Significance
As a respiratory substrate and a poison, low hydrogen sulfide concentrations can induce significant, but reversible, changes in metabolism by mechanisms that are poorly understood. In this study, we report the surprising durability of sulfide-induced changes in cellular O2 metabolism, following an acute sulfide exposure. We also furnish a quantitative analysis of sulfide preconditioning on P50(O2), the O2 pressure for half-maximal cellular respiration, revealing the underlying mechanism for flux modulation through the electron transport chain. We demonstrate the persistence of the P50(O2) effect up to 24 h after H2S exposure and downstream metabolic changes, which impact fuel utilization and storage, revealing a cellular strategy for long-lived regulation by sulfide.
Keywords: hydrogen sulfide, oxygen metabolism, bioenergetics, electron transport chain
Abstract
Hydrogen sulfide exposure in moderate doses can induce profound but reversible hypometabolism in mammals. At a cellular level, H2S inhibits the electron transport chain (ETC), augments aerobic glycolysis, and glutamine-dependent carbon utilization via reductive carboxylation; however, the durability of these changes is unknown. We report that despite its volatility, H2S preconditioning increases P50(O2), the O2 pressure for half-maximal cellular respiration, and has pleiotropic effects on oxidative metabolism that persist up to 24 to 48 h later. Notably, cyanide, another complex IV inhibitor, does not induce this type of metabolic memory. Sulfide-mediated prolonged fractional inhibition of complex IV by H2S is modulated by sulfide quinone oxidoreductase, which commits sulfide to oxidative catabolism. Since induced hypometabolism can be beneficial in disease settings that involve insufficient or interrupted blood flow, our study has important implications for attenuating reperfusion-induced ischemic injury and/or prolonging the shelf life of biologics like platelets.
Clinical outcomes of traumatic injury or diseases that result from insufficient or interrupted blood supply can be improved by decreasing metabolic demand (1, 2). The discovery of H2S as a signaling molecule (3) paved a paradigm shift in our understanding of its biology from that of a mere environmental toxin to a modulator of mammalian energy metabolism (4, 5). Moderate exposure to H2S (~80 ppm, 6 h) in mice elicits profound hypometabolism that is characterized by a drop in metabolic rate by 90% and in the core body temperature to 15 °C, which is rapidly reversed in room air (6). While the mechanism of sulfide-induced hypometabolism remains elusive, an ever-increasing span of physiological effects has been ascribed to H2S, which encompasses major organ systems, including the cardiovascular, gastrointestinal, and central nervous system (7–11). At a cellular level, the durability and multifaceted implications of “buying time” by suppressing oxidative metabolism are poorly understood, and the therapeutic promise of sulfide in clinical and experimental models of injury and disease remains to be realized.
The steady-state levels of H2S are estimated to be in the tens of nanomolar range in most tissues (12–14). Fluctuations in sulfur metabolism due to factors such as dietary sulfur intake, antibiotic use, gut microbial composition, or hypoxia can potentially disturb the delicate balance between H2S synthesis and clearance, allowing cellular levels to spike transiently. In the gut, sulfide-producing and using microbes contribute to colonic concentrations estimated to range from 0.2 to 2.4 mM H2S (15, 16). Sulfide is efficiently oxidized by sulfide quinone oxidoreductase (SQOR), an inner mitochondrial membrane enzyme (17, 18), which shields complex IV from respiratory poisoning (19). The reversible inhibition of complex IV by H2S is accompanied by induction of reductive stress, which propagates from the mitochondrion to other compartments, and serves as an important mechanism of H2S signaling (4, 5).
The sulfide oxidation pathway converts H2S through a series of oxidation and sulfur transferase steps to thiosulfate and sulfate (Fig. 1A). SQOR commits H2S to catabolism, utilizes coenzyme Q (CoQ) as an electron acceptor, and thus resides at the intersection of sulfide oxidation pathway and the electron transport chain (ETC) (20, 21). SQOR-dependent H2S oxidation not only clears this respiratory poison but also enhances proton-coupled electron transfer, fueling ATP synthesis (19, 22). However, when concentrations exceed SQOR clearance capacity, H2S inhibits complex IV (23), which manifests as a decrease in the cellular oxygen consumption rate (OCR). The twin effects of H2S as a respiratory substrate and an inhibitor influence the redox state of CoQ, which is used by multiple ETC feeders. As H2S levels rise and complex IV is inhibited, the restricted availability of CoQ causes a reductive shift in the NAD(P)+ pool (19) (Fig. 1A). The ripple effects of the reductive shift in the ETC influence both central carbon and lipid metabolism (19, 24–26). The interaction between H2S and the ETC is also postulated to underlie the profound but reversible ability of H2S to trigger hypometabolism, leading to a state of suspended animation even in a nonhibernating animal (6). Additionally, short-term H2S exposure protects mice against lethal hypoxia, suggesting that an H2S-dependent decrease in O2 metabolism is important for attenuating hypoxia-induced damage (27).
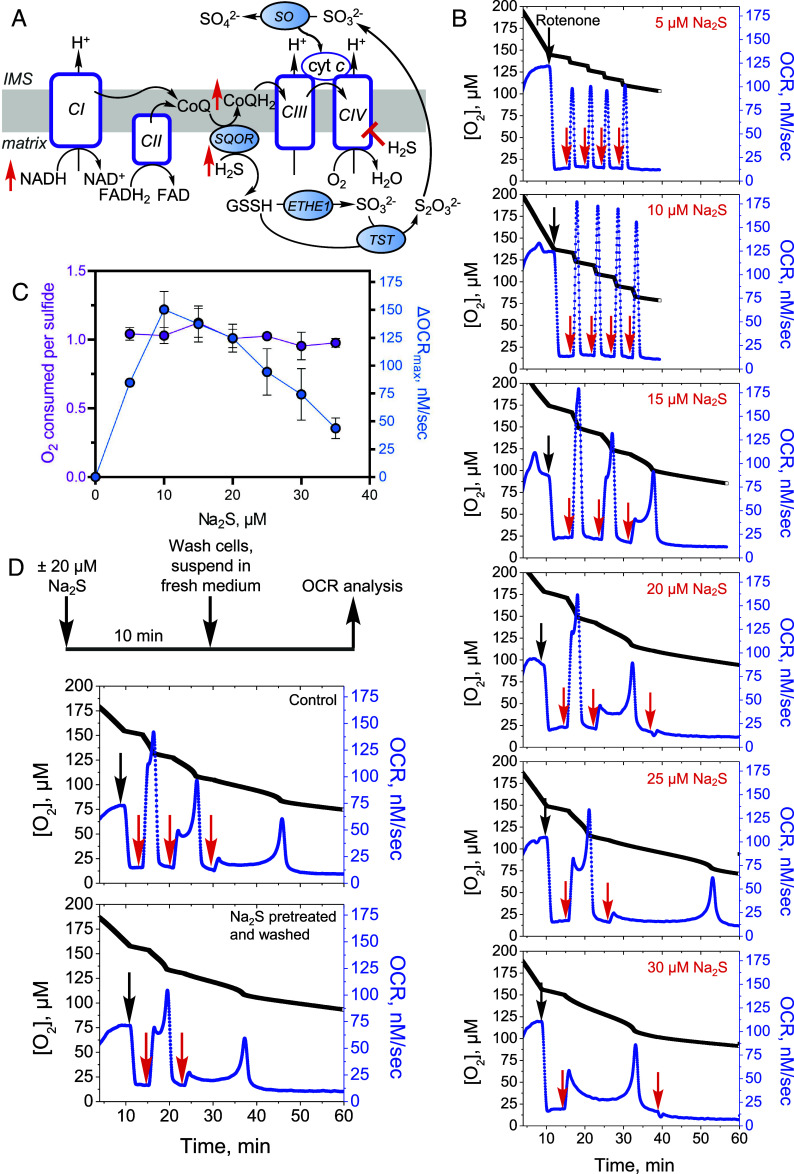
Fig. 1.
Effect of repeated sulfide exposure on ETC flux. (A) Sulfide inhibition of complex IV (CIV) causes electron acceptor insufficiency in the ETC due to a predicted build-up of NADH, CoQH2, and reduced cytochrome c (cyt c). IMS is intermembrane space, ETHE1 is persulfide dioxygenase, TST is thiosulfate sulfur transferase, and SO is sulfite oxidase. (B) Rotenone (0.5 µM)-treated HT29 cells exposed to low sulfide (5 to 10 µM) showed sharp increases in OCR with each aliquot of Na2S. At higher Na2S concentrations (15 to 30 µM), complex kinetics were observed. (C) Replotting the data in B as described under Methods showed a bell-shaped dependence of OCR on sulfide dose and an O2:sulfide ratio of 1.0 ± 0.1 between 5 and 35 µM Na2S. The data are representative of at least four independent experiments and represent the mean ± SD. (D) Experimental setup for removing residual sulfide and secreted oxidation products (Top). Washing did not impact the persistence of Na2S-triggered OCR (Bottom). Data are representative of at least three independent experiments. Black and red arrows indicate when rotenone and sulfide, respectively, were added.
In this study, we report that H2S preconditioning elicits persistent bioenergetic perturbations, which we refer to as “H2S memory.” The sustained metabolic alterations include an increase in the O2 pressure for half-maximal respiration (P50(O2)) and enhanced sensitivity to subsequent H2S exposure. The potency of this cellular memory exhibits a dose dependence on H2S, is attenuated by dissipation of the mitochondrial NADH pool, but exacerbated by attenuation of SQOR. Furthermore, SQOR expression levels across cell lines correlate inversely with the persistence of H2S memory. Collectively, these data reveal the mechanism underlying the sustained and pleiotropic effects of H2S on metabolism, including a reductive shift in cofactor pools, increased aerobic glycolysis, utilization of glutamine via reductive carboxylation and lipid accumulation, as well as changes in the pentose phosphate and pyrimidine biosynthetic pathways. These alterations result from the H2S-dependent fractional inhibition of complex IV, revealing a heretofore unappreciated cellular strategy for long-lived regulation of ETC flux.
Results
Sulfide Elicits Dose-Dependent Changes in Oxygen Consumption Kinetics.
We characterized the dose-dependent changes in respiratory rate following a single acute exposure to sulfide. Low sulfide concentrations (5 to 10 µM) elicited sharp increases in O2 consumption in HT29 cells, while mixed kinetics were seen at 20 µM Na2S (SI Appendix, Fig. S1A). The different phases at 20 µM Na2S presumably represent the sum of OCR activation and inhibition due to the dual action of H2S at this concentration. At ≥30 µM Na2S, a net decrease in OCR was observed, consistent with inhibition of respiration (SI Appendix, Fig. S1B). The protracted recovery times for establishing a new stationary OCR and the persistent fractional inhibition of OCR during the experimental time frame correlated with sulfide concentration (SI Appendix, Fig. S1 C and D).
Effect of Repeated Acute Sulfide Exposure on ETC Flux.
Next, we examined whether repeated H2S exposure elicits sustained inhibition of complex IV, which might have implications for long-term metabolic rewiring. The impact of repeated sulfide addition on OCR was more readily observed in the presence of rotenone, a complex I inhibitor that decreases competition for the CoQ pool (Fig. 1B). Under these conditions, the first injection of sulfide generally induced a net increase in OCR, although the kinetics of the response varied in a dose-dependent way. Low sulfide concentrations (<15 µM) triggered a sharp increase followed by a return to basal OCR while higher concentrations (≥20 µM) elicited complex kinetics. At low concentrations (≤10 µM), consecutive sulfide injections led to its clearance with similar kinetics, while at higher concentrations (≥15 µM), complex kinetics and longer clearance times were observed, consistent with sustained inhibition of complex IV.
The maximal change in OCR exhibited a bell-shaped dependence on sulfide concentration, and a stoichiometry of 1.0 ± 0.1 O2 consumed to sulfide added was observed across the Na2S concentration range (Fig. 1C). Washing cells to remove thiosulfate, a product of the sulfide oxidation pathway that accumulates in the conditioned medium (25), did not affect sensitivity to subsequent sulfide injections (Fig. 1D).
SQOR Increases the IC50 for Complex IV Inhibition by H2S.
We evaluated the magnitude of ETC protection conferred by SQOR by comparing the sulfide-dependent inhibition of OCR in SQOR knockdown (HT29SQOR KD) versus scrambled control (HT29Scr) cells. Sulfide inhibits complex IV in vitro with an estimated Ki of 0.2 µM and kon and koff values of 1.5 × 104 M−1 s−1 and 6 × 10−4 s−1, respectively, at 30 °C (28). Sulfide can bind to CuB (in the 1 + or 2 + oxidation states) and the heme a3 site in the MT-CO1 subunit of complex IV (Fig. 2A). shRNA-induced SQOR knockdown led to a 90 to 95% decrease in protein expression (19) (SI Appendix, Fig. S2 A and B). From the sigmoidal dependence on sulfide concentration, an IC50 value of 30 ± 1 µM was estimated (Hill coefficient = 4.8) for HT29Scr cells and 6.0 ± 0.3 µM (Hill coefficient of 5.2) for HT29SQOR KD cells (Fig. 2B). These data provide a quantitative measure of the protection afforded by SQOR against sulfide poisoning in HT29 cells.
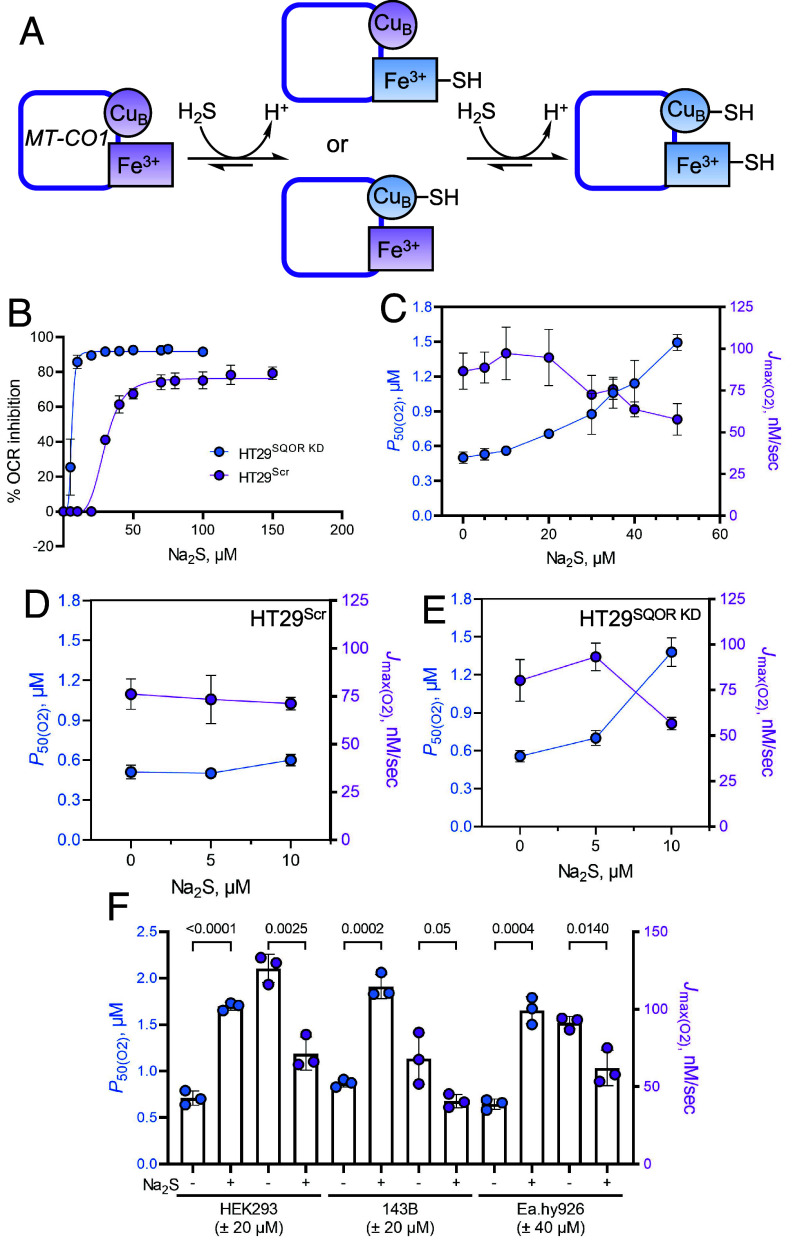
Fig. 2.
H2S increases P50(O2) for complex IV. (A) Postulated mechanism for sulfide binding to the metal sites in the MT-CO1 subunit of complex IV. (B) Inhibition of OCR as a function of sulfide concentration in HT29Scr (purple) and HT29SQOR KD (blue) cells yielded IC50 estimates of 30 ± 1 and 6.0 ± 0.3 µM, respectively. Data represent the mean ± SD of three to five independent experiments. (C) Dependence of P50(O2) (blue) and Jmax(O2) (purple) values on sulfide concentration in HT29 cells. Data represent the mean ± SD of at least three independent experiments. (D and E) Na2S increases the P50(O2) and decreases the Jmax(O2) value in HT29SQOR KD (at 10 µM) but not in HT29Scr cells (n = 4). (F) The P50(O2) and Jmax(O2) of HEK293 and 143B cells are significantly altered by 20 µM Na2S as in HT29 cells, while Ea.hy926 cells need 40 µM Na2S for similar modulation (n = 3). The two-sample unpaired t test was performed for statistical analysis.
H2S Increases P50(O2) for Complex IV.
We postulated that the sustained decrease in ETC flux after single or repeated sulfide exposure (SI Appendix, Fig. S1D and Fig. 1C) results from fractional inhibition of complex IV and assessed this by evaluating sulfide-dependent modulation of the O2 pressure at half-maximal respiration, a measure of O2 affinity. The P50(O2) value increased from 0.50 ± 0.05 to 1.49 ± 0.07 µM while Jmax(O2) decreased from 90 ± 10 to 58 ± 9 nM s−1 at 37 °C, as sulfide concentrations increased from 0 to 50 µM (Fig. 2C). Technical issues precluded determination of P50 values at higher sulfide concentrations as the dissolved O2 was depleted during the recovery time, i.e., before cells could establish a new stationary OCR. Due to the greater sensitivity of HT29SQOR KD cells, P50(O2) modulation could only be evaluated at low Na2S (5 to 10 µM) concentrations (Fig. 2 D and E). In contrast to HT29Scr cells (Fig. 2D), HT29SQOR KD cells showed a 1.3-fold increase in P50(O2) (0.56 ± 0.05 versus 0.70 ± 0.06 µM) with 5 µM Na2S, while the Jmax(O2) value was unchanged (89 ± 9 versus 93 ± 8 nM s−1) (Fig. 2E). A 2.5-fold increase in P50(O2) (1.4 ± 0.1 µM) was observed at 10 µM Na2S, which was accompanied by a 30% decrease in the Jmax(O2) value (57 ± 3 nM s−1).
The ability of sulfide to modulate O2 metabolism was compared across cell lines (Fig. 2F). In HEK293 and 143B cells, 20 µM Na2S sulfide was sufficient to achieve a similar magnitude of P50(O2) modulation as 40 µM Na2S in Ea.hy926 and HT29 cells. The P50(O2) value increased 2.4-fold (0.71 ± 0.08 to 1.70 ± 0.04 µM) in HEK293 cells while Jmax(O2) decreased from 126 ± 9 to 71 ± 11 nM s−1. In 143B cells, P50(O2) increased 2.2-fold (from 0.87 ± 0.04 to 1.91 ± 0.13 µM) while Jmax(O2) decreased from 68 ± 17 to 41 ± 4 nM s−1. In Ea.hy926 cells, P50(O2) increased 2.5-fold (from 0.64 ± 0.06 to 1.7 ± 0.1 µM) and Jmax(O2) decreased from 91 ± 4 to 60 ± 10 nM s−1.
A Single Acute H2S Exposure Elicits Long-Lived Effects on O2 Metabolism.
Unlike the short-term response of cells to acute H2S exposure (19, 24–26), the long-term impacts are less well characterized. We therefore examined the durability of metabolic effects following a single acute exposure to Na2S (100 µM) after 4, 24, or 48 h (Fig. 3 A and B). Perturbed OCR kinetics were clearly visible even 48 h later as evidenced by an increased sensitivity to subsequent Na2S exposure. A measurable increase in the P50(O2) value was seen 24 h after a single exposure to Na2S, but the effect was dissipated at 48 h (Fig. 3C). Sulfide preconditioning decreased H2S consumption and thiosulfate production (Fig. 3D).
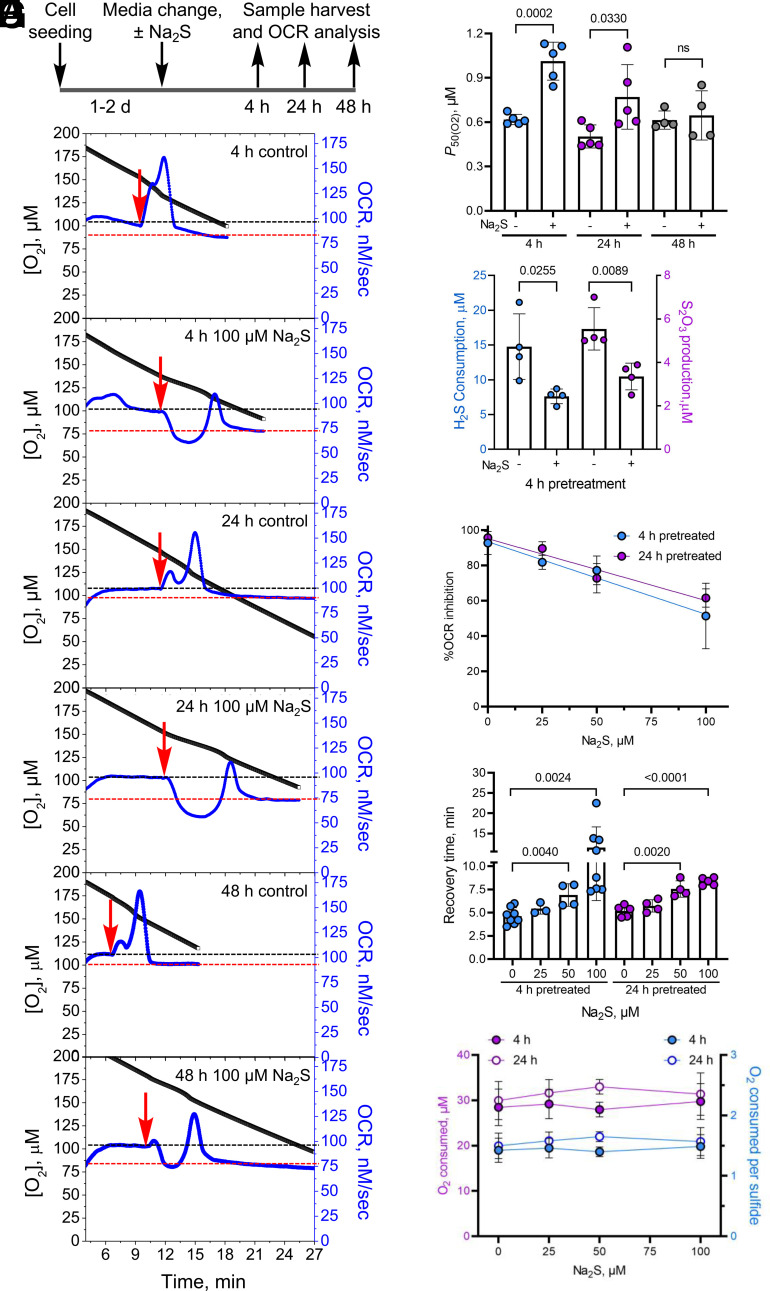
Fig. 3.
H2S memory is long-lived. (A) Scheme showing the experimental setup used to test the durability of sulfide exposure. (B) Representative OCR traces for cells ± 100 µM Na2S after 4, 24, and 48 h. While control cells rapidly oxidized 20 µM Na2S (red arrow), pretreated cells were inhibited up to 48 h. The black and red lines represent the OCR before and after recovery from 20 µM Na2S, respectively. (C) Changes in cellular P50(O2) values 4, 24, and 48 h after a single exposure to 100 µM Na2S. (D) Pretreatment with 100 µM Na2S results in lower H2S clearance and thiosulfate production 4 h later. (E–G) Sulfide (0 to 100 µM) pretreatment (4 or 24 h) resulted in a dose-dependent inhibition of OCR following Na2S reexposure (20 µM) (E), increased time to recovery of a new stationary OCR (F), but had no impact on the total O2 consumed during the recovery time (G). The average total O2 consumed per sulfide was estimated to be 1.5 ± 0.1 across all sulfide pretreatment concentrations. The two-sample unpaired t test was performed for statistical analysis (C–G).
Next, the sensitivity of preconditioning to the sulfide dose was examined. Sulfide pretreatment (4 or 24 h) at concentrations as low as 25 µM H2S increased OCR inhibition by a subsequent injection of sulfide (Fig. 3E). The recovery time to a new stationary OCR, showed significant differences with ≥50 µM sulfide pretreatment (Fig. 3F). Regardless of the duration of recovery, control and sulfide pretreated cells consumed 30 ± 1 µM O2 before establishing a new stationary OCR, corresponding to an O2:sulfide ratio of ~1.5 ± 0.1 (Fig. 3G).
Acute H2S exposure did not increase SQOR expression (SI Appendix, Fig. S2 A–C) or affect mitochondrial content as monitored by cardiolipin levels (SI Appendix, Fig. S3A). Acute sulfide exposure led to small but statistically significant decreases in MT-CO1 and MT-CO2 protein levels, which was correlated with slightly lower complex IV activity (SI Appendix, Fig. S3 B–D).
Cyanide also inhibits complex IV (29). However, unlike sulfide, cyanide only coordinates to the heme a3 iron (in the ferric or ferrous state) but not to CuB in MT-CO1 (SI Appendix, Fig. S4A). The Ki for the in vitro inhibition of complex IV by cyanide is 0.2 µM, with kon = 5 × 103 M−1s−1 and koff = 5 × 10−4 s−1 at 30 °C (28). Surprisingly, cyanide pretreatment (500 µM, 4 h) enhanced sulfide-triggered OCR and recovery, following subsequent exposure to sulfide (SI Appendix, Fig. S4 B–D). These data suggest a specific mechanism for H2S preconditioning effects that is not general to complex IV inhibition.
Durability of H2S Preconditioning Correlates with SQOR Expression Levels.
We examined whether an ~10-fold difference in SQOR expression levels across five cell lines (SI Appendix, Fig. S5A) is correlated with the durability of the preconditioning effect. HEK293 cells have very low SQOR expression and showed prolonged inhibition in response to Na2S over ~50 min (SI Appendix, Fig. S5B). Similarly, pretreatment of HT29SQOR KD but not HT29Scr control cells with sulfide for 4 or 24 h led to sustained inhibition of OCR after a subsequent injection with a low (10 µM) Na2S (SI Appendix, Fig. S2 D–G). Panc-1 cells showed the highest SQOR expression and were the least sensitive to the preconditioning effect (SI Appendix, Fig. S5C). However, HT29, LoVo, and Ea.hy926 cells showed variable sensitivity to sulfide preconditioning although they had similar SQOR levels, suggesting that additional factors influence cellular H2S memory.
H2S Induces Long-Lived Changes in Mitochondrial Function.
We evaluated which aspects of mitochondrial function are impacted by sulfide exposure and their modulation by SQOR. Mitochondrial function analysis revealed pleiotropic changes in response to H2S (100 µM, 4 h) in HT29, HT29Scr, and HT29SQOR KD cells (Fig. 4A and SI Appendix, Fig. S6A). Basal, ATP-linked, and maximal respiration as well as the proton leak rate decreased, while nonmitochondrial respiration was unaffected, except in HT29SQOR KD cells (Fig. 4 B–G and SI Appendix, Fig. S6 B–G). Each of these parameters was more impacted in HT29SQOR KD relative to HT29Scr controls.
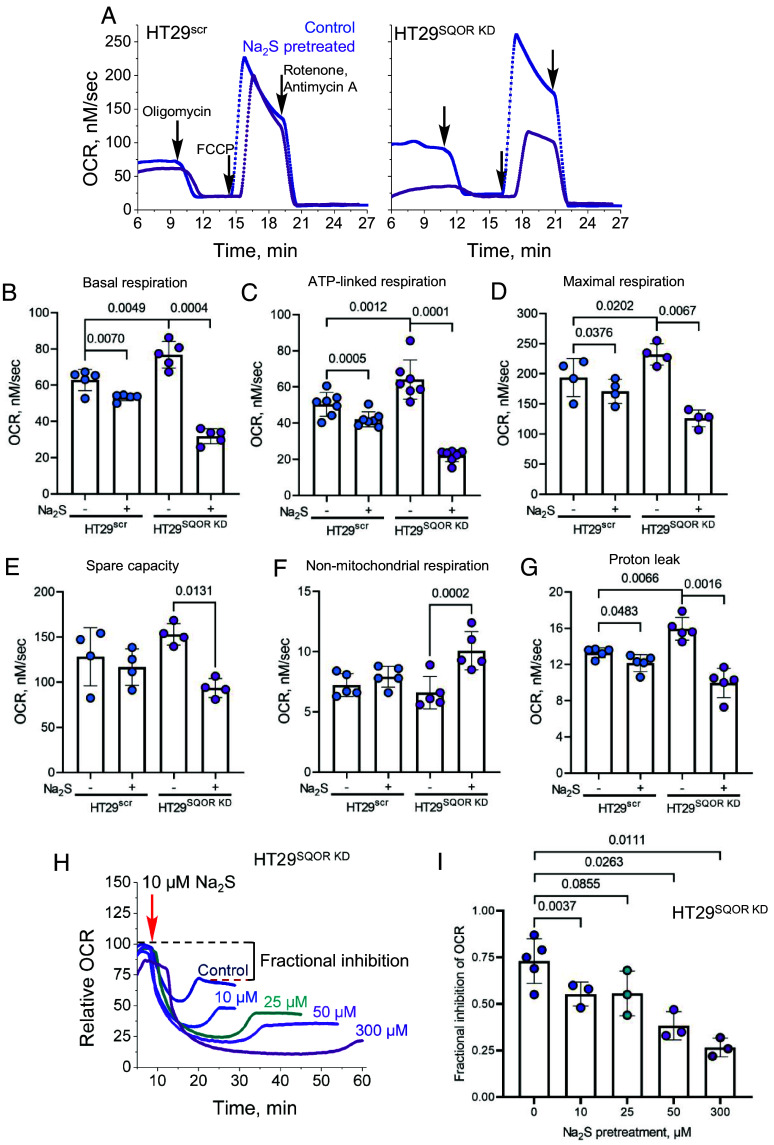
Fig. 4.
SQOR modulates the impact of H2S on mitochondrial function. (A) Representative traces of mitochondrial function profiles in HT29scr (Left) and HT29SQOR KD (Right) cells treated for 4 h ±100 µM Na2S in response to oligomycin (125 nM), FCCP (125 nM), and rotenone and antimycin A (0.5 µM each). (B–G) Quantitative analysis of the data in A reveals the effects of sulfide pretreatment on basal respiration (B), ATP-linked respiration (C), maximal respiration (D), spare capacity (E), nonmitochondrial respiration (F), and proton leak rate (G). (H and I) Dose-dependent effects of Na2S pretreatment (10 to 300 µM, 24 h) on OCR (H) and the fraction of OCR inhibited (I) in HT29SQOR KD cells following exposure to 10 µM Na2S (red arrow). The two-sample paired t test was performed for statistical analysis.
Prolonged and dose-dependent perturbations in mitochondrial bioenergetics were observed in HT29SQOR KD cells after sulfide pretreatment (10 to 300 µM, 24 h). Enhanced sensitivity to respiratory inhibition was evident at concentrations as low as 10 µM Na2S, which elicited a pronounced decrease in the stationary OCR, indicative of fractional inhibition (Fig. 4 H and I).
H2S Induces Long-Lived Metabolic Changes.
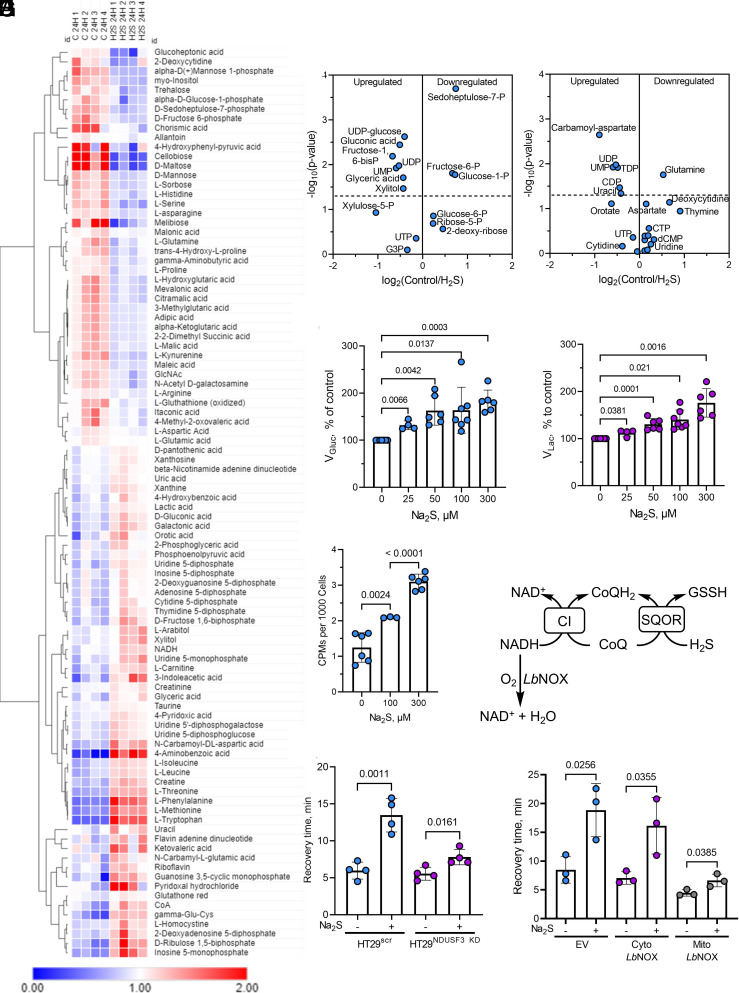
The long-lived effects of sulfide on mitochondrial function are expected to have more widespread metabolic effects. Indeed, our metabolomics analysis revealed changes in numerous metabolites (Fig. 5A) 24 h after a single exposure to H2S, including some in the pentose phosphate and pyrimidine biosynthesis pathways (Fig. 5 B and C and SI Appendix, Table S1), as well as an ~50% decrease in serine levels (SI Appendix, Fig. S7A). We have previously shown that sulfide induces an ~2-fold increase in the glycolytic rate in HT29 cells to compensate for decreased ATP-linked respiration (25). Remarkably, the dose-dependent increase in glucose consumption and lactate production persisted 24 h after an initial exposure to 25 to 300 µM sulfide (Fig. 5 D and E). We have also shown that repeated exposure to sulfide increases incorporation of glutamine carbon to lipids via reductive carboxylation (19, 24). Surprisingly, a single exposure to 100 or 300 µM Na2S was sufficient to increase [14C]-glutamine incorporation into lipids by 70% and 150%, respectively (Fig. 5F).
Fig. 5.
Sulfide preconditioning induces long-term metabolic changes implicating electron acceptor insufficiency. (A) Metabolomics analysis in HT29 cells 24 h after ±100 µM Na2S exposure. (B and C) Changes in pentose phosphate (B) and pyrimidine biosynthesis (C) pathway metabolites are among the changes observed in the metabolomics profile. (D and E) Pretreatment with sulfide (25 to 300 µM, 24 h) increased the relative rates of glucose consumption (D) and lactate production (E). Data are representative of at least four independent experiments. (F) A single bolus exposure to sulfide (100 or 300 µM) increased [U14-C]-glutamine incorporation into lipids 13 h later (n ≥ 3). (G) Scheme showing how knockdown of the complex I subunit, NDUSF3, or mitochondrial LbNOX expression can impact CoQ availability. (H and I) NDUSF3 knockdown (H) and expression of mitochondrial but not cytosolic LbNOX (I) decreased recovery time.
Electron Acceptor Insufficiency Contributes to H2S Memory.
A backup in the ETC is expected to cause an upstream reductive shift, resulting in electron acceptor insufficiency. We tested the hypothesis that CoQH2 recycling is limiting during the period that cells exhibit H2S memory. For this, electron transfer from complex I to CoQ was limited either by knockdown of the NDUFS3 subunit of complex I or via heterologous expression of LbNOX, a water-generating NADH oxidase (30), which can be targeted to the cytoplasm or mitochondria (Fig. 5G). In comparison to the HT29Scr controls, H2S memory was substantially attenuated in HT29NDUFS3 KD cells as monitored by recovery time (Fig. 5H). Mitochondrial but not cytoplasmic LbNOX expression also decreased recovery time compared to the empty vector control (Fig. 5I). Complex II reversal with fumarate, serving as an electron acceptor, can recycle CoQH2 (26) during acute H2S exposure (SI Appendix, Fig. S7B). However, neither knockdown of the SDHA subunit of complex II nor provision of dimethyl fumarate, a membrane-permeable fumarate derivative, alleviated H2S memory (SI Appendix, Fig. S7 C and D).
Discussion
The remarkable response of mice to moderate H2S exposure has the hallmarks of inducing hibernation-like behavior, including low core body temperature and depressed cardiac and metabolic function (6, 27, 31). A study using a synthetic model of cytochrome c oxidase ascribed the molecular basis of these changes to reversible sulfide coordination at ferrous heme a3 in a manner that is competitive with O2 (32). While characterizing the short-term consequences of H2S exposure in a cell culture model in which systems like SQOR that interact with H2S are present, we found that H2S-induced metabolic perturbations endure over 4 to 48 h. The durability of this cellular memory, which is sensitive to H2S dose during pretreatment, and the intrinsic capacity for sulfide oxidation by SQOR, was observed across all tested cell lines.
The persistence of cellular sulfide effects is surprising because H2S disappears rapidly under culture conditions (t1/2 ~4 min in 6-cm plates, 37 °C) (25). The durability of the preconditioning effects is also surprising since conserved stress response pathways exist to sense and alleviate acute reductive stress to preserve cellular integrity. For example, the redox status of conserved cysteines on FNIP1 (folliculin interacting protein 1) in myoblasts is sensed by the E3 ubiquitin ligase CUL2FEM1B, leading to FNIP degradation under reductive stress, with a consequent increase in mitochondrial activity (33). Despite recent advances in our understanding of the acute effects of H2S on cellular metabolism (4, 5, 34), the durability of the stress response to this volatile metabolite is not known.
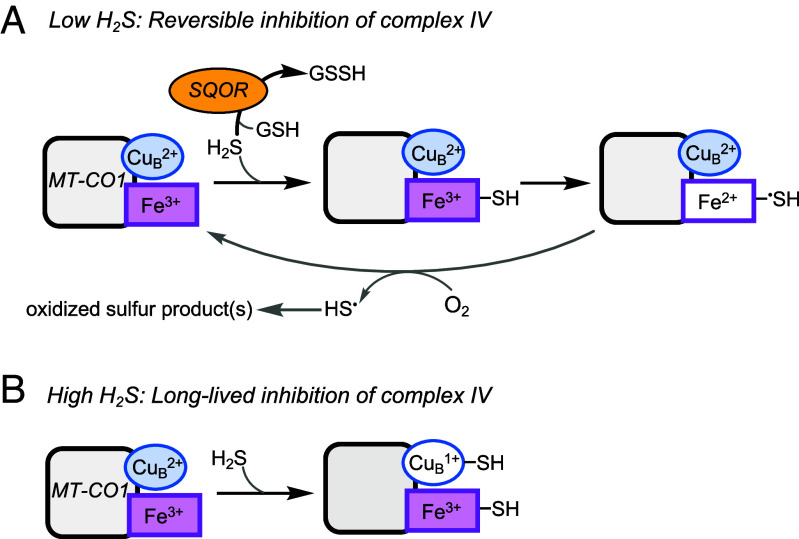
The interaction of sulfide with the MT-CO1 subunit of complex IV is complex and poorly characterized (28), and much less is known about its interaction with the dicopper-containing MT-CO2 subunit. Sulfide reacts with fully oxidized MT-CO1 to give ferric heme a3 with a sulfide ligand and cuprous CuB (35). Sulfide oxidation can occur from this mixed valence state of MT-CO1 although the product, which might be HSSH, polysulfide, S0, or S8, remains to be characterized (Fig. 6A). Sulfide binding and oxidation to catenated sulfur species has been described for other hemeproteins (36–39).
Fig. 6.
Summary of H2S interactions with MT-CO1 at low and high H2S concentrations. (A) At low sulfide concentrations, H2S is efficiently cleared by SQOR, and its interaction with MT-CO1 leads to weak and reversible inhibition that is competitive with respect to O2. (B) When SQOR capacity is exceeded and H2S levels rise, coordination to the CuB site in MT-CO1 leads to long-lived inhibition of complex IV.
In a cellular milieu, the interaction of H2S with complex IV is modulated by SQOR (19). We posit that at low concentrations, SQOR limits H2S exposure, leading to reversible complex IV inhibition via formation of ferrous heme a3 (Fig. 6A). When the capacity to clear sulfide is limiting or compromised, as in SQOR deficiency, H2S concentrations rise and MT-CO1 containing ferric heme a3 and cuprous CuB-sulfide is predicted to accumulate (Fig. 6B). Indeed, spectroscopic analysis of sulfide-treated MT-CO1 reveals that while the CuB site binds to and is reduced by sulfide, it is resistant to subsequent air oxidation, which reoxidizes the heme centers. This resistance of cuprous CuB to oxidation has been interpreted as evidence for an increase in its redox potential via direct sulfide coordination (40). Long-lived fractional inhibition of complex IV following H2S preconditioning, would explain lower basal OCR, ATP-linked respiration, and spare respiratory capacity as well as higher P50(O2) values (Figs. 3C and 4 and SI Appendix, Fig. S6). Sustained fractional inhibition of complex IV is also supported by the statistically significant decrease in its activity (SI Appendix, Fig. S3D), which could be an underestimate since ascorbate/TMPD could promote loss of the sulfide ligand. While the effect of sulfide coordination on the stability of MT-CO1 or MT-CO2 is unknown, we note small but statistically significant decreases in protein levels just 4 h after an acute sulfide exposure (SI Appendix, Fig. S3 B and C). Our model also explains why cyanide, which interacts with the heme but not the copper site, fails to elicit comparable long-lived changes (SI Appendix, Fig. S4).
The cellular response to H2S ranges from stimulation of O2 consumption at low, to inhibition at high concentrations, while complex kinetics are observed at intermediate concentrations, which reflect the dual ETC substrate and inhibitor dynamic of this metabolite. Regardless of the initial response, cells appear to return to basal OCR, superficially suggesting reversibility. However, a closer inspection reveals fractionally lower stationary OCR values after successive exposure to H2S, which is more evident at the higher concentrations (Fig. 1B) and marked in HT29SQOR KD cells (SI Appendix, Fig. S2 D and F). Persistent dampening of the ETC flux after sulfide injection is more readily seen in sulfide-pretreated cells (Fig. 3B) and particularly in HT29SQOR KD cells (Fig. 4 H and I), raising questions as to the underlying mechanism.
The observed increase in P50(O2) with a concomitant decrease in Jmax(O2) (Fig. 2C), is inconsistent with noncompetitive inhibition by sulfide with respect to O2, as reported in an in vitro steady-state kinetic analysis (29). It has been argued that the slow off-rate for sulfide (koff = 6 × 10−4 s−1) (28) likely distorted the kinetic pattern, decreasing Vmax while having a minimal effect on the KM(O2) due to an increasing fraction of the enzyme being inactive with increasing sulfide concentration. We posit that the ability of sulfide to increase P50(O2) for complex IV explains the protective effects of sulfide preconditioning on lethal hypoxia (27) as well as limiting reperfusion injury following ischemia (8). Decreased ETC flux by sulfide is expected to protect against oxidative injury by averting the buildup of a large proton motive force during the early reperfusion phase, which is one of the factors that drives reverse electron transfer and reactive oxygen species formation (41).
SQOR releases two electrons per catalytic cycle, generating one equivalent of COQH2 that leads to the reduction of 0.5 O2 per mole of sulfide oxidized. Since glutathione persulfide, the other product of the SQOR reaction, is further oxidized to sulfite, consuming one mole of O2, the predicted stoichiometry of O2 consumed:sulfide is 1.5. However, if H2S also serves as an alternate sulfane sulfur acceptor, generating HSSH (20), the expected O2:sulfide (1.5:2.0) stoichiometry is 0.75. The experimentally determined ratio was 1.0 ± 0.1 in rotenone-treated cells (Fig. 1C), and 1.5 ± 0.2 in sulfide-preconditioned cells (in the absence of rotenone) (Fig. 3G). Both values are higher than the 0.74 to 0.89 stoichiometry reported previously (42). Loss through volatilization of H2S is unlikely to be a significant factor in the closed respirometry chamber used in our study. Furthermore, a larger fractional deviation from the expected stoichiometry would be expected due to H2S loss at lower sulfide concentrations, which was not observed. Deviation from the 1.5 stoichiometry in rotenone-treated cells could be explained by i) incomplete coupling between the SQOR and ETHE1-catalyzed reactions or between SQOR and complex III activity (Fig. 1A), ii) the dual use of H2S (kcat/KM = 3.7 × 105 M−1 s−1) and GSH (kcat/KM = 1.6 × 104 M−1 s−1) (20) as sulfur acceptors during an acute H2S exposure, or iii) diversion of CoQH2 to complex II functioning in reverse (SI Appendix, Fig. S7B) (26).
We have previously reported the profound effect of SQOR in protecting against sulfide inhibition of the ETC (19) and characterized it quantitatively in this study. SQOR deficiency decreased the IC50 for H2S from 30 ± 1 to 6.0 ± 0.3 µM in HT29 cells and rendered the P50(O2) and Jmax(O2) values sensitive to low H2S concentrations (≤10 µM) in SQOR knockdown but not in control cells (Fig. 2 B, D, and E). Since the knockdown efficiency was ~90 to 95%, our study underestimates the full extent of ETC modulation by SQOR in these cells. Patients with SQOR deficiency present with Leigh disease and limited tissue analysis shows markedly lower complex IV activity (43). The protein levels of only the nuclear-encoded subunits were investigated in this study and found to be normal, leading to the conclusion that complex IV activity, but not its assembly, was affected by SQOR deficiency. Ethylmalonic encephalopathy, another inborn error of metabolism characterized by elevated H2S, results from deficiency of ETHE1, the second enzyme in the sulfide oxidation pathway (Fig. 1A) (44). The disease is characterized by severe deficiency of complex IV activity, particularly in the brain and muscle, and decreased MT-CO1 and MT-CO2 protein levels (45). These studies provide additional support of our finding that acute H2S exposure has long-lived effects on complex IV, which are exacerbated in the absence of a functional sulfide oxidation pathway.
In summary, our study provides a quantitative analysis of ETC regulation by H2S via decreased O2 affinity, sustained complex IV inhibition, and partial destabilization of the metal-containing MT-CO1 and MT-CO2 subunits. SQOR activity, which is a key determinant of cellular H2S levels (14), could represent both an “on” and an “off” switch for sulfide-dependent ETC regulation (34). The prolonged effects on oxidative metabolism provide an opportunity for extending the time window in which the potential therapeutic effects of H2S for attenuating injury or prolonging platelet shelf life might be exploited.
Methods
Materials.
Na2S, nonahydrate (99.99% purity (431648), rotenone (R8775), protease inhibitor cocktail for mammalian tissue extract (P8340), puromycin (P8833), dimethyl fumarate (242926), doxycycline (D3447), hexane (110543), RIPA lysis buffer (R0278), dimethyl sulfoxide (D2650), oligomycin A (75351), antimycin A (A8674), and ascorbate (PHR1279-1G) were from Sigma. Dulbecco’s modified Eagle’s medium (DMEM) [with 4.5 g/L glucose, 584 mg/L glutamine, and 110 mg/L sodium pyruvate (11995-065)], RPMI 1640 with glutamine (11875-093), fetal bovine serum (FBS, 10437-028), penicillin/streptomycin mixture (15140-122), 0.05% (w/v) trypsin-EDTA (25300-054), phenol red free 0.5% trypsin EDTA (15400054), PBS (10010-023), and Dulbecco’s phosphate-buffered saline medium (DPBS, 14040-133) were from Gibco. Geneticin (10131-035) was from Life Technologies. N,N,N′,N′-tetramethyl-p-phenylenediamine dihydrochloride (TPMD, T01511G), methanol (A452-4), molecular grade isopropanol (BP2618-500), molecular grade HEPES (7365-45-9), and Tween20 (BP337500) were from Fisher. [U-14C]-glutamine (281.0 mCi/mmol) was from PerkinElmer. KCl (7447-40-7) was from Acros Organics and Nonidet P40 substitute (74385) was from Fluka BioChemika. Nonyl acridine orange (NAO, A1372) was purchased from Invitrogen. The D-glucose detection kit (K-GluHK-110A) was from Megazyme, Carbonyl cyanide-p-trifluoromethoxyphenylhydrazone (FCCP, 15218) and the L-lactate assay kit (700510) were from Cayman Chemical. 10% Precast tris-glycine gels (4561033), PVDF membranes (162-0177), thick blot filter paper (1703932), and clarity ECL substrate (1705062) were purchased from Bio-Rad. Anti-SQOR antibody (17256-1-AP) was from Proteintech; anti-MTC-O1 and anti-MT-CO2 antibodies (ab14705 and ab110258, respectively) were from Abcam. Anti-rabbit horseradish peroxidase–linked IgG and anti-mouse IgG, horseradish peroxidase–linked antibodies (NA944V and NA931, respectively) were from GE Healthcare. Polystyrene round-bottom tubes (5 mL) with cell-strainer caps used for FACS analysis were from BD Biosciences (720035).
Cell Culture.
HT29 and Panc-1 cells were maintained in RPMI 1640 medium. HEK293, RKO, LoVo, and Ea.hy926 cells were maintained in DMEM medium. Both RPMI and DMEM media were supplemented with 10% FBS along with 100 units/mL penicillin and 100 µg/mL streptomycin. All cells were maintained at 37 °C with ambient O2 and 5% CO2. A separate incubator was used for cells exposed to Na2S to avoid cross-contamination of control samples with the volatile metabolite. HT29scr, HT29SQOR KD, HT29NDUSF3 KD, and HT29SDHA KD cell lines were maintained in the same medium as the parent HT29 cells but with 1 µg/mL puromycin. Puromycin was removed from the culture medium for 2 to 3 d experiments to avoid potential off target effects of the additional antibiotic. HT29 cells expressing the cytosolic or mitochondrial LbNOX were cultured as described previously (24), with geneticin antibiotic (300 µg/mL) added for selection and 300 ng/mL doxycycline used for induction 24 h before the start of the experiment.
OCR Measurements.
All OCR measurements were performed using a respirometer (Oroboros Instruments Corp) at 37 °C with a stirring rate of 750 rpm. Cells were harvested from 10 cm plates (at ~70 to 90% confluency) washed with 1 × 8 mL PBS prior to digestion with 1 mL trypsin (0.05%) at 37 °C for 5 to 10 min. Cells were collected in 7 mL of the cell culture medium and centrifuged at 1600×g for 5 min, and pellets were resuspended in 1 mL modified PBS (MPBS) or DPBS + 5 mM glucose + 20 mM HEPES, pH 7.4. Cell suspensions were transferred to preweighed tubes and centrifuged at 1600×g for 3 min, and the supernatant was carefully aspirated with a 2-µL tip fixed to a vacuum line. The wet weight of the pellet was determined to prepare 5% (w/v) cell suspensions in MPBS, which were kept on ice and used for dilution to 0.75% or 1% (w/v) suspensions for OCR experiments. Experiments in which 0.75% (w/v) cell suspensions were used included those performed in the presence of rotenone (before Na2S treatment) or with LbNOX expressing cells and in experiments in which mitochondrial function was assessed.
Analysis of OCR traces was performed using DatLab v6 (Oroboros Instruments, Austria) and replotted in Origin 7.0. Recovery time following Na2S injection is defined as the time taken by cells to return to a new stationary basal OCR. Sulfide-dependent “% OCR inhibition” refers to the maximal drop in OCR following sulfide injection and is denoted as a percent of the starting basal OCR (Figs. 2B and 3E). “Fractional inhibition of OCR” refers to the change in basal OCR following recovery from sulfide-induced inhibition and is expressed as a fractional difference between the basal OCR at the start of the experiment and the new stationary basal OCR (SI Appendix, Fig. S1D and Fig. 4I). When calculating the ratio of O2 consumed per sulfide added, the contribution of nonmitochondrial respiration, which was determined to be 10% of the basal OCR in HT29 cells, was subtracted from the total O2 consumed before dividing by the concentration of sulfide added (5 to 35 µM). The ΔOCRmax represents the maximal change in OCR in response to 5 to 35 µM Na2S in rotenone-treated cells.
Mitochondrial Function Analysis.
The two chambers in the Oroboros instrument were filled with control versus 100 µM Na2S pretreated (4 h) HT29, HT29SQOR KD, or HT29scr cell suspensions (0.75%, 2 mL total volume). Once basal OCR stabilized, 1 µL of 250 µM oligomycin (125 nM final concentration) was injected into each chamber to inhibit ATP-linked respiration. After the OCR stabilized (~3 min), 1 µL of 250 µM FCCP (125 nM) was added to elicit maximal respiration, followed 2 to 3 min later by 0.5 µL of a combined stock of 2 mM rotenone + 2 mM antimycin A (0.5 µM each, final concentration). Working stocks of oligomycin (250 µM), FCCP (250 µM) and rotenone + antimycin A (2 mM each) were prepared in cell culture grade DMSO, aliquoted, stored at −20 °C, and thawed for single use.
Complex IV Activity Measurements Using the TMPD Assay.
Complex IV activity was measured in 1% (w/v) cell suspensions of control and Na2S pretreated (100 µM, 4 h) cells in the Oroboros instrument as described previously (46). Briefly, once basal OCR stabilized, complexes I and III were blocked with rotenone and antimycin A (0.5 µM each final concentration). After a new stable OCR was established, FCCP (1 µM) was added, followed after a few minutes by ascorbate (200 µM) and then N,N,N′,N′-tetramethyl-p-phenylenediamine dihydrochloride (TMPD, 80 µM). TMPD leads to a spike in the OCR, corresponding to complex IV activity, and was recorded over at least 2 min. The ascorbate stock solution (200 mM) was prepared by diluting a freshly prepared 800 mM stock solution whose pH was adjusted to ≈6 with 5 N HCl.
P50(O2) and Jmax(O2) Analysis.
Cell suspensions (1% (w/v)) were prepared as described above for all other OCR experiments. Temperature and stirrer settings were as noted (37 °C and 750 rpm). Cells were pretreated with 100 µM Na2S (4, 24 or 48 h) and Na2S (5–50 µM) was injected into the Oroboros chamber after stabilization of basal OCR (typically, 10–15 min). O2 concentration was recorded at 0.5 s time intervals until O2 in the chamber was depleted and for an additional 5 min to determine the flux at zero O2 for background correction. DatLab v2 (Oroboros Instruments) was used to estimate the P50(O2) (or apparent KM(O2)) and Jmax(O2) (or maximum flux) values as described (47, 48). Briefly, instrumental background parameters (zero intercept, a°, and slope, b°, of the background O2 flux as a function of O2 concentration) and time constants (τ, time delay due to diffusion of O2 through sensor membrane) for each chamber were determined experimentally according to the manufacturer’s instructions in the O2k-Manual. Time constants were determined via stirrer test and DatLab v2 macro TIMECONS, and O2 traces were recorded for 100% air calibration in MPBS at the start of each day when experiments were run. DatLab v2 macro P50 was used to estimate P50(O2) and Jmax(O2) values. Volume-specific OCR (oxygen flux, JO2) was calculated as the negative slope of the recorded oxygen concentration. After interpolation of zero flux, the macro iteratively fits the data to a hyperbolic equation given by
where JO2 = volume-specific OCR and PO2 = [O2] in kPa. Oxygen partial pressure used for the fit was 0 to 1.1 kPa, and oxygen solubility was 10.56 µM/kPa.
IC50 for H2S.
The IC50 for H2S was determined with 1% (w/v) HT29SQOR KD or HT29scr cell suspensions in MPBS in the Oroboros instrument. After the basal OCR stabilized at 37 °C, Na2S (30 to 150 µM for HT29scr and 5 to 100 µM for HT29SQOR KD) was injected from a freshly prepared 10 mM stock solution. Sulfide-dependent % OCR inhibition was estimated by subtracting the lowest recorded OCR value from the basal OCR value and expressed as a percentage of the starting basal OCR. From the dependence of % OCR inhibition on H2S concentration, the IC50 was obtained by fitting the data to a four-parameter sigmoidal equation given by
where “y” represents ΔOCR, “a” is the lowest value of ΔOCR, “b” is the highest value of ΔOCR, and “n” is the Hill coefficient.
Western Blot Analysis.
SQOR.
HT29SQOR KD and HT29scr were seeded at 500,000 cells per well in 6-well plates and cultured for 24 to 48 h before changing to fresh RPMI medium ± 100 µM Na2S for 4, 24, or 48 h. Cells that would be harvested after 4 h were seeded reach ~70% confluency while cells that would be harvested after 24 to 48 h were seeded to reach ~50% confluency before sulfide treatment. Cells were washed with 2 × 2 mL PBS/well followed by addition of 0.5 mL 0.05% trypsin/well for 5 min at 37 °C and harvested in 1 mL RPMI medium. Cell suspensions were centrifuged at 1600×g for 5 min, and the pellet was washed once with 1 mL ice-cold PBS and resuspended in 200 µL RIPA lysis buffer + protease inhibitor and saved at −80 °C until lysis. For SQOR detection in different cell lines, a similar procedure was used but each cell line was cultured in 10 cm plates and grown to confluency. Cells were washed with 8 mL PBS/plate followed by addition of 1 mL 0.05% trypsin for 5 to 10 min and harvested in 7 mL RPMI. Cells were centrifuged at 1600×g for 5 min; the pellet was washed with 1 × 1 mL ice-cold PBS and resuspended in 300 µL nondenaturing lysis buffer with protease inhibitor (0.5% (v/v), Nonidet P40 substitute, 25 mM KCl, 20 mM HEPES, pH 7.4) with the exception of Ea.hy926 cells, which were suspended in 100 µL of lysis buffer, due to the smaller pellet size.
MT-CO1 and MT-CO2.
Cells (2 million per 6 cm dish) were grown for 36 h before changing to fresh RPMI medium ± 100 µM Na2S. After 4 h, the cells were harvested as described above, and the pellet was resuspended in 400 µL nondenaturing Nonidet P40 lysis buffer with protease inhibitor.
Development of western blots.
Frozen cell pellets were lysed by three freeze–thaw cycles and centrifuged at 13,000×g for 10 min. Protein content in the supernatant was determined using the Bradford reagent (Bio-Rad). Cell lysates (50 µg) were loaded on precast Bio-Rad 10% tris-glycine gels and separated by SDS-PAGE, transferred to PVDF membranes, and blocked for 1 h with 5% milk in Tris-buffered saline with 0.3% Tween 20 (TBST). Membranes were incubated overnight in TBST, 5% (w/v) milk containing diluted SQOR (1:5,000) or MTCO1 or MTCO2 (1:1,000) antibodies then washed quickly with 2 × 5 mL TBST followed by 5 × 10 min washes with 10 to 15 mL TBST. Then, the membranes were exposed for 2 h to the secondary antibody [horseradish peroxidase–linked anti-rabbit (for anti-SQOR) or anti-mouse IgG (for MTCO-1 and MTCO-2)] used at a 1:10,000 dilution in TBST, 5% milk. The membranes were washed quickly with 2 × 5 mL TBST followed by 5 × 10 min 10 to 15 mL TBST washes, before rinsing quickly with 2 × 10 mL TBS before treating with clarity ECL substrate (Bio-Rad). Signals were detected using a Bio-Rad ChemiDoc Imaging System. All images were exported as 16-bit TIF files and imported into Fiji (49) for semiquantitative analysis, which was performed by drawing equal-sized rectangles over each band to estimate its mean pixel intensity. The background was subtracted from each band.
Cardiolipin Analysis.
Mitochondrial content was estimated by staining for cardiolipin with nonyl acridine orange (NAO) as described previously (19). Briefly, HT29 cells were seeded at 700,000 cells/well in 6-well plates, and the medium was changed after 24 h before ± 100 µM Na2S treatment. After 24 h, cells were washed once with and then replaced by phenol red free RPMI and treated ±100 nM NAO for 30 min. Cells were then washed twice with 1 mL ice-cold PBS, treated with 0.5 mL phenol red free trypsin EDTA (0.05%) for 5 min, and harvested with 1 mL phenol red free RPMI. Cells were centrifuged at 700×g for 5 min, and the pellet was resuspended in 750 µL ice-cold PBS by pipetting and then filtered using 5-mL BD round-bottom falcon tubes with cell-strainer caps. FACS analysis was performed on the Bio-Rad Ze5 multilaser, high-speed cell analyzer operated with the Everest software package at the University of Michigan Flow Cytometry Core Facility. Data were analyzed using FlowJo (v10.8.1) for median fluorescence (488 excitation 525/50 emission). The median unstained fluorescence was subtracted from the value for the stained sample.
Metabolite Analyses.
Changes in glucose consumption and lactate production kinetics were measured in 5% (w/v) cell suspensions 24 h after a single bolus treatment with 25-300 µM Na2S exactly as described previously (25). Incorporation of [U-14C]-glutamine into lipids was measured 13 h after a single bolus treatment with 100 or 300 µM Na2S as described (24). H2S and thiosulfate concentrations in the conditioned medium were monitored 4 h after a single bolus treatment with 100 µM Na2S following derivatization with monobromobimane as previously described (26). Metabolomics analysis was performed on HT29 cells treated ± 100 µM Na2S for 24 h, as described previously (19).
Statistics.
Statistical analysis for the indicated pairwise comparisons of P50(O2), Jmax(O2), and protein abundance was performed using a two-sample unpaired t test. For complex IV activity, parameters of mitochondrial function, glucose consumption, lactate production, and enhanced radioactive incorporation into lipids, paired t tests were employed. Any other method used for statistical analysis is mentioned in the respective figure legend.
Supplementary Material
Appendix 01 (PDF)
Dataset S1 (XLSX)
Acknowledgments
This work was supported in part by grants from the NIH (GM130183 to R.B., R01CA248160 to C.L., and F32GM140694 to D.A.H.) and the Michigan Life Sciences Fellow Program (to J.D.).
Author contributions
D.A.H., J.D., A.G., R.K., and R.B. designed research; D.A.H., J.D., A.G., R.K., and A.A. performed research; C.L. contributed new reagents/analytic tools; D.A.H., J.D., A.G., R.K., A.A., C.L., and R.B. analyzed data; and D.A.H., J.D., A.G., R.K., and R.B. wrote the paper.
Competing interests
C.L. has received consulting fees from Astellas Pharmaceuticals, Odyssey Therapeutics, and T-Knife Therapeutics. C.L. is an inventor on patents pertaining to Kras regulated metabolic pathways, redox control pathways in pancreatic cancer, and targeting the GOT1-pathway as a therapeutic approach (US Patent No: 2015126580-A1, 05/07/2015; US Patent No: 20190136238, 05/09/2019; and International Patent No: WO2013177426-A2, 04/23/2015).
Footnotes
This article is a PNAS Direct Submission.
Data, Materials, and Software Availability
All study data are included in the article and/or SI Appendix.
Supporting Information
References
- 1.Morrison M. L., et al. , Surviving blood loss using hydrogen sulfide. J. Trauma 65, 183–188 (2008). [DOI] [PubMed] [Google Scholar]
- 2.Yenari M. A., Han H. S., Neuroprotective mechanisms of hypothermia in brain ischaemia. Nat. Rev. Neurosci. 13, 267–278 (2012). [DOI] [PubMed] [Google Scholar]
- 3.Abe K., Kimura H., The possible role of hydrogen sulfide as an endogenous neuromodulator. J. Neurosci. 16, 1066–1071 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hanna D., Kumar R., Banerjee R., A metabolic paradigm for hydrogen sulfide signaling via electron transport chain plasticity. Antioxid. Redox Signal. 38, 57–67 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kumar R., Banerjee R., Regulation of the redox metabolome and thiol proteome by hydrogen sulfide. Crit. Rev. Biochem. Mol. Biol. 56, 221–235 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Blackstone E., Morrison M., Roth M. B., H2S induces a suspended animation-like state in mice. Science 308, 518 (2005). [DOI] [PubMed] [Google Scholar]
- 7.Filipovic M. R., Zivanovic J., Alvarez B., Banerjee R., Chemical biology of H2S signaling through persulfidation. Chem. Rev. 118, 1253–1337 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Elrod J. W., et al. , Hydrogen sulfide attenuates myocardial ischemia-reperfusion injury by preservation of mitochondrial function. Proc. Natl. Acad. Sci. U.S.A. 104, 15560–15565 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Marutani E., et al. , Sulfide catabolism ameliorates hypoxic brain injury. Nat. Commun. 12, 3108 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Linden D. R., Hydrogen sulfide signaling in the gastrointestinal tract. Antioxid. Redox Signal. 20, 818–830 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li Z., et al. , Mitochondrial H(2)S regulates BCAA catabolism in heart failure. Circ. Res. 131, 222–235 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Furne J., Saeed A., Levitt M. D., Whole tissue hydrogen sulfide concentrations are orders of magnitude lower than presently accepted values. Am. J. Physiol. Regul. Integr. Comp. Physiol. 295, R1479–R1485 (2008). [DOI] [PubMed] [Google Scholar]
- 13.Levitt M. D., Abdel-Rehim M. S., Furne J., Free and acid-labile hydrogen sulfide concentrations in mouse tissues: Anomalously high free hydrogen sulfide in aortic tissue. Antioxid. Redox Signal. 15, 373–378 (2011). [DOI] [PubMed] [Google Scholar]
- 14.Vitvitsky V., Kabil O., Banerjee R., High turnover rates for hydrogen sulfide allow for rapid regulation of its tissue concentrations. Antioxid. Redox Signal. 17, 22–31 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Deplancke B., et al. , Gastrointestinal and microbial responses to sulfate-supplemented drinking water in mice. Exp. Biol. Med. (Maywood) 228, 424–433 (2003). [DOI] [PubMed] [Google Scholar]
- 16.Macfarlane G. T., Gibson G. R., Cummings J. H., Comparison of fermentation reactions in different regions of the human colon. J. Appl. Bacteriol. 72, 57–64 (1992). [DOI] [PubMed] [Google Scholar]
- 17.Landry A. P., Ballou D. P., Banerjee R., Hydrogen sulfide oxidation by sulfide quinone oxidoreductase. ChemBioChem 22, 949–960 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Landry A. P., Roman J., Banerjee R., Structural perspectives on H2S homeostasis. Curr. Opin. Struct. Biol. 71, 27–35 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Libiad M., et al. , Hydrogen sulfide perturbs mitochondrial bioenergetics and triggers metabolic reprogramming in colon cells. J. Biol. Chem. 294, 12077–12090 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Landry A. P., Ballou D. P., Banerjee R., H2S oxidation by nanodisc-embedded human sulfide quinone oxidoreductase. J. Biol. Chem. 292, 11641–11649 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Landry A. P., et al. , A catalytic trisulfide in human sulfide quinone oxidoreductase catalyzes coenzyme A persulfide synthesis and inhibits butyrate oxidation. Cell Chem. Biol. 26, 1515–1525.e1514 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Goubern M., Andriamihaja M., Nubel T., Blachier F., Bouillaud F., Sulfide, the first inorganic substrate for human cells. FASEB J. 21, 1699–1706 (2007). [DOI] [PubMed] [Google Scholar]
- 23.Nicholls P., Kim J. K., Sulphide as an inhibitor and electron donor for the cytochrome c oxidase system. Can. J. Biochem. 60, 613–623 (1982). [DOI] [PubMed] [Google Scholar]
- 24.Carballal S., et al. , Hydrogen sulfide stimulates lipid biogenesis from glutamine that is dependent on the mitochondrial NAD(P)H pool. J. Biol. Chem. 297, 100950 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vitvitsky V., et al. , The mitochondrial NADH pool is involved in hydrogen sulfide signaling and stimulation of aerobic glycolysis. J. Biol. Chem. 296, 100736 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kumar R., et al. , A redox cycle with complex II prioritizes sulfide quinone oxidoreductase-dependent H2S oxidation. J. Biol. Chem. 298, 101435 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Blackstone E., Roth M. B., Suspended animation-like state protects mice from lethal hypoxia. Shock 27, 370–372 (2007). [DOI] [PubMed] [Google Scholar]
- 28.Cooper C. E., Brown G. C., The inhibition of mitochondrial cytochrome oxidase by the gases carbon monoxide, nitric oxide, hydrogen cyanide and hydrogen sulfide: Chemical mechanism and physiological significance. J. Bioenerg. Biomembr. 40, 533–539 (2008). [DOI] [PubMed] [Google Scholar]
- 29.Petersen L. C., The effect of inhibitors on the oxygen kinetics of cytochrome c oxidase. Biochim. Biophys. Acta 460, 299–307 (1977). [DOI] [PubMed] [Google Scholar]
- 30.Titov D. V., et al. , Complementation of mitochondrial electron transport chain by manipulation of the NAD+/NADH ratio. Science 352, 231–235 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Volpato G. P., et al. , Inhaled hydrogen sulfide: A rapidly reversible inhibitor of cardiac and metabolic function in the mouse. Anesthesiology 108, 659–668 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Collman J. P., Ghosh S., Dey A., Decreau R. A., Using a functional enzyme model to understand the chemistry behind hydrogen sulfide induced hibernation. Proc. Natl. Acad. Sci. U.S.A. 106, 22090–22095 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Manford A. G., et al. , A cellular mechanism to detect and alleviate reductive stress. Cell 183, 46–61.e21 (2020). [DOI] [PubMed] [Google Scholar]
- 34.Banerjee R., Kumar R., Gas regulation of complex II reversal via electron shunting to fumarate in the mammalian ETC. Trends Biochem. Sci. 47, 689–698 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nicholls P., Marshall D. C., Cooper C. E., Wilson M. T., Sulfide inhibition of and metabolism by cytochrome c oxidase. Biochem. Soc. Trans. 41, 1312–1316 (2013). [DOI] [PubMed] [Google Scholar]
- 36.Bostelaar T., et al. , Hydrogen sulfide oxidation by myoglobin. J. Am. Chem. Soc. 138, 8476–8488 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Vitvitsky V., et al. , Structural and mechanistic insights into hemoglobin-catalyzed hydrogen sulfide oxidation and the fate of polysulfide products. J. Biol. Chem. 292, 5584–5592 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Vitvitsky V., Yadav P. K., Kurthen A., Banerjee R., Sulfide oxidation by a noncanonical pathway in red blood cells generates thiosulfate and polysulfides. J. Biol. Chem. 290, 8310–8320 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ruetz M., et al. , A distal ligand mutes the interaction of hydrogen sulfide with human neuroglobin. J. Biol. Chem. 292, 6512–6528 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hill B. C., et al. , Interactions of sulphide and other ligands with cytochrome c oxidase. An electron-paramagnetic-resonance study. Biochem. J. 224, 591–600 (1984). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chouchani E. T., et al. , Ischaemic accumulation of succinate controls reperfusion injury through mitochondrial ROS. Nature 515, 431–435 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lagoutte E., et al. , Oxidation of hydrogen sulfide remains a priority in mammalian cells and causes reverse electron transfer in colonocytes. Biochim. Biophys. Acta 1797, 1500–1511 (2010). [DOI] [PubMed] [Google Scholar]
- 43.Friederich M. W., et al. , Pathogenic variants in SQOR encoding sulfide:quinone oxidoreductase are a potentially treatable cause of Leigh disease. J. Inherit. Metab. Dis. 43, 1024–1036 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tiranti V., et al. , Loss of ETHE1, a mitochondrial dioxygenase, causes fatal sulfide toxicity in ethylmalonic encephalopathy. Nat. Med. 15, 200–205 (2009). [DOI] [PubMed] [Google Scholar]
- 45.Di Meo I., et al. , Chronic exposure to sulfide causes accelerated degradation of cytochrome c oxidase in ethylmalonic encephalopathy. Antioxid. Redox Signal. 15, 353–362 (2011). [DOI] [PubMed] [Google Scholar]
- 46.Brown G. C., Foxwell N., Moncada S., Transcellular regulation of cell respiration by nitric oxide generated by activated macrophages. FEBS Lett. 439, 321–324 (1998). [DOI] [PubMed] [Google Scholar]
- 47.Gnaiger E., Bioenergetics at low oxygen: Dependence of respiration and phosphorylation on oxygen and adenosine diphosphate supply. Respir. Physiol. 128, 277–297 (2001). [DOI] [PubMed] [Google Scholar]
- 48.Gnaiger E., Steinlechner-Maran R., Mendez G., Eberl T., Margreiter R., Control of mitochondrial and cellular respiration by oxygen. J. Bioenerg. Biomembr. 27, 583–596 (1995). [DOI] [PubMed] [Google Scholar]
- 49.Schindelin J., et al. , Fiji: An open-source platform for biological-image analysis. Nat. Methods 9, 676–682 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix 01 (PDF)
Dataset S1 (XLSX)
Data Availability Statement
All study data are included in the article and/or SI Appendix.