Pulsatile Myc induction, through epigenetic alterations, mediates several aspects of the exercise response and may restore muscle plasticity with age.
Key Words: Yamanaka factors, hypertrophy, Geroscience, DNA methylation, biological age
Abstract
Of the “Yamanaka factors” Oct3/4, Sox2, Klf4, and c-Myc (OSKM), the transcription factor c-Myc (Myc) is the most responsive to exercise in skeletal muscle and is enriched within the muscle fiber. We hypothesize that the pulsatile induction of MYC protein after bouts of exercise can serve to epigenetically reprogram skeletal muscle toward a more resilient and functional state.
KEY POINTS
Myc is a Yamanaka “epigenetic reprogramming” factor and oncogene that controls transcription, cell proliferation, ribosome biogenesis, protein synthesis, circadian rhythm, and metabolism, among other things.
Differentiated multinuclear skeletal muscle fibers are resistant to tumorigenesis, and Myc gene and protein increases acutely but dramatically in skeletal muscle after exercise across species, including in humans.
With aging, several hallmark adaptations to exercise training such as muscle hypertrophy are attenuated or delayed, as is Myc’s responsiveness to an exercise bout.
A controlled pulse of Myc in skeletal muscle fibers regulates more than 1300 genes and recapitulates molecular aspects of the skeletal muscle response to exercise.
Through its role as an epigenetic reprogramming factor, pulsatile expression of Myc in skeletal muscle throughout the lifespan and/or late in life could be leveraged to cause a shift toward a more youthful and “rejuvenated” state.
INTRODUCTION
Skeletal muscle mass and function declines with aging, leading to loss of independence and mortality. This inevitable deterioration, termed “sarcopenia,” is elusive in its precise definition and etiology, but the deleterious ramifications are well documented and understood (1). Exercise, and specifically hypertrophic resistance exercise, is the most widely accessible and effective therapy for combatting sarcopenia (1). Exercise is relatively simple to deploy and confers numerous benefits including increased muscle mass, strength, power, and quality throughout the lifespan (2). Unfortunately, aspects of the potency of exercise may decline in the later years of life. A dysregulated molecular response to resistance exercise characterizes aging (3), and the anabolic effects of resistance training may be attenuated or delayed beyond age 80 in both men and women (4,5). An improved understanding of what exercise-induced signals control muscle mass and function throughout the lifespan, and how those signals are altered by aging, could lead to therapeutic approaches that prevent sarcopenia and boost the efficacy of physical training.
The Geroscience field has matured over the last decade in conjunction with the identification of the hallmarks (or pillars) of aging (6–8). The list of hallmarks has expanded and evolved (8), but the overarching goal of Geroscience remains the same: accelerate research on the mechanisms of aging, in part by studying the hallmarks of aging, to find interventions that extend healthspan and potentially prolong lifespan (7). Numerous therapeutic strategies may accomplish these goals. Among them are caloric restriction and “antiaging” drug therapies such as senolytics, metformin, and rapamycin (9). Some of these interventions may modify the epigenome — that is, a layer of regulation that controls gene expression in the absence of a modification to the DNA sequence. Recent efforts to leverage epigenetic “rejuvenation” to slow or reverse aging in preclinical models have become increasingly popular. One key epigenetic modification is the methylation of cytosines in a CpG context that can repress or promote gene transcription, known as DNA methylation. In broad strokes, relative hypomethylation of promoter regions can promote transcription while hypermethylation can repress it. DNA methylation profiling across tissues and species reveals systematic epigenetic changes throughout the lifespan that can accurately predict chronological age (10,11). Lower DNA methylation “clock” age relative to chronological age is thought to reflect younger biological age which could translate to improved health and delayed mortality.
One intervention that reverses DNA methylation age and promotes cellular youthfulness and resiliency is the induction of Yamanaka factors to elicit partial epigenetic reprogramming (10,12). Yamanaka factors were initially discovered as a strategy to induce pluripotent stem cells from differentiated somatic cells via epigenetic reprogramming (13). This discovery earned Shinya Yamanaka the Nobel Prize in Physiology or Medicine in 2012 (shared with Sir John B. Gurdon). Consistent with their function, brief systemic induction of Yamanaka factors (Oct3/4, Sox2, Klf4, and Myc, or OSKM) in mice ameliorates several hallmarks of aging in vivo including the age-associated impairment in skeletal muscle recovery time following injury (14). Exercise training also restores muscle regenerative capacity after injury in aged animals (15,16). Exercise is arguably the most powerful anti-aging “drug” available, yet it is still underprescribed (17,18). The extent to which Yamanaka factor induction and exercise share common mechanisms for promoting muscle health is an open area of inquiry.
Essentially all forms of exercise, either acute or chronic in rodents and humans, alter the skeletal muscle DNA methylome to influence gene expression (19). Our laboratory recently showed that late-life combined resistance and endurance exercise in mice reduced DNA methylation age in skeletal muscle (20,21). Compatible conclusions were reached in human muscle tissue with aerobic, high-intensity interval, and resistance exercise training throughout the lifespan (22). Our work showed that reduced methylation age coincided with elevated Myc levels (21), an altered DNA methylome specifically in skeletal muscle fiber nuclei (myonuclei), higher muscle mass, and greater in vivo muscle function than aged (24 months) sedentary mice (23). Myc is the Yamanaka factor that is most responsive to exercise in skeletal muscle (21) and is made by myonuclei during muscle loading (24). The sustained induction of Myc is linked to tumorigenesis, but differentiated syncytial muscle fibers are resistant to developing tumors (25,26). MYC can remain elevated in hypertrophying skeletal muscle tissue of rodents for up to 14 d without overt adverse consequence (27). We hypothesize that pulses of Myc transcript and subsequent MYC protein in muscle fibers serve as an epigenetic reprogramming stimulus that facilitates adaptability and a more youthful phenotype throughout the lifespan in the muscle of aged organisms. We also hypothesize that a blunted Myc response to resistance exercise may contribute to reduced muscle plasticity with training as aging progresses.
CURRENT UNDERSTANDING OF THE ROLE OF MYC IN SKELETAL MUSCLE
MYC is a transcription factor that regulates around 15% of the genome (28). It dimerizes with MAX and other proteins to bind DNA and regulate cell cycle, apoptosis, metabolism, circadian rhythm, ribosome biogenesis, protein synthesis, miRNA and extracellular vesicle biogenesis, and a variety of other cellular functions (28,29). Recent evidence suggests that MYC is a universal amplifier of transcription (30) and can directly bind RNA to control gene expression (31). Chronic upregulation of MYC is typically oncogenic, but overexpression in certain tissues such as the liver can serve beneficial roles such as preventing obesity and insulin resistance (32). Postdevelopmental systemic knockout of Myc in mice causes a premature aging phenotype characterized by low muscle mass, albeit with extended lifespan due to lower incidence of cancer in nonmuscle tissues (33). Its role as a Yamanaka factor points to its function in regulating the epigenome, and specifically DNA methylation. MYC can influence DNA methylation via its interactions with DNA methyltransferases and ten-eleven translocase enzymes that control methylation status (34–36).
The effects of MYC are well characterized in numerous tissues and in the context of cancer, but our understanding of its role in skeletal muscle is somewhat limited. MYC protein is generally not abundant in resting adult skeletal muscle but is elevated during developmental muscle growth (37,38). MYC protein is strongly upregulated in rodent muscle during surgical mechanical overload (38) and localizes to myonuclei during muscle loading in birds and rodents (39,40). MYC transcript peaks 3 h after acute resistance exercise in skeletal muscle of humans, remains elevated by 12 h, and returns to resting levels by 24 h (41). MYC is also elevated by endurance exercise in muscle, but to a lesser extent than resistance exercise (41). MYC protein is induced by resistance exercise in human muscle (42,43) and its levels associate with the magnitude of hypertrophic adaptation to resistance training in humans (44). A disproportionately higher MYC response with hypertrophic versus endurance stimuli is likely a result of MYC’s influence on processes known to associate with growth, such as ribosome biogenesis. Enhanced ribosome biogenesis is a hallmark response to resistance exercise (41,45–47). The majority of studies on MYC in skeletal muscle focus on its role in driving ribosome biogenesis, translational capacity, and protein synthesis during loading-induced hypertrophy (e.g., synergist ablation in rodents or resistance training in humans) (48). Despite MYC’s well-documented ability to drive rDNA transcription in skeletal muscle and other tissues, it is currently unknown whether MYC induction is necessary or sufficient for adult muscle hypertrophy in vivo.
To provide a more detailed understanding of the role of MYC across skeletal muscles in vivo, our laboratory developed a genetically modified muscle-specific reverse tetracycline transactivator mouse model of pulsatile MYC induction that is controlled by the delivery of doxycycline in water (21,24). We can control the expression of MYC at will specifically in all skeletal muscle fibers in vivo at any point in the lifespan of the mouse using this tool. In our hands, overnight doxycycline in drinking water causes an accumulation of MYC protein in skeletal muscle 12 h after the removal of doxycycline. When MYC protein is high in skeletal muscle, we observe changes in the transcriptome that are greatest in the soleus (mixed slow- and fast-twitch, similar to human vastus lateralis), intermediate in the plantaris (primarily fast-twitch), and lowest in the quadriceps (mixed fiber types) (21,24). In the soleus muscle, a single pulse of MYC altered approximately 1400 genes (21). Ribosome biogenesis-related genes were most induced by MYC, and skeletal muscle identity genes were repressed; the latter is indicative of a transient cellular reprogramming response toward “stemness,” which is characteristic of Yamanaka factor expression (13).
Among ribosomal genes that were altered by MYC in muscle was Ribosomal protein large 3 (Rpl3) and muscle-specific Ribosomal protein large 3 like (Rpl3l). The induction of Rpl3 and repression of Rpl3l is characteristic of muscle growth and could be related to ribosome biogenesis as well as ribosome specialization. Both processes can support the translational and metabolic demands of muscle adaptation (24). In addition to an alteration of myosin heavy chain transcripts in all muscles by MYC, it also repressed circadian rhythm genes such as Reverbα? and Reverbβ. MYC strongly influences the circadian rhythm in cancerous cells (49). Circadian clock transcription factors bind E-boxes the same as MYC. Exercise is known to affect circadian rhythms in skeletal muscle (50). Perhaps exercise-induced MYC competes with clock genes at E-boxes to affect the muscle circadian rhythm. In the plantaris muscle, the transcriptome signature from a brief pulse of MYC overlapped with the myonuclear transcriptome response to short-term mechanical overload in mice (24). MYC was also predicted as one of the most influential transcription factors controlling the myonuclear transcriptome during mechanical overload in mice, in addition to being highly enriched in myonuclei (24). These data, in context with the literature, collectively point to MYC playing a key role in regulating exercise adaptation in skeletal muscle.
MYC AND PARTIAL EPIGENETIC REPROGRAMMING IN SKELETAL MUSCLE WITH EXERCISE DURING AGING
The beneficial effects of exercise in aged skeletal muscle are numerous and should not be underestimated (18). Exercise improves muscle strength and power-producing capacity, insulin sensitivity and metabolic health, as well as muscle mass, all of which can contribute to a more youthful phenotype. Indeed, high muscle function and health is negatively associated with mortality (17,51). The functional benefits of exercise throughout the lifespan are well understood, but the mechanisms for how these benefits are controlled is unclear. We therefore hypothesized that late-life hypertrophic/endurance exercise in aged animals would share molecular features of partial epigenetic reprogramming by Yamanaka factors in skeletal muscle, as well as the induction of Myc (21).
To begin to address our hypothesis, we first developed an exercise training model for mice called progressive weighted wheel running, or PoWeR. This approach elicits muscle hypertrophy and faster-to-slower fiber type switching across various hind limb muscles in young (4–6 months) and aged (22–24 months) mice (23,52,53). Since Yamanaka factor induction reduces DNA methylation clock age (10), we corroborated that exercise has a similar effect on the muscle DNA methylome after 2 months of late-life PoWeR in mice (20,21). We then compared the muscle transcriptome after late-life PoWeR (21,23) to the transcriptome after brief muscle fiber-specific Yamanaka factor-mediated partial reprogramming (54). A quarter of the genes that were downregulated by late-life exercise were shared with partial reprogramming (21). These data suggest that exercise and Yamanaka factors elicit a common molecular signature in skeletal muscle. Next, we overlapped the late-life exercise transcriptome with Yamanaka factor induction as well as a pulse of MYC in muscle to further define a potentially age-mitigating molecular program. Our rationale for overexpressing Myc is that it is the Yamanaka factor that is most responsive to any form of exercise, but particularly hypertrophy-inducing exercise (21,41). All three interventions were associated with lower Ndufb11 and Romo1; both are implicated in aging-associated dysregulation such as excessive reactive oxygen species (ROS) production and senescence (21). Finally, we profiled the muscle DNA methylome after a single pulse of MYC and found a shift in the epigenetic landscape (21). MYC altering the muscle methylome is consistent with its role as an epigenetic reprogramming factor in other cell types (34,35) and points to MYC as a regulator of the DNA methylation machinery in muscle with exercise. Since several metabolic intermediates are necessary for DNA methylation changes to ensue (55), epigenetic alterations may also be enabled by changes in cellular metabolism caused by MYC (56).
In the soleus muscle of aged mice, Myc is approximately 60% higher following 2 months of PoWeR when compared to the soleus of untrained controls (21,23). The mice began running 6–8 km/night beginning at 22 months of age, which is equivalent to approximately 65 y of age in humans, and the muscle was harvested 24 h after the final wheel running bout. Although Myc gene and protein is highly responsive to muscle loading (e.g., exercise) in young adult rodents and humans, there is evidence to suggest that Myc in muscle is less responsive to resistance exercise in old age. Relative to young participants, MYC gene and protein in muscle is not higher at rest (57,58) but is less responsive after a bout of resistance exercise in people approximately 70 y of age (59,60). These data dovetail with classic work by Alway showing blunted MYC with mechanical loading of wing muscles in aged versus young birds (39). Unfortunately, aspects of human skeletal muscle biology can become refractory to resistance training beyond 80 y of age (4,5). We propose that pulses of MYC in muscle is a central mediator of the exercise training response throughout the lifespan and may mimic aspects of exercise to promote muscle health and curtail sarcopenia. Furthermore, periodic expression of MYC late in life could restore muscle adaptability that is reduced in old age and promote longevity and healthspan (Fig.).
CONCLUSION
MYC is a powerful transcription factor whose role in skeletal muscle is still being defined. It is highly responsive to resistance exercise in young adult rodent and human skeletal muscle and is implicated in various facets of adaptation including ribosome biogenesis and specialization, metabolism, and circadian rhythm. In its role as a Yamanaka factor, pulses of MYC may facilitate epigenetic reprogramming and contribute to exercise’s ability to improve muscle health throughout the lifespan. MYC pulses combined with exercise may “supercharge” the adaptive response whereas depletion of MYC may blunt hypertrophic exercise adaptations, but more work is needed in this area. Furthermore, understanding whether MYC has muscle-specific, fiber type-specific, and/or sex-specific effects will provide further insights into how MYC influences muscle biology. Since MYC responsiveness to resistance exercise in muscle can decline later in life, restoring youthful regulation of MYC could improve muscle plasticity during aging. A deeper understanding of how MYC affects muscle biology will help inform therapeutic approaches that ameliorate muscle wasting with aging and other conditions and clarify the mechanisms through which exercise serves as a “fountain of youth” (18).
Figure.
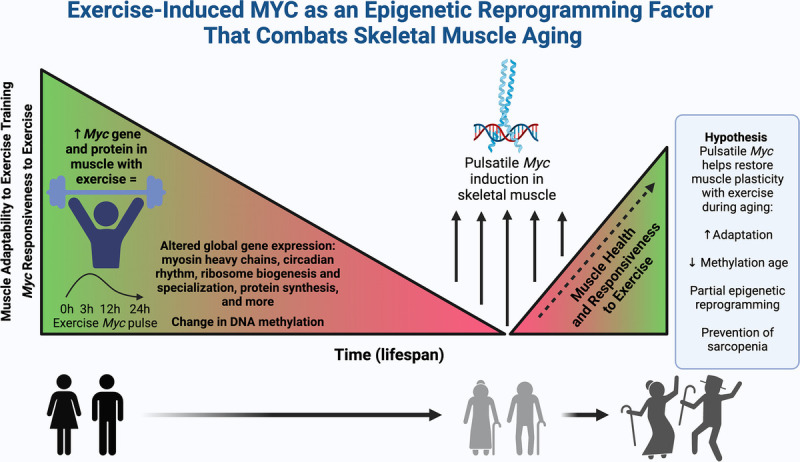
Summary of hypotheses about Myc in muscle with exercise and aging. Myc gene and protein increases acutely after exercise, and particularly hypertrophic resistance exercise, in young healthy skeletal muscle. MYC performs a variety of key functions in skeletal muscle cells and influences DNA methylation. With aging, skeletal muscle becomes smaller and weaker (i.e., sarcopenia), and exercise adaptive potential declines concomitant with reduced Myc responsiveness to exercise. Pulses of Myc/MYC throughout the lifespan and late in life may attenuate sarcopenia and/or restore skeletal muscle adaptability to training. The benefits of MYC pulses in muscle may be related to partial epigenetic reprogramming (The figure was generated using BioRender).
Acknowledgments
The authors declare that they have no conflicts of interest. This work was supported by National Institutes of Health R00 AG063994 and R01 AG080047 to KAM. This research was conducted while Kevin A. Murach, PhD, was a Glenn Foundation for Medical Research and AFAR Grant for Junior Faculty awardee. The Swedish Research Council for Sport Science #2020/3 and The Swedish Research Council #2022-01392 supported FvW. Thank you to our collaborators and friends at the University of Kentucky Center for Muscle Biology and beyond that contributed to the work summarized in this article. The figure was generated using BioRender.
Footnotes
Accepted for publication: December 14, 2023.
Editor: Kimberly Huey, Ph.D., FACSM
Contributor Information
Ronald G. Jones, III, Email: ronaldj@uark.edu.
Ferdinand von Walden, Email: ferdinand.von.walden@ki.se.
References
- 1.Coletta G, Phillips SM. An elusive consensus definition of sarcopenia impedes research and clinical treatment: a narrative review. Ageing Res. Rev. 2023; 86:101883. [DOI] [PubMed] [Google Scholar]
- 2.Smith JA, Murach KA, Dyar KA, Zierath JR. Exercise metabolism and adaptation in skeletal muscle. Nat. Rev. Mol. Cell Biol. 2023; 24:607–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jozsi AC Dupont-Versteegden EE Taylor-Jones JM, et al. Aged human muscle demonstrates an altered gene expression profile consistent with an impaired response to exercise. Mech. Ageing Dev. 2000; 120(1–3):45–56. [DOI] [PubMed] [Google Scholar]
- 4.Raue U, Slivka D, Minchev K, Trappe S. Improvements in whole muscle and myocellular function are limited with high-intensity resistance training in octogenarian women. J. Appl. Physiol. 2009; 106(5):1611–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Slivka D, Raue U, Hollon C, Minchev K, Trappe S. Single muscle fiber adaptations to resistance training in old (>80 yr) men: evidence for limited skeletal muscle plasticity. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2008; 295(1):R273–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kennedy BK Berger SL Brunet A, et al. Geroscience: linking aging to chronic disease. Cell. 2014; 159(4):709–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lebrasseur NK De Cabo R Fielding R, et al. Identifying biomarkers for biological age: geroscience and the ICFSR task force. J. Frailty Aging. 2021; 10:196–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.López-Otín C, Blasco MA, Partridge L, Serrano M, Kroemer G. Hallmarks of aging: an expanding universe. Cell. 2023; 186(2):243–78. [DOI] [PubMed] [Google Scholar]
- 9.Zhang B, Trapp A, Kerepesi C, Gladyshev VN. Emerging rejuvenation strategies—reducing the biological age. Aging Cell. 2022; 21(1):e13538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Horvath S. DNA methylation age of human tissues and cell types. Genome Biol. 2013; 14(10):R115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lu AT Fei Z Haghani A, et al. Universal DNA methylation age across mammalian tissues. Nature Aging. 2023; 3(9):1144–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Olova N, Simpson DJ, Marioni RE, Chandra T. Partial reprogramming induces a steady decline in epigenetic age before loss of somatic identity. Aging Cell. 2019; 18(1):e12877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006; 126(4):663–76. [DOI] [PubMed] [Google Scholar]
- 14.Ocampo A Reddy P Martinez-Redondo P, et al. In vivo amelioration of age-associated hallmarks by partial reprogramming. Cell. 2016; 167(7):1719–33. e12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Brett J Arjona M Ikeda M, et al. Exercise rejuvenates quiescent skeletal muscle stem cells in old mice through restoration of cyclin D1. Nat. Metab. 2020; 2(4):307–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Joanisse S, Nederveen JP, Baker JM, Snijders T, Iacono C, Parise G. Exercise conditioning in old mice improves skeletal muscle regeneration. FASEB J. 2016; 30(9):3256–68. [DOI] [PubMed] [Google Scholar]
- 17.Furrer R, Handschin C. Drugs, clocks and exercise in aging: hype and hope, facts and fiction. J. Physiol. 2022; 601(11):2057–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Joyner MJ, Barnes JN. I am 80 going on 18: exercise and the fountain of youth. J. Appl. Physiol. 2013; 114(1):1–2. [DOI] [PubMed] [Google Scholar]
- 19.Sharples AP, Seaborne RA. Exercise and DNA Methylation in Skeletal Muscle. Sports, Exercise, and Nutritional Genomics. Elsevier: Amsterdam, Netherlands; 2019. p. 211–29. [Google Scholar]
- 20.Wen Y, Dungan CM, Mobley CB, Valentino T, von Walden F, Murach KA. Nucleus type-specific DNA methylomics reveals epigenetic “memory” of prior adaptation in skeletal muscle. Function. 2021; 2:zqab038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jones RG III Dimet-Wiley A Haghani A, et al. A molecular signature defining exercise adaptation with ageing and in vivo partial reprogramming in skeletal muscle. J. Physiol. 2022; 601(4):763–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Voisin S Seale K Jacques M, et al. Exercise is associated with younger methylome and transcriptome profiles in human skeletal muscle. Aging Cell. 2023; 23:e13859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dungan CM Brightwell C Wen Y, et al. Muscle-specific cellular and molecular adaptations to late-life voluntary concurrent exercise. Function (Oxf). 2022; 3(4):zqac027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Murach KA Liu Z Jude B, et al. Multi-transcriptome analysis following an acute skeletal muscle growth stimulus yields tools for discerning global and MYC regulatory networks. J. Biol. Chem. 2022; 298:102515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Keckesova Z Donaher JL De Cock J, et al. LACTB is a tumour suppressor that modulates lipid metabolism and cell state. Nature. 2017; 543(7647):681–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Seely S. Possible reasons for the high resistance of muscle to cancer. Med. Hypotheses. 1980; 6(2):133–7. [DOI] [PubMed] [Google Scholar]
- 27.Goodman CA, Dietz JM, Jacobs BL, McNally RM, You J-S, Hornberger TA. Yes-associated protein is up-regulated by mechanical overload and is sufficient to induce skeletal muscle hypertrophy. FEBS Lett. 2015; 589(13):1491–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dang CV, O’Donnell KA, Zeller KI, Nguyen T, Osthus RC, Li F. The c-Myc target gene network. Semin. Cancer Biol. 2006; 16(4):253–64. [DOI] [PubMed] [Google Scholar]
- 29.Bui TV, Mendell JT. Myc: maestro of microRNAs. Genes Cancer. 2010; 1(6):568–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nie Z Guo C Das SK, et al. Dissecting transcriptional amplification by MYC. Elife. 2020; 9:e52483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Oksuz O Henninger JE Warneford-Thomson R, et al. Transcription factors interact with RNA to regulate genes. Molecular Cell. 2023; 83(14):2449–63. e13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Riu E Ferre T Hidalgo A, et al. Overexpression of c-myc in the liver prevents obesity and insulin resistance. FASEB J. 2003; 17(12):1715–7. [DOI] [PubMed] [Google Scholar]
- 33.Wang H Lu J Stevens T, et al. Premature aging and reduced cancer incidence associated with near-complete body-wide Myc inactivation. Cell Reports. 2023; 42(8):112830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Brenner C Deplus R Didelot C, et al. Myc represses transcription through recruitment of DNA methyltransferase corepressor. EMBO J. 2005; 24(2):336–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Poole CJ, Lodh A, Choi J-H, Van Riggelen J. MYC deregulates TET1 and TET2 expression to control global DNA (hydroxy) methylation and gene expression to maintain a neoplastic phenotype in T-ALL. Epigenetics Chromatin. 2019; 12(1):1–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chen L-L Lin H-P Zhou W-J, et al. SNIP1 recruits TET2 to regulate c-MYC target genes and cellular DNA damage response. Cell Reports. 2018; 25(6):1485–500. e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Veal E, Jackson M. C-myc is expressed in mouse skeletal muscle nuclei during post-natal maturation. Int. J. Biochem. Cell Biol. 1998; 30(7):811–21. [DOI] [PubMed] [Google Scholar]
- 38.Whitelaw PF, Hesketh JE. Expression of c-myc and c-fos in rat skeletal muscle. Evidence for increased levels of c-myc mRNA during hypertrophy. Biochem. J. 1992; 281(1):143–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Alway SE. Overload-induced C-Myc oncoprotein is reduced in aged skeletal muscle. J. Gerontol. A Biol. Sci. Med. Sci. 1997; 52(4):B203–11. [DOI] [PubMed] [Google Scholar]
- 40.Armstrong DD, Esser KA. Wnt/β-catenin signaling activates growth-control genes during overload-induced skeletal muscle hypertrophy. American Journal of Physiology-Cell Physiology. 2005; 289(4):C853–9. [DOI] [PubMed] [Google Scholar]
- 41.Figueiredo VC Wen Y Alkner B, et al. Genetic and epigenetic regulation of skeletal muscle ribosome biogenesis with exercise. J. Physiol. 2021; 599(13):3363–84. [DOI] [PubMed] [Google Scholar]
- 42.Broholm C Laye MJ Brandt C, et al. LIF is a contraction-induced myokine stimulating human myocyte proliferation. J. Appl. Physiol. 2011;111(1):251–9. [DOI] [PubMed] [Google Scholar]
- 43.Apró W, Wang L, Pontén M, Blomstrand E, Sahlin K. Resistance exercise induced mTORC1 signaling is not impaired by subsequent endurance exercise in human skeletal muscle. American Journal of Physiology-Endocrinology and Metabolism. 2013; 305(1):E22–32. [DOI] [PubMed] [Google Scholar]
- 44.Stec MJ, Kelly NA, Many GM, Windham ST, Tuggle SC, Bamman MM. Ribosome biogenesis may augment resistance training-induced myofiber hypertrophy and is required for myotube growth in vitro. American Journal of Physiology-Endocrinology and Metabolism. 2016; 310(8):E652–E661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.von Walden F, Casagrande V, Östlund Farrants A-K, Nader GA. Mechanical loading induces the expression of a pol I regulon at the onset of skeletal muscle hypertrophy. American Journal of Physiology-Cell Physiology. 2012; 302(10):C1523–30. [DOI] [PubMed] [Google Scholar]
- 46.Mori T Ato S Knudsen JR, et al. C-Myc overexpression increases ribosome biogenesis and protein synthesis independent of mTORC1 activation in mouse skeletal muscle. American Journal of Physiology-Endocrinology and Metabolism. 2020; 321(4):E551–9. [DOI] [PubMed] [Google Scholar]
- 47.Viggars MR, Sutherland H, Lanmüller H, Schmoll M, Bijak M, Jarvis JC. Adaptation of the transcriptional response to resistance exercise over 4 weeks of daily training. FASEB J. 2022; 37(1):e22686. [DOI] [PubMed] [Google Scholar]
- 48.von Walden F. Ribosome biogenesis in skeletal muscle: coordination of transcription and translation. J. Appl. Physiol. 2019; 127(2):591–8. [DOI] [PubMed] [Google Scholar]
- 49.Masri S, Sassone-Corsi P. The emerging link between cancer, metabolism, and circadian rhythms. Nat. Med. 2018; 24(12):1795–803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wolff G, Esser KA. Scheduled exercise phase shifts the circadian clock in skeletal muscle. Med. Sci. Sports Exerc. 2012; 44(9):1663–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Demontis F, Piccirillo R, Goldberg AL, Perrimon N. The influence of skeletal muscle on systemic aging and lifespan. Aging Cell. 2013; 12(6):943–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Dungan CM Murach KA Frick KK, et al. Elevated myonuclear density during skeletal muscle hypertrophy in response to training is reversed during detraining. American Journal of Physiology-Cell Physiology. 2019; 316(5):C649–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Murach KA Mobley CB Zdunek CJ, et al. Muscle memory: myonuclear accretion, maintenance, morphology, and miRNA levels with training and detraining in adult mice. Journal of Cachexia, Sarcopenia and Muscle. 2020; 11(6):1705–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wang C Ros RR Martinez-Redondo P, et al. In vivo partial reprogramming of myofibers promotes muscle regeneration by remodeling the stem cell niche. Nat. Commun. 2021; 12(1):1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.FC Lopes A. Mitochondrial metabolism and DNA methylation: a review of the interaction between two genomes. Clin. Epigenetics. 2020; 12(1):182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wu J, Ocampo A, Belmonte JCI. Cellular metabolism and induced pluripotency. Cell. 2016; 166(6):1371–85. [DOI] [PubMed] [Google Scholar]
- 57.Drummond MJ McCarthy JJ Sinha M, et al. Aging and microRNA expression in human skeletal muscle: a microarray and bioinformatics analysis. Physiol. Genomics. 2011; 43(10):595–603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Stec MJ, Mayhew DL, Bamman MM. The effects of age and resistance loading on skeletal muscle ribosome biogenesis. J. Appl. Physiol. 2015; 119(8):851–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Brook MS Wilkinson DJ Mitchell WK, et al. Synchronous deficits in cumulative muscle protein synthesis and ribosomal biogenesis underlie age-related anabolic resistance to exercise in humans. J. Physiol. 2016; 594(24):7399–417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Rivas DA Lessard SJ Rice NP, et al. Diminished skeletal muscle microRNA expression with aging is associated with attenuated muscle plasticity and inhibition of IGF-1 signaling. FASEB J. 2014; 28(9):4133–47. [DOI] [PMC free article] [PubMed] [Google Scholar]


