wild-type and double-deleted NL4-3 clones in our study may contribute to the different rates observed.
Finally, the results reported here pertain to an infectious molecular clone of HIV-1 subtype B that has been selected to replicate efficiently in T-cell lines in vitro. Differences in replication rate conferred by DIS loop variation and the effect of DIS nonhomology on recombination frequency could be more substantial in primary HIV-1 isolates replicating in peripheral blood mononuclear cells. Experiments to address this issue are ongoing. Furthermore, distal sequence elements, viral proteins, and other factors may also be variable and not fully compatible across HIV-1 subtypes; in the in vitro system developed here, these factors were exclusively from HIV-1 subtype B.
Among the intersubtype HIV-1 recombinants characterized to date, subtype A has often been observed to recombine with subtypes C, D, E, and G. Recombinants with subtype B have been observed less frequently and, when found, usually involve subtype F. Notably, subtype A/D recombinants and subtype B/F recombinants would be expected to derive from dimer formation between RNAs with different DIS loop sequences, and DIS (TG) is not an essential intermediary of recombination. Many factors, including geographic dispersal patterns of subtypes, opportunities for coinfection of individuals and susceptible cells, and functional incompatibilities that may develop with mosaic genomes from different subtypes, may influence the frequency of intersubtype recombination. The disparate DIS loop sequences found in different HIV-1 subtypes reemphasize the complex matrix of variables that contribute to the mixture of HIV-1 genetic variants in the global epidemic.
ACKNOWLEDGMENTS
This work was supported in part by cooperative agreement DAMD17-93-V-3004 between the U.S. Army Medical Research and Materiel Command and The Henry M. Jackson Foundation for the Advancement of Military Medicine.
We are grateful to Donald S. Burke for helpful discussions during the course of this work. We thank Richard C. Carroll and Nelson Micheal for helpful suggestions during preparation of the manuscript.
REFERENCES
- 1.Adachi A, Gendelman H E, Koenig S, Folks T, Willey R, Rabson A, Martin M A. Production of acquired immunodeficiency syndrome-associated retrovirus in human and nonhuman cells transfected with an infectious molecular clone. J Virol. 1986;59:284–291. doi: 10.1128/jvi.59.2.284-291.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Awang G, Sen D. Mode of dimerization of HIV-1 genomic RNA. Biochemistry. 1993;32:11453–11457. doi: 10.1021/bi00093a024. [DOI] [PubMed] [Google Scholar]
- 3.Beemon K L, Faras A, Haase A, Duesberg P H, Maisel J. Genomic complexities of murine leukemia and sarcoma, reticuloendotheliosis, and visna viruses. J Virol. 1976;17:525–537. doi: 10.1128/jvi.17.2.525-537.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bender W, Chien Y-H, Chattopadhyay S, Vogt P K, Gardner M B, Davidson N. High-molecular-weight RNAs of AKR, NZB, and wild mouse viruses and avian reticuloendotheliosis virus all have similar dimer structures. J Virol. 1978;25:888–896. doi: 10.1128/jvi.25.3.888-896.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Berkhout B, Essink B B, Schoneveld I. In vitro dimerization of HIV-2 leader RNA in the absence of PuGGAPuA motifs. FASEB J. 1993;7:181–187. doi: 10.1096/fasebj.7.1.8422965. [DOI] [PubMed] [Google Scholar]
- 6.Berkhout B, van Wamel J L B. Role of the DIS hairpin in replication of human immunodeficiency virus type 1. J Virol. 1996;70:6723–6732. doi: 10.1128/jvi.70.10.6723-6732.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bieth E, Gabus C, Darlix J L. A study of the dimer formation of Rous sarcoma virus RNA and of its effect on viral protein synthesis in vitro. Nucleic Acids Res. 1990;18:119–127. doi: 10.1093/nar/18.1.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Billeter M A, Parsons J T, Coffin J M. The nucleotide sequence complexity of avian tumor virus RNA. Proc Natl Acad Sci USA. 1974;71:4254–4258. doi: 10.1073/pnas.71.9.3560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Buchschacher G L, Jr, Panganiban A T. Human immunodeficiency virus vectors for inducible expression of foreign genes. J Virol. 1992;66:2731–2739. doi: 10.1128/jvi.66.5.2731-2739.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Carr J K, Salminen M O, Gotte D, Koch C, St. Louis D C, Burke D S, McCutchan F E. Mosaic structure of the full-length genome from a human immunodeficiency virus type 1 isolate of clade E from Thailand. J Virol. 1996;70:5935–5943. doi: 10.1128/jvi.70.9.5935-5943.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Clever J, Sassette C, Parslow T G. RNA secondary structure and binding sites for gag gene products in the 5′ packaging signal of human immunodeficiency virus type 1. J Virol. 1995;69:2101–2109. doi: 10.1128/jvi.69.4.2101-2109.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Clever J L, Parslow T G. Mutant human immunodeficiency virus type 1 genomes with defects in RNA dimerization or encapsidation. J Virol. 1997;71:3407–3414. doi: 10.1128/jvi.71.5.3407-3414.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Coffin J M. Retroviridae and their replication. In: Fields B N, Knipe D M, Chanock R M, Hirsch M S, Melnick J L, Monath T P, Roizman B, editors. Virology. 2nd ed. New York, N.Y: Raven Press; 1990. pp. 1437–1500. [Google Scholar]
- 14.Coffin J M. Structure of the retroviral genome. In: Weiss R, Teich N, Varmus H, Coffin J M, editors. RNA tumor viruses. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1984. pp. 261–368. [Google Scholar]
- 15.Coffin J M. Structure, replication and recombination of retrovirus genomes: some unifying hypotheses. J Gen Virol. 1979;42:1–26. doi: 10.1099/0022-1317-42-1-1. [DOI] [PubMed] [Google Scholar]
- 16.Darlix J L, Gabus C, Nugeyre M T, Clavel F, Barre-Sinoussi F. Cis elements and trans-acting factors involved in the RNA dimerization of the human immunodeficiency virus HIV-1. J Mol Biol. 1990;216:689–699. doi: 10.1016/0022-2836(90)90392-Y. [DOI] [PubMed] [Google Scholar]
- 17.De Rocquigny H, Gabus C, Vincent A, Fournie-Zaluski M C, Roques B, Darlix J L. Viral RNA annealing activities of human immunodeficiency virus type 1 nucleocapsid protein require only peptide domains outside the zinc fingers. Proc Natl Acad Sci USA. 1992;89:6472–6476. doi: 10.1073/pnas.89.14.6472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fu W, Gorelick R J, Rein A. Characterization of human immunodeficiency virus type 1 dimeric RNA from wild-type and protease-defective virions. J Virol. 1994;68:5013–5018. doi: 10.1128/jvi.68.8.5013-5018.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gao F, Robertson D L, Morrison S G, Huxiong H, Craig S, Decker J, Fultz P N, Girard M, Thornton C L, Shaw G M, Hahn B H, Sharp P M. The heterosexual human immunodeficiency virus 1 epidemic in Thailand is caused by an intersubtype (A/E) recombinant of African origin. J Virol. 1996;70:7013–7029. doi: 10.1128/jvi.70.10.7013-7029.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Geigenmuller U, Linial M L. Specific binding of human immunodeficiency virus type 1 (HIV-1) Gag-derived proteins to a 5′ HIV-1 genomic RNA sequence. J Virol. 1996;70:667–671. doi: 10.1128/jvi.70.1.667-671.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Harrison G P, Lever A M. The human immunodeficiency virus type 1 packaging signal and major splice donor region have a conserved stable secondary structure. J Virol. 1992;66:4144–4153. doi: 10.1128/jvi.66.7.4144-4153.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hirt B. Selective extraction of polyoma DNA from infected mouse cell cultures. J Mol Biol. 1967;26:365–369. doi: 10.1016/0022-2836(67)90307-5. [DOI] [PubMed] [Google Scholar]
- 23.Hu W S, Temin H M. Effect of gamma radiation on retroviral recombination. J Virol. 1992;66:4457–4463. doi: 10.1128/jvi.66.7.4457-4463.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hu W S, Temin H M. Genetic consequences of packaging two RNA genomes in one retroviral particle: pseudodiploidy and high rate of genetic recombination. Proc Natl Acad Sci USA. 1990;87:1556–1560. doi: 10.1073/pnas.87.4.1556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hu W S, Temin H M. Retroviral recombination and reverse transcription. Science. 1990;250:1227–1233. doi: 10.1126/science.1700865. [DOI] [PubMed] [Google Scholar]
- 26.Jones J S, Allan R W, Temin H M. Alteration of location of dimer linkage sequence in retroviral RNA: little effect on replication or homologous recombination. J Virol. 1993;67:3151–3158. doi: 10.1128/jvi.67.6.3151-3158.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Katz R A, Skalka A M. Generation of diversity in retroviruses. Annu Rev Genet. 1990;24:409–445. doi: 10.1146/annurev.ge.24.120190.002205. [DOI] [PubMed] [Google Scholar]
- 28.Kim H-J, Lee K, O’Rear J J. A short sequence upstream of the 5′ major splice site is important for encapsidation of HIV-1 genomic RNA. Virology. 1994;198:336–340. doi: 10.1006/viro.1994.1037. [DOI] [PubMed] [Google Scholar]
- 29.Kimpton J, Emerman M. Detection of replication-competent and pseudotyped human immunodeficiency virus with a sensitive cell line on the basis of activation of an integrated β-galactosidase gene. J Virol. 1992;66:2232–2239. doi: 10.1128/jvi.66.4.2232-2239.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Laughrea M, Jette L. A 19-nucleotide sequence upstream of the 5′ major splice donor is part of the dimerization domain of human immunodeficiency virus 1 genomic RNA. Biochemistry. 1994;33:13464–13474. doi: 10.1021/bi00249a035. [DOI] [PubMed] [Google Scholar]
- 31.Lever A, Gottlinger H, Haseltine W, Sodroski J. Identification of a sequence required for efficient packaging of human immunodeficiency virus type 1 RNA into virions. J Virol. 1989;63:4085–4087. doi: 10.1128/jvi.63.9.4085-4087.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Linial M, Blair D. Genetics of retroviruses. In: Weiss R, Teich N, Varmus H, Coffin J, editors. RNA tumor viruses. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1984. pp. 649–783. [Google Scholar]
- 33.Louwagie J, Delwart E L, Mullins J I, McCutchan F E, Eddy G, Burke D S. Genetic analysis of HIV-1 isolates from Brazil reveals presence of two distinct genetic subtypes. AIDS Res Hum Retroviruses. 1994;10:561–567. doi: 10.1089/aid.1994.10.561. [DOI] [PubMed] [Google Scholar]
- 34.Louwagie J, Janssens W, Mascola J, Heyndrickx L, Hegerich P, van der Groen G, McCutchan F E, Burke D S. Genetic diversity of the envelope glycoprotein from human immunodeficiency virus type 1 isolates of African origin. J Virol. 1995;69:263–271. doi: 10.1128/jvi.69.1.263-271.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Luban J, Goff S P. Mutational analysis of cis-acting packaging signals in human immunodeficiency type 1 RNA. J Virol. 1994;68:3784–3793. doi: 10.1128/jvi.68.6.3784-3793.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mann R, Mulligan R C, Baltimore D. Construction of a retrovirus packaging mutant and its use to produce helper-free defective retrovirus. Cell. 1983;33:153–159. doi: 10.1016/0092-8674(83)90344-6. [DOI] [PubMed] [Google Scholar]
- 37.Marquet R, Baudin F, Gabus C, Darlix J L, Mougel M, Ehresmann C, Ehresmann B. Dimerization of human immunodeficiency virus (type 1) RNA: stimulation by cations and possible mechanism. Nucleic Acids Res. 1991;19:2349–2357. doi: 10.1093/nar/19.9.2349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Marquet R, Paillart J C, Skripkin E, Ehresmann C, Ehresmann B. Dimerization of human immunodeficiency virus type 1 RNA involves sequences located upstream of the splice donor site. Nucleic Acids Res. 1994;22:145–151. doi: 10.1093/nar/22.2.145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.McCutchan F E, Salminen M O, Carr J K, Burke D S. HIV-1 genetic diversity. AIDS. 1996;10:S13–S20. [PubMed] [Google Scholar]
- 40.Muriaux D, Girard P M, Bonnet-Mathoniere B, Paoletti J. Dimerization of HIV-1Lai RNA at low ionic strength. An autocomplementary sequence in the 5′ leader region is evidenced by an antisense oligonucleotide. J Biol Chem. 1995;270:8209–8216. doi: 10.1074/jbc.270.14.8209. [DOI] [PubMed] [Google Scholar]
- 41.Murti K G, Boundurant M, Tereba A. Secondary structural features in the 70S RNAs of Moloney murine leukemia and Rous sarcoma viruses as observed by electron microscopy. J Virol. 1981;37:411–419. doi: 10.1128/jvi.37.1.411-419.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Myers G, Korber B, Wain-Hobson S, Smith R, Pavlakis G. Human retroviruses and AIDS 1995: a compilation and analysis of nucleic acid and amino acid sequences. Los Alamos, N.Mex: Los Alamos National Laboratory; 1995. [Google Scholar]
- 43.Paillart J C, Berthoux L, Ottmann M, Darlix J L, Marquet R, Ehresmann B, Ehresmann C. A dual role of the putative RNA dimerization initiation site of human immunodeficiency virus type 1 in genomic RNA packaging and proviral DAN synthesis. J Virol. 1996;70:8348–8354. doi: 10.1128/jvi.70.12.8348-8354.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Paillart J C, Marquet R, Skripkin E, Ehresmann B, Ehresmann C. Mutational analysis of the bipartite dimer linkage structure of human immunodeficiency virus type 1 genomic RNA. J Biol Chem. 1994;269:27486–27493. [PubMed] [Google Scholar]
- 45.Paillart J C, Skripkin E, Ehresmann B, Ehresmann C, Marquet R. A loop-loop “kissing” complex is the essential part of the dimer linkage of genomic HIV-1 RNA. Proc Natl Acad Sci USA. 1996;93:5572–5577. doi: 10.1073/pnas.93.11.5572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Parolin C, Dorfman T, Palu G, Gottlinger H, Sodroski J. Analysis in human immunodeficiency virus type 1 vectors of cis-acting sequences that affect gene transfer into human lymphocytes. J Virol. 1994;68:3888–3895. doi: 10.1128/jvi.68.6.3888-3895.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pear W S, Nolan G P, Scott M L, Baltimore D. Production of high-titer helper-free retroviruses by transient transfection. Proc Natl Acad Sci USA. 1993;90:8392–8396. doi: 10.1073/pnas.90.18.8392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Robertson D L, Sharp P M, McCutchan F E, Hahn B H. Recombination in HIV-1. Nature. 1995;374:124–126. doi: 10.1038/374124b0. [DOI] [PubMed] [Google Scholar]
- 49.Sakaguchi K, Zambrano N, Baldwin E T, Shapiro B A, Erickson J W, Omichinski J G, Clore G M, Gronenborn A M, Appella E. Identification of a binding site for the human immunodeficiency virus type 1 nucleocapsid protein. Proc Natl Acad Sci USA. 1993;90:5219–5223. doi: 10.1073/pnas.90.11.5219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Salminen M O, Koch C, Sanders B E, Ehrenberg P K, Michael N L, Carr J K, Burke D S, McCutchan F E. Recovery of virtually full-length HIV-1 provirus of diverse subtypes from primary virus cultures using the polymerase chain reaction. Virology. 1995;213:80–86. doi: 10.1006/viro.1995.1548. [DOI] [PubMed] [Google Scholar]
- 51.Shank P R, Linial M. Avian oncovirus mutant (SE21Q1b) deficient in genomic RNA: characterization of a deletion in the provirus. J Virol. 1980;36:450–456. doi: 10.1128/jvi.36.2.450-456.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Skripkin E, Paillart J C, Marquet R, Ehresmann B, Ehresmann C. Identification of the primary site of the human immunodeficiency virus type 1 RNA dimerization in vitro. Proc Natl Acad Sci USA. 1994;91:4945–4949. doi: 10.1073/pnas.91.11.4945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Stuhlmann H, Berg P. Homologous recombination of copackaged retrovirus RNAs during reverse transcription. J Virol. 1992;66:2378–2388. doi: 10.1128/jvi.66.4.2378-2388.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sundquist W I, Heaphy S. Evidence for interstrand quadruplex formation in the dimerization of human immunodeficiency virus 1 genomic RNA. Proc Natl Acad Sci USA. 1993;90:3393–3397. doi: 10.1073/pnas.90.8.3393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Temin H M. Retrovirus variation and reverse transcription: abnormal strand transfers result in retrovirus genetic variation. Proc Natl Acad Sci USA. 1993;90:6900–6903. doi: 10.1073/pnas.90.15.6900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Temin H M. Sex and recombination in retroviruses. Trends Genet. 1991;7:71–74. doi: 10.1016/0168-9525(91)90272-R. [DOI] [PubMed] [Google Scholar]
- 57.Tsuchihashi Z, Brown P O. DNA strand exchange and selective DNA annealing promoted by the human immunodeficiency virus type 1 nucleocapsid protein. J Virol. 1994;68:5863–5870. doi: 10.1128/jvi.68.9.5863-5870.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Vincenzi E, Dimitrov D S, Engelman A, Migone T-S, Purcell D F J, Leanard J, Englund G, Martin M A. An integration-defective U5 deletion mutant of human immunodeficiency virus type 1 reverts by eliminating additional long terminal repeat sequences. J Virol. 1994;68:7879–7890. doi: 10.1128/jvi.68.12.7879-7890.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Watanabe S, Temin H M. Construction of a helper cell line for avian reticuloendotheliosis virus cloning vectors. Mol Cell Biol. 1983;3:2241–2249. doi: 10.1128/mcb.3.12.2241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Weiss R A, Mason E S, Vogt P K. Genetic recombinants and heterozygotes derived from endogenous and exogenous avian RNA tumor viruses. Virology. 1973;52:535–552. doi: 10.1016/0042-6822(73)90349-8. [DOI] [PubMed] [Google Scholar]
- 61.Weiss S, Hausl G, Famulok M, Konig B. The multimerization state of retroviral RNA is modulated by ammonium ions and affects HIV-1 full-length cDNA synthesis in vitro. Nucleic Acids Res. 1993;21:4879–4885. doi: 10.1093/nar/21.21.4879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wyke J A, Beamond J A. Genetic recombination in Rous sarcoma virus: the genesis of recombinants and lack of evidence for linkage between pol, env and src genes in three factor crosses. J Gen Virol. 1979;43:349–364. doi: 10.1099/0022-1317-43-2-349. [DOI] [PubMed] [Google Scholar]
- 63.Zuker M. On finding all suboptimal foldings of an RNA molecule. Science. 1989;244:48–52. doi: 10.1126/science.2468181. [DOI] [PubMed] [Google Scholar]