Significance
Myelin is essential for the development and function of the central nervous system (CNS), and its loss or dysfunction is central to aging and neurodegenerative diseases. To build myelin, oligodendrocytes undergo dramatic cell morphology changes that are powered by actin cytoskeletal dynamics. However, how the oligodendrocyte cytoskeleton is regulated during myelination remains poorly understood. Here, we identify the transcription factor SRF (serum response factor) as essential for myelination. SRF directly regulates expression of actin regulatory genes in oligodendrocytes and, surprisingly, also inhibits disease-associated gene expression. Together with our recent discovery that SRF promotes oligodendrocyte rejuvenation, our findings uncover a pathway promoting myelin formation that may represent a therapeutic target for restoring myelin in the aged or diseased CNS.
Keywords: myelin, SRF, cytoskeleton, oligodendrocytes, neurodevelopment
Abstract
Myelination of neuronal axons is essential for nervous system development. Myelination requires dramatic cytoskeletal dynamics in oligodendrocytes, but how actin is regulated during myelination is poorly understood. We recently identified serum response factor (SRF)—a transcription factor known to regulate expression of actin and actin regulators in other cell types—as a critical driver of myelination in the aged brain. Yet, a major gap remains in understanding the mechanistic role of SRF in oligodendrocyte lineage cells. Here, we show that SRF is required cell autonomously in oligodendrocytes for myelination during development. Combining ChIP-seq with RNA-seq identifies SRF-target genes in oligodendrocyte precursor cells and oligodendrocytes that include actin and other key cytoskeletal genes. Accordingly, SRF knockout oligodendrocytes exhibit dramatically reduced actin filament levels early in differentiation, consistent with its role in actin-dependent myelin sheath initiation. Surprisingly, oligodendrocyte-restricted loss of SRF results in upregulation of gene signatures associated with aging and neurodegenerative diseases. Together, our findings identify SRF as a transcriptional regulator that controls the expression of cytoskeletal genes required in oligodendrocytes for myelination. This study identifies an essential pathway regulating oligodendrocyte biology with high relevance to brain development, aging, and disease.
Myelination is essential for rapid nerve signaling and has emerged as an important regulator of central nervous system (CNS) development, plasticity, and disease (1). The cellular mechanisms controlling myelin formation and dynamics are still incompletely understood. Oligodendrocytes form myelin by dramatically rearranging their cell morphology to ensheath and then spirally wrap dozens of individual myelin sheaths around neuronal axons. Central to the ability of an oligodendrocyte to form myelin is its actin cytoskeleton (2), which powers morphological changes in two distinct steps: first, actin assembly is required for early stages of myelination in which oligodendrocytes extend their cellular processes to make first contact with axons that they loosely ensheath (3, 4) similar to how actin assembly drives the extension of neuronal growth cones (5). Second, and unexpectedly, dramatic disassembly of the oligodendrocyte actin cytoskeleton occurs prior to the start of myelin wrapping (4, 6). The first stage—ensheathment—requires expression of proteins that promote actin filament assembly, including Arp2/3 and its regulators. In contrast, the subsequent stage—wrapping—does not require actin assembly factors, while actin disassembly factors (e.g., cofilin, ADF, gelsolin) are all highly upregulated and are required for wrapping (4, 6). Thus, precise control over actin assembly and disassembly—e.g., through tight regulation of gene expression of actin regulatory proteins—is likely to be essential to coordinate oligodendrocyte morphology changes required for these sequential steps of myelination. The mechanisms regulating the oligodendrocyte cytoskeleton during myelination remain largely unknown.
Serum response factor (SRF) is a MADS box transcription factor that functions as a major transcriptional regulator of the actin cytoskeleton in diverse cell types (7) including neurons (8, 9), muscle (10, 11), and cardiac cells (12, 13). Many of the cytoskeletal genes that are induced during oligodendrocyte differentiation (14–16) are known targets of SRF (17). In the developing nervous system, SRF is required in neural precursor cells for the specification of oligodendrocyte precursor cells (OPCs) and astrocytes (18). In addition, neuronal SRF was found to affect myelination non-cell autonomously by controlling neuronal secretion of paracrine signals that affect oligodendrocyte differentiation (19). However, whether SRF also plays a direct, cell-autonomous role in oligodendrocytes during myelination has not been directly addressed.
We recently found that infusing cerebrospinal fluid (CSF) from young mice into the brains of old mice improves memory function by a mechanism that appears to be largely dependent on the formation of new myelin (20). Young CSF increased OPC proliferation, differentiation into oligodendrocytes, and the number of myelinated axons in the hippocampus. Mechanistically, we found that the major cellular target of young CSF in oligodendrocytes is SRF. Its expression is rapidly induced in OPCs treated with young CSF, followed within hours by the induction of numerous known SRF target genes. Additionally, young CSF increased actin filament levels and growth cones in OPCs, consistent with activating SRF. In culture, the young CSF-induced increase in OPC proliferation is dependent on SRF, as SRF-KO OPCs failed to proliferate in response to young CSF. However, it is still unknown whether SRF also regulates myelination itself—for example, by controlling expression of actin genes known to be required in oligodendrocytes for ensheathment and/or wrapping of axons.
Here, we show that SRF is required cell-autonomously in oligodendrocytes for the formation of myelin during development. On the gene regulatory level, the direct gene targets of SRF in OPCs and oligodendrocytes include actin itself and several genes that regulate the formation and stability of actin filaments. Loss of SRF in cultured oligodendrocytes causes a dramatic reduction in actin filament levels early in differentiation, a time point in which actin filament assembly is required for axon ensheathment. Unexpectedly, SRF-loss also induced a gene signature associated with aging and neurodegenerative diseases such as Alzheimer’s Disease. Together, our results reveal mechanistic insight into how the oligodendrocyte cytoskeleton is regulated to promote myelination during development and dissect a unique pathway that could be targeted to boost, protect, and/or restore myelination in aging or disease.
Results
SRF Is Expressed by OPCs and Oligodendrocytes during CNS Development.
We first sought to determine when in the oligodendrocyte lineage SRF is expressed during CNS development. We used multiplexed fluorescence RNA in situ hybridization (RNAScope) to visualize and quantify expression of SRF transcripts in brain sections from postnatal day 8 (P8) and P16 mice (Fig. 1A), time points representing the start of myelination in the mouse brain. Simultaneous labeling of PDGFRα (OPCs) and Olig2 (all oligodendrocyte lineage) allowed us to quantify SRF expression in both OPCs (Olig2-expressing, PDGFRα-high) and differentiating or mature oligodendrocytes (Olig2-expressing, PDGFRα-low) while avoiding other cell types that express SRF (Fig. 1 B and C). Consistent with published transcriptomics studies (16, 21) and our prior work on SRF in the aging brain (20), SRF mRNA was expressed by both OPCs and oligodendrocytes throughout the brain at both P8 and P16 (Fig. 1D). Furthermore, immunostaining with a knockout-validated antibody confirmed SRF protein expression in oligodendrocyte nuclei (Fig. 1E), consistent with its known function as a transcription factor. We confirmed SRF’s nuclear localization by immunostaining primary cultured OPCs and oligodendrocytes using the same antibody (Fig. 1F). Thus, SRF is expressed by both OPCs and oligodendrocytes during postnatal development, positioning it to regulate gene expression during myelination.
Fig. 1.
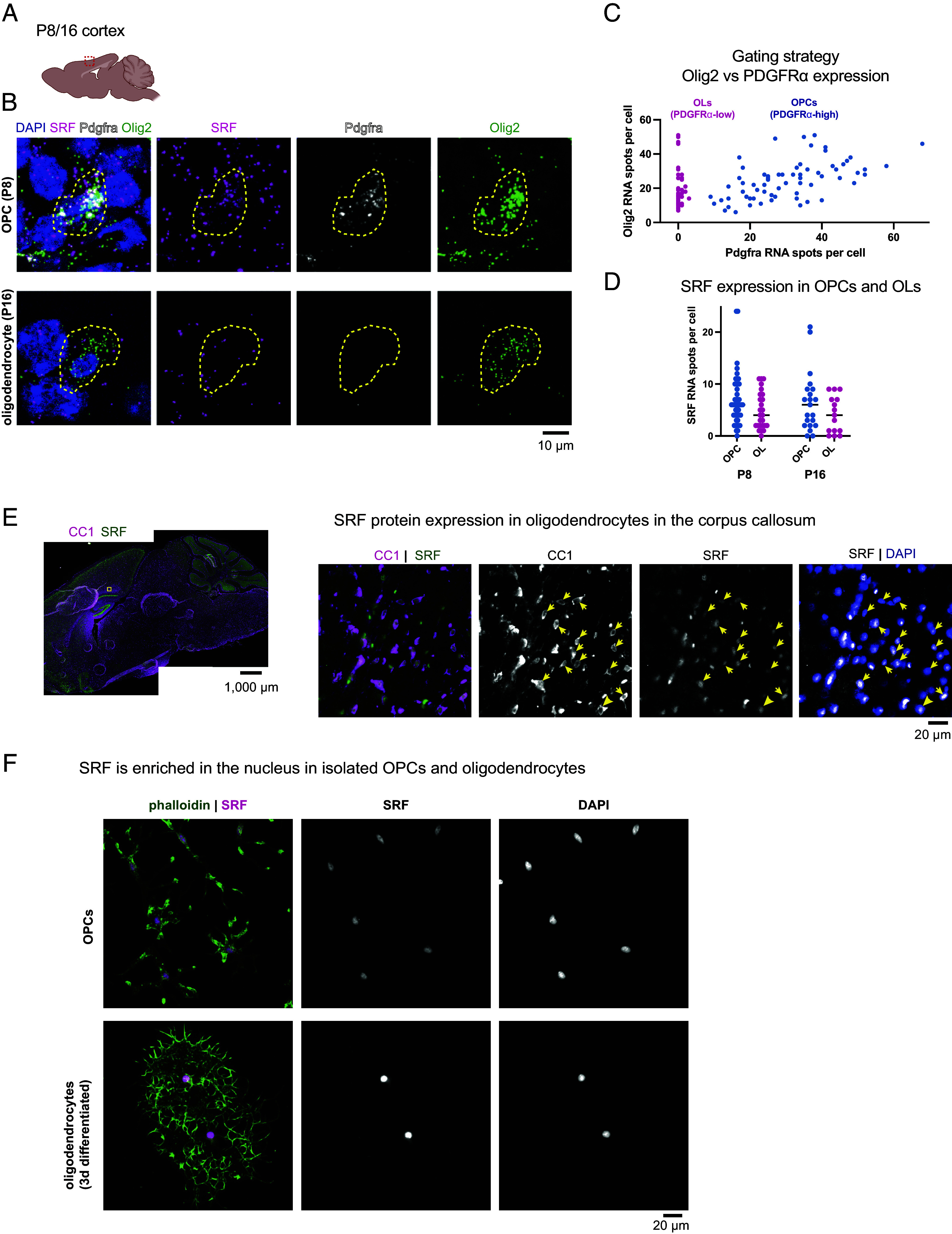
Expression of SRF in the oligodendrocyte lineage. (A) Region of interest in sagittal sections of P8 and P16 brains. (B) RNAscope for Pdgfrα, Olig2, and SRF in the cortex of P8 and P16 brains, n = 2 (Scale bar, 10 μm.) (C) Gating strategy for selection of OPCs (Pdgfrα-high) and oligodendrocytes (OLs; Pdgfrα-low) expressing cells. (D) Quantification of SRF spots per cell in OPCs and oligodendrocyte population at P8 and P16. Each dot represents a cell, bar represents the mean from all cells. n = 3 mice at each age. (E) Overview of immunostaining of a P16 mouse with CC1 (mature oligodendrocytes) and SRF and an enlargement of the corpus callosum. (Scale bar, 1,000 μm and 20 μm in the enlargement.) (F) SRF and phalloidin staining in isolated OPCs or OL differentiated for 3 d in culture. (Scale bar, 20 μm.)
Conditional Knockout of SRF Causes CNS Hypomyelination.
To test the role of SRF in myelination in vivo, we generated mice in which a floxed allele of SRF (22) was conditionally deleted from OPCs using Olig2-Cre (23) (SI Appendix, Fig. S1A). SRF-flox/flox; Olig2-Cre/+ conditional knockout mice (hereafter “SRF-cKO”) were born in Mendelian frequencies and displayed no gross behavioral or motor defects. We confirmed complete loss of SRF mRNA in OPCs using RT-PCR on purified OPCs from SRF-cKO mice and littermate controls (“SRF-Flox,” genotype: SRF-flox/flox; and “SRF-cHet,” genotype: SRF-flox/+; Olig2-Cre/+; SI Appendix, Fig. S1B). In vivo, immunostaining revealed loss of SRF protein from oligodendrocytes but not from neighboring cells (e.g., neurons) (SI Appendix, Fig. S1C).
Having confirmed loss of SRF from oligodendrocyte cells in SRF-cKO mice, we used transmission electron microscopy to test whether SRF is required in oligodendrocytes for myelination. We focused on the optic nerve and the corpus-callosum, two highly myelinated axon tracts that we and others have studied extensively as a model of CNS myelination (4, 6, 24, 25). At P18 (during active myelination), SRF-cKO optic nerves had on average half as many myelinated axons as control SRF-Flox littermates with no other abnormalities (SI Appendix, Fig. S2 A and B). This degree of hypomyelination of SRF-cKO mice persisted into adulthood in both the optic nerve (SI Appendix, Fig. S2 D and E) and the corpus callosum (Fig. 2 A–E and SI Appendix, Fig. S3 A and B), indicating that the reduction of myelinated axons in cKOs was not just due to a transient developmental delay. Focusing on only myelinated axons, morphometric analysis revealed that the average g-ratio (ratio describing myelin thickness relative to axonal diameter) of SRF-cKO myelinated axons was significantly higher than WT littermates in the adult corpus callosum (0.760 vs. 0.703; Fig. 2E), but this result did not achieve significance in the optic nerve (SI Appendix, Fig. S2 G–N). Interestingly, average myelin thickness was not significantly different between WT and SRF-cKO mice in either optic nerve or corpus callosum (SI Appendix, Figs. S2 C and F and S3C), arguing against a role of SRF in myelin wrapping. Instead, the higher g-ratios observed in SRF-cKO mice were due to significantly larger diameters of myelinated axons in cKOs (SI Appendix, Figs. S2 K and L and S3D)—suggesting a specific defect in ensheathing smaller caliber axons. Thus, while SRF is important for axonal ensheathment, it appears to be dispensable for the later stage of myelin wrapping.
Fig. 2.
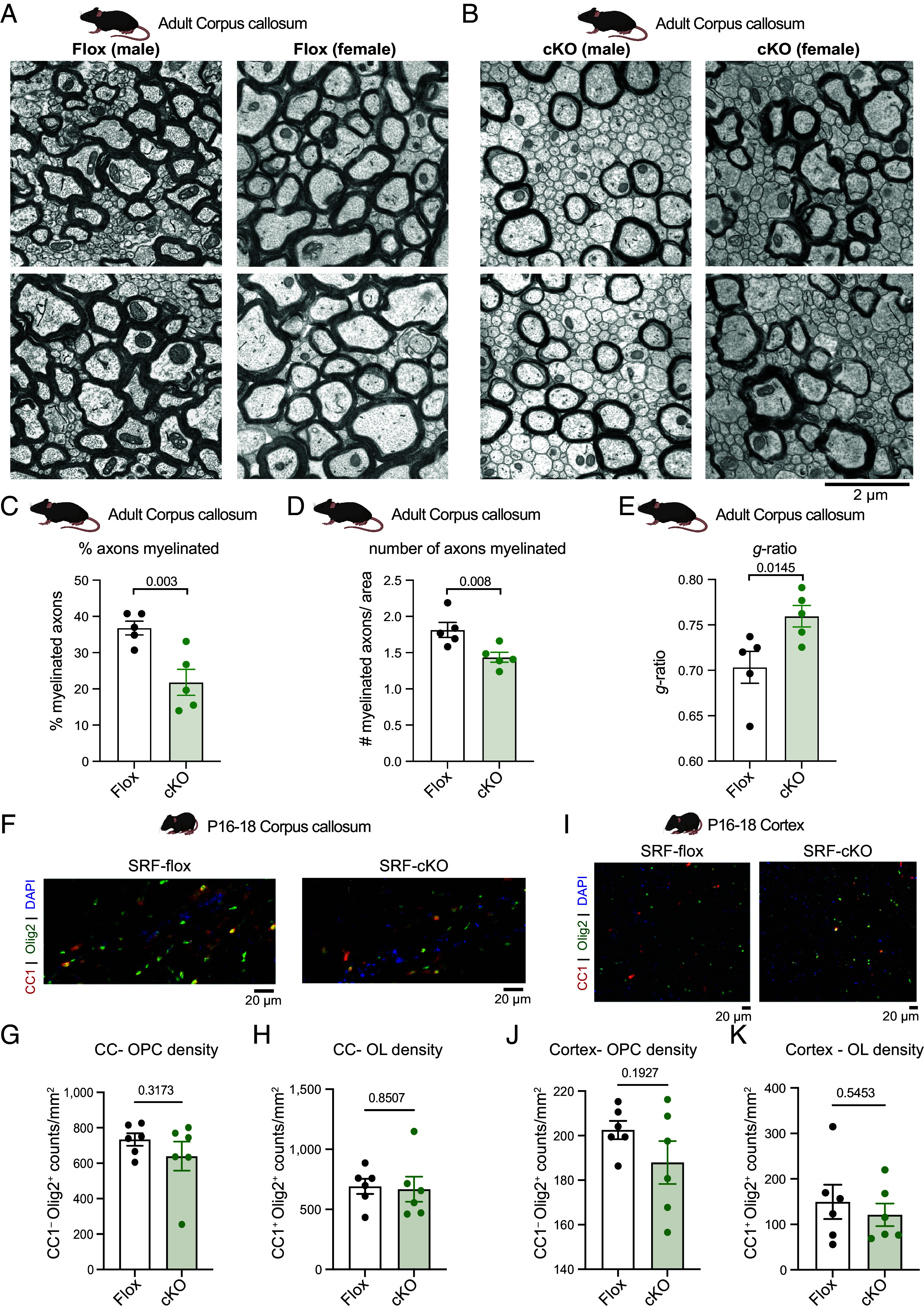
SRF is required in oligodendrocytes for myelination. (A) Representative transmission electron microscopy image of corpus callosum of adult SRF-Flox male and female mice. (Scale bar, 2 μm.) (B) Representative transmission electron microscopy image of corpus callosum of adult SRF-cKO male and female mice. (Scale bar, 2 μm.) (C) Quantification of percent axons myelinated in adult corpus callosum. Each data point is based on at least 2,500 axons counted per mouse. n = 5 animals from each genotype. Unpaired t test. (D) Quantification of number of myelinated axons per area (at least 1,000 μm2) in adult corpus callosum. Each data point is the average per mouse. n = 5 animals from each genotype. Unpaired t test. (E) Quantification of the g-ratio in adult corpus callosum. Each data point is the average of 100 myelinated axons from one animal. n = 5 animals from each genotype. Unpaired t test. (F) Representative images of CC1 (red) and Olig2 (green) staining of corpus callosum (CC) of P16–P18 SRF-Flox and SRF-cKO mice. (Scale bar, 20 μm.) (G) Quantification of OPC density (CC1−Olig2+/mm2) in CC of P16–P18 SRF-Flox and SRF-cKO mice. n = 6 animals from each genotype. Unpaired t test. (H) Quantification of oligodendrocyte density (CC1+Olig2+/mm2) in CC of P16–P18 SRF-Flox and SRF-cKO mice. n = 6 animals from each genotype. Unpaired t test. (I) Representative images of CC1 (red) and Olig2 (green) staining of cortex of P16–P18 SRF-Flox and SRF-cKO mice. (Scale bar, 20 μm.) (J) Quantification of OPC density (CC1−Olig2+/mm2) in cortex of P16–P18 SRF-Flox and SRF-cKO mice. n = 6 animals from each genotype. Student t test. (K) Quantification of oligodendrocyte density (CC1+Olig2+/mm2) in cortex of P16–P18 SRF-Flox and SRF-cKO mice. n = 6 animals from each genotype. Unpaired t test.
The hypomyelination phenotype of SRF-cKO mice could be a result of a defect in differentiation or viability of mature oligodendrocytes. We therefore quantified the cell density of OPCs (CC1−Olig2+ cells) and oligodendrocytes (CC1+Olig2+ cells) in the corpus callosum (CC, Fig. 2 F–H) and cortex (Fig. 2 I–K). We found similar densities of OPCs and oligodendrocytes in both regions with a slight trend towards fewer OPCs in the cortex (Fig. 2J), suggesting that SRF is not necessary for oligodendrocyte maturation or viability. This was in line with similar levels of MBP staining intensity in the cortex and corpus callosum (SI Appendix, Fig. S3 E–G). Together, these results indicated that the hypomyelination phenotype in SRF-cKO mice is a result of improper myelination and that SRF is required cell-autonomously within oligodendrocytes for the initial steps of myelination.
ChIP-Seq Identifies SRF Target Genes in Oligodendrocytes.
To gain a better mechanistic understanding of the role of SRF in myelination, we used chromatin immunoprecipitation-sequencing (ChIP-seq) to identify direct SRF gene targets in the oligodendrocyte lineage. Due to the high number of cells required to perform transcription factor ChIP-seq, we performed the experiments on primary rat OPC cultures under proliferation conditions (OPC) or on day 3 of differentiation (immature oligodendrocytes). We identified 848 significant SRF binding sites (peaks) in total and narrowed down the list to 329 high-confidence targets that appeared consistently across replicates within the same group and not in the IgG control (Fig. 3A and Dataset S1). Consistent with other SRF ChIP-seq datasets (10), most peaks were enriched within 2 kb of the transcription start site (TSS) (Fig. 3B). Inspection of the target sequences confirmed an enrichment for the SRF CArG consensus (Fig. 3C) as well as for Thap11 and Zic2 motifs (Dataset S1). Roughly half (n = 60) of the target genes with a confirmed CArG consensus were shared between OPCs and oligodendrocytes and included known SRF targets such as immediate early genes (Egr1, Egr2, Egr3, Fos, Junb, and Srf) and cytoskeleton genes (Actb, Actng1, Arc, and Vcl) (Fig. 3D). Among the OPC unique target genes, we found several genes with promyelinating effects such as Cardiotrophin-1 (Ctf1) (26) and involved in nervous system development such as Reticulon 4 (Rtn4/Nogo) and UTP11. Notably, among the oligodendrocyte unique target genes we found cytoskeletal proteins such as Filamin A (Flna) and Pppr12b and members of the postsynaptic density scaffold Homer1 and Dlg4.
Fig. 3.
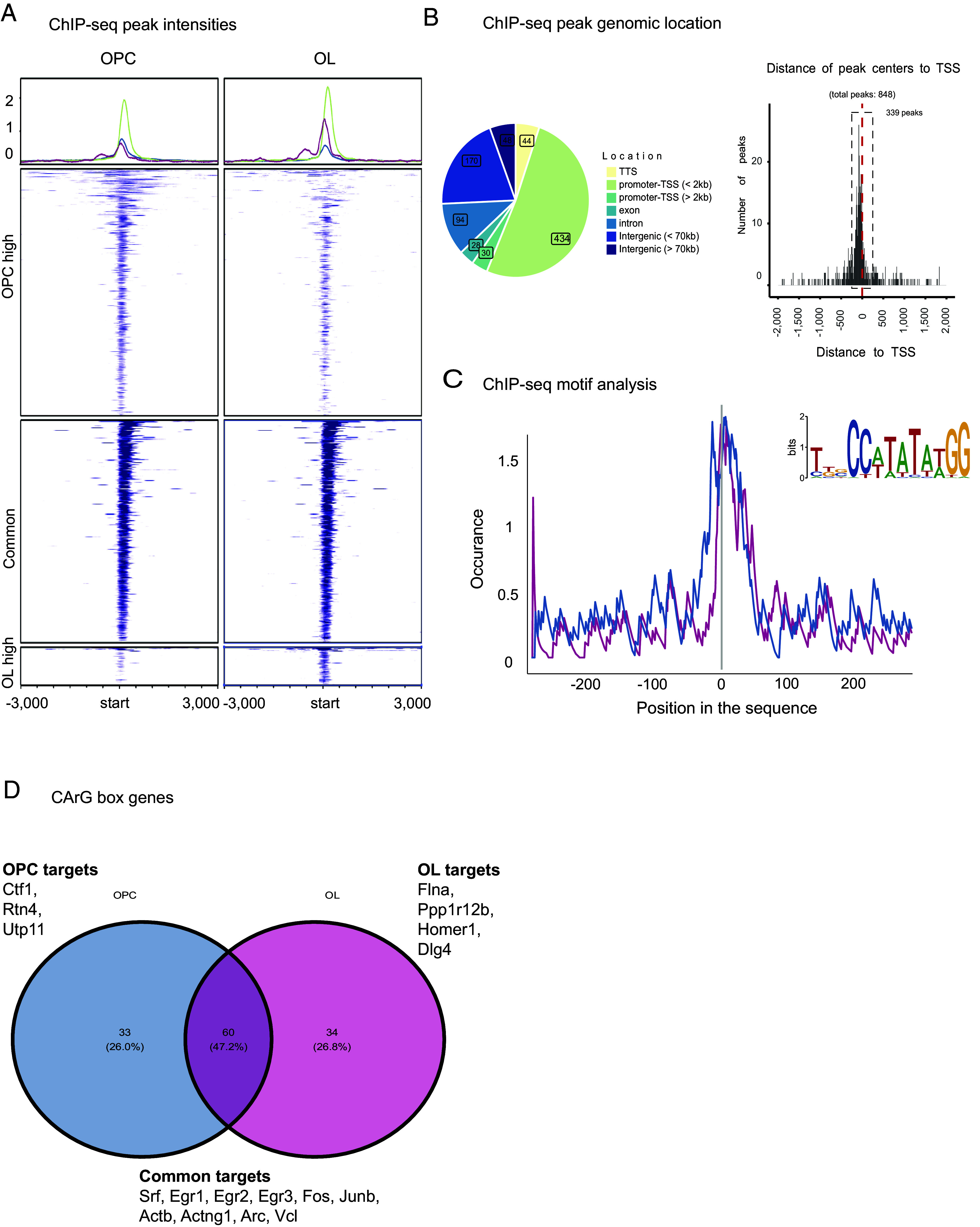
ChIP-seq identifies SRF target genes in OPCs and oligodendrocytes. (A) Heatmap of SRF ChIP-seq peak intensities in OPC and oligodendrocyte samples. OPCs n = 4, oligodendrocytes n = 3. (B) Distribution of genomic location of ChIP-seq peaks showing that most binding events localize to the promoter and TSS. (C) ChIP-seq motif analysis identifying CarG box, the SRF binding motif in OPC (blue), and oligodendrocyte (pink) peaks. (D) Venn-diagram of genes associated with the CarG box peaks in OPC and oligodendrocyte samples.
SRF Regulates Actin Cytoskeleton Gene Expression.
To test whether the ChIP-seq targets were functionally regulated by SRF in oligodendrocytes, we performed transcriptome (RNA-seq) analysis of primary OPCs purified from SRFflox/flox mice, infected with a Cre-expressing virus ex vivo and sequenced at the proliferative stage (OPC) or on differentiation day 3 (immature oligodendrocytes). We identified 157 differentially expressed genes (P.adj < 0.05) in SRF-KO OPCs vs SRF-WT and 264 in SRF-KO oligodendrocytes, including known SRF target genes (according to the TRANSFAC database) (27) (Fig. 4 A and B and Datasets S2 and S3). We further validated these findings using RNAscope in the cortex of P18 mice and found that Srf (Fig. 4 C, E, and G), Actb (Fig. 4 D, F, and G), Actg1 (SI Appendix, Fig. S4 B and D) were significantly down-regulated in our primary cells and in vivo in SRF-cKO Olig2 positive nuclei.
Fig. 4.
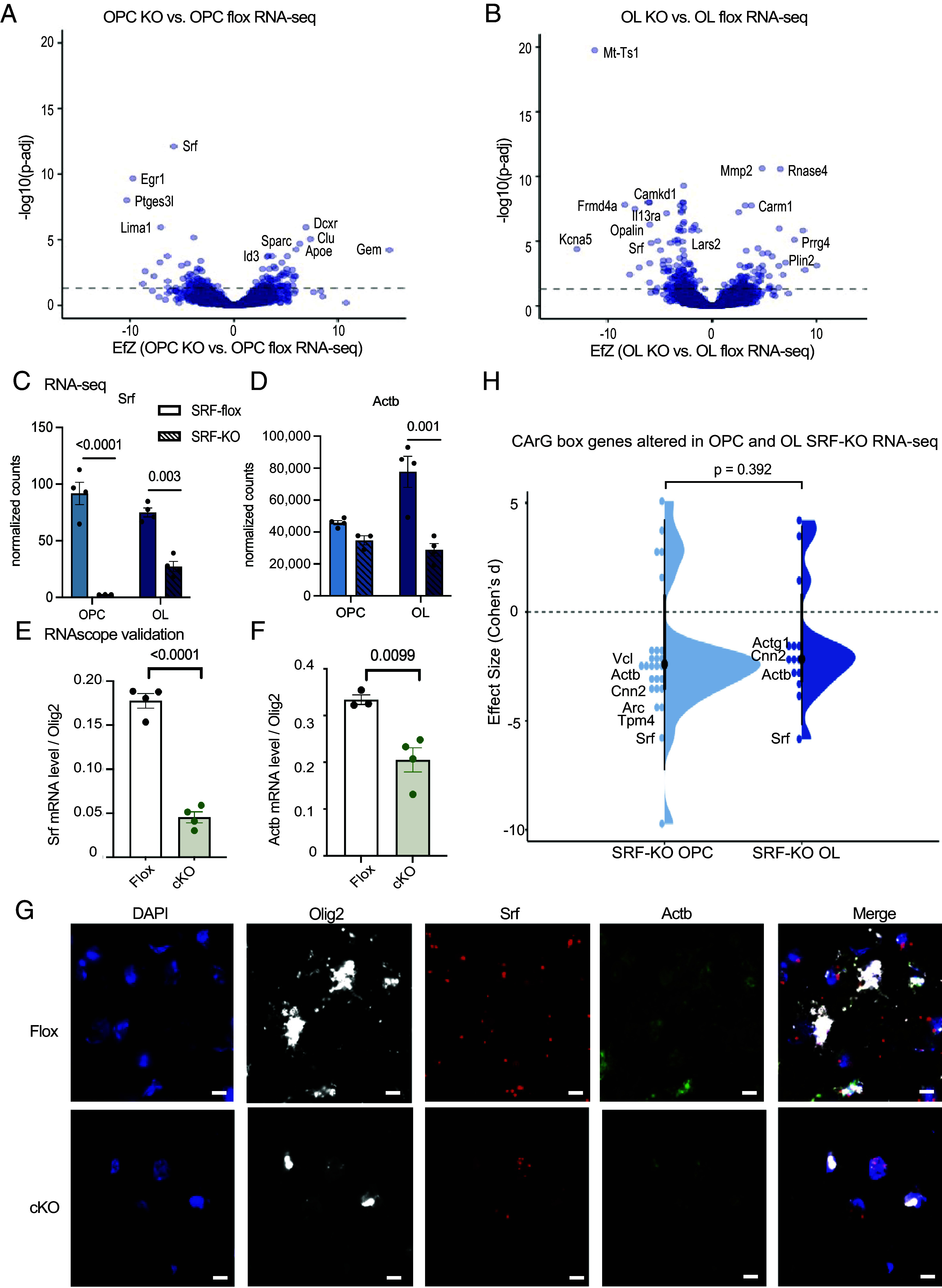
SRF regulates actin cytoskeleton gene expression. (A) Volcano plot of differentially expressed genes of SRF-KO OPCs vs. SRF-Flox OPCs. The dashed line represents Padj = 0.05. SRF-Flox n = 4, SRF-KO n = 3. (B) Volcano plot of differentially expressed genes of SRF-KO oligodendrocytes (OL) vs. SRF-Flox OLs. The dashed line represents Padj = 0.05. n = 4. (C) Normalized expression counts of Srf in SRF-Flox and SRF-KO OPCs and oligodendrocytes. OPC SRF-KO n = 3. All the rest n = 4. Statistics by DESeq2. (D) Normalized expression counts of Actb in SRF-flox and SRF-KO OPCs and oligodendrocytes. OPC SRF-KO n = 3. All the rest n = 4. Statistics by DESeq2. (E) Quantification of Srf expression levels in Olig2+ nuclei in the cortex of P16/P18 SRF-Flox and SRF-cKO mice detected by RNAscope. n = 4. (F) Quantification of Actb expression levels in Olig2+ nuclei in the cortex of P16/P18 SRF-Flox and SRF-cKO mice detected by RNAscope. n = 4. (G) Representative images of DAPI, Olig2, Srf, Actb, and merge channel in SRF-Flox and SRF-cKO mice. (Scale bar, 10 μm.) (H) Violin plot of CarG box genes in SRF-KO vs. SRF-Flox OPC and oligodendrocyte RNA-seq. Wilcoxon rank-sum test.
Next, we plotted the CarG consensus-containing genes identified by ChIP-seq as direct SRF targets that were significantly altered in SRF-KO OPCs and oligodendrocytes (Pval < 0.05, Fig. 4H). Actin genes such as beta actin (Actb) were down-regulated in SRF-KO OPCs and oligodendrocytes indicating that they are indeed directly regulated primarily by SRF and therefore down-regulated when SRF is knocked-out (SI Appendix, Fig. S4A). Interestingly, we did not identify “myelin genes” as direct SRF targets. Together, these experiments identified genes controlled by SRF in OPCs and oligodendrocytes and highlighted actin cytoskeletal regulation as a potential role of SRF during early stages of myelination.
SRF Functionally Regulates the Oligodendrocyte Actin Cytoskeleton.
Actin dynamics are critical for CNS myelination, with the early (SRF-dependent) stage of ensheathment requiring actin filament assembly (4, 6). SRF is known in other cell types to regulate actin filament assembly and disassembly by transcriptional control of numerous actin regulatory genes (10, 28, 29). Our ChIP-seq and RNA-seq results suggested that, in oligodendrocytes, SRF primarily regulates genes that promote actin assembly (including actin itself) and not genes that regulate actin disassembly (SI Appendix, Fig. S4 E and F). To test this functionally, we purified OPCs from SRF-Flox and SRF-cKO littermates and compared their actin cytoskeletons over a time course of differentiation (Fig. 5A). In OPCs and early stages of oligodendrocyte differentiation, SRF-cKO cells had dramatically reduced actin filament levels (~20% to 40% of SRF-Flox levels; Fig. 5 B, C, and E). However, both SRF-Flox and SRF-cKO oligodendrocytes underwent a similar degree of actin disassembly during late stages of differentiation, and by full maturation at 7 d of differentiation cells from both genotypes were nearly indistinguishable (Fig. 5D). Furthermore, the protein level of the differentiation marker and major myelin protein MBP was not different between SRF-WT and SRF-cKO oligodendrocytes, similar to our findings in vivo (SI Appendix, Fig. S3 E–G). These data suggest that the effect of SRF on the actin cytoskeleton is due to its direct regulation of actin gene expression rather than a more general effect on oligodendrocyte differentiation.
Fig. 5.
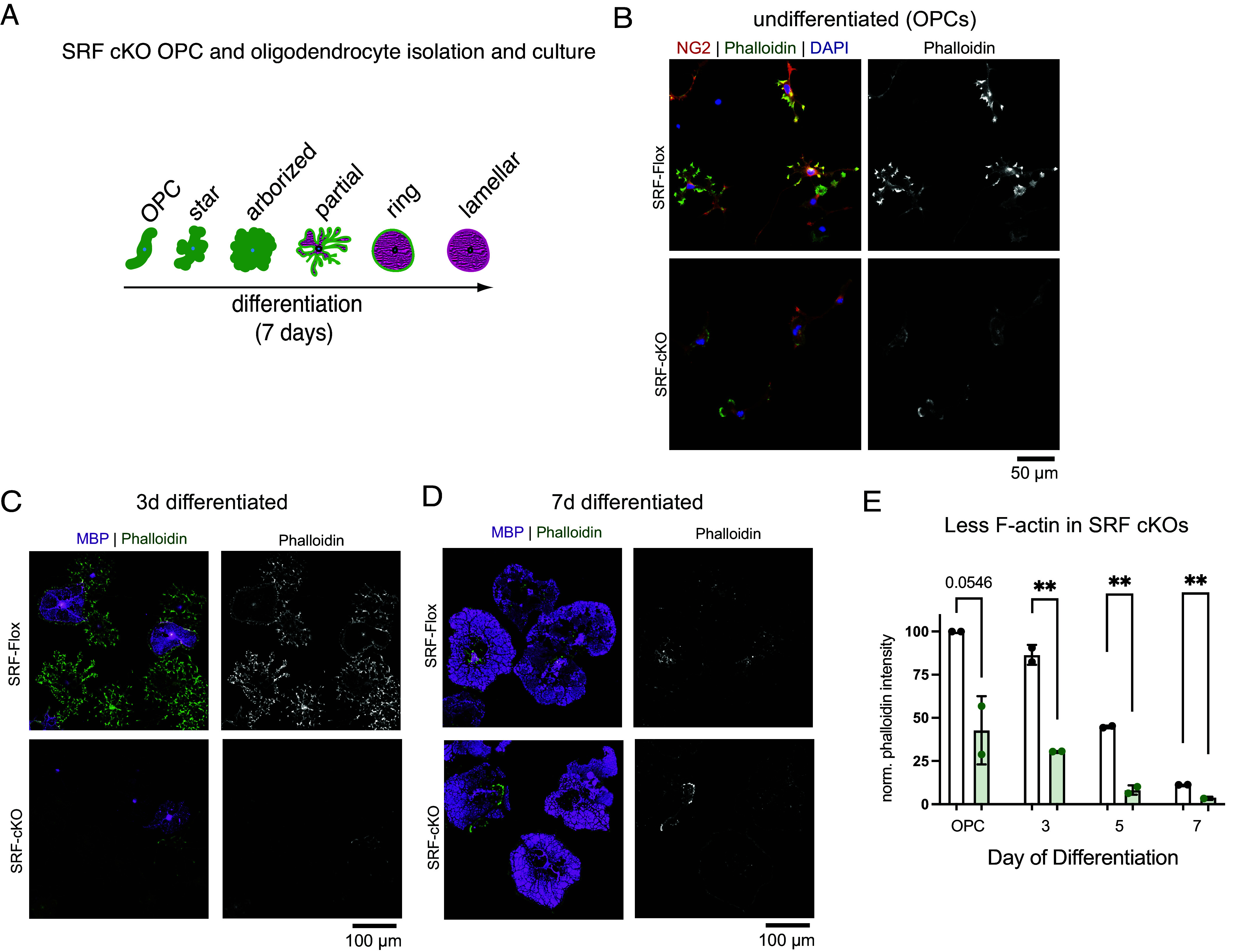
SRF regulates the oligodendrocyte cytoskeleton. (A) Diagram of OPC to oligodendrocyte morphology changes when differentiated in culture. (B) SRF-Flox and SRF-cKO OPCs stained for the OPC marker NG2 (red), phalloidin (green) and DAPI (blue). (Scale bar, 50 μm.) (C) SRF-Flox and SRF-cKO immature oligodendrocytes (3 d differentiated) stained for MBP (magenta), phalloidin (green), and DAPI (blue). (Scale bar, 100 μm.) (D) SRF-Flox and SRF-cKO mature oligodendrocytes (7 d differentiated) stained for MBP (magenta), phalloidin (green) and DAPI (blue). (Scale bar, 100 μm.) (E) Quantification of phalloidin intensity in SRF-Flox and SRF-cKO at different stages of differentiation. n = 2 preps. Unpaired t test; **P < 0.01.
Together, our results indicate that SRF promotes early stages of developmental myelination—at least in part—by controlling the expression of genes required for actin filament assembly.
SRF Loss Induces Oligodendrocyte Disease-Associated Genes.
Beyond the actin cytoskeleton, what other gene pathways does SRF control during myelination? As expected, Gene Set Enrichment Analysis (GSEA) of SRF-KO OPC genes identified depletion of pathways associated with “Transcription regulation” and “Cell cycle” (SI Appendix, Fig. S5A), and SRF-KO oligodendrocytes were depleted of pathways including “actin cytoskeleton,” “postsynaptic density,” and “myelin sheath” (SI Appendix, Fig. S5B) in accordance with the role of SRF as a transcriptional regulator driving cell proliferation (20) and regulating the actin cytoskeleton.
To our surprise, enriched pathways in SRF-KO OPC genes also consisted of genes like Apoe and Clusterin (Clu) associated with diseases such as “Alzheimer’s Disease” and “Parkinson’s Disease,” suggesting that SRF negatively regulates their expression. To validate these findings in vivo, we performed snRNA-seq of 10-mo-old SRF-Floxed and SRF-cKO pre-frontal cortex white matter. To enrich for glial cells, we sorted NeuN− nuclei and indeed captured mostly oligodendrocytes (cluster 0 and 2) and astrocytes (cluster 1) with modest numbers of OPCs (cluster 5) (Fig. 6A and SI Appendix, Fig. S5 C–E). Differential gene expression analysis of SRF-cKO over SRF-Flox oligodendrocyte clusters identified 63 significantly up-regulated and 213 down-regulated genes on both OL clusters combined (log 2FC > 0.1, P.adj < 0.05, Fig. 6B). GSEA pathway analysis identified pathways enriched with endothelial and immune genes and depletion of pathways associated with oligodendrocyte specific genes as well as “regulation of actin cytoskeleton” and “mRNA processing” (Fig. 6C). Oligodendrocyte marker genes and myelin genes were among the top down-regulated genes (Fig. 6D) suggesting reduced myelination in cKO mice, in line with the TEM hypomyelination findings in postnatal and adult cKO mice (Fig. 2 and SI Appendix, Figs. S2 and S3).
Fig. 6.
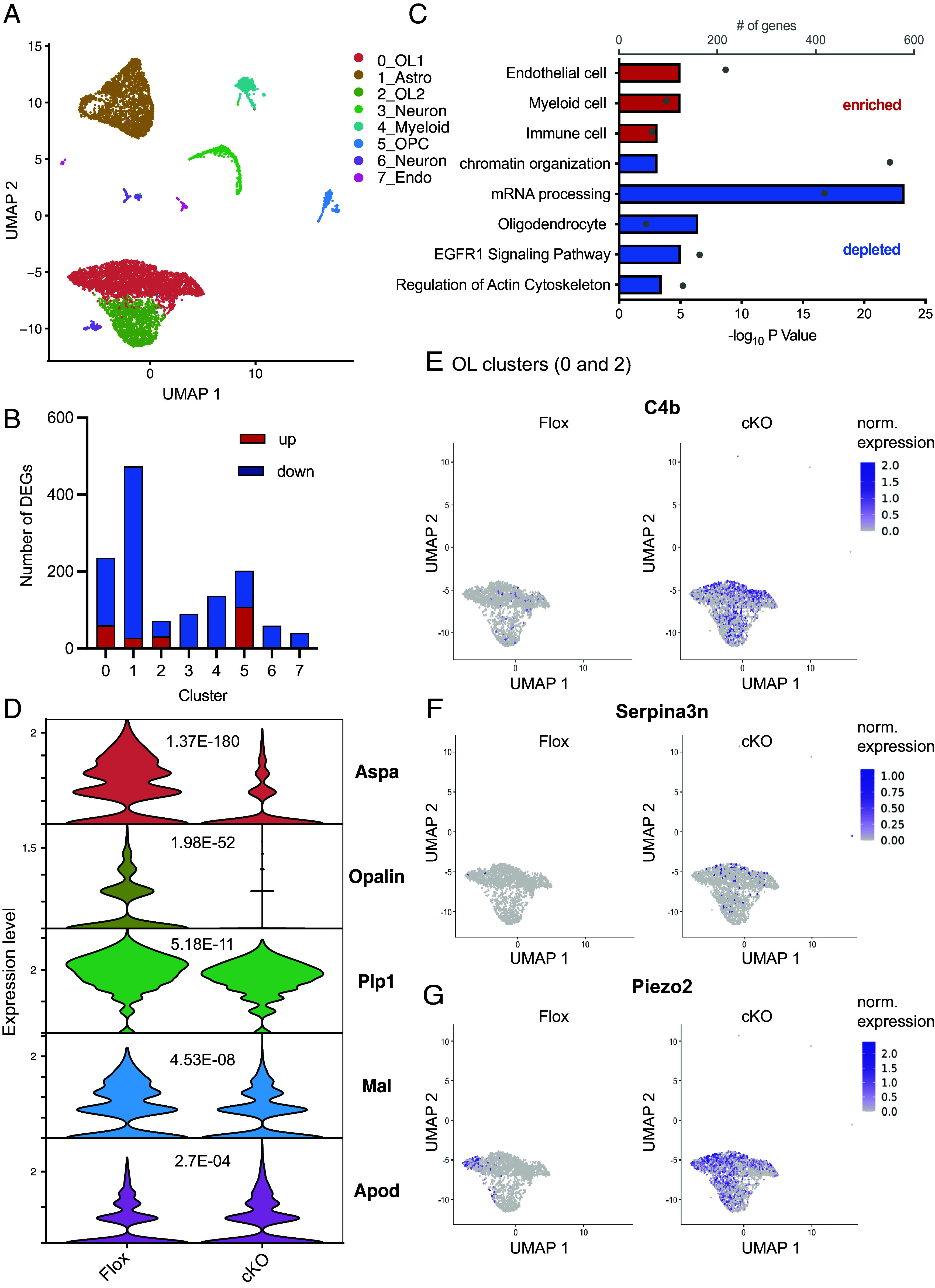
Loss of SRF induces oligodendrocyte disease–associated genes. (A) UMAP plot showing Seurat clusters and their annotation of 10-mo-old SRF-Flox and SRF-cKO NeuN− nuclei sequenced by 10× snRNA-seq. (B) Bar graph showing the number of DEGs significantly (P.adj < 0.05) up- (red) or down-regulated (blue) in each Seurat cluster. (C) Pathways enriched or depleted in SRF-cKO oligodendrocytes (clusters 0 and 2 combined) detected by Gene Set Enrichment Analysis (GSEA) of descending Log 2FC. (D) Violin plots of expression levels and P-adj. values of top differentiated mature oligodendrocyte marker genes. (E) Scatter plot showing the normalized expression of C4b overlayed over the oligodendrocyte clusters (0 and 2) split by SRF-Floxed and SRF-cKO groups. (F) Scatter plot showing the normalized expression of Serpina3n overlayed over the oligodendrocyte clusters (0 and 2) split by SRF-Floxed and SRF-cKO groups. (G) Scatter plot showing the normalized expression of Piezo2 overlayed over the oligodendrocyte clusters (0 and 2) split by SRF-Floxed and SRF-cKO groups.
Similar to our findings in culture, among the top upregulated genes in SRF-cKO oligodendrocytes were genes like C4b (Fig. 6E) and Serpina3n (Fig. 6F) previously reported as upregulated in disease associated-oligodendrocytes (30–33) and in mouse aging datasets (34), and the mechanosensitive ion channel Piezo2 (Fig. 6G) which, to our knowledge, was not reported to be an oligodendrocyte disease-associated gene. Notably, these genes were absent or very lowly expressed in middle-aged (10-mo-old) SRF-Flox mice. These findings were intriguing in light of our previous observation that SRF is down-regulated in OPCs in aged mice (20) and suggests that SRF might have an additional role in maintaining oligodendrocyte homeostatic state.
Discussion
Myelin plays essential roles in CNS development, dynamics, and disease, but the mechanisms that regulate its formation are still incompletely understood. In particular, actin cytoskeleton dynamics precisely regulate the morphological changes oligodendrocytes must undergo to build myelin sheaths, but the molecules that regulate actin in oligodendrocytes are largely unknown. Here, we show that the transcription factor SRF is required in oligodendrocytes for early stages of myelination. SRF is expressed developmentally by both OPCs and oligodendrocytes throughout the CNS. SRF-cKO mice have severe defects in numbers of axons myelinated, consistent with an ensheathment defect, while myelin sheaths that do form wrap normally. Combining ChIP-seq with RNA-seq of SRF knockout oligodendrocytes identifies SRF target genes during myelination and include genes required for the formation of actin filaments. Accordingly, SRF knockout oligodendrocytes fail to assemble actin normally, potentially explaining their inability to ensheath axons. Thus, SRF is a transcription factor that promotes myelination by regulating the expression of actin and actin-regulatory genes during the early, actin-dependent stage of myelination.
SRF is a versatile regulator of a multitude of cellular functions in various tissues like skeletal muscle (35), heart (36), and brain (8). In the developing brain, SRF modulates hippocampal neuronal growth cone integrity, migration, axon guidance, and circuit assembly (37). Mechanistically, at least half of all known SRF gene targets encode proteins that regulate actin-dependent processes in mammalian cells (38). It is interesting to speculate that SRF regulates neuronal and oligodendrocyte development by modulating their cytoskeleton to promote each cell’s unique structure and function.
How does SRF regulate myelination? Studying mice with conditional deletion of ArpC3 (essential subunit of the actin-nucleating Arp2/3 complex), we previously formulated a two-step model of myelination in which actin assembly drives oligodendrocyte process outgrowth and ensheathment at the start of myelination, while at later stages actin disassembly drives myelin wrapping (4). In the current study, SRF-cKO mice show very similar phenotypes to ArpC3-cKO mice, including hypomyelination in the optic nerve (SI Appendix, Fig. S2) and reduced actin filament levels in oligodendrocytes (Fig. 5). These similarities, along with the ChIP-seq and RNA-seq of SRF-KO OPCs and oligodendrocytes, suggest that a major role of SRF is to regulate the expression of target genes required for actin filament assembly at the actin-dependent stage of early myelination. Interestingly, both SRF-cKO and ArpC3-cKO mice only have a partial reduction of myelination. This suggests that SRF- and Arp2/3-mediated actin assembly is not absolutely required for ensheathment or that sufficient actin assembly remains in these oligodendrocytes to ensheath axons—albeit at a reduced frequency. An alternative possibility is that oligodendrocytes are heterogeneous in their requirement for SRF and/or Arp2/3, which future studies (e.g., using deeper sequencing methods) may help resolve. Our results also address an open question in the field: Is accumulation of actin filaments followed by their disassembly a required step prior to myelin wrapping (e.g., actin disassembly serving as a “checkpoint” for wrapping), or does wrapping just require the lack of actin filaments? We believe that since SRF-cKO oligodendrocytes have dramatically reduced actin filaments, were still able to differentiate and extended their membranes normally in culture (Fig. 5), and were able to wrap normally in vivo (Fig. 2), these results are consistent with the second model in which wrapping requires the lack of actin filaments—rather than accumulation and subsequent loss of actin filaments.
Of note, Knoll et al. hypothesized that SRF could also drive myelination directly in oligodendrocytes by binding to myelin gene promoters (19). This was supported by another study which showed that pharmacologic inhibition of SRF inhibited OPC differentiation (39). However, our ChIP-seq studies found no evidence that SRF directly regulates the expression of classical myelin genes but rather promotes actin formation which is required for proper axon ensheathment (Fig. 3). Based on the downregulation of mature oligodendrocyte genes (Fig. 6D) and the downregulation of Myrf, a major regulator of oligodendrocyte differentiation (40) in vitro (SI Appendix, Fig. S4G), it is possible that loss of SRF causes a downstream delay in oligodendrocyte differentiation indirectly—e.g., via other transcription factors and immediate early genes that it regulates.
Interestingly, in other cell types, SRF’s transcriptional activity is directly mediated by cofactors that respond to the balance of actin monomer/filament in the cytoplasm (41–43). In this way, it has been proposed that SRF could act as an “actin homeostat” to respond to the state of the actin cytoskeleton by inducing gene expression of proteins that, in turn, regulate actin assembly or disassembly. SRF has even been shown to shuttle from the nucleus to the cytoplasm (44–47), for example, in response to PDGF signaling (44) or the Rho kinase pathway that also regulates actin (45). This may explain why we observed a small amount of SRF immunostaining out in the processes of cultured oligodendrocytes, particularly in OPCs that are grown in the presence of PDGF (Fig. 1F). Although the actin-dependent regulation of SRF has—thus far—been only observed in cultured cells, it is tempting to speculate that such a regulatory feedback circuit could function in cells to coordinate cytoskeletal changes with gene expression. An ideal cell type to study actin-SRF feedback may be the oligodendrocyte, a cell type that undergoes dramatic cytoskeleton rearrangements during its differentiation from actin-rich to nearly devoid of actin filaments (4, 6). In future studies, it will be interesting to explore whether SRF responds to cytoskeletal changes during oligodendrocyte differentiation to coordinate gene expression with cytoskeletal remodeling.
Beyond development, myelin loss and the progressive inability of oligodendrocytes to regenerate myelin are increasingly found to underlie cognitive defects associated with aging and neurodegenerative disease. We have recently found that SRF is down-regulated in aged mouse OPCs and that infusion of young CSF promotes SRF pathway activation and actin assembly in OPCs, as well as OPC proliferation and differentiation (20). In the current study, we gained more mechanistic insight into the functional outcomes of SRF loss. We found that SRF-cKO in oligodendrocytes leads to developmental hypomyelination which persists into adulthood. Future studies focusing on adult-induced SRF-cKO in OPCs and oligodendrocytes could unravel the important roles of SRF in adaptive myelination in the context of learning and memory and cognitive aging. Furthermore, in our SRF-KO RNA-seq dataset, expectedly, SRF and many SRF targets such as Egr1 were transcriptionally down-regulated. However, to our surprise, some disease-related genes were up-regulated (Fig. 6). Combined, these results may link SRF loss to a dysfunctional oligodendrocyte disease state that appears in aging and neurodegenerative diseases.
In this study, we gained a deeper understanding of the roles of SRF in regulating oligodendrocyte maturation during development. Ultimately, a full mechanistic understanding of how oligodendrocytes undergo such dramatic morphology changes to form myelin will open the door to understanding how myelin is dynamically remodeled during learning and could reveal unique approaches for regenerating myelin in aging and disease.
Methods
Animals and Ethics Statement.
All procedures involving animals were approved by the Institutional Administrative Panel on Laboratory Animal Care (APLAC) of Stanford University and followed NIH guidelines. Mice were group-housed under standard 12:12 light–dark cycles at temperatures of 18 °C to 23 °C and 40 to 60% humidity with free access to food and water and disposable bedding in plastic cages. All mice received regular monitoring from veterinary and animal care staff and were not involved in prior procedures or testing. Sprague-Dawley rats and C57BL/6 mice were ordered from Charles River Laboratories. SRF-Flox mice (22) (Jax strain #: 006658) were a kind gift of Prof. David Ginty (Harvard University), and Olig2-Cre mice (48) were a kind gift of Prof. David Rowitch (University of Cambridge). All mouse lines were maintained by breeding with C57BL/6 mice. Both male and female mice were studied for all in vivo experiments. For cell culture studies with OPCs from mutant mice, brains of both sexes were pooled to obtain sufficient cell numbers.
Data Analysis and Statistics.
All analysis was conducted blind to the genotype and experimental condition. Data analysis and statistics were performed using GraphPad Prism 9.0 software. Descriptive statistics (mean, SEM, and N) were reported in figure legends. Statistical significance was determined between biological replicate (N = 3 to 5) by an unpaired, two-tailed t test, unless otherwise stated in the Figure Legends. For in vivo assays, N refers to the number of mice. For cellular assays, N refers to the number of independent cell purifications from 1 to 2 mouse brains. We pre-determined that using 3 to 5 biological replicates for culture or in vivo experiments were sufficient based on previously published studies (4, 49). There were no outliers/excluded data in any experiment. Detailed description of all other methods can be found in SI Appendix, Materials and Methods.
Supplementary Material
Appendix 01 (PDF)
Dataset S01 (XLSX)
Dataset S02 (XLSX)
Dataset S03 (XLSX)
Acknowledgments
We thank current and past members of the Zuchero, Wyss-Coray, and Barres labs (especially Adiljan Ibrahim, Sophia Wienbar, and Alexandra Münch) and Ben A. Barres for their discussion and support; David Ginty for SRF-Floxed mice; David Rowitch for Olig2-Cre mice. Images and diagrams were created using BioRender.com. We gratefully acknowledge the Stanford University Cell Sciences Imaging Core Facility for EM data collection, especially John Perrino and Ibanri Phanwar for processing and staining EM samples (RRID:SCR_017787: supported by ARRA Award Number 1S10RR026780-01and NIH S10 Award Number 1S10OD028536-01). We thank the staff at the University of California–Berkeley Electron Microscope Laboratory for EM sample preparation. We also thank the Stanford Neuroscience Gene Vector and Virus Core for producing the adeno-associated viruses used in this study. This project was supported by Wu Tsai Neurosciences Interdisciplinary Scholarships (to T.I., M.A.G., and M.L.); the Stanford Graduate Fellowship (M.I.); the Schaller-Nikolich Foundation (A. Keller); the Michael J. Fox Foundation for Parkinson’s Research (125491594 for A. Keller and F.K.; MJFF-021418 for T.W.-C., A. Keller, and F.K.); the NIH AG064897 (T.W.-C.), R01NS119823 (J.B.Z.), and NINDS Diversity Supplement R01NS119823-01S1 (M.A.G. and J.B.Z.); the McKnight Endowment Fund for Neuroscience (J.B.Z.); the National Multiple Sclerosis Society Harry Weaver Neuroscience Scholar Award (J.B.Z.); the Beckman Young Investigator Award (J.B.Z.); the Myra Reinhard Family Foundation (J.B.Z.); the Koret Family Foundation (J.B.Z.); and was originally launched in the lab of Ben A. Barres with support from the National MS Society RG4777-A-10. M.L. is a Merck-sponsored fellow of the Helen Hay Whitney Foundation.
Author contributions
T.I., M.A.G., A.Keller, T.W.-C., F.K., and J.B.Z. designed research; T.I., M.A.G., A. Kaur, M.A., M.I., M.L., N.A., D.M.J., and J.B.Z. performed research; T.I., M.A.G., J.A., A.Kaur, F.K., and J.B.Z. analyzed data; F.K., A.Keller, T.W.-C., and J.B.Z. supervised and funded the research; and T.I. and J.B.Z. wrote the paper.
Competing interests
The authors declare no competing interest.
Footnotes
This article is a PNAS Direct Submission.
Contributor Information
Tal Iram, Email: tal.iram@stanford.edu.
J. Bradley Zuchero, Email: zuchero@stanford.edu.
Data, Materials, and Software Availability
Raw and processed ChIP-seq, RNA-seq, and snRNA-seq data are freely accessible at NCBI GEO using Accession No. GSE241561 (50). All other data are included in the manuscript and/or supporting information.
Supporting Information
References
- 1.Monje M., Myelin plasticity and nervous system function. Annu. Rev. Neurosci. 41, 61–76 (2018), 10.1146/annurev-neuro-080317-061853. [DOI] [PubMed] [Google Scholar]
- 2.Brown L., Macklin W. B., The actin cytoskeleton in myelinating cells. Neurochem. Res. 45, 684–693 (2020), 10.1007/s11064-019-02753-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wilson R., Brophy P. J., Role for the oligodendrocyte cytoskeleton in myelination. J. Neurosci. Res. 22, 439–448 (1989), 10.1002/jnr.490220409. [DOI] [PubMed] [Google Scholar]
- 4.Zuchero J. B., et al. , CNS myelin wrapping is driven by actin disassembly. Dev. Cell 34, 152–167 (2015), 10.1016/j.devcel.2015.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fox A., Afshari F. S., Alexander J. K., Colello R. J., Fuss B., Growth conelike sensorimotor structures are characteristic features of postmigratory, premyelinating oligodendrocytes. Glia 53, 563–566 (2006), 10.1002/glia.20293. [DOI] [PubMed] [Google Scholar]
- 6.Nawaz S., et al. , Actin filament turnover drives leading edge growth during myelin sheath formation in the central nervous system. Dev. Cell 34, 139–151 (2015), 10.1016/j.devcel.2015.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Miano M., Long X., Fujiwara K., Serum response factor: Master regulator of the actin cytoskeleton and contractile apparatus. Am. J. Physiol. Cell Physiol. 292, C70–C81 (2007), 10.1152/ajpcell.00386.2006. [DOI] [PubMed] [Google Scholar]
- 8.Knoll B., Nordheim A., Functional versatility of transcription factors in the nervous system: The SRF paradigm. Trends Neurosci. 32, 432–442 (2009), 10.1016/j.tins.2009.05.004. [DOI] [PubMed] [Google Scholar]
- 9.Ghosh A., Ginty D. D., Bading H., Greenberg M. E., Calcium regulation of gene expression in neuronal cells. J. Neurobiol. 25, 294–303 (1994). 10.1002/neu.480250309. [DOI] [PubMed] [Google Scholar]
- 10.Esnault C., et al. , Rho-actin signaling to the MRTF coactivators dominates the immediate transcriptional response to serum in fibroblasts. Genes Dev. 28, 943–958 (2014), 10.1101/gad.239327.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rivera M., Sheng M., Greenberg M. E., The inner core of the serum response element mediates both the rapid induction and subsequent repression of c-fos transcription following serum stimulation. Genes Dev. 4, 255–268 (1990), 10.1101/gad.4.2.255. [DOI] [PubMed] [Google Scholar]
- 12.Hauschka D., Myocardin. A novel potentiator of SRF-mediated transcription in cardiac muscle. Mol. Cell 8, 1–2 (2001), 10.1016/s1097-2765(01)00297-0. [DOI] [PubMed] [Google Scholar]
- 13.Deshpande A., Shetty P. M. V., Frey N., Rangrez A. Y., SRF: A seriously responsible factor in cardiac development and disease. J. Biomed. Sci. 29, 38 (2022). 10.1186/s12929-022-00820-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dugas C., Ibrahim A., Barres B. A., The T3-induced gene KLF9 regulates oligodendrocyte differentiation and myelin regeneration. Mol. Cell Neurosci. 50, 45–57 (2012), 10.1016/j.mcn.2012.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cahoy D., et al. , A transcriptome database for astrocytes, neurons, and oligodendrocytes: A new resource for understanding brain development and function. J. Neurosci. 28, 264–278 (2008), 10.1523/JNEUROSCI.4178-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang Y., et al. , An RNA-sequencing transcriptome and splicing database of glia, neurons, and vascular cells of the cerebral cortex. J. Neurosci. 34, 11929–11947 (2014), 10.1523/JNEUROSCI.1860-14.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sun Q., et al. , Defining the mammalian CArGome. Genome Res. 16, 197–207 (2006), 10.1101/gr.4108706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lu P., Ramanan N., A critical cell-intrinsic role for serum response factor in glial specification in the CNS. J. Neurosci. 32, 8012–8023 (2012), 10.1523/JNEUROSCI.5633-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Stritt C., et al. , Paracrine control of oligodendrocyte differentiation by SRF-directed neuronal gene expression. Nat. Neurosci. 12, 418–427 (2009), 10.1038/nn.2280. [DOI] [PubMed] [Google Scholar]
- 20.Iram T., et al. , Young CSF restores oligodendrogenesis and memory in aged mice via Fgf17. Nature 605, 509–515 (2022), 10.1038/s41586-022-04722-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Marques S., et al. , Oligodendrocyte heterogeneity in the mouse juvenile and adult central nervous system. Science 352, 1326–1329 (2016), 10.1126/science.aaf6463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ramanan N., et al. , SRF mediates activity-induced gene expression and synaptic plasticity but not neuronal viability. Nat. Neurosci. 8, 759–767 (2005), 10.1038/nn1462. [DOI] [PubMed] [Google Scholar]
- 23.Lappe-Siefke C., et al. , Disruption of Cnp1 uncouples oligodendroglial functions in axonal support and myelination. Nat. Genet. 33, 366–374 (2003), 10.1038/ng1095. [DOI] [PubMed] [Google Scholar]
- 24.Dangata Y., Kaufman M. H., Myelinogenesis in the optic nerve of (C57BL x CBA) F1 hybrid mice: A morphometric analysis. Eur. J. Morphol. 35, 3–17 (1997), 10.1076/ejom.35.1.3.13057. [DOI] [PubMed] [Google Scholar]
- 25.Snaidero N., et al. , Myelin membrane wrapping of CNS axons by PI(3,4,5)P3-dependent polarized growth at the inner tongue. Cell 156, 277–290 (2014), 10.1016/j.cell.2013.11.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Stankoff B., et al. , Ciliary neurotrophic factor (CNTF) enhances myelin formation: A novel role for CNTF and CNTF-related molecules. J. Neurosci. 22, 9221–9227 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fu Y., Weng Z., Improvement of TRANSFAC matrices using multiple local alignment of transcription factor binding site sequences. Conf. Proc. IEEE Eng. Med. Biol. Soc. 2004, 2856–2859 (2004), 10.1109/IEMBS.2004.1403814. [DOI] [PubMed] [Google Scholar]
- 28.Sotiropoulos A., Gineitis D., Copeland J., Treisman R., Signal-regulated activation of serum response factor is mediated by changes in actin dynamics. Cell 98, 159–169 (1999). 10.1016/s0092-8674(00)81011-9. [DOI] [PubMed] [Google Scholar]
- 29.Miralles F., Posern G., Zaromytidou A. I., Treisman R., Actin dynamics control SRF activity by regulation of its coactivator MAL. Cell 113, 329–342 (2003). 10.1016/s0092-8674(03)00278-2. [DOI] [PubMed] [Google Scholar]
- 30.Kenigsbuch M., et al. , A shared disease-associated oligodendrocyte signature among multiple CNS pathologies. Nat. Neurosci. 25, 876–886 (2022), 10.1038/s41593-022-01104-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pandey S., et al. , Disease-associated oligodendrocyte responses across neurodegenerative diseases. Cell Rep. 40, 111189 (2022), 10.1016/j.celrep.2022.111189. [DOI] [PubMed] [Google Scholar]
- 32.Falcao M., et al. , Disease-specific oligodendrocyte lineage cells arise in multiple sclerosis. Nat. Med. 24, 1837–1844 (2018), 10.1038/s41591-018-0236-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.McNamara B., et al. , Microglia regulate central nervous system myelin growth and integrity. Nature 613, 120–129 (2023), 10.1038/s41586-022-05534-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hahn O., et al. , Atlas of the aging mouse brain reveals white matter as vulnerable foci. Cell 186, 4117–4133.e22 (2023), 10.1016/j.cell.2023.07.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Braun T., Gautel M., Transcriptional mechanisms regulating skeletal muscle differentiation, growth and homeostasis. Nat. Rev. Mol. Cell Biol. 12, 349–361 (2011). 10.1038/nrm3118. [DOI] [PubMed] [Google Scholar]
- 36.Guo Y., et al. , Hierarchical and stage-specific regulation of murine cardiomyocyte maturation by serum response factor. Nat. Commun. 9, 3837 (2018), 10.1038/s41467-018-06347-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Knoll B., et al. , Serum response factor controls neuronal circuit assembly in the hippocampus. Nat. Neurosci. 9, 195–204 (2006), 10.1038/nn1627. [DOI] [PubMed] [Google Scholar]
- 38.Medjkane S., Perez-Sanchez C., Gaggioli C., Sahai E., Treisman R., Myocardin-related transcription factors and SRF are required for cytoskeletal dynamics and experimental metastasis. Nat. Cell Biol. 11, 257–268 (2009). 10.1038/ncb1833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Buller B., et al. , Regulation of serum response factor by miRNA-200 and miRNA-9 modulates oligodendrocyte progenitor cell differentiation. Glia 60, 1906–1914 (2012), 10.1002/glia.22406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Emery B., et al. , Myelin gene regulatory factor is a critical transcriptional regulator required for CNS myelination. Cell 138, 172–185 (2009), 10.1016/j.cell.2009.04.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Vartiainen K., Guettler S., Larijani B., Treisman R., Nuclear actin regulates dynamic subcellular localization and activity of the SRF cofactor MAL. Science 316, 1749–1752 (2007), 10.1126/science.1141084. [DOI] [PubMed] [Google Scholar]
- 42.Knoll B., Actin-mediated gene expression in neurons: The MRTF-SRF connection. Biol. Chem. 391, 591–597 (2010). 10.1515/BC.2010.061. [DOI] [PubMed] [Google Scholar]
- 43.Connelly T., et al. , Actin and serum response factor transduce physical cues from the microenvironment to regulate epidermal stem cell fate decisions. Nat. Cell Biol. 12, 711–718 (2010), 10.1038/ncb2074. [DOI] [PubMed] [Google Scholar]
- 44.Kaplan-Albuquerque N., Garat C., Desseva C., Jones P. L., Nemenoff R. A., Platelet-derived growth factor-BB-mediated activation of Akt suppresses smooth muscle-specific gene expression through inhibition of mitogen-activated protein kinase and redistribution of serum response factor. J. Biol. Chem. 278, 39830–39838 (2003). 10.1074/jbc.M305991200. [DOI] [PubMed] [Google Scholar]
- 45.Liu W., et al. , The RhoA/Rho kinase pathway regulates nuclear localization of serum response factor. Am. J. Respir. Cell Mol. Biol. 29, 39–47 (2003), 10.1165/rcmb.2002-0206OC. [DOI] [PubMed] [Google Scholar]
- 46.Stern S., et al. , The transcription factor serum response factor stimulates axon regeneration through cytoplasmic localization and cofilin interaction. J. Neurosci. 33, 18836–18848 (2013), 10.1523/JNEUROSCI.3029-13.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kuchler O., et al. , Single-molecule tracking (SMT) and localization of SRF and MRTF transcription factors during neuronal stimulation and differentiation. Open Biol. 12, 210383 (2022), 10.1098/rsob.210383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Schuller U., et al. , Acquisition of granule neuron precursor identity is a critical determinant of progenitor cell competence to form Shh-induced medulloblastoma. Cancer Cell 14, 123–134 (2008), 10.1016/j.ccr.2008.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Harterink M., et al. , DeActs: Genetically encoded tools for perturbing the actin cytoskeleton in single cells. Nat. Methods 14, 479–482 (2017), 10.1038/nmeth.4257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Iram T., et al. , SRF transcriptionally regulates the oligodendrocyte cytoskeleton during CNS myelination. GEO. https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE241561. Deposited 23 August 2023. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix 01 (PDF)
Dataset S01 (XLSX)
Dataset S02 (XLSX)
Dataset S03 (XLSX)
Data Availability Statement
Raw and processed ChIP-seq, RNA-seq, and snRNA-seq data are freely accessible at NCBI GEO using Accession No. GSE241561 (50). All other data are included in the manuscript and/or supporting information.


