Abstract
In the western North Pacific, prominent granulomatous testes have been detected in many Brucella-infected common minke whales (Balaenoptera acutorostrata), but there have been no reports in toothed cetaceans. We found severe orchitis with granulomatous lesions in a rough-toothed dolphin (Steno bredanensis) stranded on the Pacific coast of Japan in 2011. Histopathological examination revealed leukocyte infiltration of the lesions. DNA from the lesion was analyzed by PCR and it showed molecular biological similarities with those of Brucella-infected common minke whales and Brucella ceti of sequence-type 27 (ST27). These results suggest that the type of Brucella ceti that infected the dolphin was ST27, which may have caused severe orchitis. This study adds to our understanding of Brucella infections in marine mammals.
Keywords: Brucella, cetacean, dolphin, orchitis, testis
Gram-negative intracellular bacteria, Brucella ceti, infect many cetacean species in the world’s oceans and cause cetacean brucellosis [5,6,7, 13]. Although associated pathological changes have been observed in various organs, they have been less severe and less frequently reported than in terrestrial animals infected with terrestrial Brucella species such as B. abortus or B. melitensis [5, 6]. Severe orchitis is a hallmark of brucellosis in terrestrial animals. However, it had not been reported in cetaceans until our research in the Pacific [17].
In the western North Pacific, many common minke whales (Balaenoptera acutorostrata) have been found to have orchitis with pronounced severe granulomatous lesions, suggesting possible Brucella infection [15, 17]. Pathologic and serologic studies have supported this hypothesis [15, 17]. Brucella-specific DNA sequences have been detected by PCR in DNA extracted from abnormal whale testes (DNA ID: JM13/00), although the bacteria have not been isolated [13, 16].
Anti-Brucella antibodies have been detected in serum samples from several species of toothed cetaceans in the region without gross pathologic changes [14, 18]. B. ceti was recently isolated for the first time from a bottlenose dolphin (Tursiops truncates) with osteomyelitis in the western North Pacific [20]. Therefore, B. ceti is thought to be widespread among toothed cetaceans in the region. However, no reproductive abnormalities have been reported in these animals. Here, we report a case of a rough-toothed dolphin (Steno bredanensis) stranded on the Pacific coast of Japan with a severely granulomatous testis and discuss B. ceti infection based on pathologic and molecular biological studies.
A male rough-toothed dolphin was stranded on the coast of Ichinomiya, Chiba Prefecture, Japan (35°20’09.6” N, 140°23’42.2” E) on May 12, 2011. The dolphin was thin and had some skin injuries from the cookie-cutter shark bites (Fig. 1a). It was determined to be mature based on its body length of 2.3 m. The body was transferred to National Museum of Nature and Science, Shinjuku, Tokyo, and stored at −20°C. After thawing, a postmortem examination was conducted on September 24, 2011.
Fig. 1.
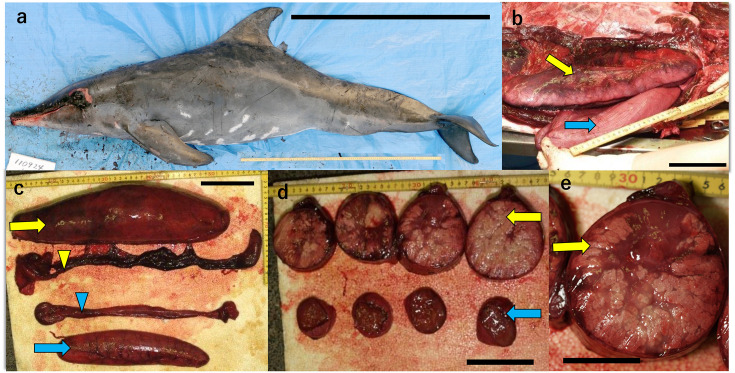
Gross pathology of the stranded rough-toothed dolphin. a. Side view of the body. Black bar: 1 m. b and c., Abnormal testes and epididymides were found during postmortem examination. Yellow arrow: left testis, blue arrow: right testis, yellow arrowhead: left epididymis, blue arrowhead: right epididymis. Black bar: 10 cm. d. Sequential cross sections of the left (lower) and right (upper) testes. Black bar: 10 cm. e. Close-up of a section of the left testis. Black bar: 5 cm.
The relevant organs were fixed in 10% neutral buffered formalin for histopathological examination. Thin tissue sections were prepared and stained with hematoxylin and eosin. Immunohistochemical analysis was also performed to identify inflammatory cells in the relevant lesions. The antibodies against ionized calcium-binding adaptor molecule 1 (lba1) for macrophages, B lymphocyte antigen 36 (BLA36) for B lymphocytes and CD3 for T lymphocytes, were used as primary antibodies (Supplementary Table 1). Color development was performed using the standard horseradish peroxidase system.
Tissue samples were collected from three spots on each testis, two spots on the left epididymis, and the spleen for molecular biological analysis by PCR. These samples were stored in 70% ethanol until further use. DNA was extracted from these tissues using a commercially available DNA extraction kit (DNeasy Tissue Kit; Qiagen, Boston, MA, USA). Brucella-specific sequences were examined using two PCR methods, each with specific primers (Supplementary Table 2) [3, 16]. Using a commercial PCR kit (Ex Taq Kit; Takara, Kyoto, Japan), amplification was performed as follows: 96°C for 1 min; 30 cycles of 96°C for 20 sec, 57°C for 30 sec, and 72°C for 1 min; followed by a final extension at 72°C for 10 min. Amplified DNA was confirmed by agarose gel electrophoresis. After purification of the amplified DNA fragments, nucleotide sequences were analyzed by the dye terminator method using a commercial kit (BigDye Terminator v3.1 Cycle sequencing kit: Thermo Scientific Inc., Waltham, MA, USA).
Gross observations at the necropsy showed that the left testis was markedly enlarged and swollen (Fig. 1b and 1c). It had severe granulomatous orchitis with caseation (Fig. 1d and 1e). In addition, the left epididymis was slightly enlarged, and dark in color (Fig. 1c). Other organs, including the brain, showed no significant changes, except that the left and right lungs showed chronic bronchopneumonia.
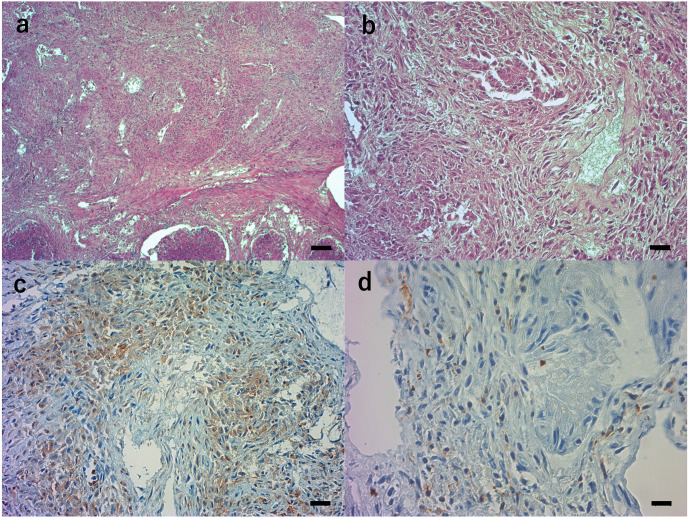
Histopathological examination showed that the parenchyma of the left testis was diffusely necrotic with granulomatous infection, consisting of many infiltrated macrophages, epithelioid cells, and some plasma cells, neutrophils and lymphocytes (Fig. 2a–d), although some postmortem changes were found. The right testis and other organs did not show any significant pathological changes related to brucellosis. These pathological observations were similar to the previously described granulomatous testis of minke whales [17]. Many large macrophages and epithelial cells were positive with the anti-lba1 antibody to detect macrophages (Fig. 2c) and some lymphocytes were positive with the anti-BLA36 antibody to detect B lymphocytes (Fig. 2d). However, no reactivity was observed with the anti-CD3 antibody to detect T lymphocytes, although this antibody has previously been shown to react with T lymphocytes from harbor porpoise (Phocoena phocoena), Dall’s porpoise (Phocoenoides dalli), and Hubbs’ beaked whale (Mesoplodon carlhubbsi) [12]. This discrepancy may be due to the epitope structure of rough-toothed dolphin CD3 being different from that of the three dolphin species. Alternatively, the epitope structure of the dolphin may be affected by postmortem changes.
Fig. 2.
Histopathological examination of the left testis of the stranded rough-toothed dolphin. Tissue sections were stained with hematoxylin and eosin (a, b) or immunostained with primary antibodies against leukocytes (c, d). a and b. The parenchyma of the left testis was diffusely necrotic with granulomatous infections consisting of many macrophages, epitheloid cells, and some plasma cells, neutrophils and lymphocytes. Black bar: 100 μm (a), 20 μm (b). c. Many macrophages and epithelioid cells were positive with the lba1 polyclonal antibody. Black bar: 20 μm. d. Some lymphocytes were positive with the monoclonal antibody BLA36. Black bar: 10 μm.
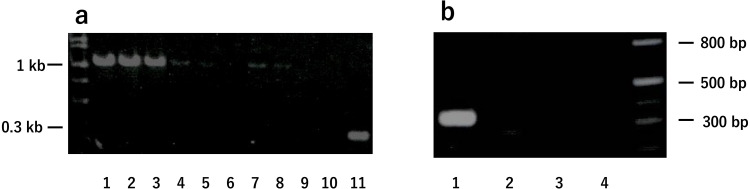
There is a unique mobile genetic element in the Brucella genus, insertion sequence 711 (IS711), which has species-specific locations [9]. We first examined the insertion of the IS711 element downstream of bp26, a specific marine Brucella marker [2, 16]. DNA from the left testis (DNA ID: JRT1/11) showed a highly amplified DNA band of 1,080 bp (lanes 1–3), which was larger than that of terrestrial Brucella (B. abortus, 250 bp on lane 11) (Fig. 3a). This indicated that JRT1/11 contained marine Brucella DNA. DNA from the right testis (lanes 4–6) and left epididymis (lanes 7 and 8) showed weaker bands at the same positions, whereas DNA from the spleen did not produce any bands (Fig. 3a, lanes 9 and 10). Next, the DNA, JRT1/11, was subjected to the infrequent restriction site-PCR (IRS-PCR) method, consisting of four PCR sets (PCR-I, -II, -III, and -IV) with four PCR primer sets (Supplementary Table 2) [3]. This method divides B. ceti into three groups based on amplification patterns; in the first group, a single amplicon is produced by PCR-I only (approximately 300 bp); in the second group, two amplicons are produced by PCR-II and -III (approximately 600 and 400 bp, respectively), and in the third group, a single amplicon is produced by PCR-IV only (approximately 300 bp). None of these bands have been reported in terrestrial Brucella. From JRT1/11, a DNA band of approximately 300 bp was amplified by PCR-I only, indicating that the dolphin B. ceti belongs to the first group (Fig. 3b). The sequences determined in the present study were deposited in DDBJ (accession numbers: LC757032 and LC757033).
Fig. 3.
Amplified DNA bands by two PCR methods targeting marine Brucella-specific sequences. a. An insertion of IS711 downstream of bp26 was analyzed using extracted DNA from three spots of left testis (lanes 1–3) and right testis (lanes 4–6), two spots of left epididymis (lanes 7, 8) and spleen (lanes 9, 10), and B. abortus as a control (lane 11). Due to the IS711 insertion in B. ceti, larger size of amplicons (1,080 bp) were produced in dolphin DNAs rather than in B. abortus DNA (250 bp). b. The JRT1/11 DNA from the left testis was analyzed by infrequent restricted site-PCR (IRS-PCR). lane 1; PCR-I, lane 2; PCR-II, lane 3; PCR-III, lane 4; PCR-IV. The dolphin testis DNA produced a single amplicon (approximately 300 bp) by PCR-I only.
A recently developed multilocus sequence typing (MLST) scheme targeting 21 housekeeping genes, is useful for classifying whole Brucella strains. It classifies B. ceti into three sequence types (ST), ST23, ST26, and ST27 [21, 22]. Each ST shows a different IRS-PCR pattern; ST23, ST 26, and ST27 produce two amplicons by PCR-II and -III, a single amplicon by PCR-IV only, and a single amplicon by PCR-I only, respectively [22]. In Table 1, we have summarized the characteristics of JRT1/11, together with ST23, ST26 and ST27 of B. ceti, B. melitensis, and JM13/00 [13, 16, 20, 22]. The JRT1/11 shows the same IRS-PCR amplification pattern as B. ceti of ST27 and JM13/00. The pathologic and molecular biological examinations in the present study strongly suggested that B. ceti belonging to ST27 infected the rough-toothed dolphin and induced the severe granulomatous lesions in the testis, although the bacterial antigens were not examined histochemically. The MLST sequence type of JRT1/11 should be confirmed in the future.
Table 1. Comparison of JRT1/11 with other strains in ST27, 23, 26 and Brucella melitensis 16M.
| Strain | Host animal | Sea area | IS711 post bp26 | IRS-PCRa) |
MLST 21b) |
|---|---|---|---|---|---|
| I / II / III / IV | |||||
| JRT1/11c) | Rough-toothed dolphin | Japan, Pacific | + | + / − / − / − | NA |
| JM13/00c) | Common minke whale | Japan, Pacific | + | + / − / − / − | NA |
| BD1442 | Bottlenose dolphin | Japan, Pacific | + | + / − / − / − | ST27 |
| F5/99 | Bottlenose dolphin | USA, Pacific | + | NA | ST27 |
| F8/08-1 | Bottlenose dolphin | USA, Pacific | + | + / − / − / − | ST27 |
| F8/08-24 | California sea lion | USA, Pacific | + | + / − / − / − | ST27 |
| 85A05748 | Human | Peru, Pacific | + | NA | ST27 |
| 01A09163 | Human | Peru, Pacific | + | NA | ST27 |
| 02/611 | Human | New Zealand, Pacific | + | + / − / − / − | ST27 |
| B1/94 | Harbor porpoise | UK, Atlantic | + | − / + / + / − | ST23 |
| B14/94 | Common dolphin | UK, Atlantic | + | − / − / − / + | ST26 |
| B. melitensis, 16M | Caprine | USA | – | − / − / − / − | ST73 |
+; PCR positive, –; PCR megative, NA; not available. a) Infrequent restricted site-PCR, ST27 produces a single amplicon by PCR-I only, while ST23, ST 26, produce two amplicons by PCR-II and -III, or a single amplicon by PCR-IV only, respectively. No amplicon is produced from terrestrial-originated Brucella, such as B. melitensis. b) Multilocus sequence typing scheme targeting 21 genes, c) The ID of bacterial DNA. This table was created based on the references [16, 20, 22].
The ST27 is reported to have unique characteristics. It is found primarily in Pacific waters. In addition to cetaceans, which are the primary hosts of ST27, it has also been detected in pinnipeds and humans. Importantly, it has been reported that ST27 strains are often associated with severe disease. Reproductive abnormalities, including abortion, have been reported in dolphins associated with ST27 strains [1, 4, 8, 10, 22]. Strains possibly belonging to ST27 have also been detected in JRT1/11 and JM13/00 from cetaceans with severe orchitis [16]. In addition, an ST27 strain was recently isolated from a bottlenose dolphin with osteomyelitis in the sea around Japan [20]. Neurobrucellosis has also been reported in humans infected with ST27 strains, although there is no evidence of contact between the patients and marine mammals [11, 19].
The present study adds to our understanding that B. ceti, possibly the ST27 strain, causes a severe male reproductive disorder in toothed cetaceans, as well as in common minke whales, which are baleen cetaceans. This severe orchitis may affect reproduction or be involved in the sexual transmission of bacteria from males to females. Further studies using pathology and molecular biology are needed to understand the relationship between the ST and related diseases, as well as their impact on the overall health of cetaceans.
CONFLICTS OF INTEREST
All authors have no conflicts of interest to disclose.
Supplementary
Acknowledgments
The authors would like to thank their gratitude to all the individuals who participated in the stranding field work. We also express our appreciation to Dr. Y. Nagai and Ms. M. Tsuda of the Japan Agency for Marine-Earth Science and Technology for their technical advice on molecular biology. In addition, we would like to thank Dr. M. Mitishita of Nippon Veterinary and Life Science University for providing us with valuable technical advice on immunohistochemistry.
REFERENCES
- 1.Buckle K, Roe WD, Howe L, Michael S, Duignan PJ, Burrows E, Ha HJ, Humphrey S, McDonald WL. 2017. Brucellosis in endangered Hector’s dolphins (Cephalorhynchus hectori). Vet Pathol 54: 838–845. doi: 10.1177/0300985817707023 [DOI] [PubMed] [Google Scholar]
- 2.Cloeckaert A, Grayon M, Grepinet O. 2000. An IS711 element downstream of the bp26 gene is a specific marker of Brucella spp. isolated from marine mammals. Clin Diagn Lab Immunol 7: 835–839. doi: 10.1128/CDLI.7.5.835-839.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cloeckaert A, Grayon M, Grépinet O, Boumedine KS. 2003. Classification of Brucella strains isolated from marine mammals by infrequent restriction site-PCR and development of specific PCR identification tests. Microbes Infect 5: 593–602. doi: 10.1016/S1286-4579(03)00091-1 [DOI] [PubMed] [Google Scholar]
- 4.Ewalt DR, Payeur JB, Martin BM, Cummins DR, Miller WG. 1994. Characteristics of a Brucella species from a bottlenose dolphin (Tursiops truncatus). J Vet Diagn Invest 6: 448–452. doi: 10.1177/104063879400600408 [DOI] [PubMed] [Google Scholar]
- 5.Foster G, MacMillan AP, Godfroid J, Howie F, Ross HM, Cloeckaert A, Reid RJ, Brew S, Patterson IA. 2002. A review of Brucella sp. infection of sea mammals with particular emphasis on isolates from Scotland. Vet Microbiol 90: 563–580. doi: 10.1016/S0378-1135(02)00236-5 [DOI] [PubMed] [Google Scholar]
- 6.Guzmán-Verri C, González-Barrientos R, Hernández-Mora G, Morales JA, Baquero-Calvo E, Chaves-Olarte E, Moreno E. 2012. Brucella ceti and brucellosis in cetaceans. Front Cell Infect Microbiol 2: 3. doi: 10.3389/fcimb.2012.00003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hernández-Mora G, Palacios-Alfaro JD, González-Barrientos R. 2013. Wildlife reservoirs of brucellosis: Brucella in aquatic environments. Rev Sci Tech 32: 89–103. doi: 10.20506/rst.32.1.2194 [DOI] [PubMed] [Google Scholar]
- 8.Hernández-Mora G, González-Barrientos R, Víquez-Ruíz E, Palacios-Alfaro JD, Bettoni-Rodríguez G, Gendre M, Vincent C, Roca-Monge K, Ruiz-Villalobos N, Suárez-Esquivel M, Cordero-Chavarría M, Chaves-Olarte E, Thompson NR, Barquero-Calvo E, Moreno E, Guzmán-Verri C. 2021. Brucella sp. sequence-type 27 associated with abortion in dwarf sperm whale Kogia sima. Eur J Wildl Res 67: 63. doi: 10.1007/s10344-021-01502-5 [DOI] [Google Scholar]
- 9.López-Goñi I, García-Yoldi D, Marín CM, de Miguel MJ, Muñoz PM, Blasco JM, Jacques I, Grayon M, Cloeckaert A, Ferreira AC, Cardoso R, Corrêa de Sá MI, Walravens K, Albert D, Garin-Bastuji B. 2008. Evaluation of a multiplex PCR assay (Bruce-ladder) for molecular typing of all Brucella species, including the vaccine strains. J Clin Microbiol 46: 3484–3487. doi: 10.1128/JCM.00837-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mackie JT, Blyde D, Harris L, Roe WD, Keyburn AL. 2020. Brucellosis associated with stillbirth in a bottlenose dolphin in Australia. Aust Vet J 98: 92–95. doi: 10.1111/avj.12903 [DOI] [PubMed] [Google Scholar]
- 11.McDonald WL, Jamaludin R, Mackereth G, Hansen M, Humphrey S, Short P, Taylor T, Swingler J, Dawson CE, Whatmore AM, Stubberfield E, Perrett LL, Simmons G. 2006. Characterization of a Brucella sp. strain as a marine-mammal type despite isolation from a patient with spinal osteomyelitis in New Zealand. J Clin Microbiol 44: 4363–4370. doi: 10.1128/JCM.00680-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nakagun S, Kobayashi Y. 2020. Histochemical and immunohistochemical characterizations of hepatic trematodiasis in odontocetes. Front Vet Sci 7: 336. doi: 10.3389/fvets.2020.00336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ohishi K, Fujise Y, Maruyama T. 2008. Brucella spp. in the western North Pacific and Antarctic cetaceans: A review. J Cetacean Res Manag 10: 67–72. doi: 10.47536/jcrm.v10i1.661 [DOI] [Google Scholar]
- 14.Ohishi K, Katsumata E, Uchida K, Maruyama T. 2007. Two stranded pygmy sperm whales (Kogia breviceps) with anti-Brucella antibodies in Japan. Vet Rec 160: 628–629. doi: 10.1136/vr.160.18.628 [DOI] [PubMed] [Google Scholar]
- 15.Ohishi K, Bando T, Abe E, Kawai Y, Fujise Y, Maruyama T. 2016. Long-term and large-scale epidemiology of Brucella infection in baleen whales and sperm whales in the western North Pacific and Antarctic Oceans. J Vet Med Sci 78: 1457–1464. doi: 10.1292/jvms.16-0076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ohishi K, Takishita K, Kawato M, Zenitani R, Bando T, Fujise Y, Goto Y, Yamamoto S, Maruyama T. 2004. Molecular evidence of new variant Brucella in North Pacific common minke whales. Microbes Infect 6: 1199–1204. doi: 10.1016/j.micinf.2004.06.006 [DOI] [PubMed] [Google Scholar]
- 17.Ohishi K, Zenitani R, Bando T, Goto Y, Uchida K, Maruyama T, Yamamoto S, Miyazaki N, Fujise Y. 2003. Pathological and serological evidence of Brucella-infection in baleen whales (Mysticeti) in the western North Pacific. Comp Immunol Microbiol Infect Dis 26: 125–136. doi: 10.1016/S0147-9571(02)00036-X [DOI] [PubMed] [Google Scholar]
- 18.Ohishi K, Amano M, Nakamatsu K, Miyazaki N, Tajima Y, Yamada TK, Matsuda A, Ochiai M, Matsuishi TF, Taru H, Iwao H, Maruyama T. 2020. Serologic survey of Brucella infection in cetaceans inhabiting along the coast of Japan. J Vet Med Sci 82: 43–46. doi: 10.1292/jvms.19-0481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sohn AH, Probert WS, Glaser CA, Gupta N, Bollen AW, Wong JD, Grace EM, McDonald WC. 2003. Human neurobrucellosis with intracerebral granuloma caused by a marine mammal Brucella spp. Emerg Infect Dis 9: 485–488. doi: 10.3201/eid0904.020576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ueno Y, Yanagisawa M, Kino S, Shigeno S, Osaki M, Takamatsu D, Katsuda K, Maruyama T, Ohishi K. 2020. Molecular characterization of Brucella ceti from a bottlenose dolphin (Tursiops truncatus) with osteomyelitis in the western Pacific. J Vet Med Sci 82: 754–758. doi: 10.1292/jvms.20-0015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Whatmore AM, Koylass MS, Muchowski J, Edwards-Smallbone J, Gopaul KK, Perrett LL. 2016. Extended multilocus sequence analysis to describe the global population structure of the genus Brucella: Phylogeography and relationship to biovars. Front Microbiol 7: 2049. doi: 10.3389/fmicb.2016.02049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Whatmore AM, Dawson C, Muchowski J, Perrett LL, Stubberfield E, Koylass M, Foster G, Davison NJ, Quance C, Sidor IF, Field CL, St Leger J. 2017. Characterisation of North American Brucella isolates from marine mammals. PLoS One 12: e0184758. doi: 10.1371/journal.pone.0184758 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.