Significance
Plant and fungal viruses have divergently evolved to adapt to the distinct biological characteristics of plant and fungal kingdoms. While cross-infection of plant viruses in fungi has been observed, the question of whether a virus could naturally spread to both plant and fungal populations remains unclear. An RNA virus, Valsa mali negative-strand RNA virus 1 (VmNSRV1) was identified from fungus Valsa mali infecting apple trees. Aside from the prevalence of VmNSRV1 in V. mali population, VmNSRV1 was found experimentally to be able to infect plants. More importantly, VmNSRV1 was detected in apple trees grown in the fields. VmNSRV1 could be considered a virus with natural plant and fungal hosts. This finding redefines the conventional concept of plant and fungal viruses.
Keywords: cross-kingdom infection, negative-strand RNA virus, mycovirus, movement protein, hypovirulence
Abstract
The presence of viruses that spread to both plant and fungal populations in nature has posed intriguingly scientific question. We found a negative-strand RNA virus related to members of the family Phenuiviridae, named Valsa mali negative-strand RNA virus 1 (VmNSRV1), which induced strong hypovirulence and was prevalent in a population of the phytopathogenic fungus of apple Valsa canker (Valsa mali) infecting apple orchards in the Shaanxi Province of China. Intriguingly, VmNSRV1 encodes a protein with a viral cell-to-cell movement function in plant tissue. Mechanical leaf inoculation showed that VmNSRV1 could systemically infect plants. Moreover, VmNSRV1 was detected in 24 out of 139 apple trees tested in orchards in Shaanxi Province. Fungal inoculation experiments showed that VmNSRV1 could be bidirectionally transmitted between apple plants and V. mali, and VmNSRV1 infection in plants reduced the development of fungal lesions on leaves. Additionally, the nucleocapsid protein encoded by VmNSRV1 is associated with and rearranged lipid droplets in both fungal and plant cells. VmNSRV1 represents a virus that has adapted and spread to both plant and fungal hosts and shuttles between these two organisms in nature (phyto-mycovirus) and is potential to be utilized for the biocontrol method against plant fungal diseases. This finding presents further insights into the virus evolution and adaptation encompassing both plant and fungal hosts.
Certain viruses have narrow host ranges, infecting only particular species within a genus, but the host ranges of viruses could span higher taxonomic ranks such as class, phylum, and kingdom (1). Although some virus groups are exceptions, virus infection across kingdoms is generally thought of as uncommon (2); therefore, viruses can be broadly categorized according to their host kingdoms, such as bacterial, animal, plant, and fungal viruses. In fact, many viruses possess morphological and genetic characteristics that are unique to each virus category, implying that these viruses have evolved to adapt to the distinct structural, physiological, genetic, and ecological characteristics of the kingdoms. For instance, many animal viruses are enveloped by a lipid bilayer decorated with viral envelope glycoprotein(s), which facilitates viral entry into cells via the fusion of the envelope with cell membranes (3, 4). On the other hand, the majority of plant viruses are not enveloped. They are commonly introduced to plant cells through the breaking of the rigid cell walls and cell membranes by biological vectors or mechanical injuries (5–7). In addition, plant viruses usually encode the movement protein (8) to facilitate the transport of the viral genome or virion to adjacent cells through plasmodesmata, membranous conduits that connect plant cells (9). In the case of fungal viruses (mycoviruses), a considerable proportion lacks genome encapsidation (10). This is likely related to the adaptation of mycoviruses to the nonextracellular route of virus transmission through hyphal anastomosis or sporulation in fungi (11).
Several viruses can replicate in hosts belonging to two different kingdoms and shuttle between the two host kingdoms in nature, but in this case, one of the hosts is usually an arthropod that also serves as the virus vector. Plant RNA viruses belonging to the families Reoviridae, Rhabdoviridae, Tospoviridae, and Phenuiviridae infect plants as well as insect vectors (12). Additionally, a DNA mycovirus was acquired by a fungus-feeding insect and replicated in the insect. Furthermore, the insect could transmit this virus to the fungus (13). Thus, these viruses may have adapted to the hosts of two kingdoms during long-term association.
In the natural environment, plants are often colonized by various fungi or fungus-like organisms (such as oomycetes) (14, 15). The possibility of virus transmission between plants and fungi was initially highlighted by the finding that several plant and fungal viruses share a high degree of genetic similarity (2, 16, 17). Importantly, a plant RNA virus, cucumber mosaic virus (CMV, family Bromoviridae), was found infecting the phytopathogenic fungus Rhizoctonia solani (18), providing evidence of naturally occurring cross-kingdom infection of a plant virus in a fungus. The bidirectional transmission of CMV and other plant and fungal viruses, including plant viroids, between plants and fungi was also demonstrated by fungal inoculation experiments in the laboratory (18–21). In a subsequent study, many of the fungal strains isolated from various vegetable plants infected with various plant viruses were found to carry at least 10 plant RNA virus species, but the presence of plant viruses in these fungal strains was not persistent (22). This suggests that the transmission of plant viruses to fungi may be common in the natural environment, but not all plant viruses can adapt subsequently and become native to the fungal hosts. Given that plants and fungi are structurally and genetically very divergent organisms, it remains an open question whether a virus can adapt to both organisms and spread to plant and fungal populations in nature.
The family Phenuiviridae is an expanding and highly diverse virus family belonging to Bunyavirales, an order of negative-strand segmented RNA viruses (8, 23). Phenuivirids have 2 to 8 genomic RNA segments with negative-sense or negative- and ambi-sense coding strategies, depending on the genus (24, 25). Most animal phenuivirid genomes encode a glycoprotein precursor, whereas plant phenuivirid genomes likely lack structural glycoproteins and encode a nonstructural protein that allows cell-to-cell movement in plant hosts. The phenuiviral virions are mostly enveloped spherical particles (26), while those of the known plant-infecting phenuiviruses are nonenveloped filamentous particles (27–29). Phenuiviruses infect a very broad range of hosts, including humans, vertebrates, arthropods, fungi, and plants, and many can replicate in two distinct hosts belonging to distant taxa (insects and plants or vertebrates), reflecting the high host adaptability of this virus group (25). Notably, many phenuiviruses are known to cause medically and economically important diseases (30–34). The highly adaptative and arthropod-borne nature of phenuiviruses is considered a factor in their continuing potential as emerging viral pathogens that pose a great threat to humans, animals, and agriculture (25, 35). In this study, we identified a virus related to phenuiviruses that has adapted and spread to both plant and fungal hosts.
Results
A Hypovirulence-Inducing Negative-Strand RNA Virus that Spreads in the Valsa mali Population.
Apple Valsa canker caused by the plant pathogenic fungus V. mali var. mali (order Diaporthales) has affected apple orchards in the Shaanxi Province of China (36, 37). V. mali primarily infects wounded parts of the tree, such as pruning cuts or frost-damaged bark, leading to the formation of cankers (38). To investigate the virome of V. mali strains infecting apple trees, we isolated V. mali strains from 167 stem tissue samples with visible Valsa canker symptoms obtained from apple trees grown in several orchards located in different areas belonging to Xianyang City, Shaanxi Province, China (SI Appendix, Fig. S1A). Out of a total of 1,110 V. mali strains isolated, 91 fungal strains with dsRNA accumulation were detected by the dsRNA isolation method. The isolated fungal strains had diverse double-stranded RNA (dsRNA) profiles and showed varying phenotypic growth on PDA (potato dextrose agar) medium (SI Appendix, Fig. S1 B and C). The dsRNA fractions extracted from the 91 fungal strains were subjected to high-throughput sequencing and subsequent bioinformatic analysis for virus identification.
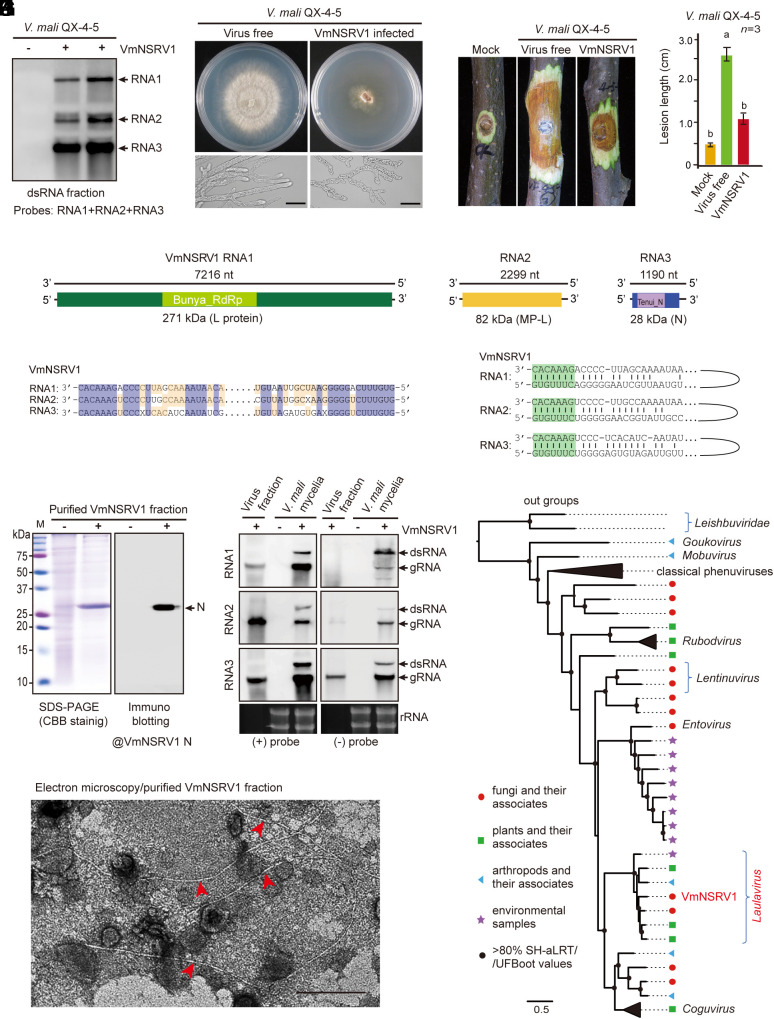
Numerous virus candidates were identified that were available in the database or had not been reported previously. In this study, we were particularly interested in a negative-strand RNA virus candidate that had genetic similarities to the members of the family Phenuiviridae and was thus tentatively named Valsa mali negative-strand RNA virus 1 (VmNSRV1). The VmNSRV1 genome consists of three RNA segments. RT-PCR analysis indicated that VmNSRV1 was present in 72.5% of 91 strains carrying dsRNA and in around 36.4% of 19 randomly selected fungal strains that were not previously detected to have dsRNA accumulation (SI Appendix, Fig. S2). These results indicate that VmNSRV1 is widespread in the V. mali population infecting apple trees in Xianyang City. Following the RT-PCR detection of VmNSRV1 and other viruses, we identified a fungal strain (QX-4-5) that was infected with VmNSRV1 but not with other viruses. Virus-free isogenic strains were obtained from this strain via single-spore isolation. RNA blot analysis using dsRNA fractions isolated from VmNSRV1-infected and virus-free strains confirmed the presence of three RNA segments corresponding to the VmNSRV1 genome in virus-infected strains (Fig. 1A).
Fig. 1.
Identification of a hypovirulence-inducing negative-strand RNA virus from V. mali. (A) Detection of three RNA segments corresponding to the RNA1, 2, and 3 genomes of VmNSRV1 in a V. mali strain by RNA blotting with dsRNA fractions. (B) Phenotypic growth and mycelial morphology of a V. mali strain (VmNSRV1-free and -infected, QX-4-5) on PDA medium. The fungi were photographed at 3 d after culturing. (Scale bar, 20 µm.) (C) Virulence assay of the V. mali (QX-4-5) strain on apple twigs. Fungal disease lesions (peeled twigs) were photographed at 5 d after inoculation. (D) Measurement of fungal disease lesions observed in the experiment described in (C). Different letters indicate significant differences (P < 0.05, one-way ANOVA). (E) A schematic genome structure of VmNSRV1. Black lines represent negative-strand genomic RNAs. Colored boxes represent open reading frames (ORFs) encoded by positive-strand genomic RNAs. The estimated molecular weight of each encoded protein (L[RdRp], MP-L, and N) is indicated. The relative position of conserved domains in the encoded RdRp and N proteins is presented (small colored boxes). (F) Sequence similarities of the 5′- and 3′-terminal regions of VmNSRV1 RNA1, 2, and 3 segments. (G) Sequence complementarity of the 5′- and 3′-terminal regions of each VmNSRV1 RNA1, 2, and 3 segment. (H) SDS-PAGE and immunoblot analyses of the protein associated with VmNSRV1 purified fractions. Immunoblotting was carried out using an antibody specific to the VmNSRV1 N protein. (I) Electron microscopy of a VmNSRV1 purified fraction from a virus-infected V. mali (QX-4-5) strain. Red arrowheads mark the potential VmNSRV1 particles. (Scale bar, 200 nm.) (J) Detection of positive- and negative-strand RNA of VmNSRV1 in purified virus fraction and mycelia of a virus-infected V. mali (QX-4-5) strain by RNA blotting. (K) Phylogenetic relationships of VmNSRV1 with phenuiviruses or other selected phenui-like viruses. The maximum likelihood tree was based on multiple sequence alignment of the RNA1-encoded L protein (RdRp). For the virus names and details of the tree, see SI Appendix, Fig. S6 and its legend.
On PDA medium, the VmNSRV1-infected V. mali strain showed drastically reduced growth, stunted and irregularly shaped mycelia, and reduced pycnidia (fruiting bodies) development compared to the virus-free strain (Fig. 1B and SI Appendix, Fig. S3 A and B). A fungal virulence test using apple twigs, fruits, and leaves showed that VmNSRV1 infection significantly reduced the pathogenicity of the fungal host (Fig. 1 C and D and SI Appendix, Fig. S3 C–F). VmNSRV1 was transmitted horizontally through hyphal anastomosis between vegetatively compatible strains but not between vegetatively incompatible strains (SI Appendix, Fig. S4 A, B, and E) and vertically through spores produced by pycnidia (SI Appendix, Fig. S4 C–E). The coinoculation of virus-free and VmNSRV1-infected V. mali strains on apple leaves and twigs produced smaller lesions than inoculation with the virus-free strain alone (SI Appendix, Fig. S5 A–D). In addition, the direct application of purified VmNSRV1 preparations to V. mali inoculated on apple twigs also resulted in smaller fungal lesions due to virus infection in the fungi (SI Appendix, Fig. S5 E–H).
VmNSRV1 Is Closely Related to Viruses Belonging to the Family Phenuiviridae.
The complete sequences of VmNSRV1 RNA1, 2, and 3 (Acc. Nos. OR209170–2) were 7,216 nt, 2,299 nt, and 1,190 nt, respectively (Fig. 1E). The 5′- and 3′-terminal regions of the RNA1, 2, and 3 genome segments (viral negative-strand) shared identical nucleotide stretches, and the termini of each RNA segment were complementary to each other (Fig. 1 F and G), a common characteristic of viruses in the order Bunyavirales (39). The complementary strand of RNA1 encoded an RNA-dependent RNA polymerase (RdRp) protein (~271 kDa) containing a conserved domain for the bunyavirus RdRp superfamily (E-value 7.6e−38). TBLASTN results revealed its identities with the RdRp encoded by phenuiviruses, with the highest protein identity (62.8%) to the RdRp encoded by grapevine-associated cogu-like virus 3, a phenuivirus (genus Laulavirus) associated with grapevine samples (40). The complementary strand of RNA2 encoded a protein (~82 kDa; hereafter referred to as MP-like protein or MP-L) with no known viral conserved domain; however, it had 27.0 to 47.9% identity with MP-like proteins of phenuiviruses belonging to the genera Coguvirus and Laulavirus, such as grapevine-associated cogu-like virus 2 to 4, watermelon crinkle leaf-associated virus, citrus virus A, citrus concave gum-associated virus (29, 40–42), and other unclassified phenui-like viruses. The complementary strand of RNA3 encoded a protein (~28 kDa) containing a conserved domain for the nucleocapsid (N) protein superfamily (E-value 2.9e−15). It had the highest protein identity (75.1%) with the N protein of Laurel Lake virus (genus Laulavirus), a phenuivirus identified in ticks (43).
Sodium dodecyl sulfate-polyacrylamide gel electrophoresis analysis of purified VmNSRV1 showed a major band above 25 kDa, and immunoblotting analysis using a polyclonal antibody raised against the VmNSRV1 N protein confirmed that this major band was the N protein (Fig. 1H). Electron microscopy of purified VmNSRV1 showed the presence of long, thread-like, filamentous particles of 1 to 2 nm width and heterogenous length (100 to >2,000 nm; Fig. 1I). RNA blot analysis using total RNA extracted from the purified viral fractions revealed the predominant presence of minus-strand RNA1, 2, and 3, while only small or lower amounts of positive-strand RNA2 and 3 were detected (Fig. 1J), confirming the potential minus-strand nature of the VmNSRV1 genome. In total RNA extracted from mycelia, both strands of RNA1, 2, and 3 and their potential dsRNA forms were detected (Fig. 1J).
In a phylogenetic tree constructed using L protein sequences derived from representative members of established genera in the family Phenuiviridae and several unclassified phenui-like viruses, VmNSRV1 was placed in a well-supported clade together with members of the genus Laulavirus, which consists of viruses derived from arthropods, plants, fungi, and environmental samples (Fig. 1K and SI Appendix, Fig. S6), suggesting that VmNSRV1 is a candidate member of this genus.
VmNSRV1 RNA2 Encodes a Protein with Plant Viral MP-Like Function.
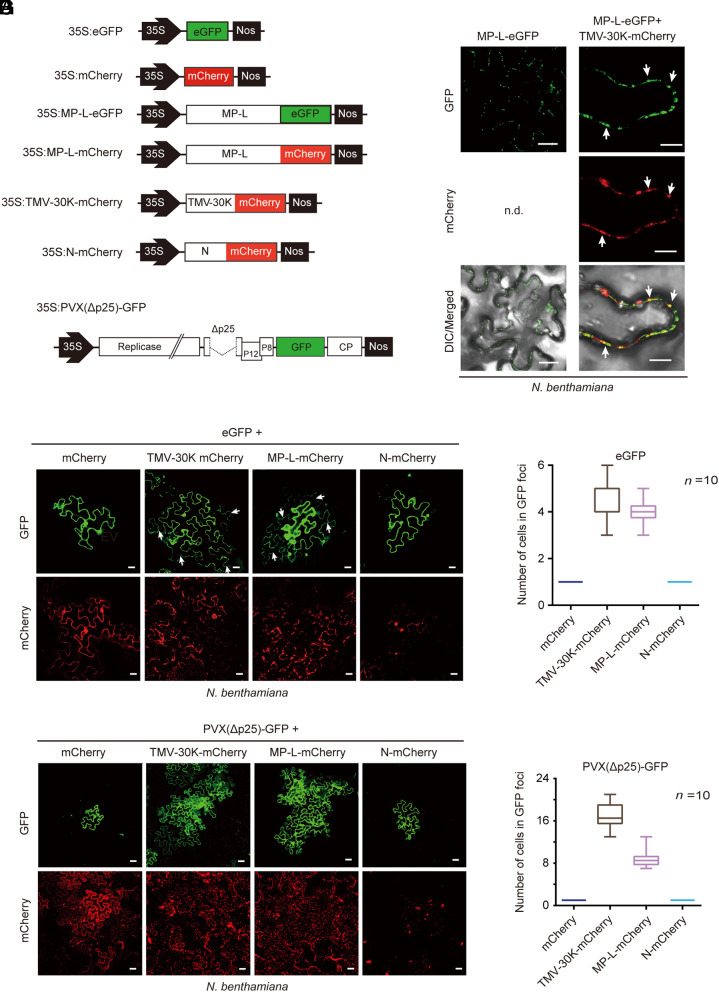
Since the protein encoded by VmNSRV1 RNA2 shares identity with the putative MP-like gene products of plant-infecting phenuiviruses, we investigated whether VmNSRV1 MP-L protein has characteristics similar to those of plant viral MPs. First, to examine the subcellular localization of the protein in the plant, MP-L was fused to eGFP (enhanced green fluorescent protein) (MP-L-eGFP, Fig. 2A) and transiently expressed in leaves of Nicotiana benthamiana by agroinfiltration. Confocal laser scanning microscopy (CLSM) showed that in epidermal cells, MP-L-eGFP was localized in punctate spots along the cell periphery (Fig. 2B), suggesting protein localization in plasmodesmata, which is commonly observed for plant viral MPs such as the 30K MP of tobacco mosaic virus (TMV, genus Tobamovirus) (44). To verify this, MP-L-eGFP was coexpressed with TMV 30K fused to mCherry (TMV-30K-mCherry, Fig. 2A). MP-L-eGFP was colocalized with 30K-mCherry in punctate spots along the cell periphery (Fig. 2C), indicating that MP-L localizes to plasmodesmata.
Fig. 2.
Viral plant MP function of the MP-L protein encoded by RNA2 of VmNSRV1. (A) A schematic diagram of the constructs used to assess the ability of MP-L to increase the SEL of plasmodesmata and complement the cell-to-cell movement of movement-defective potato virus X expressing GFP [PVX(Δp25)-GFP]. TMV 30K fused to mCherry (TMV-30K-mCherry) was used as a plasmodesmatal marker. 35S and Nos represent the cauliflower mosaic virus (CaMV) 35S promoter and nopaline synthase terminator sequences, respectively. (B) Subcellular localization of MP-L protein fused to eGFP (MP-L-eGFP) in the epidermal cells of N. benthamiana plants transiently expressed by Agrobacterium infiltration. Fluorescence was observed by CLSM 3 d after inoculation. (Scale bar, 25 µm.) (C) Coexpression of MP-L-eGFP with TMV-30K-mCherry in epidermal cells. Arrows indicate the representative overlapping fluorescence signals. (Scale bar, 10 µm.) (D) Coexpression of the tag-free eGFP with TMV-30K-mCherry, MP-L-mCherry, N-mCherry, or unfused mCherry in epidermal cells. An Agrobacterium culture harboring 35S: eGFP (OD600 = 1) was diluted 10,000-fold and mixed (1:1) with an Agrobacterium culture harboring one of the mCherry expression constructs. Arrows indicate cells with diffused eGFP. (Scale bar, 20 µm.) (E) Quantification of the GFP-expressing cells in GFP foci observed in the experiment described in (D). (F) Coexpression of PVX(Δp25)-GFP with TMV-30K-mCherry, MP-L-mCherry, N-mCherry, or unfused mCherry in epidermal cells. (Scale bar, 40 µm.) (G) Quantification of the GFP-expressing cells in GFP foci observed in the experiment described in (F).
To examine whether VmNSRV1 MP-L has the ability to increase the size exclusion limit (SEL) of plasmodesmata (9), the effect of MP-L expression on the cell-to-cell diffusion of an unfused eGFP in plant cells was observed. An Agrobacterium culture (OD600 = 1) harboring an eGFP expression vector (35S: eGFP) was highly diluted (10,000-fold) and used for coexpression with mCherry, TMV-30K-mCherry, MP-L-mCherry, or N-mCherry (Fig. 2A). eGFP expression was found to be restricted to a single cell in unfused mCherry or N-mCherry protein-expressing leaf tissue, whereas it was diffused to 2 to 4 adjacent cells in TMV-30K-mCherry or MP-L-mCherry-expressing tissue (Fig. 2 D and E), showing that MP-L has the ability to increase the plasmodesmata SEL. Based on predicted structural similarity, MP or putative MP encoded by plant-infecting phenuiviruses belong to members of the 30K superfamily of plant virus MPs (29, 45, 46). Previous reports demonstrated that the cell-to-cell movement of an MP-defective mutant of potato virus X (PVX, genus Potexvirus) was restored by the expression of MPs belonging to the 30K superfamily (47–49). Similarly, in a trans-complementation experiment using an MP-defective mutant of (PVX) expressing GFP (PVX(Δp25)-GFP, Fig. 2A), the expression of MP-L-mCherry or TMV-30K-mCherry but not N-mCherry or unfused mCherry promoted the cell-to-cell movement of PVX(Δp25)-GFP (Fig. 2 F and G), demonstrating a potential MP function of VmNSRV1 MP-L. The observation that a mycovirus encodes a protein with the MP function is intriguing. Further studies using different experimental assays are necessary to verify the MP-L function in plant viral movement in plants.
VmNSRV1 Can Infect Plants and Was Detected in Orchard Apples.
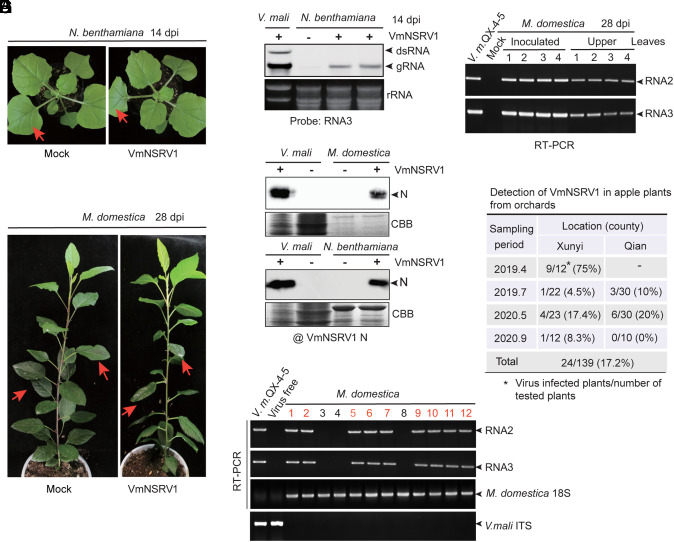
Next, we tested whether VmNSRV1 can infect plants. Purified VmNSRV1 preparations were used as an inoculum for mechanical rub-inoculation on the leaves of N. benthamiana plants. Up to 14 days postinoculation (dpi), the inoculated plants did not develop any visible symptoms of viral disease (Fig. 3A). However, in noninoculated upper leaves, VmNSRV1 RNA3 and N protein accumulation were detected by RNA blot and immunoblot analyses (Fig. 3 B and E). Purified VmNSRV1 preparations were also inoculated on young apple trees. No virus symptoms were observed up to 28 dpi (Fig. 3C), but in noninoculated upper leaves, VmNSRV1 RNA2 and RNA3 and N protein accumulation were detected by RT-PCR and immunoblotting (Fig. 3 D and E).
Fig. 3.
Infectivity of VmNSRV1 in plants. (A) N. benthamiana plant mechanically inoculated with purified VmNSRV1 fractions. Plants were photographed at 14 dpi. Arrows indicate the inoculated leaves. (B) RNA blot detection of VmNSRV1 RNA3 accumulation in the upper leaves of N. benthamiana plants described in (A). Leaves were harvested at 14 dpi. (C) Apple plant (Malus domestica) mechanically inoculated with purified VmNSRV1 fraction. Plants were photographed at 28 dpi. Arrows indicate the inoculated leaves. (D) RT-PCR detection of VmNSRV1 RNA2 and RNA3 accumulation in the upper leaves of apple plants described in (C). Leaves were harvested at 28 dpi. (E) Immunoblot analysis of the protein fractions from noninoculated upper leaves of virus-free and VmNSRV1-infected N. benthamiana and M. domestica. Immunoblotting was carried out using an antibody specific to the VmNSRV1 N protein. (F) RT-PCR detection of VmNSRV1 RNA2 and RNA3 accumulation in the upper leaves of field-grown apple trees. Primers specific for M. domestica 18S rRNA and V. mali ITS were used as a plant reference gene or to confirm the absence of V. mali in leaf samples, respectively. (G) Detection rate of VmNSRV1 in apple trees grown in orchards located in two counties of Shaanxi Province, China. VmNSRV1 detection was performed as described in (F).
We then investigated whether VmNSRV1 infects apple trees grown in orchards. Apple leaf samples were collected from orchards in two counties of Xianyang City, during four sampling periods in 2019 and 2020 and subjected to VmNSRV1 detection by RT-PCR. VmNSRV1 was detected in 24 out of 139 plants tested (17.2% of tested samples) grown in orchards located in both counties (Fig. 3 F and G).
VmNSRV1 Can Be Transmitted between Plants and Fungi.
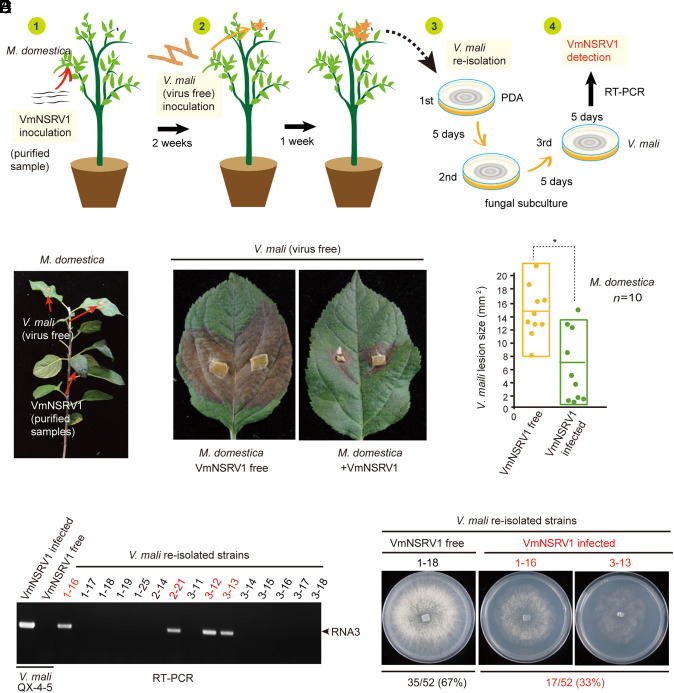
Previously, we showed experimentally under laboratory conditions that some plant RNA viruses can be transmitted from a plant to a fungus and vice versa during fungal colonization of the plants (18, 19). We performed a similar virus and fungal inoculation experiment to investigate whether VmNSRV1 undergoes cross-kingdom transmission during colonization of the apple plant by V. mali (Fig. 4A and SI Appendix, Fig. S7A). Two weeks after the mechanical inoculation of apple plants with VmNSRV1, a virus-free V. mali strain was inoculated on the upper leaves of VmNSRV1-infected plants (Fig. 4B). Remarkably, V. mali developed significantly smaller lesions on the leaves of VmNSRV1-infected apple plants than on those of virus-free apple plants (Fig. 4 C and D). Among 52 V. mali strains obtained by the reisolation of fungi from VmNSRV1-infected apple plants, 17 fungal strains were infected with VmNSRV1, as indicated by RT-PCR (Fig. 4 E and F). Accordingly, VmNSRV1-infected fungal strains showed slower growth on PDA medium compared to VmNSRV1-free fungal strains (Fig. 4F). These observations indicate that V. mali acquires VmNSRV1 from apple plants and that cross-infection with VmNSRV1 suppresses the growth and pathogenicity of V. mali. In an experiment to assess the transmission of VmNSRV1 from V. mali to the apple plant, VmNSRV1 RNA3 was detected by RT-PCR in the upper leaves of apple plants inoculated 4 wk earlier with a VmNSRV1-infected V. mali strain on the lower leaves (SI Appendix, Fig. S7B), although V. mali was not present in these leaves (SI Appendix, Fig. S7C), showing that VmNSRV1 had moved systemically in these plants but that the fungus had not moved systemically. These experiments demonstrate the bidirectional transmission of VmNSRV1 between V. mali and the apple plant.
Fig. 4.
Transmission of VmNSRV1 from the plant to fungus. (A) Diagram describing the experimental procedure used to study VmNSRV1 acquisition by V. mali. (1) Mechanical inoculation of purified VmNSRV1 fractions on apple plants. (2) Inoculation of upper leaves with mycelia of a virus-free V. mali (QX-4-5) strain 2 wk after VmNSRV1 inoculation. (3) Reisolation of V. mali from fungal lesions on leaves 1 wk after inoculation with V. mali mycelia. (4) RT-PCR detection of VmNSRV1 in reisolated V. mali strains after at least three subcultures. (B) A young apple plant inoculated with purified VmNSRV1 fractions on the lower leaves and a virus-free V. mali (QX-4-5) strain on the noninoculated upper leaves. Arrows indicate the inoculated leaves. (C) Representative V. mali lesions on the leaves of virus-free and VmNSRV1-infected apple plants. Fungal disease lesions were photographed at 5 d after inoculation. (D) Lesion areas of V. mali measured on leaves described in (C). “*” indicates significant differences (P < 0.05, Student’s t test). (E) RT-PCR detection of VmNSRV1 in V. mali strains reisolated from lesions on leaves described in (C). (F) Phenotypic growth of representative reisolated V. mali strains with VmNSRV1 infection or without virus infection. The fungi were photographed at 3 d after culturing. The numbers of VmNSRV1-free and VmNSRV1-infected V. mali strains are shown below the images.
The VmNSRV1 N Protein Rearranges Lipid Droplets in Host Cells.
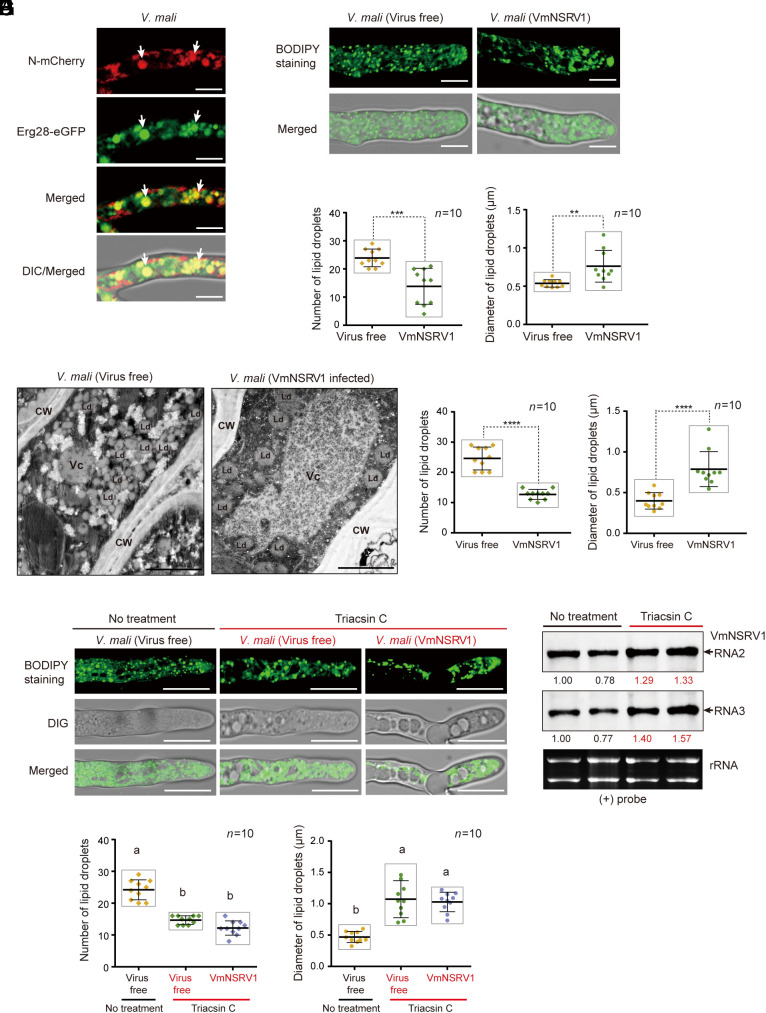
To further characterize the VmNSRV1-encoded proteins, MP-L-mCherry and N-mCherry were expressed in V. mali. MP-L-mCherry was diffusely distributed throughout the fungal cells, while N-mCherry localized in aggregate structures of heterogenous size (SI Appendix, Fig. S8A), suggesting the possibility that N protein associates with membranous structures. To investigate whether the VmNSRV1 N protein is associated with the fungal endomembrane system, N-mCherry was coexpressed with eGFP-tagged endoplasmic reticulum (SecE), trans-Golgi network (KEX), peroxisome (PER4), and lipid droplet (Erg28) markers. Fluorescence observation showed that N-mCherry consistently colocalized with Erg28-eGFP but not with other organelle markers (Fig. 5A and SI Appendix, Fig. S8B), suggesting that the N protein mainly associates with lipid droplets. Lipid droplets are cellular organelles involved in the storage and metabolism of neutral lipids such as triglycerides and cholesterol esters. They are spherical structures composed of lipids enclosed by a phospholipid monolayer containing integral and peripheral proteins (50). Numerous studies have indicated the involvement of lipid droplets in virus infection and host immune responses (51–53). To further confirm our observation, MP-L-mCherry- or N-mCherry-expressing fungi were treated with BODIPY 493/503, which is commonly used to stain lipid droplets in fungi. N-mCherry-expressing aggregates colocalized with BODIPY-stained fluorescent signals regardless of the presence of VmNSRV1 (SI Appendix, Fig. S8C). We also observed that in VmNSRV1-infected or N-mCherry-expressing cells, BODIPY-stained signals showed larger but fewer aggregates or granule-like structures compared to those observed in virus- or N-mCherry-free cells (Fig. 5 B and C and SI Appendix, Fig. S8C). Likewise, in the VmNSRV1-infected cells, Erg28-eGFP localized mainly to strikingly larger aggregate structures than in virus-free cells (SI Appendix, Fig. S9A). Moreover, sucrose gradient fractions showed that in the presence of VmNSRV1, some of the Erg28-eGFP was distributed in the upper phases, suggesting an increased structural size and possibly reduced density of some fractions of lipid droplets (SI Appendix, Fig. S9B). Electron microscopy of ultrathin sections prepared from V. mali mycelia revealed that fungal cells infected with VmNSRV1 had enlarged but reduced numbers of lipid droplets compared to virus-free cells (Fig. 5 D and E). Notably, VmNSRV1 infection also drastically increased the size of vacuoles (Fig. 5D), similar to what was seen in the fluorescence observation of VmNSRV1-infected mycelial cells (SI Appendix, Fig. S8C, large hollow structures). Intriguingly, treatment of V. mali with triacsin C, which blocks the accumulation of triacylglycerol in lipid droplets by inhibiting long-chain acyl-CoA synthetase (54), resulted in enlarged but reduced numbers of lipid droplets (Fig. 5 F and G) and appeared to be associated with the increased accumulation of VmNSRV1 genomic RNA (Fig. 5H), suggesting that the rearrangement of lipid droplets by the N protein may promote virus multiplication in the fungal cells.
Fig. 5.
Association of the VmNSRV1 N protein with lipid droplets. (A) Coexpression of N protein fused to mCherry (N-mCherry) with Erg28-eGFP (a lipid droplet marker) in V. mali (QX-4-5) strain. Fluorescence in mycelial cells was observed by CLSM. Arrows indicate representative overlapping fluorescence signals. (Scale bar, 10 µm.) (B) Staining of lipid droplets with BODIPY 493/503. (Scale bar, 10 µm.) (C) Quantification and measurement of the number and size of lipid droplets in the area of 10 µm2 per cell observed in the staining experiment described in (B). “**” or “***” indicates significant differences (P < 0.01 or 0.001, Student’s t test). (D) Electron microscopy observations of ultrathin sections prepared from the mycelia of virus-free and VmNSRV1-infected V. mali (QX-4-5) strains. CW, Vc, and Ld indicate cell wall, vacuolar, and lipid droplet, respectively. (Scale bar, 2.0 μm.) (E) Quantification and measurement of the number and size of lipid droplets in the area of 10 µm2 per cell observed in the staining experiment described in (D). “****” indicates significant differences (P < 0.0001, Student’s t test). (F) Treatment of virus-free and virus-infected V. mali (QX-4-5) strains with triacsin C (acetyl-CoA synthetase inhibitor). Lipid droplets were stained with BODIPY 493/503. (Scale bar, 10 µm.) (G) Quantification and measurement of the number and size of lipid droplets in the area of 10 µm2 per fungal cell observed in the staining experiment described in (F). Different letters indicate significant differences (P < 0.01, one-way ANOVA). (H) RNA blot analysis of VmNSRV1 negative-strand genomic RNA in triacsin C-treated and non-treated V. mali (QX-4-5) strains obtained from the experiment described in (F).
Expression of N-eGFP in N. benthamiana cells also showed accumulation in granule-like or large aggregate structures that colocalized with lipid droplet staining signals of Nile red, which is usually used to visualize lipid droplets in plants (SI Appendix, Fig. S10A). Moreover, N-eGFP expression in the cells also enlarged the size of aggregates visualized by Nile red staining as compared to those observed in eGFP-expressing cells (SI Appendix, Fig. S10B). These observations revealed that the VmNSRV1 N protein rearranges lipid droplets in both fungal and plant cells.
Discussion
While cross-infection of plant viruses in fungi has been documented, it is still unclear whether a virus could naturally spread to both plant and fungal populations. In this study, VmNSRV1 was found to be present in many V. mali strains (more than 30%) isolated from apple trees grown in numerous apple orchards distributed in a wide area of Shaanxi Province, suggesting that VmNSRV1 is widespread in the V. mali population in the field. VmNSRV1 had the typical characteristics of a mycovirus; it stably infected V. mali strains, transmitted through hyphal anastomosis and spores, and reduced the virulence of the fungal host. Interestingly, VmNSRV1 could also infect plants, and to some extent, was distributed in apple trees in the field. Thus, VmNSRV1 can be considered a virus with dual natural fungal and plant hosts. The transmissibility of VmNSRV1 between the plant and fungus suggests that VmNSRV1 shuttles between plants and fungi in nature and that V. mali may serve as a biological vector for the spread of VmNSRV1 among apple plants, although VmNSRV1 may also be transmitted by other common vectors for plant viruses. On the other hand, apple plants may serve as a biological reservoir for VmNSRV1, wherein diverse V. mali strains colonizing the plants can acquire VmNSRV1, leading to the spread of VmNSRV1 to various vegetatively incompatible fungal strains in V. mali populations. As VmNSRV1 cannot be typically defined as either a mycovirus or a plant virus, we propose the concept or term “phyto-mycovirus” to refer to a virus that has spread in nature between plant and fungal hosts. A phenuivirus (genus Lentinuvirus) found in the shiitake mushroom and a phenui-like virus infecting Trichoderma gamsii also encode a plant MP-like protein (46, 55). Phenuiviruses belonging to the genus Laulavirus have been identified in grapevines infected with downy mildew (40). Further studies are needed to investigate whether other phenui- or phenui-like viruses spread in nature to both plant and fungal hosts.
As VmNSRV1 infection causes mycelial malformations, slower growth, lower spore production, and hypovirulence to V. mali, it has potential as a biological control agent for apple Valsa canker disease. We have previously proposed using mycoviruses as biocontrol agents for fungal diseases of crops by establishing mycovirus infection in the plant; when the phytopathogenic fungus colonizes the plant, the mycovirus is expected to cross-infect the fungus and reduce the pathogenicity of the fungus (16). Systemic infection of a mycovirus, Cryphonectria hypovirus 1 (CHV1, family Hypoviridae), throughout the plant, can be achieved by coinfection with a plant virus that facilitates the cell-to-cell and systemic movement of CHV1 (19). Unlike conventional fungal viruses, VmNSRV1 alone was competent for systemic infection in the plant, and VmNSRV1 infection in the plant could confer protection against severe fungal diseases (Fig. 4 B–D). Moreover, the direct application of purified VmNSRV1 preparations to V. mali mycelia could reduce fungal lesions on apple twigs (SI Appendix, Fig. S5 E and F). The efficacy of VmNSRV1 infection in protecting apple plants against V. mali needs to be further evaluated under field conditions.
Lipid droplets are organelles that were initially thought to function mainly as neutral lipid storage; however, it is now established that lipid droplets are also involved in many physiological and pathological processes in the cell (56). Numerous studies have implicated lipid droplets in the replication of RNA viruses (53). For example, the core and NS5A proteins of hepatitis C virus colocalize on cytoplasmic lipid droplets, and this association is likely to influence viral particle assembly (57). The association of proteins encoded by plant and fungal viruses with lipid droplets has been unclear. Lipid droplet rearrangement by the N protein may be critical for the ability of VmNSRV1 to infect both fungal and plant hosts, and further detailed studies are required to confirm this view.
Materials and Methods
Sample Collection, Fungal Isolation, and Fungal Culture.
The stem tissue samples (consisting of bark and vascular cambium tissues) were collected from around the junctions of Valsa canker lesions and healthy parts of the tissues on the trunks of apple trees (>10 y old) grown in orchards located in Yangling District, Wugong County, Qianxian County, Liquan County, and Xunyi County in the Shaanxi Province of China in the spring of 2018 and 2019. For virus detection in apple trees, young, newly emerged leaves on the tops of shoots were collected from apple trees grown in orchards in Xunyi County and Qian County in the Shaanxi Province of China in 2019 and 2020.
For the isolation of V. mali strains, about 5 mm2 of stem tissue samples were soaked in 75% alcohol for 5 min and then washed twice with sterile water for 5 min each. The tissue samples were placed on sterile absorbent papers to remove excess water and then placed on PDA culture medium supplemented with streptomycin (20 μg/mL) at 25 °C. After 3 to 5 d, the tips of the growing hyphae were collected and transferred to fresh PDA medium. Generally, fungi were cultured and maintained in PDA medium at 25 °C for observation or in PDA medium layered with cellophane for nucleic acid extraction.
RNA Extraction, High-Throughput Sequencing, RT-PCR, and Rapid Amplification of cDNA Ends (RACE).
dsRNA and total RNA were extracted from fungal mycelia as described previously (58). The RNA was treated with RQ1 RNase-Free DNase (Promega, USA) to remove fungal genomic DNA. The dsRNA was used as a template for high-throughput sequencing analysis on the Illumina HiSeq 4000 platform (Illumina, San Diego, CA, United States) carried out by Hanyu Biotechnology Co., Ltd. (Shanghai, China) as described previously (59), and subsequent bioinformatic analyses were performed as described previously (22).
Based on the assembled virus-like contig sequences from high-throughput sequencing, VmNSRV1-specific primers were designed for RT-PCR detection. First-strand cDNA was synthesized using EasyScript® reverse transcriptase (TransGen Biotech, China) and amplified using DNA polymerase (Kangwei, China) with VmNSRV1-specific primers.
RACE of the 5′- or 3′-terminal sequences of VmNSRV1 genome sequences was carried out as described previously (60). The PCR products were purified and subjected to Sanger sequencing. All primers used in this study are listed in SI Appendix, Table S1.
Plant Expression Vectors and Transient Expression in Plants by Agroinfiltration.
The coding sequences of the VmNSRV1 N (Acc. No. OR209172) and MP-L (Acc. No. OR209171) genes were fused to the eGFP gene and inserted into the binary vector pBIN41 (61). The coding regions of the MP-L, NP, and TMV 30K (J02412.1) genes were inserted into the pBI121-mCherry vector (62). The potato virus X (PVX) vector with an internal deletion in the triple gene block 1 (p25), named PVX(Δp25)-GFP, has been described previously (63) and was coexpressed with a p19 RNA silencing suppressor-carrying plasmid (pBIN-p19) (64). All plasmids generated in this study are described in SI Appendix, Table S2.
Agrobacterium infiltration was performed as previously described (65). The binary vector construct plasmids were introduced to Agrobacterium tumefaciens (strain GV3101). Agrobacterium cultures were resuspended in infiltration buffer (10 mM MES, pH 5.7, 10 mM MgCl2, and 150 mM acetosyringone) and injected into the leaves of N. benthamiana plants (six-leaf stage).
Fungal Expression Vectors and Transformation of V. mali.
The coding sequences of the VmNSRV1 N and MP-L genes were fused with the mCherry gene and inserted into a fungal expression plasmid vector pCPXHY2 (66), carrying a hygromycin B phosphotransferase gene (hph) as a selectable marker. The coding sequences of the KEX (Protease KEX1, KUI70994.1), PER4 (Peroxisomal membrane protein 4, KUI68068.1), SecE (Putative protein transport protein Sec61 subunit gamma, KUI57675.1) and Erg28 (Ergosterol biosynthetic protein 28, KUI63417.1) genes were fused with the eGFP gene and inserted into the pCPXHY2 plasmid.
V. mali protoplasts were prepared as previously described (58). The recombinant plasmid was transfected into fungal protoplasts using polyethylene glycol (PEG 4000), and the fungal transformants were screened based on resistance to Hygromycin B. The fungal transformants expressing two different fusion proteins were obtained by hyphal fusion.
BODIPY and Nile Red Staining.
BODIPY 493/503 dye (difluoro {2- [1-(3, 5-dimethyl-2H-pyrrol-2-ylidene-N) ethyl]-3, 5-dimethyl-1H-pyrrolato-N}, Sigma-Aldrich, USA) was dissolved in dimethyl sulfoxide (DMSO) and further diluted in 0.01 M PBS. Fungal mycelia were treated with 1 µM BODIPY 493/503 for 10 min at room temperature and then washed with 0.01 M PBS. Mycelia were mounted on slides and observed with a laser scanning confocal microscope. Nile Red dye (Solarbio, China) was dissolved in DMSO and further diluted in 0.01 M PBS. Leaves (0.5 × 0.5 cm) were soaked in 1 µM Nile Red solution (500 μL) and vacuum-infiltrated for 2 h at room temperature. Leaves were washed with 0.01 M PBS before observation.
Triacsin C Inhibitor Treatment.
Fungus was cultured for 4 d in PDA medium containing 5 μM Triacsin C (MedChemExpress, USA) layered with cellophane. Each treatment group had two replicates. The virus-free and virus-infected fungal mycelia treated with Triacsin C were stained using BODIPY 493/503, and LDs were observed.
Observation of Fluorescent Proteins.
An Olympus FV3000 confocal laser scanning microscope was used to visualize the signals of eGFP/BODIPY 493/503 dye (excitation, 488 nm; emission, 510 to 550 nm) and mCherry/Nile Red staining (excitation, 587 nm; emission, 560 to 610 nm). The number of LDs in single fungal cell within 10 μm2 observed area and single plant cell was quantified, and the diameter of LDs was measured with ImageJ software version 1.54t.
The methods for phylogenetic analysis, fungal and viral inoculations, pathogenicity assays, prokaryotic protein expression and antibody preparation, western blot and northern blot analyses, virus particle purification, lipid droplet fractionation, and transmission electron microscopy observation were described in SI Appendix.
Supplementary Material
Appendix 01 (PDF)
Acknowledgments
We thank the High-Performance Computing Center of Northwest A&F University for providing computing resources. We thank Zhen Wang and Lei Chen (Life Science Research Core Services, Northwest A&F University, Yangling, China) for technical supports. This research was supported by the National Natural Science Foundation of China (31970163), Natural Science Foundation of Xinjiang Uygur Autonomous Region (2021D01D12) to L.S., and JSPS/MEXT of Japan (KAKENHI, 23H02214, 23K18029, and 21K18222) to H.K.
Author contributions
L.S. designed research; R.D., S.Y., M.T., and H.W. performed research; D.Z. and Y.W. contributed to research materials; Z.K., T.P., H.K., I.B.A., and L.S. analyzed data; and I.B.A. and L.S. wrote the paper.
Competing interests
The authors declare no competing interest.
Footnotes
This article is a PNAS Direct Submission.
Contributor Information
Ida Bagus Andika, Email: idabagusyf@yahoo.com.
Liying Sun, Email: sunliying@nwsuaf.edu.cn.
Data, Materials, and Software Availability
All of the materials, protocols, and data that were used or generated in this study are described and available in the article and/or SI Appendix.
Supporting Information
References
- 1.Pommerville J. C., Alcamo’s Fundamentals of Microbiology: Body Systems (Jones & Bartlett Publishers, 2012). [Google Scholar]
- 2.Sun L., Kondo H., Andika I. B., “Cross-kingdom virus infection” in Encyclopedia of Virology, Bamford D. H., Zuckerman M., Eds. (Elsevier, Amsterdam, 2020), pp. 443–449. [Google Scholar]
- 3.Navaratnarajah C., Warrier R., Kuhn R. J., “Assembly of viruses: Enveloped particles” in Encyclopedia of Virology, van Regenmortel M. H. V., Mahy B. W. J., Eds. (Elsevier, Amsterdam, 2008), pp. 193–200, 10.1016/B978-012374410-4.00667-1. [DOI] [Google Scholar]
- 4.Buchmann J. P., Holmes E. C., Cell walls and the convergent evolution of the viral envelope. Microbiol. Mol. Biol. Rev. 79, 403–418 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Whitfield A. E., Falk B. W., Rotenberg D., Insect vector-mediated transmission of plant viruses. Virology 479, 278–289 (2015). [DOI] [PubMed] [Google Scholar]
- 6.Bragard C., et al. , Status and prospects of plant virus control through interference with vector transmission. Annu. Rev. Phytopathol. 51, 177–201 (2013). [DOI] [PubMed] [Google Scholar]
- 7.Hull R., Plant Virology (Academic, 2013). [Google Scholar]
- 8.Kuhn J. H., et al. , 2022 taxonomic update of phylum Negarnaviricota (Riboviria: Orthornavirae), including the large orders Bunyavirales and Mononegavirales. Arch. Virol. 167, 2857–2906 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schoelz J. E., Harries P. A., Nelson R. S., Intracellular transport of plant viruses: Finding the door out of the cell. Mol. Plant 4, 813–831 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kondo H., Botella L., Suzuki N., Mycovirus diversity and evolution revealed/inferred from recent studies. Annu. Rev. Phytopathol. 60, 307–336 (2022). [DOI] [PubMed] [Google Scholar]
- 11.Ghabrial S. A., Castón J. R., Jiang D., Nibert M. L., Suzuki N., 50-plus years of fungal viruses. Virology 479, 356–368 (2015). [DOI] [PubMed] [Google Scholar]
- 12.Hogenhout S. A., Ammar E.-D., Whitfield A. E., Redinbaugh M. G., Insect vector interactions with persistently transmitted viruses. Annu. Rev. Phytopathol. 46, 327–359 (2008). [DOI] [PubMed] [Google Scholar]
- 13.Liu S., et al. , Fungal DNA virus infects a mycophagous insect and utilizes it as a transmission vector. Proc. Natl. Acad. Sci. U.S.A. 113, 12803–12808 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Derevnina L., et al. , Emerging oomycete threats to plants and animals. Philos. Trans. R. Soc. B: Biol. Sci. 371, 20150459 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Buckley M., The Fungal Kingdom: Diverse and essential roles in earth’s ecosystem: This report is based on a colloquium, sponsored by the American academy of microbiology. Convened November 2–4, 2007 in Tucson, Arizona (American Society for Microbiology, Washington, DC, 2008). https://www.ncbi.nlm.nih.gov/books/NBK559443/. Accessed 20 July 2020. [PubMed]
- 16.Andika I. B., et al. , Cross-kingdom interactions between plant and fungal viruses. Annu. Rev. Virol. 10, 119–138 (2023). [DOI] [PubMed] [Google Scholar]
- 17.Roossinck M. J., Lifestyles of plant viruses. Philos. Trans. R. Soc. B: Biol. Sci. 365, 1899–1905 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Andika I. B., et al. , Phytopathogenic fungus hosts a plant virus: A naturally occurring cross-kingdom viral infection. Proc. Natl. Acad. Sci. U.S.A. 114, 12267–12272 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bian R., et al. , Facilitative and synergistic interactions between fungal and plant viruses. Proc. Natl. Acad. Sci. U.S.A. 117, 3779–3788 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wei S., et al. , Symptomatic plant viroid infections in phytopathogenic fungi. Proc. Natl. Acad. Sci. U.S.A. 116, 13042–13050 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Afanasenko O. S., Khiutti A. V., Mironenko N. V., Lashina N. M., Transmission of potato spindle tuber viroid between Phytophthora infestans and host plants. Vavilov J. Genet. Breed. 26, 272–280 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cao X., et al. , Common but nonpersistent acquisitions of plant viruses by plant-associated fungi. Viruses 14, 2279 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wichgers Schreur P. J., Kormelink R., Kortekaas J., Genome packaging of the Bunyavirales. Curr. Opin. Virol. 33, 151–155 (2018). [DOI] [PubMed] [Google Scholar]
- 24.Sasaya T., et al. , ICTV virus taxonomy profile: Phenuiviridae 2023. J. Gen. Virol. 104, 001893 (2023). [DOI] [PubMed] [Google Scholar]
- 25.Sun M. H., et al. , Highly adaptive Phenuiviridae with biomedical importance in multiple fields. J. Med. Virol. 94, 2388–2401 (2022). [DOI] [PubMed] [Google Scholar]
- 26.Koch J., Xin Q., Tischler N. D., Lozach P.-Y., Entry of phenuiviruses into mammalian host cells. Viruses 13, 299 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Toriyama S., Characterization of rice stripe virus: A heavy component carrying infectivity. J. Gen. Virol. 61, 187–195 (1982). [Google Scholar]
- 28.Hibino H., et al. , Rice grassy stunt virus: A planthopper-borne circular filament. Phytopathology 75, 894–899 (1985). [Google Scholar]
- 29.Navarro B., et al. , The first phlebo-like virus infecting plants: A case study on the adaptation of negative-stranded RNA viruses to new hosts. Mol. Plant Pathol. 19, 1075–1089 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yu X.-J., et al. , Fever with thrombocytopenia associated with a novel bunyavirus in China. N. Engl. J. Med. 364, 1523–1532 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lancelot R., et al. , Drivers of Rift Valley fever epidemics in Madagascar. Proc. Natl. Acad. Sci. U.S.A. 114, 938–943 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Baudin M., et al. , Association of Rift Valley fever virus infection with miscarriage in Sudanese women: A cross-sectional study. Lancet Glob. Health 4, e864–e871 (2016). [DOI] [PubMed] [Google Scholar]
- 33.Baba M., Masiga D. K., Sang R., Villinger J., Has Rift Valley fever virus evolved with increasing severity in human populations in East Africa? Emerg. Microbes Infect. 5, 1–10 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Xu Y., Fu S., Tao X., Zhou X., Rice stripe virus: Exploring molecular weapons in the arsenal of a negative-sense RNA virus. Annu. Rev. Phytopathol. 59, 351–371 (2021). [DOI] [PubMed] [Google Scholar]
- 35.Elliott R. M., Brennan B., Emerging phleboviruses. Curr. Opin. Virol. 5, 50–57 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li Z., et al. , Survey of apple Valsa canker in Weibei area of Shaanxi province. Acta Agric. Boreali-Occident. Sin. 1, 029 (2013). [Google Scholar]
- 37.Wang X., Shi C.-M., Gleason M. L., Huang L., Fungal species associated with apple Valsa canker in East Asia. Phytopathol. Res. 2, 1–14 (2020). [Google Scholar]
- 38.Feng H., et al. , Apple Valsa canker: Insights into pathogenesis and disease control. Phytopathol. Res. 5, 45 (2023). [Google Scholar]
- 39.Ferron F., Weber F., de la Torre J. C., Reguera J., Transcription and replication mechanisms of Bunyaviridae and Arenaviridae L proteins. Virus Res. 234, 118–134 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chiapello M., et al. , Putative new plant viruses associated with Plasmopara viticola-infected grapevine samples. Ann. Appl. Biol. 176, 180–191 (2020). [Google Scholar]
- 41.Xin M., et al. , Two negative-strand RNA viruses identified in watermelon represent a novel clade in the order Bunyavirales. Front. Microbiol. 8, 1514 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Navarro B., et al. , A negative-stranded RNA virus infecting citrus trees: The second member of a new genus within the order Bunyavirales. Front. Microbiol. 9, 2340 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tokarz R., et al. , Identification of novel viruses in Amblyomma americanum, Dermacentor variabilis, and Ixodes scapularis ticks. mSphere 3, e00614-17 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tomenius K., Clapham D., Meshi T., Localization by immunogold cytochemistry of the virus-coded 30K protein in plasmodesmata of leaves infected with tobacco mosaic virus. Virology 160, 363–371 (1987). [DOI] [PubMed] [Google Scholar]
- 45.Mushegian A. R., Elena S. F., Evolution of plant virus movement proteins from the 30K superfamily and of their homologs integrated in plant genomes. Virology 476, 304–315 (2015). [DOI] [PubMed] [Google Scholar]
- 46.Lin Y.-H., et al. , Two novel fungal negative-strand RNA viruses related to mymonaviruses and phenuiviruses in the shiitake mushroom (Lentinula edodes). Virology 533, 125–136 (2019). [DOI] [PubMed] [Google Scholar]
- 47.SYu M., et al. , Complementation of a potato virus X mutant mediated by bombardment of plant tissues with cloned viral movement protein genes. J. Gen. Virol. 78, 2077–2083 (1997). [DOI] [PubMed] [Google Scholar]
- 48.Andika I. B., et al. , Endoplasmic reticulum export and vesicle formation of the movement protein of Chinese wheat mosaic virus are regulated by two transmembrane domains and depend on the secretory pathway. Virology 435, 493–503 (2013). [DOI] [PubMed] [Google Scholar]
- 49.Xiong R., Wu J., Zhou Y., Zhou X., Identification of a movement protein of the tenuivirus rice stripe virus. J. Virol. 82, 12304–12311 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kory N., Farese R. V. Jr., Walther T. C., Targeting fat: Mechanisms of protein localization to lipid droplets. Trends Cell Biol. 26, 535–546 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Farias M. A., Diethelm-Varela B., Navarro A. J., Kalergis A. M., Gonzalez P. A., Interplay between lipid metabolism, lipid droplets, and DNA virus infections. Cells 11, 2224 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Monson E. A., Trenerry A. M., Laws J. L., Mackenzie J. M., Helbig K. J., Lipid droplets and lipid mediators in viral infection and immunity. FEMS Microbiol. Rev. 45, fuaa066 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Farias M. A., Diethelm-Varela B., Kalergis A. M., Gonzalez P. A., Interplay between lipid metabolism, lipid droplets and RNA virus replication. Crit. Rev. Microbiol., in press. [DOI] [PubMed] [Google Scholar]
- 54.Dechandt C. R. P., et al. , Triacsin C reduces lipid droplet formation and induces mitochondrial biogenesis in primary rat hepatocytes. J. Bioenerg. Biomembr. 49, 399–411 (2017). [DOI] [PubMed] [Google Scholar]
- 55.Pagnoni S., et al. , A collection of Trichoderma isolates from natural environments in Sardinia reveals a complex virome that includes negative-sense fungal viruses with unprecedented genome organizations. Virus Evol. 9, vead042 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Welte M. A., Gould A. P., Lipid droplet functions beyond energy storage. Biochim. Biophys. Acta Mol. Cell Biol. Lipids 1862, 1260–1272 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Galli A., et al. , Analysis of hepatitis C virus core/NS5A protein co-localization using novel cell culture systems expressing core–NS2 and NS5A of genotypes 1–7. J. Gen. Virol. 94, 2221–2235 (2013). [DOI] [PubMed] [Google Scholar]
- 58.Yang S., et al. , Infection of two heterologous mycoviruses reduces the virulence of Valsa mali, a fungal agent of apple Valsa canker disease. Front. Microbiol. 12, 659210 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Li H., et al. , Identification of a novel hypovirulence-inducing hypovirus from Alternaria alternata. Front. Microbiol. 10, 1076 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Mu F., et al. , Nine viruses from eight lineages exhibiting new evolutionary modes that co-infect a hypovirulent phytopathogenic fungus. PLoS Pathog. 17, e1009823 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Voinnet O., Vain P., Angell S., Baulcombe D. C., Systemic spread of sequence-specific transgene RNA degradation in plants is initiated by localized introduction of ectopic promoterless DNA. Cell 95, 177–187 (1998). [DOI] [PubMed] [Google Scholar]
- 62.Zhang X., et al. , Acidovorax citrulli type III effector AopP suppresses plant immunity by targeting the watermelon transcription factor WRKY6. Front. Plant Sci. 11, 579218 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sun L., Andika I. B., Kondo H., Chen J., Identification of the amino acid residues and domains in the cysteine-rich protein of Chinese wheat mosaic virus that are important for RNA silencing suppression and subcellular localization. Mol. Plant Pathol. 14, 265–278 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Voinnet O., Rivas S., Mestre P., Baulcombe D., Retracted: An enhanced transient expression system in plants based on suppression of gene silencing by the p19 protein of tomato bushy stunt virus. Plant J. 33, 949–956 (2003). [DOI] [PubMed] [Google Scholar]
- 65.Sun L., Andika I. B., Shen J., Yang D., Chen J., The P2 of Wheat yellow mosaic virus rearranges the endoplasmic reticulum and recruits other viral proteins into replication-associated inclusion bodies. Mol. Plant Pathol. 15, 466–478 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Craven M., Pawlyk D., Choi G., Nuss D., Papain-like protease p29 as a symptom determinant encoded by a hypovirulence-associated virus of the chestnut blight fungus. J. Virol. 67, 6513–6521 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix 01 (PDF)
Data Availability Statement
All of the materials, protocols, and data that were used or generated in this study are described and available in the article and/or SI Appendix.