Abstract
Island organisms often evolve phenotypes divergent from their mainland counterparts, providing a useful system for studying adaptation under differential selection. In the white-winged fairywren (Malurus leucopterus), subspecies on two islands have a black nuptial plumage whereas the subspecies on the Australian mainland has a blue nuptial plumage. The black subspecies have a feather nanostructure that could in principle produce a blue structural color, suggesting a blue ancestor. An earlier study proposed independent evolution of melanism on the islands based on the history of subspecies divergence. However, the genetic basis of melanism and the origin of color differentiation in this group are still unknown. Here, we used whole-genome resequencing to investigate the genetic basis of melanism by comparing the blue and black M. leucopterus subspecies to identify highly divergent genomic regions. We identified a well-known pigmentation gene ASIP and four candidate genes that may contribute to feather nanostructure development. Contrary to the prediction of convergent evolution of island melanism, we detected signatures of a selective sweep in genomic regions containing ASIP and SCUBE2 not in the black subspecies but in the blue subspecies, which possesses many derived SNPs in these regions, suggesting that the mainland subspecies has re-evolved a blue plumage from a black ancestor. This proposed re-evolution was likely driven by a preexisting female preference. Our findings provide new insight into the evolution of plumage coloration in island versus continental populations, and, importantly, we identify candidate genes that likely play roles in the development and evolution of feather structural coloration.
Keywords: white-winged fairywren, male ornamentation, color genetics, feather coloration, structural color, avian plumage color
Introduction
Studies of the genetic basis underlying phenotypic variation are crucial to understanding processes leading to evolutionary adaptation. The “island syndrome”, which describes the divergence of organisms on islands from their continental counterparts (Baeckens and Van Damme 2020), has provided model systems ideal for studying adaptive phenotypes under differential selective pressures (Lamichhaney et al. 2016; Cerca et al. 2023). For example, insular populations of birds frequently have evolved plumage coloration differing from related continental populations (Uy et al. 2016; Romano et al. 2021). One well-known example is that of island melanisms, where populations of birds (Driskell et al. 2002; Uy et al. 2016, 2019; Campagna et al. 2022) and other taxa (Ritchie 1978; Castilla 1994; Buades et al. 2013) on islands evolve melanic coloration that is speculated to confer an advantage (Bonser 1995; Rathburn and Montgomerie 2003; Goldstein et al. 2004; Margalida et al. 2008; Roulin 2014; Surmacki et al. 2021). The repeated evolution of melanic coloration in island populations provides an opportunity to understand the genetic basis of adaptive evolutionary convergence, yet knowledge of the underlying molecular mechanisms of this convergence remains limited to a few case studies (Uy et al. 2016, 2019; Campagna et al. 2022).
An intriguing case of island melanism occurs in the white-winged fairywren (Malurus leucopterus), which has intraspecific variation in plumage structural color rather than pigment-based coloration (Driskell et al. 2002; Doucet et al. 2004). The color polymorphism in this species could originate due to loss or gain of structural coloration (Schodde 1982; Ford 1987; Driskell et al. 2002). Although we have made good progress in identifying genes responsible for pigment-based plumage coloration in birds in recent years, such as pigmentation due to melanins (Mundy 2005), carotenoids (Toomey et al. 2022), and psittacofulvins (Cooke et al. 2017), the genetic basis of feather structural colors is still largely unknown (Price-Waldman and Stoddard 2021). Structural colors are produced by coherent scattering or interference of light by nanostructures with periodic variation in refractive index (Prum and Torres 2003). Many birds have iridescent (changing in hue depends on the viewing or illumination angle) or noniridescent structurally based plumage colors (Eliason et al. 2015, 2023; Shawkey and D’Alba 2017; Nordén et al. 2019, 2021; Eliason and Clarke 2020; Saranathan and Finet 2021). Noniridescent structural plumage colors are produced by coherent light scattering due to quasi-ordered β-keratin nanostructures and air in the medullary spongy layer of feather barbs (Prum et al. 2009), which generates blue, violet, and ultraviolet feather colors. The presence of melanosomes (melanin-containing organelles) beneath the spongy layer is also essential for the production of structural color as it absorbs incoherently scattered light (Saranathan and Finet 2021).
Malurus fairywrens (Maluridae) are passerines displaying considerable variation in ornamental structural colors of males, from blue, indigo, violet, to almost pure UV hues (Fan et al. 2019). Most species exhibit a high degree of sexual dichromatism, and breeding males have striking ornamentation, whereas females of most populations are cryptically brown (Enbody et al. 2022). Structural color variation in fairywrens is associated with the thickness of the keratin cortex on top of the spongy layer, the size and density of the spongy layer, the relative amount of melanosomes underneath the spongy layer, and their interactions (Fan et al. 2019). This variation in different microstructural components contributing to the diversity of structural colors in fairywren nuptial plumages makes them an ideal model system to study mechanisms of structural color evolution.
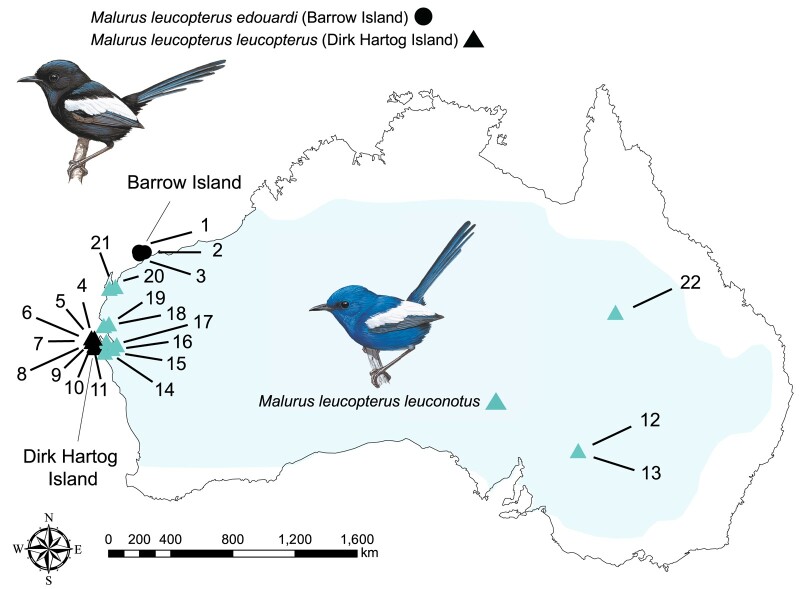
The white-winged fairywren (M. leucopterus) is particularly well suited to studying the genetic basis of structural coloration because males in this species have either blue or black nuptial plumage. Malurus leucopterus is distributed across most of Australia, with males of the mainland subspecies (Malurus leucopterus leuconotus) having a bright cobalt blue nuptial plumage with white wings, whereas the two island subspecies in Western Australia (M. l. leucopterus on Dirk Hartog Island [DHI] and M. l. edouardi on Barrow Island [BI]) exhibit a black male nuptial plumage with white wings (Driskell et al. 2002; Doucet et al. 2004) (Fig. 1). DHI and BI are far (∼600 km) from each other and separated from continental Australia by ∼2 and ∼56 km, respectively. The feather barbs of blue males on the mainland are composed of a spongy layer with a keratin matrix and air space, with melanin granules beneath the spongy layer around the central vacuole, a feather nanostructure typically producing blue colors (Fig. 2; Doucet et al. 2004; Driskell et al. 2010). Surprisingly, feather barbs of black males on both DHI and BI also have a spongy layer that could in principle produce a blue color similar to that on the mainland (Doucet et al. 2004; Driskell et al. 2010), but the feather barbs of these males have several features that render them black instead of blue in coloration: they have a thicker cortex with more melanin; a spongy layer with more and larger holes; and melanin granules that are distributed both within and beneath the spongy layer (Fig. 2; Doucet et al. 2004; Driskell et al. 2010). Because black feather barbs in other species do not normally possess a spongy layer, the presence of a spongy layer that could produce blue color in black feathers might suggest that the black M. leucopterus island populations evolved from blue ancestors (Doucet et al. 2004; Driskell et al. 2010). The low divergence between M. leucopterus populations (FST = 0.04 to 0.18) (Walsh et al. 2021) indicates that the switch of male nuptial plumage color occurred quickly and in the recent past (<2.5M generations) (Walsh et al. 2021), suggesting a simple genetic change (Driskell et al. 2002).
Fig. 1.
Geographical distribution and sampling sites of M. leucopterus. Malurus leucopterus males from BI (M. l. edouardi, denoted by black circles on the map) and DHI (M. l. leucopterus, denoted by black triangles) have black nuptial plumage. Malurus leucopterus males distributed across the Australian continent (M. l. leuconotus, denoted by blue triangles) are with blue plumage. Light blue color indicates the range of M. leucopterus. Refer to supplementary table S1, Supplementary Material online for detailed information of sampled individuals. The map was generated using QGIS 3.24. Illustrations of birds were reproduced with the permission of Lynx Edicions.
Fig. 2.
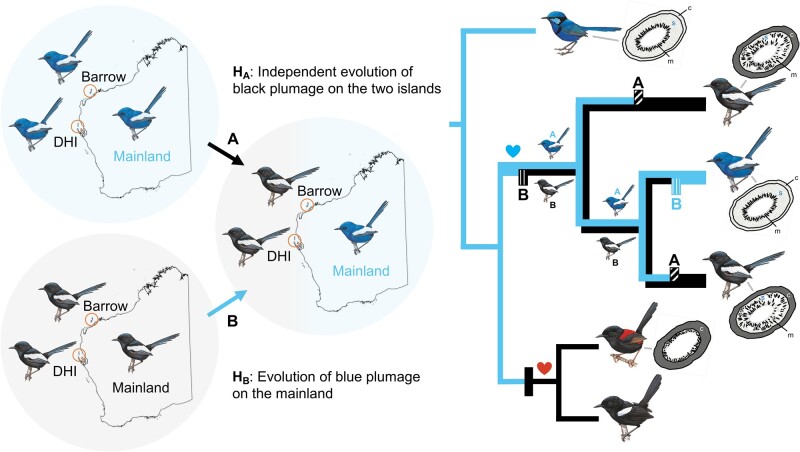
Hypotheses of plumage color evolution in the white-winged fairywren (M. leucopterus). Left side of the figure shows the two hypotheses: hypothesis A (HA)—independent evolution of black plumage on DHI and BI; hypothesis B (HB)—evolution of blue plumage on the mainland from birds with black plumage. Right side shows the corresponding positions of the plumage color change on the phylogenetic tree predicted by the two hypotheses. In the M. leucopterus clade, the upper half of the branches leading to the subspecies indicates scenario under hypothesis A, and the lower half of the branches represents hypothesis B. Hypothesized color changes are indicated by boxes (HA: with diagonal lines and “A” above, HB: with vertical lines and “B” below, black: change from blue to black, blue: change from black to blue). The heart symbol denotes proposed female color preference based on male petal carrying behavior. Female preference for blue color likely maintained in all M. leucopterus subspecies, and female preference for red color might have evolved in the common ancestor of M. melanocephalus and M. alboscapulatus. Diagrams of feather barb cross-section are illustrated based on Driskell et al. (2010). c, cortex; s, spongy layer, m, melanosome. Illustrations of birds were reproduced with the permission of Lynx Edicions.
Malurus leucopterus subspecies on DHI and mainland have been suggested based on reduced representation sequencing data to be more closely related to each other than either is to the subspecies on BI (Walsh et al. 2021). The emergence of color polymorphism in M. leucopterus therefore occurred either due to convergent melanism on the two islands or because the mainland subspecies re-evolved blue plumage from a black ancestor that in turn was originally blue (Fig. 2) (Schodde 1982; Ford 1987; Driskell et al. 2002). The two sister species in the bi-colored wren clade, Malurus melanocephalus and Malurus alboscapulatus, found on mainland Australia and New Guinea, respectively, also have black male nuptial plumage, which could imply a black ancestor to the bi-colored wren clade, but feather microstructure suggests this plumage pattern arose independent of the switch to a melanic form in M. leucopterus (Driskell et al. 2010). Based on the independent colonization events of DHI and BI, and the presence of spongy layer in black subspecies suggesting a blue ancestor, Walsh et al. (2021) concluded that there was independent evolution of melanic plumage in island M. leucopterus subspecies. However, the genetic basis of melanism, both for M. leucopterus and for other species in the bi-colored clade, is still an unknown. Accordingly, the alternative hypothesis is still plausible, such that melanism on the islands could occur with or without convergence, even if colonization occurred independently (Fig. 2), and understanding the genetic basis of black plumage in this clade will help clarify mechanisms of plumage color evolution generally (Robert et al. 2021). For convergent evolution of island melanism, the presence of signatures of selective sweep in genomic regions associated with relevant genes in both black subspecies would indicate independent evolution of melanism (Campagna et al. 2022). Conversely, the presence of signatures of selective sweep and derived SNPs in regions of relevant genes in the blue subspecies would suggest the evolution of blue plumage on the mainland.
Here we used whole-genome sequencing to study the genetic basis of melanism in M. leucopterus. We compared the blue and black M. leucopterus subspecies to identify highly divergent genomic regions between subspecies and candidate genes associated with plumage color differentiation. We further looked for signatures of selective sweeps at these loci and also examined genotypes in M. leucopterus subspecies and other Malurus species to test hypotheses of plumage color evolution in M. leucopterus.
Results
Phylogeny of the Bi-colored Clade and M. Leucopterus Subspecies
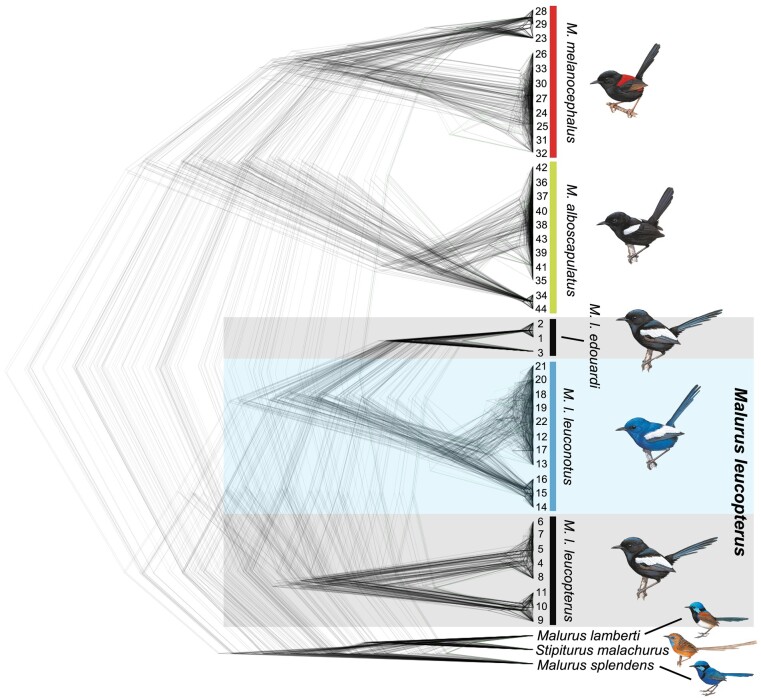
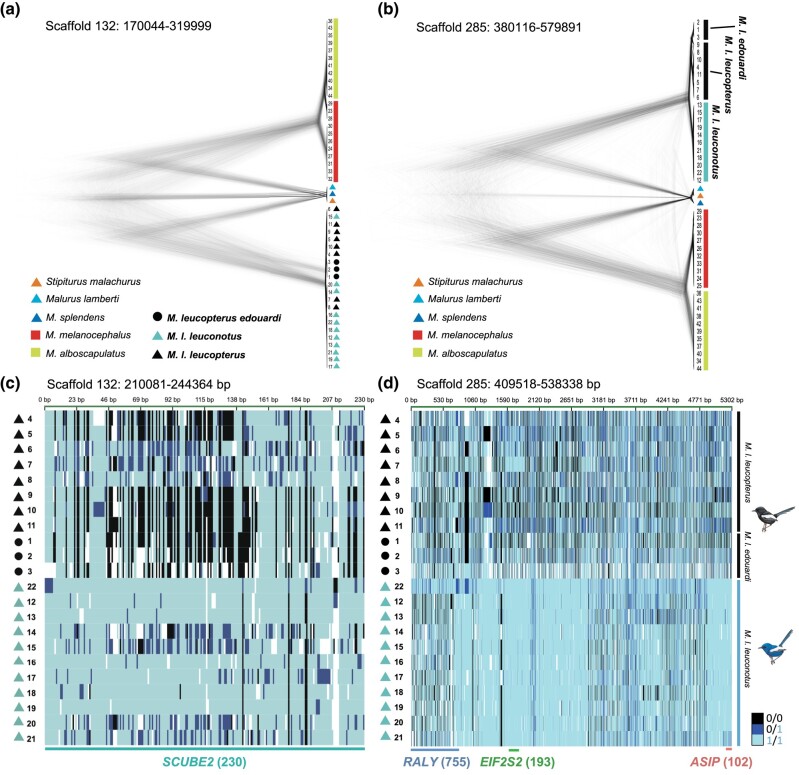
Phylogenomic analysis identified a monophyletic clade of M. leucopterus (Fig. 3 and supplementary fig. S1, Supplementary Material online), sister to the clade formed by two other bi-colored wrens, M. melanocephalus and M. alboscapulatus. Within M. leucopterus, the two black-plumaged insular populations did not form a monophyletic clade (Fig. 3). The maximum-likelihood (ML) tree based on 168,378 concatenated loci (supplementary fig. S1A, Supplementary Material online) shows that M. l. edouardi from BI formed a monophyletic clade that was sister to the intermixed cluster of individuals from the mainland (M. l. leuconotus) and DHI (M. l. leucopterus). The coalescent-based SNAPP (Fig. 3) and SVDquest (supplementary fig. S1B, Supplementary Material online) trees instead show that M. l. edouardi with black plumage was more closely related to blue-plumaged M. l. leuconotus than to black-plumaged M. l. leucopterus. Coalescent-based methods outperform concatenation in species-level inference, especially when gene flow is present (Long and Kubatko 2018), and should be more accurate given the gene flow between mainland and island populations (Walsh et al. 2021). Nevertheless, both trees indicate that the two insular populations were divergent from each other (Fig. 3, supplementary figs. S1 and S2, Supplementary Material online), and one of the black-plumaged insular populations was more closely related to the blue-plumaged mainland population than to another black-plumaged population, suggesting at least two color change events predicted by both hypotheses (Fig. 2). The reconstruction of ancestral plumage color suggested that the common ancestor of M. l. leuconotus and M. l. edouardi could have had either blue or black plumage, and the common ancestor of all subspecies could have also had either blue or black plumage (supplementary results; supplementary fig. S3, Supplementary Material online), a result that is consistent with the predictions of the two hypotheses.
Fig. 3.
Phylogenetic positions of M. leucopterus subspecies. SNAPP tree based on 6,790 nuclear SNPs. Illustrations of birds were reproduced with the permission of Lynx Edicions.
Genomic Regions of Divergence Reveal Possible Targets of Selection for Male Plumage Color
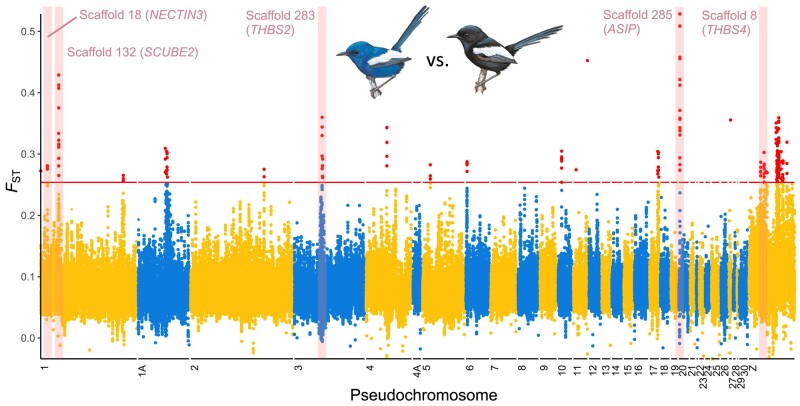
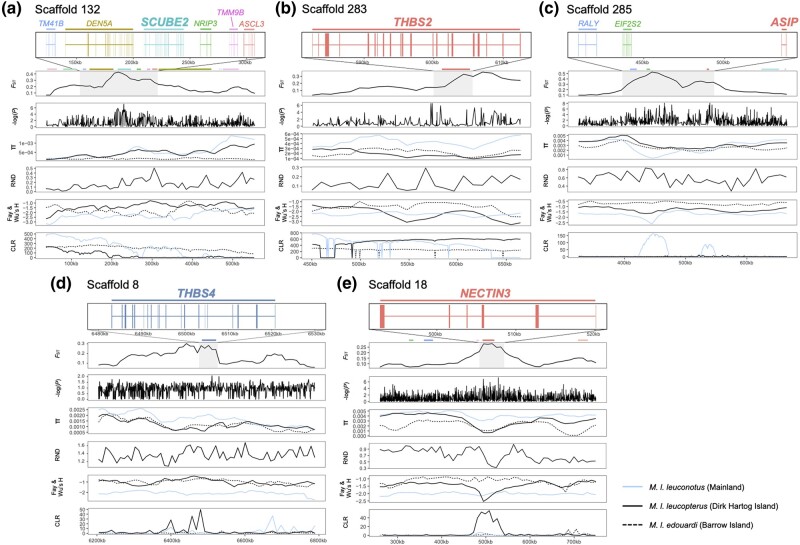
Genome-wide divergence (mean autosomal FST) between M. l. leuconotus (blue plumage) and M. l. leucopterus (black plumage) was 0.119 (supplementary fig. S4, Supplementary Material online), between M. l. leuconotus and M. l. edouardi (black plumage) was 0.274, and between M. l. leucopterus and M. l. edouardi was 0.369. In addition to comparison between subspecies, combining the two subspecies with black plumage (i.e. M. l. leucopterus and M. l. edouardi) in the divergence analysis facilitated identification of divergent genomic regions between blue and black birds, lowering the divergence (mean autosomal FST) between black insular and blue mainland subspecies to 0.081 (Fig. 4). Using windowed FST estimates, we identified 42 highly divergent genomic regions (exceeding 99.9th percentile) relative to the background (Fig. 4; supplementary table S2, Supplementary Material online), of which 20 were autosomal. These divergent genomic regions contained 90 annotated genes (supplementary table S2, Supplementary Material online) enriched for gene ontology categories of osteoblast and muscle cell differentiation and migration (supplementary fig. S5, Supplementary Material online) and not showing any obvious link to plumage color development. Enrichment of these gene ontology categories offers insights for future studies on the difference in muscle and bone development between island and mainland populations. However, five genes—ASIP, SCUBE2, THBS2, THBS4, and NECTIN3—were plausibly associated with plumage color differentiation (Fig. 5). ASIP has a known regulatory role in vertebrate melanogenesis. SCUBE2, THBS2, THBS4, and NECTIN3 all play roles in keratinocyte development and cell-to-cell/matrix interactions. When we separated the two black subspecies for the FST analysis, the comparison between M. l. leuconotus and M. l. leucopterus shows that the same regions harboring these five genes were all above the 99.9th percentile (supplementary fig. S4B, Supplementary Material online), whereas only the regions containing ASIP (scaffold 285) and SCUBE2 (scaffold 132) out of the five regions passed the cut-off in the comparison between M. l. leuconotus and M. l. edouardi (supplementary fig. S4C, Supplementary Material online).
Fig. 4.
Genome-wide differentiation between black and blue M. leucopterus. Points indicate overlapping sliding window FST values in 50 kb windows. Red points above red horizontal line are above 99.9th percentile. Five divergent regions with genes that are potentially relevant to plumage color development are highlighted in pink, with scaffold and gene identities labeled. Scaffolds are reordered as pseodochromosomes according to the T. guttata genome. Illustrations of birds were reproduced with the permission of Lynx Edicions.
Fig. 5.
The genomic landscape of the five divergent regions between black and blue M. leucopterus. The five regions are: a) scaffold 132 containing SCUBE2, b) scaffold 283 containing THBS2, c) scaffold 285 containing ASIP, d) scaffold 8 containing THBS4, and e) scaffold 18 containing NECTIN3. FST, SNP associations, genetic diversity (π), RND, Fay and Wu's H, and CLR are shown in different panels for each region. Blue, and black solid and dotted lines in the same panel of π, Fay and Wu's H, and CLR represent mainland, DHI, and BI populations, respectively. Annotated genes in those regions are depicted using blocks and lines to represent exons and introns, respectively, in the top panel.
In the five highly divergent regions, SNPs strongly associated with plumage color were identified by genome-wide association analysis (GWAS) in the loci and/or in the regions flanking those loci (Fig. 5). The region with the ASIP gene was lower in nucleotide diversity in the blue birds on mainland (π < 0.001; Fig. 5) than black birds on the two islands (both π > 0.002), which was in turn lower than the global π (autosomal; mainland population = 0.0034; DHI population = 0.0022; BI population = 0.0019). No introgression was identified from blue wrens to the blue M. leucopterus in those regions based on the low relative node depth (RND) values (Fig. 5). Signatures of selective sweep were detected in the ASIP and SCUBE2 regions in the blue birds, indicated by the significant negative Fay and Wu's H values (<99th percentile [Sterken et al. 2009], i.e. −2.59) and higher SweepFinder2 composite likelihood ratio (CLR) values in blue than in black birds (Fig. 5). The signature of selective sweep was less apparent in the SCUBE2 region (Fig. 5a) but particularly clear in the ASIP region in blue birds (Fig. 5c). Strikingly, the phylogeny of these two genomic regions differed from the species tree (Shimodaira-Hasegawa test, P < 0.001; Fig. 6a and b, supplementary fig. S6, Supplementary Material online): in these trees individuals clustered by plumage color. This could be due to the selective sweep in the blue birds. The genotype matrix of those two regions also show a pattern of selective sweep in the blue birds (Fig. 6c and d), which was not detected in the birds with black plumage.
Fig. 6.
Phylogeny and individual genotypes of the two regions of divergence. Phylogenetic tree reconstructed using SNAPP based on SNPs from the whole divergence peak region contains a) SCUBE2 or b) ASIP. c, d) Genotypes at SNPs between black and blue Malurus leucopterus populations in the (c) SCUBE2 and (d) ASIP regions. Each row represents one individual. Black triangles and circles denote M. leucopterus from DHI and BI, respectively. Light blue, black, and dark blue indicate positions homozygous for the allele that was the same as the reference genome (from a M. l. leuconotus sampled in Queensland, Australia), homozygous for the allele different from the reference, and heterozygous for both alleles, respectively. Missing values are represented by white. Number in parentheses indicates the polymorphic site number of each gene. Illustrations of birds were reproduced with the permission of Lynx Edicions.
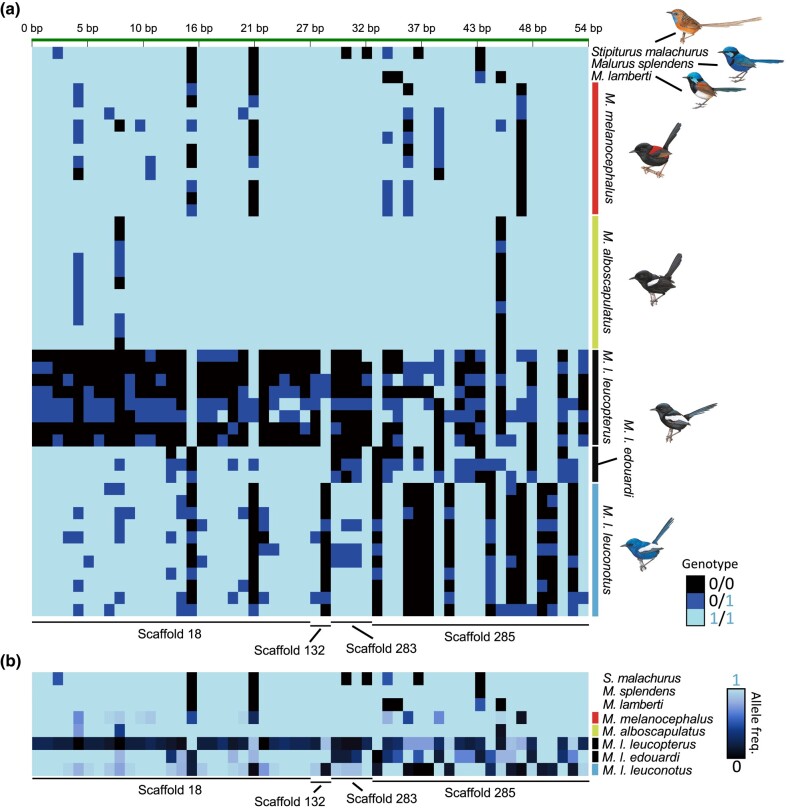
When we extracted those loci that were highly associated with plumage color differentiation between DHI and mainland populations in the five highly divergent genomic regions and compared the genotypes and allele frequencies between BI and DHI, the pattern for scaffold 285, 283, and 132 (one out of two SNPs) was similar between the two insular populations (Fig. 7). The genotypes and allele frequencies of the mainland population were different in these regions, especially for the region in scaffold 285, which was consistent with the selective sweep signature in the blue birds. The comparison of genotypes among all sequenced bi-colored wrens and outgroup individuals showed that the selective sweep regions of the blue M. leucopterus (i.e. scaffold 285 and 132) contain many derived SNPs that are different from the other two bi-colored wrens and outgroups (Fig. 7). Moreover, the SNPs in black M. leucopterus were not found in the other two bi-colored wrens that were also black in male nuptial plumage color, suggesting that the evolution of black plumage in M. leucopterus was independent from that in M. melanocephalus and M. alboscapulatus.
Fig. 7.
a) Genotypes and b) allele frequencies at 54 SNPs in the five highly divergent genomic regions (see Fig. 5) that were strongly associated (−log[P] > 6) with plumage color differentiation between M. l. leucopterus from DHI and M. l. leuconotus from the mainland. a) Genotypes of M. l. edouardi from BI, other bi-colored wrens, and outgroups are also shown. Each row represents one individual. Light blue, black, and dark blue indicate positions homozygous for the allele that was the same as the reference genome (from a M. l. leuconotus sampled in Queensland, Australia), homozygous for the allele different from the reference, and heterozygous for both alleles, respectively. Each column represents one SNP position. The scaffolds where the SNPs originated from are indicated at the bottom. b) Each row represents the allele frequencies of one species or subspecies of M. leucopterus, ranging from 0 (all alleles were different from the reference) to 1 (all alleles were the same as the reference). Illustrations of birds were reproduced with the permission of Lynx Edicions.
Discussion
Genetic Basis of Plumage Color Difference in M. Leucopterus
We have identified highly divergent genomic regions between blue and black M. leucopterus subspecies containing candidate genes associated with plumage color development (Figs. 4 and 5). The candidate genes include an important pigmentation gene and genes that may play a role in keratinocyte and feather nanostructure development. Variation of structural color in fairywrens is related to multiple independent features of the feather barb rather than a single factor (Fan et al. 2019). Both the density and distribution of melanin and thickness and nanostructure of cortex and spongy layer contribute to chromatic and achromatic variation in structural color (Fan et al. 2019). The gene encoding the ASIP (agouti signaling protein), which is an antagonistic ligand of MC1R, is in the most divergent genomic region between blue and black M. leucopterus and is the only well-studied pigmentation gene identified in the divergent regions. ASIP is an important melanin production modifier that can switch the synthesis of eumelanin to pheomelanin and regulate melanocyte precursor behavior in hair follicles (Gantz and Fong 2003).
In birds, ASIP was found to be upregulated in peripheral pulp adjacent to apigmented feather barbs, and it suppresses melanocyte differentiation from progenitor cells during feather growth (Lin et al. 2013). Differential melanocytes are required to produce and transport melanosomes to keratinocytes in developing feathers (Lin et al. 2013; D’Alba and Shawkey 2019; Saranathan and Finet 2021); expression of ASIP would therefore inhibit the formation and deposition of melanosomes. ASIP expression has been shown to affect body color and pattern in birds and other vertebrates (Nadeau et al. 2008; Norris and Whan 2008; Toews et al. 2016; Kratochwil 2019; Robic et al. 2019; Wang et al. 2020; Semenov et al. 2021; Goutte et al. 2022). Mutation of this gene has also been reported to link to island melanism in Monarcha castaneiventris, which has an insular population possessing a nonsynonymous substitution in the coding sequence (CDS) of ASIP (Campagna et al. 2022). In contrast, no mutations in the ASIP CDS were fixed in M. leucopterus subspecies (supplementary results, Supplementary Material online). The upstream region of ASIP is highly divergent between blue and black subspecies and likely regulates the expression of this gene. Mutations in the genomic region containing ASIP, EIF2S2, and RALY are also associated with pigmentation change in other avian species (Nadeau et al. 2008; Wang et al. 2020). We predict ASIP to be underexpressed in developing feathers of black M. leucopterus relative to the blue form, which leads to more differential melanocytes in developing feathers and hence more melanosomes in both the cortex and spongy layer of the feather barbs, resulting in the black color of the feather (Lin et al. 2013).
Another candidate gene also in a divergent genomic region with a selective sweep signature and that may link to structural plumage color development is SCUBE2 (signal peptide, cubulin [CUB] domain, epidermal growth factor [EGF]-like protein 2). This gene plays important roles in Sonic Hedgehog (SHH)-related biology such as organ development (Tsai et al. 2009), including feather development (Cooper and Milinkovitch 2023). SHH induces feather development and mediates programmed cell death in the feather barb marginal plate epithelium during keratinization of the barb plate epithelium (Ting-Berreth and Chuong 1996; Jung et al. 1998), which regulates barb and barbule formation (Chuong et al. 2000; Yu et al. 2002). SCUBE2 may play a role in developing feather barb nanostructure required for structural color production, and accounting for the difference in the nanostructure between blue and black subspecies (e.g. a more degraded spongy layer in black feathers). The peak of the genomic island of divergence is in the upstream region of SCUBE2, suggesting mutations in a cis-regulatory region may affect gene expression. Here, a protein difference could also be linked to the differentiation in phenotype, since a nearly fixed nonsynonymous substitution between blue and black subspecies has also been identified (supplementary results, Supplementary Material online).
Other candidate genes in divergent genomic regions that may link to feather development and plumage color differences are NECTIN3, thrombospondin-2 (THBS2), and thrombospondin-4 (THBS4). Nectin-3 is a cell–cell adhesion molecule that plays an important role in the formation of cell–cell adhesion junctions in keratinocytes (Tanaka et al. 2003) and is required for cell adhesion and differentiation in hair follicles and for proper hair formation in mice (Yoshida et al. 2014). Thrombospondins are extracellular glycoproteins participate in cell–cell and cell–extracellular matrix interaction (Adams and Lawler 2004, 2011) and are important for skin keratinocyte development (Adams and Lawler 2004, 2011; Mäemets-Allas et al. 2023). In particular, THBS4 orthologs are related to skin pigmentation in pearl danio (Danio albolineatus) and may play a general role in body color development across vertebrates (Huang et al. 2021). These three genes are divergent between DHI and mainland populations but not between BI and mainland populations (supplementary fig. S4, Supplementary Material online), a result that could be caused by the small sample size from BI or independent evolution of these genes on the two islands (e.g. the DHI population has a signature of selective sweep in the NECTIN3 region; Fig. 5e). Including more samples from BI in future studies would reduce the variance and may reveal a clearer pattern. With their associations with keratinocyte development, these genes, plus SCUBE2, may play a role in developing the feather nanostructure required for structural color, which is an important step in unravelling the molecular mechanism and evolution of structural coloration in birds. Future studies may perform functional analysis to confirm the roles that these candidate genes play in structural coloration.
Re-evolution of Blue Plumage in Mainland M. Leucopterus
We identified genomic islands of divergence that show a signature of selective sweep (Figs. 5 and 6). Surprisingly, selective sweeps occurred in the genomic regions containing ASIP and SCUBE2 in the blue mainland subspecies but not the black subspecies on islands, in contrast to signatures of selective sweep found in insular M. castaneiventris populations showing convergent evolution of island melanism (Campagna et al. 2022). Individual genotypes and allele frequencies in these two sweep regions in the blue subspecies are different from the two island subspecies (Fig. 7), which are more similar to each other even though they are more divergent genome-wide. Given the similarity of the two black subspecies in these two regions, the ancestor of M. leucopterus subspecies could have already become melanic before they arrived DHI and BI, i.e. black plumage evolved from a blue ancestor was ancestral for all the subspecies. The two island subspecies possess derived SNPs in these two regions not found in M. melanocephalus and M. alboscapulatus, which also have evolved black male nuptial plumage. If these are, or are closely linked to, the causal mutations render the plumage black in color in the two M. leucopterus island subspecies, the switch to melanism in the other two bi-colored wrens is an independent event. This independent origin of melanism is supported by the different extents of feather nanostructure organization—M. melanocephalus has a high degree of structural degradation (over evolutionary time) of the spongy layer in the black feather barb, whereas black M. leucopterus subspecies possess a spongy layer that is similar in nanostructure to those of blue M. leucopterus and Malurus splendens (Driskell et al. 2010), suggesting the switch to melanic form happened later in M. leucopterus than the other two melanic bi-colored species.
The degradation of feather nanostructure in these black fairywrens is consistent with an initial hypermelanization, followed by subsequent changes in spongy layer keratin organization and ramus shape for pigment presentation. The common ancestor of all M. leucopterus subspecies was suggested to have blue plumage and a functional spongy layer and feather barb nanostructure as in other blue wrens (Doucet et al. 2004; Driskell et al. 2010). The hypermelanization proposed to occur in the common ancestor of all M. leucopterus subspecies could be due to mutations in the ASIP region, which were likely inherited by all subspecies. This explains the sharing of SNPs in that region by the two island populations but no other species. Hypermelanization would then have relaxed selection to maintain the feather nanostructure needed for structural color, hence genes contributing to the nanostructure might accumulate mutations that affect the phenotype, which was shown by the spongy layer degraded with more holes (Driskell et al. 2010). If the melanic form was at the same time favored by natural selection, feather nanostructure would be selected for features that consolidate the black color, such as the thicker cortex and rami observed (Driskell et al. 2010). Alternatively, nanostructure could be selected for other nonsignaling functions. These changes could be due to mutations in SCUBE2 or other candidate genes relevant to feather nanostructure development, and could occur before or after the divergence of the subspecies. However, in contrast to the other two black bi-colored wren species, black M. leucopterus still have a functional spongy layer that can produce a blue color by coherent light scattering (Doucet et al. 2004), which means only a reversal of hypermelanization was required to switch the black plumage to blue again. The strong selective sweep signature in the genomic region containing ASIP supports this hypothesis of re-evolution of blue plumage on the mainland by mutations that reversed hypermelanization, in contrast to the hypothesis of convergent melanism on islands (Walsh et al. 2021). This sweep region in the blue subspecies contains many derived SNPs associated with the plumage color difference between blue and black subspecies (Fig. 7). These derived SNPs are not only different from the two island subspecies but also from the other two bi-colored wrens and outgroups, including one of the blue wrens, M. splendens. We, therefore, propose that the mainland subspecies likely re-evolved the mechanism to develop blue plumage again, from a black M. leucopterus ancestor that itself evolved from a blue ancestor. Assuming SCUBE2 is linked to the development of feather nanostructure, the selective sweep in the genomic region containing this gene in the blue subspecies likely postdated the dehypermelanization, since selection could only favor the development of optimal feather nanostructure for blue structural color (Igic et al. 2016) after the density and distribution of melanosome were altered. The mutations of the SCUBE2 region might facilitate the development of feather nanostructure similar to its blue ancestor or the sister blue wren species, e.g. a more structured and intact spongy layer (Driskell et al. 2010), in order to increase light scattering and interference and produce a brighter blue hue (Fan et al. 2019).
Therefore, instead of a case of convergent evolution of melanism on island populations (Walsh et al. 2021), the differentiation in plumage color between the island and mainland populations could be a case of re-evolution of blue plumage in the mainland population. The mutations and selective sweep in these two genomic regions containing important candidate genes, especially the pigmentation gene ASIP, in M. l. leuconotus was likely the mechanism of re-evolution of blue plumage on the mainland. Although hybridizations between fairywren species have been reported (Haines 2014; Ross and Briggs 2022; Welklin et al. 2022), including between M. l. leuconotus and the Superb fairywren M. cyaneus (Welklin et al. 2022), re-evolution of blue plumage in mainland M. leucopterus was not a result of genetic introgression from blue wrens (Fig. 5). The high diversity in feather plumage color among Malurus species and body regions within a species suggest that plumage color is a very labile trait during evolution of these birds (Fan et al. 2019), and re-evolution of blue plumage from a black one is feasible. A substantial difference in plumage color can be due to minor changes in the size and structure of light-scattering elements or pigment concentration or distribution (Prum and Torres 2003; Shawkey et al. 2005; Fan et al. 2019). Convergent regulatory changes in melanin synthesis pathway have also led to repeated evolution of structural coloration in birds (Saranathan and Finet 2021). In Malurus, Fan et al. (2019) showed that black feather barbs from species with both black and blue plumage patches, such as M. splendens and M. lamberti, contain a spongy layer that could produce a blue color, but the degradation and high density of melanosome in the spongy layer plus the thicker and more melanized cortex make them appear black. This suggests that evolutionary changes in modular regulatory control of color for blue and black plumage patches are common and relatively feasible in this clade, which likely also applies to the switches between blue and black whole body plumages in M. leucopterus. The mainland population of M. leucopterus diverged from the island populations in the late Pleistocene (Walsh et al. 2021), when the population size began to expand on the mainland (supplementary results; supplementary fig. S7, Supplementary Material online). This rapid population expansion might increase dispersal and gene flow on mainland and facilitate the spread of mutations for a “novel” phenotype, i.e. the blue plumage, especially if the trait was under strong selection.
Selective Pressure for Plumage Color Differentiation Between Islands and Mainland
Malurus fairywrens are one of the birds with the highest rates of extra-pair paternity (Karubian 2002; Colombelli-Négrel et al. 2009; Cockburn et al. 2013), and their high sexual dichromatism and very colorful male plumages indicate that their plumage is under strong sexual selection (Lindsay et al. 2011; Friedman and Remeš 2015). Males with brighter plumage are more attractive to females and have higher within- and extra-pair paternity (Karubian 2002; Webster et al. 2008). In M. leucopterus, female preference for bright blue color is likely retained from their ancestor in all descendant subspecies. Males of many fairywren species often carry colorful petals in their bills during courtship toward females (Karubian and Alvarado 2003; Montgomerie 2006). The black male M. leucopterus on the islands have higher incidence of petal carrying than mainland males and show a strong preference to carry blue petals (Rathburn and Montgomerie 2003), suggesting female's preference for males carrying blue petals. Female's preference for bright blue color has likely been maintained from an ancestor with a blue male nuptial plumage, even though it became black (Fig. 2), and could be the selective force driving the proposed re-evolution of blue male nuptial plumage in the mainland M. leucopterus population. If males carrying the mutations for a bluer plumage have a higher reproductive success, then the haplotype would spread quickly in the population, leaving a signature of selective sweep (Hejase et al. 2020) in M. l. leuconotus. We therefore propose that the preexisting female preference for blue color in M. leucopterus drove the re-evolution of blue nuptial plumage.
In contrast to the mainland subspecies, black plumage on the two islands is maintained, even though there is gene flow between mainland and island subspecies (Walsh et al. 2021). The maintenance of black plumage on islands could be due to weaker sexual selection on islands and/or natural selection. Both sexual dichromatism and dimorphism of body size in this species are reduced on islands (Endler 1990; Rathburn and Montgomerie 2003). Small population size, low genetic diversity, and limited choice of mates are believed to reduce sexual selection in isolated island populations compared to mainland counterparts (Frankham 1997; Omland 1997; Griffith 2000; Rathburn and Montgomerie 2003). The effective population size on the islands was estimated to be much smaller than that on the mainland (Walsh et al. 2021). In this study, the autosomal π were 0.0030 and 0.0019 for the DHI and BI subspecies, respectively, which were lower than the 0.0034 of the mainland subspecies. In addition to the difference in sexual dichromatism, mainland birds have bigger clutches, shorter incubation periods, and higher realized reproductive success (Rathburn and Montgomerie 2003). Island birds also have fewer helpers and are largely socially monogamous, and, importantly, appear to have lower levels of extra-pair paternity (Rathburn and Montgomerie 2003). These differences in reproductive behavior and social structure could further result in reduced strength of sexual selection on islands compared to mainland. In parallel, black plumage may confer an advantage under natural selection through providing solar radiation protection (Roulin 2014), bacterial degradation resistance (Goldstein et al. 2004), abrasion resistance (Bonser 1995), thermoregulation (Margalida et al. 2008), and crypsis against predation and brood parasitism (Rathburn and Montgomerie 2003; Surmacki et al. 2021). However, since black plumage might have evolved on the mainland but not small islands, similar to what likely took place in the other two bi-colored wrens, natural selection specific to insular environments alone cannot explain the evolution of black plumage. Alternatively, indirect selection through pleiotropic effects was proposed to be the evolutionary driver of melanism (Walsh et al. 2021). It is worth noting that the estimated size of the ancestral M. leucopterus population before splitting into three subspecies is much smaller than the current mainland population and comparable to the current DHI population (Walsh et al. 2021), suggesting that the strength of sexual selection could be lower in the ancestral population than the later expanded mainland population and might have led to a reduced sexual dichromatism in the ancestral form.
Conclusions
Feather barb nanostructure and melanosome features are crucial component in avian structural coloration. Their nature of being independent and evolutionary labile are likely to facilitate the evolution of a high diversity of plumage structural color, both within and between species (Maia et al. 2013; Fan et al. 2019). Feather structural colors have independently evolved many times in more than 45 avian families (Saranathan and Finet 2021). The convergent evolution of similar feather barb nanostructure and melanosome distribution in many avian lineages shows that the genetic basis to develop the medullary spongy layer and change melanosome density and distribution in keratinocytes is simple, and feather structural color can evolve relatively easily (Saranathan and Finet 2021). The transition to blue structural color, from the black plumage M. leucopterus that already possess a partly degraded spongy layer interrupted by melanin granules, appears to be an example of this evolutionary lability. We show that a selective sweep in the genomic region containing the pigmentation gene ASIP was likely the cause of a change in the melanism state of the feather barb leading to the proposed re-evolution of the blue plumage color. Any changes in the molecular mechanisms that affect melanin synthesis and deposition would influence the potential to develop plumage structural color. In M. l. leuconotus, genetic changes related to feather nanostructure development likely happened following the mutations in the ASIP region to consolidate the blue structural color. The parallel evolution of noniridescent structural color in many avian lineages (Saranathan and Finet 2021) could follow the same order of evolutionary changes. Future studies of the genetic basis in different species and how much it is shared among birds with similar feather structural color will increase our understanding of the evolution of avian phenotypic diversity and its interaction with sexual and natural selection.
Materials and Methods
Sample Collection and DNA Extraction
Samples from three subspecies of M. leucopterus were collected, including M. l. leuconotus on mainland Australia (n = 11), M. l. leucopterus on DHI (n = 8), and M. l. edouardi on BI (n = 3) (Fig. 1; supplementary table S1, Supplementary Material online). In total, 22 M. leucopterus males were included in this study, with 11 mainland individuals displayed a blue plumage and 11 insular individuals on island with black plumage. The two sister species M. melanocephalus (n = 11) and M. alboscapulatus (n = 11) were also sampled (supplementary table S1, Supplementary Material online). One individual of M. splendens, a member of blue wrens that are sister to the bi-colored wrens, were included. Malurus lamberti and Stipiturus malachurus were also included as outgroups (supplementary table S1, Supplementary Material online). Samples were collected from the field or requested from museum collections (supplementary table S1, Supplementary Material online). We isolated genomic DNA using DNeasy Blood and Tissue Kit (Qiagen, Hilden, Germany) following the manufacturer's protocol. We confirmed the sex of all individuals to be male using published primers targeting CHD1 genes (2550F and 2718R) (Fridolfsson and Ellegren 1999) for PCR and measured DNA concentration with a Qubit dsDNA HS Assay Kit (Invitrogen, Carlsbad, USA). Sample collection of M. leucopterus were approved by the Cornell University Institutional Animal Care and Use Committee (IACUC 2009 to 0105) and the James Cook University Animal Ethics Committee (A2100), and was performed with authorization of the Australian Bird and Bat Banding Scheme (#1965) and the Western Australia Department of Parks and Wildlife (BB003372). Sample collection of M. alboscapulatus was conducted under the approval of IACUC #0395 and ASAF #04573 and the Australian Bird and Bat Banding Scheme, and with annual permits from the Papua New Guinea Department of Environment and Conservation, permissions from the provincial governments of Milne Bay, Western, and Madang Provinces.
Data Generation and Processing
Refecrence Genome Sequencing and Assembly
The genome of one individual of each species was sequenced and assembled. We performed whole-genome library preparation, sequencing, and assembly following Sin et al. (2020), Huynh et al. (2023), and Grayson et al. (2017). In brief, a DNA fragment library of 220 bp insert size was prepared for M. l. leuconotus (from Queensland, Australia; supplementary table S1, Supplementary Material online), M. splendens, M. lamberti, and S. malachurus using the PrepX ILM 32i DNA Library Kit (Takara), and mate-pair libraries of 3 kb insert size were prepared for M. leucopterus and M. splendens using the Nextera Mate Pair Sample Preparation Kit (cat. No. FC-132-1001, Illumina). Mate-pair library of 6 kb insert size were also prepared for M. leucopterus. We performed DNA shearing for the fragment and mate-pair library preparations using Covaris S220. We used the 0.75% agarose cassette in the Pippin Prep (Sage Science) for size selection of the mate-pair library (target size 3 or 6 kb, “Tight” mode). We then assessed fragment and mate-pair library qualities using the High Sensitivity D1000 ScreenTape for the Tapestation (Agilent) and High Sensitivity DNA Kit for the Bioanalyzer (Agilent), respectively, and quantified the libraries with qPCR (KAPA library quantification kit) prior to sequencing. We sequenced the libraries on an Illumina HiSeq 2500 instrument (High Output 250 kit, PE 125 bp reads) at the Bauer Core facility at Harvard University to ∼65× coverage. We assessed the quality of the sequencing data using FastQC and removed adapters using Trimmomatic (Bolger et al. 2014). We performed de novo genome assembly using AllPaths-LG v52488 (Gnerre et al. 2011) for M. leucopterus and M. splendens. We estimated the completeness of the assembled genomes with BUSCO v2.0 (Simão et al. 2015) (supplementary table S3, Supplementary Material online). The de novo genomes of M. alboscapulatus (GCA_025434525.1) (Enbody et al. 2022) and M. melanocephalus (GCA_030028575.1) (Khalil et al. 2023) included in the analysis of this study were sequenced and assembled using the same strategies.
Genome Annotation
We annotated the M. leucopterus genome following Sin et al. (2022). We used MAKER v2.31.8 (Holt and Yandell 2011), combining ab initio gene prediction with protein-based evidence from 16 other vertebrates (10 birds, 3 reptiles, 2 mammals, and 1 fish species) and gene predictions from Gallus gallus. The genome annotation using MAKER identified a total of 17,443 gene models. We functionally annotated the genome to identify putative gene function and protein domains using NCBI BLAST+ and the UniProt/Swiss-Prot set of proteins. We used BLASTP on the list of proteins identified by MAKER with an e-value of 1e−6.
Pseudochromosome Reconstruction
We used Satsuma (Grabherr et al. 2010) and MUMmer (Kurtz et al. 2004) to align all assembled scaffolds in the M. leucopterus genome against the zebra finch (Taeniopygia guttata) genome (bTaeGut1_v1; GCF_003957565.1). A total of 312 sex-linked scaffolds—accounting for ∼9.08% of M. leucopterus draft genome—aligned to the Z chromosome. The aligned scaffolds of the M. leucopterus draft genome were reordered to form pseudochromosomes.
Whole-genome Resequencing Library Preparation
To determine the genomic region(s) associated with male ornamentation polymorphism, we performed whole-genome resequencing at ∼4× depth-of-coverage for the 22 M. leucopterus. We also re-sequenced 11 samples of each of the two sister species, M. melanocephalus and M. alboscapulatus. DNA fragment libraries of 220 bp insert size were prepared using the protocol mentioned in the whole-genome library preparation section. All libraries were multiplexed in equimolar ratio and sequenced using an Illumina HiSeq instrument (High Output 250 kit, PE 125 bp reads). Preliminary quality assessment was performed using FastQC (www.bioinformatics.babraham.ac.uk/projects/fastqc).
Data Analysis
Whole-genome Resequencing Data Preprocessing
We followed GATK best practices (Van der Auwera et al. 2013) using Picard Tools v. 2.20.6 (broadinstitute.github.io/picard/) and Genome Analysis Toolkit (GATK) 3.8 (McKenna et al. 2010) to preprocess the data. We removed adapters of the resequencing data using NGmerge v0.3 (Gaspar 2018) and Picard Tools. We used BWA-MEM (0.7.13; Li and Durbin 2009) to map reads to the M. leucopterus genome assembly. The BAM files were merged, duplicates marked, sorted, and validated using Picard Tools. The reads were realigned around indels using GATK and underwent an initial variant calling using GATK HaplotypeCaller. We then applied a base quality score recalibration (BQSR) on SNPs and indels. The recalibrated BAM files were then passed to ANGSD v0.921 (Korneliussen et al. 2014) to estimate genotype likelihood for final SNP calling, by first calculating the site allele frequency for each species/subspecies with the following settings: -skipTriAllelic 1 -remove_bads 1 -trim 0 -minMapQ 20 -minQ 20 -uniqueOnly 1 -baq 1 -C 50 -only_proper_pairs 0 -doCounts 1 -GL 1 -doMaf 1 -doPost 1 -doMajorMinor 1 -dobcf 1 –ignore-RG 0 -dogeno 1 -doSaf 1, and the output was converted to VCF format using a custom R script (github.com/rcristofari/RAD-Scripts/blob/master/angsd2vcf.R). The VCF files from ANGSD were used for summary statistic calculations unless otherwise specified.
Inference of Demographic History
To investigate the historical change in effective population size (Ne) in M. leucopterus we used the Pairwise Sequential Markovian Coalescent (PSMC) model (Li and Durbin 2011) based on the diploid whole-genome sequence to reconstruct the population history. We filtered the raw sequencing data of the M. l. leuconotus individual that we used for whole-genome assembly using fastp (Chen et al. 2018) and removed sex-linked scaffolds. The settings for the PSMC atomic time intervals were “4 + 25*2 + 4 + 6”. We performed 100 bootstraps to compute the variance in estimate of Ne. We used the estimated mutation rate of 3.3 × 10−3 substitutions per site per million years (Passeriformes; Zhang et al. 2014). The estimated generation time is 2 yr (Rowley and Russell 1995; Garnett 2021).
Phylogenomic Analysis and PCA
This VCF file was first formatted using a script vcf2phylip.py (https://github.com/edgardomortiz/vcf2phylip) for phylogenetic reconstruction using IQTREE 2.2.0 (Minh et al. 2020). Bi-allelic SNPs thinned by intervals of 5 kb, resulting in 168,378 polymorphic sites, were concatenated and used for ML tree reconstruction with 1,000 bootstraps. The best substitution model was selected using ModelFinder (Kalyaanamoorthy et al. 2017). We also employed + ASC model to correct ascertainment bias as SNP data did not contain invariant sites, and we appended -bnni to the regular UFBoot command to reduce the risk of overestimating branch support (Hoang et al. 2018). Coalescent-based methods were shown to outperform concatenation in inferring species-level phylogeny, even when gene flow is present (Long and Kubatko 2018). Phylogeny was therefore also inferred using the coalescent-based SNAPP package (Bryant et al. 2012) in BEAST2 (Bouckaert et al. 2019) and SVDquest (Vachaspati and Warnow 2018) based on 6,790 genome-wide SNPs, with one SNP sampled from each 200 kb window. For SNAPP, 2 millions Monte Carlo Markov Chain (MCMC) generations were run with trees sampled every 1,000 generations. The resulting tree was visualized using DensiTree (Bouckaert 2010). For SVDquest, 1,000 replicates were conducted. We also used ngsTools (Fumagalli et al. 2014) to generate a covariance matrix based on genotype-likelihoods for PCA.
Ancestral Plumage Color Reconstruction
We inferred the ancestral state of plumage color (i.e. black or blue) using PastML v1.9.42 (Ishikawa et al. 2019), with the SVDquest tree (supplementary fig. S1B, Supplementary Material online) as the input tree topology. We used the ML algorithm with the probabilistic model of marginal posterior probabilities approximation (MPPA) and the F81 model.
Ancestral State Inference
The ancestral genome of M. leucopterus, M. melanocephalus, and M. alboscapulatus was reconstructed using the software est-sfs v2.0395 (Keightley and Jackson 2018) based on resequencing data, by setting these species in the bi-colored clade as ingroups and M. splendens, M. lamberti, and S. malachurus as outgroups, ordered from the most to the least closely related based on our inferred species trees. The software was designed to infer the unfolded site frequency spectrum (SFS) but it also output the allelic state probabilities of each site, which we further used to reconstruct the ancestral genome sequence. The inferred ancestral genome was then used in downstream genomic analyses of the re-sequenced samples to polarize alleles.
Summary Statistic Calculations and Characterization of Regions of Divergence
We used the output from ANGSD and generated the site allele frequencies (SAF) (with -doSaf), which was then used to compute the unfolded SFS and to estimate various statistics. Alleles were polarized using the ancestral genome. We then obtained joint frequency spectrums for each between-subspecies comparison using realSFS, and then estimated FST. We calculated FST for the blue and black M. leucopterus comparison. We calculated FST in overlapping sliding windows of 50 kb with 10 kb steps. Windows with FST above the 99.9th percentile of autosomal values were considered regions of divergence and further examined for gene content.
Genetic diversity (π) and Fay and Wu's H (Fay and Wu 2000) were also calculated for sliding windows across the genome using ANGSD. A significant negative value of Fay and Wu's H (<99th percentile) (Sterken et al. 2009) indicates an excess of high-frequency derived SNPs due to selective sweep. We used SweepFinder2 (DeGiorgio et al. 2016) to detect signature of selective sweep in the regions of divergence within M. leucopterus subspecies with blue or black plumage. Stipiturus malachurus was used as the outgroup for this analysis, and the allele frequency within target group was calculated using VCFtools (Danecek et al. 2011) including both variant and invariant sites but separately for autosomal or Z-linked scaffolds. We estimated the CLR statistics every 1,000 bp. Elevated CLR values estimated by SweepFinder2 indicates a signature of selection sweep. We also conducted GWAS based on allelic counts of each locus in Plink v1.90b6.26 (Purcell et al. 2007) to identify the SNPs associated with the plumage color differentiation between black and blue M. leucopterus. Loci with a minor allele frequency lower than 0.05 and violated Hardy–Weinberg equilibrium (P ≤ 1e−6) were excluded for this analysis. To test if the candidate genomic regions underlying plumage color difference between blue and black M. leucopterus were introgressed from blue fairywren species that are sister to the bi-colored wrens, we calculated the RND (Feder et al. 2005) between blue and black M. leucopterus, using M. alboscapulatus as the outgroup. The RND was calculated as absolute nucleotide divergence (DXY)(between black and blue M. leucopterus)/([DXY(between black M. leucopterus and M. alboscapulatus) + DXY(between blue M. leucopterus and M. alboscapulatus)]/2). DXY was calculated using pixy (Korunes and Samuk 2021) with VCF files generated by GATK's GenotypeGVCFs, including variant and invariant sites. RND values are expected to lie between 0 and 1 for nonintrogressed genomic regions, while for introgressed regions RND values should be well above 1 (Lopes et al. 2016).
In the genomic regions of divergence, we examined all the annotated genes and identified those that are potentially relevant to plumage color development, such as genes related to pigment development or feather/keratinocyte development. For each highly divergent region with relevant candidate genes and a signature of selective sweep, we reconstructed the gene tree based on concatenated SNPs (2,355 and 666 SNPs for the ASIP and SCUBE2 regions, respectively) using IQTREE 2.2.0 (Minh et al. 2020), with 1,000 bootstraps. The best substitution model was selected using ModelFinder (Kalyaanamoorthy et al. 2017). We also inferred the phylogeny using SNAPP (Bryant et al. 2012). Two millions Monte Carlo Markov Chain (MCMC) generations were run with trees sampled every 1,000 generations. Topology discordance between the species tree and trees of the regions with a selective sweep signature was tested using the Shimodaira–Hasegawa test (SH test) (Shimodaira and Hasegawa 1999) with 1,000 bootstrap replicates. We also visualized the genotypes using RectChr (github.com/BGI-shenzhen/RectChr). To perform gene enrichment analysis, we first extracted protein sequences from the genome annotation file and functionally annotated the sequences using eggNOG-mapper v2 (Cantalapiedra et al. 2021). The genes in the top 99.9th percentile FST peaks in the black and blue subspecies comparison were used to search for enriched pathways using clusterProfiler 4.0 (Wu et al. 2021).
To study the evolution of plumage color across Malurus, we investigated the SNP loci that had strong association with the plumage color difference between black and blue M. leucopterus in all studied species, including other bi-colored wrens and outgroup species. We first performed association analysis (Purcell et al. 2007) between the blue M. l. leuconotus and black M. l. leucopterus from DHI, excluding those black individuals from the BI, and extracted those SNPs with strong association (i.e. −log[P] > 6) with the color polymorphism. We then used RectChr to plot the genotype of these SNP loci in all individuals and species, including M. l. edouardi from BI. We also visualized the allele frequencies of these loci in all M. leucopterus subspecies and other species.
We also examined the coding sequence (CDS) of the candidate genes for fixed or nearly fixed nucleotide substitutions. We used SnpEff (Cingolani et al. 2012) to annotate the variants of the candidate genes and to differentiate synonymous and nonsynonymous substitutions. Allele frequency at each site was calculated using VCFtools (Danecek et al. 2011).
Supplementary Material
Acknowledgments
This research was supported by the University of Hong Kong start-up granted to S.Y.W.S. and funds from Harvard University. We thank Museum of Comparative Zoology, University of Washington Burke Museum, University of Kansas Biodiversity Institute, and American Museum of Natural History for providing the samples; and Tim Sackton, Leonardo Campagna, Jennifer Walsh, and Michaël Nicolaï for helpful discussion. The computations were performed using research computing facilities offered by the Information Technology Services at the University of Hong Kong, and the Odyssey cluster supported by the FAS Division of Science, Research Computing Group at Harvard University. We thank the Wetmore Colles Fund of the Museum of Comparative Zoology for offsetting costs of publishing and open access for this paper.
Contributor Information
Simon Yung Wa Sin, School of Biological Sciences, The University of Hong Kong, Hong Kong, China; Department of Organismic and Evolutionary Biology, Harvard University, Cambridge, MA 02138, USA.
Fushi Ke, School of Biological Sciences, The University of Hong Kong, Hong Kong, China.
Guoling Chen, School of Biological Sciences, The University of Hong Kong, Hong Kong, China.
Pei-Yu Huang, School of Biological Sciences, The University of Hong Kong, Hong Kong, China.
Erik D Enbody, Department of Ecology and Evolutionary Biology, Tulane University, New Orleans, LA 70118, USA.
Jordan Karubian, Department of Ecology and Evolutionary Biology, Tulane University, New Orleans, LA 70118, USA.
Michael S Webster, Cornell Lab of Ornithology and Department of Neurobiology and Behavior, Cornell University, Ithaca, NY 14853, USA.
Scott V Edwards, Department of Organismic and Evolutionary Biology, Harvard University, Cambridge, MA 02138, USA.
Supplemental Material
Supplementary material is available at Molecular Biology and Evolution online.
Author Contributions
Conceptualization, S.Y.W.S. and S.V.E.; sample collection, J.K., E.E., M.S.W., and S.V.E.; methodology, S.Y.W.S.; analysis, S.Y.W.S., F.K., G.C., and P.Y.H.; writing—original draft, S.Y.W.S.; writing—review and editing, all authors; supervision, S.Y.W.S. and S.V.E.
Conflict of Interest
The authors declare no competing interests.
Data Availability
The sequencing data and genome assemblies have been archived in NCBI under the BioProject accession number PRJNA1046240, PRJNA1046245, and PRJNA1046643. All software are cited in the Materials and methods and are publicly available.
References
- Adams JC, Lawler J. The thrombospondins. Int J Biochem Cell Biol. 2004:36(6):961–968. 10.1016/j.biocel.2004.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adams JC, Lawler J. The thrombospondins. Cold Spring Harb Perspect Biol. 2011:3(10):a009712. 10.1101/cshperspect.a009712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baeckens S, Van Damme R. The island syndrome. Curr Biol. 2020:30(8):R338–R339. 10.1016/j.cub.2020.03.029. [DOI] [PubMed] [Google Scholar]
- Bolger A, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014:30(15):2114–2120. 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonser RH. Melanin and the abrasion resistance of feathers. Condor. 1995:97(2):590–591. 10.2307/1369048. [DOI] [Google Scholar]
- Bouckaert RR. DensiTree: making sense of sets of phylogenetic trees. Bioinformatics. 2010:26(10):1372–1373. 10.1093/bioinformatics/btq110. [DOI] [PubMed] [Google Scholar]
- Bouckaert R, Vaughan TG, Barido-Sottani J, Duchêne S, Fourment M, Gavryushkina A, Heled J, Jones G, Kühnert D, De Maio N. BEAST 2.5: an advanced software platform for Bayesian evolutionary analysis. PLoS Comput Biol. 2019:15(4):e1006650. 10.1371/journal.pcbi.1006650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryant D, Bouckaert R, Felsenstein J, Rosenberg NA, RoyChoudhury A. Inferring species trees directly from biallelic genetic markers: bypassing gene trees in a full coalescent analysis. Mol Biol Evol. 2012:29(8):1917–1932. 10.1093/molbev/mss086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buades JM, Rodríguez V, Terrasa B, Perez-Mellado V, Brown RP, Castro JA, Picornell A, Ramon M. Variability of the mc1r gene in melanic and non-melanic Podarcis lilfordi and Podarcis pityusensis from the balearic archipelago. PLoS One. 2013:8(1):e53088. 10.1371/journal.pone.0053088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campagna L, Mo Z, Siepel A, Uy JAC. Selective sweeps on different pigmentation genes mediate convergent evolution of island melanism in two incipient bird species. PLoS Genet. 2022:18(11):e1010474. 10.1371/journal.pgen.1010474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cantalapiedra CP, Hernández-Plaza A, Letunic I, Bork P, Huerta-Cepas J. eggNOG-mapper v2: functional annotation, orthology assignments, and domain prediction at the metagenomic scale. Mol Biol Evol. 2021:38(12):5825–5829. 10.1093/molbev/msab293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castilla A. A case of melanism in a population of the insular lizard podarcis hispanica atrata. Boll Soc d'Hist Nat Balears. 1994:37:175–179. [Google Scholar]
- Cerca J, Cotoras DD, Bieker VC, De-Kayne R, Vargas P, Fernández-Mazuecos M, López-Delgado J, White O, Stervander M, Geneva AJ. Evolutionary genomics of oceanic island radiations. Trends Ecol Evol (Amst). 2023:3(7):631–642. 10.1016/j.tree.2023.02.003. [DOI] [PubMed] [Google Scholar]
- Chen S, Zhou Y, Chen Y, Gu J. Fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics. 2018:34(17):i884–i890. 10.1093/bioinformatics/bty560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuong C-M, Chodankar R, Widelitz RB, Jiang T-X. Evo-devo of feathers and scales: building complex epithelial appendages. Curr Opin Genet Dev. 2000:10(4):449–456. 10.1016/S0959-437X(00)00111-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cingolani P, Platts A, Wang L, Coon M, Nguyen T, Wang L, Land S, Lu X, Ruden DM. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly. 2012:6(2):80–92. 10.4161/fly.19695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cockburn A, Brouwer L, Double MC, Margraf N, van de Pol M. Evolutionary origins and persistence of infidelity in Malurus: the least faithful birds. Emu. 2013:113(3):208–217. 10.1071/MU12094. [DOI] [Google Scholar]
- Collins P. Variation in the plumage of the White-winged fairy-wren ‘Malurus leucopterus’. Aust Bird Watch. 1995:16:130–131. [Google Scholar]
- Colombelli-Négrel D, Schlotfeldt BE, Kleindorfer S. High levels of extra-pair paternity in superb fairy-wrens in South Australia despite low frequency of auxiliary males. Emu. 2009:109(4):300–304. 10.1071/MU09035. [DOI] [Google Scholar]
- Cooke TF, Fischer CR, Wu P, Jiang T-X, Xie KT, Kuo J, Doctorov E, Zehnder A, Khosla C, Chuong C-M. Genetic mapping and biochemical basis of yellow feather pigmentation in budgerigars. Cell. 2017:171(2):427–439.e421. 10.1016/j.cell.2017.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper RL, Milinkovitch MC. Transient agonism of the sonic hedgehog pathway triggers a permanent transition of skin appendage fate in the chicken embryo. Sci Adv. 2023:9(20):eadg9619. 10.1126/sciadv.adg9619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Alba L, Shawkey MD. Melanosomes: biogenesis, properties, and evolution of an ancient organelle. Physiol Rev. 2019:99(1):1–19. 10.1152/physrev.00059.2017. [DOI] [PubMed] [Google Scholar]
- Danecek P, Auton A, Abecasis G, Albers C, Banks E, DePristo M, Handsaker R, Lunter G, Marth G, Sherry S, et al. The variant call format and VCFtools. Bioinformatics. 2011:27(15):2156–2158. 10.1093/bioinformatics/btr330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeGiorgio M, Huber CD, Hubisz MJ, Hellmann I, Nielsen R. SweepFinder2: increased sensitivity, robustness and flexibility. Bioinformatics. 2016:32(12):1895–1897. 10.1093/bioinformatics/btw051. [DOI] [PubMed] [Google Scholar]
- Doucet S, Shawkey M, Rathburn M, Mays H Jr, Montgomerie R. Concordant evolution of plumage colour, feather microstructure and a melanocortin receptor gene between mainland and island populations of a fairy–wren. Proc R Soc London Ser B Biol Sci. 2004:271(1549):1663–1670. 10.1098/rspb.2004.2779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Driskell AC, Pruett-Jones S, Tarvin KA, Hagevik S. Evolutionary relationships among blue-and black-plumaged populations of the white-winged fairy-wren (Malurus leucopterus). Aust J Zool. 2002:50(6):581–595. 10.1071/ZO02019. [DOI] [Google Scholar]
- Driskell AC, Prum RO, Pruett-Jones S. The evolution of black plumage from blue in Australian fairy-wrens (Maluridae): genetic and structural evidence. J Avian Biol. 2010:41(5):505–514. 10.1111/j.1600-048X.2009.04823.x. [DOI] [Google Scholar]
- Eliason CM, Clarke JA. Cassowary gloss and a novel form of structural color in birds. Sci Adv. 2020:6(20):eaba0187. 10.1126/sciadv.aba0187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eliason CM, Clarke JA, Kane SA. Wrinkle nanostructures generate a novel form of blue structural color in great argus flight feathers. iScience. 2023:26(1):105912. 10.1016/j.isci.2022.105912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eliason CM, Maia R, Shawkey MD. Modular color evolution facilitated by a complex nanostructure in birds. Evolution. 2015:69(2):357–367. 10.1111/evo.12575. [DOI] [PubMed] [Google Scholar]
- Enbody ED, Sin SYW, Boersma J, Edwards SV, Ketaloya S, Schwabl H, Webster MS, Karubian J. The evolutionary history and mechanistic basis of female ornamentation in a tropical songbird. Evolution. 2022:76(8):1720–1736. 10.1111/evo.14545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endler JA. On the measurement and classification of colour in studies of animal colour patterns. Biol J Linn Soc. 1990:41(4):315–352. 10.1111/j.1095-8312.1990.tb00839.x. [DOI] [Google Scholar]
- Fan M, D’alba L, Shawkey MD, Peters A, Delhey K. Multiple components of feather microstructure contribute to structural plumage colour diversity in fairy-wrens. Biol J Linn Soc. 2019:128(3):550–568. 10.1093/biolinnean/blz114. [DOI] [Google Scholar]
- Fay JC, Wu C-I. Hitchhiking under positive Darwinian selection. Genetics. 2000:155(3):1405–1413. 10.1093/genetics/155.3.1405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feder JL, Xie X, Rull J, Velez S, Forbes A, Leung B, Dambroski H, Filchak KE, Aluja M. Mayr, Dobzhansky, and Bush and the complexities of sympatric speciation in Rhagoletis. Proc Natl Acad Sci USA. 2005:102(suppl_1):6573–6580. 10.1073/pnas.0502099102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ford J. Minor isolates and minor geographical barriers in avian speciation in continental Australia. Emu. 1987:87(2):90–102. 10.1071/MU9870090. [DOI] [Google Scholar]
- Frankham R. Do island populations have less genetic variation than mainland populations? Heredity. 1997:78(3):311–327. 10.1038/hdy.1997.46. [DOI] [PubMed] [Google Scholar]
- Fridolfsson AK, Ellegren H. A simple and universal method for molecular sexing of non-ratite birds. J Avian Biol. 1999:30(1):116–121. 10.2307/3677252. [DOI] [Google Scholar]
- Friedman N, Remeš V. Rapid evolution of elaborate male coloration is driven by visual system in Australian fairy-wrens (Maluridae). J Evol Biol. 2015:28(12):2125–2135. 10.1111/jeb.12737. [DOI] [PubMed] [Google Scholar]
- Fumagalli M, Vieira FG, Linderoth T, Nielsen R. ngsTools: methods for population genetics analyses from next-generation sequencing data. Bioinformatics. 2014:30(10):1486–1487. 10.1093/bioinformatics/btu041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gantz I, Fong TM. The melanocortin system. Am J Physiol Endocrinol Metab. 2003;284(3):E468–E474. 10.1152/ajpendo.00434.2002. [DOI] [PubMed] [Google Scholar]
- Garnett ST. The action plan for Australian birds 2020. Clayton, Victoria, Australia: CSIRO publishing; 2021. [Google Scholar]
- Gaspar JM. NGmerge: merging paired-end reads via novel empirically-derived models of sequencing errors. BMC Bioinformatics. 2018:19(1):1–9. 10.1186/s12859-018-2579-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gnerre S, MacCallum I, Przybylski D, Ribeiro F, Burton J, Walker B, Sharpe T, Hall G, Shea T, Sykes S, et al. High-quality draft assemblies of mammalian genomes from massively parallel sequence data. Proc Natl Acad Sci USA. 2011:108(4):1513–1518. 10.1073/pnas.1017351108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldstein G, Flory KR, Browne BA, Majid S, Ichida JM, Burtt EH Jr. Bacterial degradation of black and white feathers. Auk. 2004:121(3):656–659. 10.1642/0004-8038(2004)121[0656:BDOBAW]2.0.CO;2. [DOI] [Google Scholar]
- Goutte S, Hariyani I, Utzinger KD, Bourgeois Y, Boissinot S. Genomic analyses reveal association of ASIP with a recurrently evolving adaptive color pattern in frogs. Mol Biol Evol. 2022:39(11): msac235. 10.1093/molbev/msac235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grabherr MG, Russell P, Meyer M, Mauceli E, Alföldi J, Palma FD, Lindblad-Toh K. Genome-wide synteny through highly sensitive sequence alignment: Satsuma. Bioinformatics. 2010:26(9):1145–1151. 10.1093/bioinformatics/btq102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grayson P, Sin SYW, Sackton TB, Edwards SV. Comparative genomics as a foundation for evo-devo studies in birds. New York, NY: Humana Press; 2017. [DOI] [PubMed] [Google Scholar]
- Griffith SC. High fidelity on islands: a comparative study of extrapair paternity in passerine birds. Behav Ecol. 2000:11(3):265–273. 10.1093/beheco/11.3.265. [DOI] [Google Scholar]
- Haines P. Hybrid fairywren at hart lagoon, river Murray, South Australia. South Aust Ornithol. 2014:39:87–89. [Google Scholar]
- Hejase HA, Salman-Minkov A, Campagna L, Hubisz MJ, Lovette IJ, Gronau I, Siepel A. Genomic islands of differentiation in a rapid avian radiation have been driven by recent selective sweeps. Proc Natl Acad Sci USA. 2020:117(48):30554–30565. 10.1073/pnas.2015987117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoang DT, Chernomor O, Von Haeseler A, Minh BQ, Vinh LS. UFBoot2: improving the ultrafast bootstrap approximation. Mol Biol Evol. 2018:35(2):518–522. 10.1093/molbev/msx281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holt C, Yandell M. MAKER2: an annotation pipeline and genome-database management tool for second-generation genome projects. BMC Bioinformatics. 2011:12(1):491. 10.1186/1471-2105-12-491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang D, Lewis VM, Foster TN, Toomey MB, Corbo JC, Parichy DM. Development and genetics of red coloration in the zebrafish relative Danio albolineatus. Elife. 2021:10:e70253. 10.7554/eLife.70253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huynh S, Cloutier A, Chen G, Chan DTC, Lam DK, Huyvaert KP, Sato F, Edwards SV, Sin SYW. Whole-genome analyses reveal past population fluctuations and low genetic diversities of the North Pacific albatrosses. Mol Biol Evol. 2023:40(7):msad155. 10.1093/molbev/msad155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Igic B, D’Alba L, Shawkey MD. Manakins can produce iridescent and bright feather colours without melanosomes. J Exp Biol. 2016:219(12):1851–1859. 10.1242/jeb.137182. [DOI] [PubMed] [Google Scholar]
- Ishikawa SA, Zhukova A, Iwasaki W, Gascuel O. A fast likelihood method to reconstruct and visualize ancestral scenarios. Mol Biol Evol. 2019:36(9):2069–2085. 10.1093/molbev/msz131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung H-S, Francis-West PH, Widelitz RB, Jiang T-X, Ting-Berreth S, Tickle C, Wolpert L, Chuong C-M. Local inhibitory action of BMPs and their relationships with activators in feather formation: implications for periodic patterning. Dev Biol. 1998:196(1):11–23. 10.1006/dbio.1998.8850. [DOI] [PubMed] [Google Scholar]
- Kalyaanamoorthy S, Minh BQ, Wong TK, Von Haeseler A, Jermiin LS. ModelFinder: fast model selection for accurate phylogenetic estimates. Nat Methods. 2017:14(6):587–589. 10.1038/nmeth.4285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karubian J. Costs and benefits of variable breeding plumage in the red-backed fairy-wren. Evolution. 2002:56(8):1673–1682. 10.1111/j.0014-3820.2002.tb01479.x. [DOI] [PubMed] [Google Scholar]
- Karubian J, Alvarado A. Testing the function of petal-carrying in the red-backed fairy-wren (Malurus melanocephalus). Emu. 2003:103(1):87–92. 10.1071/MU01063. [DOI] [Google Scholar]
- Keightley PD, Jackson BC. Inferring the probability of the derived vs. the ancestral allelic state at a polymorphic site. Genetics. 2018:209(3):897–906. 10.1534/genetics.118.301120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khalil S, Enbody ED, Frankl-Vilches C, Welklin JF, Koch RE, Toomey MB, Sin SYW, Edwards SV, Gahr M, Schwabl H. Testosterone coordinates gene expression across different tissues to produce carotenoid-based red ornamentation. Mol Biol Evol. 2023:40(4):msad056. 10.1093/molbev/msad056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korneliussen T, Albrechtsen A, Nielsen R. ANGSD: analysis of next generation sequencing data. BMC Bioinformatics. 2014:15(1):356. 10.1186/s12859-014-0356-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korunes KL, Samuk K. pixy: unbiased estimation of nucleotide diversity and divergence in the presence of missing data. Mol Ecol Resour. 2021:21(4):1359–1368. 10.1111/1755-0998.13326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kratochwil CF. Molecular mechanisms of convergent color pattern evolution. Zoology. 2019:134:66–68. 10.1016/j.zool.2019.04.004. [DOI] [PubMed] [Google Scholar]
- Kurtz S, Phillippy A, Delcher AL, Smoot M, Shumway M, Antonescu C, Salzberg SL. Versatile and open software for comparing large genomes. Genome Biol. 2004:5(2):R12. 10.1186/gb-2004-5-2-r12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamichhaney S, Han F, Berglund J, Wang C, Almén MS, Webster MT, Grant BR, Grant PR, Andersson L. A beak size locus in Darwin's finches facilitated character displacement during a drought. Science. 2016:352(6284):470–474. 10.1126/science.aad8786. [DOI] [PubMed] [Google Scholar]
- Li H, Durbin R. Fast and accurate short read alignment with burrows–wheeler transform. Bioinformatics. 2009:25(14):1754–1760. 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Durbin R. Inference of human population history from individual whole-genome sequences. Nature. 2011:475(7357):493–496. 10.1038/nature10231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin S, Foley J, Jiang T, Yeh C, Wu P, Foley A, Yen C, Huang Y, Cheng H, Chen C. Topology of feather melanocyte progenitor niche allows complex pigment patterns to emerge. Science. 2013:340(6139):1442–1445. 10.1126/science.1230374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindsay WR, Webster MS, Schwabl H. Sexually selected male plumage color is testosterone dependent in a tropical passerine bird, the red-backed fairy-wren (Malurus melanocephalus). PLoS One. 2011:6(10):e26067. 10.1371/journal.pone.0026067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long C, Kubatko L. The effect of gene flow on coalescent-based species-tree inference. Syst Biol. 2018:67(5):770–785. 10.1093/sysbio/syy020. [DOI] [PubMed] [Google Scholar]
- Lopes RJ, Johnson JD, Toomey MB, Ferreira MS, Araujo PM, Melo-Ferreira J, Andersson L, Hill GE, Corbo JC, Carneiro M. Genetic basis for red coloration in birds. Curr Biol. 2016:26(11):1427–1434. 10.1016/j.cub.2016.03.076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mäemets-Allas K, Klaas M, Cárdenas-León CG, Arak T, Kankuri E, Jaks V. Stimulation with THBS4 activates pathways that regulate proliferation, migration and inflammation in primary human keratinocytes. Biochem Biophys Res Commun. 2023:642:97–106. 10.1016/j.bbrc.2022.12.052. [DOI] [PubMed] [Google Scholar]
- Maia R, Rubenstein DR, Shawkey MD. Key ornamental innovations facilitate diversification in an avian radiation. Proc Natl Acad Sci USA. 2013:110(26):10687–10692. 10.1073/pnas.1220784110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margalida A, Negro JJ, Galván I. Melanin-based color variation in the bearded vulture suggests a thermoregulatory function. Comp Biochem Physiol Part A Mol Integr Physiol. 2008:149(1):87–91. 10.1016/j.cbpa.2007.10.017. [DOI] [PubMed] [Google Scholar]
- McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, Garimella K, Altshuler D, Gabriel S, Daly M, et al. The genome analysis toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010:20(9):1297–1303. 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minh BQ, Schmidt HA, Chernomor O, Schrempf D, Woodhams MD, Von Haeseler A, Lanfear R. IQ-TREE 2: new models and efficient methods for phylogenetic inference in the genomic era. Mol Biol Evol. 2020:37(5):1530–1534. 10.1093/molbev/msaa015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montgomerie R. Cosmetic and adventitious colors. Bird Coloration. 2006:1:399–427. 10.2307/j.ctv22jnscm. [DOI] [Google Scholar]
- Mundy NI. A window on the genetics of evolution: MC1R and plumage colouration in birds. Proc R Soc B Biol Sci. 2005:272(1573):1633–1640. 10.1098/rspb.2005.3107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nadeau NJ, Minvielle F, Shin’ichi I, Inoue-Murayama M, Gourichon D, Follett SA, Burke T, Mundy NI. Characterization of Japanese quail yellow as a genomic deletion upstream of the avian homolog of the mammalian ASIP (agouti) gene. Genetics. 2008:178(2):777–786. 10.1534/genetics.107.077073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nordén KK, Eliason CM, Stoddard MC. Evolution of brilliant iridescent feather nanostructures. Elife. 2021:10:e71179. 10.7554/eLife.71179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nordén KK, Faber JW, Babarović F, Stubbs TL, Selly T, Schiffbauer JD, Peharec Štefanić P, Mayr G, Smithwick FM, Vinther J. Melanosome diversity and convergence in the evolution of iridescent avian feathers—implications for paleocolor reconstruction. Evolution. 2019:73(1):15–27. 10.1111/evo.13641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norris BJ, Whan VA. A gene duplication affecting expression of the ovine ASIP gene is responsible for white and black sheep. Genome Res. 2008:18(8):1282–1293. 10.1101/gr.072090.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Omland KE. Examining two standard assumptions of ancestral reconstructions: repeated loss of dichromatism in dabbling ducks (Anatini). Evolution. 1997:51(5):1636–1646. 10.2307/2411215. [DOI] [PubMed] [Google Scholar]
- Price-Waldman R, Stoddard MC. Avian coloration genetics: recent advances and emerging questions. J Hered. 2021:112(5):395–416. 10.1093/jhered/esab015. [DOI] [PubMed] [Google Scholar]
- Prum RO, Dufresne ER, Quinn T, Waters K. Development of colour-producing β-keratin nanostructures in avian feather barbs. J R Soc Interface. 2009:6(suppl_2):S253–S265. 10.1098/rsif.2008.0466.focus. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prum RO, Torres RH. A Fourier tool for the analysis of coherent light scattering by bio-optical nanostructures. Integr Comp Biol. 2003:43(4):591–602. 10.1093/icb/43.4.591. [DOI] [PubMed] [Google Scholar]
- Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira M, Bender D, Maller J, Sklar P, Bakker PD, Daly M, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Human Genet. 2007:81(3):559–575. 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rathburn MK, Montgomerie R. Breeding biology and social structure of white-winged fairy-wrens (Malurus leucopterus): comparison between island and mainland subspecies having different plumage phenotypes. Emu. 2003:103(4):295–306. 10.1071/MU03011. [DOI] [Google Scholar]
- Ritchie J. Melanism in Oedaleus senegalensis and other oedipodines (Orthoptera, acrididae). J Nat Hist. 1978:12(2):153–162. 10.1080/00222937800770041. [DOI] [Google Scholar]
- Robert A, Peona V, Ottenburghs J. Digest: population genomics reveals convergence toward melanism in different island populations. Evolution. 2021:75(6):1582–1584. 10.1111/evo.14242. [DOI] [PubMed] [Google Scholar]
- Robic A, Morisson M, Leroux S, Gourichon D, Vignal A, Thebault N, Fillon V, Minvielle F, Bed’Hom B, Zerjal T. Two new structural mutations in the 5′ region of the ASIP gene cause diluted feather color phenotypes in Japanese quail. Genet Sel Evol. 2019:51(1):1–10. 10.1186/s12711-019-0458-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romano A, Séchaud R, Roulin A. Evolution of wing length and melanin-based coloration in insular populations of a cosmopolitan raptor. J Biogeogr. 2021:48(4):961–973. 10.1111/jbi.14053. [DOI] [Google Scholar]
- Ross M, Briggs A. Behaviour of probable hybrid red-backed x superb fairy-wrens at gladstone, Queensland. Austra Field Ornithol. 2022:39:76–81. 10.20938/afo39076081. [DOI] [Google Scholar]
- Roulin A. Melanin-based colour polymorphism responding to climate change. Glob Chang Biol. 2014:20(11):3344–3350. 10.1111/gcb.12594. [DOI] [PubMed] [Google Scholar]
- Rowley I, Russell E. The breeding biology of the white-winged fairy-wren Malurus leucopterus leuconotus in a western Australian coastal heathland. Emu. 1995:95(3):175–184. 10.1071/MU9950175. [DOI] [Google Scholar]
- Saranathan V, Finet C. Cellular and developmental basis of avian structural coloration. Curr Opin Genet Dev. 2021:69:56–64. 10.1016/j.gde.2021.02.004. [DOI] [PubMed] [Google Scholar]
- Schodde R. The Fairy-wrens: a monograph of the Maluridae. Melbourne: Lansdowne Editions; 1982. [Google Scholar]
- Semenov GA, Linck E, Enbody ED, Harris RB, Khaydarov DR, Alström P, Andersson L, Taylor SA. Asymmetric introgression reveals the genetic architecture of a plumage trait. Nat Commun. 2021:12(1):1019. 10.1038/s41467-021-21340-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shawkey MD, D’Alba L. Interactions between colour-producing mechanisms and their effects on the integumentary colour palette. Philos Trans R Soc B Biol Sci . 2017:372(1724):20160536. 10.1098/rstb.2016.0536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shawkey MD, Estes AM, Siefferman L, Hill GE. The anatomical basis of sexual dichromatism in non-iridescent ultraviolet-blue structural coloration of feathers. Biol J Linn Soc. 2005:84(2):259–271. 10.1111/j.1095-8312.2005.00428.x. [DOI] [Google Scholar]
- Shimodaira H, Hasegawa M. Multiple comparisons of log-likelihoods with applications to phylogenetic inference. Mol Biol Evol. 1999:16(8):1114–1116. 10.1093/oxfordjournals.molbev.a026201. [DOI] [Google Scholar]
- Simão FA, Waterhouse RM, Ioannidis P, Kriventseva EV, Zdobnov EM. BUSCO: assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics. 2015:31(19):3210–3212. 10.1093/bioinformatics/btv351. [DOI] [PubMed] [Google Scholar]
- Sin SYW, Cloutier A, Nevitt G, Edwards SV. Olfactory receptor subgenome and expression in a highly olfactory procellariiform seabird. Genetics. 2022:220(2):iyab210. 10.1093/genetics/iyab210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sin SYW, Lu L, Edwards SV. De novo assembly of the northern cardinal (Cardinalis cardinalis) genome reveals candidate regulatory regions for sexually dichromatic red plumage coloration. G3. 2020:10(10):3541–3548. 10.1534/g3.120.401373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sterken R, Kiekens R, Coppens E, Vercauteren I, Zabeau M, Inze D, Flowers J, Vuylsteke M. A population genomics study of the Arabidopsis core cell cycle genes shows the signature of natural selection. Plant Cell. 2009:21(10):2987–2998. 10.1105/tpc.109.067017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Surmacki A, Minias P, Kudelska K. Occurrence and function of melanin-based grey coloration in western palaearctic songbirds (Aves: Passeriformes). Ibis. 2021:163(2):390–406. 10.1111/ibi.12878. [DOI] [Google Scholar]
- Tanaka Y, Nakanishi H, Kakunaga S, Okabe N, Kawakatsu T, Shimizu K, Takai Y. Role of nectin in formation of E-cadherin-based adherens junctions in keratinocytes: analysis with the N-cadherin dominant negative mutant. Mol Biol Cell. 2003:14(4):1597–1609. 10.1091/mbc.e02-10-0632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ting-Berreth SA, Chuong CM. Sonic Hedgehog in feather morphogenesis: induction of mesenchymal condensation and association with cell death. Dev Dyn. 1996:207(2):157–170. 10.1002/(SICI)1097-0177(199610)207:23.0.CO;2-G. [DOI] [PubMed] [Google Scholar]
- Toews DPL, Taylor SA, Vallender R, Brelsford A, Butcher BG, Messer PW, Lovette IJ. Plumage genes and little else distinguish the genomes of Hybridizing Warblers. Curr Biol. 2016:26(17):2313–2318. 10.1016/j.cub.2016.06.034. [DOI] [PubMed] [Google Scholar]
- Toomey MB, Marques CI, Araújo PM, Huang D, Zhong S, Liu Y, Schreiner GD, Myers CA, Pereira P, Afonso S. A mechanism for red coloration in vertebrates. Curr Biol. 2022:32(19):4201–4214.e12. 10.1016/j.cub.2022.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai MT, Cheng CJ, Lin YC, Chen CC, Wu AR, Wu MT, Hsu CC, Yang RB. Isolation and characterization of a secreted, cell-surface glycoprotein SCUBE2 from humans. Biochem J. 2009:422(1):119–128. 10.1042/BJ20090341. [DOI] [PubMed] [Google Scholar]
- Uy JAC, Cooper EA, Chaves JA. Convergent melanism in populations of a Solomon island flycatcher is mediated by unique genetic mechanisms. Emu. 2019:119(3):242–250. 10.1080/01584197.2019.1586446. [DOI] [Google Scholar]
- Uy JAC, Cooper EA, Cutie S, Concannon MR, Poelstra JW, Moyle RG, Filardi CE. Mutations in different pigmentation genes are associated with parallel melanism in island flycatchers. Proc R Soc B Biol Sci. 2016:283(1834):20160731. 10.1098/rspb.2016.0731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vachaspati P, Warnow T. SVDquest: improving SVDquartets species tree estimation using exact optimization within a constrained search space. Mol Phylogenet Evol. 2018:124:122–136. 10.1016/j.ympev.2018.03.006. [DOI] [PubMed] [Google Scholar]
- Van der Auwera GA, Carneiro MO, Hartl C, Poplin R, Del Angel G, Levy-Moonshine A, Jordan T, Shakir K, Roazen D, Thibault J. From FastQ data to high-confidence variant calls: the genome analysis toolkit best practices pipeline. Curr Protoc Bioinformatics. 2013:43(1):11.10. 11–11.10. 33. 10.1002/0471250953.bi1110s43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh J, Campagna L, Feeney WE, King J, Webster MS. Patterns of genetic divergence and demographic history shed light on island-mainland population dynamics and melanic plumage evolution in the white-winged fairywren. Evolution. 2021:75(6):1348–1360. 10.1111/evo.14185. [DOI] [PubMed] [Google Scholar]
- Wang S, Rohwer S, de Zwaan DR, Toews DP, Lovette IJ, Mackenzie J, Irwin D. Selection on a small genomic region underpins differentiation in multiple color traits between two warbler species. Evol Lett. 2020:4(6):502–515. 10.1002/evl3.198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webster MS, Varian CW, Karubian J. Plumage color and reproduction in the red-backed fairy-wren: why be a dull breeder? Behav Ecol. 2008:19(3):517–524. 10.1093/beheco/arn015. [DOI] [Google Scholar]
- Welklin JF, Johnson AE, Black A, Nye G, Sramek P, Michalek B, Ross M, Newport A, Walker D, Welburn T. New records of hybridisation in Australian fairy-wrens’ Malurus’ spp. Austr Field Ornithol. 2022:39:63–75. 10.20938/afo39063075. [DOI] [Google Scholar]
- Wu T, Hu E, Xu S, Chen M, Guo P, Dai Z, Feng T, Zhou L, Tang W, Zhan L. clusterProfiler 4.0: a universal enrichment tool for interpreting omics data. The Innovation. 2021:2(3):100141. 10.1016/j.xinn.2021.100141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida T, Takai Y, Thesleff I. Cooperation of Nectin-1 and Nectin-3 is required for maintenance of epidermal stratification and proper hair shaft formation in the mouse. Dev Biol J. 2014:2014:1–12. 10.1155/2014/432043. [DOI] [Google Scholar]
- Yu M, Wu P, Widelitz RB, Chuong C-M. The morphogenesis of feathers. Nature. 2002:420(6913):308–312. 10.1038/nature01196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang G, Li C, Li Q, Li B, Larkin D, Lee C, Storz J, Antunes A, Greenwold M, Meredith R, et al. Comparative genomics reveals insights into avian genome evolution and adaptation. Science. 2014:346(6215):1311–1320. 10.1126/science.1251385. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The sequencing data and genome assemblies have been archived in NCBI under the BioProject accession number PRJNA1046240, PRJNA1046245, and PRJNA1046643. All software are cited in the Materials and methods and are publicly available.