Significance
Mitotic progression into anaphase is monitored by the spindle assembly checkpoint that is poorly understood in plants. Using Arabidopsis thaliana as a model system, we identified an interaction pattern centered at the BUB1 and MAD3 protein BMF3 at kinetochores. A noncanonical isoform of the evolutionarily conserved BUB3 family protein BUB3.3 interacted with two internal repeats in BMF3 for recruiting the CDC20 protein to unattached kinetochores in order to inhibit its function in anaphase onset. Hence, our work sheds light on how the spindle assembly checkpoint operates in flowering plants that produce the highly conserved BUB3.3 protein but divergent BMF proteins.
Keywords: Arabidopsis, kinetochore, spindle assembly checkpoint, BUB3, CDC20
Abstract
The spindle assembly checkpoint (SAC) ensures faithful chromosome segregation during cell division by monitoring kinetochore-microtubule attachment. Plants produce both sequence-conserved and diverged SAC components, and it has been largely unknown how SAC activation leads to the assembly of these proteins at unattached kinetochores to prevent cells from entering anaphase. In Arabidopsis thaliana, the noncanonical BUB3.3 protein was detected at kinetochores throughout mitosis, unlike MAD1 and the plant-specific BUB1/MAD3 family protein BMF3 that associated with unattached chromosomes only. When BUB3.3 was lost by a genetic mutation, mitotic cells often entered anaphase with misaligned chromosomes and presented lagging chromosomes after they were challenged by low doses of the microtubule depolymerizing agent oryzalin, resulting in the formation of micronuclei. Surprisingly, BUB3.3 was not required for the kinetochore localization of other SAC proteins or vice versa. Instead, BUB3.3 specifically bound to BMF3 through two internal repeat motifs that were not required for BMF3 kinetochore localization. This interaction enabled BMF3 to recruit CDC20, a downstream SAC target, to unattached kinetochores. Taken together, our findings demonstrate that plant SAC utilizes unconventional protein interactions for arresting mitosis, with BUB3.3 directing BMF3’s role in CDC20 recruitment, rather than the recruitment of BUB1/MAD3 proteins observed in fungi and animals. This distinct mechanism highlights how plants adapted divergent versions of conserved cell cycle machinery to achieve specialized SAC control.
Cell division results in the production of two daughter cells, each inheriting an equal amount of genetic material. This is achieved by aligning all chromosomes at the metaphase plate, a process facilitated by the attachment of sister kinetochores to microtubule fibers that originate from opposite spindle poles prior to anaphase onset. The spindle assembly checkpoint (SAC) monitors this amphitelic chromosome attachment or biorientation, and its functionality relies on proteins known as Budding Uninhibited by Benzimidazoles (BUB) and Mitotic Arrest Defective (MAD) (1). In fungi and animals, SAC activation triggers the kinase-dependent recruitment of MAD1 and MAD2 to kinetochores, causing a conformational change in MAD2. Activated or conformationally closed MAD2 is joined by BUBR1/MAD3 which is docked on the kinetochores by interaction with the WD40 repeat protein BUB3. BUB3, BUBR1/MAD3, and MAD2 form the mitotic checkpoint complex (MCC) with CDC20 which is the activator of the Anaphase Promoting Complex/Cyclosome (APC/C). MCC formation prevents anaphase onset, resulting in mitotic arrest at prometaphase due to SAC activation (1).
The genomes of flowering plants like Arabidopsis thaliana contain genes encoding BUB3, MAD1, and MAD2 homologs resembling their counterparts in fungi and animals, as well as three proteins distantly related to BUB1 and BUBR1/MAD3 (2). These three BUB1/MAD3 family proteins (BMFs) all possess a BUB1/MAD3-type tetratricopeptide repeat (TPR) domain but otherwise share little if any homology outside this N-terminal domain (2). Only BMF1, but not BMF2 and BMF3, has a C-terminal kinase domain, and none of the BMF proteins possess the BUB3-interacting Gle2-binding sequence (GLEBS) domain found in fungal and animal BUB1 and BUBR1/MAD3 proteins (3, 4). Therefore, it is unknown whether the BUB3 protein interacts with BMFs or whether it is a bona fide SAC component acting at kinetochores that trigger SAC activation.
A characteristic feature of SAC components in animal cells is their presence at the kinetochores of unattached chromosomes, with dissociation occurring once chromosomes align at the metaphase plate (1). In A. thaliana, MAD1 and BMF3 exhibit this typical kinetochore localization before reaching metaphase but BMF1 resides at kinetochores throughout mitosis (4). Conversely, other SAC proteins exhibit varied localization patterns during mitosis. For example, functional BMF2-GFP (green fluorescent protein) and MAD2-GFP fusions displayed a predominantly diffuse pattern in the cytoplasm (4). There are three BUB3 proteins, with BUB3.1 and BUB3.2 closely related to the fungal and animal BUB3, associating with phragmoplast microtubules by direct interaction with the microtubule-associated protein MAP65-3 for cytokinesis (5). However, the location and function of the less conserved, noncanonical BUB3.3 protein during mitosis remain unclear.
The characteristic phenotype caused by the loss of an essential SAC component is hypersensitivity to microtubule-depolymerizing drugs like the synthetic herbicide oryzalin (4) which is highly specific and potent against microtubules in plant cells. For example, mutants like bub3.3, mad1, mad2, and bmf3 grow indistinguishably from the wild-type control under unchallenged conditions but had their root growth severely inhibited by 150 nM oryzalin that does not obviously affect the growth of wild-type plants (4). Interestingly, the bmf1 mutant displays insensitivity to oryzalin, while the bmf2 mutant shows less sensitivity compared to bmf3. This suggests that BMF1-dependent phosphorylation is likely not essential for SAC regulation, and BMF2 may either play a minor role or not be involved in the SAC in A. thaliana (2). Thus, the differences in the protein architecture, localization, and function between plant MAD and BMF proteins and those in fungi and animals raise the question of how plants engage necessary SAC components in the formation of the MCC-like complex when the SAC is activated.
The oryzalin hypersensitivity phenotype of bub3.3 prompted us to test whether the function of MAD1 and BMF3 at kinetochores upon SAC activation was dependent on BUB3.3, as what has been described in fungi and animals. Fungal and animal BUB3 proteins typically recognize the GLEBS motif in their interacting partners (6). But none of the BMF proteins contains GLEB motifs, leading to the question of whether BUB3.3 directly interacts with one or more BMFs. In A. thaliana, we identified a mode of interaction between BUB3.3 and BMF3 for the recruitment of CDC20 to unattached kinetochores, thereby preventing anaphase onset in mitotic cells.
Results
BUB3.3 Associates with Kinetochores throughout Mitotic Cell Division.
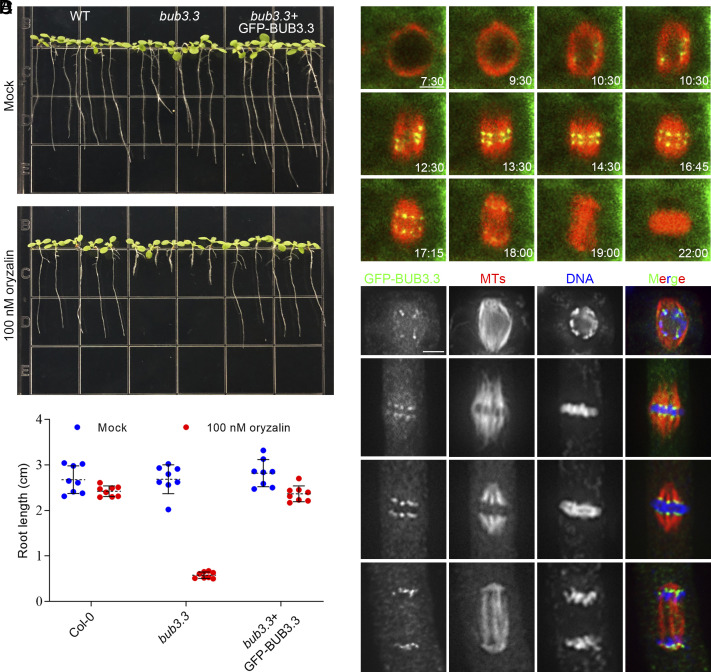
To investigate the function of BUB3.3 during mitosis, we introduced a construct for GFP-BUB3.3 expression under the control of the BUB3.3 promoter into a bub3.3 mutant carrying a T-DNA insertion (SI Appendix, Fig. S1). Upon testing the transformants together with the wild-type control and the bub3.3 mutant, we observed significant growth retardation in the bub3.3 mutant when exposed to 100 nM oryzalin, as quantified by the root lengths (Fig. 1 A and B). This oryzalin hypersensitivity phenotype was suppressed when the BUB3.3(p)::GFP-BUB3.3 transgene was expressed in the homozygous mutant (Fig. 1 A and B), indicating that the phenotype was caused by the inactivation of the BUB3.3 gene.
Fig. 1.
BUB3.3 associates with kinetochores throughout mitotic cell division. (A) Ten-day-old seedlings grown in the absence or presence of 100 nM oryzalin. The wild-type (WT) control, bub3.3 mutant, and bub3.3 mutant expressing GFP-BUB3.3 are included. (B) Quantification of root lengths of the seedlings with or without oryzalin treatment (n = 8 for each sample). (C) Live imaging of GFP-BUB3.3 and mCherry-TUB6 (for microtubules) in a mitotic cell. Snapshots are taken from Movie S1. (D) Triple localization of BUB3.3, microtubules, and DNA mitosis. The merged images have GFP-BUB3.3 detected by the anti-GFP antibody pseudocolored in green, microtubules in red, and DNA in blue. (Scale bars: 5 μm.)
Because the GFP-BUB3.3 fusion protein was functional as indicated by genetic suppression of the bub3.3 mutation, we used the transgenic line to examine subcellular localization of GFP-BUB3.3 during mitosis. We introduced an mCherry-TUB6 (β-tubulin 6) constrcut into the transgenic plants to monitor mitotic progression in live cells (Movie S1). During prophase, GFP-BUB3.3 was detected as paired dots inside the nucleus surrounded by a bipolar microtubule array (Fig. 1C). The signal then aligned in the middle of the spindle before segregating into two groups toward opposite spindle poles in the cell. The prominent signal persisted as bright dots during telophase (Fig. 1C). To explore the association of the GFP-BUB3.3 signal with kinetochores/chromosomes, we conducted immunofluorescence experiments in fixed cells. At late prophase when a bipolar spindle array was formed, paired GFP-BUB3.3 dots were associated with chromosomes in the nucleus (Fig. 1D). At metaphase when chromosomes were aligned at the metaphase plate flanked by mirrored kinetochore fibers, GFP-BUB3.3 appeared at the two edges of aligned chromosomes and the end of kinetochore fibers (Fig. 1D). As kinetochore fibers shortened, GFP-BUB3.3 tracked the shortening fibers until reaching the two spindle poles at telophase (Fig. 1D). Therefore, these data led to the conclusion that BUB3.3 associates with kinetochores throughout mitosis.
The Loss of BUB3.3 Causes Frequent Chromosome Misalignment.
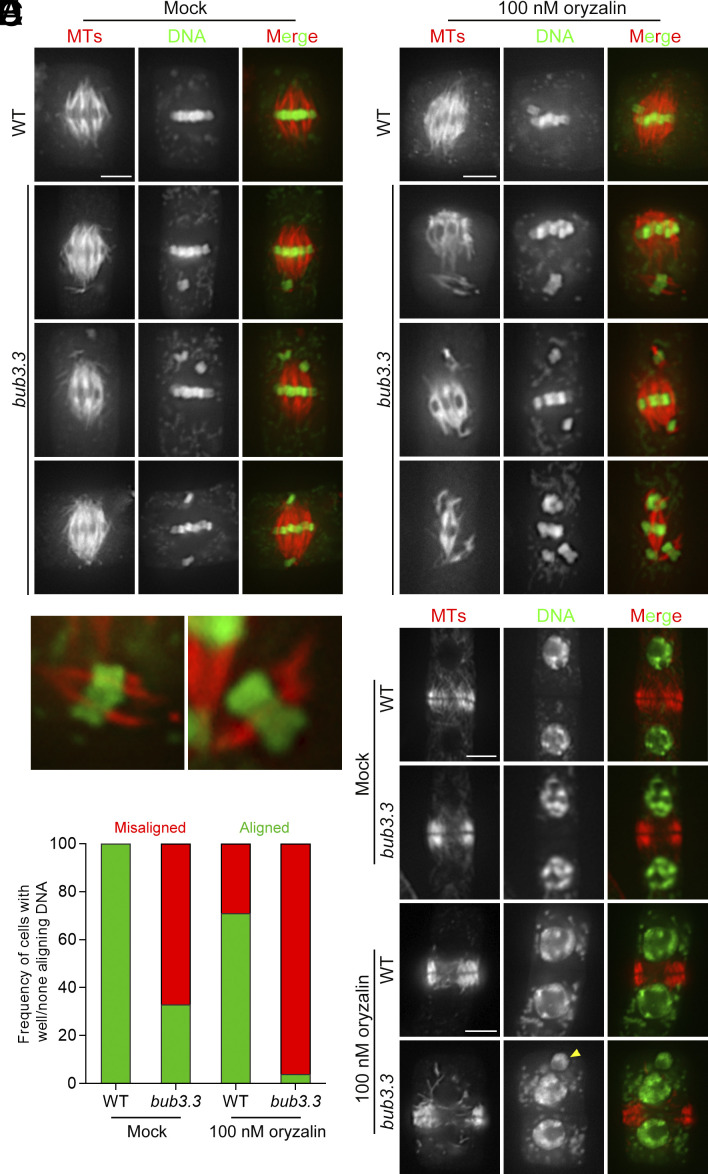
As BUB3.3 is the sole BUB3 protein localized at kinetochores in A. thaliana, we asked whether its loss led to mitotic defects by examining meristematic cells. Despite the bub3.3 mutant producing healthy plants, its mitotic cells often exhibited chromosome misalignment (Fig. 2A). In wild-type cells producing typical metaphase spindles with paired kinetochore–microtubule fibers, chromosomes were always aligned at the metaphase plate (Fig. 2A). In the bub3.3 mutant cells, however, mitotic cells bearing similar spindle microtubule arrays frequently had one or two chromosomes positioned away from the metaphase plate, either displaced to one side of the cell or misaligned on both sides near spindle poles (Fig. 2A). In fact, over 60% of the bub3.3 mutant cells at mitosis showed misaligned chromosomes outside the metaphase plate where the majority of chromosomes had been congressed (Fig. 2D). After treatment with 100 nM oryzalin, over 95% (62/65) of bub3.3 mutant cells displayed exacerbated chromosome misalignment, with misaligned chromosomes sometimes outnumbered ones at the metaphase plate (Fig. 2 B and D). Furthermore, the misaligned chromosomes typically had microtubules attached as if a “minispindle” was formed outside the main spindle that had already congressed chromosomes (Fig. 2C). This challenge also caused less frequent chromosome misalignment phenomena in the control cells (Fig. 2 B and D). We then examined whether the misaligned chromosomes were engaged in chromosome segregation or gave birth to a nucleus-like structure by examining cytokinetic cells. In wild-type cells, cytokinesis consistently resulted in the birth of two identical nuclei, regardless of oryzalin treatment (Fig. 2 E and F), suggesting that chromosome misalignment was corrected before anaphase. In bub3.3 mutant cells, however, micronuclei were formed toward the end of cytokinesis after oryzalin treatment, an occurrence not observed without oryzalin (Fig. 2 E and F). Therefore, the results indicated that BUB3.3 functioned in chromosome alignment at prometaphase and prevented the formation of micronuclei when chromosomes are misaligned.
Fig. 2.
Chromosome misalignment in the absence of BUB3.3. (A and B) Chromosome alignment in wild-type (WT) control and bub3.3 mutant cells at late stages of prometaphase in the absence (A) or presence (B) of oryzalin. (C) Enlarged views of kinetochore fibers/minispindles with misaligned chromosomes in bub3.3 mutant cells. (D) Quantitative assessment of abnormal cells exhibiting misaligned chromosomes in bub3.3 mutant cells compared to WT cells with or without oryzalin treatment (n = 65). (E and F) Comparative views of cytokinetic cells in WT and bub3.3 mutant plants in the absence (E) or presence (F) of oryzalin. The yellow arrowhead points at a representative micronucleus formed after oryzalin treatment in a bub3.3 mutant cell. Merged images have microtubules in red and DNA in green. (Scale bars: 5 μm.)
The bub3.3 Mutant Cells Show Enhanced SAC Defects in the Presence of Oryzalin.
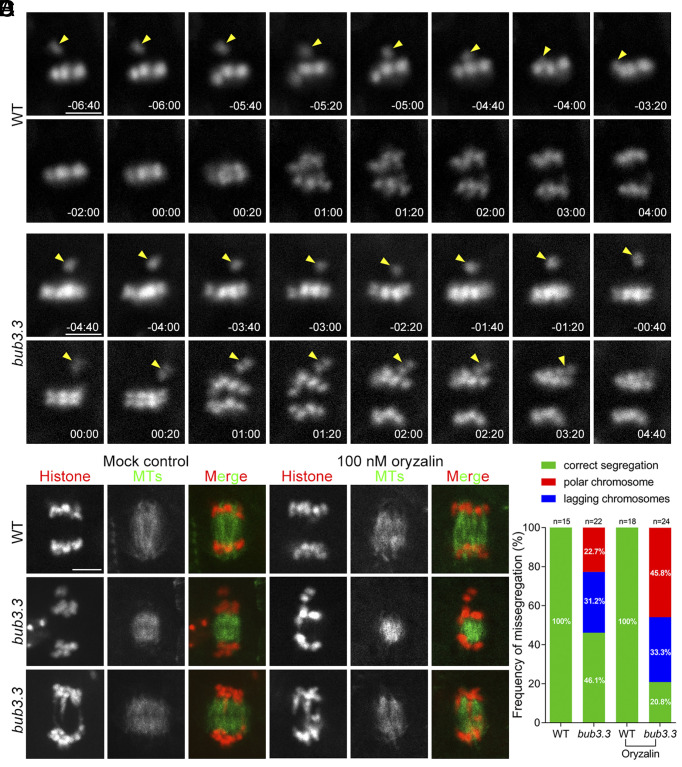
The oryzalin hypersensitivity phenotype prompted us to test whether the loss of BUB3.3 led to errors in mitosis. To monitor mitotic progression in living cells, we used an NDC80-TagRFP fusion protein to mark kinetochores after transformation, as previously reported (7). When wild-type roots were exposed to 100 nM oryzalin, we observed the NDC80-TagRFP signal outside the metaphase plate, representing a misaligned chromosome (Fig. 3A). Anaphase took place after this misaligned chromosome was brought to the metaphase plate (arrowheads, Fig. 3A and Movie S2). In oryzalin-treated bub3.3 mutant cells, however, mitotic cells entered anaphase without having the misaligned chromosome congressed to the metaphase plate (arrowheads, Fig. 3B and Movie S3). In such cells, unaligned chromosome(s) joined the segregated sister chromatids, leading to erroneous mitosis due to unequal chromosome segregation (Fig. 3B). Therefore, our result provided evidence showing that the loss of this noncanonical BUB3.3 led to defective SAC upon oryzalin treatment.
Fig. 3.
BUB3.3 monitors chromosome alignment at the metaphase plate to prevent premature sister chromatid segregation. (A and B) Mitotic progression is monitored by live-cell imaging of the NDC80-TagRFP fusion protein in WT (A) and bub3.3 mutant (B) cells treated with 100 nM oryzalin. Snapshots are taken from Movies S2 and S3. In the WT cell, anaphase onset takes place after the misaligned chromosome (arrowhead) is brought to the metaphase plate. In bub3.3 mutant cells, however, the cell ignores the misaligned chromosome (arrowhead) and enters anaphase. (C) Live-cell imaging shows examples of bub3.3 mutant cells that have polar/misaligned chromosomes (middle cells) or lagging chromosomes (bottom cells) at late anaphase/telophase in the absence (Movies S4–S6) or presence of 100 nM oryzalin (Movies S7 and S8). (D) Quantitative assessment of polar/misaligned chromosomes and lagging chromosomes in control and bub3.3 mutant cells without or with 100 nM oryzalin treatment. (Scale bars: 5 μm.)
To monitor chromosome and spindle dynamics, we had both histone H1-TagRFP and visGreen-TUB6 expressed (Fig. 3 C and D). We found that anaphase cells of the bub3.3 mutant also showed a phenotype of lagging chromosomes in addition to having misaligned chromosomes outside of the primary chromatid mass (Fig. 3C). Such a phenotype became more obvious when the microtubule array of the developing phragmoplast was viewed simultaneously. Again, treatment with 100 nM oryzalin further enhanced both phenotypes (Fig. 3C). To quantitatively evaluate the phenotypes, we followed mitotic progression by imaging both chromosomes and microtubules in both control and bub3.3 mutant cells. First, we examined chromosome misalignment phenotypes prior to anaphase onset and found that 63.6% (14/22) of bub3.3 cells displayed this phenotype, which increased to 70.8% (17/24) after oryzalin treatment. Additionally, we assessed the frequency of anaphase progression with polar/misaligned or lagging chromosomes. While the control cells never showed such phenotypes in the absence or presence of oryzalin (SI Appendix, Fig. S2 and Movie S4), the bub3.3 mutant cells had 22.7% and 31.2% showing polar and lagging chromosomes, respectively (Fig. 3D and SI Appendix, Fig. S2 and Movies S5 and S6). These percentages increased to 45.8% and 33.3%, respectively, after oryzalin treatment. Chromosome misalignment was also observed in wild-type cells after oryzalin treatment but always corrected by becoming engaged in the spindle microtubule array before anaphase onset (SI Appendix, Fig. S3 and Movie S7). In contrast, bub3.3 mutant cells entered anaphase with misaligned chromosomes that did not undergo segregation, and lagging chromosomes were found during anaphase (SI Appendix, Fig. S3 and Movie S8). Collectively, these data suggest that BUB3.3 is required for mitotic arrest when cells experienced microtubule depolymerization challenges.
Independent Kinetochore Localization of BUB3.3 and Other SAC Proteins.
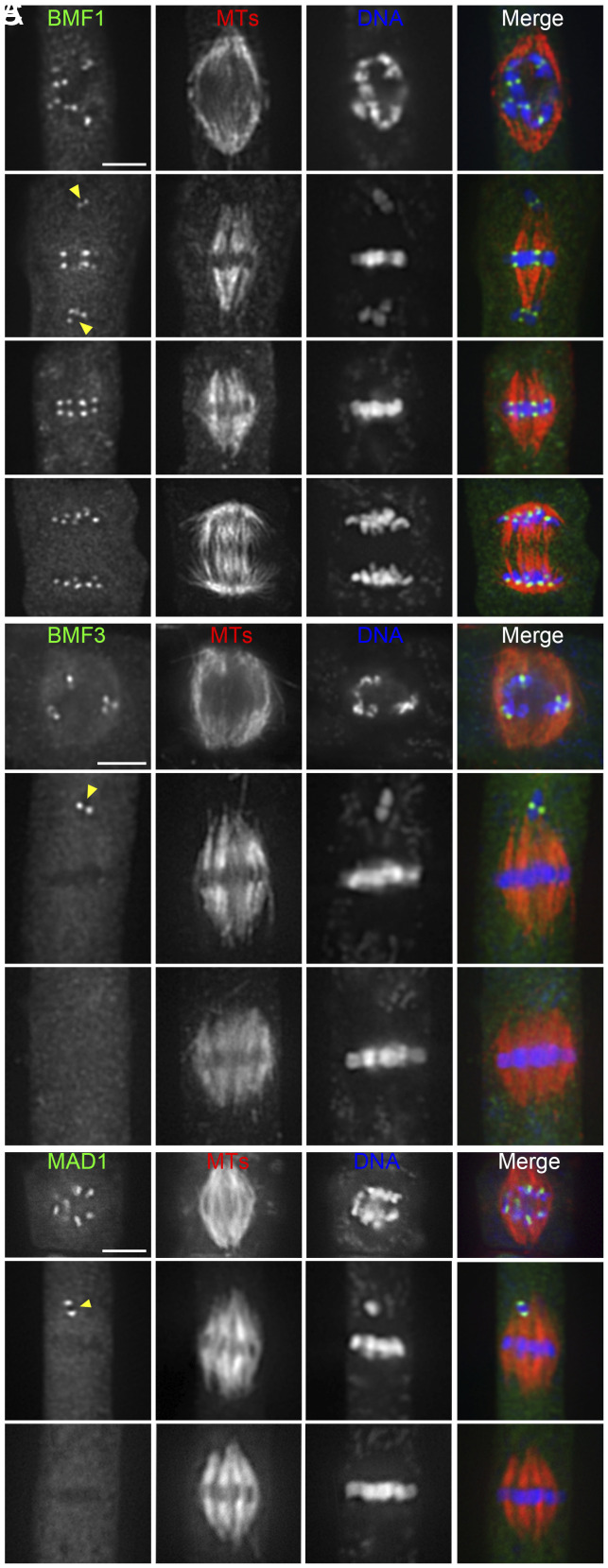
Because BUB3 showed continuous kinetochore localization during mitosis, we tested whether it still functioned in the recruitment of SAC proteins in A. thaliana. To do so, we transformed constructs, which have been demonstrated to be functional GFP fusions of BMF1, BMF3, and MAD1 proteins (4), into the bub3.3 mutant. In unchallenged wild-type cells expressing BUB3.3, BMF1-GFP fusion protein decorated kinetochores throughout mitosis while GFP-MAD1 and BMF3-GFP were detected at the kinetochores prior to chromosome alignment or specifically at those of frequently misaligned chromosomes, while aligned ones lacked the signals (SI Appendix, Fig. S4). Such localization patterns were unaltered in the bub3.3 mutant, where GFP-MAD1 and BMF3-GFP selectively decorated kinetochores of misaligned chromosomes (arrowheads. Fig. 4 A–C).
Fig. 4.
BUB3.3 is not required for the kinetochore localization of other SAC proteins. (A) As in the control cells, the BMF1-GFP fusion protein localizes to kinetochores of both aligned and misaligned chromosomes in bub3.3 mutant cells. (B and C) BMF3-GFP and MAD1-GFP decorate kinetochores of unattached/misaligned chromosomes similarly in bub3.3 mutant cells. Note that the BMF3 and MAD1 signals are no longer detected at the kinetochores when chromosomes arrive at the metaphase plate. Merged images have GFP-tagged proteins in green, microtubules in red, and DNA in blue. (Scale bars: 5 μm.)
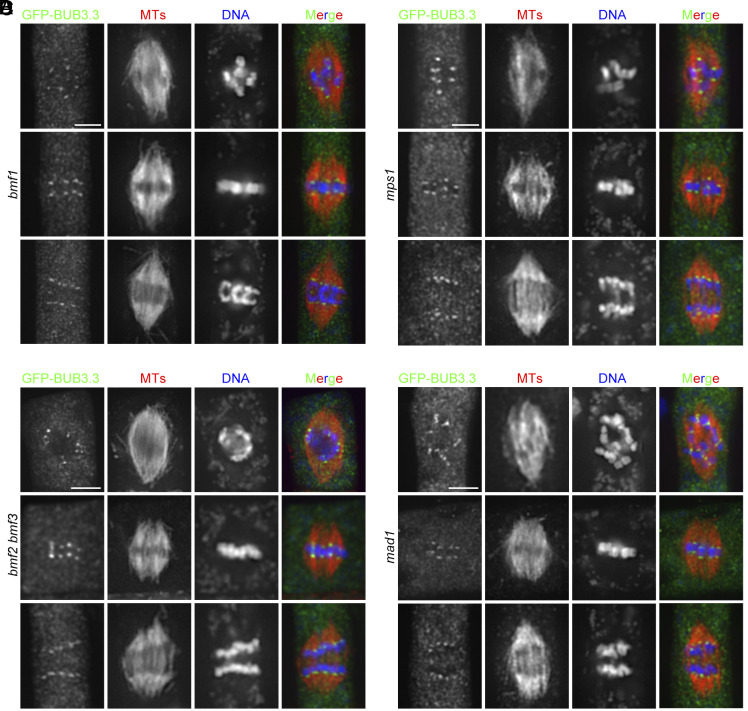
Conversely, we asked whether any of the MAD and BMF proteins were required for BUB3.3 localization. When GFP-BUB3.3 was expressed in the bmf1 and mad1 mutant, it associated with kinetochores at all stages of mitosis (Fig. 5 A and D), consistent with our earlier observations (Fig. 1C). Because BMF2 and BMF3 but not BMF1 have been detected as functional SAC components based on the oryzalin hypersensitivity test (4), we generated a bmf2; bmf3 double mutant in which the GFP-BUB3.3 fusion was expressed. Again, GFP-BUB3.3 remained undisturbed at kinetochores throughout mitosis (Fig. 5B). Because of the essential role of the MPS1 kinase in SAC signaling, we also examined GFP-BUB3.3 localization in the mps1 mutant cells and found it was undisturbed (4). Therefore, our results indicated that BUB3.3 localization is independent of the other critical SAC proteins of MPS1, MAD1, BMF2, and BMF3, and vice versa.
Fig. 5.
The kinetochore localization of BUB3.3 is independent to other SAC proteins. The GFP-BUB3.3 fusion protein was expressed and detected by immunostaining at kinetochores during mitosis in the bmf1 (A), bmf2 bmf3 (B), mps1 (C), and mad1 (D) mutant cells at prometaphase (Top), metaphase (Middle), and anaphase. Merged images have GFP-tagged proteins in green, microtubules in red, and DNA in blue. (Scale bars: 5 μm.)
BUB3.3 Recognizes Two Internal Repeat Motifs in BMF3.
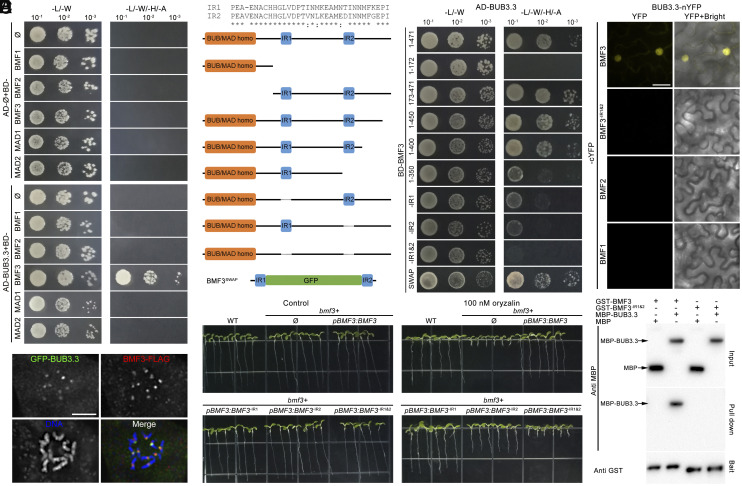
We then tested whether BUB3.3 interacted with one or more proteins described above. To do so, we employed yeast two-hybrid assays to examine potential interactions between BUB3.3 and BMF1, BMF2, BMF3, MAD1, and MAD2. We detected interaction only between BUB3.3 and BMF3, with no interactions detected with other proteins (Fig. 6A). The two proteins also colocalized at kinetochores of the prometaphase chromosomes (Fig. 6B).
Fig. 6.
BUB3.3 recognizes two internal repeat (IR) motifs in BMF3. (A) BUB3.3 interacts with BMF3 but not other Arabidopsis SAC components by the yeast two-hybrid (Y2H) assay. The empty vector is used as a negative control (Ø). The yeast cultures are spotted on vector-selective (-L/-W, left column) and interaction-selective (-L/-W/-H/-A, right column) media and photographed after incubation at 30 °C for 2 d. (B) Colocalization of GFP-BUB3.3 and BMF3-FLAG in a mitotic cell. The merged image has BUB3.3 pseudocolored in green, BMF3 in red, and DNA in blue. (C) Schematic representations of full-length and truncated versions of BMF3 used for mapping the BUB3.3-binding domains by Y2H assay. The IR1 (amino acids 196 to 228) and IR2 (amino acids 354 to 387) peptides show high degrees of amino acid identity to each other. The BMF3SWAP fusion protein has the region between IR1 and IR2 replaced by GFP. Note the deletion of either IR1 or IR2 significantly reduces the interaction strength and BMF3SWAP interacts with BUB3.3 similarly to the full-length protein. (D) In BiFC assays, YFP fluorescence is established when BUB3.3-nYFP and BMF3-cYFP, but not BMF1 or BMF2, are coexpressed. Deletion of the two internal repeats abolishes the interaction. (E) Purified MBP-BUB3.3 but not MBP alone is pulled down or cosedimented with GST-BMF3 but not the truncated version missing the repeats. The proteins are detected by either anti-GST or anti-MBP antibodies. (F) Growth phenotypes of 7-d-old plants associated with the expression of various BMF3 derivatives on media lacking or including 100 nM oryzalin. (Scale bars: 5 μm.)
To learn how the BUB3.3–BMF3 interaction was established, we made truncations of BMF3 for the yeast two-hybrid assays. First, we separated BMF3 into two parts, the N-terminal BUB1/MAD3 characteristic TPR domain (BMF31–172) and the remaining fragment with two internal repeats (IR1 and IR2) (BMF3173–471) (SI Appendix, Fig. S5). It was found that the BMF3173–471 fragment was sufficient for the interaction with BUB3.3 (Fig. 6C). Subsequent truncation of the BMF3 protein from the C terminus revealed that deletions of peptides after the IR2 motif did not affect the interaction (Fig. 6C). However, removal of the IR2 motif in BMF31–350 weakened the interaction, though it was not completely abolished (Fig. 6C), indicating a role for IR2 in the interaction. Consequently, we made a truncated BMF3 with the IR1 deleted and found that this BMF3-IR1(197–229) fragment also exhibited a partially compromised interaction with BUB3.3 (Fig. 6C), similar to the result obtained with the truncated BMF3-IR2(355–388) fragment lacking the IR2 motif (Fig. 6C). Simultaneous deletions of both IR1 and IR2 motifs completely abolished the interaction (Fig. 6C), suggesting the necessity of these two internal repeats for the interaction. The question then arose as whether the two IR motifs alone were sufficient for BUB3.3 interaction. To do so, we designed an artificial IR1-GFP-IR2 fusion protein and found that this BMF3SWAP protein interacted with BMF3 in yeast cells (Fig. 6C). We also conducted interaction tests using bimolecular fluorescence complementation (BiFC) of the yellow fluorescent protein (YFP) and in vitro protein cosedimentation. In tobacco cells as the expression host, YFP fluorescence was established only when BUB3.3-nYFP and BMF3-cYFP were coexpressed (Fig. 6D). In contrast, truncated BMF3 lacking IR1 and IR2, as well as BMF1 and BMF2 did not show positive BiFC. For the in vitro protein cosedimentation assay, glutathione S-transferase (GST)-BMF3 and maltose-binding protein (MBP)-BUB3.3 fusion proteins, purified from bacterial host cells, were incubated together. Capture of GST-BMF3 by glutathione beads had MBP-BUB3.3 cosedimented, while the truncated GST-BMF3-IR1&2 or MBP alone did not show such cosedimentation results (Fig. 6E).
We then examined the in vivo functionality of the aforementioned BMF3 truncations after having them expressed under their native promoter in the bmf3 homozygous mutant background. The bmf3 mutant grew indistinguishably from the wild-type control under unchallenged conditions but exhibited hypersensitivity to 100 nM oryzalin (Fig. 6F), similar to what has been reported previously (4). We found that the BMF3-IR1 version without the IR1 motif was largely functional as the full-length protein and the BMF3-IR2 fragment without IR2 partially suppressed the bmf3 mutation (Fig. 6F). The removal of both IR1 and IR2 abolished the functionality of BMF3 in this oryzalin assay (Fig. 6F). Therefore, we concluded that both IR1 and IR2 contributed to the interaction with BUB3.3 but IR2 probably played a more critical role than IR1 for the functionality of BMF3 when SAC is activated.
The BUB3.3–BMF3 Interaction Is Required for the Recruitment of CDC20 to Unattached Kinetochores.
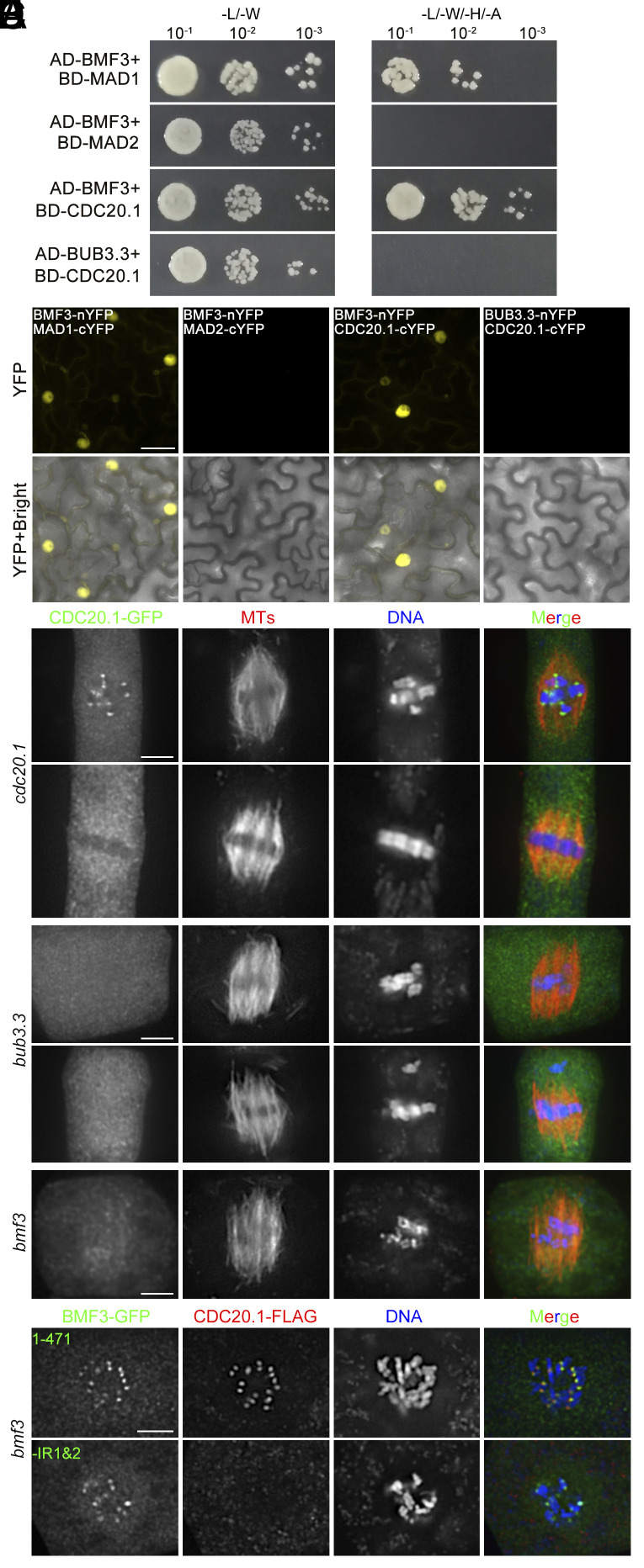
Because BUB3.3 interacted with BMF3 without affecting its kinetochore localization, we proceeded to investigate whether the interaction with BUB3.3 impacted other targets of BMF3. We first screened BMF3 binding proteins among putative MCC components via yeast two-hybrid assay. Indeed, BMF3 interacted with MAD1 (Fig. 7A), consistent with previous reports (4). We also found BMF3 interacted with CDC20.1 but not MAD2 by the yeast two-hybrid assay (Fig. 7A). The BMF3-MAD1 and BMF3-CDC20 interactions were complementarily verified by the BiFC assay (Fig. 7B). Using bacterially expressed GST-BUB3.3, GST-BMF3, MBP-CDC20 fusion proteins, we also tested their interaction and found that MBP-CDC20, but not MBP alone, was cosedimented with GST-BMF3 but not GST-BUB3.3 when using immobilized glutathione (SI Appendix, Fig. S6).
Fig. 7.
BUB3.3–BMF3 interaction is required for the recruitment of CDC20 to unattached kinetochores. (A) BMF3 interacts with MAD1 and CDC20.1 in a Y2H assay. But BUB3.3 does not interact with CDC20.1. (B) BiFC assays report the interaction between BMF3 and MAD1 as well as BMF3 and CDC20.1, but not between BMF3 and MAD2 or BUB3.3 and CDC20.1. (C–E) CDC20.1-GFP localization in different genetic backgrounds. When it is expressed in the cdc20.1 mutant (C), the fusion protein is detected at kinetochores of all chromosomes at prometaphase and becomes cytosolic upon chromosome congression at the metaphase plate. In the bub3.3 (D) and bmf3 (E) mutant cells, however, CDC20.1-GFP is no longer detected at kinetochores at any stages of mitosis. (F) When the CDC20.1-FLAG fusion protein is expressed in the bmf3 mutant cells coexpressing either the full-length BMF3-GFP (Top rows) or truncated BMF3ΔIR1&2-GFP (Bottom rows) fusion proteins, it colocalizes with BMF3-GFP at kinetochores but not BMF3ΔIR1&2-GFP which is still detected at kinetochores in prometaphase cells. In the merged images, the GFP fusion proteins are pseudocolored in green, microtubules by anti-tubulin (C–E) or CDC20.1-FLAG by anti-FLAG (F) in red, and DNA by DAPI in blue. (Scale bars: 5 μm.)
Given that CDC20 is a crucial MCC component and the key target upon SAC activation, and it is structurally and functionally conserved in plants (1, 8), we then examined the CDC20.1 protein in mitotic cells after expressing a CDC20.1-GFP fusion under the control of the CDC20.1 promoter in the null cdc20.1 mutant. CDC20.1-GFP was detected at kinetochores at prometaphase prior to chromosome congression to the metaphase plate (Fig. 7C). The signal disappeared from there after chromosomes were amphitellicaly attached and aligned (Fig. 7C). To determine whether such cell cycle-dependent localization was dependent on BUB3.3 and/or BMF3, the fusion protein was expressed in the bub3.3 and bmf3 mutants, respectively. In the bub3.3 mutant, the CDC20.1-GFP signal was no longer detected at kinetochores following nuclear envelope breakdown or those of misaligned chromosomes (Fig. 7D). Similarly, CDC20.1-GFP was not observed at kinetochores during prometaphase either in the bmf3 mutant (Fig. 7D). Therefore, these results revealed that the kinetochore localization of CDC20 was a response to SAC activation and was dependent on both BUB3.3 and BMF3 in A. thaliana.
To further verify the significance of the internal repeats, we had the truncated BMF3ΔIR1&2 protein lacking both IR1 and IR2 expressed in the bmf3 mutant. In comparison to cells expressing the full-length BMF3-GFP which exhibited kinetochore association and restored the localization of CDC20.1 in a fusion with the FLAG tag, mutant cells expressing the BMF3ΔIR1&2-GFP fusion no longer had CDC20.1-FLAG detected at kinetochores although the mutant BMF3 protein remained localized there (Fig. 7F). These results collectively led to the conclusion that BUB3.3 is essential for BMF3 to recruit CDC20 through its two internal repeats.
Discussion
Our results revealed a mode of action underlying plant SAC regulation, centered on the noncanonical BUB3 family protein, BUB3.3, in A. thaliana. Specifically, BUB3.3 functioned in recruiting CDC20 to kinetochores of unattached chromosomes through its interaction with BMF3. These findings bring insights into how plant cells respond to SAC activation, uncovering a mechanism that employs the evolutionarily conserved BUB3 family protein to trigger SAC-dependent mitotic arrest without having to dissociate from the kinetochores to assemble the equivalent of the MCC found in vertebrates.
BUB3.3-Interaction Domains in the Plant-Specific BMF3 Protein.
BUB3 family proteins are highly conserved and made of mostly WD40 repeats. In animal cells, they recognize the GLEBS domain which is commonly found in BUB1/BUBR1 proteins (6). In mammals, BUB3 also interacts with both the BuGZ protein bearing the GLEBS domain and the kinetochore scaffold protein KNL1 by recognizing the phosphorylated MELT motif (9–11). However, plant BMF proteins, which are distantly related to the animal BUB1 family proteins with isolated functional domain(s), lack such interaction motifs. Therefore, it has been unclear how BUB3.3 could be coupled with BMF family proteins to prevent anaphase onset upon SAC activation (2, 4). Here we provided direct evidence showing that BUB3.3 interacted with two internal repeats within the BMF3 protein. We did not find any vertebrate proteins that share significant sequence homology with such internal repeats. Therefore, these two repeats represent a mode of interaction with the evolutionarily conserved BUB3 family proteins in A. thaliana.
Among three BUB3 isoforms in A. thaliana, BUB3.1 and BUB3.2 are nearly identical and show a closer relationship with animal and fungal BUB3 proteins than BUB3.3 (12). Interestingly, only BUB3.3 exhibited exclusive localization at the kinetochores and played a role in SAC regulation, despite being more divergent from the fungal or animal BUB3 proteins. Collectively, these findings support the notion that the classical interaction modules of BUB3-GLEBS/MELT perhaps do not apply to flowering plants. Instead, plants seem to have established an alternative interaction module involving BUB3.3 and the internal repeats of BMF3, following the diversification of functions among BUB3 isoforms.
BUB3.3 Exhibits SAC Activation–Independent Kinetochore Localization.
In human cells, BUB3 localization at kinetochores shows a drastic reduction from prometaphase to metaphase, suggesting a great deal of BUB3 delocalization from there after SAC satisfaction (13). The kinetochore localization of BUB3.3, however, did not show obvious fluctuations throughout mitosis. In fact, different SAC-implicated proteins have different dynamic patterns during mitosis in A. thaliana and only BMF3 and MAD1 exhibit clear SAC activation-dependent localization patterns (4). Similar to BUB3.3, both BMF1 and the pivotal SAC kinase MPS1 are associated with kinetochores throughout mitosis. In contrast to the MAD2 homolog in maize which localizes to kinetochores of unaligned chromosomes, the Arabidopsis counterpart seems to be predominantly cytosolic (4, 14). These phenomena raised the questions about whether the functions of these proteins converge toward SAC regulation and, if so, how these proteins are functionally linked in plant cells.
In animal cells, a crucial role of BUB3 is to recruit BUB1 and BUBR1/MAD3 to kinetochores (15). In contrast, BUB3.3 and MAD/BMF proteins in A. thaliana exhibited independent localization. Such independence was further supported by the kinetochore localization of the truncated BMF3 lacking the internal repeats responsible for BUB3.3 interaction.
Our results raise the question of how SAC proteins including BUB3.3 achieve kinetochore localization. In animal cells, the kinetochore scaffold protein KNL1 presents the phosphorylated MELT motifs to recruit BUB3 (6). Plants produce proteins that share homologies in two functional domains with animal KNL1 proteins but lack obvious MELT motifs and localize to kinetochore in mitotic and meiotic cells (3, 16, 17). Furthermore, the maize KNL1 homolog directly interacts with the BMF1 and BMF2 homologs (3). Therefore, it is possible that the classical KMN network is established by KNL1, along with the Mis12 and NDC80 complexes, in plants (18). Although it has been shown that inactivation of the KNL1 gene in maize leads to defects in chromosome congression during mitosis (3), it is yet to be tested whether the plant KNL1 homologs are required for the kinetochore localization of either BMF1, BMF2, or BUB3 during mitosis.
It is intriguing to speculate that BUB3.3–BMF3 interaction-dependent SAC regulation is conserved among plants. However, studies in maize reported only one BUB3 protein and indicated that its BUB3 and BMF3 proteins did not interact with each other (3). This result aligns with the observation that the maize BMF3 lacks the two internal repeats found in its Arabidopsis counterpart. Furthermore, the maize BUB3 and BMF3 do not interact with KNL1 by yeast two-hybrid assay, leaving uncertainty about whether these two proteins function similarly to their counterparts in A. thaliana. Functional dissections of these genes and the interrogation of their interacting partners in maize and other plants would be very informative although technically challenging.
Inhibition of CDC20 upon SAC Activation.
The ultimate consequence of SAC activation is the inhibition of the APC/C activator CDC20 via the formation of the MCC complex as demonstrated in animal cells (15). In A. thaliana, the function of CDC20 in spindle assembly and chromosome segregation has been demonstrated in male meiotic cells (8). Here, we demonstrated that a direct interaction between BMF3 and BUB3.3 was required for CDC20 localization to kinetochores. This finding suggests that BUB3.3 may play a role in recruiting CDC20 to kinetochores via BMF3, leading to the inhibition of APC/C and prevention of anaphase onset. This function may be realized by augmenting BMF3-CDC20 interaction or enhancing CDC20 residence at kinetochores. On the one hand, this is different from animal cells in which BUB3 is required for BUB1 and BUBR1 localization to kinetochores (19). On the other hand, such an action may partly resemble the interaction of BUBR1 with BUB3 in human cells in which BUB3 enhances the tethering of BUBR1 at kinetochores for CDC20 recruitment (20). It is unclear whether plant cells assemble the equivalent of the animal MCC complex and, if so, what such an MCC complex is made of (4). Among the three BMF proteins, only BMF1 possesses a kinase domain but is likely dispensable for the SAC because its mutant lacks the oryzalin hypersensitivity phenotype; and BMF2 is cytosolic but not detectable at kinetochores (4). Therefore, if the MCC exists in A. thaliana, its assembly probably involves BUB3.3, BMF3, and CDC20 but excludes BMF1 and BMF2 due to their distinct localization patterns. This notion was also supported by the data that BUB3.3 did not interact with BMF1 or BMF2 in our experiments.
However, the oryzalin hypersensitivity phenotype of the bmf2 mutant and the predominant cytosolic localization of the BMF2 protein suggest that it may contribute to SAC signaling in the cytosol. However, such a cytosolic SAC signaling scheme must be BUB3.3 independent, unlike the scenario in animal cells, again because BUB3.3 only interacted with BMF3 but not BMF2. It is interesting that MAD1 interacts with both MAD2 and BMF3 in A. thaliana (4). However, unlike MAD1 which exhibits a typical SAC activation-dependent kinetochore localization, MAD2 shows striking cytosolic localization. Because BMF3 is required for the kinetochore localization of MAD1, upon SAC activation, the MCC might be assembled in the following sequential steps. At first, BMF3 interacts with BUB3.3 at kinetochores while recruited MAD1 catalyzes the conformational change of MAD2. When CDC20 is recruited to unattached kinetochores following BUB3.3–BMF3 interaction, it then complexes with MAD2 in the closed conformation. The catalysis of MAD2-CDC20 complex formation might dissociate from unattached kinetochores and associated with BMF2 in the cytosol to form the MCC complex. Alternatively, MCC formation may be independent of kinetochore localization. This hypothesis was supported by at least two lines of evidence from studies in animal cells. For example, it was suggested that the interaction between BUB3 and CDC20, MAD2, and MAD3 does not require intact kinetochores (21). An independent work also led to the hypothesis that CDC20–MAD2 association may be established in a kinetochore-independent manner (22).
BUB3.3 and Chromosome Congression/Alignment.
Besides exhibiting phenotypes of lagging chromosomes during anaphase and the formation of micronuclei after telophase, the bub3.3 mutant cells also displayed frequent chromosome misalignment during mitosis which was not a typical SAC-related phenotype. BUB3.3 function in chromosome congression was also detected in an independent study recently (23). Impressively, this chromosome congression phenotype was drastically enhanced when the cells experienced mild microtubule depolymerization challenges. As a result, micronuclei were born from the misaligned chromosomes, as the bub3.3 mutant cells entered anaphase prematurely without having the error corrected. This phenotype suggested that BUB3.3 might possess a function in chromosome congression during prometaphase. In animal mitosis, chromosome congression to the metaphase plate is dependent on various microtubule-associated factors, like the Kinesin-7 motor CENP-E (24). The possibility of a BUB3.3–microtubule interaction was partly inspired by findings in vertebrate cells that demonstrate interactions between Kinesin-7/CENP-E and BUBR1, as well as between Kinesin-5 and MAD1 (25, 26). There are 15 Kinesin-7 and 4 Kinesin-5 paralogs in A. thaliana (27), but to date, it is unclear whether any of them function similarly as CENP-E and whether they are functionally linked to SAC proteins like BUB3.3. It would be interesting to learn whether any of these motors play a role in chromosome alignment and whether BUB3.3 or MAD and BMF proteins interact with them or other microtubule-associated proteins.
Materials and Methods
Materials and Methods are summarized here and described in detail in SI Appendix.
Plant Materials and Growth Conditions.
The A. thaliana control, mutant, and transgenic plants of the Columbia (Col-0) background were grown either in soil or on solid Murashige-Skoog (MS) medium. Oryzalin treatment was carried out by including it in the MS medium.
Recombinant DNA Manipulations.
DNA fragments were amplified by standard PCR reactions and cloned into pDONR vectors by Gateway-based cloning methods to generate entry clones and migrated into specific destination vectors by clonase-based reactions. Primers used in this study are summarized in SI Appendix, Table S1.
Yeast Two-Hybrid Assays.
Yeast cells were transformed with plasmids containing cDNA fragments to be tested. Interactions were reported by yeast growth on control and selection media lacking corresponding amino acids.
In Vitro Protein–Protein Interaction Assay.
The coding sequences of target proteins were cloned into the bacterial expression plasmids for the expression of GST or MBP fusion proteins. Fusion proteins were purified using immobilized glutathione or maltose prior to being used in the cosedimentation assay by using glutathione resin.
Transient Expression in Tobacco Leaf Cells and Transformation in A. thaliana.
Transient expression in tobacco (Nicotiana benthamiana) was carried out by agrobacterial infiltration for the BiFC assays. Stable transformation was carried out by floral dipping in A. thaliana and transformants were selected by selection markers carried by the corresponding destination vectors.
Microscopic Observation and Imaging.
Live-cell imaging was carried out under a laser scanning confocal microscope, and immunolocalization results were acquired under a standard wide-field microscope coupled with a cMOS digital camera. Images were analyzed in the Image J/Fiji software package prior to being assembled into figure plates.
Supplementary Material
Appendix 01 (PDF)
Live-cell imaging of GFP-BUB3.3 and mCherry-labelled microtubules in the bub3.3 background. Images have intervals of 15 s.
Live-cell imaging of WT cells expressing NDC80-TagRFP following the treatment with 100 nM oryzalin. Images have intervals of 20 s.
Live-cell imaging of bub3.3 cells expressing NDC80-TagRFP following the treatment with 100 nM oryzalin. Images have intervals of 20 s.
Live-cell imaging of VisGreen-TUB6 and histone H1-TagRFP in control cells without oryzalin treatment.
Live-cell imaging of VisGreen-TUB6 and histone H1-TagRFP in bub3.3 cells without oryzalin treatment.
Live-cell imaging of VisGreen-TUB6 and histone H1-TagRFP in bub3.3 cells without oryzalin treatment.
Live-cell imaging of VisGreen-TUB6 and histone H1-TagRFP in control cells treated with 100 nM oryzalin.
Live-cell imaging of VisGreen-TUB6 and histone H1-TagRFP in bub3.3 cells treated with 100 nM oryzalin.
Acknowledgments
We want to thank Dr. A. Schnittger and Dr. S. Komaki for sharing unpublished data and their patience and cooperation throughout the work. We are very grateful to Prof. T. Nakagawa for sharing the pGWB vectors. We also thank the anonymous reviewers and Dr. T. Rost for careful examination of the text and findings. This work was supported by the NSF grant MCB-1920358 to Y.-R.J.L. and B.L., the National Natural Science Foundation of China grants 32270354 and U22A20494 to X.D. and H.-H. Lin, and the Institutional Research Fund of Sichuan University grant 2020SCUNL212 to X.D. B.L. is supported by the U.S. Department of Agriculture (USDA)-the National Institute of Food and Agriculture (NIFA) under an Agricultural Experiment Station (AES) hatch project (CA-D-PLB-2536-H).
Author contributions
X.D., Y.-R.J.L., H.-H.L., and B.L. designed research; X.D., F.L.P., X.T., and Y.-R.J.L. performed research; X.D. and Y.-R.J.L. contributed new reagents/analytic tools; X.D., Y.-R.J.L., H.-H.L., and B.L. analyzed data; and X.D. and B.L. wrote the paper.
Competing interests
The authors declare no competing interest.
Footnotes
This article is a PNAS Direct Submission.
Contributor Information
Hong-Hui Lin, Email: hhlin@scu.edu.cn.
Bo Liu, Email: bliu@ucdavis.edu.
Data, Materials, and Software Availability
All study data are included in the article and/or supporting information.
Supporting Information
References
- 1.Lara-Gonzalez P., Pines J., Desai A., Spindle assembly checkpoint activation and silencing at kinetochores. Semin. Cell Dev. Biol. 117, 86–98 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Komaki S., Schnittger A., The spindle checkpoint in plants-a green variation over a conserved theme? Curr. Opin. Plant Biol. 34, 84–91 (2016). [DOI] [PubMed] [Google Scholar]
- 3.Su H., et al. , Knl1 participates in spindle assembly checkpoint signaling in maize. Proc. Natl. Acad. Sci. U.S.A. 118, e2022357118 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Komaki S., Schnittger A., The spindle assembly checkpoint in Arabidopsis is rapidly shut off during severe stress. Dev. Cell 43, 172–185.e175 (2017). [DOI] [PubMed] [Google Scholar]
- 5.Zhang H., et al. , Role of the BUB3 protein in phragmoplast microtubule reorganization during cytokinesis. Nat. Plants 4, 485–494 (2018). [DOI] [PubMed] [Google Scholar]
- 6.Kops G., Snel B., Tromer E. C., Evolutionary dynamics of the spindle assembly checkpoint in eukaryotes. Curr. Biol. 30, R589–R602 (2020). [DOI] [PubMed] [Google Scholar]
- 7.Shin J., Jeong G., Park J. Y., Kim H., Lee I., MUN (MERISTEM UNSTRUCTURED), encoding a SPC24 homolog of NDC80 kinetochore complex, affects development through cell division in Arabidopsis thaliana. Plant J. 93, 977–991 (2018). [DOI] [PubMed] [Google Scholar]
- 8.Niu B. X., et al. , Arabidopsis cell division cycle 20.1 is required for normal meiotic spindle assembly and chromosome segregation. Plant Cell 27, 3367–3382 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jiang H., et al. , A microtubule-associated zinc finger protein, BuGZ, regulates mitotic chromosome alignment by ensuring Bub3 stability and kinetochore targeting. Dev. Cell 28, 268–281 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Toledo C. M., et al. , BuGZ is required for Bub3 stability, Bub1 kinetochore function, and chromosome alignment. Dev. Cell 28, 282–294 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vleugel M., et al. , Sequential multisite phospho-regulation of KNL1-BUB3 interfaces at mitotic kinetochores. Mol. Cell 57, 824–835 (2015). [DOI] [PubMed] [Google Scholar]
- 12.Lermontova I., Fuchs J., Schubert I., The Arabidopsis checkpoint protein Bub3.1 is essential for gametophyte development. Front Biosci. 13, 5202–5211 (2008). [DOI] [PubMed] [Google Scholar]
- 13.Shirnekhi H. K., Herman J. A., Paddison P. J., DeLuca J. G., BuGZ facilitates loading of spindle assembly checkpoint proteins to kinetochores in early mitosis. J. Biol. Chem. 295, 14666–14677 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yu H. G., Muszynski M. G., Kelly Dawe R., The maize homologue of the cell cycle checkpoint protein MAD2 reveals kinetochore substructure and contrasting mitotic and meiotic localization patterns. J. Cell Biol. 145, 425–435 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Barisic M., Rajendraprasad G., Steblyanko Y., Spindle assembly checkpoint activation and silencing at kinetochores. Semin Cell Dev. Biol. 117, 99–117 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kozgunova E., Nishina M., Goshima G., Kinetochore protein depletion underlies cytokinesis failure and somatic polyploidization in the moss Physcomitrella patens. eLife 8, e43652 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.van Hooff J. J. E., Tromer E., van Wijk L. M., Snel B., Kops G. J. P. L., Evolutionary dynamics of the kinetochore network in eukaryotes as revealed by comparative genomics. EMBO Rep. 18, 1559–1571 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liu B., Lee Y. R. L., Spindle assembly and mitosis in plants. Annu. Rev. Plant Biol. 73, 227–254 (2022). [DOI] [PubMed] [Google Scholar]
- 19.Taylor S. S., Ha E., McKeon F., The human homologue of Bub3 is required for kinetochore localization of Bub1 and a Mad3/Bub1-related protein kinase. J. Cell Biol. 142, 1–11 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Han J. S., Vitre B., Fachinetti D., Cleveland D. W., Bimodal activation of BubR1 by Bub3 sustains mitotic checkpoint signaling. Proc. Natl. Acad. Sci. U.S.A. 111, E4185–E4193 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fraschini R., et al. , Bub3 interaction with Mad2, Mad3 and Cdc20 is mediated by WD40 repeats and does not require intact kinetochores. EMBO J. 20, 6648–6659 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li J., Dang N., Wood D. J., Huang J. Y., The kinetochore-dependent and -independent formation of the CDC20-MAD2 complex and its functions in HeLa cells. Sci. Rep. 7, 41072 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lampou K., Böwer F., Komaki S., Köhler M., Schnittger A., A cytological and functional framework of the meiotic spindle assembly checkpoint in Arabidopsis thaliana. bioRxiv [Preprint] (2023). 10.1101/2023.05.26.542430 (Accessed 27 February 2024). [DOI] [Google Scholar]
- 24.Maiato H., Gomes A. M., Sousa F., Barisic M., Mechanisms of chromosome congression during mitosis. Biology 6, E13 (2017), 10.3390/biology6010013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mao Y. H., Desai A., Cleveland D. W., Microtubule capture by CENP-E silences BubR1-dependent mitotic checkpoint signaling. J. Cell Biol. 170, 873–880 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Akera T., Goto Y., Sato M., Yamamoto M., Watanabe Y., Mad1 promotes chromosome congression by anchoring a kinesin motor to the kinetochore. Nat. Cell Biol. 17, 1124–1133 (2015). [DOI] [PubMed] [Google Scholar]
- 27.Richardson D. N., Simmons M. P., Reddy A. S., Comprehensive comparative analysis of kinesins in photosynthetic eukaryotes. BMC Genomics 7, 18 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix 01 (PDF)
Live-cell imaging of GFP-BUB3.3 and mCherry-labelled microtubules in the bub3.3 background. Images have intervals of 15 s.
Live-cell imaging of WT cells expressing NDC80-TagRFP following the treatment with 100 nM oryzalin. Images have intervals of 20 s.
Live-cell imaging of bub3.3 cells expressing NDC80-TagRFP following the treatment with 100 nM oryzalin. Images have intervals of 20 s.
Live-cell imaging of VisGreen-TUB6 and histone H1-TagRFP in control cells without oryzalin treatment.
Live-cell imaging of VisGreen-TUB6 and histone H1-TagRFP in bub3.3 cells without oryzalin treatment.
Live-cell imaging of VisGreen-TUB6 and histone H1-TagRFP in bub3.3 cells without oryzalin treatment.
Live-cell imaging of VisGreen-TUB6 and histone H1-TagRFP in control cells treated with 100 nM oryzalin.
Live-cell imaging of VisGreen-TUB6 and histone H1-TagRFP in bub3.3 cells treated with 100 nM oryzalin.
Data Availability Statement
All study data are included in the article and/or supporting information.