Abstract
One group of sequence variants of Epstein-Barr virus is characterized by a 10-amino-acid deletion within the CTAR-2 functional domain of the latent membrane protein, LMP1. A role for this deletion in enhancing the tumorigenicity of the viral oncogene in rodent fibroblasts was recently demonstrated. We examined the effect of this deletion upon LMP1 function in four human lymphoid cell lines by using three natural variants of LMP1: the prototype B95.8 gene and the CAO and AG876 genes, both of which have codons 343 to 352 of the B95.8-LMP1 deleted. These experiments revealed that LMP1-mediated upregulation of CD40 and CD54 was markedly impaired (by 60 to 90%) with CAO-LMP1 compared with B95.8-LMP1. In contrast, the function of AG876-LMP1 was indistinguishable from that of B95.8-LMP1 in two lines and was only slightly impaired in the other two lines. Activation of NF-κB by CAO-LMP1 was not impaired in any of the lines; rather, activation of an NF-κB reporter by CAO-LMP1 was consistently about twofold greater than the activation with B95.8- or AG876-LMP1. Therefore, while the CAO-LMP1 is functionally distinct from the prototype B95.8-LMP1 in human lymphocytes, the 10-amino-acid deletion appears not to be directly responsible. This conclusion was confirmed by using a B95.8-LMP1 mutant with codons 343 to 352 deleted and chimerae of CAO- and B95.8-LMP1 in which the CTAR-2 domains of these genes were exchanged. Sequences outside the CTAR-2 domain were implicated in the distinct functional characteristics of CAO-LMP1 in human lymphoid cells.
Epstein-Barr virus (EBV) is a human gammaherpesvirus that contributes to the development of several malignant diseases of both the lymphoid and the epithelial cell compartments (53). The virus is a potent transforming agent for human B lymphocytes, a property that is readily demonstrated in vitro by experimental infection of resting B cells, which results in the outgrowth of permanent lymphoblastoid cell lines (LCLs). In these LCLs, the virus is maintained as a predominantly nonproductive infection by the expression of a number of latent infection proteins (35). Of the four or more viral gene products known to be essential for B-cell transformation (35), latent membrane protein 1 (LMP1) is of particular interest because it is a classical oncogene by virtue of its ability to transform rodent fibroblasts (5, 33, 63). Furthermore, gene transfection experiments suggest that LMP1 is a major effector of EBV-induced human B-cell transformation since its expression in B-cell lines leads to the induction of many of the cell phenotypic changes observed following infection with EBV. These changes include the upregulation of a number of lymphocyte activation markers and adhesion molecules (52, 64, 65), as well as enhancement of the survival capacity of the B cells by the induction of anti-apoptotic genes such as Bcl-2, A20, and Mcl-1 (24, 41, 55, 66). Expression of LMP1 in epithelial cells has also profound effects, including upregulation of CD40 and epidermal growth factor receptor, secretion of interleukin-6, and inhibition of cellular differentiation (12, 15, 17, 44). In some circumstances, LMP1 can also be oncogenic in human epithelial cells (27, 51).
LMP1 activates NF-κB (22, 41), and many of the LMP1-induced genes appear to be regulated by this transcription factor. Members of the NF-κB family, which include p50 (NFKB1), p52 (NFKB2), p65 (RelA), c-Rel, and RelB, can form both homodimers and heterodimers (3). An active NF-κB moiety generally comprises NFKB/Rel heterodimers, the prototype NF-κB being p50/p65. The heterodimers are normally held in the cytoplasm as inactive complexes with an inhibitory component, IκB. Activation appears to involve phosphorylation and subsequent release and degradation of the IκB component, thus permitting translocation of the active heterodimers to the nucleus, where they bind to specific DNA sequences in the promoters of the genes they activate. There are a number of different types of IκB, and recent evidence suggests that LMP1 activates NF-κB through phosphorylation of IκBα (25).
The LMP1 gene from the prototype B95.8 virus is a 386-amino-acid (aa) protein comprising a short N-terminal cytoplasmic domain, six putative membrane-spanning helices, and a large cytoplasmic C-terminal domain of approximately 200 aa (18, 43). Two regions within the C-terminal domain have previously been identified to be vital in the function of the molecule (29, 47). Designated CTAR-1 (C-terminal activating region 1) and CTAR-2, these domains comprise residues 194 to 232 immediately proximal to the membrane and the last 55 aa (332 to 386) respectively. Both CTAR-1 and CTAR-2 are able to activate NF-κB independently in certain cell backgrounds, but the major NF-κB-activating region of LMP1 appears to be within CTAR-2 (8, 29, 47). CTAR-1 can bind four members of a family of tumor necrosis factor (TNF) receptor-associated factors (TRAF1, TRAF2, TRAF3, and TRAF5) that are involved in NF-κB activation pathways in other members of the TNF receptor family (8, 13, 32, 50, 58). While there is no evidence for direct association of any TRAF species with CTAR-2, direct binding of the TNF receptor-associated death domain protein (TRADD) to CTAR-2 was recently reported (30). Studies with a dominant negative mutant of TRAF2 (15, 30, 32) have implicated TRAF2 as a common mediator of NF-κB activation from the CTAR-1 and CTAR-2 domains, although the mechanism presumably differs for each domain since TRAF2 can bind directly only to CTAR-1. Therefore, the precise nature of TRAF involvement in LMP1-mediated signaling remains unclear (8, 46, 58). Analysis of laboratory mutants of LMP1 reveals an incomplete correlation between LMP1-mediated activation of NF-κB and changes in cell phenotype (29, 46, 47), and these data point to a separate signalling pathway(s) being activated by LMP1 along with NF-κB to effect full downstream functions. One such pathway was recently identified as the SEK/JNK kinase cascade that leads to activation of the AP-1 transcription factor (36).
Although the LMP1 gene is relatively well conserved in its protein-coding region, typically showing greater than 95% amino acid sequence identity among different EBV isolates (45, 57), there has been considerable interest in the possibility that natural sequence variation affects the function of the LMP1 and influences the development of EBV-associated disease. In particular, attention has focused on a deletion variant (del-LMP1) that was originally identified in tumors of Chinese patients with undifferentiated nasopharyngeal carcinoma (NPC) and that is characterized by a 30-bp deletion corresponding to codons 343 to 352 of the B95.8-LMP1, together with other hot spots of point mutations (9, 28, 38, 60). The geographic distribution of this variant means that the majority of Chinese NPC patients are infected with EBV carrying the del-LMP1 gene. LMP1 is expressed in about 65% of undifferentiated NPCs (16, 67) and appears to influence the growth and clinical course of the cancer (26). In addition, the two prototype del-LMP1 variants, CAO and 1510, are widely accepted as being more oncogenic than the B95.8 gene in rodent fibroblasts and a human epithelial cell line (9, 27, 68), although these results have recently been questioned by one group (51).
The del-LMP1 gene has also been identified in a number of EBV-associated diseases other than NPC, including Hodgkin’s disease and lymphomas, and here also it has been suggested that the LMP1 sequence variations may be contributing to the malignant process (37, 39, 40). In this study, we investigated whether the del-LMP1 genes have different functional properties in human lymphoid cells and whether any differences were due to the 10-aa deletion itself or to other sequence differences in LMP1. Our structure-function analysis was performed on a panel of four human lymphoid lines, and activation of NF-κB and induction of cell surface markers were used as functional readouts.
MATERIALS AND METHODS
Cell lines.
DG75 is an EBV-negative Burkitt’s lymphoma (BL) cell line derived from a patient with a sporadic case of BL (6). Eli-BL is an EBV-positive BL cell line retaining a latency I pattern of gene expression in which EBNA1 is the only nuclear antigen expressed and the LMP1 gene is repressed (56). Daudi is an EBV-positive BL tumor line which carries an EBV genome with a deletion that removes the EBNA2 gene, which in turn prevents the EBNA2-regulated expression of the intact LMP1 gene (11, 31). Jurkat is an EBV-negative T-cell line from an acute lymphocytic leukemia patient (7). All these lines were maintained as suspension cultures in growth medium, consisting of bicarbonate-buffered RPMI 1640 supplemented with 10% fetal bovine serum and antibiotics (200 U of penicillin per ml and 200 mg of streptomycin per ml) at 37°C in a humidified atmosphere containing 5% CO2.
DNA expression vectors.
All LMP1-expressing constructs were based on the pSG5 vector in which the inserted genes are placed downstream of a β-globin intron and a simian virus 40 promoter (20). The pSG5-LMP1 plasmid containing a cDNA of B95.8 has been described previously (29). The pSG5.CAO-LMP1 plasmid was prepared by excising the LMP1 gene from J124-Cao5 (28) as a BamHI fragment and ligating it into the BamHI site of pSG5. The pSG5.AG876-LMP1 plasmid was generated by inserting an AG876 cDNA into the BamHI site of pSG5; mRNA of the AG876 cell line was used to produce cDNA from which the LMP1 gene was amplified as an EcoRI fragment by using the following pair of primers: 5′-ATA GAA TTC CTG AGG ATG GAA CAC-3′ and 3′-TGG TCG GTC GCT GAC TTA AGA TA-5′.
The chimeric genes B95.8 × CAO and CAO × B95.8 were created from pSG5-LMP1 and pSG5.CAO-LMP1 by swapping the BstEII-SmaI fragments containing the CTAR-2 domain from codon 333 (B95.8 sequence) to the C terminus. Plasmid pSG5-LMP1Δ(343–352) was generated by using PCR mutagenesis to introduce three point mutations into the chimera B95.8 × CAO to revert the three amino acid changes found in the CAO sequence to the B95.8 sequence. The primers used to introduce these mutations were 5′-ATA GGT GAC CAG GGC CCG CCT TTT GA-3′ (R334→Q, S338→L) and 3′-ATA AAC CGG AAC CAG AAG AAC CCA A-5′ (T356→S). Underlined bases indicate the introduction of point mutations. The primers were used to amplify a BsaWI-BstEII fragment of B95.8 × CAO, generating an 85-bp product that was then inserted in B95.8 × CAO. Plasmid pSG5-LMP1.Δ(343–352) was identified by the loss of the three restriction sites NciI, TaqI, and RsaI, and its identity was confirmed by sequencing. Plasmid pSG5-LMP1.Δ(352–360) was generated by use of the transformer site-directed mutagenesis kit (Clontech) with the selection primer 5′-CAT TGG GAA AAC GCT CTT CGG to destroy a unique Asp 700 site within the ampicillin resistance gene and the primer 5′-AGT CAT GAT TCC GGC ACG CTG CTT TTG GGT to introduce the deletion into pSG5.LMP1. The deletion mutant was screened for by loss of a BglI restriction site, and its identity was confirmed by sequencing.
Plasmid RSV-IκB, which encodes IκBα driven from a Rous sarcoma virus long terminal repeat (LTR) promoter (14), was a kind gift from Colin Duckett (Howard Hughes Medical Institute, University of Michigan Medical Center).
DNA reporter plasmids.
Plasmid pSG5-rCD2, containing a nonfunctional truncated rat CD2 gene under the control of a constitutively active simian virus 40 promoter, was constructed as described elsewhere (19) and was used in phenotype assays as a marker of transfected cells. The luciferase reporter constructs 3Enh.κB-ConALuc, human immunodeficiency virus (HIV) LTR, and HIV LTRΔκB were kindly supplied by F. Arenzana-Seisdedos (Institut Pasteur, Paris, France). The 3Enh.κB-ConALuc reporter contains three tandem repeats of the NF-κB-binding sites from the Igκ promoter located upstream of a minimal conalbumin promoter controlling a luciferase gene (1). The HIV LTR luciferase reporter plasmid contains HIV LTR sequences controlling a luciferase gene, and HIV LTRΔκB is the HIV LTR with the κB motifs deleted (2). The cytomegalovirus CMV-βGal construct, in which a bacterial β-galactosidase gene is constitutively expressed from the human CMV immediate-early promoter, was obtained from D. McCance (University of Rochester).
Transfection of lymphoid cell lines.
For electroporation of suspension cell cultures, 107 cells in 500 μl of HEPES-buffered RPMI 1640 containing 10% fetal calf serum and DNA were placed in electroporation cuvettes with 0.4-mm spacing between electrodes. For cells intended for luciferase reporter assays, 3 μg of NF-κB reporter plasmid and 2 μg of CMV-βGal plasmid were added to the cell suspension, together with 4 μg of pSG5 vector or LMP1 construct plasmid DNA. For cells intended for fluorescence-activated cell sorter analysis, 3 μg of pSG5-rCD2 plasmid was added to the cell suspension, together with 4 μg of pSG5 vector or LMP1 construct plasmid DNA. The cells were then pulsed with a Bio-Rad Gene Pulser at 960 μF and either 270 V (for DG75 and Eli-BL cells) or 280 V (for Daudi and Jurkat cells) and were subsequently transferred to 8 ml of RPMI 1640 growth medium in 3.5-cm-diameter culture dishes.
Detection of LMP1 expression by Western blotting.
Electroporated cells were harvested 40 h posttransfection, washed in phosphate-buffered saline, solubilized in gel sample buffer, and separated by Laemmli discontinuous sodium dodecyl sulfate-polyacrylamide gel electrophoresis before being electroblotted onto nitrocellulose filters essentially as described previously (55). Nitrocellulose transfers were blocked with 5% (wt/vol) dried skimmed milk in Tris-buffered saline (pH 8.0) (TBS) for 1 h before being probed overnight at 4°C with CS.1–4, a pool of murine monoclonal antibodies reactive with epitopes within the C-terminal cytosolic domain of LMP1 (54), at 1 μg/ml in TBS-milk. The blots were then incubated for 1 h with rabbit anti-mouse immunoglobulin G (Dako Z0259) diluted 1:2,000 in TBS plus 0.1% Tween 20, and specifically bound antibody was detected by incubation for 2 h with rabbit anti-mouse-conjugated alkaline phosphatase (Sigma; A-3812) diluted 1:10,000 in TBS-Tween 20. Antibody-protein complexes were detected with a 5-bromo-4-chloro-3-indolylphosphate–nitroblue tetrazolium (BCIP-NBT) chromogenic substrate kit (Bio-Rad; 170-6432).
Flow cytometry assay for surface phenotype induction.
Induction of CD40 and CD54 expression by LMP1 was analyzed by flow cytometry as described previously (55). Briefly, cells were harvested 48 h posttransfection, and the live cells were isolated by isopycnic centrifugation over Ficoll-Hypaque. The cells were dually stained with biotinylated OX34 anti-rat CD2 followed by phycoerythrin (PE)-streptavidin and fluorescein isothiocyanate (FITC)-conjugated antibodies to CD40 or CD54. Fluorescent staining of cell surface markers was analyzed by flow cytometry with a Becton-Dickinson FACScalibur; the transfected-cell population identified by PE staining was gated and analyzed for green fluorescence. Induction of CD40 or CD54 was measured by determining a gate for positive FITC staining on cells transfected with SG5 vector plus the rCD2 reporter and determining the percentage of positive cells induced in cells transfected with the experimental LMP1 vectors plus the rCD2 reporter. All the results were expressed relative to B95.8 LMP1 (100%) after subtraction of the percentage of FITC-stained cells in the SG5 vector control transfection.
Assay for NF-κB activity.
NF-κB activity was determined by quantitating the luciferase expressed from the cotransfected NF-κB reporter plasmid at 24 h posttransfection as described previously (29). Briefly, the cells were washed in phosphate-buffered saline and then lysed at approximately 100 μl/106 cells in reporter lysis buffer (Promega) and clarified by centrifugation. A 50-μl aliquot of lysate was analyzed in a Berthold LB 9501 luminometer following injection of 100 μl of assay reagent buffer (20 mM glycylglycine, 5 mM MgCl2, 40 μM EDTA, 3 mM dithiothreitol, 27 μM coenzyme A (lithium salt), 0.5 mM luciferin) and integration of light release for 10 s. The luciferase activity in each sample was normalized for variations in transfection efficiency by measuring the β-galactosidase enzyme expressed from the cotransfected CMV-βGal plasmid, as described previously (29). Briefly, endogenous β-galactosidase activity was inactivated by heating the lysate to 50°C for 1 h, then 20 μl of the lysate was mixed with 200 μl of 35 mM AMPGD chemiluminescent substrate (Tropix) dissolved in 100 mM sodium phosphate (pH 8.0)–1 mM magnesium chloride and allowed to react for 30 min at room temperature. Light release was integrated in a Berthold LB9501 luminometer for precisely 5 s after injection of 300 μl of a 10% solution of Emerald luminescence amplifier (Tropix) in 0.1 M NaOH.
RESULTS
LMP1 variants.
A sequence comparison of the three natural LMP1 gene variants used in this study is shown in Fig. 1A. It should be noted that in contrast the original published sequence (28) of the Chinese NPC-derived gene, CAO-LMP1, our sequence data demonstrated that the N-terminal sequence of CAO-LMP1 shows fewer differences from the B95.8 prototype sequence; specifically, we did not find changes in codons 7, 11, 12, or 13. The reason for this discrepancy is unclear, but the differences have no bearing upon our functional analysis of the C-terminal region. Otherwise, we confirmed that CAO-LMP1 contains a 10-aa deletion corresponding to aa 343 to 352 of B95.8-LMP1, as well as an increased number of repeats and several point mutations compared to B95.8, as shown in Fig. 1A. Many of the point mutations lie within the transmembrane loops (aa 19 to 182). However, one point mutation (G212→S) is found within the CTAR-1 region of the molecule (aa 194 to 232) in close proximity to the PxQxT motif (aa 204 to 208) reported to bind members of the TRAF family. Other point mutations in the C-terminal cytoplasmic domain include three in the CTAR-2 region (aa 332-386 in B95.8-LMP1), within which also lies the 10-aa deletion (aa 343 to 352 in B95.8-LMP1). AG876 is a prototype type 2 EBV, and its LMP1 gene displays a higher degree of sequence identity to B95.8 LMP1 (57). It does, however, contain a number of point mutations, and it has the 30-bp deletion detected in the NPC isolate. Many of the point mutations are conservative and are found in regions outside those thought to be of functional significance (Fig. 1A). The CTAR-2 region of AG876 is identical to that of CAO.
FIG. 1.
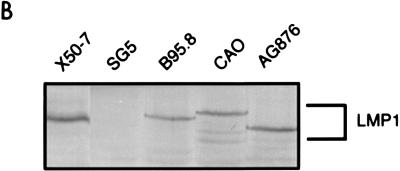
(A) Amino acid sequence variation between the natural variant LMP1 genes derived from B95.8, CAO, and AG876. Dots represent areas of amino acid identity, and dashes represent deleted amino acids. Shaded areas depict the previously defined CTAR-1 (amino acids 194 to 232 of B95.8-LMP1) and CTAR-2 (amino acids 332 to 386). The boxed area within CTAR-1 at aa 204 to 208 denotes the PxQxT TRAF binding motif, and the boxed area within CTAR-2 at aa 343 to 352 (B95.8) indicates the 10-aa deletion in AG876-LMP1 and CAO-LMP1. Note that the N-terminal sequence of CAO-LMP1 shown here differs from the previously published sequence (see the text). (B) Immunoblot showing the expression of the LMP1 variants from pSG5 vectors in transiently transfected Jurkat cells. The EBV-transformed B-cell line X50/7 is a positive control for LMP1, and SG5-transfected Jurkat is a negative control. Approximately 1.5 × 105 cells were separated on a 7.5% polyacrylamide gel and transferred to nitrocellulose. LMP1 expression was detected by probing blots with the CS1–4 anti-LMP MAbs and using an alkaline phosphatase-based chromogenic protocol.
The three natural variant LMP1 genes were placed into the pSG5 vector, and expression from each plasmid was verified by transfection into the Jurkat cell line and assaying by Western blotting at 40 h posttransfection. Blots were probed with a cocktail of four anti-LMP1 antibodies (CS1–4) recognizing different epitopes on the C-terminal cytoplasmic domain. As illustrated in Fig. 1B, expression of LMP1 polypeptides was detected for all the variants at similar intensity, suggesting similar expression levels. Also, as expected, CAO-LMP1 had a higher apparent molecular weight and AG876-LMP1 had a lower apparent molecular weight than did B95.8-LMP1.
Function of LMP1 variants in lymphoid cells. (i) Induction of CD54 and CD40.
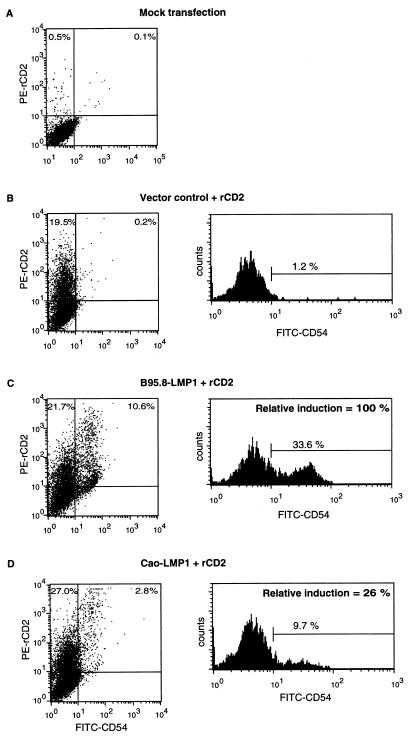
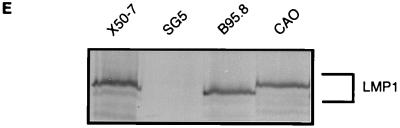
Cell surface phenotype changes induced by the LMP1 variants were measured by a flow cytometry assay in which the LMP1 expression vector was electroporated into the target cell together with a marker plasmid encoding an inactive derivative of the rat CD2 protein that acts as cell surface marker of transfected cells. Since LMP1 and rCD2 are expressed coincidentally in the transfected subpopulation, alterations of the cell surface phenotype in the LMP1-expressing population can be detected and quantified by the application of two-color flow cytometry, e.g., gating for cells stained for rCD2 with PE-tagged antibodies, and analyzing the expression of CD40 or CD54 stained with FITC-conjugated monoclonal antibodies. Figure 2 illustrates the results obtained in a representative experiment with the Eli-BL B-cell line 48 h after transfection with different combinations of plasmid DNA. Mock-transfected cells (SG5 vector, but no rCD2 or LMP1 plasmid DNA) were stained with fluorescent-antibody reagents to establish the background staining levels (Fig. 2A), and cells transfected with SG5 vector plus the rCD2 plasmid DNA were similarly stained so that the transfected subpopulation of cells (PE-rCD2 positive) in the upper left and upper right quadrants of the two-colour plot (Fig. 2B, left) could be gated and analyzed separately for CD54 expression detected with FITC-conjugated CD54 antibodies (Fig. 2B, right histogram). This CD54 expression histogram of the transfected-cell subpopulation was used as a reference for analysis of parallel transfections with LMP1 genes to quantify the percentage of LMP1-transfected cells that showed increased expression of CD54 (Fig. 2C, B95.8-LMP1; Fig. 2D, CAO-LMP1). It is clear from this experiment that the CAO-LMP1 gene was markedly less efficient at inducing CD54 expression than was the B95.8 gene. Western blot analysis (Fig. 2E) revealed that expression levels of B95.8-LMP1 and CAO-LMP1 in the transfectants were similar and that the levels of expression at the single-cell level were comparable to those observed in the reference normal EBV-transformed LCLs, allowing for the transfection efficiency of about 30% (Fig. 2C and D).
FIG. 2.
Flow cytometry assay for the induction of cell surface CD54 expression 40 h after transfection of Eli-BL cells with B95.8-LMP1 or CAO-LMP1 genes. (A to D) Two-color flow cytometry analysis of four transfections. The cells were stained for rCD2 with PE-conjugated antibody and for CD54 with FITC-conjugated antibody; the left-hand panels show the two-color plots of the total viable population, while the right-hand panels show histograms of CD54 staining of the subpopulation of cells gated for PE-rCD2 positivity. The four transfections were as follows: mock-transfected cells with no rCD2 or LMP1 DNA (A), control transfected cells with pSG5 vector plus rCD2 DNA (B), B95.8-LMP1 plus rCD2 DNA (C), and CAO-LMP1 plus rCD2 DNA (D). An arbitrary gate was set on the control transfection CD54 histogram (B) so that 1.2% of the transfected cells were classified as positive for CD54. The same gate was applied to the CD54 histograms of the B95.8-LMP1 (C) and CAO-LMP1 (D) transfections, which gave 33.6 and 9.7% CD54-positive cells, respectively. (E) Western blot illustrating the expression of LMP1 in the transfected Eli-BL cultures. The blot was probed with CS.1–4 anti-LMP1 MAbs, and the samples (left to right) were equivalent amounts of whole-cell protein from the reference X50-7 EBV-transformed LCLs, the vector control-transfected Eli-BL cells from panel B, the B95.8-LMP1-transfected Eli-BL cells from panel C, and the CAO-LMP1-transfected cells from panel D.
To facilitate a comparison of results from different experiments and with different cell lines where the transfection efficiency may range from 5 to 45%, the phenotype assay results were calculated as the induction relative to the vector control transfection (designated 0%) and the B95.8-LMP1 reference transfection (designated 100%). By this calculation, the example shown in Fig. 2 produced an induction of CD54 expression by CAO-LMP1 that was 26% of that detected with B95.8-LMP1. The reproducibility of this result in replicate experiments is illustrated by the standard deviations shown in Fig. 3A.
FIG. 3.
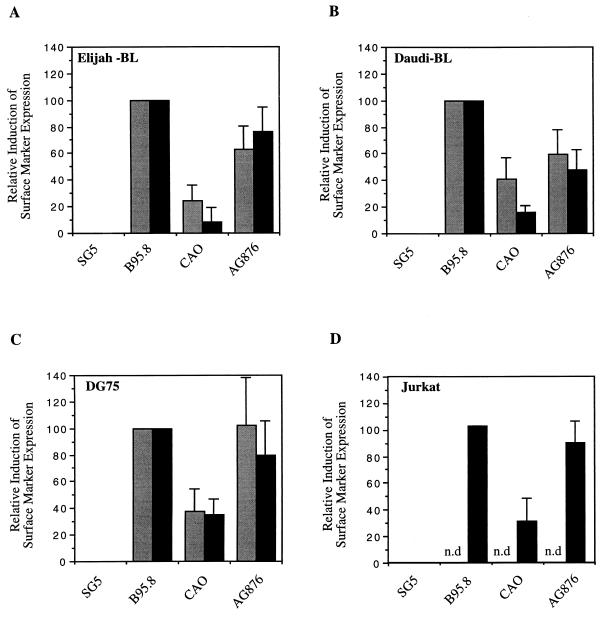
Effects of the LMP1 variants on the induction of cell surface markers CD40 (░⃞) and CD54 (▪) on the B-cell lines Eli-BL (A), Daudi (B), and DG75 (C) and the T-cell line Jurkat (D). Phenotypic changes in the transfected subpopulation of cells were measured by two-color flow cytometry, as in Fig. 2. The measured induction of surface markers was expressed relative to the degree of induction achieved with the B95.8 LMP1 gene (100%) and the pSG5 vector control (0%). The data for each cell line are the mean and standard deviation of at least three separate experiments. Induction of CD40 was not determined (n.d.) for the Jurkat cell line, since it is documented that CD40 cannot be upregulated in this cell line.
This phenotype assay was then applied to four different lymphoid cell lines of human origin, and the relative induction of CD54 and CD40 by the three natural LMP1 variants (B95.8-LMP1, CAO-LMP1, and AG876-LMP1) was determined. The target lines for transfection included three B-lymphoid lines: Eli-BL, an EBV-positive BL line in the latency I state where EBNA1 is the only viral protein expressed; Daudi, another EBV-positive BL line in which EBNA2 is deleted and which therefore does not express the endogenous EBNA2-regulated LMP1 gene; and DG75, an EBV-negative BL line. The EBV-negative T-lymphoid line, Jurkat, was also examined, since it is apparent that a number of T-cell lymphomas are associated with EBV.
Figure 3 shows the results obtained by expression of the LMP1 variants in these four lymphoid cell lines. Several points can be made from these data. First, for each of the B-cell lines, induction of the cell surface markers CD40 and CD54 gave broadly similar results (Fig. 3A through C); whereas CD40 could not be induced by LMP1 in the Jurkat T-cell line, apparently because of an inherent inability of this line to express CD40, the induction of CD54 showed a similar pattern of results to those obtained with the B-cell lines (Fig. 3D). Second, in all the lines, CAO-LMP1 consistently showed a marked impairment of CD54 and CD40 induction relative to that shown by B95.8-LMP1. Third, the results obtained with AG876-LMP1 more closely mirrored the results obtained with B95.8-LMP1. In DG75 and Jurkat cells, the effects of AG876-LMP1 were indistinguishable from those of B95.8 LMP1 (Fig. 3C and D). In the Daudi and Eli-BL lines, AG876-LMP1 was clearly less efficient than B95.8-LMP1 at inducing the cell surface phenotype but was consistently better than CAO-LMP1 (Fig. 3A and B). Since the sequence of the CTAR-2 domains of AG876-LMP1 and CAO-LMP1 are identical (Fig. 1), these results suggested that the more substantial phenotypic differences observed between CAO-LMP1 and B95.8-LMP1 may be due largely to sequences outside the CTAR-2 domain. However, the modest impairment of AG876-LMP1 function in the Daudi and Eli-BL lines does not allow us at this stage to rule out the possibility that the 10-aa deletion has some effect upon the induction of surface phenotype.
(ii) Activation of NF-κB.
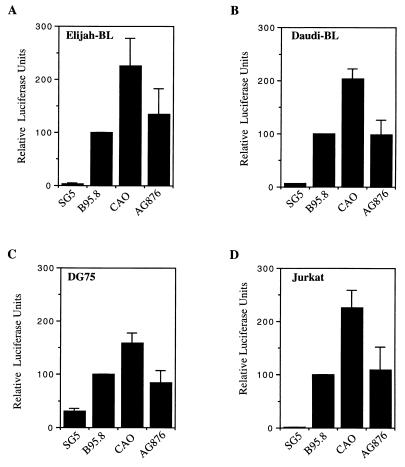
In parallel with the phenotype assays, we investigated the upregulation of NF-κB by the LMP1 variants by using a reporter plasmid (3Enh.κB-ConALuc) which contains a triplet of NF-κB binding motifs regulating a luciferase gene. The constructs expressing the LMP1 variants were cotransfected with the 3Enh.κB-ConALuc construct into the target cells, and cell lysates were assayed for luciferase enzyme activity at 24 h posttransfection. Variations in transfection efficiency were corrected for by measuring the levels of β-galactosidase constitutively expressed from a cotransfected CMV early promoter-driven expression plasmid; these β-galactosidase data were used to normalize the NF-κB results. The results obtained in this transient-transfection assay with the four lymphoid lines are shown in Fig. 4. Unexpectedly, we consistently observed the CAO-LMP1-induced NF-κB activation to be about twofold greater than that of B95.8-LMP1 in all four lines. Furthermore, AG876-LMP1 activated NF-κB to levels similar to those achieved by activation by B95.8. While the increased NF-κB-activating function of CAO-LMP1 contrasted with the decreased CD54/40-inducing function, the results obtained with AG876-LMP1 again implicated sequences outside the CTAR-2 domain as being responsible for the distinct functional characteristics of CAO-LMP1.
FIG. 4.
Effects of the LMP1 variants on the activation of NF-κB on the cell lines Eli-BL (A), Daudi (B), DG75 (C), and Jurkat (D), as determined by quantitation of the luciferase produced from a cotransfected reporter plasmid, 3Enh.κB-ConALuc, regulated by NF-κB. The data were normalized for transfection efficiency by measuring the β-galactosidase enzyme produced from a cotransfected constitutively active reporter plasmid. The data were then expressed relative to the activity obtained with the B95.8 LMP1 gene (100%) without subtracting the basal activity in control pSG5-transfected cells. The data for each cell line are the mean and standard deviation of at least four separate experiments.
Analysis of NF-κB activation by CAO-LMP1.
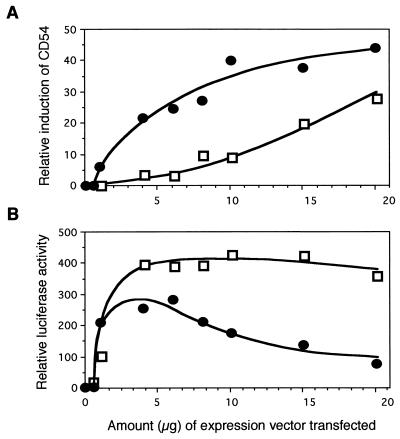
It could be argued that the increased NF-κB activation by CAO-LMP1 is relatively small and might be due to small differences in levels of LMP1 expression that are not reliably quantified by Western blotting because of the possibility that the monoclonal antibodies bind with different affinities to the two forms of LMP1. This was rebutted by the titer determination experiment (Fig. 5), in which different amounts of B95.8-LMP1 and CAO-LMP1 vector DNA (0.5 to 20 μg) were transfected into Jurkat cells and the effects upon CD54 induction and NF-κB activation were measured as before. The induction of CD54 by different amounts of the two LMP1 genes, as shown in Fig. 5A, revealed two distinct dose-response curves that cannot be due simply to different amounts of functionally identical proteins being produced from the two vectors. Likewise, the activation of NF-κB by different amounts of the two LMP1 genes also produced two distinct dose-response curves (Fig. 5B). In fact, by increasing the amount of B95.8-LMP1 beyond 5 μg, the activation of NF-κB gradually decreased, whereas the activation of NF-κB by CAO-LMP1 remained constantly high over the range from 4 to 20 μg DNA. Indeed, these data suggest that CAO-LMP1 may activate NF-κB up to fivefold more than B95.8-LMP1 does.
FIG. 5.
Determination of the effects of B95.8-LMP1 and CAO-LMP1 upon induction of CD54 (A) and activation of NF-κB (B) by measurement of titers. Jurkat cells were transfected with different amounts of either B95.8-LMP1 plasmid DNA (•) or CAO-LMP1 DNA (□) over a range of 0.5 to 20 μg of DNA per transfection. Induction of CD54 and activation of NF-κB were assayed as in the experiments in Fig. 3 and 4.
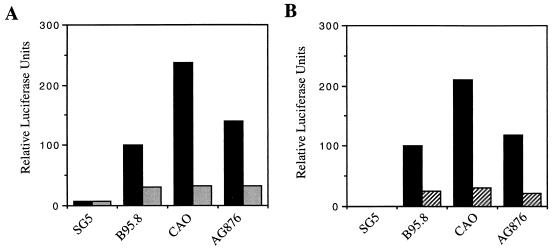
One important point about the CAO-LMP1 function is that by one readout (CD54 induction) it is impaired relative to B95.8-LMP1 while by another readout (activation of NF-κB) it is enhanced relative to B95.8-LMP1. The lack of correlation between NF-κB activation and surface phenotype induction prompted us to reconfirm the specificity of the NF-κB reporter assay. We had previously shown that the ConALuc parental reporter plasmid, containing the minimal conalbumin promoter but lacking the three κB enhancer elements, does not produce luciferase in response to LMP1 expression (data not shown). To establish the involvement of NF-κB in the induction of the reporter, we cotransfected Jurkat cells with the 3Enh.κB-ConALuc reporter and the pSG5-LMP1 plasmids together with a plasmid encoding the physiological inhibitor, IκBα. As shown in Fig. 6A, expression of IκBα substantially reduced the reporter activity induced by all three LMP1 genes to the same low basal level. Further confirmation of the NF-κB results was obtained with a second luciferase reporter regulated by the natural HIV LTR enhancer sequences; it is well documented that LMP1 upregulates the HIV LTR by induction of NF-κB (22). Figure 6B shows that this reporter produced similar results to those obtained with the 3Enh.κB-ConALuc reporter. Furthermore, CAO-LMP1 did not activate an HIV LTR reporter in which the NF-κB binding sites were deleted (Fig. 6B).
FIG. 6.
(A) Effects of IκBα upon the NF-κB activity induced by the LMP1 variants in Jurkat cells transfected as in Fig. 3, but with (▪) or without (░⃞) the expression vector for IκBα. The data were normalized for transfection efficiency and expressed relative to the activity obtained with the B95.8-LMP1 gene without the inhibitor (100%). Data shown are from one representative experiment. (B) Regulation of the HIV LTR by the LMP1 variants in Jurkat cells. Effects of the variants were determined by cotransfection of a luciferase reporter plasmid regulated by the HIV LTR and quantitation of the luciferase produced (▪). To show the specificity of the induction, a derivative of the HIV LTR reporter plasmid mutated at the NF-κB sites at the HIV LTR was also used (▨). Data were normalized for transfection efficiency and expressed relative to the activity obtained with the B95.8 LMP1 on the wild-type HIV LTR. The data shown are from one representative experiment.
Sequences responsible for the CAO-LMP1 functional differences.
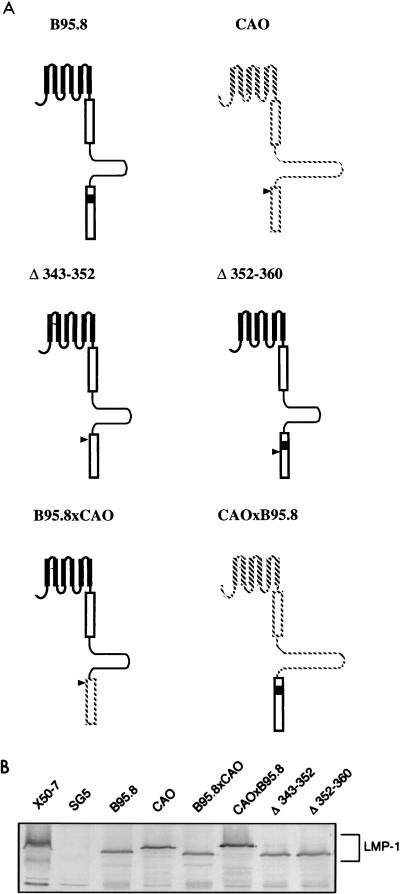
While not conclusive, the results in Fig. 3 and 4 suggest that the 10-aa deletion itself has little effect upon LMP1 function in human lymphocytes and is not responsible for the marked difference between CAO-LMP1 and B95.8-LMP1. To investigate further the sequences responsible for the functional properties of CAO-LMP1, we constructed two deletion mutants of B95.8 LMP1. One mutant, LMP1.Δ[343–352], had a deletion of the 10 codons that are missing in the CTAR-2 region of CAO-LMP1; the other control mutant, LMP1.Δ[352–360], had the 3′ adjacent 8 codons deleted. In addition, we constructed chimerae of the B95.8-LMP1 and CAO-LMP1 genes by swapping the BstEII-SmaI restriction fragments spanning the CTAR-2 region. As illustrated in Fig. 7A, the chimeras B95.8 × CAO and CAO × B95.8 express LMP1 molecules in which the CTAR-2 domains are exchanged between B95.8-LMP1 and CAO-LMP1. The B95.8 × CAO-LMP1 chimera contains sequences from B95.8-LMP1 to aa 333, and the remaining sequence is derived from CAO-LMP1; the chimera CAO × B95.8-LMP1 contains sequences from CAO-LMP1 to aa 359, and the remainder of the sequence is from B95.8-LMP1. Expression of each of the constructs was confirmed by Western blotting, and similar levels of expression were obtained for all mutants following transient transfection into Jurkat cells (Fig. 7B).
FIG. 7.
(A) Schematic representation of the LMP1 mutants generated for further analysis of CAO-LMP1 function. The LMP1 chimeras B95.8 × CAO and CAO × B95.8 were made by swapping the BstEII-SmaI fragments spanning the CAO deletion, thus exchanging sequences downstream of glycine 333 in B95.8 (glycine 373 in CAO). This effectively exchanges the CTAR-2 between B95.8-LMP1 and CAO-LMP1. B95.8 sequences are represented by solid lines, and CAO sequences are represented by broken lines. Solid boxes show the 10-aa sequence in B95.8-LMP1 that is deleted in CAO-LMP1, whereas the arrows indicate the position of this deletion in CAO. The Δ[343–352] mutant is a B95.8-LMP1 gene from which codons 343 to 352 were deleted (see Materials and Methods). The Δ[352–360] mutant is also derived from B95.8 LMP1. (B) Immunoblot showing the expression of the LMP1 mutants in transiently transfected Jurkat cells. The EBV-transformed B-cell line X50/7 is a positive control for LMP1, and SG5 vector-transfected Jurkat is a negative control. LMP1 expression was detected with CS1–4 antibodies as described in the legend to Fig. 1.
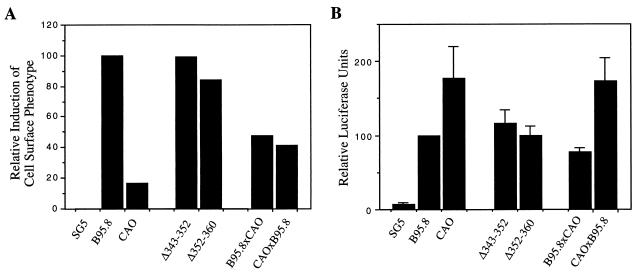
Using the same flow cytometric method as above, we investigated the effects of the LMP1 mutants on the cell surface phenotype in the Jurkat cell line. The results shown in Fig. 8A were obtained in the same representative experiment from which the Western blots were prepared in Fig. 7. As shown above, the induction of CD54 by CAO-LMP1 was markedly impaired relative to that by B95.8 LMP1. Both deletion mutants of B95.8-LMP1 (i.e., Δ[343–352] and Δ[352–360]) induced CD54 expression as efficiently as did the parental B95.8-LMP1 gene, confirming that the deletion alone is not responsible for the functional properties of CAO-LMP1. Surprisingly, the induction of CD54 by both chimeras (B95.8 × CAO and CAO × B95.8) was impaired relative to B95.8-LMP1, although neither chimera was as functionally impaired as CAO-LMP1. The result with the CAO × B95.8 chimera supports the conclusion that regions outside of CTAR-2 contribute to the loss of phenotype induction with CAO-LMP1. However, the result with B95.8 × CAO suggests either (i) that the other sequence changes within the CAO CTAR-2 are important or (ii) that the CAO CTAR-2 domain (which is identical to AG876) is not fully compatible with the B95.8 sequences in codons 1 to 332.
FIG. 8.
(A) Analysis of the effects of LMP1 mutants on cell surface phenotype. The T-cell line Jurkat was transfected with the indicated expression plasmids, and the relative induction of CD54 was measured by two-color flow cytometry as described in the legend to Fig. 2. Data shown are from one representative experiment from which the Western blot in Fig. 7B was obtained. (B) Effects on NF-κB activation by the LMP1 mutants. Jurkat cells were transfected with the indicated expression plasmids together with the 3Enh.κB-ConALuc reporter plasmid, and NF-κB activation was measured as described in the legend to Fig. 3. Data shown are the mean and standard deviation of at least four separate experiments.
In parallel with the phenotype assays, we also investigated the effects of the LMP1 mutants on NF-κB activation by using the same luciferase-based reporter assay as used above, and a summary of the results obtained following transfection of Jurkat cells is shown in Fig. 8B. As observed in earlier experiments, CAO-LMP1 activated NF-κB about twofold more than did B95.8-LMP1. As with the surface phenotype assays, the deletion mutants of B95.8-LMP1 (i.e., Δ[343–352] and Δ[352–360]) both behaved similarly to the parental B95.8-LMP1 with respect to the degree of activation of NF-κB. In contrast to the results obtained in the cell surface phenotype assays, the two chimeras B95.8 × CAO and CAO × B95.8 showed quite distinct properties with respect to activation of NF-κB. Thus, whereas the B95.8 × CAO chimera activated NF-κB to levels similar to those induced by B95.8, the CAO × B95.8 chimera behaved indistinguishably from CAO-LMP1 (Fig. 8B). These results clearly implicate sequences outside the CTAR-2 domain as being responsible for the elevated NF-κB-activating function of CAO-LMP1.
DISCUSSION
In this study, we show that the CAO-LMP1 gene differs markedly from the prototype B95.8-LMP1 gene with respect to the ability to activate NF-κB and to induce cell surface expression of CD40 and CD54 in human lymphoid cells. This observation extends the results of previous studies on CAO-LMP1 function in human cells, which demonstrated that the NPC-derived gene has greater transforming activity in the Rhek-1 human epithelial line as measured in vivo by increased tumor formation in SCID mice (27) and in vitro by differences in cell morphology and colony growth pattern (68). Taken together with the recent observation of Li et al., who showed that deletion of the 10-aa sequence (codons 343 to 352) from B95.8-LMP1 conferred a more tumorigenic phenotype to rodent fibroblasts when inoculated into nude mice (42), these data lend credence to the possibility that deletion variant LMP1 genes also influence the development of EBV-positive lymphoid tumors (37, 38, 40).
Against this background, it was surprising that our structure-function experiments ruled out a direct causative role for the 10-aa deletion in determining the distinct functional characteristics of CAO-LMP1 in lymphoid cells (Fig. 8). Equivalent structure-function studies of deletion variant LMP1 genes, using tumorigenicity and/or colony growth assays as a readout, have not been reported for human epithelial cell targets. Furthermore, while the experiments of Li et al. clearly showed that the 10-aa deletion was responsible for LMP1-induced tumorigenic transformation of BALB/c3T3 fibroblasts inoculated into nude mice (42), earlier studies demonstrated inconsistent results when comparing transformation of BALB/c3T3 and Rat-1 fibroblasts (4, 48) and demonstrated a lack of correlation between LMP1 function in rodent fibroblasts and human cells (29, 47). Therefore, our present data with human lymphoid cells should not be construed as contradictory; rather, they reflect the fact that we are using different functional readouts in different target cell types. In fact, our results are more consistent with those of a recent study in which human B-cell lines transformed with different EBV isolates, some of which expressed deletion variants of LMP1, were tested for the ability to form tumors in SCID and nude mice (59). In that study, the LCLs whose EBV expressed deletion variant LMP1 genes were, like those with undeleted LMP1 genes, unable to establish tumors in nude mice; only one LCL expressing a variant LMP1 gene with a larger (23-aa) deletion was tumorigenic in that assay. The observations of Sandvej et al. (59), together with our present results, appear to be at variance with one clinical report showing a correlation of the presence of LMP1 deletion variants with cancer in patients with lymphoproliferative disease (37). However, another clinical study published during the preparation of this paper found that the presence of del-LMP1 among 58 lesions from 36 heart and kidney organ transplant patients with lymphoproliferative disorders did not correlate with the aggressiveness of the lesions or with the progression of disease (61). Therefore, the hypothesis that the 10-aa deletion in LMP1 might influence the development of lymphoproliferative diseases is not convincingly supported by the evidence. Nevertheless, if sequence changes in the LMP1 gene do influence the development and clinical course of lymphoproliferations, our data would point to a role for sequence variations other than, or in addition to, the 10-aa deletion itself.
Our first indication that the 10-aa deletion itself was not the prime cause of the functional differences in our present study came from comparison with the function of the AG876-LMP1 gene, which displays the deletion but is otherwise more similar to the B95.8-LMP1 sequence (Fig. 1). With respect both to activation of NF-κB and to induction of CD40 and CD54, the AG876-LMP1 gene behaved similarly to the B95.8-LMP1 gene rather than to CAO-LMP1 (Fig. 3 and 4). Since the entire CTAR-2 sequence of AG876-LMP1 is identical to that of CAO-LMP1, these data would argue against a role for any of the CTAR-2 sequence in determining the distinct functional characteristics of CAO-LMP1. This is supported by the recent mapping of the NF-κB-activating domain of CTAR-2 to a highly conserved stretch of 6 aa comprising residues 379 to 384 at the far C terminus of B95.8-LMP1 (8, 19). Ruling out CTAR-2 turns the focus of attention to the CTAR-1 domain, which is known to bind TNF receptor-associated factors (TRAFs) that are involved in NF-κB activation pathways in other members of the TNF receptor family (50). A TRAF-binding motif (PxQxT) at codons 204 to 208 within CTAR-1 has been identified in B95.8-LMP1 (13), and this sequence is conserved in all published LMP1 gene sequences (45, 57). However, the precise role of TRAFs in activating LMP1-mediated activation of NF-κB is unclear (8, 32, 58). Furthermore, while mutation of sequences in the PxQxT motif undoubtedly impairs both TRAF-binding and NF-κB-activating functions (13, 15, 58), flanking sequences are also clearly important (13, 58). At this stage, therefore, the possibility cannot be excluded that sequence changes on either side of the PxQxT motif, e.g., the Q→P (codon 189 B95.8 LMP1), S→T (codon 192), or G→S (codon 212) changes, account for the functional differences between B95.8-LMP1 and CAO-LMP1 genes.
Notwithstanding the above discussion, our experiments with chimeric B95.8 × CAO and CAO × B95.8 LMP1 genes suggest that it may be naive to attempt to attribute all the functional differences of the CAO-LMP1 to a specific sequence variation in one functional domain. Thus, while these two chimeras showed properties consistent with the elevated NF-κB-activating function of CAO-LMP1 residing outside the CTAR-2 domain (Fig. 8B), the ability to induce cell surface CD54 was unexpectedly impaired in the B95.8 × CAO chimera as well as in the CAO × B95.8 chimera (Fig. 8A). Furthermore, neither chimera showed as severe an impairment of CD54-inducing function as did the parental CAO gene (Fig. 8). Our data with chimeric LMP1 genes are compatible with a scenario in which the signaling events arising from a physical interaction between CTAR-1 and CTAR-2 may differ qualitatively or quantitatively from the sum of the signalling events that can be induced separately from CTAR-1 and CTAR-2. Recent work in our laboratory, which involved cotransfection of a series of mutated B95.8-LMP1 genes that formed either mixed oligomeric complexes or separate homogeneous oligomers, demonstrated that CTAR-1 and CTAR-2 do indeed cooperate with each other to affect the nature of the signals generated by LMP1 complexes (19a). The recent data of other groups showing that TRAF1 and TRAF2 bind more efficiently to CTAR-2-deleted LMP1 sequences than they do to the whole carboxy region containing both CTAR-1 and CTAR-2 (13, 58) is also consistent with the possibility that CTAR-1 and CTAR-2 cooperate to bind a different set of TRAF signaling molecules than would be predicted from the factors known to bind CTAR-1 and CTAR-2 in isolation. Against this background, it is not surprising that swapping of the CTAR-1 and CTAR-2 domains between B95.8-LMP1 and CAO-LMP1 does not necessarily produce simple results. In addition to the likelihood that the CTAR-1 and CTAR-2 domains of these two LMP1 genes may not be fully compatible with each other, it is possible to envisage a more subtle role for intervening sequences such as the number of repeats and the phosphorylation sites (49), which have received little attention in recent structure-function investigations. Further work is in progress to elucidate the sequence changes responsible for the distinct functional properties of CAO, but the evidence to date points to a cooperative effect of more than one sequence variation in different parts of the molecule.
There remains the paradox of why the CAO-LMP1 gene causes a two- to threefold-greater activation of NF-κB than the B95.8-LMP1 or Ag876-LMP1 genes and yet is impaired in its ability to induce more downstream functions such as expression of cell surface CD40 and CD54 in lymphoid cells (Fig. 3 and 4). Both the CD40 and CD54 genes have NF-κB binding regulatory sequences in their promoter regions, and they do appear to be regulated by NF-κB activation in response to cytokines (21, 62). We have confirmed the specificity of our regular luciferase reporter assay by testing a second luciferase reporter construct based on an HIV promoter construct, together with a control reporter in which the NF-κB binding site is mutated; furthermore, the LMP1-mediated luciferase activity was inhibited by cotransfection with a gene expressing the IκBα inhibitory protein (Fig. 5). One explanation for the paradox is that CAO-LMP1 and B95.8-LMP1 genes differ in their ability to activate signalling pathways other than NF-κB. This is supported by many published examples where LMP1-mediated NF-κB activation does not correlate with other phenotypic readouts of LMP1 function (29, 46, 47), and the nature of the second signaling pathway may have been revealed by a report published during the preparation of this paper which showed that LMP1 can activate the AP-1 transcription factor via the SEK/JNK kinase cascade (36). Another explanation, which is not mutually exclusive, is that the CAO-LMP1 gene activates different species of NF-κB or NF-κB-like transcription factors that may bind with higher affinity to the reporter constructs but do not efficiently activate the endogenous CD40 or CD54 genes.
The recent literature has been inundated with papers describing the presence of del-LMP1 in various EBV-associated tumors. To some extent, these data may simply reflect the incidence of variant LMP1 in the healthy population in the geographical location studied (10, 23, 34). However, this does not preclude a role for sequence variants in promoting lymphoproliferative disease in certain circumstances such as immunosuppression. If distinct functional properties of del-LMP1 genes are important in EBV-associated lymphoproliferations, our results suggest that attention should be paid to the role of sequence variations other than the 10-aa deletion itself.
ACKNOWLEDGMENTS
This work was funded jointly by a Cancer Research Campaign studentship to R.J.J. and by a Leukemia Research Fund grant to M.S. and M.R. The contributions of S.A.H. and C.G.B. were sponsored by the Wellcome Trust. L.-F.H. was supported by the Swedish Cancerfonden.
REFERENCES
- 1.Arenzana-Seisdedos F, Fernandez B, Dominguez I, Jacqué J M, Thomas D, Diaz-Meco M T, Moscat J, Virelizier J L. Phosphatidylcholine hydrolysis activates NF-κB and increases human immunodeficiency virus replication in human monocytes and T lymphocytes. J Virol. 1993;67:6596–6604. doi: 10.1128/jvi.67.11.6596-6604.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bachelerie F, Alcami J, Arenzana-Seisdedos F, Virelizler J-L. HIV enhancer activity perpetuated by NF-κB induction on infection of monocytes. Nature. 1991;350:709–712. doi: 10.1038/350709a0. [DOI] [PubMed] [Google Scholar]
- 3.Baeuerle P, Baltimore D. NF-κB: ten years after. Cell. 1996;87:13–20. doi: 10.1016/s0092-8674(00)81318-5. [DOI] [PubMed] [Google Scholar]
- 4.Baichwal V, Sugden B. The multiple membrane-spanning segments of the BNLF-1 oncogene from Epstein-Barr virus are required for transformation. Oncogene. 1989;4:67–74. [PubMed] [Google Scholar]
- 5.Baichwal V R, Sugden B. Transformation of Balb 3T3 cells by the BNLF-1 gene of Epstein-Barr virus. Oncogene. 1988;2:461–467. [PubMed] [Google Scholar]
- 6.Ben-Bassat H, Goldblum N, Mitrani S, Goldblum T, Yoffey J M, Cohen M M, Bentwitch Z, Ramot B, Klein E, Klein G. Establishment in culture of a new type of lymphocyte from a “Burkitt-like” lymphoma (line D.G.-75) Int J Cancer. 1977;19:27–33. doi: 10.1002/ijc.2910190105. [DOI] [PubMed] [Google Scholar]
- 7.Brattsand G, Cantrell D A, Ward S, Ivars F, Gulberg M. Signal transduction through the T-cell receptor-CD3 complex: evidence for heterogeneity in receptor coupling. J Immunol. 1990;144:3651–3658. [PubMed] [Google Scholar]
- 8.Brodeur S R, Cheng G, Baltimore D, Thorley-Lawson D A. Localization of the major NF-κB-activating site and the sole TRAF3 binding site of LMP1 defines two distinct signalling motifs. J Biol Chem. 1997;272:19777–19784. doi: 10.1074/jbc.272.32.19777. [DOI] [PubMed] [Google Scholar]
- 9.Chen M L, Tsai C N, Liang C L, Shu C H, Huang C R, Sulitzeanu D, Liu S T, Chang Y S. Cloning and characterisation of the latent membrane protein (LMP) of a specific Epstein-Barr virus variant derived from the nasopharyngeal carcinoma in the Taiwanese population. Oncogene. 1992;7:2131–2140. [PubMed] [Google Scholar]
- 10.Chen W G, Chen Y Y, Bacchi M M, Bacchi C E, Alvarenga M, Weiss L M. Genotyping of Epstein-Barr-Virus in Brazilian Burkitts lymphoma and reactive lymphoid tissue-type A with a high prevalence of deletions within the latent membrane protein gene. Am J Pathol. 1996;148:17–23. [PMC free article] [PubMed] [Google Scholar]
- 11.Contreras-Salazar B, Ehlin-Henriksson B, Klein G, Masucci M G. Up regulation of the Epstein-Barr virus (EBV)-encoded membrane protein LMP in the Burkitt’s lymphoma line Daudi after exposure to n-butyrate and after EBV superinfection. J Virol. 1990;64:5441–5447. doi: 10.1128/jvi.64.11.5441-5447.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dawson C W, Rickinson A B, Young L S. Epstein-Barr virus latent membrane protein inhibits human epithelial cell differentiation. Nature. 1990;344:777–780. doi: 10.1038/344777a0. [DOI] [PubMed] [Google Scholar]
- 13.Devergne O, Hatzivassiliou E, Izumi K M, Kaye K M, Kleijnen M F, Kieff E, Mosialos G. Association of TRAF1, TRAF2, and TRAF3 with an Epstein-Barr virus LMP1 domain important for B-lymphocyte transformation: role in NF-κB activation. Mol Cell Biol. 1996;16:7098–7108. doi: 10.1128/mcb.16.12.7098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Duckett C S, Perkins N D, Kowalik T F, Schmid R M, Huang E-S, Baldwin A S, Jr, Nabel G J. Dimerization of NF-κB2 with RelA(p65) regulates DNA binding, transcriptional activation, and inhibition by an IκB-α (MAD-3) Mol Cell Biol. 1993;13:1315–1322. doi: 10.1128/mcb.13.3.1315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Eliopoulos A G, Stack M, Dawson C W, Kaye K M, Rowe M, Young L S. Epstein Barr virus-encoded LMP1 and CD40 mediate IL6 production in epithelial cells via an NF-κB pathway involving TNF receptor-associated factors. Oncogene. 1997;14:2899–2916. doi: 10.1038/sj.onc.1201258. [DOI] [PubMed] [Google Scholar]
- 16.Fåhraeus R, Hu L-F, Ernberg I, Finke J, Rowe M, Klein G, Falk K, Nilsson E, Yadav M, Busson P, Tursz T, Kallin B. Expression of the Epstein-Barr virus genome in nasopharyngeal carcinoma. Int J Cancer. 1988;42:329–338. doi: 10.1002/ijc.2910420305. [DOI] [PubMed] [Google Scholar]
- 17.Fåhraeus R, Rymo L, Rhim J S, Klein G. Morphological transformation of human keratinocytes expressing the LMP gene of Epstein-Barr virus. Nature. 1990;345:447–449. doi: 10.1038/345447a0. [DOI] [PubMed] [Google Scholar]
- 18.Fennewald S, van Santen V, Kieff E. Nucleotide sequence of an mRNA transcribed in latent growth-transforming virus indicates that it may encode a membrane protein. J Virol. 1984;51:411–419. doi: 10.1128/jvi.51.2.411-419.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Floettmann J E, Rowe M. Epstein-Barr virus latent membrane protein-1 (LMP1) C-terminus activation region-2 (CTAR-2) maps to the far C-terminus and requires oligomerisation for NF-κB activation. Oncogene. 1997;15:1851–1858. doi: 10.1038/sj.onc.1201359. [DOI] [PubMed] [Google Scholar]
- 19a.Floettman, J. E., et al. Unpublished data.
- 20.Green S, Issemann I, Sheer E. A versatile in vivo and in vitroexpression vector for protein engineering. Nucleic Acids Res. 1988;16:369. doi: 10.1093/nar/16.1.369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Grimaldi J C, Torres R, Kozak C A, Chang R, Clark E A, Howard M, Cockayne D A. Genomic structure and chromosomal mapping of the murine CD40 gene. J Immunol. 1991;149:3921–3926. [PubMed] [Google Scholar]
- 22.Hammarskjöld M-L, Simurda M C. Epstein-Barr virus latent membrane protein transactivates the human immunodeficiency virus type 1 long terminal repeat through induction of NF-κB activity. J Virol. 1992;66:6496–6501. doi: 10.1128/jvi.66.11.6496-6501.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hayashi K, Chen W G, Bacci C E, Alvarenga M, Abreau E S, Chang K L, Weiss L M. Deletion of Epstein-Barr virus latent membrane protein 1 gene in United States and Brazilian Hodgkin’s disease and reactive lymphoid tissue: high frequency of a 30-bp deletion. Hum Pathol. 1997;28:1408–1414. doi: 10.1016/s0046-8177(97)90231-8. [DOI] [PubMed] [Google Scholar]
- 24.Henderson S, Rowe M, Gregory C, Wang F, Kieff E, Rickinson A. Induction of bcl-2expression by Epstein-Barr virus latent membrane protein-1 protects infected B cells from programmed cell death. Cell. 1991;65:1107–1115. doi: 10.1016/0092-8674(91)90007-l. [DOI] [PubMed] [Google Scholar]
- 25.Herrero J A, Mathew P, Paya C V. LMP-1 activates NF-κB by targeting the inhibitory molecule IκBα. J Virol. 1995;69:2168–2174. doi: 10.1128/jvi.69.4.2168-2174.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hu L-F, Chen F, Zhen Q-F, Zhang Y-W, Luo Y, Zheng X, Winberg G, Ernberg I, Klein G. Differences in the growth pattern and clinical course of EBV-LMP1 expressing and non-expressing nasopharyngeal carcinomas. Eur J Cancer. 1995;31A:658–660. doi: 10.1016/0959-8049(94)00468-k. [DOI] [PubMed] [Google Scholar]
- 27.Hu L-F, Chen F, Zheng X, Ernberg I, Cao S-L, Christensson B, Klein G, Winberg G. Clonability and tumorigenicity of human epithelial cells expressing the EBV encoded membrane protein LMP1. Oncogene. 1993;8:1575–1583. [PubMed] [Google Scholar]
- 28.Hu L-F, Zabarovsky E R, Chen F, Cao S-L, Ernberg I, Klein G, Winberg G. Isolation and sequencing of the Epstein-Barr virus BNLF-1 gene (LMP1) from a Chinese nasopharyngeal carcinoma. J Gen Virol. 1991;72:2399–2409. doi: 10.1099/0022-1317-72-10-2399. [DOI] [PubMed] [Google Scholar]
- 29.Huen D S, Henderson S A, Croom-Carter D, Rowe M. The Epstein-Barr-virus latent membrane protein-1 (LMP1) mediates activation of NF-κB and cell surface phenotype via 2 effector regions in its carboxy-terminal cytoplasmic domain. Oncogene. 1995;10:549–560. [PubMed] [Google Scholar]
- 30.Izumi K M, Kaye K M, Kieff E D. The Epstein-Barr virus LMP1 amino acid sequence that engages tumor necrosis factor associated factors is critical for primary B lymphocyte growth transformation. Proc Natl Acad Sci USA. 1997;94:1447–1452. doi: 10.1073/pnas.94.4.1447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jones M D, Foster L, Sheedy T, Griffin B E. The EB virus genome in Daudi Burkitt’s lymphoma cells has a deletion similar to that observed in a non-transforming strain of the virus. EMBO J. 1984;3:813–821. doi: 10.1002/j.1460-2075.1984.tb01890.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kaye K M, Devergne O, Harada J N, Izumi K M, Yalamanchilli R, Kieff E, Mosialos G. Tumor necrosis factor receptor associated factor 2 is a mediator of NF-κB by latent infection membrane protein 1, the Epstein-Barr virus transforming protein. Proc Natl Acad Sci USA. 1996;93:11085–11090. doi: 10.1073/pnas.93.20.11085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kaye K M, Izumi K M, Kieff E. Epstein-Barr virus latent membrane protein 1 is essential for B-lymphocyte growth transformation. Proc Natl Acad Sci USA. 1993;90:9150–9154. doi: 10.1073/pnas.90.19.9150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Khanim F, Yao Q-Y, Niedobitek G, Sihota S, Rickinson A B, Young L S. Analysis of Epstein-Barr virus gene polymorphisms in normal donors and in virus associated tumors from different geographic locations. Blood. 1996;88:3491–3501. [PubMed] [Google Scholar]
- 35.Kieff E. Epstein-Barr virus and its replication. In: Fields B N, Knipe D M, Howley P M, editors. Fields virology. 3rd ed. Vol. 2. Philadelphia, Pa: Lippincott-Raven Publishers; 1996. pp. 2343–2396. [Google Scholar]
- 36.Kieser A, Kilger E, Giles O, Ueffing M, Kolch W, Hammerschmidt W. AP-1 induction by Epstein-Barr virus latent membrane protein-1 via the SEK/JNK cascade. EMBO J. 1997;16:6478–6485. doi: 10.1093/emboj/16.21.6478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kingma D W, Weiss W B, Jaffe E S, Kumar S, Frekko K, Raffeld M. Epstein-Barr virus latent membrane protein-1 oncogene deletions—correlations with malignancy in Epstein-Barr virus-associated lymphoproliferative disorders and malignant lymphomas. Blood. 1996;88:242–251. [PubMed] [Google Scholar]
- 38.Knecht H, Bachmann E, Brousset P, Rothenberger S, Einsele H, Lestou V S, Delsol G, Bachmann F, Ambros P F, Odermatt B F. Mutational hot spots within the carboxy terminal region of the LMP1 oncogene of Epstein-Barr virus are frequent in lymphoproliferative disorders. Oncogene. 1995;10:523–528. [PubMed] [Google Scholar]
- 39.Knecht H, Bachmann E, Brousset P, Sandvej K, Nadal D, Bachmann F, Odermatt B F, Delsol G, Pallesen G. Deletions within the LMP1 oncogene of Epstein-Barr virus are clustered in Hodgkins disease and are identical to those observed in nasopharyngeal carcinoma. Blood. 1993;82:2937–2942. [PubMed] [Google Scholar]
- 40.Knecht H, Raphael M, McQuain C, Rothenberger S, Pihan G, Camilleri-Broet S, Bachmann E, Kershaw G, Ryan S, Kittler E, Quesenberry P, Schlaifer D, Woda B, Brousset P. Deletion variants within the NF-κB activation domain of the LMP1 oncogene prevail in acquired immunodeficiency syndrome-related large cell lymphomas and human immunodeficiency virus-negative atypical lymphoproliferations. Blood. 1996;87:876–881. [PubMed] [Google Scholar]
- 41.Laherty C D, Hu H M, Opipari A W, Wang F, Dixit V M. Epstein-Barr virus LMP1 gene product induces A20 zinc finger protein expression by activating nuclear factor κB. J Biol Chem. 1992;267:24157–24160. [PubMed] [Google Scholar]
- 42.Li S-N, Chang Y-S, Liu S-H. Effect of a 10-amino-acid deletion on the oncogenic activity of latent membrane 1 of Epstein-Barr virus. Oncogene. 1996;12:2129–2135. [PubMed] [Google Scholar]
- 43.Mann K P, Staunton D, Thorley-Lawson D A. Epstein-Barr virus-encoded protein found in plasma membranes of transformed cells. J Virol. 1985;55:710–720. doi: 10.1128/jvi.55.3.710-720.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Miller W E, Earp H S, Raab-Traub N. The Epstein-Barr virus latent membrane protein 1 induces expression of the epidermal growth factor receptor. J Virol. 1995;69:4390–4398. doi: 10.1128/jvi.69.7.4390-4398.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Miller W E, Edwards R H, Walling D M, Raab-Traub N. Sequence variation in the Epstein-Barr virus latent membrane protein-1. J Gen Virol. 1994;75:2729–2740. doi: 10.1099/0022-1317-75-10-2729. [DOI] [PubMed] [Google Scholar]
- 46.Miller W E, Mosialos G, Kieff E, Raab-Traub N. Epstein-Barr virus LMP1 induction of the epidermal growth factor receptor is mediated through a TRAF signaling pathway distinct from NF-κB activation. J Virol. 1996;71:586–594. doi: 10.1128/jvi.71.1.586-594.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mitchell T, Sugden B. Stimulation of NF-κB-mediated transcription by mutant derivatives of the latent membrane protein of Epstein-Barr virus. J Virol. 1995;69:2968–2976. doi: 10.1128/jvi.69.5.2968-2976.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Moorthy R K, Thorley-Lawson D A. All three domains of the Epstein-Barr virus-encoded latent membrane protein LMP-1 are required for transformation of Rat-1 fibroblasts. J Virol. 1993;67:1638–1646. doi: 10.1128/jvi.67.3.1638-1646.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Moorthy R K, Thorley-Lawson D A. Biochemical, genetic, and functional analyses of the phosphorylation sites on the Epstein-Barr virus-encoded oncogenic latent membrane protein LMP-1. J Virol. 1993;67:2637–2645. doi: 10.1128/jvi.67.5.2637-2645.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mosialos G, Birkenbach M, Yalamanchili R, VanArsdale T, Ware C, Kieff E. The Epstein-Barr virus transforming protein LMP1 engages signalling proteins for the tumor necrosis factor receptor family. Cell. 1995;80:389–399. doi: 10.1016/0092-8674(95)90489-1. [DOI] [PubMed] [Google Scholar]
- 51.Nicholson L J, Hopwood P, Johannessen I, Salisbury J R, Codd J, Thorley-Lawson D, Crawford D H. Epstein-Barr virus latent membrane protein does not inhibit differentiation and induces tumorigenicity of human epithelial cells. Oncogene. 1997;15:275–283. doi: 10.1038/sj.onc.1201187. [DOI] [PubMed] [Google Scholar]
- 52.Peng M, Lundgren E. Transient expression of the Epstein Barr virus LMP1 gene in B-cell chronic lymphocytic leukemia cells, T cells, and hematopoietic cell lines: cell type independent induction of CD23, CD21 and ICAM-1. Leukemia. 1993;7:104–112. [PubMed] [Google Scholar]
- 53.Rickinson A B, Kieff E. Epstein-Barr virus. In: Fields B N, Knipe D M, Howley P M, editors. Fields virology. 3rd ed. Vol. 2. Philadelphia, Pa: Lippincott-Raven; 1996. pp. 2397–2446. [Google Scholar]
- 54.Rowe M, Evans H S, Young L S, Hennessy K, Kieff E, Rickinson A B. Monoclonal antibodies to the latent membrane protein of Epstein-Barr virus reveal heterogeneity of the protein and inducible expression in virus-transformed cells. J Gen Virol. 1987;68:1575–1586. doi: 10.1099/0022-1317-68-6-1575. [DOI] [PubMed] [Google Scholar]
- 55.Rowe M, Peng-Pilon M, Huen D S, Hardy R, Croom-Carter D, Lundgren E, Rickinson A B. Up-regulation of Bcl-2 by the Epstein-Barr virus latent membrane protein LMP1: a B-cell-specific response that is delayed relative to NF-κB activation and to induction of cell surface markers. J Virol. 1994;68:5602–5612. doi: 10.1128/jvi.68.9.5602-5612.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Rowe M, Rowe D T, Gregory C D, Young L S, Farrell P J, Rupani H, Rickinson A B. Differences in B cell growth phenotype reflect novel patterns of Epstein-Barr virus latent gene expression in Burkitt’s lymphoma. EMBO J. 1987;6:2743–2751. doi: 10.1002/j.1460-2075.1987.tb02568.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sample J, Kieff E F, Kieff E D. Epstein-Barr virus type-1 and type-2 have nearly identical LMP-1 transforming genes. J Gen Virol. 1994;75:2741–2746. doi: 10.1099/0022-1317-75-10-2741. [DOI] [PubMed] [Google Scholar]
- 58.Sandberg M, Hammerschmidt W, Sugden B. Characterization of LMP1 association with TRAF1, TRAF2, and TRAF3. J Virol. 1997;71:4649–4656. doi: 10.1128/jvi.71.6.4649-4656.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sandvej K, Munch M, Hamilton-Dutoit S. Mutations in the Epstein-Barr virus latent membrane protein-1 (BNLF-1) gene in spontaneous lymphoblastoid cell lines: effect on in vitro transformation associated parameters and tumorigenicity in SCID and nude mice. J Clin Pathol. 1996;49:M290–M297. doi: 10.1136/mp.49.5.m290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sandvej K, Peh S C, Andresen B S, Pallesen G. Identification of potential hotspots in the carboxy terminal part of the Epstein-Barr virus (EBV) BNLF-1 gene in both malignant and benign EBV-associated diseases: high frequency of a 30bp deletion in Malaysian and Danish T-cell lymphomas. Blood. 1994;84:4053–4060. [PubMed] [Google Scholar]
- 61.Scheinfeld A G, Nador R G, Cesarman E, Chadburn A, Knowles D M. Epstein-Barr virus latent membrane protein-1 oncogene deletion in post-transplantation lymphoproliferative disorders. Am J Pathol. 1997;151:805–812. [PMC free article] [PubMed] [Google Scholar]
- 62.Voraberger G, Schäfer R, Stratowa C. Cloning of the human gene for intracellular adhesion molecule 1 and analysis of its 5′-regulatory region. Induction by cytokines and phorbol ester. J Immunol. 1991;147:2777–2786. [PubMed] [Google Scholar]
- 63.Wang D, Liebowitz D, Kieff E. An EBV membrane protein expressed in immortalized lymphocytes transforms established rodent cells. Cell. 1985;43:831–840. doi: 10.1016/0092-8674(85)90256-9. [DOI] [PubMed] [Google Scholar]
- 64.Wang D, Liebowitz D, Wang F, Gregory C, Rickinson A, Larson R, Springer T, Kieff E. Epstein-Barr virus latent infection membrane protein alters the human B-lymphocyte phenotype: deletion of the amino terminus abolishes activity. J Virol. 1988;62:4173–4184. doi: 10.1128/jvi.62.11.4173-4184.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wang F, Gregory C D, Sample C, Rowe M, Liebowitz D, Murray R, Rickinson A B, Kieff E. Epstein-Barr virus latent membrane protein (LMP1) and nuclear proteins 2 and 3C are effectors of phenotypic changes in B lymphocytes: EBNA-2 and LMP1 cooperatively induce CD23. J Virol. 1990;64:2309–2318. doi: 10.1128/jvi.64.5.2309-2318.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wang S, Rowe M, Lundgren E. Expression of the Epstein-Barr virus transforming protein LMP1 causes a rapid and transient stimulation of the Bcl-2 homologue Mcl-1 levels in B cell lines. Cancer Res. 1996;56:4610–4613. [PubMed] [Google Scholar]
- 67.Young L, Dawson C, Clark D, Rupani H, Busson P, Tursz T, Johnson A, Rickinson A. Epstein-Barr virus gene expression in nasopharyngeal carcinoma. J Gen Virol. 1988;69:1051–1065. doi: 10.1099/0022-1317-69-5-1051. [DOI] [PubMed] [Google Scholar]
- 68.Zheng N, Yuan F, Hu L, Chen F, Klein G, Christensson B. Effect of a B-lymphocyte and NPC derived EBV-LMP1 gene expression on in vitro growth and differentiation of human epithelial cells. Int J Cancer. 1994;57:747–753. doi: 10.1002/ijc.2910570523. [DOI] [PubMed] [Google Scholar]