Abstract
Epstein-Barr virus (EBV) is a ubiquitous herpesvirus associated with infectious mononucleosis and several tumors. The BARF1 gene is transcribed early after EBV infection from the BamHI A fragment of the EBV genome. Evidence shown here indicates that the BARF1 protein is secreted into the medium of transfected cells and from EBV-carrying B cells induced to allow lytic replication of the virus. Expression cloning identified colony-stimulating factor-1 (CSF-1) as a ligand for BARF1. Computer-assisted analyses indicated that subtle amino acid sequence homology exists between BARF1 and c-fms, the cellular proto-oncogene that is the receptor for CSF-1. Recombinant BARF1 protein was found to be biologically active, and it neutralized the proliferative effects of human CSF-1 in a dose-dependent fashion when assayed in vitro. Since CSF-1 is a pleiotropic cytokine best known for its differentiating effects on macrophages, these data suggest that BARF1 may function to modulate the host immune response to EBV infection.
Recent studies have indicated that many DNA viruses encode proteins that function to modulate the host immune response to infection. A variety of mechanisms are exploited by viruses, including interference with antigen presentation, synthesis of cytokines and cytokine receptors, and inhibition of apoptosis (for a review, see reference 44 and references therein).
Epstein-Barr virus (EBV) is a ubiquitous human herpesvirus whose tissue tropism is typically restricted to B lymphocytes and epithelial cells. Although EBV infection is not usually life threatening, it is the causative agent of infectious mononucleosis, and it is closely associated with at least three forms of cancer: Burkitt’s lymphoma, Hodgkin’s lymphoma, and nasopharyngeal carcinoma (NPC). The EBV-specific genes necessary for cellular transformation have been characterized, but it is believed that the virus encodes other gene products that may contribute either directly or indirectly to EBV oncogenicity. Candidates include some of the virus-encoded immunomodulators such as BHRF-1, which exhibits structural and functional homology to Bcl-2 (21, 51), a cellular gene product that functions to inhibit apoptotic cell death, and BCRF-1 (viral interleukin-10 [IL-10]), a cytokine that stimulates B lymphocytes (22). In an attempt to identify other EBV-encoded immunomodulators and to assess their role in immune evasion or oncogenicity, we expressed the EBV BARF1 gene, an early gene reported to confer tumorigenic capacity onto human B-cell lines (55), in mammalian cells and analyzed its biological activity in vitro. Two forms of recombinant BARF1 protein, (i) a tagged version that contains an eight-amino-acid flag residue at the amino-proximal end of the putative mature protein (BARF1.flag) and (ii) an immunoglobulin (Ig) fusion protein that contains the CH2 and CH3 domains of the heavy chain of human IgG1 fused to the carboxy-terminal amino acids of the mature BARF1 protein (BARF1.Fc), were purified. These proteins were used to identify the α, β, and γ forms of human colony-stimulating factor-1 (hCSF-1) as ligands for the BARF1 protein and to produce a BARF1-specific monoclonal antibody (MAb). This MAb was used to immunoprecipitate BARF1 protein from the medium of B95-8 cells that had been induced to allow lytic replication of EBV, indicating that BARF1 is naturally secreted from infected B cells during the lytic cycle. The biological activity of this protein was tested in bone marrow macrophage nonadherent (BMMNA) proliferation assays, where BARF1 protein efficiently neutralized the proliferative effects of hCSF-1 but not mouse CSF-1. Computer-assisted amino acid sequence analyses indicate a subtle, highly localized region of homology is shared between BARF1 and several members of the tyrosine kinase receptor family. This family includes the cellular receptor for CSF-1, the cellular proto-oncogene c-fms product.
MATERIALS AND METHODS
Protein production and purification.
DEAE transfections were performed in COS-7 cells as described previously (29) except that the cells were fed with medium containing 0.5% low-IgG fetal bovine serum (FBS; HyClone, Logan, Utah) and supernatants were harvested 7 days posttransfection. Supernatants from transfected cells were passed through a 0.45-μm-pore-size filter. BARF1.flag was affinity purified on a column of Affigel-10 (Bio-Rad, Hercules, Calif.) coupled to the anti-flag antibody (4E11) as described previously (23). BARF1.Fc-containing supernatants were passed through a 0.45-μm-pore-size filter and applied to a 0.5-ml protein A/G (Pierce, Rockford, Ill.) affinity column (1.5 by 12 cm) (Bio-Rad) at 4°C with a flow rate of 80 ml/h. The column was washed with 60 ml of pyrogen-free phosphate-buffered saline (PBS). Bound BARF1.Fc was eluted with 12.5 mM citrate (pH 2.8) and collected in 1.0-ml fractions by gravity flow into polypropylene tubes containing 100 μl of 500 mM HEPES (pH 9.0). Fractions were examined by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), and BARF1.Fc-containing fractions were pooled and concentrated. Purified proteins for biological assays were screened for low endotoxin levels (1 pg/ml, final concentration) in the Limulus amoebocyte assay (Whittaker M.A. Bioproducts, Walkersville, Md.).
Isotopic labeling and immunoprecipitation.
CV1/EBNA cells (29) were transfected as described above. Cells were labeled 48 h after DNA transfection with 100 μCi of l-[35S]methionine/cysteine (Amersham, Arlington Heights, Ill.) for 3 h at 37°C. The supernatants and cell lysates were harvested and clarified at 14,000 × g for 30 min. For immunoprecipitation of the α, β, and γ forms of CSF-1, supernatants or cell lysates were incubated with 1 μg of BARF1.Fc followed by the addition of protein A-Sepharose beads (Pharmacia, Piscataway, N.J.). For metabolically labeled BARF1.Fc, protein A-Sepharose beads alone were incubated with supernatants. For EBV-encoded BARF1, supernatants from transfected CV1/EBNA cells or from B95-8 cells induced to express EBV lytic antigens with 5 ng phorbol 12,13-dibutyrate (Sigma, St. Louis, Mo.) per ml were incubated with a MAb directed against BARF1 or with an anti-flag MAb (International Biotechnologies, Inc., New Haven, Conn.), followed by incubation with a rabbit anti-mouse antiserum and the addition of protein A-Sepharose beads. Immunoprecipitates were analyzed by SDS-PAGE on 8 to 16% gels under reducing conditions.
Plasmid construction.
The α, β, and γ forms of hCSF-1 were expressed by using plasmid constructs as described previously (10). All three forms were cloned into the expression plasmid pDC201 (42). A cDNA encoding a soluble flag-tagged BARF1 gene was constructed by PCR using the BamHI A fragment of EBV genomic DNA in plasmid pBR322 as a template for amplification. The oligonucleotides used were 5′-TGTCACTAGTTCTGATTACAAAGATGACGATGATAAAGTCACCGCTTTCTTGGGTGAGCGA-3′, which introduces a SpeI site and flag moiety (23) downstream of the predicted signal cleavage site after amino acid 20 of the BARF1 protein sequence, and 5′-GACAGCGGCCGCCTATTATTGCGACAAGTATCCAGA-3′, which encodes the 3′ end of the BARF1 protein including the native termination codon followed by a NotI site. The amplified DNA was digested with SpeI and NotI and cloned into pDC206 (26) for expression under control of the IL-7 secretory leader sequence to construct pDC206/BARF1.flag. A cDNA encoding BARF1.Fc was constructed by PCR amplification from the same template. The oligonucleotides used were 5′-GATCGGTACCATGGCCAGGTTCATCGCT-3′, which introduces a KpnI site upstream of the initiator methionine of BARF1, and 5′-GCTCTACGTAGGAACCCCCACCGCCTGAACCGCCTCCTCCTGACCCTCCGCCACCTTGCGACAAGTATCCAGAAAC-3′, encoding a ([Gly4]Ser)2 repeat that serves as a flexible linker domain after amino acid 221 of BARF1, followed by a SnaBI site. The amplified DNA was digested with KpnI and SnaBI, and the SnaBI site was used to fuse BARF1 to a modified cDNA copy of the human IgG1 Fc region as described previously (17). This cDNA copy of human IgG1 Fc was mutated to minimize binding to the Fc receptor (7, 8). The fusion gene was cloned into pDC304 (31) to construct pDC304/BARF1.Fc.
Cell separation and flow cytometry.
Peripheral blood T (PBT) cells were isolated exactly as described previously (46). T cells were incubated on two changes of plastic tissue culture flasks to remove adherent cells and then cultured in the presence of phorbol myristate acetate (10 ng/ml; Sigma) plus ionomycin (500 ng/ml; Calbiochem, San Diego, Calif.) for 16 h. After incubation, nonadherent cells were removed for flow cytometric analysis or RNA isolation. Purity was typically greater than 95%. PBT were washed twice in fluorescence-activated cell sorting (FACS) buffer (PBS, 1% FBS, and 0.02% NaN3), and then 106 cells were incubated on ice for 30 min in 100 μl of FACS buffer with 10 μg of BARF1.Fc per ml, whole human IgG, or a control Fc protein. After thorough washing, the cells were stained with phycoerythrin-conjugated anti-human IgG (Fc specific) (Rockland, Gilbertsville, Pa.) in 100 μl of FACS buffer. Cells were then washed thoroughly in FACS buffer. A minimum of 5,000 cells were analyzed in a FACScan (Becton Dickinson, San Jose, Calif.).
Activated primary PBT cDNA expression library.
Random hexamer-primed cDNA was generated from oligo(dT)-selected activated primary PBT mRNA and cloned into the expression vector pDC410 (1), using BglII adapters. The library was screened as described previously (29). Briefly, CV1/EBNA cell monolayers on chamber slides were transfected with plasmid DNA derived from pooled transformants. Cell monolayers were incubated 2 days after transfection with 1 μg of BARF1.Fc per ml and then washed and incubated with 125I-labeled mouse anti-human Ig. After extensive washing, cells were fixed, dipped in photographic emulsion, and then developed. Positive pools of cDNAs were identified and subdivided into smaller cDNA pools until single-positive clones were isolated.
BMMNA proliferation assays.
BMMNA proliferation assays were performed essentially as described previously (10). Briefly, bone marrow was flushed from the femurs of C57BL/6 mice (12 to 20 weeks of age) and drawn through a 22-gauge needle to disperse the cells. Pelleted cells were suspended in BMMNA medium (alpha minimal essential medium, 15% FBS, 20 μg of asparagine per ml) and seeded at 5 × 107 per T175 flask in a 1:25 dilution of L929 medium. Flasks were incubated at 37°C with 6.5% CO2, and after 72 h, nonadherent cells were removed from the flask. A standard proliferation assay was performed in a 96-well flat-bottom microtiter plate. hCSF-1 was added at a final concentration of 1 μg/ml and titrated with a serial twofold dilution to 11 wells or was preincubated for 30 min at 37°C with a titration of BARF1.Fc, BARF1.flag, appropriate control proteins, a blocking polyclonal antiserum to hCSF-1, or a normal rabbit control serum. The heterologous Fc-containing control proteins used in these studies contain the extracellular domain of the p35 protein of vaccinia virus fused to the Fc portion of human IgG1 (45). Cells were seeded at 4 × 105/ml and incubated at 37°C with 6.5% CO2 for 5 h. Cells were pulsed with 10 μCi of [3H]thymidine per ml. One percent antifoam and 5% Nonidet P-40 were added after 19 h at the time of cell harvest.
Solid-phase binding assay for CSF-1.
Ninety-six-well enzyme immunoassay plates (Dynatech, Chantilly, Va.) were coated with BARF1.Fc (5 μg/ml in PBS) for 16 h at 4°C, washed thoroughly (three times with PBS–0.05% Tween 20 and three times with PBS alone), blocked with PBS–1% nonfat dry milk for 1 h at room temperature, and then washed thoroughly. The wells were then incubated with a titration of biotinylated CSF-1, Flt3L (fms-like tyrosine kinase receptor 3 ligand), or human or mouse MGF (mast cell growth factor) (all four provided by Immunex, Seattle, Wash.), beginning at a concentration of 0.3 μg/ml with serial twofold dilutions out to eight wells (in PBS with 10% normal goat serum). Cytokines were biotinylated essentially as described previously (4). The plates were then washed thoroughly. Bound biotinylated CSF-1 was detected with streptavidin-horseradish peroxidase (Zymed, San Francisco, Calif.), washed, and then developed with the use of the TMB Microwell peroxidase substrate system (Kirkegaard & Perry, Gaithersburg, Md.). Data reduction was accomplished by using the Deltasoft 3.1 ELISA analysis program for the Macintosh (Biometallics, Inc., Princeton, N.J.). Binding of BARF1.Fc to human granulocyte-macrophage colony-stimulating factor (Immunex) was tested by adding 10 μg of recombinant human GMCSF (R&D Systems, Minneapolis, Minn.) per ml to the titration of hCSF-1-biotin before addition to the wells.
RESULTS
The BARF1 gene encodes a secreted protein.
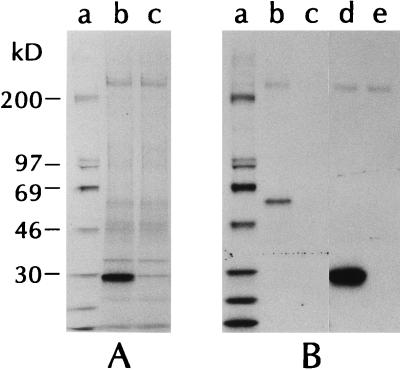
Computer-assisted analyses of the putative BARF1 amino acid sequence using two separate algorithms (16, 27) suggested that this protein contains an N-terminal signal sequence (predicted cleavage is following the alanine at position 20) and contains no transmembrane domain (data not shown). This predicted topology suggests that BARF1 is a secreted protein, which would be consistent with both cytosolic and membrane immunofluorescent staining patterns previously seen in cells transfected with the BARF1 gene (14, 50, 54, 55). Based on these analyses, a cDNA containing the complete coding region of the BARF1 gene was amplified by PCR from a plasmid containing the EBV BamHI A genomic fragment. The resultant DNA product was subcloned into a mammalian expression vector which was then transfected into CV1/EBNA cells, and supernatants from metabolically labeled cells were examined for the presence of the BARF1 protein. Cells transfected with the BARF1-containing plasmid secreted a protein of approximately 29 kDa (Fig. 1A, lane b).
FIG. 1.
Recombinant and native BARF1 are secreted proteins. (A) CV1/EBNA cells were transfected with plasmids containing the full-length, unmodified coding region of the BARF1 gene (lane b) or empty plasmid (lane c) and radiolabeled as described in Materials and Methods. Supernatants were harvested 3 h after the addition of label and analyzed directly by SDS-PAGE on 8 to 16% gradient gels. Molecular weight markers are shown in lane a. (B) CV1/EBNA cells were transfected with plasmids containing the BARF1.Fc coding sequence (lane b), the BARF1.flag coding sequence (lane d), or empty vector (lanes c and e) and then radiolabeled as for panel A. Supernatants were collected, and proteins were precipitated with protein A-Sepharose alone for BARF1.Fc and empty vector-containing samples (lanes b and c) or with anti-flag MAb M1 and protein A-Sepharose for the BARF1.flag protein and empty vector-containing samples (lanes d and e). Molecular weight markers are shown in lane a.
The predicted molecular mass for BARF1 in the supernatant is approximately 23 kDa. The difference between the observed and predicted values likely arises as a result of posttranslational modifications, such as the addition of carbohydrate to the single N-linked attachment site. Additional recombinant forms of the BARF1 protein were constructed and tested for the ability to be secreted into the medium of transfected CV1/EBNA cells. Figure 1B, lane b, shows a protein of ∼60 kDa precipitated from metabolically labeled cells that were transfected with a BARF1.Fc-containing plasmid. The apparent molecular mass of this protein is in good agreement with the size predicted as a result of combining BARF1 (∼29 kDa) and the Fc moiety (∼30 kDa). Lane d shows a 29-kDa non-Fc, flag-tagged version of the BARF1 protein that has been immunoprecipitated with an anti-flag MAb from BARF1.flag-transfected cell supernatants.
The secretion of all three forms of recombinant BARF1 protein strongly suggested that naturally occurring BARF1 protein also would be secreted from EBV-infected cells. This was confirmed in an assay using a BARF1-specific MAb (M71) that had been raised against purified BARF1.Fc protein and medium collected from metabolically radiolabeled B95-8 cells that had been induced to undergo lytic infection. Induced B95-8 cells, but not uninduced cells, secreted a 29-kDa protein that was immunoprecipitated with the BARF1 MAb (Fig. 2), confirming that the BARF1 protein is normally secreted into the extracellular milieu during lytic replication of EBV in B cells.
FIG. 2.
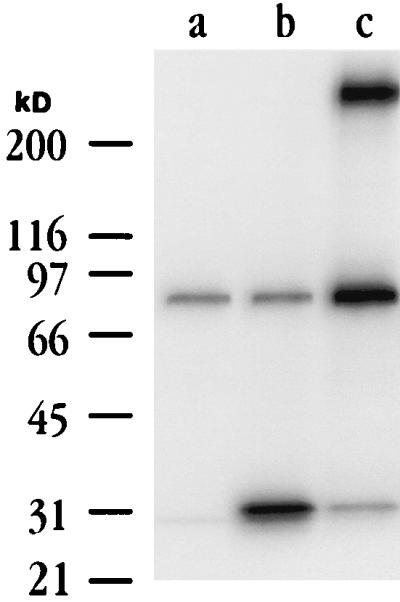
BARF1 is secreted from EBV-infected cells. CV1/EBNA cells transfected with empty vector (lane a) or vector containing a nonmodified BARF1 gene (lane b), and B95-8 cells induced to express EBV lytic antigens (lane c) were metabolically labeled as described in Materials and Methods. Cell supernatants were immunoprecipitated with a BARF1 MAb, and precipitates were analyzed by SDS-PAGE on 8 to 16% gradient gels. Induced B95-8 cell supernatants contained a BARF1 MAb-reactive protein of approximately 29 kDa that comigrated with that found in cells transfected with BARF1 expression plasmids. A very large (>200-kDa) protein was detected in immunoprecipitations from B95-8 cells (lane c) but not from EBV-negative B cells (12).
Identification of a ligand for BARF1.
Purified BARF1.Fc protein was used in flow cytometric analyses of a variety of cell lines to determine a potential cell source for a counterstructure for BARF1. These experiments indicated significant binding of BARF1.Fc to PBT activated either with phorbol myristate acetate and ionomycin or with an anti-CD3 MAb (data not shown). An activated, human PBT cDNA library was generated and used in an expression cloning strategy that has successfully identified a number of membrane-bound ligands for soluble molecules (5, 29, 45). As a result of these experiments, a single cDNA capable of conferring binding of BARF1.Fc to transfected mammalian cells was isolated and found to encode the β form of hCSF-1.
CSF-1 is a cytokine that traditionally has been considered a macrophage growth and differentiation factor, owing in large part to work performed in the mouse system (47). Remarkably little is known about the biological activity of hCSF-1; however, its expression by fibroblasts, endothelial cells, monocytes/macrophages (reference 41 and references therein), and activated human T cells and T-cell lines (25, 59) as well as the expression of the CSF-1 receptor (CSF-1R) on osteoclasts and on myeloid, epithelial, and B cells strongly suggest that hCSF-1 is a pleiotropic cytokine with many as yet undetermined activities.
hCSF-1 is encoded by a single gene; however, alternative splicing gives rise to multiple transcripts, and cDNAs for three different forms of hCSF-1 (α, β, and γ) have been identified. The α form of hCSF-1 comprises 256 amino acids; the β form contains an 894-bp insert within the coding region and comprises 554 amino acids; and the γ form contains a 546-bp insertion in the same position (relative to the α form) and comprises 438 amino acids (10). All three forms retain their transmembrane domain, and the soluble form of the cytokine is generated by proteolytic cleavage.
BARF1 binds the α, β, and γ forms of hCSF-1.
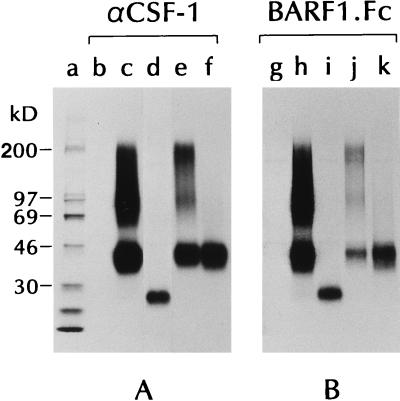
Flow cytometric analysis and the identification of β hCSF-1 as a ligand for BARF1.Fc indicated that BARF1.Fc could bind to cell surface hCSF-1. To confirm the recognition of β hCSF-1 by BARF1.Fc and to test whether BARF1.Fc bound to other forms of hCSF-1, immunoprecipitations of supernatants from cells transfected with cDNAs encoding either the α, β, or γ form of hCSF-1 were performed (Fig. 3). Radiolabeled, soluble hCSF-1 of the α, β, or γ form was precipitated with BARF1.Fc, whereas no proteins were precipitated from supernatants derived from cells transfected with vector alone. Identical results were obtained when a similar experiment was performed with cell lysates (data not shown). These data indicate that BARF1.Fc binds efficiently to all forms of hCSF-1.
FIG. 3.
Immunoprecipitation of the α, β, and γ forms of hCSF-1 with BARF1.Fc protein. CV1/EBNA cells were transfected with empty plasmids (lanes b and g), with plasmids containing the α (lanes d and i), β (lanes e and j), or γ (lanes f and k) form of hCSF-1, or with the plasmid (containing a β form) isolated by expression cloning (lanes c and h) and then metabolically labeled, and the cell supernatants were precipitated with a commercially available pan-hCSF-1 MAb (A) or recombinant BARF1.Fc protein (B).
CSF-1 belongs to a family of related ligands that includes Flt3L and MGF (also known as c-kit ligand and stem cell factor), all of which bind to specific receptor tyrosine kinase molecules. The specificity of the interaction between hCSF-1 and BARF1.Fc was examined by testing these molecules for their ability to bind to recombinant BARF1 protein in solid-phase binding assays. Flt3L, MGF, and hCSF-1 were directly biotinylated and assayed for binding to immobilized BARF1.Fc. In the case of granulocyte-macrophage colony-stimulating factor, which is an unrelated colony-stimulating factor, a titration of unlabeled cytokine was used in an attempt to inhibit biotinylated hCSF-1 binding to BARF1.Fc protein in similar assays. Only hCSF-1 showed binding to BARF1.Fc, whereas all other cytokines failed to bind (data not shown). This finding specificity strongly argues for CSF-1 playing an important, and at present, unappreciated, role in the response of the host to EBV infection.
BARF1 can function as an antagonist for hCSF-1.
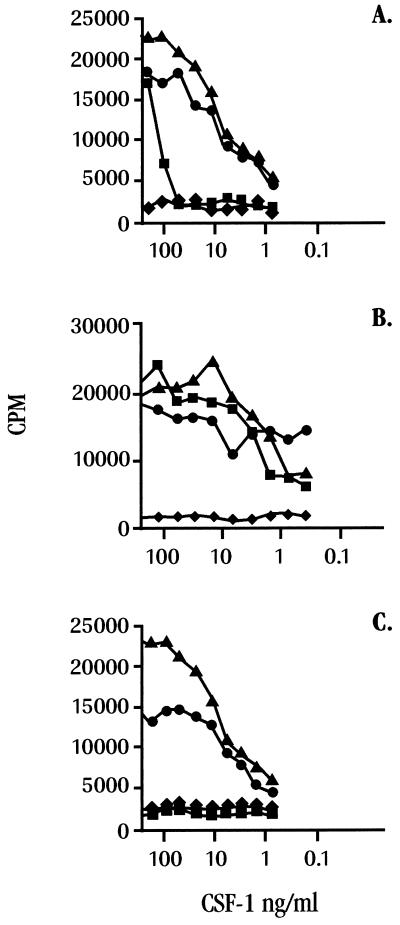
The identification of BARF1.Fc as a soluble hCSF-1-specific binding protein suggested that this viral molecule may act as an hCSF-1 antagonist. To examine this, the BARF1.Fc protein was tested for its ability to inhibit the proliferative effects of hCSF-1 in a mouse BMMNA proliferation assay. In this assay, nonadherent macrophage progenitor cells were cultured in the presence of hCSF-1, and their proliferation was measured by [3H]thymidine uptake. Cultures contained either a titration of hCSF-1 and a constant amount of BARF1.Fc protein or a heterologous control Fc protein (Fig. 4A). Although some diminution in proliferation was typically seen when the control Fc protein was included in these cultures, the BARF1.Fc protein effectively neutralized hCSF-1 proliferative effects on mouse macrophage precursors at all but the highest concentration (100 ng/ml) of cytokine. Similar experiments were performed with the BARF1.flag (non-Fc-tagged version), and the results were indistinguishable from those in Fig. 4A, indicating that possible Fc receptor interactions arising from the use of the BARF1.Fc chimeric protein do not play a role in the observed inhibitory effect.
FIG. 4.
Recombinant BARF1 protein neutralizes hCSF-1. BMMNA proliferation assays were performed with a titration of hCSF-1 (A and C, triangles) or mouse CSF-1 (B, triangles). Parallel cultures contained the same titration of CSF-1 and either BARF1.Fc protein at 2.5 μg/ml (A and B, squares) or a heterologous Fc-containing protein at 2.5 μg/ml (A and B, circles). The control consisted of similar cultures (C) that included a titration of hCSF-1 and a 1:350 dilution of a CSF-1 neutralizing polyclonal serum (squares) or normal rabbit serum (circles). Cultures containing medium only are indicated by diamonds. The results shown in all panels are representative of four independent experiments.
Flow cytometric data had indicated that the BARF1.Fc protein could bind to mouse 3T3 fibroblast cells (data not shown), which can constitutively express mouse CSF-1 (41). The ability of BARF1.Fc protein to neutralize mouse CSF-1 was tested in a BMMNA proliferation assay as described above. Interestingly, the inclusion of either BARF1.Fc protein (Fig. 4B) or the BARF1.flag protein (data not shown) had no effect on the proliferative capacity of mouse CSF-1 on mouse macrophage precursors. The ability of BARF1 to bind to mouse CSF-1 (as indicated by flow cytometry studies) but not block its biological activity underscores the fine specificity of this immunomodulator, but perhaps it is not surprising because BARF1 is derived from a virus whose host range is limited to humans.
BARF1 shows limited sequence similarity to the cellular receptor for CSF-1.
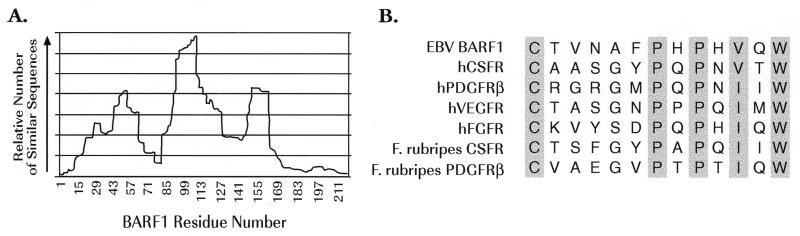
Computer-assisted analyses of the predicted amino acid sequence of the BARF1 open reading frame by using BLAST (basic local alignment search tool) (2) against the nonredundant protein database indicated three regions within BARF1 that exhibited similarity to other proteins (Fig. 5A). The first region, comprising residues 10 to 70, exhibited similarity to numerous unrelated proteins, which contain stretches of hydrophobic amino acids that align either with the N-terminal signal sequence of BARF1 or with the isolated hydrophobic residues that occur in BARF1 between residues 48 and 60. The second region that showed similarity to a number of proteins spanned residues between positions 85 and 127 of the BARF1 sequence, and again, all of these peptides appear unrelated to one another except that they all contain an Ig-like domain. The BARF1 protein was not predicted to contain an Ig-like domain based on conformity with the defining consensus sequence (the sequence surrounding the C-terminal cysteine residue of an Ig domain is [F,Y]XCX[V,A]XH, where X is any amino acid) (6, 9); however, BARF1 does contain four cysteine residues, and two of the four (at positions 14 and 104 or positions 104 and 201) are separated by 90 to 100 amino acids, the distance typically found in an Ig domain loop. This similarity likely explains the sequence relationships found within this second region. In contrast to regions 1 and 2, a third region of BARF1, comprising amino acids 146 to 158, exhibited homology to a group of proteins consisting almost exclusively of the extracellular domains of members of the tyrosine kinase receptor family. A representative sequence alignment including the hCSF-1R and a few of the receptors closely related to it is shown in Fig. 5B. This alignment indicates an absolute conservation of a cysteine residue (located in the BARF1 protein at position 146), a proline at position 7 of this motif, a second proline at position 9, and a tryptophan at position 13. Several other positions (2, 3, 5, and 11) contain similar amino acids. This short stretch of sequences is the only region of apparent homology between BARF1 and its cellular counterpart, the CSF-1R. Included in this alignment are the amino acid sequences for the CSF-1R and the platelet-derived growth factor receptor β homologs that have been sequenced from the Japanese puffer fish, Fugu rubripes (24). The sequence homology of these teleost receptors indicates a broad evolutionary conservation of this motif.
FIG. 5.
(A) Histogram indicating the relative number of proteins (vertical axis) similar to BARF1 at each amino acid residue along the BARF1 sequence (horizontal axis), as identified by BLAST searches against a nonredundant protein database. Three discrete regions of similarity to a large number of proteins are identifiable. (B) The amino acid motif comprising residues 146 to 158 in region 3 of panel A, which is conserved between the BARF1 sequence and the indicated receptor tyrosine kinases.
DISCUSSION
The data presented here indicate that the BARF1 protein can bind and neutralize soluble hCSF-1. The conserved block of amino acid sequences present in both BARF1 and many members of the hCSF-1R family supports the idea that the BARF1 gene most likely originated as a cellular gene. Thus, the BARF1 gene joins several other EBV-encoded genes whose primary function may be to modulate the host response to initial infection, and may also contribute to EBV pathogenicity.
The BARF1 gene is transcribed before the onset of DNA replication (58) in a rightward direction from the BamHI A EBV genome fragment. In prior studies, this region was found to give rise to numerous rightward transcripts that were believed to be expressed in NPC tissues but not in tissues from other EBV-associated tumors, such as Burkitt’s lymphoma (18, 33). More recent studies of the transcripts from this region, using cell lines derived from NPC tissue or from primary NPC cells, have indicated that while NPC cells contain numerous rightward transcripts from the BamHI A region, these transcripts terminate prior to the BARF1 gene (11, 39). This finding suggests that transcription of the BARF1 gene does not play a singular role in NPC compared to other forms of EBV-associated tumor (40). Consistent with this conclusion, immunoprecipitations of radiolabeled, nontagged, recombinant BARF1 protein with serum from healthy EBV-seropositive donors or from age- and race-matched NPC patients clearly indicate that all seropositive serum tested contained antibodies capable of reacting with the BARF1 protein (47a).
A study by Wei and Ooka (56) has suggested that the overexpression of the BARF1 gene by retrovirus expression vectors in mouse fibroblast cells rendered these cells oncogenic, as assayed by tumor formation, after injection into newborn rats. Similar results were obtained by this group when BARF1-expressing retroviruses were used to transform the EBV-negative, human Louckes B-cell line (55). In these latter experiments, BARF1-expressing B cells were tumorigenic when injected into newborn rats. Such results are provocative because CSF-1R was originally identified as the product of c-fms, a cellular tyrosine kinase proto-oncogene whose overexpression can lead to cell transformation.
Although it has been studied for some years, the exact role played by CSF-1 and CSF-1R in human tumor establishment and/or maintenance is unknown; however, several postulated mechanisms may be relevant to the understanding of the oncogenic potential of overexpressed BARF1 (for a review, see references 20 and 41). For example, NIH 3T3 cells expressing hCSF-1R can undergo transformation and proliferate in serum-free medium in the presence of human CSF-1 (35–37). This transformation is presumably a result of the kinase activity of the CSF-1R that occurs following binding of CSF-1 and the subsequent activation of downstream mitogenic signals. Support view for this comes from studies in which mutagenized CSF-1R was examined for its ability to induce transcription of other proto-oncogenes such as c-myc. In these studies, mutant receptors, which were found to be mitogenically inactive, also failed to induce c-myc, suggesting that c-myc induction is required for CSF-1-induced mitogenesis (34, 38). In the study by Wei et al. (55), overexpression of BARF1 protein in Louckes B cells activated the c-myc proto-oncogene. Thus, it is possible that the expression of BARF1 protein may, in rare instances associated with EBV infection, result in activation of downstream oncogenic proteins, contributing to the formation of tumors in some patients. While BARF1 could have a role for some tumors in vivo, the protein is not expressed during latent infection of EBV-transformed B cells in vitro (50). Preliminary experiments using an EBV mutant in which the BARF1 gene is disrupted confirm that this gene is not required for maintenance of transformation by EBV (12).
It is likely that the primary role of the BARF1 protein during EBV infection is to act as an antagonist for hCSF-1. As stated previously, CSF-1 is a pleiotropic cytokine whose receptor is expressed on many cell types. Although little is known about the biology of hCSF-1, and relatively little is known about mouse CSF-1 outside of its role as a macrophage growth and differentiation factor, there are some studies that clearly suggest a role for hCSF-1 in modulating the innate and cell-mediated immune responses. The expression of hCSF-1 on the surface of activated, but not resting, human T cells, as indicated by our expression cloning experiments, is consistent with other reports (19, 49, 57, 59). Zisman et al. (59) performed proliferation assays using antigen-specific human T-cell clones, irradiated syngeneic spleen cells, and antigen and found that the inclusion of hCSF-1 blocking antibodies inhibited T-cell proliferation. This work, then, suggests a role for hCSF-1 as a costimulatory molecule. The biological activities of hCSF-1 are better understood on monocytes, where it has been shown to enhance complement component C3 production (3), regulate integrin expression (15), and induce interferon and tumor necrosis factor alpha (53).
It is worth noting that EBV encodes a viral homolog of the cytokine IL-10. IL-10 also has profound immunosuppressive effects on monocytes and macrophages, inhibiting their ability to secrete cytokines and to serve as accessory cells for T cells and natural killer cells (22). IL-10 also inhibits superoxide anion production (32), but interestingly, viral IL-10 does not upregulate major histocompatibility complex class II expression on monocyte cell surfaces, although cellular IL-10 does (30). The inclusion of two gene products within the EBV genome that are involved with the inhibition of monocyte/macrophage functions clearly points to an important role for these cells in controlling EBV infection.
The BARF1 gene joins a growing family of virus-encoded immunomodulators that have little or no detectable sequence similarity to their cellular counterparts, yet seem to function at least as efficiently. Two striking examples of this have been reported recently. The B18R gene of poxviruses shows clear homology to the IL-1 receptor family of proteins yet binds alpha/beta interferon with high affinity (13, 48). A soluble poxvirus protein with no apparent sequence similarity to proteins in the nonredundant database has been shown to bind and neutralize chemokines (28, 43), an impressive fact considering all known mammalian chemokine receptors are multiple membrane spanners. The relationship of BARF1 to the CSF-1R is similar in that although there is a highly conserved amino acid motif, it does not occur in the region of the CSF-1R previously reported to be critical for hCSF-1 binding (52); in addition, this motif is found in several other members of the tyrosine kinase receptor family. These observations argue that this stretch of conserved sequence might be more important in the preservation of some secondary structure common to members of this family than in the formation of the binding site of hCSF-1. It is conceivable that the study of these nonconserved virus immunomodulators will allow new insight into the development of computer-assisted structure-function programs.
The identification of a novel, soluble CSF-1 receptor encoded by EBV implies a previously unrecognized importance for the role of hCSF-1 in controlling EBV infection, and it raises many questions regarding the potential link between BARF1, hCSF-1, and CSF-1R in EBV-associated tumors.
REFERENCES
- 1.Alderson M R, Smith C A, Tough T W, Davis-Smith T, Armitage R J, Falk B, Roux E, Baker E, Sutherland G R, Din W S, Goodwin R G. Molecular and biological characterization of human 4-1BB and its ligand. Eur J Immunol. 1994;24:2219–2227. doi: 10.1002/eji.1830240943. [DOI] [PubMed] [Google Scholar]
- 2.Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 3.Andoh A, Fujiyama Y, Kitoh K, Niwakawa M, Hodohara K, Bamba T, Hosoda S. Macrophage colony-stimulating factor (M-CSF) enhances complement component C3 production by human monocytes/macrophages. Int J Hematol. 1993;57:53–59. [PubMed] [Google Scholar]
- 4.Armitage R J, Beckmann M P, Idzerda R L, Alpert A, Fanslow W C. Regulation of interleukin 4 receptors on human T cells. Int Immunol. 1990;2:1039–1045. doi: 10.1093/intimm/2.11.1039. [DOI] [PubMed] [Google Scholar]
- 5.Armitage R J, Fanslow W C, Strockbine L, Sato T A, Clifford K N, Macduff B M, Anderson D M, Gimpel S D, Davis-Smith T, Maliszewski C R, Clark E A, Smith C A, Grabstein K H, Cosman D, Spriggs M K. Molecular and biological characterization of a murine ligand for CD40. Nature. 1992;357:80–82. doi: 10.1038/357080a0. [DOI] [PubMed] [Google Scholar]
- 6.Bairoch, A. The PROCITE dictionary of protein sites and patterns. University of Geneva, Geneva, Switzerland.
- 7.Baum P R, Gayle III R B, Ramsdell F, Srinivasan S, Sorensen R A, Watson M L, Seldin M F, Baker E, Sutherland G R, Clifford K N, Alderson M R, Goodwin R G, Fanslow W C. Molecular characterization of murine and human OX40/OX40 ligand systems: identification of a human OX40 ligand as the HTLV-1-regulated protein gp34. EMBO J. 1994;13:3992–4001. doi: 10.1002/j.1460-2075.1994.tb06715.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Baum P R, Gayle III R B, Ramsdell F, Srinivasan S, Sorensen R A, Watson M L, Seldin M F, Clifford K N, Grabstein K, Alderson M R, Goodwin R G, Fanslow W C. Identification of OX40 ligand and preliminary characterization of its activities on OX40 receptor. Circ Shock. 1994;44:30–34. [PubMed] [Google Scholar]
- 9.Bork P, Holm L, Sander C. The immunoglobulin fold. Structural classification, sequence patterns and common core. J Mol Biol. 1994;242:309–320. doi: 10.1006/jmbi.1994.1582. [DOI] [PubMed] [Google Scholar]
- 10.Cerretti D P, Wignall J, Anderson D, Tushinski R J, Gallis B M, Stya M, Gillis S, Urdal D L, Cosman D. Human macrophage-colony stimulating factor: alternate RNA and protein processing from a single gene. Mol Immunol. 1988;25:761–770. doi: 10.1016/0161-5890(88)90112-5. [DOI] [PubMed] [Google Scholar]
- 11.Chen H-L, Lung M M L, Sham J S T, Choy D T K, Griffin B E, Ng M H. Transcription of BamHI-A region of the EBV genome in NPC tissues and B cells. Virology. 1992;191:193–201. doi: 10.1016/0042-6822(92)90181-n. [DOI] [PubMed] [Google Scholar]
- 12.Cohen, J. I. Unpublished data.
- 13.Colamonici O R, Domanski P, Sweitzer S M, Larner A, Buller R M L. Vaccinia virus B18R gene encodes a type I interferon-binding protein that blocks interferon α transmembrane signaling. J Biol Chem. 1995;270:15974–15978. doi: 10.1074/jbc.270.27.15974. [DOI] [PubMed] [Google Scholar]
- 14.Decaussin G, Leclerc V, Ooka T. The lytic cycle of Epstein-Barr virus in the nonproducer Raji line can be rescued by the expression of a 135-kilodalton protein encoded by the BALF2 open reading frame. J Virol. 1995;69:7309–7314. doi: 10.1128/jvi.69.11.7309-7314.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.De Nichilo M O, Burns G F. Granulocyte-macrophage and macrophage colony-stimulating factors differentially regulate αv integrin expression on cultured human macrophages. Proc Natl Acad Sci USA. 1993;90:2517–2521. doi: 10.1073/pnas.90.6.2517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Engelman D M, Steitz T A, Goldman A. Identifying nonpolar transbilayer helices in amino acid sequences of membrane proteins. Annu Rev Biophys Biophys Chem. 1986;15:321–353. doi: 10.1146/annurev.bb.15.060186.001541. [DOI] [PubMed] [Google Scholar]
- 17.Fanslow W C, Anderson D, Grabstein K H, Clark E A, Cosman D, Armitage R J. Soluble forms of CD40 inhibit biologic responses of human B cells. J Immunol. 1992;149:655–660. [PubMed] [Google Scholar]
- 18.Gilligan K J, Rajadurai P, Lin J-C, Busson P, Abdel-Hamid M, Prasad U, Tursz T, Raab-Traub N. Expression of the Epstein-Barr virus BamHI A fragment in nasopharyngeal carcinoma: evidence for a viral protein expressed in vivo. J Virol. 1991;65:6252–6259. doi: 10.1128/jvi.65.11.6252-6259.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hallet M-M, Praloran V, Vié H, Peyrat M-A, Wong G, Witek-Giannotti J, Soulillou J-P, Moreau J-F. Macrophage colony-stimulating factor (CSF-1) gene expression in human T-lymphocyte clones. Blood. 1991;77:780–786. [PubMed] [Google Scholar]
- 20.Hamilton J A. CSF-1 signal transduction: what is of functional significance. Immunol Today. 1997;18:313–317. doi: 10.1016/s0167-5699(97)01084-0. [DOI] [PubMed] [Google Scholar]
- 21.Henderson S, Huen D, Rowe M, Dawson C, Jonhson G, Rickinson A. Epstein-Barr virus-encoded BHRF1 protein, a viral homologue of Bcl-2, protects human B cells from programmed cell death. Proc Natl Acad Sci USA. 1993;90:8479–8483. doi: 10.1073/pnas.90.18.8479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ho A S-Y, Moore K W. Interleukin-10 and its receptor. Ther Immunol. 1994;1:173–185. [PubMed] [Google Scholar]
- 23.Hopp T P, Prickett K S, Price V L, Liggy R T, March C J, Cerretti D P, Urdal D L, Conlon P J. A short polypeptide marker sequence useful for recombinant protein identification and purification. Bio/Technology. 1988;6:1204–1210. [Google Scholar]
- 24.How G-F, Venkatesh B, Brenner S. Conserved linkage between the puffer fish (Fugu rubripes) and human genes for platelet-derived growth factor receptor and macrophage colony-stimulating factor receptor. Genome Res. 1996;6:1185–1191. doi: 10.1101/gr.6.12.1185. [DOI] [PubMed] [Google Scholar]
- 25.Kawasaki C, Okamura S, Omori F, Shimoda K, Hayashi S, Kondo S, Yamada M, Niho Y. Both granulocyte-macrophage colony-stimulating factor and monocytic colony-stimulating factor are produced by the human T-cell line, HUT 102. Exp Hematol. 1990;18:1090–1093. [PubMed] [Google Scholar]
- 26.Kozlosky C J, Maraskovsky E, McGrew J T, VandenBos T, Teepe M, Lyman S D, Srinivasan S, Fletcher F A, Gayle III R B, Cerretti D P, Beckmann M P. Ligands for the receptor tyrosine kinases hek and elk: isolation of cDNAs encoding a family of proteins. Oncogene. 1995;10:299–306. [PubMed] [Google Scholar]
- 27.Kyte J, Doolittle R F. A simple method for displaying the hydropathic character of a protein. J Mol Biol. 1982;157:105–132. doi: 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]
- 28.Lalani A S, Graham K, Mossman K, Rajarathnam K, Clark-Lewis I, Kelvin D, McFadden G. The purified myxoma virus gamma interferon receptor homolog M-T7 interacts with the heparin-binding domains of chemokines. J Virol. 1997;71:4356–4363. doi: 10.1128/jvi.71.6.4356-4363.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.McMahan C J, Slack J L, Mosley B, Cosman D, Lupton S D, Brunton L L, Grubin C E, Wignall J M, Jenkins N A, Brannan C I, Copeland N G, Huebner K, Croce C M, Cannizzarro L A, Benjamin D, Dower S K, Spriggs M K, Sims J E. A novel IL-1 receptor, cloned from B cells by mammalian expression, is expressed in many cell types. EMBO J. 1991;10:2821–2832. doi: 10.1002/j.1460-2075.1991.tb07831.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Moore K W, O’Garra A, de Waal Malefyt R, Vieira P, Mosmann T R. Interleukin-10. Annu Rev Immunol. 1993;11:165–190. doi: 10.1146/annurev.iy.11.040193.001121. [DOI] [PubMed] [Google Scholar]
- 31.Mosley B, Beckmann M P, March C J, Idzerda R L, Gimpel S D, VandenBos T, Friend D, Alpert A, Anderson D, Jackson J, Wignall J M, Smith C, Gallis B, Sims J E, Urdal D, Widmer M B, Cosman D, Park L S. The murine interleukin-4 receptor: molecular cloning and characterization of secreted and membrane bound forms. Cell. 1989;59:335–348. doi: 10.1016/0092-8674(89)90295-x. [DOI] [PubMed] [Google Scholar]
- 32.Niiro H, Otsuka T, Abe M, Satoh H, Ogo T, Nakano T, Furukawa Y, Niho Y. Epstein-Barr virus BCRF1 gene product (viral interleukin 10) inhibits superoxide anion production by human monocytes. Lymphokine Cytokine Res. 1992;11:209–214. [PubMed] [Google Scholar]
- 33.Raab-Traub N, Hood R, Yang C-S, Henry II B, Pagano J S. Epstein-Barr virus transcription in nasopharyngeal carcinoma. J Virol. 1983;48:580–590. doi: 10.1128/jvi.48.3.580-590.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Roussel M F, Cleveland J L, Shurtleff S A, Sherr C J. Myc rescue of a mutant CSF-1 receptor impaired in mitogenic signalling. Nature. 1991;353:361–363. doi: 10.1038/353361a0. [DOI] [PubMed] [Google Scholar]
- 35.Roussel M F, Downing J R, Rettenmier C W, Sherr C J. A point mutation in the extracellular domain of the human CSF-1 receptor (c-fms proto-oncogene product) activates its transforming potential. Cell. 1988;55:979–988. doi: 10.1016/0092-8674(88)90243-7. [DOI] [PubMed] [Google Scholar]
- 36.Roussel M F, Dull T J, Rettenmier C W, Ralph P, Ullrich A, Sherr C J. Transforming potential of the c-fms proto-oncogene (CSF-1 receptor) Nature. 1987;325:549–552. doi: 10.1038/325549a0. [DOI] [PubMed] [Google Scholar]
- 37.Roussel M F, Sherr C J. Mouse NIH 3T3 cells expressing human colony-stimulating factor 1 (CSF-1) receptors overgrow in serum-free medium containing human CSF-1 as their only growth factor. Proc Natl Acad Sci USA. 1989;86:7924–7927. doi: 10.1073/pnas.86.20.7924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Roussel M F, Shurtleff S A, Downing J R, Sherr C J. A point mutation at tyrosine-809 in the human colony-stimulating factor 1 receptor impairs mitogenesis without abrogating tyrosine kinase activity, association with phosphatidylinositol 3-kinase, or induction of c-fos and junB genes. Proc Natl Acad Sci USA. 1990;87:6738–6742. doi: 10.1073/pnas.87.17.6738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sadler R H, Raab-Traub N. Structural analyses of the Epstein-Barr virus BamHI A transcripts. J Virol. 1995;69:1132–1141. doi: 10.1128/jvi.69.2.1132-1141.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sbih-Lammali F, Djennaoui D, Belaoui H, Bouguermouh A, Decaussin G, Ooka T. Transcriptional expression of Epstein-Barr virus genes and proto-oncogenes in north African nasopharyngeal carcinoma. J Med Virol. 1996;49:7–14. doi: 10.1002/(SICI)1096-9071(199605)49:1<7::AID-JMV2>3.0.CO;2-A. [DOI] [PubMed] [Google Scholar]
- 41.Sherr C J. Colony-stimulating factor-1 receptor. Blood. 1990;75:1–12. [PubMed] [Google Scholar]
- 42.Sims J E, March C J, Cosman D, Widmer M B, MacDonald H R, McMahan C J, Grubin C E, Wignall J M, Call S M, Friend D, Alpert A R, Gillis S R, Urdal D L, Dower S K. cDNA expression cloning of the IL-1 receptor, a member of the immunoglobulin superfamily. Science. 1988;241:585–589. doi: 10.1126/science.2969618. [DOI] [PubMed] [Google Scholar]
- 43.Smith C A, Smith T D, Smolak P J, Friend D, Hagen H, Gerhart M, Park L, Pickup D J, Torrance D, Mohler K, Schooley K, Goodwin R G. Poxvirus genomes encode a secreted, soluble protein that completely inhibits β chemokine activity yet lacks sequence homology to known chemokine receptors. Virology. 1997;236:316–327. doi: 10.1006/viro.1997.8730. [DOI] [PubMed] [Google Scholar]
- 44.Spriggs M K. One step ahead of the game: viral immunomodulatory molecules. Annu Rev Immunol. 1996;14:101–130. doi: 10.1146/annurev.immunol.14.1.101. [DOI] [PubMed] [Google Scholar]
- 45.Spriggs M K, Armitage R J, Comeau M R, Strockbine L, Farrah T, Macduff B, Ulrich D, Alderson M R, Müllberg J, Cohen J I. The extracellular domain of the Epstein-Barr virus BZLF2 protein binds the HLA-DR β chain and inhibits antigen presentation. J Virol. 1996;70:5557–5563. doi: 10.1128/jvi.70.8.5557-5563.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Spriggs M K, Armitage R J, Strockbine L, Clifford K N, Macduff B M, Sato T A, Maliszewski C R, Fanslow W C. Recombinant human CD40 ligand stimulates B cell proliferation and immunoglobulin E secretion. J Exp Med. 1992;176:1543–1550. doi: 10.1084/jem.176.6.1543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Stanley E R, Berg K L, Einstein D B, Lee P S, Pixley F J, Wang Y, Yeung Y G. Biology and action of colony-stimulating factor-1. Mol Reprod Dev. 1997;46:4–10. doi: 10.1002/(SICI)1098-2795(199701)46:1<4::AID-MRD2>3.0.CO;2-V. [DOI] [PubMed] [Google Scholar]
- 47a.Strockbine, L. D., and M. K. Spriggs. Unpublished observations.
- 48.Symons J A, Alcamí A, Smith G L. Vaccinia virus encodes a soluble type I interferon receptor of novel structure and broad species specificity. Cell. 1995;81:551–560. doi: 10.1016/0092-8674(95)90076-4. [DOI] [PubMed] [Google Scholar]
- 49.Takahashi M, Hong Y-M, Yasuda S, Takano M, Kawai K, Nakai S, Hirai Y. Macrophage colony-stimulating factor is produced by human T lymphoblastoid cell line, CEM-ON: identification by amino-terminal amino acid sequence analysis. Biochem Biophys Res Commun. 1988;152:1401–1409. doi: 10.1016/s0006-291x(88)80441-8. [DOI] [PubMed] [Google Scholar]
- 50.Tanner J E, Wei M X, Alfieri C, Ahmad A, Taylor P, Ooka T, Menezes J. Antibody and antibody-dependent cellular cytotoxicity responses against the BamHI A rightward open-reading frame-1 protein of Epstein-Barr virus (EBV) in EBV-associated disorders. J Infect Dis. 1997;175:38–46. doi: 10.1093/infdis/175.1.38. [DOI] [PubMed] [Google Scholar]
- 51.Tarodi B, Subramanian T, Chinnadurai G. Epstein-Barr virus BHRF1 protein protects against cell death induced by DNA-damaging agents and heterologous viral infection. Virology. 1994;201:404–407. doi: 10.1006/viro.1994.1309. [DOI] [PubMed] [Google Scholar]
- 52.Wang Z E, Myles G M, Brandt C S, Lioubin M N, Rohrschneider L. Identification of the ligand-binding regions in the macrophage colony-stimulating factor receptor extracellular domain. Mol Cell Biol. 1993;13:5348–5359. doi: 10.1128/mcb.13.9.5348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Warren M K, Ralph P. Macrophage growth factor CSF-1 stimulates human monocyte production of interferon, tumor necrosis factor, and colony stimulating activity. J Immunol. 1986;137:2281–2285. [PubMed] [Google Scholar]
- 54.Wei M X, de Turenne-Tessier M, Decaussin G, Benet G, Ooka T. Establishment of a monkey kidney epithelial cell line with the BARF1 open reading frame from Epstein-Barr virus. Oncogene. 1997;14:3073–3081. doi: 10.1038/sj.onc.1201128. [DOI] [PubMed] [Google Scholar]
- 55.Wei M X, Moulin J-C, Decaussin G, Berger F, Ooka T. Expression and tumorigenicity of the Epstein-Barr virus BARF1 gene in human Louckes B-lymphocyte cell line. Cancer Res. 1994;54:1843–1848. [PubMed] [Google Scholar]
- 56.Wei M X, Ooka T. A transforming function of the BARF1 gene encoded by Epstein-Barr virus. EMBO J. 1989;8:2897–2903. doi: 10.1002/j.1460-2075.1989.tb08438.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wong G G, Temple P A, Leary A C, Witek-Giannotti J S, Yang Y C, Ciarletta A B, Chung M, Murtha P, Kriz R, Kaufman R J, Ferenz C R, Sibley B S, Turner K J, Hewick R M, Clark S C, Yanai N, Yokota H, Yamada M, Saito M, Motoyoshi K, Takaku F. Human CSF-1: molecular cloning and expression of 4-kb cDNA encoding the human urinary protein. Science. 1987;235:1504–1508. doi: 10.1126/science.3493529. [DOI] [PubMed] [Google Scholar]
- 58.Zhang C X, Decaussin G, Daillie J, Ooka T. Altered expression of two Epstein-Barr virus early genes localized in BamHI-A in nonproducer Raji cells. J Virol. 1988;62:1862–1869. doi: 10.1128/jvi.62.6.1862-1869.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zisman E, Waisman A, Ben-Yair E, Tartakovsky B. Production of colony-stimulating factor 1 by T cells: possible involvement in their interaction with antigen-presenting cells. Cytokine. 1993;5:309–318. doi: 10.1016/1043-4666(93)90062-a. [DOI] [PubMed] [Google Scholar]