Significance
Cortical neurogenesis is essential for the generation of neuronal networks and brain function. KCTD10 belongs to the KCTD family, many members of which are associated with neuropsychiatric disorders. However, the biological function of KCTD10 in brain development remains unexplored. Here, we show that KCTD10 is critical for cortical neurogenesis, and loss of KCTD10 leads to severe neural developmental defects and motor deficits. Mechanistically, we identify KCTD13 as a new KCTD10-interacting protein and reveal that KCTD10 regulates cortical neurogenesis by mediating the degradation of KCTD13. Given that both KCTD13 and CUL3 are high-confidence neuropsychiatric risk factors, our study strongly advocates for the screening of KCTD10 mutations in patients with neurodevelopmental disorders, including autism.
Keywords: KCTD10, cortical neurogenesis, KCTD13, ubiquitination-dependent degradation, motor deficits
Abstract
KCTD10 belongs to the KCTD (potassiumchannel tetramerization domain) family, many members of which are associated with neuropsychiatric disorders. However, the biological function underlying the association with brain disorders remains to be explored. Here, we reveal that Kctd10 is highly expressed in neuronal progenitors and layer V neurons throughout brain development. Kctd10 deficiency triggers abnormal proliferation and differentiation of neuronal progenitors, reduced deep-layer (especially layer V) neurons, increased upper-layer neurons, and lowered brain size. Mechanistically, we screened and identified a unique KCTD10-interacting protein, KCTD13, associated with neurodevelopmental disorders. KCTD10 mediated the ubiquitination-dependent degradation of KCTD13 and KCTD10 ablation resulted in a considerable increase of KCTD13 expression in the developing cortex. KCTD13 overexpression in neuronal progenitors led to reduced proliferation and abnormal cell distribution, mirroring KCTD10 deficiency. Notably, mice with brain-specific Kctd10 knockout exhibited obvious motor deficits. This study uncovers the physiological function of KCTD10 and provides unique insights into the pathogenesis of neurodevelopmental disorders.
Mammalian cortex development begins with the proliferation of neuroepithelial cells, which expand the neuronal progenitor cell (NPC) pool through symmetric divisions. As the neural epithelium thickens into the pseudostratified ventricular wall, neuroepithelial cells in the ventricular zone (VZ) transform into radial glial cells (RGCs) or neural stem cells. RGCs generate almost all excitatory neurons in the cerebral cortex, either directly through asymmetric division or indirectly through basal progenitor cells (BPCs) (1, 2). Newly generated neurons migrate along glial fibers to the cortical plate after undergoing a multi-to-bipolar transition in the upper intermediate zone (IZ) (3). Upon arrival at the cortical plate, neurons undergo further differentiation and maturation, establishing synaptic connections and forming intricate neural circuits. Compromised cortical development can cause a variety of neurological disorders, including autism spectrum disorder (ASD), intellectual disability, autosomal recessive primary microcephaly, epilepsy, and schizophrenia (4–7).
The KCTD (potassium channel tetramerization domain-containing) family consists of 25 soluble proteins that share a conserved N-terminal potassium (K+) channel tetramerization domain (a BTB domain subtype) but have diverse C-termini. Many KCTD-encoding genes have been linked with neurodevelopmental and neuropsychiatric disorders, with varying degrees of genetic evidence implicating KCTD genes in neurocognitive disorders (KCTD3) (8–10); progressive myoclonus epilepsy (KCTD7) (11–19); bipolar disorder, depression, and schizophrenia (KCTD12) (20–22); autism and schizophrenia (KCTD13) (23–26); movement disorders (KCTD17) (27–30); epilepsy and autism (KCTD18) (31); cerebral visual impairment (KCTD19) (32); Alzheimer’s disease (KCTD2) (33); and sporadic amyotrophic lateral sclerosis (BTBD10) (34).
KCTD10, a member of the KCTD family, functions as a substrate recognition receptor within the RING-type ubiquitin ligase complex (35–37). It serves as a bridge between substrates and CUL3, a high-confidence ASD risk protein (38–40). Multiple KCTD10 substrates, including RhoB, CEP97, EIF3D, and TRIF, are degraded by the CUL3/KCTD10 E3 complex. The targets of KCTD10 highlight the diverse function of KCTD10 in regulating biological processes including endothelial barrier formation, primary cilium formation, plasma membrane dynamics, cell proliferation, and immune responses (41–44). Kctd10-knockout mice exhibited developmental delay by embryonic day 9 and embryonic lethality by E10.5, indicating a critical role in embryonic development (45).
KCTD13, also known as BACURD1, POLDIP1, PDIP1, and TNFAIP1-like, is found at chromosome 16 region 16p11.2. Copy-number variants of KCTD13 are closely associated with neuropsychiatric disorders and are frequently found in patients with ASD (24, 25, 46, 47). Moreover, KCTD13 has been identified as a predominant driver of neurodevelopmental disorders associated with 16p11.2 copy-number variation. Overexpressing KCTD13 in zebrafish results in microcephaly (also associated with 16p11.2 duplication), due to reduced NPC proliferation and a concomitant increase in apoptosis in the developing brain (24). In addition, Kctd13 knockdown induces a macrocephalic phenotype in zebrafish, and increased NPC proliferation is observed in zebrafish and mouse brains (24). Patients with ASD or schizophrenia have been found to possess mutations in the KCTD13 (26, 48, 49), although the underlying mechanism of pathogenesis remains ambiguous.
In this study, we show that the conditional knockout (cKo) of Kctd10 in mice results in diminished NPC proliferation, enhanced differentiation, reduced brain size, and motor impairment. Similarly, KCTD13 overexpression in the mouse embryonic cortex also reduced NPC proliferation and disturbed cellular distribution. We demonstrate that KCTD10 promotes the degradation of KCTD13 via the ubiquitin/proteasome pathway throughout cortical development.
Results
cKo of Kctd10 in the Brain Causes Severe Developmental Defects.
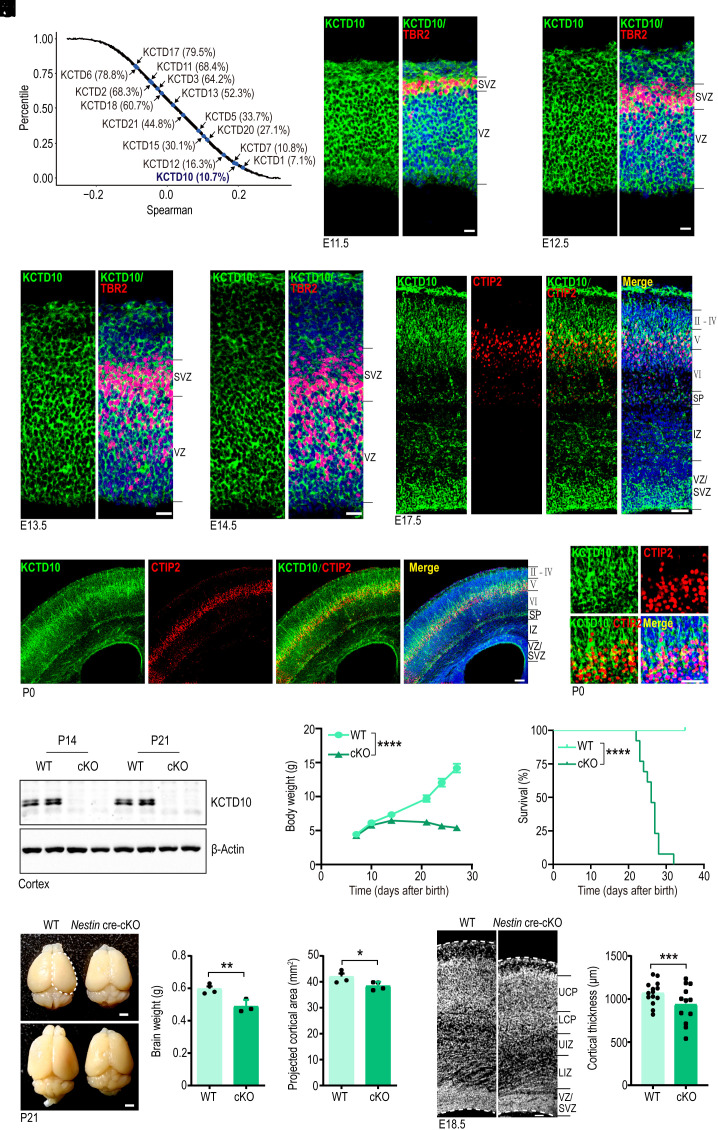
Many genes in the KCTD family are associated with neurodevelopmental and neuropsychiatric disorders. However, the relationship between KCTD10 and brain disorders such as NDD (neuronal developmental disorder) remains uncertain. We, therefore, explored the potential biological connection of KCTD10 with NDD genes by examining their co-expression. We computed the Spearman’s correlation coefficient of all protein-coding genes with a well-established NDD gene set (Fig. 1A). Within the KCTD family, KCTD10 was highly correlated with the known NDD gene set (Percentile = 10.8%, top 10.8%) (Fig. 1A), indicating that the expression of KCTD10 is more highly correlated with NDD genes compared to most other cortex-expressed genes. This analysis revealed 88 genes highly correlated with KCTD10 (|Spearman’s correlation coefficient| > 0.7). To support this analysis, we inspected the expression of KCTD10 during mouse brain development, revealing significant expression in the cortex that was highest on embryonic day 15.5 (E15.5) and gradually decreased during development (SI Appendix, Fig. S1B). Meanwhile, KCTD10 expression in the hippocampus was relatively low during embryogenesis and gradually increased after birth, reaching maximal levels on postnatal day 28 (P28) (SI Appendix, Fig. S1C). Immunofluorescence staining of rostral cortices revealed that KCTD10 was expressed in specific cell populations at various developmental stages, with robust KCTD10 expression found in the VZ/SVZ of the cerebral cortex at E11.5 to E14.5 (Fig. 1 B–E and SI Appendix, Fig. S1A), At E17.5, KCTD10 expression dropped significantly in the VZ/SVZ and was highest in cortical layer V in CTIP2+ (a marker of Layer V) cells (Fig. 1F). At P0, KCTD10 was mainly expressed around cortical layer V and CTIP2+ cells displayed strong KCTD10 costaining (Fig. 1 G and H and SI Appendix, Fig. S1D). There was also weak expression in the IZ, subplate, and other layers of the cortex as that at E17.5 (Fig. 1F). At P7, P21 and P26, KCTD10 was primarily expressed in layer V (CTIP2+ cells) of the neocortex, although at gradually decreased levels (SI Appendix, Fig. S1 E and F). These results demonstrate that KCTD10 is highly and dynamically expressed during brain development.
Fig. 1.
Kctd10 cKO in the brain leads to severe developmental defects. (A) Co-expression analyses of KCTD family genes with NDD genes. The percentile of the average correlation coefficient for all developing cortex-expressed genes with NDD genes. The average percentile of KCTD family genes was marked. (B–E) Representative images of E11.5 (B), E12.5 (C), E13.5 (D), and E14.5 (E) cortices co-stained for KCTD10 (green), TBR2 (red), and DAPI (blue). (F) Representative images of E17.5 cortices stained for KCTD10 (green), CTIP2 (red), and DAPI (blue). (G) Representative images of P0 cortices stained for KCTD10 (green), CTIP2 (red), and DAPI (blue). (H) Magnifying images of P0 cortices stained for KCTD10 (green), CTIP2 (gray), and DAPI (blue). (I) Western blot confirms KCTD10 loss in the cortex of Nestin-cKO mice at P14 and P21. (J) Body weight of WT and cKO mice from P7 to P28. WT and cKO: n = 10. (K) Survival curve of WT and Kctd10 cKO mice. WT and cKO: n = 13. (L) WT and cKO brains at P21 (Left). Brain weight (Middle) and projected cortical area (Right) of WT and cKO mice. WT and cKO: n = 5. (M) Representative images of WT and cKO cortex stained with DAPI at E18.5 (Left). Quantification of cortical thickness (Right). WT and cKO: n = 9/3 (n, slice numbers/brain numbers). All data are means ± SEM (error bars). Student’s t test. *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001. [Scale bars: 20 μm (B–E), 50 μm (F, H, and M), 100 μm (G) and 2 mm (L).]
The expression pattern of KCTD10 suggested a role in regulating brain development. To determine the function of KCTD10, four short hairpin RNAs (shRNAs) targeting Kctd10 were designed. Among them, K10 sh1 and K10 sh4 effectively suppressed the overexpression of KCTD10 in HEK293 cells (SI Appendix, Fig. S2 A and B). We then used in utero electroporation to introduce scrambled shRNA (shNC) or K10 sh4 into cortical neuronal progenitors at E12.5 to investigate their impact on cortical development at E17.5. A disruption in the distribution of transfected cells with green fluorecent protein (GFP+) was observed in cortices expressing K10 sh4, characterized by the significant reduction in GFP+ cells in the VZ/SVZ and lower CP, and a notable increase in GFP+ cells located in the upper CP (SI Appendix, Fig. S2 C and D). Similar observations were made in cortices electroporated at E13.5 and inspected at E17.5 (SI Appendix, Fig. S2 E and F). This indicates that KCTD10 has a functional role in neuronal development.
We went on to establish a brain-specific Kctd10 cKO mouse line by crossing Kctd10fl/fl mice (45) with Nestin-Cre transgenic mice, circumventing the lethality associated with Kctd10 null mutations. The presence of Cre recombinase led to the deletion of the second exon of Kctd10, resulting in a frameshift mutation (SI Appendix, Fig. S1G). Western blot and immunostaining analyses confirmed the efficient deletion of KCTD10 in the cortex and/or hippocampus of Kctd10-cKO mice at P14 and P21, respectively (Fig. 1I and SI Appendix, Fig. S1 H–J). Compared to wild-type (WT) mice, Kctd10-cKO mice were significantly smaller at P21 and P26 (SI Appendix, Fig. S1K), indicating severe growth defects. The body weight of Kctd10-cKO mice steadily declined after P10 (Fig. 1J), with most dying at or around P25, and the remainder before P31 (Fig. 1H). Kctd10-cKO mice exhibited markedly reduced brain sizes and overall projected cortical areas compared to controls at P21 (Fig. 1L), characterized by a significantly decreased thickness of cortical layers V–VI and increased thickness of cortical layers II–IV (SI Appendix, Fig. S1L). Kctd10-cKO mice exhibited significantly decreased cortical thickness at E18.5 (Fig. 1M), with significantly reduced thickness of the VZ/SVZ, lower IZ, and lower CP (SI Appendix, Fig. S1M). Analysis of motor function further revealed that Kctd10-cKO mice had abnormal gait in the hind limbs around P24 (Movie S1). The abnormal gait consisted of shorter stride lengths than WT mice at both P19 and P23 (SI Appendix, Fig. S3A). We also noticed a concurrent decrease in both forearm and forelimb grip strength (SI Appendix, Fig. S3B). In the cylinder assay, the rearing duration and the number of rearing events of Kctd10 cKO mice were significantly reduced compared to WT mice (SI Appendix, Fig. S3C). Open field tests revealed significant differences in spontaneous motor activity including movement distance and/or resting duration between Kctd10 cKO and WT mice (SI Appendix, Fig. S3D).
Kctd10 cKO Results in Reduced Proliferation and Premature Differentiation of NPC in the Neocortex.
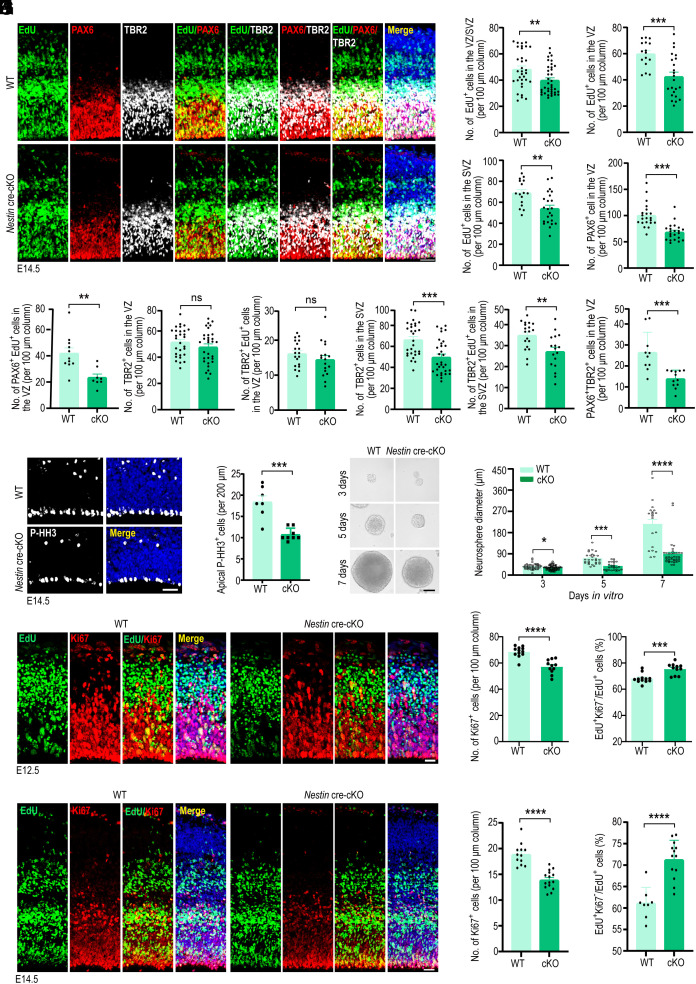
Reduced NPC proliferation is the primary cause of microcephaly. To determine whether the smaller brain sizes and thinner cerebral cortices observed in Kctd10-cKO mice were due to reduced progenitor cell proliferation, intraperitoneal injection of EdU into pregnant mice was performed at E11.5 to label cells in the S phase. Brains were collected 24 h later and subjected to immunofluorescence staining for PAX6 (a marker for RGCs or APCs), TBR2 (a marker for BPCs or IPCs), and EdU (SI Appendix, Fig. S4 A and B). EdU+ cells density was decreased in both VZ and SVZ of the dorsal cortex of Kctd10 cKO mice, indicating lowered NPC proliferation. In line with this observation, Kctd10 deletion substantially reduced the total number of PAX6+ cells in the VZ and TBR2+ cells located in the SVZ at E12.5. Meanwhile, there was a notable decrease in the density of PAX6+ EdU+ cells in the VZ and TBR2+ EdU+ cells in the SVZ in Kctd10-cKO mice at E12.5 (SI Appendix, Fig. S4 A and B). EdU labeling was also performed at E13.5. Similarly, there was a significant reduction in the density of EdU+ cells in both VZ and SVZ of the dorsal cortex, and a significant decreased in both the density of PAX6+ cells in the VZ and of TBR2+ cells in the SVZ at E14.5 (Fig. 2 A–C). However, no noticeable difference in the number of TBR2+ cells in the VZ was detected between Kctd10-cKO and WT mice (Fig. 2 A–C). Meanwhile, the density of PAX6+ EdU+ cells in the VZ and TBR2+ EdU+ cells in the SVZ, but not that of TBR2+ EdU+ cells in the VZ, was significantly reduced in Kctd10-cKO mice at E14.5 (Fig. 2 A–C). These data indicate that APC and BPC cycling was reduced. Given that TBR2+ BPCs are typically generated either through BPC self-renewal within the SVZ or by the transition of APCs in the VZ, we quantified the number of PAX6/TBR2 double-positive cells in the VZ region. As expected, the density of PAX6+ TBR2+ cells was significantly reduced in the cortex of Kctd10-cKO mice at E14.5 (Fig. 2 A–C). Similarly, in brains labeled with EdU at E15.5, we observed a noticeable reduction in the numbers of EdU+ cells in the VZ/SVZ, PAX6+ cells in the VZ, and TBR2+ cells in the SVZ in the cortex of Kctd10-cKO animals at E16.5 (SI Appendix, Fig. S4 C and D). Moreover, the number of PAX6+ EdU+ cells in the VZ region was also reduced, whereas no difference in the number of TBR2+ EdU+ cells in the SVZ was detected between the two groups at E16.5 (SI Appendix, Fig. S4 C and D). To repeat these observations in a distinct genetic model of conditional Kctd10 inactivation, thereby confirming the results are due to the inactivation of Kctd10 and not from any technical issue with the Nestin cre alle, Kctd10fl/fl mice were crossed with Emx1-Cre transgenic mice. Similar to results obtained using Nestin cre, we found that in brains labeled with EdU at E12.5, there was a noticeable reduction in the numbers of EdU+ cells in the VZ/SVZ, PAX6+ cells, and EdU+PAX6+ cells in the VZ, and TBR2+ cells as well as EdU+TBR2+ cells in the SVZ in the cortex of Kctd10 Emx1-Cre cKO animals at E13.5 (SI Appendix, Fig. S4 E and F).
Fig. 2.
KCTD10 regulates NPC development in the cortex. (A) Representative images of E14.5 WT and Kctd10 cKO mouse cortices stained for EdU (green), PAX6 (red), TBR2 (white), and DAPI (blue). EdU was injected at E13.5. (B) Quantification of the total number of EdU+ cells in the VZ and SVZ, and PAX6+ cells in the VZ per 100 µm column in A. WT: n = 33/8, cKO: n = 37/10. (C) Quantification of EdU+; PAX6+ in the VZ (WT: n = 11/3, cKO: n = 8/3), TBR2+ cells in the VZ and SVZ (WT: n = 19/3, cKO: n = 17/5), EdU+;TBR2+ in the VZ and SVZ (WT: n = 29/4, cKO: n = 31/5) and PAX6+;TBR2+ cells (WT: n = 11/3, cKO: n = 9/3) per 100 μm column. (D) Representative images of E14.5 WT and cKO cortices stained for P-HH3 (gray). (E) Quantification of apical P-HH3+ cells per 200 µm, WT: n = 15/3, cKO: n = 15/3. (F) Representative images of second-generation neurospheres derived from WT and cKO cortices. (G) Quantification of the diameters of neurospheres in F. Day 3 (WT: n = 46/3, cKO: n = 72/3), Day 5 (WT: n = 29/3, cKO: n = 26/3), Day 7 (WT: n = 20/3, cKO: n = 38/3), (n, neurosphere numbers/brain numbers). (H) Representative images of WT and cKO cortices stained for EdU (green), Ki67 (red), and with DAPI (blue). (I) Quantification of the number of Ki67+ cells per 100 μm column, and the percentage of EdU+; Ki67−/EdU+ cells represented exiting the cell cycle. WT: n = 12/3, cKO: n = 11/3. (J) Representative fluorescent images of E14.5 WT and cKO cortices stained for EdU (green), Ki67 (red), and with DAPI (blue). (K) Determination of the number of Ki67+ cells per 100 μm column (WT: n = 12/3, cKO: n = 15/4), and the percentage of EdU+; Ki67−/EdU+ cells (WT: n = 8/3, cKO: n = 13/4). All data are means ± SEM. Student’s t test. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001. n in B, C, E, I, and K = slice/brain numbers. [Scale bars: 50 μm (A, D, and F), 20 μm (H), 25 μm (J).]
In agreement with the altered densities of PAX6+ APCs in the VZ and TBR2+ IPCs in the SVZ, there was a substantial decrease in the density of mitotic cells (labeled with phosphorylated histone H3, P-HH3) in the VZ of Kctd10-cKO mice (Fig. 2 D and E). To further verify the effect of Kctd10 deletion on NPC proliferation, cortical neural stem cells were isolated and grown in suspension culture to form neurospheres. We found that neurospheres formed by Kctd10-cKO cells were smaller than those formed by WT cells (Fig. 2 F and G). These results demonstrate that the deletion of Kctd10 in the brain disrupts NPC proliferation.
Premature cell cycle exit and differentiation may also contribute to reduced NPCs. To test this hypothesis, the cell cycle exit index and neurogenesis were examined at E12.5, E14.5, and E16.5. EdU labeling was performed as described previously at E11.5, and brains were isolated at E12.5 and co-stained for the proliferation marker Ki67 to determine the numbers of EdU+, Ki67+, or EdU+Ki67− cells. The fraction of proliferating cells (EdU+Ki67+/EdU+) was significantly reduced in Kctd10-cKO brains. In contrast, the number of precursors that had exited the cell cycle (EdU+Ki67−/EdU+, cell cycle exit index) was markedly increased (Fig. 2 H and I). Similar results were obtained by repeating the experiment with brains labeled with EdU at E13.5 or E15.5 and collected at E14.5 or E16.5 (Fig. 2 J and K and SI Appendix, Fig. S5 A and B). Using a distinct genetic model of KCTD10 inactivation, we found that the cell cycle exit index was significantly elevated in the Kctd10 Emx1-cre cKO cortex at E13.5, when EdU labeling was performed at E12.5 (SI Appendix, Fig. S5 C and D). The observed changes occurred with a concomitant rise in the proportion of cells expressing TUJ1 (a marker of immature neurons) in the Kctd10-cKO brains on E15.5 (SI Appendix, Fig. S5 E and F).
To further examine the effects of KCTD10 level, we overexpressed KCTD10 in cortical neural progenitors at E13.5 using in utero electroporation. In contrast to Kctd10 knockdown (SI Appendix, Fig. S2 C–F), overexpression of KCTD10 resulted in a significantly increased fraction of GFP+ cells in the VZ/SVZ (SI Appendix, Fig. S5 G and H). Consistently, the percentage of GFP+SOX2+/GFP+ cells was significantly elevated in the VZ/SVZ of cortices overexpressing KCTD10 (SI Appendix, Fig. S5 I and J).
Overall, these results indicate that Kctd10 deletion results in a notable reduction of APCs and BPCs due to decreased cell proliferation and an increase in the number of cells exiting the cell cycle.
Kctd10 Deletion Results in Abnormal Lamination of the Cortex.
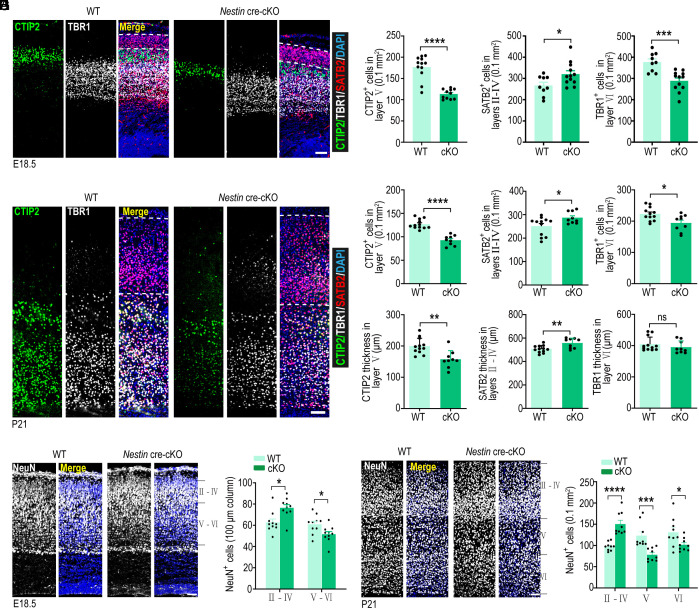
Because Kctd10 cKO results in impaired NPC self-renewal and smaller brain size, we examined whether Kctd10 deletion disturbed neuron generation and organization in the cortex. Brain sections from E18.5 and P21 were stained for TBR1 (a marker for deep layer VI neurons), CTIP2 (a marker of deep layer V–VI neurons) (50), and Satb2 (a marker of superficial layer II–IV neurons) (50). Interestingly, we observed a substantially lower TBR1+ and CTIP2+ cell density but an elevated SATB2+ cell density in the Kctd10-cKO cortex at both stages (Fig. 3 A–D). The thickness of the CTIP2+ cell layer was dramatically decreased at E18.5 and P21, while the thickness of the SATB2+ cells layer was found to be increased at P21 (Fig. 3 A–D and SI Appendix, Fig. S6A). Similar results were observed in E17.5 coronal cortices (rostral to caudal) and sagittal sections of Kctd10 Emx1-cre cKO mice (SI Appendix, Fig. S6 B–D). To determine whether Kctd10 loss affected the total number of neurons in the dorsal cortex, we stained sections from E18.5, P7, and P21 brains for NeuN. The density of NeuN+ cells in deep layers (layers V and VI) was substantially decreased in the Kctd10-cKO cortex at these three stages, accompanied by a significant increase in superficial layer (layer II–IV) neurons, changes that were consistent with the changes of marker gene expression from each layer (Fig. 3 E–H and SI Appendix, Fig. S6 E and F).
Fig. 3.
Kctd10 deficiency leads to abnormal neuron production. (A) Representative images of E18.5 WT and cKO cortical sections stained with SATB2 (red), CTIP2 (green), TBR1 (gray), and DAPI (blue). (B) Quantification of the number of SATB2+, CTIP2+, and TBR1+ neurons per 0.1 mm2 in A. CTIP2 (WT: n = 12/4, cKO: n = 9/3), TBR1, and SATB2 (WT: n = 9/3, cKO: n = 12/4). (C) Representative images of P21 WT and cKO cortical sections stained for SATB2 (red), CTIP2 (green), TBR1 (gray), and DAPI (blue). (D) Quantification of the number of CTIP2+, SATB2+, and TBR1+ neurons and their thickness in C. CTIP2, SATB2, and TBR1 (WT: n = 12/4, cKO: n = 9/3). (E) Representative coronal sections of E18.5 WT and cKO brains stained for NeuN (gray). (F) Quantification of NeuN+ cells per 100 μm column in E. WT: n = 12/4, cKO: n = 9/3. (G) Representative coronal sections of P21 WT and cKO brains stained for NeuN (gray). (H) Quantification of NeuN+ cells per 0.1 mm2 in G. WT and cKO: n = 9/3. All data are presented as means ± SEM. *P < 0.05, **P < 0.01, ns: no significance. Student’s t test. Slice thickness: 20 μm (A, C, and G), 30 μm (E). n in B, D, F, and H = slice/brain numbers. [Scale bars: 50 μm (A and E), 100 μm (C and G).]
Identification of KCTD13 as a KCTD10-Interacting Protein.
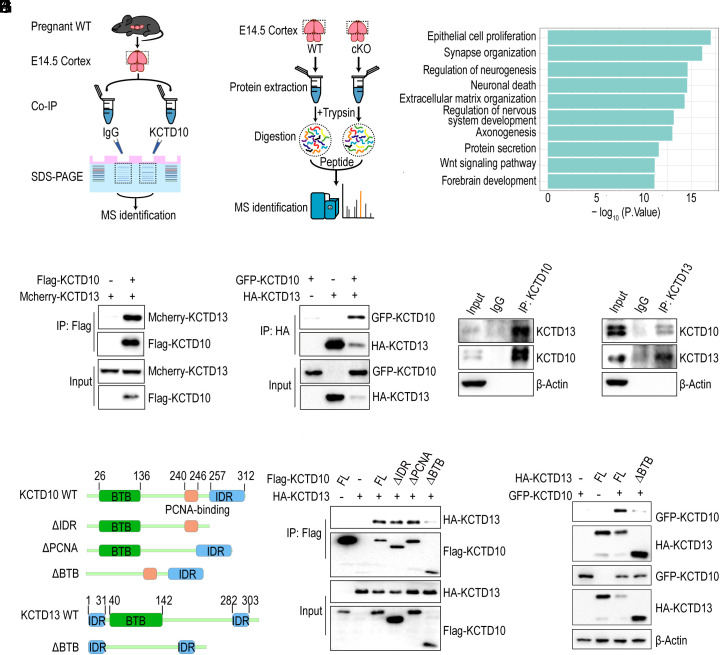
To determine the mechanism for how Kctd10 deficiency impacts brain development, co-immunoprecipitation (co-IP) was performed to identify KCTD10-interacting proteins from the E14.5 cerebral cortex using a KCTD10 antibody, followed by mass spectrometric analysis (Fig. 4A). In total, 143 proteins uniquely precipitated by the KCTD10 antibody were detected (SI Appendix, Table S1). Next, quantitative mass spectrometry (MS) comparing E14.5 WT and Kctd10-cKO cortical samples was performed to identify the functional consequence of KCTD10 loss on protein abundance (Fig. 4B). A total of 3,442 differentially expressed proteins (DEPs) were identified using three or more biological replicates. Gene Ontology (GO) analysis of the DEPs demonstrated their involvement in neurogenesis, nervous system development, forebrain development, synapse organization, neuronal death, axonogenesis, and protein transport (Fig. 4C). Interestingly, 27 of the 3,442 DEPs were significantly up-regulated or down-regulated in the cKO cortices and found to interact with KCTD10 by immunoprecipitation (SI Appendix, Tables S1 and S2). Among these 27 proteins, the level of KCTD13, which has been associated with ASD and schizophrenia and has been shown to regulate neurogenesis (24, 26, 48, 49), was elevated. These observations led us to investigate the relationship between KCTD10 and KCTD13.
Fig. 4.
KCTD10 interacts with KCTD13. (A) Schematic diagram illustrating the procedure of IP-MS assay. IP: immunoprecipitation, MS: mass spectrum. (B) Schematic plot illustrating the procedure of quantitative MS assay of E14.5 WT and cKO cortex. (C) GO enrichment analysis of DEPs identified in cKO cortices. (D) KCTD10 interacts with KCTD13. Co-IP with anti-Flag beads was performed with cell lysates from HEK293 cells transfected with Mcherry-KCTD13 alone or together with Flag-KCTD10, and probed with Flag and RFP antibodies. (E) The interaction of KCTD13 with KCTD10. Co-IP with an anti-HA antibody was performed with cell lysates from HEK293 cells transfected constructs as indicated and analyzed with HA and GFP antibodies. (F and G) Interaction between endogenous KCTD10 and KCTD13 in the cortex was confirmed by co-IP with KCTD10 (F) and KCTD13 (G) antibodies. (H) Schematic representation of KCTD10 and KCTD13 as well as their truncations. (I and J) The BTB domains of KCTD10 and KCTD13 are essential for their interaction. Co-IP was performed with HEK293 cells transfected with the indicated constructs using anti-Flag or HA antibodies and probed with Flag, HA, or GFP antibodies.
The interaction between KCTD10 and KCTD13 was first confirmed through co-IP assays involving both exogenous and endogenous proteins. To examine the exogenous interaction, Flag-KCTD10 was expressed either alone or in combination with mCherry-KCTD13 in HEK293 cells. The ensuing co-IP using anti-Flag beads demonstrated an interaction between KCTD10 and KCTD13 (Fig. 4D). In a reverse assay, HA-KCTD13 was expressed either alone or in combination with GFP-KCTD10. Again, co-IP with anti-HA beads demonstrated an interaction between KCTD13 and KCTD10 (Fig. 4E). To examine the endogenous interaction, the reciprocal co-IP experiments were performed using E14.5 cerebral cortex cell lysates and both KCTD10 and KCTD13 antibodies, demonstrating that these proteins physically interact in the developing cortex (Fig. 4 F and G).
Both KCTD10 and KCTD13 contain an N-terminal BTB domain. KCTD10 features an intrinsically disordered region (IDR) along with a PCNA-binding motif in the C-terminus, while KCTD13 contains an IDR on both the N and C termini (Fig. 4H). To pinpoint the domain(s) of KCTD10 involved in its interaction with KCTD13, several KCTD10 deletion products were constructed and examined for their capacity to bind KCTD13 via co-IP assays. Only the removal of the KCTD10 BTB domain, but not the other domains, dramatically reduced the interaction with KCTD13, indicating the importance of the BTB domain in facilitating the interaction between the two proteins (Fig. 4I). Similarly, the deletion of the KCTD13 BTB domain also significantly impeded its interaction with KCTD10, suggesting that the BTB domains of both proteins mediate their interaction (Fig. 4J). Taken together, these results indicate that KCTD10 and KCTD13 physically interact through the association of their BTB domains.
KCTD10 Negatively Regulates KCTD13 Protein Stability through Proteasomal Degradation.
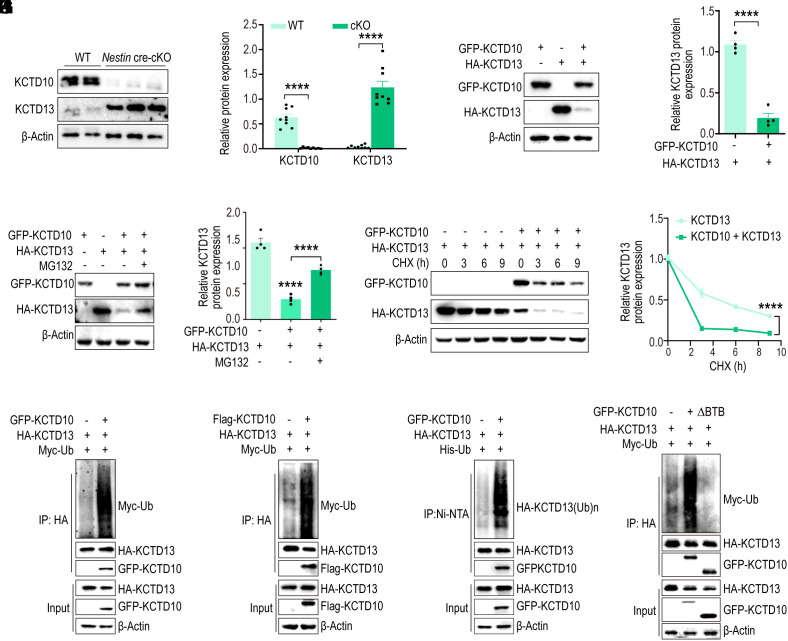
To decipher the functional impact of the KCTD10/KCTD13 interaction, we examined the expression of KCTD13 during mouse brain development. In contrast to the expression of KCTD10, the expression of KCTD13 increased gradually from E11.5 to P7 (SI Appendix, Fig. S7A). Subsequently, we assessed KCTD13 expression at protein and mRNA levels in the cortex of Kctd10-cKO mice. There was a striking elevation in KCTD13 protein levels in the Kctd10-cKO cortex at P14 (Fig. 5 A and B), whereas KCTD13 mRNA levels were unaltered (SI Appendix, Fig. S7 B and C). This implied that KCTD10 regulates KCTD13 protein levels at the post-translational level. Considering that KCTD10 functions as a substrate recognition receptor and regulates the ubiquitination of proteins (36, 37, 44), we hypothesized that KCTD10 might regulate KCTD13 stability via the proteasomal degradation pathway. To test this, HA-KCTD13 was transfected into HEK293 cells alone or in combination with GFP-KCTD10, resulting in decreased KCTD13 protein abundance (Fig. 5 C and D). However, the decrease in KCTD13 protein levels observed when KCTD10 was overexpressed could be abrogated by proteasome inhibitor MG132 (Fig. 5 E and F).
Fig. 5.
KCTD10 regulates the stability of KCTD13 through the proteasomal pathway. (A) P14 cortices from cKO and WT littermates were analyzed by immunoblotting for the expression of KCTD13 and KCTD10, with β-Actin as a loading control. (B) Quantification of relative levels normalized with β-Actin. N = 9 from > 3 independent experiments. (C) Overexpression of KCTD10 in HEK293 cells results in decreased KCTD13 levels. Cell lysates were analyzed by immunoblotting with GFP, HA, and β-Actin antibodies. (D) Quantification of relative levels of KCTD13 normalized with β-Actin. n = 4 from 4 independent experiments. (E) HEK293 cells were transfected with constructs as indicated and treated with MG132 for 4 h prior to analyzing with GFP and HA antibodies. (F) Quantification of relative levels of KCTD13 normalized with β-Actin. n = 4 from 4 independent experiments. (G) KCTD13 was transfected alone or together with KCTD10 and the half-life of KCTD13 was inspected by CHX chase assay by western blot analysis. (H) The relative level of KCTD13 was quantified and normalized against β-Actin. n = 3 independent experiments. (I–K) KCTD13 ubiquitination levels in HEK293 cells co-transfected with indicated constructs, together with or without GFP-KCTD10, and treated with MG132 for 4 h. KCTD13 was immunoprecipitated from cell lysates with HA antibody (I and J), and ubiquitinated proteins were pulled down by Ni-NTA beads from cell lysates under denaturing conditions (K) and immunoblotted with antibodies as indicated. (L) KCTD10 without the BTB domain cannot induce ubiquitination of KCTD13. HA-KCTD13 and Myc-Ub were co-transfected with KCTD10, immunoprecipitated with HA antibody, and probed with HA, GFP, and Myc antibodies. All data are presented as means ± SEM. ****P < 0.0001. Student’s t test (B and D), One-way ANOVA (F), Two-way ANOVA (H).
To further validate KCTD13 as a KCTD10-targeted substrate for proteasomal degradation, we found that the stability of KCTD13 diminished rapidly in the presence of KCTD10, as discerned by cycloheximide-based protein chase assays (Fig. 5 G and H). To determine whether KCTD10 mediates the ubiquitination-dependent degradation of KCTD13, we transfected HA-KCTD13 into HEK293 cells either alone or in combination with GFP-KCTD10 or Flag-KCTD10 and evaluated KCTD13 ubiquitination levels in vivo. Co-expression with either GFP-KCTD10 or Flag-KCTD10 increased KCTD13 ubiquitination (Fig. 5 I and J). To exclude the possibility that the observed diffuse ubiquitination signals originated from KCTD10-associated proteins, reciprocal His-Ub immunoprecipitation also demonstrated markedly increased ubiquitination signals after blotting with an HA antibody in cells co-expressing Flag-KCTD10, HA-KCTD13, and His-Ub (Fig. 5K). Moreover, the removal of the BTB domain of KCTD10 resulted in almost the complete loss of KCTD13 ubiquitination (Fig. 5L). Collectively, these results indicate that KCTD10 regulates the stability of KCTD13 via the proteasomal degradation pathway.
KCTD13 Overexpression Phenocopies the Defects in NPC Development Caused by Kctd10 cKO.
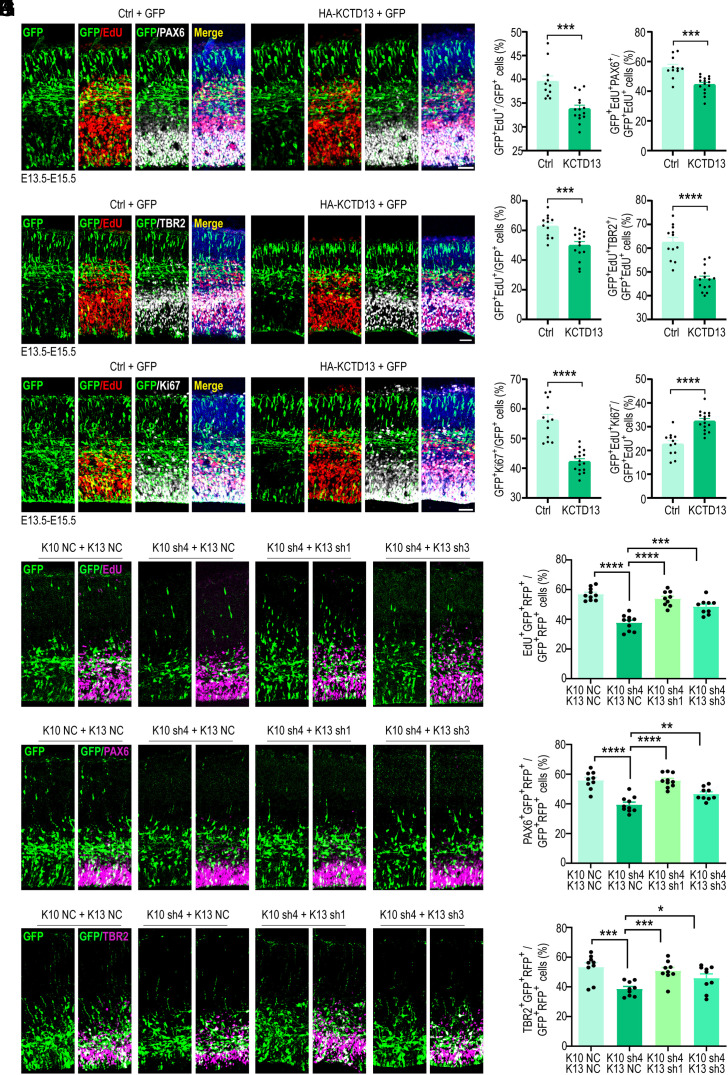
Downregulation of KCTD13 expression in zebrafish has been shown to stimulate the proliferation of NPCs (24). Consequently, we speculated that Kctd10 cKO results in a substantially elevated KCTD13 level, leading to the suppression of NPC proliferation. To test this, we overexpressed KCTD13 in cortical neuronal progenitors at E13.5 using in utero electroporation and evaluated the subsequent impact on cortical development at E15.5. EdU pulse labeling was conducted 24 h before analysis (E14.5), and brain sections were immunolabelled for GFP and PAX6. The overexpression of KCTD13 resulted in a significant reduction in EdU incorporation within the GFP+ cells, compared to overexpressing GFP alone. Additionally, we observed a reduction in the population of dividing APCs in cells overexpressing KCTD13, characterized by a prominent decrease in the ratio of EdU+ Pax6+ GFP+/EdU+ GFP+ cells (Fig. 6 A and B). This is similar to the result observed after inactivating Kctd10. Co-immunostaining for GFP, TBR2, and EdU further revealed a notable decrease in GFP+ EdU+ cells in NPCs overexpressing KCTD13. As anticipated, the GFP+ TBR2+ cell population was also greatly reduced in NPCs overexpressing KCTD13, signaling a reduction of IPCs (Fig. 6 C and D). The quantity of GFP+ EdU+ TBR2+ cells was also significantly reduced, further signifying a reduced number of cycling IPCs. Consistent with this decline, the proportion of GFP+ Ki67+/GFP+ cells was also significantly decreased (Fig. 6 E and F). To determine whether the reduction in NPC proliferation was due to premature cell cycle exit, cortical sections were stained for GFP, Ki67, and EdU. KCTD13 overexpression resulted in increased cell cycle exit (Fig. 6 E and F). Additionally, the overexpression of KCTD13 led to an abnormal distribution of GFP+ cells, exemplified by a prominent decrease in the VZ/SVZ and IZ, accompanying a significant increase in the CP, results that further imitate the Kctd10 knock-out or -down phenotypes (SI Appendix, Fig. S8A).
Fig. 6.
Overexpression of KCTD13 inhibits NPC proliferation and promotes NPC differentiation. (A–F) All images are from E15.5 cortices electroporated at E13.5 with empty vector (Ctrl) or HA-KCTD13 together with a GFP-expressing construct and stained with indicated antibodies 24 h after EdU labeling at E14.5. (A) Representative images stained for GFP (green), EdU (red), PAX6 (gray), and DAPI (blue). (B) Quantification of transfected cells positive for EdU or PAX6. Ctrl: n = 11/3, HA-KCTD13: n = 15/4. (C) Representative images stained for GFP (green), EdU (red), TBR2 (gray), and DAPI (blue). (D) Quantification of transfected cells positive for EdU or TBR2. Ctrl: n = 12/3, HA-KCTD13: n = 15/4. (E) Representative images stained for GFP (green), EdU (red), Ki67 (gray), and DAPI (blue). (F) Quantification of transfected cells positive for EdU or Ki67. Ctrl: n = 12/3, HA-KCTD13: n = 16/4. (G–L) All images depict E15.5 cortices electroporated at E13.5 with the following constructs: empty vector (K10 NC+K13 NC), K10 sh4 + K13 NC, K10 sh4 + K13 sh1, or K10 sh4 + K13 sh3. The brain sections were stained with the indicated antibodies 24 h after EdU labeling at E14.5. (G) Representative images depicting GFP (green) and EdU (magenta) staining. (H) Quantification of transfected cells positive for EdU. K10 NC + K13 NC and K10 sh4 + K13 NC: n = 10/3, K10 sh4 + K13 sh1 and K10 sh4 + K13 sh3: n = 9/3. (I) Representative images depicting GFP (green) and PAX6 (magenta) staining. (J) Quantification of transfected cells expressing PAX6. K10 NC + K13 NC: n = 9/3, K10 sh4 + K13 NC and K10 sh4 + K13 sh1: n = 10/3, K10 sh4 + K13 sh3: n = 9/3. (K) Representative images stained for GFP (green) and TBR2 (magenta). (L) Quantification of transfected cells expressing TBR2. K10 NC + K13 NC, K10 sh4 + K13 NC, K10 sh4 + K13 sh1, K10 sh4 + K13 sh3: n = 9/3. All data are presented as means ± SEM. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001, Student’s t test. n = slice/brain numbers. [Scale bars: 50 μm (A, C, E, G, I, and K).]
To investigate whether the knockdown of Kctd13 can rescue the NPC proliferation defects caused by Kctd10 knockdown, we constructed four shRNAs fused with RFP targeting Kctd13. Among them, K13 sh1 and K13 sh4 effectively down-regulated KCTD13 overexpression in HEK293 cells (SI Appendix, Fig. S8 B and C). We then introduced scrambled shRNA (NC) or K10 sh4 as well as K13 sh1 or K13 sh3 into cortical neuronal progenitors at E13.5 using in utero electroporation, performed EdU pulse labeling at E14.5, and harvested tissue at E15.5 for analysis. E15.5 brain sections were stained for GFP, RFP, and EdU, PAX6, or TBR2. Nearly all of transfected cells exhibited positivity for both GFP and RFP (SI Appendix, Fig. S8D). Knockdown of KCTD13 was shown to rescue decreased NPCs proliferation induced by KCTD10 knockdown (Fig. 6 G–L). These results supported the model that KCTD10 regulates neuronal development by controlling the level of KCTD13 during cortical development.
Discussion
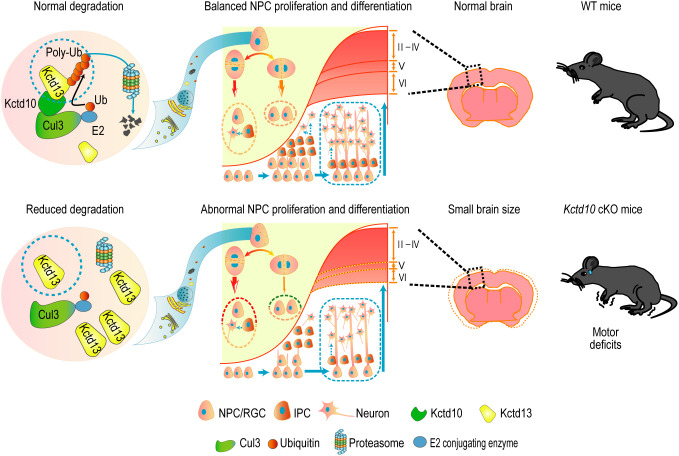
Our research elucidates a critical role for KCTD10 in regulating NPC proliferation and differentiation during brain development. We demonstrate that Kctd10 cKO results in reduced brain size and motor deficits. Mechanistically, KCTD10 orchestrated NPC development by interacting with KCTD13 to regulate KCTD13 protein stability by facilitating KCTD13 degradation via the proteasomal pathway (Fig. 7).
Fig. 7.
The regulatory mechanism of KCTD10 in the brain. Schematic model illustrating the regulation of cortical neurogenesis by KCTD10 through priming KCTD13 for degradation via the proteasomal pathway, preserving NPC proliferation and preventing premature differentiation. Kctd10 cKO in the brain results in smaller brain size and motor deficits.
KCTD10 is highly expressed in embryonic and neonatal mouse cortices but gradually decreases in expression as corticogenesis progresses. Compared to the cerebral cortex, KCTD10 expression in the hippocampus escalates from P7 and peaks at P28, suggesting a role for KCTD10 in hippocampal development. Our findings show that Kctd10 deletion results in a substantial decrease in the abundance of cortical layer V and VI neurons, with an increase in the abundance of layer II–IV neurons. During early neurogenesis, RGCs self-renew and give rise to neurons destined for deep layers of the cortex. As neurogenesis continues, RGCs (or APCs) produce IPCs, which undergo one or two rounds of division before giving rise to pairs of neurons destined for the upper cortical layers (51, 52). Therefore, the decreased production of deep-layer neurons in Kctd10 cKO cortices likely stems from reduced APC proliferation. Kctd10 deficiency increased the abundance of layer II–IV neurons, likely downstream of multiple contributing factors. First, premature cell cycle exit at early brain development stages might directly lead to the expanded generation of layer II–IV neurons. Second, the increased abundance of layer II–IV neurons may be due to a compensatory effect resulting from the reduction of layer V and VI neurons downstream of Kctd10 cKO. Finally, Kctd10 deficiency might alter the neuronal identity of layer V and VI neurons to layer II–IV neuronal fates. The reduction observed in layer V projection neurons is consistent with the high KCTD10 expression in layer V CTIP2+ neurons. As a transcription factor, CTIP2 (BCL11b) might function not only in developing layer V neurons but also in fate determination. CTIP2 variants have been identified in patients with neurodevelopmental disorders (53, 54). Notably, CTIP2 is expressed in cortico-spinal motor neurons and is essential for cortico-spinal connections (55). Motor deficits in the hind limbs of the Kctd10-cKO mice suggest that Kctd10 deficiency might result in the loss of cortico-spinal projections.
We identified KCTD13 as a unique KCTD10-interacting protein. KCTD10 was shown to facilitate the ubiquitination-dependent degradation of KCTD13 by intercting with the two proteins through their respective BTB domains, which was required for KCTD13 ubiquitination. Deletions and duplications of 16p11.2, the genomic locus containing KCTD13, are recognized as drivers of genetic risk for neuropsychiatric disorders. Among the 29 genes present within this locus, KCTD13 is considered the primary contributor to neurodevelopmental disorders by regulating brain development (24, 46). However, the role of KCTD13 in regulating NPC proliferation and differentiation remains inconsistent. Discrepancies in previous reports examining KCTD13 function might reflect knockdown and/or off-target effects related to the method of KCTD13 inactivation (24, 56). Our functional screening suggests that KCTD13 overexpression phenocopies Kctd10 deficiency, supporting the model of KCTD10 regulating cortical development by controlling KCTD13 protein levels. Nevertheless, it is possible that other KCTD10 substrates also contribute to brain development. Given the close relationship between KCTD10 and two other high-confidence ASD risk proteins, KCTD13 and CUL3, our work suggests screening for KCTD10 mutations in patients with brain developmental disorders would likely result in the identification of causal genetic mutations.
In summary, our study demonstrates that KCTD10 negatively regulates KCTD13 stability through the ubiquitin/proteasome degradation pathway within the developing mouse brain to control NPC proliferation and differentiation. We propose that the motor deficits in Kctd10 cKO mice may stem from abnormal KCTD13 accumulation, leading to perturbed brain development. This research provides unique insights into the potential pathogenesis of brain developmental disorders.
Materials and Methods
Animals.
The Kctd10fl/fl mice used in this study were purchased from GemPharmatech (Nanjing, China). To obtain Kctd10-cKO mice, Kctd10fl/fl homozygous mice were crossed with Nestin-Cre mice [Jackson Laboratory, B6. Cg (SJL)-TgN (Nes-Cre)1Kln] and Emx1-Cre mice (Gift from Kun Xia, Central South University, Changsha, Hunan, China).
Antibodies.
The following antibodies were used for western blotting: anti-Human KCTD10 (1:1,000, PA553138, Invitrogen), anti-Human KCTD13 (1:1,000, PA541082, Invitrogen; 1:1,000, HPA043524, Sigma), anti-Flag (1:1,000, M185, MBL), anti-Human Myc (1:5,000, 9E10, Santa Cruz), anti-HA (1:5,000, M180-3, MBL), and anti-β-actin (1:10,000, A5441, Sigma). The antibodies used for immunostaining were as follows: anti-KCTD10 (1:500, generated by collaboration with Huaan Biotechnology Co., Ltd., not commercially available), anti-HA (1:1,000, 3724S, CST), anti-Myc (1:1,000, 2272S, CST), anti-Flag (1:5,000, F1804, Sigma), anti-PAX6 (1:400, PRB-278P, Covance), anti-TBR2 (1:500, 14-4875-82, Invitrogen), anti-phosphorylated (p)-histone H3 (1:500, ab10543, Abcam), anti-GFP (1:1,000, ab13970, Abcam), anti-Ki67 (1:1,000, ab15580, Abcam), anti-SATB2 (1:400, ab51502, Abcam), anti-TBR1 (1:500, ab31940, Abcam), anti-NeuN (rabbit polyclonal, 1:1,000; ab104225, Abcam), and anti-NeuN (mouse monoclonal, 1:1,000; ab104224, Abcam). EdU staining was performed using the Click-iT EdU Cell Proliferation Kit for Imaging (C10338, Invitrogen) following the manufacturer’s instructions. DAPI was used for nuclear counterstaining (Invitrogen).
Assay of Genic Co-Expression.
An evaluation of co-expression was carried out employing RNA-sequencing data across multiple stages of development (ranging from 8 post-conception weeks to 40 y) for various brain regions, as sourced from Brainspan (http://www.brainspan.org/). Specifically, a gene exhibiting an RPKM value greater than 0.5 in 80% of all developing cortex samples was deemed to be cortex-expressed. A set of 102 confidently identified ASD-risk genes from prior research were designated as the known ASD gene set (57). Then, 411 recognized NDD genes, inclusive of the aforementioned 102 ASD-risk genes, were identified in published studies (57–59). These gene sets were utilized to compute the Spearman's correlation coefficient with all cortex-expressed genes. Following this, the mean of the Spearman's correlation coefficient for each cortex-expressed gene with the known NDD gene set was calculated. The percentile of the average correlation coefficient between KCTD10 and NDD gene set was determined.
IP–MS.
First, appropriate amounts of protein A/G beads were washed three times in 1 mL of PBS and then centrifuged at 500 rpm for 20 s at 4 °C. The beads were subsequently incubated with 1 μg of anti-KCTD10 or anti-IgG antibody at 4 °C for 1.5 h. Meanwhile, mouse embryonic (E15) brain cortices were isolated and dissociated in cell lysis buffer (50 mM NaCl, 0.5% NP-40, 10 mM HEPES, 0.5 mM EDTA, 20 mM Tris, pH 7.4) supplemented with Complete Protease Inhibitor Tablets (Roche Applied Science) and 1 mM phenylmethanesulfonyl fluoride (PMSF). The protein concentrations were measured by bicinchoninic acid assay (BCA). After incubation, the supernatant was removed, and 2 mg of protein samples were, respectively, added to the beads, followed by incubation overnight at 4 °C with rotation. The next day, the immunoprecipitates were washed five to six times with cell lysis buffer, eluted with 2× SDS sample buffer, and analyzed by sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS–PAGE). After dyeing and decolorizing, a 1 × 1 cm gel slice was placed in a 1.5 mL tube. A small amount of deionized water was added to the gel to keep it moist. The LC–MS/MS analysis was performed by Jingjie Biological Co., LTD using a Thermo QE PLUS OBRITRAP high-resolution mass spectrometer. The proteomics data obtained from IP-MS have been successfully deposited to the Proteo-meXchange Consortium through the PRIDE partner repository. The dataset identifier assigned to this deposition is PXD050135 (60).
Quantitative LC–MS/MS Analysis.
WT and Kctd10 cKO cortices at E14.5 were extracted from −80 °C, weighed into a pre-chilled mortar, and thoroughly pulverized in liquid nitrogen. Sample groups were, respectively, added with quadruple the volume of the powder lysis buffer (comprising 8 M urea and 1% protease inhibitor), followed by ultrasonication and centrifugation at 12,000 g for 10 min at 4 °C. Upon centrifugation, cellular debris was discarded and the supernatant was moved into a fresh centrifuge tube. The protein concentration was measured using the BCA method. For each sample, protein was digested in equal quantities, the volume was adjusted with lysate, an equal volume of pre-chilled acetone was added, and subsequently, four times the volume of pre-chilled acetone was added post-vortex mixing to precipitate for 2 h at −20 °C. The sediments were procured by centrifugation at 4,500 g for 5 min and washed with pre-chilled acetone 2 to 3 times. Once dried, TEAB with a final concentration of 200 mM was employed to resuspend the precipitate using ultrasonication. Subsequently, trypsin was added at a 1:50 ratio (protease: protein, w/w) to enzymatically hydrolyze overnight. Dithiothreitol was added to a final concentration of 5 mM to deoxidize at 56 °C for 30 min. Then, iodoacetamide was incorporated to achieve a final concentration of 11 mM and incubated at room temperature for 15 min in a dark environment. Peptides for proteome analysis were dissolved in buffer A (comprising 0.1% formic acid and 2% acetonitrile), which is an aqueous solution, and were separated using the EASY-nLC 1200 ultra-high performance liquid chromatography system. The segmented gradient setting was as follows: 0 to 68 min, 6 to 23% solvent B (0.1% formic acid and 90% acetonitrile); 68 to 82 min, 23 to 32% solvent B; 82 to 86 min, 32 to 80% solvent B; 86 to 90 min, 80% solvent B. After separation, peptides were injected into an nanospray flex (NSI) ion source for electrification and then loaded into the Orbitrap Exploris 480 mass spectrometer for analysis. The results were filtered by medium and high confidence peptides with a false discovery rate below 5%. The MS proteomics data have been deposited to the Proteo-meXchange Consortium via the PRIDE partner repository with the dataset identifier PXD050077 (60).
Statistical Analysis.
All experiments were repeated at least three times. All data were analyzed with GraphPad Prism 8 (GraphPad Software, Inc.) and are presented as means ± SEM. The Student’s t test and two-way ANOVA were used for comparisons between and among groups, respectively. A P-value < 0.05 was considered significant.
Other methods and materials are shown in SI Appendix, Supplemental Methods and Materials.
Supplementary Material
Appendix 01 (PDF)
Kctd10-cKO mice exhibited motor deficits.
Acknowledgments
We thank Dr. Qingfeng Wu and our colleagues from the QinXueHaoWen forum for their constructive comments. This work was partially supported by National Natural Science Foundation of China (32070980, 32271026, 81901168, 32330038 and 32394030), Huxiang Young Talents Program (2020RC3006, 2022RC1163), Natural Science Foundation of Hunan Province (2021JJ20075, 2020JJ4689, 2018DK201, 2022JJ20056 and 2019JJ40408), Innovation Driven Project of Central South University (2020CX022), Open Project of Chinese Academy of Sciences (2022-MDB-KF-24); This work was also supported by the High Performance Computing Center of Central South University.
Author contributions
Z.X. and L.Y. designed research; J.C., Z.W., M.T., W.Z., G.L., C.M., M.H., and X.J. performed research; S.T., D.Z., Y.W., H.G., D.X., L.L., J.L., K.X., F.L., and R.D. analyzed data; and L.Y. wrote the paper.
Competing interests
The authors declare no competing interest.
Footnotes
This article is a PNAS Direct Submission.
Contributor Information
Zhiheng Xu, Email: zhxu@genetics.ac.cn.
Ling Yuan, Email: yuanling@sklmg.edu.cn.
Data, Materials, and Software Availability
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository (accession no. PXD050135 and PXD050077) (60). And this paper does not report original code. All study data are included in the article and/or supporting information.
Supporting Information
References
- 1.Haubensak W., Attardo A., Denk W., Huttner W. B., Neurons arise in the basal neuroepithelium of the early mammalian telencephalon: A major site of neurogenesis. Proc. Natl. Acad. Sci. U.S.A. 101, 3196–3201 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mihalas A. B., et al. , Intermediate progenitor cohorts differentially generate cortical layers and require Tbr2 for timely acquisition of neuronal subtype identity. Cell Rep. 16, 92–105 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ayala R., Shu T., Tsai L. H., Trekking across the brain: The journey of neuronal migration. Cell 128, 29–43 (2007). [DOI] [PubMed] [Google Scholar]
- 4.Dong Z., et al. , CUL3 deficiency causes social deficits and anxiety-like behaviors by impairing excitation-inhibition balance through the promotion of cap-dependent translation. Neuron 105, 475–490.e6 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kaindl A. M., et al. , Many roads lead to primary autosomal recessive microcephaly. Prog. Neurobiol. 90, 363–383 (2010). [DOI] [PubMed] [Google Scholar]
- 6.Moon H. M., Wynshaw-Boris A., Cytoskeleton in action: Lissencephaly, a neuronal migration disorder. Wiley Interdiscip. Rev. Dev. Biol. 2, 229–245 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sahin M., Sur M., Genes, circuits, and precision therapies for autism and related neurodevelopmental disorders. Science 350, aab3897 (2015), 10.1126/science.aab3897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Alazami A. M., et al. , Accelerating novel candidate gene discovery in neurogenetic disorders via whole-exome sequencing of prescreened multiplex consanguineous families. Cell Rep. 10, 148–161 (2015). [DOI] [PubMed] [Google Scholar]
- 9.Trujillano D., et al. , Clinical exome sequencing: results from 2819 samples reflecting 1000 families. Eur. J. Hum. Genet. 25, 176–182 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Faqeih E. A., et al. , Phenotypic characterization of KCTD3-related developmental epileptic encephalopathy. Clin. Genet. 93, 1081–1086 (2018). [DOI] [PubMed] [Google Scholar]
- 11.Metz K. A., et al. , KCTD7 deficiency defines a distinct neurodegenerative disorder with a conserved autophagy-lysosome defect. Ann. Neurol. 84, 766–780 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Van Bogaert P., et al. , Mutation of a potassium channel-related gene in progressive myoclonic epilepsy. Ann. Neurol. 61, 579–586 (2007). [DOI] [PubMed] [Google Scholar]
- 13.Farhan S. M., et al. , Linkage analysis and exome sequencing identify a novel mutation in KCTD7 in patients with progressive myoclonus epilepsy with ataxia. Epilepsia 55, e106-11 (2014). [DOI] [PubMed] [Google Scholar]
- 14.Krabichler B., et al. , Novel mutation in potassium channel related gene KCTD7 and progressive myoclonic epilepsy. Ann. Hum. Genet. 76, 326–331 (2012). [DOI] [PubMed] [Google Scholar]
- 15.Kousi M., et al. , Novel mutations consolidate KCTD7 as a progressive myoclonus epilepsy gene. J. Med. Genet. 49, 391–399 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vairo F. P., et al. , The prevalence of diseases caused by lysosome-related genes in a cohort of undiagnosed patients. Mol. Genet. Metab. Rep. 13, 46–51 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ebrahimi-Fakhari D., et al. , The spectrum of movement disorders in childhood-onset lysosomal storage diseases. Mov. Disord. Clin. Pract. 5, 149–155 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lindy A. S., et al. , Diagnostic outcomes for genetic testing of 70 genes in 8565 patients with epilepsy and neurodevelopmental disorders. Epilepsia 59, 1062–1071 (2018). [DOI] [PubMed] [Google Scholar]
- 19.Mastrangelo M., et al. , Progressive myoclonus epilepsy and ceroidolipofuscinosis 14: The multifaceted phenotypic spectrum of KCTD7-related disorders. Eur. J. Med. Genet. 62, 103591 (2019). [DOI] [PubMed] [Google Scholar]
- 20.Lee M. T., et al. , Genome-wide association study of bipolar I disorder in the Han Chinese population. Mol. Psychiatry 16, 548–556 (2011). [DOI] [PubMed] [Google Scholar]
- 21.Sibille E., et al. , A molecular signature of depression in the amygdala. Am. J. Psychiatry 166, 1011–1024 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Benes F. M., Amygdalocortical circuitry in schizophrenia: From circuits to molecules. Neuropsychopharmacology 35, 239–257 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Madison J. M., et al. , Regulation of purine metabolism connects KCTD13 to a metabolic disorder with autistic features. iScience 24, 101935 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Golzio C., et al. , KCTD13 is a major driver of mirrored neuroanatomical phenotypes of the 16p11.2 copy number variant. Nature 485, 363–367 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Crepel A., et al. , Narrowing the critical deletion region for autism spectrum disorders on 16p11.2. Am. J. Med. Genet. B Neuropsychiatr. Genet. 156, 243–245 (2011). [DOI] [PubMed] [Google Scholar]
- 26.Degenhardt F., et al. , Identification of rare variants in KCTD13 at the schizophrenia risk locus 16p11.2. Psychiatr. Genet. 26, 293–296 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mencacci N. E., et al. , A missense mutation in KCTD17 causes autosomal dominant myoclonus-dystonia. Am. J. Hum. Genet. 96, 938–947 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mencacci N. E., Bruggemann N., KCTD17 is a confirmed new gene for dystonia, but is it responsible for SGCE-negative myoclonus-dystonia? Parkinsonism Relat. Disord. 61, 1–3 (2019). [DOI] [PubMed] [Google Scholar]
- 29.Graziola F., et al. , A novel KCTD17 mutation is associated with childhood early-onset hyperkinetic movement disorder. Parkinsonism Relat. Disord. 61, 4–6 (2019). [DOI] [PubMed] [Google Scholar]
- 30.Marce-Grau A., et al. , Childhood onset progressive myoclonic dystonia due to a de novo KCTD17 splicing mutation. Parkinsonism Relat. Disord. 61, 7–9 (2019). [DOI] [PubMed] [Google Scholar]
- 31.Usui D., et al. , Interstitial duplication of 2q32.1-q33.3 in a patient with epilepsy, developmental delay, and autistic behavior. Am. J. Med. Genet. A 161A, 1078–1084 (2013). [DOI] [PubMed] [Google Scholar]
- 32.Bosch D. G., et al. , Novel genetic causes for cerebral visual impairment. Eur. J. Hum. Genet. 24, 660–665 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Boada M., et al. , ATP5H/KCTD2 locus is associated with Alzheimer’s disease risk. Mol. Psychiatry 19, 682–687 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Furuta N., et al. , Reduced expression of BTBD10 in anterior horn cells with Golgi fragmentation and pTDP-43-positive inclusions in patients with sporadic amyotrophic lateral sclerosis. Neuropathology 33, 397–404 (2013). [DOI] [PubMed] [Google Scholar]
- 35.Rodriguez-Perez F., et al. , Ubiquitin-dependent remodeling of the actin cytoskeleton drives cell fusion. Dev. Cell 56, 588–601.e9 (2021). [DOI] [PubMed] [Google Scholar]
- 36.Kovacevic I., et al. , The Cullin-3-Rbx1-KCTD10 complex controls endothelial barrier function via K63 ubiquitination of RhoB. J. Cell Biol. 217, 1015–1032 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tong X., et al. , Kctd10 regulates heart morphogenesis by repressing the transcriptional activity of Tbx5a in zebrafish. Nat. Commun. 5, 3153 (2014). [DOI] [PubMed] [Google Scholar]
- 38.Geschwind D. H., State M. W., Gene hunting in autism spectrum disorder: On the path to precision medicine. Lancet Neurol. 14, 1109–1120 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Iossifov I., et al. , The contribution of de novo coding mutations to autism spectrum disorder. Nature 515, 216–221 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.De Rubeis S., et al. , Synaptic, transcriptional and chromatin genes disrupted in autism. Nature 515, 209–215 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Murakami A., et al. , Cullin-3/KCTD10 E3 complex is essential for Rac1 activation through RhoB degradation in human epidermal growth factor receptor 2-positive breast cancer cells. Cancer Sci. 110, 650–661 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nagai T., Mukoyama S., Kagiwada H., Goshima N., Mizuno K., Cullin-3-KCTD10-mediated CEP97 degradation promotes primary cilium formation. J. Cell Sci. 131, jcs219527 (2018). [DOI] [PubMed] [Google Scholar]
- 43.Maekawa M., et al. , Cullin-3/KCTD10 complex is essential for K27-polyubiquitination of EIF3D in human hepatocellular carcinoma HepG2 cells. Biochem. Biophys. Res. Commun. 516, 1116–1122 (2019). [DOI] [PubMed] [Google Scholar]
- 44.Wu X., et al. , Regulation of TRIF-mediated innate immune response by K27-linked polyubiquitination and deubiquitination. Nat. Commun. 10, 4115 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ren K., et al. , KCTD10 is involved in the cardiovascular system and Notch signaling during early embryonic development. PLoS One 9, e112275 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lin G. N., et al. , Spatiotemporal 16p11.2 protein network implicates cortical late mid-fetal brain development and KCTD13-Cul3-RhoA pathway in psychiatric diseases. Neuron 85, 742–754 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Howrigan D. P., et al. , Exome sequencing in schizophrenia-affected parent-offspring trios reveals risk conferred by protein-coding de novo mutations. Nat. Neurosci. 23, 185–193 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.An J. Y., et al. , Genome-wide de novo risk score implicates promoter variation in autism spectrum disorder. Science 362, eaat6576 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rk C. Y., et al. , Whole genome sequencing resource identifies 18 new candidate genes for autism spectrum disorder. Nat. Neurosci. 20, 602–611 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Greig L. C., Woodworth M. B., Galazo M. J., Padmanabhan H., Macklis J. D., Molecular logic of neocortical projection neuron specification, development and diversity. Nat. Rev. Neurosci. 14, 755–769 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Noctor S. C., Martinez-Cerdeno V., Ivic L., Kriegstein A. R., Cortical neurons arise in symmetric and asymmetric division zones and migrate through specific phases. Nat. Neurosci. 7, 136–144 (2004). [DOI] [PubMed] [Google Scholar]
- 52.Kwan K. Y., Sestan N., Anton E. S., Transcriptional co-regulation of neuronal migration and laminar identity in the neocortex. Development (Cambridge, England) 139, 1535–1546 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yu Y., et al. , A novel variant in BCL11B in an individual with neurodevelopmental delay: A case report. Mol. Genet. Genomic Med. 11, e2132 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Prasad M., et al. , BCL11B-related disorder in two canadian children: Expanding the clinical phenotype. Eur. J. Med. Genet. 63, 104007 (2020). [DOI] [PubMed] [Google Scholar]
- 55.Arlotta P., et al. , Neuronal subtype-specific genes that control corticospinal motor neuron development in vivo. Neuron 45, 207–221 (2005). [DOI] [PubMed] [Google Scholar]
- 56. C. O., et al. , Kctd13 deletion reduces synaptic transmission via increased RhoA. Nature 551, 227–231 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Satterstrom F. K., et al. , Large-scale exome sequencing study implicates both developmental and functional changes in the neurobiology of autism. Cell 180, 568–584.e23 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kaplanis J., et al. , Evidence for 28 genetic disorders discovered by combining healthcare and research data. Nature 586, 757–762 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Coe B. P., et al. , Neurodevelopmental disease genes implicated by de novo mutation and copy number variation morbidity. Nat. Genet. 51, 106–116 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Perez-Riverol Y., et al. , The PRIDE database resources in 2022: A hub for mass spectrometry-based proteomics evidences. Nucleic Acids Res. 50, D543–D552 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix 01 (PDF)
Kctd10-cKO mice exhibited motor deficits.
Data Availability Statement
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository (accession no. PXD050135 and PXD050077) (60). And this paper does not report original code. All study data are included in the article and/or supporting information.