Abstract
The intracellular assembly of the transmissible gastroenteritis coronavirus (TGEV) was studied in infected swine testis (ST) cells at different postinfection times by using ultrathin sections of conventionally embedded infected cells, freeze-substitution, and methods for detecting viral proteins and RNA at the electron microscopy level. This ultrastructural analysis was focused on the identification of the different viral components that assemble in infected cells, in particular the spherical, potentially icosahedral internal core, a new structural element of the extracellular infectious coronavirus recently characterized by our group. Typical budding profiles and two types of virion-related particles were detected in TGEV-infected cells. While large virions with an electron-dense internal periphery and a clear central area are abundant at perinuclear regions, smaller viral particles, with the characteristic morphology of extracellular virions (exhibiting compact internal cores with polygonal contours) accumulate inside secretory vesicles that reach the plasma membrane. The two types of virions coexist in the Golgi complex of infected ST cells. In nocodazole-treated infected cells, the two types of virions coexist in altered Golgi stacks, while the large secretory vesicles filled with virions found in normal infections are not detected in this case. Treatment of infected cells with the Golgi complex-disrupting agent brefeldin A induced the accumulation of large virions in the cisternae that form by fusion of different membranous compartments. These data, together with the distribution of both types of virions in different cellular compartments, strongly suggest that the large virions are the precursors of the small viral particles and that their transport through a functional Golgi complex is necessary for viral maturation.
Coronaviruses are a group of large, enveloped RNA viruses involved in a number of serious diseases that affect mammals and birds (15, 43). These viruses have a simple protein composition. Basically, four to five protein species make up the infectious viral particle, with some variation in the different members of the family: S and M glycoproteins are inserted in the viral envelope (36, 56), as well as the small membrane E protein, a minor component recently identified (20, 41, 67). Some members of the Coronaviridae family contain an additional envelope protein, the hemagglutinin esterase (HE) (32, 57). The nucleocapsid (N) protein is located inside the virion and forms a complex with the viral RNA (29). Molecules of N protein also seem to be part of the internal core shell recently found in coronaviruses (52).
Morphogenesis of coronaviruses takes place in the cytoplasm of the infected cell, with the participation of different membranous compartments, as established from the early ultrastructural studies of coronavirus-infected cells (2, 12–14, 24). Early in infection, shortly after the appearance of M and S proteins, the first progeny virions are seen by electron microscopy (EM) in the perinuclear region of the cell (62–64). Viral morphogenesis starts with the assembly of a helical nucleocapsid, made up of viral RNA and the nucleocapsid N protein (13, 58, 59). It is not known whether this nucleocapsid is preformed within the cytoplasm and rapidly transported to the membranes where assembly occurs or whether it is assembled in association with cellular membranes. The nucleocapsid associates with smooth membranes belonging to the endoplasmic reticulum-Golgi intermediate compartment (ERGIC) that constitute the “budding compartment” for coronaviruses (35). This compartment, which ultrastructurally resembles the transitional elements at the rough endoplasmic reticulum now identified in a variety of cells (23), contains both perinuclear and peripheral elements (55). A structural analysis of cells infected with infectious bronchitis virus, transmissible gastroenteritis coronavirus (TGEV), or feline infectious peritonitis virus revealed that these viruses also use the smooth perinuclear membranes (33), although some budding in the Golgi complex has been occasionally detected in mouse hepatitis virus (MHV)-infected cells. Although molecular details of the budding process have not yet been elucidated, it has been demonstrated that the M protein, which is essential for assembly, accumulates locally in the ERGIC membranes and probably reaches a concentration high enough to be recognized by the nucleocapsids (53). This association would trigger the budding process, which most probably involves the collective incorporation of all other viral envelope proteins. The minor envelope E protein, which is not fully characterized, also plays a crucial role in assembly (6), although its nature has not been yet elucidated. This budding process gives rise to virions that are transported to and through the Golgi complex by vesicular carriers (35). In the Golgi complex, the viral particles are usually seen only at the rims of the stacks and at the trans side, and the virions are collected into vesicles of the constitutive exocytic pathway and released from the cell. Tooze et al. (63) described the existence of two different morphologies for MHV-A59 viral particles in infected AtT20 cells. Virions in the trans-Golgi network were often kidney shaped and had a fairly uniform and high electron density, while virions in the Golgi cisternae and pre-Golgi compartments were spherical and exhibited an “empty” center. Once released from the cell, coronavirions tend to attach to the plasma membrane, forming dense mats of adsorbed virions.
Our group has recently identified a new structural element in purified virions from two coronaviruses, TGEV and MHV (52). An internal core, potentially icosahedral, that contains the helical nucleocapsid, was visualized in intact virions, and after a purification procedure for the cores was established, their structure and protein composition were characterized (52). Since the internal core shell represents a new structural element in the sequence of coronavirus assembly, its formation in infected cells must involve some as yet undefined steps in coronavirus morphogenesis that need to be revisited. In the last 10 years, methods that preserve cellular and viral ultrastructure and that detect macromolecules at the EM level have been developed and proving to be very useful tools in understanding viral structure and morphogenesis (5, 9, 21, 22, 26, 27, 49). We have studied the morphogenesis of the TGEV and started to define the different types of viral particles that assemble inside infected cells, as well as the cellular elements potentially involved in viral maturation.
MATERIALS AND METHODS
Cells, viruses, and antibodies.
TGEV virions (PUR46-MAD strain) were purified from infected cultures of swine testis (ST) cells. Cell culture, virus infections, and purification of TGEV were performed as previously described (28). Monolayers of ST cells were infected with TGEV at a multiplicity of infection of 10 PFU/cell and collected at different times (1, 2, 3, 4, 5, 6, 8, 10, 12, and 24 h) after fixation for EM (see below). The murine monoclonal antibodies specific for TGEV proteins have been described previously (18, 51, 54). Hyperimmune serum against the PUR46-MAD strain of TGEV was raised in rabbits (28). Colloidal gold conjugates of goat anti-mouse and goat anti-rabbit immunoglobulin G were provided by Biocell Research Laboratory (Cardiff, United Kingdom).
Electron microscopy. (i) Processing of infected ST cells for embedding in EML-812.
Monolayers of control ST cells and cells infected with TGEV (at the postinfection (p.i.) times indicated in the previous paragraph) were fixed in situ with a mixture of 2% glutaraldehyde and 2% tannic acid in 0.4 M HEPES buffer (pH 7.5) for 1 h at room temperature. Fixed monolayers were removed from the dishes in the fixative and transferred to Eppendorf tubes. After centrifugation in a microcentrifuge and washes with HEPES buffer, the pellets were processed for embedding in EML-812 (EML Laboratories, Berks, United Kingdom), an epoxy resin adequate for ultrastructural studies, by methods previously established (48) with some modifications, such as two postfixation steps to stabilize the lipids and short dehydration at 4°C to minimize the extraction of soluble components. Postfixation of the cells was done with a mixture of 1% osmium tetroxide and 0.8% potassium ferricyanide in distilled water for 1 h at 4°C. This mixture has been reported to provide high levels of preservation and contrast of membranous organelles (45, 48). After washes with HEPES buffer, samples were treated with 2% uranyl acetate, which protects membranes from extraction during dehydration (22), washed again, and dehydrated in increasing concentrations of acetone (50, 70, 90, and 100%, 10 min each, at 4°C), and EML-812 was allowed to infiltrate the cells at room temperature. Polymerization of the infiltrated samples was done at 60°C for 3 days. Ultrathin (20- to 30-nm-thick) sections of the samples were stained with saturated uranyl acetate and lead citrate by standard procedures. For RNA detection, infected cells were subjected to a milder fixation (1% glutaraldehyde in phosphate-buffered saline [PBS]) before being embedded in EML-812. No postfixation was applied in this case. All samples were studied with a JEOL 1200 EX II electron microscope.
(ii) Treatment of infected ST cells with nocodazol and BFA.
Confluent monolayers of ST cells were infected with TGEV at a multiplicity of infection of 10 PFU/cell, and at 4 h p.i., cultures were treated with the microtubular disrupting agent nocodazole (Sigma) at different concentrations (1, 3, 10, and 20 μM) and incubated for 4 h at 37°C. Cells were then fixed in situ with a mixture of 2% glutaraldehyde and 2% tannic acid in HEPES buffer, washed, and kept at 4°C until use. Other cultures were treated for 2 or 4 h with the Golgi complex-disrupting agent brefeldin A (BFA) (Sigma), which was added to the cultures at 4 h p.i. and at a concentration of 1, 5, or 10 μg/ml. Some of these cultures were incubated for 2 or 4 h more after BFA was removed. Cells were then fixed with glutaraldehyde-tannic acid or maintained at 37°C for 2 or 4 h after replacing the culture medium by a fresh one without the drugs. At the times indicated, cells were fixed and processed for embedding in EML-812, as described above.
(iii) Quick-freezing and freeze-substitution of fixed infected ST cells.
Small pellets of fixed cells were cryoprotected with glycerol, applied on small pieces of filter paper (1 mm2), blotted for 15 s, and quick-frozen in liquid propane at an approximate speed of 104°C/s. Vitrified specimens were stored in liquid nitrogen until use. For freeze-substitution, samples were transferred to a Reichert Jung AFS freeze-substitution unit (Leica, Vienna, Austria), and maintained for 24 h at −90°C in a mixture of pure acetone and 0.5% (wt/vol) osmium tetroxide for a complete substitution of the water of the sample by established procedures (22, 46). After a controlled gradual increase of the temperature, the cell samples were infiltrated with the epoxy resin EML-812 and polymerized. Ultrathin sections were either stained with uranyl acetate and lead citrate or processed for immunodetection of viral proteins.
(iv) Detection of viral proteins and RNA.
Immunogold labeling of TGEV proteins was done on ultrathin EML-812 sections of infected ST cells by using monoclonal or polyclonal antibodies and conjugates of secondary antibodies and 5- or 10-nm-diameter colloidal gold particles, according to general procedures previously described in detail (49–51). RNA from cellular and viral structures was detected as previously described (3, 4) by using a conjugate of RNase and colloidal gold with a diameter of 10 nm provided by EY Laboratories (San Diego, Calif.). Sections were incubated with the RNase-gold conjugate diluted 1:40 in PBS (pH 7.5) for 30 min at room temperature, washed with PBS and distilled water, and stained with uranyl acetate and lead citrate. For a cytochemical control, some sections were preincubated with a solution of nonconjugated RNase (20 μg/ml) before treating with the RNase-gold conjugate.
(v) Quantitative studies.
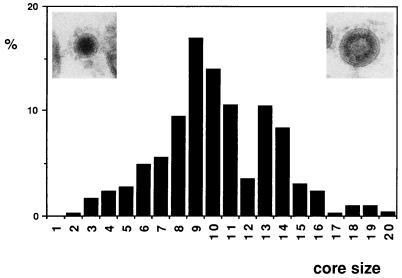
The size of the viral cores (the dense internal component) was measured from electron micrographs of freeze-substituted infected ST monolayers (6 h p.i.) (see Fig. 4). The distribution of the different types of virions in different cellular compartments (perinuclear area, Golgi complex, large secretory vesicles, and extracellular cell surface) at different p.i. times was also studied. The results are presented as the percentage of each viral morphology in the indicated compartment (see Table 1). The effect of nocodazol and BFA treatments on the intracellular accumulation of the different types of virions was also studied, and the results are summarized in Table 2. A total of 3,500 virions were included in these quantitative studies.
FIG. 4.
Size distribution of TGEV cores in viral particles from ultrathin sections of TGEV-infected cells. Only intracellular virions from freeze-substituted samples were included in the quantification. The percentage of viral cores with a defined diameter is represented. Core size (diameter [in nanometers]) classes were established as follows: 1, <36; 2, >36 to 38; 3, >38 to 41; 4, >41 to 44; 5, >44 to 47; 6, >47 to 50; 7, >50 to 53; 8, >53 to 56; 9, >56 to 59; 10, >59 to 62; 11, >62 to 65; 12, >65 to 68; 13, >68 to 71; 14, >71 to 74; 15, >74 to 77; 16, >77 to 80; 17, >80 to 83; 18, >83 to 86; 19, >86 to 89; 20, >89. Virions belonging to the two extremes of the core size distribution are shown in the two micrographs: the small virion on the left corresponds to class 8, while the large virion on the right corresponds to class 19. A total of 505 virions were included in the measurements.
TABLE 1.
Distribution of large and small virions in different compartments of infected ST cellsa
| Time or treatment | Virion typeb | % of virions in compartment
|
|||
|---|---|---|---|---|---|
| Perinuclear | Golgi complex | Secretory vesicles | Extracellularc | ||
| Time (h p.i.) | |||||
| 5 | LV | 84 | 51 | 34 | 12 |
| SV | 16 | 49 | 66 | 88 | |
| 6 | LV | 83 | 45 | 14 | 8 |
| SV | 17 | 55 | 86 | 92 | |
| 8 | LV | 85 | 46 | 34 | 5 |
| SV | 15 | 54 | 66 | 95 | |
| 10 | LV | 84 | 37 | 9 | 2 |
| SV | 16 | 63 | 91 | 98 | |
| 24 | LV | 56 | 33 | 11 | 5 |
| SV | 44 | 67 | 89 | 95 | |
| NOC (8 h p.i.)d | LV | 82 | 62 | ||
| SV | 18 | 38 | |||
A total of 2,400 virions (at least 100 per compartment and treatment) were included in this quantification.
LV, large virions; SV, small virions.
Virions attached to the cell surface.
Infected cells (4 h p.i.) were incubated for 4 h in the presence of 20 μM nocodazole (NOC).
TABLE 2.
Percentages of large and small virions in infected ST cells at different times and with different treatmentsa
| Time or treatmentb | % of virionsc
|
BPd | |
|---|---|---|---|
| LV | SV | ||
| Time (h p.i.) | |||
| 5 | 49 | 51 | 5 |
| 6 | 31 | 69 | 5 |
| 8 | 28 | 72 | 3 |
| 10 | 27 | 73 | 6 |
| 24 | 18 | 82 | 1 |
| Treatment | |||
| NOC (8 h p.i.) | 61 | 39 | 5 |
| BFA (8 h p.i.) | 88 | 12 | 22 |
A total of 2,995 virions (at least 400 per treatment) were included in this quantification. For details of the different treatments, see the text.
Infected cells (4 h p.i.) were incubated for 4 h in the presence of either 20 μM nocodazole (NOC) or 5 μg of BFA per ml.
LV, large virions; SV, small virions.
Number of budding profiles (BP) per 10 infected cells.
RESULTS
Different TGEV assemblies in infected ST cells.
ST cells infected with TGEV were studied at different p.i. times to understand the early events in morphogenesis. At 4 h p.i., the earliest p.i. time rendering sufficient amounts of virions to be studied by ultrathin sections and EM, very few virions accumulate inside infected cells. At 5 and 6 h p.i., a progressive accumulation of intracellular viral particles takes place, although the ultrastructure of the cells is still indistinguishable from that of noninfected cells. At 8 and 10 h p.i., large amounts of intracellular and extracellular virions can be seen. At longer times (24 h), large secretory vesicles with many viruses and many extracellular virions are seen, together with cellular alterations and cytoplasmic inclusions typical of late p.i. times (12, 61). Five and 6 h p.i. were then selected as the best times to analyze the early assembly events, well before the overproduction of viral components and viral particles makes interpretation more complicated.
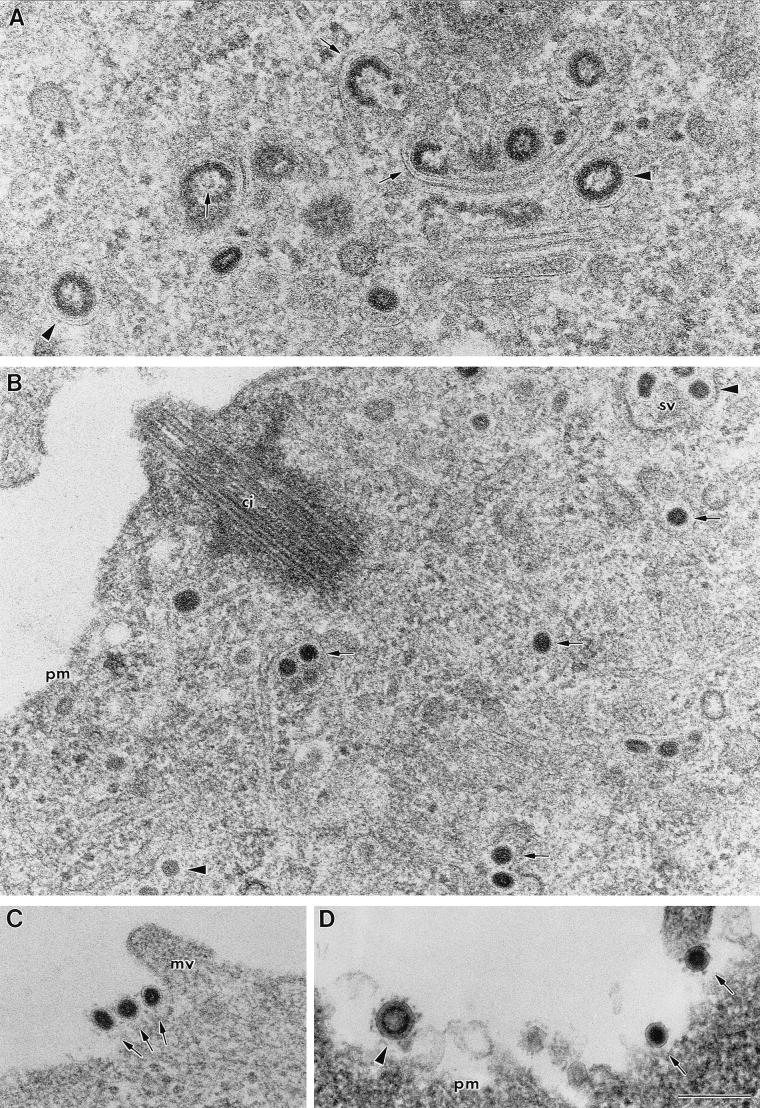
Our analysis of the different viral assemblies that form in infected cells has been done with ultrathin sections of conventionally embedded infected cells and cells processed by freeze-substitution, a method that allows a high level of preservation of fine structural details. Certain fine details, such as the shape and size of cellular organelles, are better preserved by freeze-substitution, although conventional embedding methods provide better contrast of cellular membranes. This combination of data showed the existence of virions of two different sizes and morphologies, both in freeze-substituted and conventionally embedded samples (Fig. 1). The earliest viral assembly distinguished inside infected cells consisted of characteristic budding profiles or “maturation arcs” (Fig. 1A). The budding profiles give rise to large virions that exhibit an electron-dense internal periphery and a less dense central zone (Fig. 1A). These virions, together with the budding profiles, are more abundant in the perinuclear area of the cell but are also found in smaller amounts at other locations. A different type of viral particle accumulates in regions closer to the plasma membrane. These particles are smaller and contain a dense core (Fig. 1B). The structure and dimensions of the internal core of these virions (diameter of around 60 nm) are identical to the corresponding cores found in extracellular viral particles (Fig. 1C). Only a small amount of large viral particles exit the cells (Fig. 1D); these virions constitute a minor component of the extracellular population of virions (see below).
FIG. 1.
TGEV virions in different regions of infected ST cells. Sections of freeze-substituted cells (A to C) and a conventionally embedded cell (D) are shown. (A) Certain areas of the infected cell, mainly close to the nucleus, accumulate characteristic budding profiles (arrows) and large virions with an internal electron-dense periphery and clear center (arrowheads). (B) Closer to the plasma membrane, virions exhibit a smaller size and a homogeneously dense internal core (arrows). Some of these viral cores clearly exhibit polygonal contours (arrowheads). (C and D) Most of the extracellular virions are small particles with dense cores (arrows), although a few large viral particles are also seen in the extracellular environment (arrowhead in panel D). Abbreviations: sv, secretory vesicle; ci, cilium; pm, plasma membrane; mv, microvilli. Bar, 200 nm.
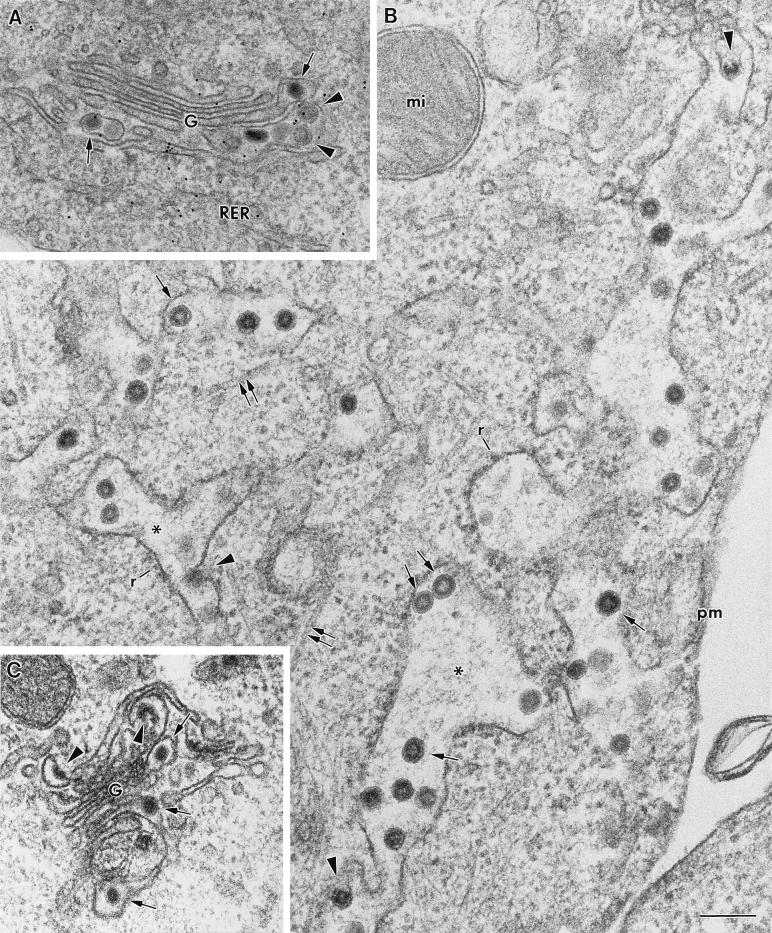
Figure 2 is a summary of the different TGEV-related assemblies detected in infected ST cells. Budding profiles contain a dense cytoplasmic element (presumably the helical nucleocapsid) and exhibit different stages from flat forms (Fig. 2A) to curved structures (Fig. 2B), some with cytoplasmic “tails” (Fig. 2C), to almost spherical (Fig. 2D). Large spherical viral particles originate from the budding profiles (Fig. 2E and F). On the other hand, smaller viral particles contain homogeneously dense cores (Fig. 2G and H), characteristic of extracellular virions. We obtained the first evidence of a potentially icosahedral internal core in the small virions with freeze-substituted cells, since a polygonal periphery is distinguished in some of the sectioned particles (Fig. 1B and 2G and H). All the described viral assemblies were unequivocally identified as related to TGEV, since they all reacted with polyclonal anti-TGEV antibodies (Fig. 2I and J), and they were always associated with the membrane. Several monoclonal antibodies specific for the TGEV M, N, and S proteins also reacted with the two different types of viral particles (not shown).
FIG. 2.
Collection of images corresponding to the different TGEV-related assemblies detected in infected ST cells. Sections of conventionally embedded cells (A, C, D, and I) and fixed, freeze-substituted cells (B, E, F, G, H, and J) are shown. (A to D) Different stages of the budding profiles. All stages present a dense cytoplasmic structure (arrows) interacting with smooth-walled membranes (arrowheads) and a variable curvature, from flat (A) to almost spherical (D). In panel C, part of the dense material is apparently not interacting with the membrane and looks like a cytoplasmic tail (arrow). Budding profiles originate large spherical viral particles with an electron-dense internal periphery and a clear center (E and F). Smaller virions have an internal dense core that frequently exhibit polygonal contours (arrows in panels G and H). Budding profiles (arrow in panel I) and both large and small viral particles (I and J) react with polyclonal anti-TGEV antibodies, as shown by immunogold labeling. Bar, 100 nm.
Large and small viral particles also bound a conjugate of RNase-gold (Fig. 3), used to detect RNA-containing structures at the EM level (3, 4). This conjugate reacted with RNA-containing cellular structures, such as ribosomes (Fig. 3A) and nucleoli (not shown). The conjugate reacted with the small viral particles and the dense periphery of large virions (Fig. 3B). Pretreatment of the sections with nonconjugated RNase totally inhibited the binding of the RNase-gold complex to viral particles and to RNA-containing cellular components (Fig. 3C and D).
FIG. 3.
Binding of an RNase-gold complex to sections of infected ST cells. (A) The signal concentrates on RNA-containing cellular structures, such as, for example, the rough endoplasmic reticulum (RER)-associated ribosomes and free ribosomes. Binding to individual ribosomes is distinguished (arrows). Some signal on mitochondria (mi) is also detected. (B) This RNase-gold complex also reacts with both small (arrows) and large (arrowheads) virions. The RNA-gold probe binds to the dense periphery of large virions. Pretreatment of the sections with nonconjugated RNase (20 μg/ml) abolished the subsequent binding of the RNase-gold complex to both RNA-containing cellular structures (C) and to TGEV particles (labeled v in panel D). r, ribosome; SV, secretory vesicle. Bar, 200 nm.
Using freeze-substituted infected ST cells, we measured the size of the dense internal component of the viral particles (the structures contrasted better in these samples) and the size distribution obtained is shown in Fig. 4. Considering the small differences in diameter caused by the plane of section, two main sizes account for more than 80% of the whole population of virions. In extracellular virions, most of the viral cores correspond to the smaller size (diameter, 50 to 65 nm), while a population of large particles (68 to 80 nm) is found in intracellular virions. Also, minor components appeared after this quantitative study: a small percentage (4%) of bigger particles (>80- to 92-nm diameter) and a small percentage of virions (1 to 5%, depending on the p.i. time) that did not correspond to the two morphologies described. We found large particles with a homogeneous, highly dense content, small virions with an annular, dense internal structure, and virions with an undefined internal structure of apparently low density. They could constitute potential intermediary assemblies, although their significance remains to be established.
The distribution of large and small virions in different cellular regions and compartments was analyzed (Table 1). This quantification indicated that large virions are more abundant in the perinuclear area, known to be rich in ERGIC-related elements, while both large and small virions coexist in the Golgi complex (Table 1 and Fig. 5A). Post-Golgi complex large secretory vesicles contain mainly small dense virions, the morphology that corresponds to that of the majority of the extracellular population of virions (Table 1). These data suggest that the large annular virions are precursors of the small viral particles and that the major structural transformation that yields the small virions could depend on the transport of TGEV particles through the Golgi complex of ST cells.
FIG. 5.
Effect of BFA and nocodazole treatment on TGEV assembly. (A) Large virions (arrowheads) and small dense viral particles (arrows) coexist within the Golgi complex (G) of normal infected cells. (B) BFA causes the disappearance of the Golgi complex as a distinguishable structure, together with the formation of large cisternae (asterisks), some of them apparently derived from the rough endoplasmic reticulum (RER), since they have ribosomes (r) attached. TGEV virions assemble in association with these cisternae (arrowheads point to budding profiles). Large viral particles with an electron-dense internal periphery and clear center (arrows) accumulate in these conditions. Some ERGIC-like tubular membranes are visible in these cells (pairs of arrows). (C) Abnormal Golgi stack (G) from a nocodazole-treated infected ST cell. Both budding profiles (arrowheads) and small dense virions (arrows) are seen within the altered membranes of the stack. mi, mitochondria; pm, plasma membrane. Bar, 200 nm.
Effects of BFA and nocodazole on TGEV assembly.
The intracellular distribution of the two types of TGEV particles strongly suggests an association between intracellular transport through the constitutive secretory pathway and the structural maturation of virions. To test this possibility, we analyzed the effects of two drugs that disrupt the exocytic pathway (BFA and nocodazole) on viral assembly. BFA acts within minutes, inducing a rapid redistribution of Golgi cisternae into the endoplasmic reticulum, leaving no definable Golgi structure (17, 39, 40). This dramatic effect on the endoplasmic reticulum and Golgi systems occurs without affecting other cellular processes, including protein synthesis. At 4 h p.i., when few virions accumulate, infected ST cells were treated with BFA as described in Materials and Methods. The disappearance of the Golgi complex was associated with the formation of large cisternae that most probably result from the fusion between different membranous compartments (Fig. 5). While both large and small virions coexist in the Golgi apparatus from normal infected cells (Fig. 5A), most of the TGEV particles visualized inside the large cisternae of BFA-treated infected cells are large virions with a very electron-dense periphery and a less dense center (Fig. 5B and Table 2). A significantly higher amount of budding profiles was seen in these cells than in normally infected cell cultures (Table 2). Removal of BFA from these cultures did not lead to a recovery of cellular internal organization and TGEV release when cells were maintained 2 or 4 h more in the absence of the drug. Large annular virions kept accumulating inside large cisternae under these conditions (not shown).
The addition of nocodazole to infected ST cells (4 h p.i.) apparently arrested the transport of viral particles beyond the Golgi apparatus. The cytoplasm of cells affected by the nocodazole treatment was less organized, as shown by EM; no microtubules were distinguished, and there were smaller and dispersed Golgi stacks (not shown). TGEV virions were seen in these disrupted Golgi stacks, and they exhibited the two types of morphologies described above (Fig. 5C and Table 1). Large secretory vesicles with numerous virions were not seen, and very few virions were detected on the extracellular surfaces of these cells (not shown). No differences in structure were observed in large and small TGEV virions assembled in the presence of nocodazole compared with virions visualized in normally infected cells.
These results strongly suggest that precursor virions need to pass through a functional Golgi complex to complete the structural transformation that yields the small, infectious virions.
DISCUSSION
The morphogenesis of coronaviruses has been defined as a complex process that involves different elements of the constitutive exocytic pathway of the cell. Special interest resides in the events that take place during the budding process and the subsequent construction of the spherical, most probably icosahedral, core shell. In this study, we have shown that two types of virus-related particles can be found in TGEV-infected cells, large virions that form from budding profiles and small viral particles that are identical to the extracellular infectious virions characterized in our previous report (52). The optimal structural preservation obtained in this study and the confirmation provided by freeze-substitution have been decisive in identifying the two types of TGEV virions in infected ST cells. Moreover, our images strongly suggest that the morphology of the small viral particles is closely related to the formation of the internal core shell.
The morphological maturation described for TGEV represents a dramatic decrease in the volume of the viral particle. Significant changes in virion volume during maturation have been extensively described in the morphogenesis of bacteriophages (8, 10, 31), although it does not seem to be so frequent among animal viruses. Nevertheless, there are cases in which a similar process has been also described. The human immunodeficiency virus, for example, assembles large virions that later transform into smaller viral particles with condensed internal cores (19, 22). The process also represents a considerable change in virion volume that must take place after the activation of the viral protease. For TGEV, we have seen that the change in volume is accompanied by a major redistribution of the dense intravirion material that first occupies a peripheral position and after maturation becomes homogeneously distributed inside the smaller condensed core. This dense material most probably contains the viral RNA, as suggested by the signal provided by the RNase-gold complex and the data obtained in another study, in which phosphorous elemental maps have been proposed to represent the direct locations of nucleic acids in viruses (47).
What are the stimuli that trigger TGEV structural maturation inside ST cells? Our data point to a close association between intracellular transport through the exocytic constitutive pathway and viral maturation. In this sense, Tooze et al. (63) reported the existence of two types of virions in AtT20 cells infected by the MHV-A59 coronavirus. While spherical large virions were visualized in pre-Golgi compartments and in Golgi cisternae, smaller, kidney-shaped, very electron-dense viral particles were detected in the trans-Golgi network. Although the mentioned kidney shape could be the result of a deformation suffered by the small viral particles, the morphological maturation described for MHV seems to be very similar to the process in TGEV that we detected. The data we have obtained in infected cells treated with BFA or nocodazole also support the possibility that viral maturation is taking place in the Golgi complex. Nocodazole treatment was chosen to study the effects that a blockade of the exocytic pathway at a post-Golgi level could have on viral morphology. The results of experiments with live cells and microtubule inhibitors (1, 7) suggest that microtubules facilitate the transport of vesicles between the Golgi apparatus and the plasma membrane, while the movement of membrane out of the endoplasmic reticulum toward the Golgi complex is believed to occur by a microtubule-independent mechanism (39). It is also known that nocodazole treatment causes a reversible fragmentation of the Golgi complex into numerous dispersed units that remain functional (65). In nocodazole-treated, infected ST cells, TGEV virions are able to reach these modified Golgi stacks and undergo structural maturation. The assembly seems, however, to be retarded, since fewer small virions accumulate with time (Table 1). On the other hand, BFA was used to analyze the morphology of TGEV particles assembled in the absence of a functional Golgi complex. Large “precursor” virions accumulated in these cells. Although it has been reported that cells appear to metabolize BFA (11), at the lower BFA concentration used in our experiments (1 μg/ml), the effects of BFA treatment on the organization of ST cells and release of TGEV were not reversible 4 h after the drug was removed. Large annular virions kept accumulating inside the large cisternae under these conditions. It is likely that the prolonged treatment with BFA needed to accumulate virions makes the effect of the drug irreversible in ST cells. We have observed (52a) that the effects on cellular ultrastructure were less dramatic when BFA was added after a 30-min incubation with nocodazole, probably because BFA-induced fusion between different membranous compartments partly depends on microtubules, as proposed by Lippincott-Schwartz et al. (39). Four hours after both drugs were removed, a partial recovery of intracellular organization was seen, together with an increase in the amount of small dense virions (up to 30% of the total population of virions). Virus secretion, however, was not recovered.
To this point we have no data on whether cellular factors are responsible for viral morphological maturation or whether specific virus-associated elements are activated during intracellular transport. A combination of both is also possible, as in the case of alphaviruses. One of the viral polyproteins of Sindbis virus is processed by the combined action of an autoproteolytic activity in the capsid protein, a cellular signal peptidase, and an enzyme thought to be a component of the Golgi apparatus (60). Maturation through a proteolytic processing has the advantage of being an irreversible process, as extensively documented, for example, in bacteriophages (8) and retroviruses (30, 34). In coronaviruses, no enzymatic activity associated to purified coronavirus particles has been reported to date. However, the large intracellular precursor virions have not been purified and characterized in vitro. Since new virus-associated proteins are being reported (16) and still several open reading frames remain to be defined (56), minor, yet unidentified virus-associated proteins could exist and be involved in the maturation process. The E envelope protein is, for example, a key element in assembly, being synthesized in ample amounts in infected cells but incorporated into extracellular virions in only minor amounts (53).
It has been proposed that proteolytic processing (25) and dephosphorylation (37, 59) of the nucleocapsid (N) protein could play an important role in assembly. For TGEV M and S proteins, glycosylation is the only posttransductional modification known to take place. Glycosylation of M is not a requirement for assembly, since a mutant TGEV defective for glycosylation in M was able to assemble normally and produce infectious virions (38). Correct glycosylation of S is necessary for its folding and incorporation into infectious virions, but not for the assembly of viral particles (42). Glycosylation is not complete for all the protein molecules within the viral particles, probably due to accessibility problems for the enzymes in reaching all the modification sites, since they have to act on whole viral particles. Some similar mechanism could explain why a small amount of virions “escape” from the maturation process and exit the cell, constituting a minor component of the extracellular population of virions. The participation of these or some other processes in the morphogenesis of coronaviruses should be studied in detail, and to this end, the purification of the large precursor virions will be of great interest.
Recent studies focused on the association between the coronavirus envelope proteins and the formation of virus-like particles in vitro have identified some of the molecular requirements for building pseudo-viral envelopes (44, 66). Lateral interactions between the M and E envelope proteins within the lipid bilayer seem to constitute the driving force for the assembly of these particles. Although they can contain variable amounts of the different envelope proteins, the sizes of the envelopes assembled under these conditions are surprisingly similar to those of whole virions (66). Although these processes are far from the natural morphogenetic pathway, they can be of interest in understanding the particular roles and assembly capabilities of the isolated structural components.
The detailed structural analysis presented here shows new aspects of the morphogenesis of coronaviruses. Further studies are necessary for defining its molecular basis and its potential application in the in vitro manipulation of assembly. The potential existence of other intermediates in the sequence of assembly should also be defined. These studies are presently under way.
ACKNOWLEDGMENTS
This work was partly supported by grant PB96-0818 from the Comisión Interministerial de Ciencia y Tecnología to J.L.C. and by grants from the Comisión Interministerial de Ciencia y Tecnología and the European Union (projects Science and Biotech) to L.E. C. Risco is the recipient of a contract from the C.S.I.C.-Fundación Ramón Areces.
REFERENCES
- 1.Arnheiter H, Dubois-Dalcq M, Lazzarini R A. Direct visualization of protein transport and processing in the living cell by microinjection of specific antibodies. Cell. 1984;39:99–109. doi: 10.1016/0092-8674(84)90195-8. [DOI] [PubMed] [Google Scholar]
- 2.Becker W B, McIntosh K, Dees J H, Chanock R M. Morphogenesis of avian infectious bronchitis virus and a related human virus (strain 229E) J Virol. 1967;1:1019–1027. doi: 10.1128/jvi.1.5.1019-1027.1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bendayan M. Ultrastructural localization of nucleic acids by the use of enzyme-gold complexes. J Histochem Cytochem. 1981;29:531–541. doi: 10.1177/29.4.6265546. [DOI] [PubMed] [Google Scholar]
- 4.Bendayan M. The enzyme-gold cytochemical approach: a review. In: Hayat M A, editor. Colloidal gold: principles, methods, and applications. Vol. 2. San Diego, Calif: Academic Press, Inc.; 1989. pp. 117–147. [Google Scholar]
- 5.Booy F P. Cryoelectron microscopy. In: Bentz J, editor. Viral fusion mechanisms. Boca Raton, Fla: CRC Press, Inc.; 1993. pp. 21–54. [Google Scholar]
- 6.Bos E C W, Luytjes W, Van der Meulen H, Koerten H K, Spaan W J M. The production of recombinant infectious DI-particles of a murine coronavirus in the absence of helper virus. Virology. 1996;218:52–60. doi: 10.1006/viro.1996.0165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Burguess T L, Kelly R B. Constitutive and regulated secretion of proteins. Annu Rev Cell Biol. 1987;3:243–293. doi: 10.1146/annurev.cb.03.110187.001331. [DOI] [PubMed] [Google Scholar]
- 8.Carrascosa J L. Bacteriophage morphogenesis. In: Harris J R, Horne R W, editors. Electron microscopy of proteins. Vol. 5. London, England: Academic Press; 1986. pp. 37–70. [Google Scholar]
- 9.Carrascosa J L. Immunoelectron microscopical studies on viruses. Electron Microsc Rev. 1988;1:1–16. doi: 10.1016/s0892-0354(98)90003-x. [DOI] [PubMed] [Google Scholar]
- 10.Casjens S, Hendrix R W. Control mechanisms in dsDNA bacteriophage assembly. In: Calendar R, editor. The bacteriophages. Vol. 1. New York, N.Y: Plenum Press; 1988. pp. 15–90. [Google Scholar]
- 11.Chen S-Y, Matsuoka Y, Compans R W. Assembly and polarized release of Punta Toro virus and effects of brefeldin A. J Virol. 1991;65:1427–1439. doi: 10.1128/jvi.65.3.1427-1439.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.David-Ferreira J F, Manaker R A. An electron microscope study on the development of a mouse hepatitis virus in tissue cells. J Cell Biol. 1965;24:57–78. doi: 10.1083/jcb.24.1.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dubois-Dalcq M E, Doller E W, Haspel M V, Holmes K V. Cell tropism and expression of mouse hepatitis viruses (MHV) in mouse spinal cord cultures. Virology. 1982;119:317–331. doi: 10.1016/0042-6822(82)90092-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dubois-Dalcq M E, Holmes K V, Rentier B. Assembly of coronaviruses. In: Kingsbury D W, editor. Assembly of RNA viruses. New York, N.Y: Springer-Verlag; 1984. pp. 100–119. [Google Scholar]
- 15.Enjuanes L, Van der Zeijst B A M. Molecular basis of the transmissible gastroenteritis virus epidemiology. In: Siddell S G, editor. The Coronaviridae. New York, N.Y: Plenum Press; 1995. pp. 337–376. [Google Scholar]
- 16.Fischer F, Peng D, Hingley S T, Weiss S R, Masters P S. The internal open reading frame within the nucleocapsid gene of mouse hepatitis virus encodes a structural protein that is not essential for viral replication. J Virol. 1997;71:996–1003. doi: 10.1128/jvi.71.2.996-1003.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fujiwara T, Oda K, Yokota S, Takatsuki A, Ikehara Y. Brefeldin A causes disassembly of the Golgi complex and accumulation of secretory proteins in the endoplasmic reticulum. J Biol Chem. 1988;263:18545–18552. [PubMed] [Google Scholar]
- 18.Gebauer F, Posthumus W A P, Correa I, Suñé C, Sánchez C M, Smerdou C, Lenstra J A, Meloen R, Enjuanes L. Residues involved in the formation of the antigenic sites of the S protein of transmissible gastroenteritis coronavirus. Virology. 1991;183:225–238. doi: 10.1016/0042-6822(91)90135-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gelderblom H R, Özel M, Pauli G. Morphogenesis and morphology of HIV. Structure-function relations. Arch Virol. 1989;106:1–13. doi: 10.1007/BF01311033. [DOI] [PubMed] [Google Scholar]
- 20.Godet M, L’Haridon R, Vautherot J-F, Laude H. TGEV coronavirus ORF4 encodes a membrane protein that is incorporated into virions. Virology. 1992;188:666–675. doi: 10.1016/0042-6822(92)90521-P. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Granzow H, Weiland F, Jöns A, Klupp B G, Karger A, Mettenleiter T C. Ultrastructural analysis of the replication cycle of pseudorabies virus in cell culture: a reassessment. J Virol. 1997;71:2072–2082. doi: 10.1128/jvi.71.3.2072-2082.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Grief C, Nermut M V, Hockley D J. A morphological and immunolabeling study of freeze-substituted human and simian immunodeficiency viruses. Micron. 1994;25:119–128. doi: 10.1016/0968-4328(94)90036-1. [DOI] [PubMed] [Google Scholar]
- 23.Hauri H P, Schweizer A. The endoplasmic reticulum-Golgi intermediate compartment. Curr Opin Cell Biol. 1992;4:600–608. doi: 10.1016/0955-0674(92)90078-Q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Holmes K V, Behnke J N. Evolution of a coronavirus during persistent infection in vitro. Adv Exp Med Biol. 1981;142:287–299. doi: 10.1007/978-1-4757-0456-3_23. [DOI] [PubMed] [Google Scholar]
- 25.Holmes K V, Boyle J F, Williams R K, Stephensen C B, Robbins S G, Bauer E C, Duchala C S, Frana M F, Weismiller D G, Compton S, McGowan J J, Sturman L S. Processing of coronavirus proteins and assembly of virions. In: Brinton M A, Rueckert R R, editors. Positive strand RNA viruses. New York, N.Y: Alan R. Liss; 1987. pp. 339–349. [Google Scholar]
- 26.Hyatt A D, Eaton B T. Virological applications of the grid-cell-culture technique. Electron Microsc Rev. 1990;3:1–27. doi: 10.1016/0892-0354(90)90011-G. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jäntti J, Hildén P, Rönkä H, Mäkiranta V, Keränen S, Kuismanen E. Immunocytochemical analysis of Uukuniemi virus budding compartments: role of the intermediate compartment and the Golgi stack in virus maturation. J Virol. 1997;71:1162–1172. doi: 10.1128/jvi.71.2.1162-1172.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jiménez G, Correa I, Melgosa M P, Bullido M J, Enjuanes L. Critical epitopes in transmissible gastroenteritis virus neutralization. J Virol. 1986;60:131–139. doi: 10.1128/jvi.60.1.131-139.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kapke P A, Brian D A. Sequence analysis of the porcine transmissible coronavirus nucleocapsid protein gene. Virology. 1986;151:41–49. doi: 10.1016/0042-6822(86)90102-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Katoh I, Yoshinaka Y, Rein A, Shibuya M, Odaka T, Oroszlan S. Murine leukemia virus maturation: protease region required for conversion from immature to mature core form and for virus infectivity. Virology. 1985;145:280–292. doi: 10.1016/0042-6822(85)90161-8. [DOI] [PubMed] [Google Scholar]
- 31.Kellenberger E. Form determination of the heads of bacteriophages. Eur J Biochem. 1990;190:233–248. doi: 10.1111/j.1432-1033.1990.tb15568.x. [DOI] [PubMed] [Google Scholar]
- 32.King B, Brian D A. Bovine coronavirus structural proteins. J Virol. 1982;42:700–707. doi: 10.1128/jvi.42.2.700-707.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Klumperman J, Krijnse Locker J, Meijer A, Horzinek M C, Geuze H J, Rottier P J M. Coronavirus M proteins accumulate in the Golgi complex beyond the site of virion budding. J Virol. 1994;68:6523–6534. doi: 10.1128/jvi.68.10.6523-6534.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kohl N E, Emini E A, Schleif W A, Davis L J, Heimbach J C, Dixon R A F, Scolnick E M, Sigal I S. Active human immunodeficiency virus protease is required for viral infectivity. Proc Natl Acad Sci USA. 1988;85:4686–4690. doi: 10.1073/pnas.85.13.4686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Krijnse-Locker J, Ericsson M, Rottier P J, Griffiths G. Characterization of the budding compartment of mouse hepatitis virus: evidence that transport from the RER to the Golgi complex requires only one vesicular transport step. J Cell Biol. 1994;124:55–70. doi: 10.1083/jcb.124.1.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lai M M C. Coronavirus. Organization, replication, and expression of genome. Annu Rev Microbiol. 1990;44:303–333. doi: 10.1146/annurev.mi.44.100190.001511. [DOI] [PubMed] [Google Scholar]
- 37.Laude H, Masters P S. The coronavirus nucleocapsid protein. In: Siddell S G, editor. The Coronaviridae. New York, N.Y: Plenum Press; 1995. pp. 141–163. [Google Scholar]
- 38.Laude H, Gelfi J, Lavenant L, Charley B. Single amino acid changes in the viral glycoprotein M affect induction of alpha interferon by the coronavirus transmissible gastroenteritis virus. J Virol. 1992;66:743–749. doi: 10.1128/jvi.66.2.743-749.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lippincott-Schwartz J, Donaldson J G, Schweizer A, Berger E G, Hauri H-P, Yuan L C, Klausner R D. Microtubule-dependent retrograde transport of proteins into the ER in the presence of brefeldin A suggests an ER recycling pathway. Cell. 1990;60:821–836. doi: 10.1016/0092-8674(90)90096-w. [DOI] [PubMed] [Google Scholar]
- 40.Lippincott-Schwartz J, Yuan L C, Bonifacino J S, Klausner R D. Rapid redistribution of Golgi proteins into the ER in cells treated with brefeldin A: evidence for membrane cycling from Golgi to ER. Cell. 1989;56:801–813. doi: 10.1016/0092-8674(89)90685-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Liu D X, Inglis S C. Association of the infectious bronchitis virus 3c protein with the virion envelope. Virology. 1991;185:911–917. doi: 10.1016/0042-6822(91)90572-S. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Luytjes W, Gerritsma H, Bos E, Spaan W. Characterization of two temperature-sensitive mutants of coronavirus mouse hepatitis virus strain A59 with maturation defects in the spike protein. J Virol. 1997;71:949–955. doi: 10.1128/jvi.71.2.949-955.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.MacIntosh K. Coronaviruses. In: Fields B N, Knipe D M, Howley P M, et al., editors. Fields virology. Philadelphia, Pa: Lippincott-Raven; 1996. pp. 1095–1133. [Google Scholar]
- 44.Opsteltein D-J, Raamsman M J B, Wolfs K, Horzinek M C, Rottier P J. Envelope glycoprotein interactions in coronavirus assembly. J Cell Biol. 1995;131:339–349. doi: 10.1083/jcb.131.2.339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pimenta P F P, de Souza W. Fine structure and cytochemistry of the endoplasmic reticulum and its association with the plasma membrane of Leishmania mexicana. J Submicrosc Cytol. 1985;17:413–419. [PubMed] [Google Scholar]
- 46.Quintana C. Cryofixation, cryosubstitution, cryoembedding for ultrastructural, immunocytochemical, and microanalytical studies. Micron. 1994;25:63–99. doi: 10.1016/0968-4328(94)90056-6. [DOI] [PubMed] [Google Scholar]
- 47.Quintana, C., S. Marco, N. Bonnet, C. Risco, M. L. Gutiérrez, A. Guerrero, and J. L. Carrascosa. Phosphorous localization by EELS and image filtering in viruses. Submitted for publication.
- 48.Risco C, Romero C, Bosch M A, Pinto da Silva P. Type II pneumocytes revisited: intracellular membranous systems, surface characteristics, and lamellar body secretion. Lab Invest. 1994;70:407–417. [PubMed] [Google Scholar]
- 49.Risco C, Menéndez-Arias L, Copeland T D, Pinto da Silva P, Oroszlan S. Intracellular transport of the murine leukemia virus during acute infection of NIH 3T3 cells: nuclear import of the nucleocapsid protein and integrase. J Cell Sci. 1995;108:3039–3050. doi: 10.1242/jcs.108.9.3039. [DOI] [PubMed] [Google Scholar]
- 50.Risco C, Carrascosa J L, Pedregosa A M, Humphrey C D, Sánchez-Fauquier A. Ultrastructure of the human astrovirus serotype 2. J Gen Virol. 1995;76:2075–2080. doi: 10.1099/0022-1317-76-8-2075. [DOI] [PubMed] [Google Scholar]
- 51.Risco C, Antón I M, Suñé C, Pedregosa A M, Martín-Alonso J M, Parra F, Carrascosa J L, Enjuanes L. Membrane protein molecules of transmissible gastroenteritis coronavirus also expose the carboxy-terminal region on the external surface of the virion. J Virol. 1995;69:5269–5277. doi: 10.1128/jvi.69.9.5269-5277.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Risco C, Antón I M, Enjuanes L, Carrascosa J L. The transmissible gastroenteritis coronavirus contains a spherical core shell consisting of M and N proteins. J Virol. 1996;70:4773–4777. doi: 10.1128/jvi.70.7.4773-4777.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52a.Risco, C., et al. Unpublished results.
- 53.Rottier P J M. The coronavirus membrane glycoprotein. In: Siddell S G, editor. The Coronaviridae. New York, N.Y: Plenum Press; 1995. pp. 115–139. [Google Scholar]
- 54.Sánchez C M, Jiménez G, Laviada M D, Correa I, Suñé C, Bullido M J, Gebauer F, Smerdou C, Callebaut P, Escribano J M, Enjuanes L. Antigenic homology among coronaviruses related to transmissible gastroenteritis virus. Virology. 1990;174:410–417. doi: 10.1016/0042-6822(90)90094-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Saraste J, Palade G E, Farquhar M G. Antibodies to rat pancreas Golgi subfractions: identification of a 58-kD cis-Golgi protein. J Cell Biol. 1987;105:2021–2029. doi: 10.1083/jcb.105.5.2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Siddell S G. The Coronaviridae. An introduction. In: Siddell S G, editor. The Coronaviridae. New York, N.Y: Plenum Press; 1995. pp. 1–10. [Google Scholar]
- 57.Spaan W, Cavanagh D, Horzinek M C. Coronaviruses: structure and genome expression. J Gen Virol. 1988;69:2939–2952. doi: 10.1099/0022-1317-69-12-2939. [DOI] [PubMed] [Google Scholar]
- 58.Spaan W J M, Rottier P J, Horzinek M C, Van der Zeijst B A M. Isolation and identification of virus-specific mRNAs in cells infected with mouse hepatitis virus (MHV-A59) Virology. 1981;108:424–434. doi: 10.1016/0042-6822(81)90449-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Stohlman S A, Fleming J O, Patton C D, Lai M M C. Synthesis and subcellular localization of the murine coronavirus nucleocapsid protein. Virology. 1983;130:527–532. doi: 10.1016/0042-6822(83)90106-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Strauss J H, Calisher C H, Dalgarno L, Dalrymple J M, Frey T K, Pettersson R F, Rice C M, Spaan W J M. Togaviridae. In: Murphy F A, Fauquet C M, Bishop D H L, et al., editors. Virus taxonomy. New York, N.Y: Springer-Verlag; 1995. pp. 428–433. [Google Scholar]
- 61.Sturman I, Holmes K V. The molecular biology of coronaviruses. Adv Virus Res. 1983;28:35–112. doi: 10.1016/S0065-3527(08)60721-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Tooze J, Tooze S, Warren G. Replication of coronavirus MHV-A59 in sac− cells: determination of the first site of budding of progeny virions. Eur J Cell Biol. 1984;33:281–293. [PubMed] [Google Scholar]
- 63.Tooze J, Tooze S A, Fuller S D. Sorting of progeny coronavirus from condensed secretory proteins at the exit from the trans-Golgi network of AtT20 cells. J Cell Biol. 1987;105:1215–1226. doi: 10.1083/jcb.105.3.1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Tooze S A, Tooze J, Warren G. Site of addition of N-acetyl-galactosamine to the E1 glycoprotein of mouse hepatitis virus-A59. J Cell Biol. 1988;106:1475–1487. doi: 10.1083/jcb.106.5.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Turner J R, Tartakoff A M. The response of the Golgi complex to microtubule alterations: the roles of metabolic energy and membrane traffic in Golgi complex organization. J Cell Biol. 1989;109:2081–2088. doi: 10.1083/jcb.109.5.2081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Vennema H, Godeke G-J, Rossen J W A, Voorhout W F, Horzinek M C, Opsteltein D-J E, Rottier P J M. Nucleocapsid-independent assembly of coronavirus-like particles by co-expression of viral envelope protein genes. EMBO J. 1996;15:2020–2028. doi: 10.1002/j.1460-2075.1996.tb00553.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Yu X, Bi W, Weiss S R, Leibowitz J L. Mouse hepatitis virus gene 5b protein is a new virion envelope protein. Virology. 1994;202:1018–1023. doi: 10.1006/viro.1994.1430. [DOI] [PubMed] [Google Scholar]