Summary
In pancreatic ductal adenocarcinoma (PDAC), no recurrent metastasis-specific mutation has been found, suggesting that epigenetic mechanisms, such as DNA methylation, are the major contributors of late-stage disease progression. Here, we performed the first whole-genome bisulfite sequencing (WGBS) on mouse and human PDAC organoid models to identify stage-specific and molecular subtype-specific DNA methylation signatures. With this approach, we identified thousands of differentially methylated regions (DMRs) that can distinguish between the stages and molecular subtypes of PDAC. Stage-specific DMRs are associated with genes related to nervous system development and cell-cell adhesions, and are enriched in promoters and bivalent enhancers. Subtype-specific DMRs showed hypermethylation of GATA6 foregut endoderm transcriptional networks in the squamous subtype and hypermethylation of EMT transcriptional networks in the progenitor subtype. These results indicate that aberrant DNA methylation contributes to both PDAC progression and subtype differentiation, resulting in significant and reoccurring DNA methylation patterns with diagnostic and prognostic potential.
Subject areas: genomics, Epigenetics, cancer systems biology, cancer
Graphical abstract

Highlights
-
•
PDAC WGBS shows distinct methylation in stages and molecular subtypes
-
•
Late-stage PDAC has major DNA methylome shifts
-
•
Squamous PDAC subtype shows hypermethylation of GATA6 transcriptional network
Genomics; Epigenetics; Cancer systems biology; Cancer
Introduction
Cancer cells accumulate numerous genetic and epigenetic alterations to proliferate and survive. While genetic contributions to cancer initiation and progression have long been studied, it has recently become clear that epigenetic regulation of gene expression is also critical for malignant transformation. Next-generation sequencing has made it much easier to profile the epigenome of cancer cells and its correlation with gene expression and patient prognosis. DNA methylation is one such epigenetic mechanism that occurs at cytosines of 5′-CG-3′ (CpG) dinucleotides and has been shown to be extensively altered in every cancer type through genome-wide microarray studies.1 Hypermethylation of promoters and CpG dense regions, called CpG islands, has been especially well studied in cancer and linked to silencing of tumor suppressor genes and DNA repair genes to promote cancer cell survival and genomic instability, respectively.2,3 Meanwhile, the mechanisms by which CpG hypomethylation promotes cancer progression have been largely overlooked even though genome-wide loss of methylation has been a known characteristic of cancers for over 40 years4 While DNA methylation is generally associated with gene downregulation at promoters and gene upregulation at gene bodies, numerous counterexamples have been observed,5,6,7 highlighting a complex relationship between DNA methylation and gene expression.
Pancreatic ductal adenocarcinoma (PDAC) accounts for over 90% of pancreatic cancer cases and is the 3rd-leading cause of cancer-related deaths in the United States.8 The genetic underpinnings of PDAC progression from normal ductal cells to pancreatic intraepithelial neoplasms (PanINs) and primary tumors have been uncovered through the identification of common somatic point mutations and copy number variants in exome sequencing studies.9,10 However, attempts to identify recurrent metastasis-specific genetic mutations have been unsuccessful,11 suggesting that late-stage disease progression is predominantly driven by epigenetic alterations. In fact, early genome-wide DNA methylation studies have made it increasingly clear that DNA methylation is an important regulator of PDAC molecular pathobiology with impacts on clinical phenotypes and patient survival.12,13,14,15,16,17 While providing invaluable insights into functional roles of aberrant DNA methylation in PDAC, these studies were limited due to use of microarrays, which assay less than 5% of CpG sites in the human genome,18 use of primary tumor samples with a high stromal content that can interfere with detection of tumor cell-specific DNA methylation changes, and/or a limited sample size (n < 10).
Recently, Espinet et al. performed whole-genome bisulfite sequencing (WGBS) of fluorescence-activated cell sorting (FACS)-isolated epithelial cells (EpCAM+/CD45-) from resected PDAC specimens, identifying a unique DNA methylation profile associated with interferon signaling and poor prognosis in a subset of PDAC tumors.19 Analysis of late-stage PDAC was lacking in this study, likely because late-stage PDAC patients are largely ineligible for surgical resection. Thus, we reasoned that WGBS analysis on PDAC patient-derived organoids (PDOs), which can be generated from resectable or biopsy-obtained late-stage specimens, would provide additional insight into how the DNA methylation landscape is altered in aggressive PDAC and throughout PDAC progression.
To assay the PDAC methylome without stromal interference, we performed the first WGBS analysis on PDOs and mouse organoid models that span each PDAC stage. In this study, over 75% of the CpGs in the human and mouse genome were assayed and used to identify differentially methylated regions (DMRs) between the stages and two major molecular subtypes of PDAC: progenitor and squamous. Characterization of these DMRs with functional enrichment testing identified pathways, transcriptional networks, and gene regulatory regions associated with aberrant DNA methylation while integrative transcriptome analysis identified DMRs significantly correlated with gene expression and poor patient survival. As a result, we have identified several pathways and transcriptional networks whose aberrant methylation may be impacting PDAC progression and DMRs that may serve as predictors of PDAC aggressiveness and treatment responsiveness.
Results
Global DNA methylation is altered at each stage of PDAC
To assess DNA methylation differences between normal pancreas tissue, precursor lesions, primary tumors, and metastases, we performed WGBS on 12 mouse organoids derived from each stage of PDAC (normal [mN], PanIN [mP], tumor [mT], and metastasis [mM]). mN organoids were derived from normal pancreata of C57BL/6J mice, mP organoids were derived from PanIN lesions of the KC (Kras+/LSL-G12D; Pdx1-Cre) mouse model of PDAC, and mT and mM organoids were derived from paired tumor and metastatic tissue of the KPC (Kras+/LSL-G12D; Trp53+/LSL-R172H; Pdx1-Cre) mouse model of PDAC (Figure 1A; Table S1). Principal component analysis (PCA) of the 16,031,225 CpGs assayed revealed that mN and mP organoids cluster together while mT and mM each form a distinct cluster. mM organoids clustered far away from all other stages of PDAC (Figure 1B), suggesting that DNA methylation is most dysregulated in distant metastases. Global methylation levels in mouse organoids were lower in neoplastic samples as mN and mP organoids had greater global methylation (66% and 67%, respectively) than mT and mM (61% and 63%, respectively) (Figure 1C; Table S1).
Figure 1.
Global DNA methylation is altered at each stage of PDAC
(A) Experimental design. Created with BioRender.com. FNA, Fine needle aspiration.
(B) Principal component analysis (PCA) of WGBS data from mouse organoids.
(C) Density plot of smoothed single CpG methylation values from mouse organoids.
(D) PCA of WGBS data from patient-derived organoids (PDOs).
(E) Density plot of smoothed single CpG methylation values from PDOs.
To profile genome-wide DNA methylation changes during PDAC progression in the patient setting, we performed WGBS on 35 PDOs from different stages of disease. 31 primary tumor PDOs (hT) were derived from tumor tissue obtained by surgical resection of patients with early-stage disease (resectable or borderline) or fine-needle aspiration of patients with late-stage disease (locally advanced or metastatic). Two normal PDOs (hN) were derived from surgical resection of adjacent normal pancreatic tissues of PDAC patients and an additional two hN organoids were previously characterized20 (Figure 1A; Table S1). PCA of the 21,818,517 CpGs assayed revealed that the global methylome of hN organoids is distinct from that of hT organoids (Figure 1D). While hT organoids from different stages do not form distinct clusters, the hT organoids from patients with metastatic disease have the most variable methylome (Figure 1D), suggesting that DNA methylation becomes more dysregulated as PDAC progresses. When comparing global methylation levels, hN organoids had the lowest global methylation (62%) of all PDOs (Figure 1E; Table S1). Global methylation levels tended to decrease with disease aggressiveness as hT organoids from patients with resectable disease had the greatest global methylation (73%), followed by borderline and locally advanced (67% and 69%, respectively), and lastly, metastatic (62%; Figure 1E; Table S1). Thus, in both mouse organoid models of PDAC and PDOs, aberrant DNA methylation is observed at each stage of PDAC and average global methylation levels decrease with disease aggressiveness.
DMR analysis of mouse organoids identifies a distinct DNA methylation signature in metastatic organoids
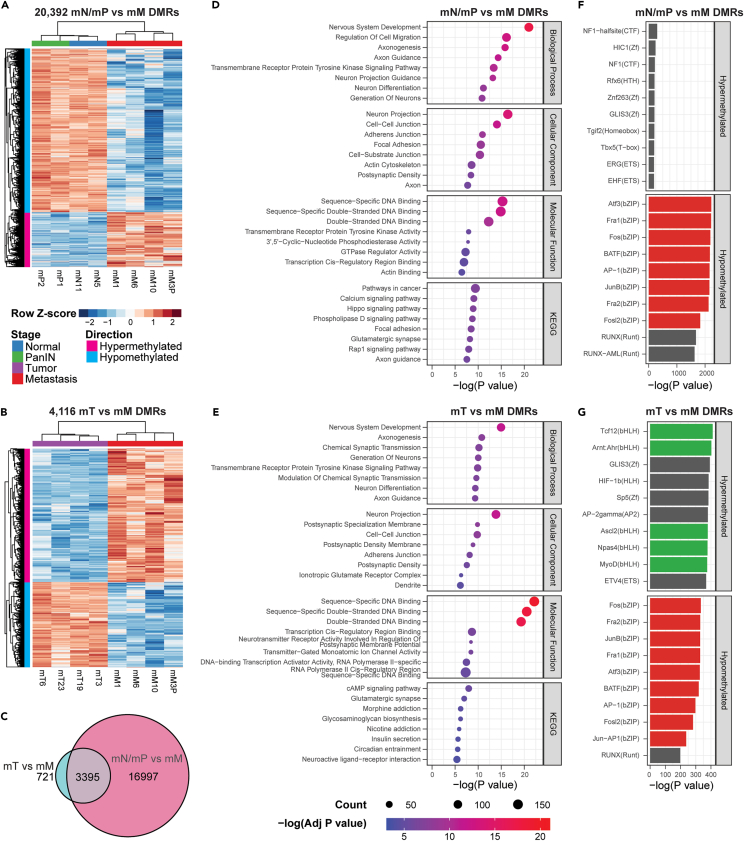
To identify DMRs that distinguish between metastatic tissue and primary tumor or pre-neoplastic tissue, we performed two comparisons: mN/mP versus mM and mT versus mM. In the mN/mP versus mM comparison, we identified 20,392 significant (q < 0.01) DMRs that completely distinguished mN/mP organoids from mM organoids in hierarchical clustering analysis (Figure 2A). 75% of the DMRs were hypomethylated in mM and 25% were hypermethylated (Table S2). In the mT versus mM comparison, we identified 4,116 significant (q < 0.01) DMRs that completely distinguished mT organoids from mM organoids in hierarchical clustering analysis (Figure 2B). 39% of the DMRs were hypomethylated in mM and 61% were hypermethylated (Table S3). Of the mT versus mM DMRs, 82% overlapped with mN/mP versus mM DMRs (Figure 2C). For both sets of DMRs, mN/mP organoids cluster with mT organoids in hierarchical clustering analysis (Figures S1A and S1B), and DMRs were evenly distributed throughout the genome (Figures S1C and S1D). In addition, no significant DMRs were detected between mN/mP and mT organoids. Using machine learning feature selection on the mN/mP versus mM DMRs, we identified a minimal set of 5 DMRs that distinguish mM from mN, mP, and mT organoids (Figure S1E). The genes associated with these DMRs (Fam20b,21 Mrpl1,22 Rab31,23 Man2a1,24 and Cited225) are primarily involved in metabolic signaling and have all been previously implicated in cancer progression and were all differentially expressed between mN and mM organoids (Figures S1F–S1J). Taken together, DMR calling of mouse organoids identified a DNA methylation signature in mM organoids that separates distant metastases from normal pancreatic tissue, PanINs, and primary tumors.
Figure 2.
DMR analysis of mouse organoids identifies a distinct DNA methylation signature in metastatic organoids
(A) Heatmap of 20,392 significant (q < 0.01) DMRs from mN/mP versus mM comparison.
(B) Heatmap of 4,116 significant (q < 0.01) DMRs from mT versus mM comparison.
(C) Euler diagram of sequence overlaps for DMRs.
(D and E) Top 10 KEGG and GO enrichments for (D) mN/mP versus mM DMRs and (E) mT versus mM DMRs.
(F and G) Top 10 transcription factor motif enrichments for (F) mN/mP versus mM DMRs and (G) mT versus mM DMRs.
To identify the pathways and ontologies impacted by these DNA methylation changes, we performed gene ontology (GO) enrichment analysis on the genes mapping to both DMR sets. In both sets of DMRs, we observed significant (p < 0.05) enrichment of biological processes related to neurodevelopment and cell migration, cellular components related to cell-cell junctions and actin cytoskeleton, and molecular functions related to DNA and actin binding (Figures 2D and 2E; Tables S2 and S3). Interestingly, cancer signaling pathways were more significantly enriched in mN/mP versus mM DMRs as several Ras-associated pathways (Rap1, PI3K/Akt, Hippo) were among the top 10 Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways and pathways in cancer were the top enriched KEGG term (Figure 2D; Table S2). In mT versus mM DMRs, the top 10 KEGG terms predominantly related to neuronal signaling, with cAMP signaling being the top enriched term (Figure 2E; Table S3). To identify transcriptional networks impacted by DNA methylation changes in PDAC progression, we performed HOMER motif analysis on DMRs relative to their background regions. DMRs that were hypomethylated in mM organoids of both mN/mP versus mM DMRs and mT versus mM DMRs were enriched in bZIP transcription factors (TFs) (Figures 2F and 2G; Tables S2 and S3), including those that form the AP-1 transcription factor complex which controls the expression of genes involved in proliferation, apoptosis, and oncogenic transformation.26 DMRs that were hypermethylated in mM compared to mT were enriched in bHLH TFs (Figure 2G; Table S3). TFs in this family included MyoD1, which is involved in suppression of cell migration in gastric cancers,27 and ASCL2, which is associated with immune infiltration in colorectal cancer.28 Enrichment of bHLH TF-binding motifs was not observed in mN/mP versus mM DMRs (Figure 2E; Table S3), suggesting that hypermethylation of bHLH TFs occurs late in PDAC progression. Overall, ontology analysis of DMRs revealed enrichment of neurodevelopment and cancer signaling pathways while HOMER motif analysis revealed that bZIP TF target genes are hypomethylated in mM organoids.
DMR analysis of PDOs identifies a DNA methylation signature associated with late-stage disease
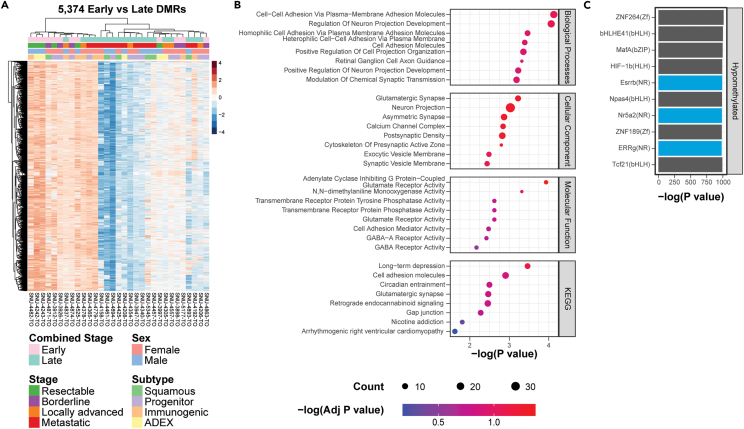
Due to presence of a distinct DNA methylation signature in mM organoids, we hypothesized that a unique DNA methylation signature of late-stage PDAC could similarly be identified in PDOs. DMR calling between early-stage organoids and late-stage organoids identified 5,374 significant (q < 0.05) DMRs that were nearly all hypomethylated in late-stage organoids (Figure 3A; Table S4). Genes associated with these DMRs were significantly enriched in biological processes related to cell adhesion, cellular components related to synapse signaling, and molecular functions related to neurotransmitter activity (Figure 3B; Table S4). In HOMER motif analysis of the regions hypomethylated in late-stage organoids, several nuclear receptor (NR) TF motifs were enriched, such as NR5A2 and ESRRB (Figure 3C). NR5A2 overexpression has been previously shown to promote proliferation and migration in pancreatic cancer cells29 while ESRRB promotes stem cell self-renewal by inducing loss of DNA methylation and recruitment of core pluripotency factors to inactive enhancers.30 Expression of these pluripotency factors was associated with worse survival outcomes and treatment resistance in several cancers,31 and may be performing a similar function in late-stage PDAC. No TF motifs were enriched in hypermethylated regions due to the small number of DMRs.
Figure 3.
DMR analysis of PDOs identifies a DNA methylation signature associated with late-stage disease
(A) Heatmap of 5,374 significant (q < 0.05) DMRs for early- versus late-stage organoid comparison.
(B) Top 10 KEGG and GO enrichments for early- versus late-stage DMRs.
(C) Top 10 transcription factor motif enrichments for DMRs hypomethylated in late-stage organoids.
To identify differential methylation between normal pancreatic tissue and primary PDAC tumors, we performed DMR calling on hN compared to hT organoids. This analysis identified 374 significant (q < 0.05) DMRs where 13% of DMRs were hypomethylated in hT and 87% were hypermethylated (Table S5). These DMRs completely separated hN organoids from hT organoids in hierarchical clustering analysis and were evenly distributed across chromosomes (Figures S2A and S2E). Early- versus late-stage DMRs overlapped minimally with the hN versus hT DMRs (Figure S2B), suggesting that most of the DNA methylation changes that occur during tumorigenesis differ from those that occur in the progression from primary tumor to metastasis. GO analysis revealed enrichment of biological processes associated with transcription, cellular components associated with ion channels and membranes, and molecular functions associated with DNA binding (Figure S2C; Table S5). Similar to HOMER motif analysis of the mT versus mM DMRs, bHLH TF motifs were enriched in regions that are hypermethylated in hT organoids (Figure S2D). No TF motifs were enriched in hypomethylated regions due to the small number of DMRs. Overall, DMR calling of PDOs identified a DNA methylations signature associated with late-stage disease that is enriched for NR TF-binding motifs in hypomethylated regions and ontologies related to cell adhesion and neurodevelopment.
Human and mouse DMRs are enriched in promoter regions and bivalent enhancers
To assess the genomic regions most impacted by differential methylation, we tested the DMRs for enrichment of CpG and gene region annotations relative to their background regions. For all DMR sets, CpG islands overlapped more with hypermethylated DMRs and open sea regions overlapped more with hypomethylated DMRs (Figure S3A; Table S6), matching previous evidence that global hypomethylation and promoter hypermethylation are common events in cancers.2,4 In addition, each hypermethylated DMR set was most highly enriched in CpG islands and the hypomethylated early- vs. late-stage DMRs were uniquely depleted in CpG islands, shores, and shelves and enriched in open seas (Figure 4A). In gene annotation testing, all DMR sets had significant (q < 0.05) enrichment of promoters and depletion of intergenic regions (Figure 4B). Most DMR sets also had significant (q < 0.05) enrichment of 5′ UTRs, exons, and 3′ UTRs (Figure 4B) even though less than 15% of DMRs were in exons and less than 5% are in 5′ UTRs and 3′ UTRs (Figure S3B). The enrichment of DMRs in CpG islands and promoters indicates that recurrent DNA methylation changes often affect genetic regions that regulate initiation of transcription.
Figure 4.
Human and mouse DMRs are enriched in promoter regions and bivalent enhancers
(A) CpG annotation enrichments. ∗FDR < 0.05.
(B) Gene region annotation enrichments. ∗FDR < 0.05.
(C) Heatmap of log(fold enrichment) for overlaps between DMR sets and Roadmap Epigenomics reference datasets in pancreas cells. ∗q < 0.05, ∗∗q < 0.01, ∗∗∗q < 0.001.
To understand the epigenetic landscape at these DMRs, we used the chromHMM core 15-state model (based on 127 epigenomes from the NIH Roadmap Epigenomics Project) to identify pancreas-specific chromatin state enrichments in the DMRs. Apart from the early- vs. late-stage DMRs, hypermethylated DMRs showed significant (q < 0.01) enrichment in active transcription start sites (TSS), flanking TSS, transcription at gene 5′ and 3′, weak transcription, genic enhancers, enhancers, and ZNF genes. In early- versus late-stage hypomethylated DMRs, the top enrichment was repressed polycombs (Figure 4C; Table S7). In mT versus mM DMRs, mN/mP versus mM DMRs, and hypermethylated hN versus hT DMRs, the top enrichment was either flanking bivalent TSS/enhancer or bivalent enhancer (Figure 4C; Table S7). Previously, it was reported that bivalent enhancers are closely associated with developmental genes that are poised for activation during development.32 Thus, the enrichment of hypermethylated regions at enhancers suggests that developmentally poised genes are more prone to methylation in late-stage PDAC and confer survival benefits and aggressive characteristics.
DNA methylation correlates with gene expression changes that have prognostic value
To estimate the impact of DNA methylation on gene expression in PDAC, we investigated correlations between DNA methylation and gene expression in mT versus mM DMRs and early- versus late-stage DMRs. 33% of mT versus mM DMRs and 10% of early- vs. late-stage DMRs had a significant (p < 0.05) correlation between DNA methylation and gene expression (Figure 5A). This smaller proportion of correlated DMRs in the patient setting is likely due to the greater genetic variation in patients compared to mice as well as the effects of variable environments and cancer treatments. When investigating the genetic regions associated with significant correlations between DNA methylation and gene expression, we found that the distribution of gene regions was largely consistent between positive and negative correlations. Exceptions to this were the enrichment of promoters in negatively correlated mT vs. mM DMRs and the enrichment of introns in positively correlated early- vs. late-stage DMRs (Figure 5B; Table S8). To assess the relationship between methylation and histone modifications at DMRs, we profiled active histone marks at mT versus mM DMRs with chromatin immunoprecipitation sequencing (ChIP-seq) (GSE99311).33 Regions that were hypermethylated in mM organoids had reduced occupancy of H3K4me1 and H3K27ac while hypomethylated regions had increased occupancy (Figures S4A–S4D). Together, these results indicate that DMRs have coordinated differences in their epigenetic landscapes that impact gene expression.
Figure 5.
DNA methylation correlates with gene expression changes that have prognostic value
(A) Percent of DMRs with correlations between DNA methylation and gene expression. RNA-seq of mT and mM organoids (GSE142467)34 and RNA-seq of early-stage and late-stage organoids.
(B) Distribution of gene region annotations in DMRs where DNA methylation levels correlate with expression of the associated gene for mT versus mM DMRs and early- versus late-stage DMRs.
(C and D) Low methylation in late-stage organoids (left graph) and positive correlation between DNA methylation and normalized gene expression (right graph) in DMRs associated with glutamatergic synapse-related genes: (C) DLGAP1 and (D) SH3GL2. Normalized gene expression calculated with DESeq2.35
(E and F) Kaplan-Meier survival showing poor survival in patients with low mRNA expression of (E) DLGAP1 and (F) SH3GL2 in TCGA-PAAD data.
Given that early- vs. late-stage DMRs are enriched in glutamatergic synapse components (Figure 4B), we sought to discern whether the associated genes have significant correlation between DNA methylation and gene expression as well as clinical significance. We found that DMRs associated with DLGAP1 and SH3GL2 were all hypomethylated in late-stage organoids, methylation at these DMRs was positively correlated with gene expression, and low expression of these glutamatergic synapse-related genes was associated with worse survival outcomes in The Cancer Genome Atlas data (Figures 5C–5F). Because glutamate is a key substrate of several metabolic pathways, metabolic reprogramming with adaptations to glutamate metabolism is a common event in cancers, including PDAC, that sustains rapid cell proliferation.36,37 These results highlight a previously underappreciated role of DNA methylation in regulation of glutamate metabolism with a close correlation to survival outcomes.
Progenitor transcriptional networks are hypermethylated in squamous PDOs
Transcriptional profiling of resected PDAC specimens has identified two main molecular subtypes, called progenitor and squamous, that have distinct molecular features and prognoses.38,39,40 The progenitor subtype is characterized by a favorable prognosis and the upregulation of transcriptional networks that are crucial for pancreatic endoderm cell-fate determination.39,41 The squamous subtype is characterized by loss of these endoderm specification TFs, metabolic reprogramming, and a worse prognosis.39 Importantly, changes in gene expression between the progenitor and squamous subtype are epigenetically regulated as evidenced by chromatin remodeling and changes in histone modifications.12,39,42,43 To assess the extent of DNA methylation-mediated regulation on the molecular subtypes, we first stratified PDOs into two major molecular subtypes (progenitor, n = 12 and squamous, n = 6) using RNA sequencing (RNA-seq) data as described previously (Figure S5A)39 and then performed DMR analysis on the hT organoids. DMR calling identified 3,751 significant (p < 0.05) DMRs that completely separated the two subtypes in hierarchical clustering analysis (Figures 6A and S5B). Machine learning identified a minimal set of 17 DMRs that distinguished squamous from progenitor PDOs (Figure S5C). All 17 of the DMRs were hypermethylated in squamous PDOs and 7 of the genes associated with these DMRs (KALRN, LINC00501, EPCAM, ACSL5, ADAP1, PAQR8, and PRR15L) were downregulated in squamous compared to progenitor patient specimens.39 GO terms enriched in subtype DMRs were associated with cell migration, cytoskeleton rearrangement, and signal transduction while pathways in cancer were among the top enriched KEGG pathways (Figure 6B; Table S9).
Figure 6.
Progenitor transcriptional networks are hypermethylated in squamous PDOs
(A) Heatmap of 3,751 significant (p < 0.05) DMRs for progenitor versus squamous subtype comparison.
(B) Top 10 KEGG and GO enrichments for subtype DMRs.
(C) HOMER motifs enriched in hypermethylated regions of squamous subtype organoids.
(D) Overlap of subtype DMRs with GATA6 peaks.
(E) GATA6 occupancy in hypermethylated DMRs that overlap with GATA6 peaks using GATA6 ChIP-seq data in progenitor subtype, PATU8988S cells (GSE47535).44
(F) Gene set enrichment analysis (GSEA) of human embryonic stem cell-derived pancreatic progenitor cells with TET1/2/3 triple knockout (TET TKO) compared to wild-type (WT) RNA-seq (GSE146486)45 using the progenitor subtype gene signature from Bailey et al.39
The pancreatic progenitor subtype of PDAC is defined by expression of transcription factors involved in pancreatic endoderm identity, such as HNF4A and GATA6.39,42,44 In the squamous subtype, these TFs are epigenetically silenced by DNA methylation, resulting in loss of endodermal identity and expression of squamous-associated transcriptional profiles.39,42,44,46 In this study, not only did we see hypermethylation of HNF4A and GATA6 (Figures 6A and 6B; Table S9), but also hypermethylation of their target genes as evidenced by significant (P < 1E-150) enrichment of their binding motifs within DMRs that were hypermethylated in the squamous subtype (Figure 6C). The enrichment of GATA6-binding motif in hypermethylated DMRs prompted us to hypothesize that GATA6 is critical to maintain hypomethylated status in the progenitor subtype-associated genes. To this end, we compared the known GATA6-binding sites in the progenitor PDAC cell line, PATU8988S, with the hypermethylated regions in the squamous subtype. GATA6 peaks were enriched in the hypermethylated regions while hypomethylated regions showed no enrichment (Figures 6D and 6E). Genes associated with these hypermethylated GATA6 peaks were downregulated in the squamous subtype (Figures S6C–S6E), indicating that GATA6 target genes are silenced by DNA methylation. Recently, GATA6 has been shown to interact with demethylating TET enzymes, resulting in demethylation of its binding sites.47 These results suggest that interactions with TET enzymes help pancreatic endoderm TFs maintain the progenitor subtype in PDAC by preventing DNA methylation-mediated silencing of their target genes. To test this hypothesis, we first investigated whether TET enzymes are necessary for progenitor gene expression using publicly available RNA-seq dataset (GSE146486) of pancreatic progenitor cells with triple knockout of TET1/2/3 (TET TKO).45 Gene set enrichment analysis of this RNA-seq data revealed that TET TKO cells are significantly depleted in the progenitor gene signature compared to wild-type cells (Figure 6F), indicating that TET enzymes are required to maintain the progenitor subtype. On the other hand, no significant difference in the squamous gene signature was observed (Figure S6F), indicating that loss of TET enzymes is not sufficient to induce the squamous subtype. With the ChIP-seq and WGBS data from the same dataset, we then tested whether TET depletion promotes epigenetic remodeling of GATA6-regulated progenitor genes to an inactive state. As expected, the TET TKO resulted in increased DNA methylation, reduced chromatin accessibility, and reduced H3K27ac occupancy (Figures S6G–S6J). In conclusion, the two main molecular subtypes of PDAC have distinct DNA methylation signatures, progenitor transcriptional networks are hypermethylated in squamous PDOs, and TET enzymes are involved in epigenetic remodeling of GATA6 target genes to promote the progenitor subtype in PDAC.
Discussion
Most genome-wide DNA methylation studies of PDAC relied on microarray platforms that assay a very small percentage of CpGs and resected primary tumor specimens that exhibit variable tumor cellularity due to the extensive desmoplastic stroma characteristic of PDAC.12,13,14,15,16,17 This high stromal content complicates characterization of molecular mechanisms specific to epithelium-derived malignant cells and reduces the sensitivity of detecting DMRs.15 Recent WGBS studies employed microdissection and FACS of epithelial cells (EpCAM+/CD45-) to achieve high purity of resected primary tumor samples and provide insights into the cell of origin debate in human PDAC and aberrant DNA methylation patterns in resectable disease.19,48 However, the role of DNA methylation in late-stage disease progression is poorly understood as most patients with late-stage disease are not eligible for tumor resection. Use of PDAC organoids provides a unique opportunity to investigate DNA methylation changes across all stages of PDAC as they can be derived from resectable tissue as well as fine-needle biopsy. Furthermore, PDAC organoids preserve tumor heterogeneity, exhibit disease stage-specific characteristics, and are derived from neoplastic ductal cells, eliminating the cellularity issues posed by primary specimens.20 Thus, we provide novel insight into the role of DNA methylation in PDAC progression and subtype transition with the first comprehensive genome-wide DNA methylation study using PDAC organoids. First, we uncovered DMRs that distinguish metastases from primary tumors and preneoplastic tissue, early-stage primary tumors from late-stage primary tumors, and primary tumors from normal pancreata. Second, we showed that that these DMRs mapped to genes that are enriched for neurodevelopment, focal adhesion, and glutamatergic signaling processes. Third, we identified DMRs whose DNA methylation correlates with expression of genes with prognostic value. Lastly, we demonstrated that squamous and progenitor core gene programs are differentially methylated and separate the two subtypes based on their DNA methylation profiles.
The DMRs identified in this study validate previously observed impacts of DNA methylation on gene expression and signaling pathways while also providing new insights. Pathways that were enriched in differentially methylated genes of previous studies as well as this study include axon guidance,15 neuroactive ligand-receptor interaction,13,14 focal adhesion,13 circadian entrainment,14 and many critical cancer-related pathways.13,14,15 To our knowledge, differential methylation of glutamate signaling has not been previously highlighted but was significantly enriched in hypomethylated early- vs. late-stage DMRs in this study. Glutamate signaling components have previously been shown to regulate PDAC metabolic reprogramming, growth, and metastasis.36,49 In addition, we found that hypomethylation was associated with low gene expression and poor survival outcomes, suggesting that DNA methylation-mediated glutamate adaptations promote PDAC progression and contribute to poor prognosis.
While TP63 expression is sufficient to induce squamous subtype transition, not all squamous subtype PDOs express TP63, suggesting that p63 is not required for the squamous identity.43 Likewise, we did not see significant enriched TP63-binding motif in the hypomethylated regions, while the significant enrichments in GATA6 and other endoderm lineage TFs were found in the hypermethylated regions. Instead, we found that the binding motifs of Snail1 and Slug are enriched in DMRs that are hypomethylated in the squamous subtype (Figure S5D). Overexpression of Snail1 in PDAC has been shown to promote many of the characteristics associated with the squamous subtype, including epithelial-to-mesenchymal transition (EMT), metabolic reprogramming, and chemoresistance.50,51 This suggests that the loss of endoderm lineage TFs (e.g., GATA6, HNF4A) is a prerequisite for squamous subtype transition and other contributing factors such as TP63 and EMT TFs can further promote squamous identity. Nonetheless, our data suggest that GATA6-mediated recruitment of TET enzymes allows PDAC cells to maintain the hypomethylated status of endodermal lineage-related genes. HNF4A may also play a role in this progenitor subtype maintenance as we found that HNF4A ChIP-seq peaks overlap with GATA6 peaks in progenitor PDAC cell lines (Figure S6K). However, it remains to be determined whether DNA methylation actively evicts GATA6 binding or GATA6 loss passively allows DNA methylation in these regions.
Taken together, our study provides analysis of the global DNA methylation landscape in murine models and PDOs for PDAC progression, revealing stage- and subtype-specific DMRs in PDAC. The identification of these DMRs indicates that recurrent DNA methylation changes underlie the disease stages and molecular subtypes of PDAC. Furthermore, these DNA methylation alterations can be utilized as functional biomarkers for PDAC progression and molecular subtyping.
Limitations of the study
One limitation in this study is the absence of human metastasis-derived organoids for stage-specific DMR analysis. Consequently, this study may be missing DNA methylation changes that occur in metastatic disease. In addition, the PDAC organoid model may have epigenetic differences from primary tumor specimens that have yet to be explored. This may especially be the case for hN organoids which had unexpectedly low DNA methylation levels relative to hT organoids. We hypothesize that this global hypomethylation of hN organoids may be the result of epigenetic changes that enable human normal samples to proliferate in cell culture conditions. Thus, the hN versus hT DMR analysis may be reflective of proliferation-independent differences between neoplastic and non-neoplastic cells, but will require further investigation.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Biological samples | ||
| Human normal pancreas | University of California, Davis Comprehensive Cancer Center | N/A |
| Critical commercial assays | ||
| Qiagen DNEasy Blood and Tissue Kit | Qiagen | Cat#69504 |
| Accel-NGS Methyl-Seq DNA Library Kit | Swift Biosciences | Cat#30096 |
| Deposited data | ||
| WGBS raw data and processed bedgraph files | This paper | GEO: GSE243528 |
| Custom code | This paper | https://github.com/swang52/pdac_wgbs.git |
| Mouse reference genome NCBI37/mm9 | Genome Reference Consortium | https://www.ncbi.nlm.nih.gov/grc/mouse |
| Human reference genome GRCh38/hg38 | Genome Reference Consortium | https://www.ncbi.nlm.nih.gov/grc/human |
| mT and mM organoid RNA-seq data | Oni et al.34 | GEO: GSE142467 |
| mT and mM H3K4me1 and H3K27ac ChIP-seq data | Roe et al.33 | GEO: GSE99311 |
| GATA6 ChIP in PATU8988S cells | Kloesch et al.44 | GEO: GSE47535 |
| TET TKO WGBS, ATAC-seq, RNA-seq, and ChIP-seq data | Li et al.45 | GEO: GSE146486 |
| HNF4A ChIP in PDAC cells | Camolotto et al.52 | GEO: GSE138452 |
| Biological samples | ||
| Mouse: mN5 Normal pancreatic organoid | Tuveson Laboratory, Boj et al.20 | NA |
| Mouse: mN11 Normal pancreatic organoid | Tuveson Laboratory, Boj et al.20 | NA |
| Mouse: mP1 PanIN organoid | Tuveson Laboratory, Boj et al.20 | NA |
| Mouse: mP2 PanIN organoid | Tuveson Laboratory, Boj et al.20 | NA |
| Mouse: mT3 Tumor organoid | Tuveson Laboratory, Boj et al.20 | NA |
| Mouse: mT6 Tumor organoid | Tuveson Laboratory, Boj et al.20 | NA |
| Mouse: mT19 Tumor organoid | Tuveson Laboratory, Roe et al.33 | NA |
| Mouse: mT23 Tumor organoid | Tuveson Laboratory, Roe et al.33 | NA |
| Mouse: mM1 Metastatic organoid (liver, paired tumor is mT3) | Tuveson Laboratory, Boj et al.20 | NA |
| Mouse: mM3P Metastatic organoid (peritoneum, paired tumor is mT6) | Tuveson Laboratory, Boj et al.20 | NA |
| Mouse: mM6 Metastatic organoid (peritoneum, paired tumor is mT19) | Tuveson Laboratory, Roe et al.33 | NA |
| Mouse: mM10 Metastatic organoid (peritoneum, paired tumor is mT23) | Tuveson Laboratory, Roe et al.33 | NA |
| Human: hN1 Normal pancreatic organoid (female, 59 years old) | This paper | NA |
| Human: hN4 Normal pancreatic organoid (female, unknown age) | Tuveson Laboratory, Boj et al.20 | NA |
| Human: hN9 Normal pancreatic organoid (female, unknown age) | Tuveson Laboratory, Boj et al.20 | NA |
| Human: hN18 Normal pancreatic organoid (male, 69 years old) | This paper | NA |
| Human: SNU-3898-TO Tumor organoid (female, 61 years old) | Korean Cell Line Bank | NA |
| Human: SNU-3912-TO Tumor organoid (female, 58 years old) | Korean Cell Line Bank | NA |
| Human: SNU-3926-TO Tumor organoid (female, 52 years old) | Korean Cell Line Bank | NA |
| Human: SNU-3947-TO Tumor organoid (female, 64 years old) | Lee et al.53 | NA |
| Human: SNU-3997-TO Tumor organoid (male, 54 years old) | Korean Cell Line Bank | NA |
| Human: SNU-4158-TO Tumor organoid (female, 54 years old) | Lee et al.53 | NA |
| Human: SNU-4192-TO Tumor organoid (female, 56 years old) | Lee et al.53 | NA |
| Human: SNU-4206-TO Tumor organoid (female, 81 years old) | Lee et al.53 | NA |
| Human: SNU-4208-TO Tumor organoid (male, 45 years old) | Lee et al.53 | NA |
| Human: SNU-4242-TO Tumor organoid (male, 74 years old) | Korean Cell Line Bank | NA |
| Human: SNU-4243-TO Tumor organoid (male, 63 years old) | Korean Cell Line Bank | NA |
| Human: SNU-4305-TO Tumor organoid (female, 65 years old) | Lee et al.53 | NA |
| Human: SNU-4340-TO Tumor organoid (male, 66 years old) | Lee et al.53 | NA |
| Human: SNU-4354-TO Tumor organoid (female, 64 years old) | Lee et al.53 | NA |
| Human: SNU-4365-TO Tumor organoid (female, 55 years old) | Lee et al.53 | NA |
| Human: SNU-4378-TO Tumor organoid (female, 69 years old) | Korean Cell Line Bank | NA |
| Human: SNU-4425-TO Tumor organoid (female, 60 years old) | Lee et al.53 | NA |
| Human: SNU-4457-TO Tumor organoid (female, 58 years old) | Korean Cell Line Bank | NA |
| Human: SNU-4461-TO Tumor organoid (female, 67 years old) | Korean Cell Line Bank | NA |
| Human: SNU-4482-TO Tumor organoid (female, 65 years old) | Korean Cell Line Bank | NA |
| Human: SNU-4525-TO Tumor organoid (male, 56 years old) | Korean Cell Line Bank | NA |
| Human: SNU-4557-TO Tumor organoid (female, 63 years old) | Korean Cell Line Bank | NA |
| Human: SNU-4779-TO Tumor organoid (female, 62 years old) | Korean Cell Line Bank | NA |
| Human: SNU-4837-TO Tumor organoid (male, 67 years old) | Korean Cell Line Bank | NA |
| Human: SNU-4863-TO Tumor organoid (female, 69 years old) | Korean Cell Line Bank | NA |
| Human: SNU-4871-TO Tumor organoid (female, 58 years old) | Korean Cell Line Bank | NA |
| Human: SNU-4874-TO Tumor organoid (female, 54 years old) | Korean Cell Line Bank | NA |
| Human: SNU-4893-TO Tumor organoid (male, 69 years old) | Korean Cell Line Bank | NA |
| Human: SNU-4894-TO Tumor organoid (male, 61 years old) | Korean Cell Line Bank | NA |
| Human: SNU-5177-TO Tumor organoid (female, 57 years old) | Korean Cell Line Bank | NA |
| Human: SNU-5345-TO Tumor organoid (male, 54 years old) | Korean Cell Line Bank | NA |
| Software and algorithms | ||
| CpG_Me | Laufer et al.54 | https://github.com/ben-laufer/CpG_Me |
| Trim Galore | Babraham Bioinformatics | https://www.bioinformatics.babraham.ac.uk/projects/trim_galore/ |
| Cutadapt | Martin55 | https://cutadapt.readthedocs.io/en/stable/ |
| FastQ Screen | Wingett et al.56 | https://www.bioinfromatics.babraham.ac.uk/projects/fastq_screen/ |
| Bismark | Krueger and Andrews57 | https://www.bioinformatics.babraham.ac.uk/projects/Bismark |
| Bowtie 2 | Langmead and Salzberg58 | https://bowtie-bio.sourceforge.net/bowtie2/index.shtml |
| SAMtools | Li et al.59 | https://www.htslib.org |
| Picard | Broad Institute | https://broadinstitute.github.io/picard/ |
| MultiQC | Ewels et al.60 | https://multiqc.info |
| R | https://www.r-project.org/ | |
| DMRichR | Laufer et al.54 | https://github.com/ben-laufer/DMRichR |
| Dmrseq | Korthauer et al.61 | https://bioconductor.org/packages/release/bioc/html/dmrseq.html |
| Bsseq | Hansen et al.62 | https://bioconductor.org/packages/release/bioc/html/bsseq.html |
| ComplexHeatmap | Gu et al.63 | https://jokergood.github.io/ComplexHeatmap-reference/book/ |
| enrichR | Chen et al.64 | https://cran.r-project.org/web/packages/enrichR/index.html |
| HOMER | Heinz et al.65 | htttps://homer.ucsd.edu/homer |
| LOLA | Sheffield and Bock66 | https://bioconductor.org/packages/release/bioc/html/LOLA.html |
| liftOver | UCSC | https://genome.ucsc.edu/cgi-bin/hgLiftOver |
| Boruta | Kursa and Rudnicki67 | https://cran.r-project.org/web/packages/Boruta/index.html |
| sigFeature | Das et al.68 | https://www.bioconductor.org/packages/release/bioc/html/sigFeature.html |
| DESeq2 | Love et al.35 | https://bioconductor.org/packages/release/bioc/html/DESeq2.html |
| ggplot2 | Wickham69 | https://cran.r-project.org/web/packages/ggplot2/index.html |
| Graphpad Prism | GraphPad Software | https://www.graphpad.com/features |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Dr. Chang-il Hwang (cihwang@ucdavis.edu).
Materials availability
This study did not generate new unique reagents.
Data and code availability
-
•
Raw and processed sequencing data has been deposited at GEO and is publicly available as of the date of publication under accession number GSE243528. The accession number is also listed in the key resources table.
-
•
All original code has been deposited at GitHub and is publicly available as of the date of publication at https://github.com/swang52/pdac_wgbs.git. The URL is also listed in the key resources table.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
Experimental model and study participant details
Human specimens
Specimens used to generate two normal organoids (hN4 and hN9) and specimens used to generate ten late-stage primary tumor organoids (SNU-3947-TO, SNU-4158-TO, SNU-4192-TO, SNU-4206-TO, SNU-4208-TO, SNU-4305-TO, SNU-4340-TO, SNU-4354-TO, and SNU-4365-TO, SNU-4425-TO) were obtained and characterized previously.20,53 An additional two normal organoids (hN1 and hN18) were derived from the adjacent normal tissue of patients undergoing surgical resection at University of California, Davis Medical Center (UCDMC). An additional twenty-one primary tumor specimens were obtained from surgical resection of PDAC patients with early-stage disease or fine-needle aspiration of PDAC patients with late-stage disease at Seoul National University Hospital. All tissue donations and experiments were reviewed and approved by the Internal Review Boards of each participating institution in compliance with the Declaration of Helsinki. Written informed consent was received from each donor and/or their legal representative prior to specimen acquisition. Samples were confirmed to be tumor or normal based on pathologic assessment. For patients included in this study, 31% were male and 69% were female and the median age was 61 years old. There was no significant difference in median age of early- versus late-stage patients or progenitor versus squamous subtype patients as determined by t-test. Information on sex, age, and treatment for each sample can be found in Table S1.
Organoid culture
All murine organoids were established and characterized previously.20 In brief, normal pancreatic ductal organoids (mN) were derived from C57BL/6 mice. Low-grade murine PanIN organoids (mP) were derived from KC mice (Kras+/LSL-G12D; Pdx1-Cre) while murine primary tumor organoids (mP) and metastatic organoids (mM) were derived from tumor-bearing KPC mice (Kras+/LSL-G12D; Trp53+/LSL-R172H ; Pdx1-Cre). Murine pancreatic cancer organoids were cultured in Matrigel with murine organoid culture media: advanced DMEM/F12, HEPES 10 mM, GlutaMAX 1X, A83-01 500 nM, mEGF 50 ng/ul, mNoggin 100 ng/ul, hFGF10 100 ng/ul, hGastrin I 10 nM, N-acetylcysteine 1.25 mM, nicotinamide 10 mM, B27 supplement (2% final), and R-spondin conditioned media (10% final). Primary tumor organoids derived from surgically resected tissue and FNA of PDAC patients as well as normal organoids derived from adjacent normal tissue of surgically resected tissue of PDAC patients were established and cultured as previously described.20,53 PDOs were cultured in Matrigel with human organoid culture media: advanced DMEM/F12, HEPES 10mM, Glutamax 1X, A83-01 500 nM, hEGF 50 ng/ml, mNoggin 100ng/mL, hFGF10 100 ng/mL, hGastrin I 10 nM, N-acetylcysteine 1.25 mM, nicotinamide 10 mM, PGE2 1uM, Primocin 100 ug/mL B27 supplement 1X, R-spondin1 conditioned media (10% final), Afamin/Wnt3A conditioned media (50% final).
Method details
DNA isolation and WGBS library preparation
Genomic DNA was extracted from murine organoids and hN organoids using the DNeasy Blood and Tissue Kit (Qiagen) with RNaseA to digest RNA. Genomic DNA from EUS-FNA-derived human organoids (SNU-) were obtained from the Korean Cell Line Bank and extracted as previously described.53 Digestion of RNA was confirmed with a 1% agarose gel and genomic DNA was quantified using a Nanodrop 2000 Spectrophotometer (Thermo Scientific). For each sample, 200 ng of genomic DNA was bisulfite converted using the EZ DNA Methylation-Lightning kit (Zymo). WBGS sequencing libraries were constructed from 50 ng of converted DNA per sample using the TruSeq DNA Methylation Kit (Illumina). Library quality was checked with Bioanalyzer (Agilent), quantified, indexed, and pooled prior to 150 bp paired-end sequencing for ∼5X coverage. All murine samples were sequenced on one lane of a single NovaSeq 6000 flow cell (Illumina) and all human samples were sequenced on two lanes of a single NovaSeq 6000 flow cell.
WGBS data processing
Raw WGBS fastq files were aligned using the CpG_Me pipeline.54 Briefly, reads were chastity filtered and trimmed to remove adapters and sequences with methylation bias using Trim Galore (https://www.bioinformatics.babraham.ac.uk/projects/trim_galore) and Cutadapt.55 Bismark,57 Bowtie 2,58 and SAMtools59 were then used to align reads to the mouse genome (mm9) for murine samples and the human genome (hg38) for human samples, remove PCR duplicates, extract CpG methylation, generate CpG count matrices, and calculate coverage. Insert size metrics were calculated using Picard (https://broadinstitute.github.io/picard). Quality control and assurance was performed using Trim Galore, Bismark, FastQ Screen,56 and MultiQC.60
Differential methylation analysis
DMR calling and most downstream analyses were performed using the DMRichR R package.54 DMRichR uses dmrseq61 and bsseq62 algorithms to perform local likelihood smoothing, which infers methylation levels from CpG count matrices where CpGs with higher coverage are given greater weight. Thus, the statistical approaches benefit from more biological replicates rather than deeper sequencing and reduces the coverage needed to achieve accurate results from 30X coverage or deeper to as little as 4X.62 Briefly, symmetric CpG sites were merged across strands and CpGs were filtered for at least 1X coverage in 100% of murine samples or at least 75% of human samples with sex adjustment. CpGs from chromosome X and Y were removed from analysis to eliminate sex-dependent DMRs. Candidate background regions with greater than 10% methylation difference were assembled and subject to permutation testing (8 permutations for murine samples and 10 permutations for human samples) to find DMRs with at least 5 CpGs and FDR corrected q-value < 0.01 for mouse organoid comparisons, FDR corrected q-value < 0.05 for stage-stratified PDO comparisons, and empirical p-value < 0.05 for the subtype-stratified PDO comparison. Individual smoothened methylation values for DMRs were generated with bsseq62 and used for heatmap visualizations with ComplexHeatmap63 and principle component analysis (PCA). Annotatr70 and ChIPseeker71 packages were used for gene region, gene symbol, and CpG annotations where promoters were defined as regions within 3 Kb of the TSS and DMRs were annotated with the following prioritization order: Promoter > 5’UTR > 3’UTR > Exon > Intron > Downstream > Intergenic.
Enrichment testing
enrichR64 was used for gene ontology (GO) and KEGG pathway enrichment testing of DMRs between 3Kb upstream of a TSS and 3 Kb downstream of a 3’ UTR. HOMER65 was used to identify transcription factor binding motifs enriched in DMRs through the findMotifsGenome.pl script with CpG% normalization (-cpg) and given region size (-size given). Annotation based enrichment was performed using two-sided Fisher’s exact tests. Locus Overlap Analysis (LOLA)66 was used to identify chromatin enrichments in pancreas cells from the ChromHMM 15-state model.72 All enrichment testing was done relative to the background regions assembled for DMR calling and odds ratios were converted to fold enrichment for data visualization. For all genes annotated to DMRs, Pearson’s correlation coefficient was calculated to identify genes whose methylation changes significantly correlate to gene expression (p < 0.05).
Machine learning
The random forest algorithm in Boruta67 and the support vector machine algorithm in sigFeature68 were used to build binary classification models and create two lists of DMRs ranked by variable importance for mN/mP vs mM DMRs and subtype DMRs. Common DMRs in the top 1% of each list were selected as minimal DMRs. Machine learning analyses were then performed to predict PDAC stage (tumor or metastatic) and PDAC subtype (progenitor or squamous) from the identified set of minimal DMRs. The ntree parameter (number of trees) was set to 500 and the mtry (number of predictors sampled at each tree split) parameter was set to 2.
Analyses of public datasets
Normalized RNA-seq expression data for mouse organoids were downloaded from the NCBI Gene Expression Omnibus (GEO) with accession number GSE142467.34 The Cancer Genome Atlas (TCGA) normalized samples from PDAC samples were downloaded from the TCGA database and used to calculate Kaplan-Meier survival curves with GraphPad Prism. H3K4me1 and H3K27ac for mT and mM organoids were obtained from accession number GSE99311 33 and used to generate mean signal plots at DMRs with deepTools73 functions computeMatrix and plotProfile. Similarly, mean signal plots at DMRs with GATA6 occupancy were generated from WGBS, ATAC-seq, H3K4me1 ChIP-seq, and H3K27ac ChIP-seq data in TET1/2/3 triple knockout pancreatic progenitor cells (TET TKO) obtained from accession number GSE146486.45 GATA6 occupancy was determined from GATA6 ChIP-seq in PATU8988S cells (progenitor subtype) from GSE47535.44 HNF4A occupancy was determined from HNF4A ChIP-seq in HC596a8 and HC800a8 PDAC cell lines (progenitor subtype) from GSE138452.52
Quantification and statistical analysis
Statistical details of experiments can be found in the figure legends. Statistical analysis of gene expression data was conducted using Dunnett’s multiple comparison test when comparing three or more groups or student t-test when comparing two groups in Graphpad Prism. All other statistical analysis was conducted in R and described above. In all figures ∗P < 0.05; ∗∗P < 0.01; ∗∗∗P < 0.001. Graphs were drawn with ggplot2,69 Graphpad Prism, or ComplexHeatmap.63
Acknowledgments
The authors would like to thank all members of the C.-I.H. laboratory for helpful discussions throughout the course of this study. The authors thank Dr. Janine LaSalle for guidance on WGBS analysis and Jihao Xu for organoid culture. The results shown here are in part based upon data generated by the TCGA Research Network: https://www.cancer.gov/tcga. S.S.W. is supported by the NIH-funded UC Davis Molecular and Cell Biology Training Program (T32 GM-007377). C.-I.H. is supported by the NCI (K22CA226037 and R37CA249007), University of California Research Initiatives, Cancer Research Coordinating Committee, Pilot Grant (C21CR2020), and the UC Davis Comprehensive Cancer Center Pilot Grant (NCI P30CA093373). Two hN organoids (hN4 and hN9) were provided by Tuveson laboratory. UC Davis Comprehensive Cancer Center Biorepository is funded by the NCI P30CA093373.
Author contributions
Conceptualization, S.S.W. and C.-I.H.; writing – original draft, S.S.W.; writing – review and editing, S.S.W. and C.-I.H.; formal analysis, S.S.W. and C.-I.H.; investigation, S.S.W., M.L.H., N.R., E.L., S.-C.K., and C.-I.H.; resources, S.H.L., J.-Y.J., R.J.B., and J.-L.K.; funding acquisition, C.-I.H.; supervision, C.-I.H. All authors read and approved the final manuscript.
Declaration of interests
The authors declare no competing interests.
Published: March 4, 2024
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2024.109414.
Supplemental information
(A) Sequencing alignment and DNA methylation extraction metrics for WGBS libraries. (B) Patient information for PDOs. (C) Average global CpG methylation for mouse organoids. (D) Average global CpG methylation for PDOs.
(A) DMR enrichment testing results. (B) Gene ontology enrichment results. (C) Known HOMER motif results.
(A) DMR enrichment testing results. (B) Gene ontology enrichment results. (C) Known HOMER motif results.
(A) DMR enrichment testing results. (B) Gene ontology enrichment results. (C) Known HOMER motif results
(A) DMR enrichment testing results. (B) Gene ontology enrichment results. (C) Known HOMER motif results.
(A) mN/mP versus mM DMRs. (B) mT versus mM DMRs. (C) early- versus late-stage DMRs. (D) hN versus hT DMRs.
(A) mN/mP versus mM DMRs. (B) mT versus mM DMRs. (C) early- versus late-stage DMRs. (D) hN versus hT DMRs.
(A) mT versus mM DMRs. (B) early- versus late-stage DMRs.
(A) DMR enrichment testing results. (B) Gene ontology enrichment results. (C) Known HOMER motif results.
References
- 1.Weisenberger D.J. Characterizing DNA methylation alterations from the cancer genome atlas. J. Clin. Invest. 2014;124:17–23. doi: 10.1172/JCI69740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.De Carvalho D.D., Sharma S., You J.S., Su S.F., Taberlay P.C., Kelly T.K., Yang X., Liang G., Jones P.A. DNA Methylation screening identifies driver epigenetic events of cancer cell survival. Cancer Cell. 2012;21:655–667. doi: 10.1016/j.ccr.2012.03.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sriraman A., Debnath T.K., Xhemalce B., Miller K.M. Making it or breaking it: DNA methylation and genome integrity. Essays Biochem. 2020;64:687–703. doi: 10.1042/EBC20200009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Feinberg A.P., Vogelstein B. Hypomethylation distinguishes genes of some human cancers from their normal counterparts. Nature. 1983;301:89–92. doi: 10.1038/301089a0. [DOI] [PubMed] [Google Scholar]
- 5.Jones P.A. Functions of DNA methylation: Islands, start sites, gene bodies and beyond. Nat. Rev. Genet. 2012;13:484–492. doi: 10.1038/nrg3230. [DOI] [PubMed] [Google Scholar]
- 6.Spainhour J.C., Lim H.S., Yi S.V., Qiu P. Correlation patterns between DNA methylation and gene expression in The Cancer Genome Atlas. Cancer Inform. 2019;18 doi: 10.1177/1176935119828776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Smith J., Sen S., Weeks R.J., Eccles M.R., Chatterjee A. Promoter DNA hypermethylation and paradoxical gene activation. Trends Cancer. 2020;6:392–406. doi: 10.1016/j.trecan.2020.02.007. [DOI] [PubMed] [Google Scholar]
- 8.Siegel R.L., Miller K.D., Wagle N.S., Jemal A. Cancer statistics, 2023. CA. Cancer J. Clin. 2023;73:17–48. doi: 10.3322/caac.21763. [DOI] [PubMed] [Google Scholar]
- 9.Biankin A.V., Waddell N., Kassahn K.S., Gingras M.C., Muthuswamy L.B., Johns A.L., Miller D.K., Wilson P.J., Patch A.M., Wu J., et al. Pancreatic cancer genomes reveal aberrations in axon guidance pathway genes. Nature. 2012;491:399–405. doi: 10.1038/nature11547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jones S., Zhang X., Parsons D.W., Lin J.C.-H., Leary R.J., Angenendt P., Mankoo P., Carter H., Kamiyama H., Jimeno A., et al. Core Signaling Pathways in Human Pancreatic Cancers Revealed by Global Genomic Analyses. Science. 2008;321:1801–1806. doi: 10.1126/science.1164368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yachida S., Jones S., Bozic I., Antal T., Leary R., Fu B., Kamiyama M., Hruban R.H., Eshleman J.R., Nowak M.A., et al. Distant metastasis occurs late during the genetic evolution of pancreatic cancer. Nature. 2010;467:1114–1117. doi: 10.1038/nature09515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lomberk G., Blum Y., Nicolle R., Nair A., Gaonkar K.S., Marisa L., Mathison A., Sun Z., Yan H., Elarouci N., et al. Distinct epigenetic landscapes underlie the pathobiology of pancreatic cancer subtypes. Nat. Commun. 2018;9:1978. doi: 10.1038/s41467-018-04383-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mishra N.K., Guda C. Genome-wide DNA methylation analysis reveals molecular subtypes of pancreatic cancer. Oncotarget. 2017;8:28990–29012. doi: 10.18632/oncotarget.15993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mishra N.K., Southekal S., Guda C. Survival analysis of multi-omics data identifies potential prognostic markers of pancreatic ductal adenocarcinoma. Front. Genet. 2019;10:624. doi: 10.3389/fgene.2019.00624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nones K., Waddell N., Song S., Patch A.M., Miller D., Johns A., Wu J., Kassahn K.S., Wood D., Bailey P., et al. Genome-wide DNA methylation patterns in pancreatic ductal adenocarcinoma reveal epigenetic deregulation of SLIT-ROBO, ITGA2 and MET signaling. Int. J. Cancer. 2014;135:1110–1118. doi: 10.1002/ijc.28765. [DOI] [PubMed] [Google Scholar]
- 16.Thompson M.J., Rubbi L., Dawson D.W., Donahue T.R., Pellegrini M. Pancreatic Cancer Patient Survival Correlates with DNA Methylation of Pancreas Development Genes. PLoS One. 2015;10 doi: 10.1371/journal.pone.0128814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vincent A., Omura N., Hong S.M., Jaffe A., Eshleman J., Goggins M. Genome-wide analysis of promoter methylation associated with gene expression profile in pancreatic adenocarcinoma. Clin. Cancer Res. 2011;17:4341–4354. doi: 10.1158/1078-0432.CCR-10-3431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sun Z., Cunningham J., Slager S., Kocher J.-P. Base resolution methylome profiling: considerations in platform selection, data preprocessing and analysis. Epigenomics. 2015;7:813–828. doi: 10.2217/epi.15.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Espinet E., Gu Z., Imbusch C.D., Giese N.A., Büscher M., Safavi M., Weisenburger S., Klein C., Vogel V., Falcone M., et al. Aggressive PDACs show hypomethylation of repetitive elements and the execution of an intrinsic IFN program linked to a ductal cell-of-origin. Cancer Discov. 2021;11:638–659. doi: 10.1158/2159-8290.CD-20-1202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Boj S.F., Hwang C.I., Baker L.A., Chio I.I.C., Engle D.D., Corbo V., Jager M., Ponz-Sarvise M., Tiriac H., Spector M.S., et al. Organoid models of human and mouse ductal pancreatic cancer. Cell. 2015;160:324–338. doi: 10.1016/j.cell.2014.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ma P., Yan W., Tian Y., Wang J., Feng J.Q., Qin C., Cheng Y.-S.L., Wang X. Inactivation of Fam20B in joint cartilage leads to chondrosarcoma and postnatal ossification defects. Sci. Rep. 2016;6 doi: 10.1038/srep29814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lin X., Guo L., Lin X., Wang Y., Zhang G. Expression and prognosis analysis of mitochondrial ribosomal protein family in breast cancer. Sci. Rep. 2022;12 doi: 10.1038/s41598-022-14724-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen K., Xu J., Tong Y.L., Yan J.-F., Pan Y., Wang W.J., Zheng L., Zheng X.X., Hu C., Hu X., et al. Rab31 promotes metastasis and cisplatin resistance in stomach adenocarcinoma through Twist1-mediated EMT. Cell Death Dis. 2023;14:115. doi: 10.1038/s41419-023-05596-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang Y., Zhao J., Fu G., Sheng C., Zhu J., Zhong T., Yang F., Jiang Z. MAN2A1 predicts prognosis and progression through cancer-related pathways in colorectal cancer. Transl. Cancer Res. 2022;11:3686–3697. doi: 10.21037/tcr-22-629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shin S.-H., Lee G.Y., Lee M., Kang J., Shin H.-W., Chun Y.-S., Park J.-W. Aberrant expression of CITED2 promotes prostate cancer metastasis by activating the nucleolin-AKT pathway. Nat. Commun. 2018;9:4113. doi: 10.1038/s41467-018-06606-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Eferl R., Wagner E.F. AP-1: a double-edged sword in tumorigenesis. Nat. Rev. Cancer. 2003;3:859–868. doi: 10.1038/nrc1209. [DOI] [PubMed] [Google Scholar]
- 27.Wu F., Qin Y., Jiang Q., Zhang J., Li F., Li Q., Wang X., Gao Y., Miao J., Guo C., et al. MyoD1 suppresses cell migration and invasion by inhibiting FUT4 transcription in human gastric cancer cells. Cancer Gene Ther. 2020;27:773–784. doi: 10.1038/s41417-019-0153-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wu L., Sun S., Qu F., Liu X., Sun M., Pan Y., Zheng Y., Su G. ASCL2 affects the efficacy of immunotherapy in colon adenocarcinoma based on single-cell RNA sequencing analysis. Front. Immunol. 2022;13 doi: 10.3389/fimmu.2022.829640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Guo F., Zhou Y., Guo H., Ren D., Jin X., Wu H. NR5A2 transcriptional activation by BRD4 promotes pancreatic cancer progression by upregulating GDF15. Cell Death Discov. 2021;7:78. doi: 10.1038/s41420-021-00462-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Adachi K., Kopp W., Wu G., Heising S., Greber B., Stehling M., Araúzo-Bravo M.J., Boerno S.T., Timmermann B., Vingron M., Schöler H.R. Esrrb unlocks silenced enhancers for reprogramming to naive pluripotency. Cell Stem Cell. 2018;23:266–275.e6. doi: 10.1016/j.stem.2018.05.020. [DOI] [PubMed] [Google Scholar]
- 31.Hepburn A.C., Steele R.E., Veeratterapillay R., Wilson L., Kounatidou E.E., Barnard A., Berry P., Cassidy J.R., Moad M., El-Sherif A., et al. The induction of core pluripotency master regulators in cancers defines poor clinical outcomes and treatment resistance. Oncogene. 2019;38:4412–4424. doi: 10.1038/s41388-019-0712-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bernhart S.H., Kretzmer H., Holdt L.M., Jühling F., Ammerpohl O., Bergmann A.K., Northoff B.H., Doose G., Siebert R., Stadler P.F., Hoffmann S. Changes of bivalent chromatin coincide with increased expression of developmental genes in cancer. Sci. Rep. 2016;6 doi: 10.1038/srep37393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Roe J.S., Hwang C.I., Somerville T.D.D., Milazzo J.P., Lee E.J., Da Silva B., Maiorino L., Tiriac H., Young C.M., Miyabayashi K., et al. Enhancer Reprogramming Promotes Pancreatic Cancer Metastasis. Cell. 2017;170:875–888.e20. doi: 10.1016/j.cell.2017.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Oni T.E., Biffi G., Baker L.A., Hao Y., Tonelli C., Somerville T.D.D., Deschênes A., Belleau P., Hwang C.I., Sánchez-Rivera F.J., et al. SOAT1 promotes mevalonate pathway dependency in pancreatic cancer. J. Exp. Med. 2020;217 doi: 10.1084/jem.20192389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Love M.I., Huber W., Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Herner A., Sauliunaite D., Michalski C.W., Erkan M., De Oliveira T., Abiatari I., Kong B., Esposito I., Friess H., Kleeff J. Glutamate increases pancreatic cancer cell invasion and migration via AMPA receptor activation and Kras-MAPK signaling. Int. J. Cancer. 2011;129:2349–2359. doi: 10.1002/ijc.25898. [DOI] [PubMed] [Google Scholar]
- 37.García-Gaytán A.C., Hernández-Abrego A., Díaz-Muñoz M., Méndez I. Glutamatergic system components as potential biomarkers and therapeutic targets in cancer in non-neural organs. Front. Endocrinol. 2022;13 doi: 10.3389/fendo.2022.1029210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Moffitt R.A., Marayati R., Flate E.L., Volmar K.E., Loeza S.G.H., Hoadley K.A., Rashid N.U., Williams L.A., Eaton S.C., Chung A.H., et al. Virtual microdissection identifies distinct tumor- and stroma-specific subtypes of pancreatic ductal adenocarcinoma. Nat. Genet. 2015;47:1168–1178. doi: 10.1038/ng.3398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bailey P., Chang D.K., Nones K., Johns A.L., Patch A.M., Gingras M.C., Miller D.K., Christ A.N., Bruxner T.J.C., Quinn M.C., et al. Genomic analyses identify molecular subtypes of pancreatic cancer. Nature. 2016;531:47–52. doi: 10.1038/nature16965. [DOI] [PubMed] [Google Scholar]
- 40.Collisson E.A., Sadanandam A., Olson P., Gibb W.J., Truitt M., Gu S., Cooc J., Weinkle J., Kim G.E., Jakkula L., et al. Subtypes of pancreatic ductal adenocarcinoma and their differing responses to therapy. Nat. Med. 2011;17:500–503. doi: 10.1038/nm.2344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tiyaboonchai A., Cardenas-Diaz F.L., Ying L., Maguire J.A., Sim X., Jobaliya C., Gagne A.L., Kishore S., Stanescu D.E., Hughes N., et al. GATA6 plays an important role in the induction of human definitive endoderm, development of the pancreas, and functionality of pancreatic β cells. Stem Cell Rep. 2017;8:589–604. doi: 10.1016/j.stemcr.2016.12.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Brunton H., Caligiuri G., Cunningham R., Upstill-Goddard R., Bailey U.M., Garner I.M., Nourse C., Dreyer S., Jones M., Moran-Jones K., et al. HNF4A and GATA6 Loss Reveals Therapeutically Actionable Subtypes in Pancreatic Cancer. Cell Rep. 2020;31 doi: 10.1016/j.celrep.2020.107625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Somerville T.D.D., Xu Y., Miyabayashi K., Tiriac H., Cleary C.R., Maia-Silva D., Milazzo J.P., Tuveson D.A., Vakoc C.R. TP63-Mediated Enhancer Reprogramming Drives the Squamous Subtype of Pancreatic Ductal Adenocarcinoma. Cell Rep. 2018;25:1741–1755.e7. doi: 10.1016/j.celrep.2018.10.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kloesch B., Ionasz V., Paliwal S., Hruschka N., Martinez De Villarreal J., Öllinger R., Mueller S., Dienes H.P., Schindl M., Gruber E.S., et al. A GATA6-centred gene regulatory network involving HNFs and "np63 controls plasticity and immune escape in pancreatic cancer. Gut. 2022;71:766–777. doi: 10.1136/gutjnl-2020-321397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Li J., Wu X., Ke J., Lee M., Lan Q., Li J., Yu J., Huang Y., Sun D.-Q., Xie R. TET1 dioxygenase is required for FOXA2-associated chromatin remodeling in pancreatic beta-cell differentiation. Nat. Commun. 2022;13:3907. doi: 10.1038/s41467-022-31611-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Eyres M., Lanfredini S., Xu H., Burns A., Blake A., Willenbrock F., Goldin R., Hughes D., Hughes S., Thapa A., et al. TET2 Drives 5hmc Marking of GATA6 and Epigenetically Defines Pancreatic Ductal Adenocarcinoma Transcriptional Subtypes. Gastroenterology. 2021;161:653–668.e16. doi: 10.1053/j.gastro.2021.04.044. [DOI] [PubMed] [Google Scholar]
- 47.Suzuki T., Furuhata E., Maeda S., Kishima M., Miyajima Y., Tanaka Y., Lim J., Nishimura H., Nakanishi Y., Shojima A., Suzuki H. GATA6 is predicted to regulate DNA methylation in an in vitro model of human hepatocyte differentiation. Commun. Biol. 2022;5:414. doi: 10.1038/s42003-022-03365-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lo E.K.W., Mears B.M., Maurer H.C., Idrizi A., Hansen K.D., Thompson E.D., Hruban R.H., Olive K.P., Feinberg A.P. Comprehensive DNA Methylation Analysis Indicates That Pancreatic Intraepithelial Neoplasia Lesions Are Acinar-Derived and Epigenetically Primed for Carcinogenesis. Cancer Res. 2023;83:1905–1916. doi: 10.1158/0008-5472.CAN-22-4052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Li F., He C., Yao H., Zhao Y., Ye X., Zhou S., Zou J., Li Y., Li J., Chen S., et al. Glutamate from nerve cells promotes perineural invasion in pancreatic cancer by regulating tumor glycolysis through HK2 mRNA-m6A modification. Pharmacol. Res. 2023;187 doi: 10.1016/j.phrs.2022.106555. [DOI] [PubMed] [Google Scholar]
- 50.Yin T., Wang C., Liu T., Zhao G., Zha Y., Yang M. Expression of Snail in pancreatic cancer promotes metastasis and chemoresistance. J. Surg. Res. 2007;141:196–203. doi: 10.1016/j.jss.2006.09.027. [DOI] [PubMed] [Google Scholar]
- 51.Liu M., Hancock S.E., Sultani G., Wilkins B.P., Ding E., Osborne B., Quek L.-E., Turner N. Snail-overexpression induces epithelial-mesenchymal transition and metabolic reprogramming in human pancreatic ductal adenocarcinoma and non-tumorigenic ductal cells. J. Clin. Med. 2019;8:822. doi: 10.3390/jcm8060822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Camolotto S.A., Belova V.K., Torre-Healy L., Vahrenkamp J.M., Berrett K.C., Conway H., Shea J., Stubben C., Moffitt R., Gertz J., et al. Reciprocal regulation of pancreatic ductal adenocarcinoma growth and molecular subtype by HNF4α and SIX1/4. Gut. 2021;70:900–914. doi: 10.1136/gutjnl-2020-321316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lee J.H., Kim H., Lee S.H., Ku J.L., Chun J.W., Seo H.Y., Kim S.C., Paik W.H., Ryu J.K., Lee S.K., et al. Establishment of patient-derived pancreatic cancer organoids from endoscopic ultrasound-guided fine-needle aspiration biopsies. Gut Liver. 2022;16:625–636. doi: 10.5009/gnl210166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Laufer B.I., Neier K., Valenzuela A.E., Yasui D.H., Schmidt R.J., Lein P.J., Lasalle J.M. Placenta and fetal brain share a neurodevelopmental disorder DNA methylation profile in a mouse model of prenatal PCB exposure. Cell Rep. 2022;38:110442. doi: 10.1016/j.celrep.2022.110442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J. 2011;17:5–7. doi: 10.14806/ej.17.1.200. [DOI] [Google Scholar]
- 56.Wingett S.W., Andrews S., Hamilton R., Donaldson I.J. FastQ Screen: A tool for multi-genome mapping and quality control. F1000Res. 2018;7:1338. doi: 10.12688/f1000research.15931.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Krueger F., Andrews S.R. Bismark: a flexible aligner and methylation caller for Bisulfite-Seq applications. Bioinformatics. 2011;27:1571–1572. doi: 10.1093/bioinformatics/btr167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Langmead B., Salzberg S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N., Marth G., Abecasis G., Durbin R. The sequence alignment/map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ewels P., Magnusson M., Lundin S., Käller M. Data and text mining MultiQC: summarize analysis results for multiple tools and samples in a single report. Bioinformatics. 2016;32:3047–3048. doi: 10.1093/bioinformatics/btw354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Korthauer K., Chakraborty S., Benjamini Y., Irizarry R.A. Detection and accurate false discovery rate control of differentially methylated regions from whole genome bisulfite sequencing. Biostatistics. 2019;20:367–383. doi: 10.1093/biostatistics/kxy007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hansen K.D., Langmead B., Irizarry R.A. BSmooth: from whole genome bisulfite sequencing reads to differentially methylated regions. Genome Biol. 2012;13:R83. doi: 10.1186/gb-2012-13-10-R83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Gu Z., Eils R., Schlesner M. Complex heatmaps reveal patterns and correlations in multidimensional genomic data. Bioinformatics. 2016;32:2847–2849. doi: 10.1093/bioinformatics/btw313. [DOI] [PubMed] [Google Scholar]
- 64.Chen E.Y., Tan C.M., Kou Y., Duan Q., Wang Z., Meirelles G.V., Clark N.R., Ma'ayan A. Enrichr: interactive and collaborative HTML5 gene list enrichment analysis tool. BMC Bioinf. 2013;14:128. doi: 10.1186/1471-2105-14-128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Heinz S., Benner C., Spann N., Bertolino E., Lin Y.C., Laslo P., Cheng J.X., Murre C., Singh H., Glass C.K. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol. Cell. 2010;38:576–589. doi: 10.1016/j.molcel.2010.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Sheffield N.C., Bock C. LOLA: Enrichment analysis for genomic region sets and regulatory elements in R and Bioconductor. Bioinformatics. 2016;32:587–589. doi: 10.1093/bioinformatics/btv612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kursa M.B., Rudnicki W.R. Feature selection with the boruta package. J. Stat. Softw. 2010;36:1–13. doi: 10.18637/jss.v036.i11. [DOI] [Google Scholar]
- 68.Das P., Roychowdhury A., Das S., Roychoudhury S., Tripathy S. sigFeature: Novel significant feature selection method for classification of gene expression data using support vector machine and t statistic. Front. Genet. 2020;11:247. doi: 10.3389/fgene.2020.00247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wickham H. 2016. Elegant Graphics for Data Analysis: Ggplot2. [Google Scholar]
- 70.Cavalcante R.G., Sartor M.A. Annotatr: Genomic regions in context. Bioinformatics. 2017;33:2381–2383. doi: 10.1093/bioinformatics/btx183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Yu G., Wang L.G., He Q.Y. ChIP seeker: An R/Bioconductor package for ChIP peak annotation, comparison and visualization. Bioinformatics. 2015;31:2382–2383. doi: 10.1093/bioinformatics/btv145. [DOI] [PubMed] [Google Scholar]
- 72.Ernst J., Kellis M. Chromatin-state discovery and genome annotation with ChromHMM. Nat. Protoc. 2017;12:2478–2492. doi: 10.1038/nprot.2017.124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ramírez F., Ryan D.P., Grüning B., Bhardwaj V., Kilpert F., Richter A.S., Heyne S., Dündar F., Manke T. deepTools2: a next generation web server for deep-sequencing data analysis. Nucleic Acids Res. 2016;44:W160–W165. doi: 10.1093/nar/gkw257. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(A) Sequencing alignment and DNA methylation extraction metrics for WGBS libraries. (B) Patient information for PDOs. (C) Average global CpG methylation for mouse organoids. (D) Average global CpG methylation for PDOs.
(A) DMR enrichment testing results. (B) Gene ontology enrichment results. (C) Known HOMER motif results.
(A) DMR enrichment testing results. (B) Gene ontology enrichment results. (C) Known HOMER motif results.
(A) DMR enrichment testing results. (B) Gene ontology enrichment results. (C) Known HOMER motif results
(A) DMR enrichment testing results. (B) Gene ontology enrichment results. (C) Known HOMER motif results.
(A) mN/mP versus mM DMRs. (B) mT versus mM DMRs. (C) early- versus late-stage DMRs. (D) hN versus hT DMRs.
(A) mN/mP versus mM DMRs. (B) mT versus mM DMRs. (C) early- versus late-stage DMRs. (D) hN versus hT DMRs.
(A) mT versus mM DMRs. (B) early- versus late-stage DMRs.
(A) DMR enrichment testing results. (B) Gene ontology enrichment results. (C) Known HOMER motif results.
Data Availability Statement
-
•
Raw and processed sequencing data has been deposited at GEO and is publicly available as of the date of publication under accession number GSE243528. The accession number is also listed in the key resources table.
-
•
All original code has been deposited at GitHub and is publicly available as of the date of publication at https://github.com/swang52/pdac_wgbs.git. The URL is also listed in the key resources table.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.