Abstract
The adenovirus gene regulatory program occurs in two distinct phases, as defined by the onset of DNA replication. During the early phase, the E1A, E1B, E2, E3, and E4 genes are maximally expressed, while the major late promoter (MLP) is minimally expressed and transcription is attenuated. After the onset of DNA replication, the IVa2 and pIX genes are expressed at high levels, transcription from the MLP is unattenuated and fully activated, and early gene expression is repressed. Although the cis elements and trans-acting factors responsible for the late-phase activation of the MLP have been identified and characterized and the role of DNA replication in activation has been established, the mechanism(s) underlying the commensurate decrease in early gene expression has yet to be elucidated. The results of this study demonstrate that this decrease depends on a fully functional MLP. Specifically, virus mutants with severely deficient transcription from the MLP exhibit a marked increase in expression of the E1A, E1B, and E2 early genes. These increases were observed at the level of transcription initiation, mRNA accumulation, and protein production. In addition, expression from the late gene pIX, which is not contained within the major late transcription unit (MLTU), is also markedly increased. To begin the analysis of the mechanisms underlying these late-phase effects, mixed-infection experiments with mutant and wild-type viruses were performed. The results show that the effects on early gene expression, as measured both at the protein and RNA levels, are mediated in trans and not in cis. These observations are consistent either with a model in which one or more late protein products encoded by the MLTU acts as a repressor of early gene expression or with one in which the wild-type MLP competes with early promoters for limiting transcription factors.
Extensive studies of adenovirus gene regulation have uncovered the complexity of the gene expression program during the course of infection. Not only are adenovirus genes regulated in a temporal manner, but viral gene products also exert control at every step in the gene expression pathway, from transcription initiation to protein modification. Both early and late gene products are involved in transcriptional activation (E1A, E2A, E4, IX, and IVa2 [10, 21, 24, 35, 36, 44, 55, 59]), various stages of RNA processing (E1B, E2, and E4 [1, 2, 6, 25, 42, 43, 45, 51]), and translational efficiency (L4 100-kDa [100K] protein [22]). In addition, VA RNA, a short (157-nucleotide) RNA polymerase III transcript, also has a regulatory role, as it blocks global shutdown of protein synthesis in the host cell by interfering with the inhibitory action of DAI kinase (37). cis-acting elements also have a wide range of effects beyond the binding of transcription factors, as they are involved in determination of splice site choice (29), polyadenylation site choice (20, 46–48), and mRNA translational efficiency (15).
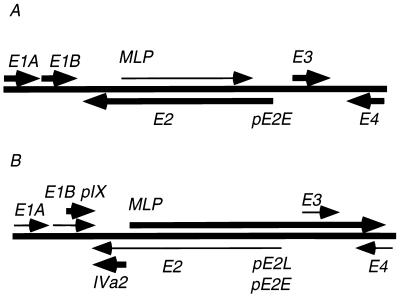
Within this diversity of levels and mechanisms of gene expression control, the most prominent regulatory event in the adenovirus life cycle is the early-to-late switch in infection (Fig. 1). At early times (from the time of adsorption to about 6 h postinfection [p.i.]), the early genes E1A, E1B, E2, E3, and E4, all of which are activated by the immediate early gene E1A (4, 26), are maximally expressed, and the major late transcription unit (MLTU) is expressed at very low levels and is attenuated (41, 53). At the onset of viral DNA replication (∼6 h p.i.), a marked switch in gene expression occurs. At this time, transcription is activated at three different transcription units: IVa2, located at 16 map units (m.u.), pIX, located at 10 m.u., and the MLTU, located at 17 m.u. The activation of late genes is accompanied by a reduction in the expression of early genes, as measured per genome (reviewed in references 39 and 52). Whether this reduction is a passive consequence of high levels of gene activity from the late promoters or is an active repression has not been conclusively determined.
FIG. 1.
Transcriptional map of adenovirus, depicting the early-to-late switch in infection. (A) At early times (0 to 6 h p.i.), transcription of early genes E1A to E4 is at a maximum, as represented by heavy arrows, and MLP transcription is at a minimum, as represented by a lighter arrow, and attenuated. (B) At later times (after the onset of DNA replication at about 6 h p.i.), MLP transcription is fully activated and unattenuated and early gene expression per genome is reduced. Late genes pIX and IVa2 are also expressed at high levels.
Studies of late gene expression from the major late promoter (MLP) have implicated both trans-acting factors and cis effects in the late-phase-specific activation of the MLP. Not only has it been known for many years that the E1A 289R protein can transactivate the MLP (38), but recent data show that the late-phase-specific protein IVa2, expressed from a gene lying outside the MLTU, up-regulates the MLP through binding to the DEF sites downstream of the initiation site (30, 32, 35, 59). The cis-acting elements required for maximum MLP activation have also been identified and extensively studied both in vitro and in vivo (7, 33, 34, 49, 50). In addition, the studies of Thomas and Mathews (56) have demonstrated that the initiation, but not the maintenance, of DNA replication is mandatory for the activation of the MLP during a viral infection.
Although it was soon recognized that the late phase of infection is accompanied by a general down-regulation of early gene expression, initial studies of the mechanism(s) underlying control of early gene expression were conducted under conditions in which entry into the late phase was inhibited. The primary tools employed were inhibitors of DNA replication (5, 11, 12, 31, 40), inhibitors of protein synthesis (5, 11, 12, 27, 31, 40), and a temperature-sensitive mutant in the single-stranded DNA binding protein (DBP) (1, 5, 8), which is essential for viral DNA replication (16). The results of these various studies indicated that lowered expression of some early genes occurs prior to the onset of the late phase of infection and therefore is not contingent upon entry into the late phase. Furthermore, they suggested that a balance of protein factors was involved in both the repression and activation of early genes, implying a complex network of regulatory interactions during the early phase of infection. Studies with H5ts125, a virus with a temperature-sensitive lesion in DBP, suggested a role of DBP in the destabilization of early mRNAs (1) and indicated that a block in DNA replication, especially when coupled with a block in protein synthesis, allowed for an enhanced accumulation of early viral RNAs (5). Taking all of these previous results together, we can conclude that some form of down-regulation of early gene expression does indeed take place during adenovirus infection, but significant questions concerning the mechanism remain, particularly concerning any late-phase-specific repression of early gene expression.
Previous work from our laboratory has examined the structure and function of the MLP in the context of the complete viral genome by the creation of mutations in known or suspected transcription elements (34, 49, 50). Several of these mutant viruses were deficient in viral growth and had a commensurate decrease in late RNA and protein due to the expected decrease in MLP activity. It was also apparent that there were other contingent effects on at least one early gene, namely E1B, and alterations to protein accumulation over and above those expected from the lowered expression of late gene products. In this study, we have used these mutants to demonstrate that the expression patterns of all early genes and of one late gene, pIX, are significantly altered. Cells infected with MLP mutant viruses overexpress the early genes E1A, E1B, and E2 and the late gene pIX, they have quantitative and qualitative changes in expression of the E4 gene, and the late-phase-specific increase in expression of E3 is not observed. These results establish that there are mechanisms of early gene control that are dependent upon entry into the late phase and that the previously documented down-regulation observed at late times is not a mere temporal coincidence. Furthermore, mixed-infection experiments provide evidence for a trans-acting mechanism of repression of E1A, E1B, E2, and pIX. In the Discussion we consider possible models and mutagenic strategies to explain the trans-acting nature of early gene regulation.
MATERIALS AND METHODS
Cell culture and viral infection methods.
All analyses were conducted with monolayer cultures of A549 human lung carcinoma cells (19). The cells were grown in Dulbecco modified Eagle medium with 10% supplemental calf serum (HyClone, Logan, Utah) and antibiotics, as described previously (60). Cells to be infected were plated in culture dishes of various sizes 1 or 2 days prior to the addition of virus and were used when the monolayers were confluent. Cells were infected at the multiplicities of infection (MOI) indicated for the individual experiments.
Labeling of infected cell proteins.
A549 cell monolayers in 35-mm-diameter dishes were infected with different virus strains at the indicated MOI. After various incubation periods, the original medium was removed and replaced with methionine-free medium (Gibco 11970-027) and then incubated further for 1 h. Following depletion of the intracellular methionine pools, the medium was replaced with fresh methionine-free medium containing 10 to 100 μCi of a mixture of [35S]methionine and [35S]cysteine (Dupont NEN Express NEG-072). After a 1-h incubation, cells were washed once with 2.5 ml of phosphate-buffered saline (PBS) and then resuspended in 1 ml of PBS. Samples were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and autoradiography as described below.
SDS-PAGE.
SDS-acrylamide gels for protein electrophoresis were formed by using premade Protogel solutions (National Diagnostics, Atlanta, Ga.). The separating gel contained a 12% solution of acrylamide–bis-acrylamide at a ratio of 37.5:1. Protein samples were mixed with an equal amount of 2× loading buffer (1.51% Tris, 20% glycerol, 4% SDS, 10% 2-mercaptoethanol, and 0.002% bromphenol blue [BPB]) and boiled for 2 min before loading. Samples were electrophoresed for approximately 3 h, until the BPB dye front reached the bottom of the separating gel. Gels were then stained for 30 min with Coomassie brilliant blue (0.25% in fixing solution of 40% methanol and 7% glacial acetic acid) and destained for at least 2 h in fixing solution. Gels used for the resolution of radiolabelled proteins were dried by a gel drier prior to exposure to autoradiography film (Fuji-RX).
Western blotting.
A549 cells in 35-mm-diameter dishes were infected with TATA0 or wild-type MLP (MLP-WT) virus. At the indicated times p.i., infected cells were washed once in cold PBS and then harvested by scraping into 1 ml of cold PBS. Cell resuspensions (0.1 ml) were electrophoresed and stained as described (see “SDS-PAGE” above) and then equilibrated in transfer buffer (39 mM glycine, 48 mM Tris, 0.037% SDS, 20% methanol) for 60 min. Proteins were transferred for 2 h to a nitrocellulose filter by using a transfer apparatus set at 30 mA. The filter was then blocked in blocking solution (5% nonfat dry milk in PBS) for 90 min before primary antibody exposure. The primary antibody was an anti-E2A monoclonal antibody (courtesy of D. F. Klessig), which was diluted 1:1,000 in blocking solution. Filters were incubated for 90 min at room temperature in 20 ml of diluted antibody. The filter was washed once in TBST (10 mM Tris-HCl [pH 8.0], 150 mM NaCl, 0.05% Tween 20) and three times in PBS. Horseradish peroxidase-conjugated goat-generated anti-mouse immunoglobulin A (IgA), IgG, and IgM (Sigma A 0412) antiserum was used as secondary antibody. Filters were incubated for 30 to 60 min in 15 ml of a 1:600 dilution of this antibody in blocking solution. The filters were then washed once in TBST and three times in PBS. Blots were developed by chemiluminescence with the Dupont Renaissance kit (Dupont NEL-100) and exposed to autoradiography film.
Northern blot analysis.
Total cell RNA was isolated by the RNA STAT-60 protocol (Tel-Test “B,” Inc.). Methods for Northern blot analysis were based on a published protocol. Total infected cell RNA (5 μg) was denatured at 55°C for 15 min in denaturation solution (50% formamide, 18% 12.3 M [37%, wt/wt] formaldehyde, 40 mM MOPS [morpholinepropanesulfonic acid] [pH 7.0], 10 mM sodium acetate, 1 mM EDTA). RNA loading buffer (10 μl of a solution containing 1 mM EDTA [pH 8.0], 0.25% BPB, 0.25% xylene cyanol, and 50% glycerol) and 1 μl of ethidium bromide (10 mg/ml) were added to each sample prior to electrophoresis. Samples were loaded onto a formaldehyde-agarose gel (1% agarose, 18% 12.3 M [37%, wt/wt] formaldehyde, 40 mM MOPS [pH 7.0], 10 mM sodium acetate, 1 mM EDTA) and electrophoresed for approximately 3 h (30 mA, 80V) in a buffer containing 40 mM MOPS (pH 7.0), 10 mM sodium acetate, and 1 mM EDTA. Gels were washed for 15 min in deionized H2O and equilibrated for 45 min in 10× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate). RNA was transferred via capillary action overnight to a Genescreen Plus (Dupont NEN) filter in 10× SSC. Filters were then either baked for 2 h under vacuum at 80°C or UV cross-linked to immobilize transferred nucleic acids. Filters were blocked in hybridization solution (50% formamide, 4× Denhardt’s solution, 2% SDS, 0.1 mg of salmon sperm DNA per ml, 750 mM NaCl, 150 mM Tris [pH 7.0], 18 mM NaH2PO4, 28 mM Na2HPO4) overnight at 42°C. Samples were then hybridized overnight at 42°C to a probe prepared by random priming of the indicated gel-purified restriction fragment. Filters were then washed twice at 25°C in 2× SSC, once at 42°C in 0.1× SSC–0.5% SDS, and once at 42°C in 0.1× SSC. Filters were exposed and analyzed by autoradiography and quantitated by either a densitometer or a PhosphorImager.
Nuclear run-on analysis.
Nuclear run-on assays were performed essentially as described previously (34). Between 2 × 107 and 6 × 107 cells were infected with TATA0 or MLP-WT virus at an MOI of 10. At 22 h p.i., cells were washed twice with ice-cold PBS and scraped into 4 ml of PBS. Cells were pelleted at 1,500 rpm, and PBS was removed. Cells were vortexed briefly and resuspended during vortexing in 4 ml of Nonidet P-40 (NP-40) lysis buffer (10 mM Tris-HCl [pH 7.4], 10 mM NaCl, 3 mM MgCl2, 0.5% NP-40). Resuspended cells were incubated for 5 min on ice to allow lysis for isolation of intact nuclei. Nuclei were then pelleted at 1,500 rpm. The cytoplasmic supernatant was removed, and nuclei were resuspended in NP-40 lysis buffer and pelleted again. Nuclear pellets were then resuspended in 200 μl of glycerol storage buffer (50 mM Tris-HCl [pH 8.3], 40% glycerol, 5 mM MgCl2, 0.1 mM EDTA) before the transcription reaction. An equal volume of transcription buffer (10 mM Tris-HCl [pH 8], 5 mM MgCl2, 0.3 M KCl, 1 mM ATP, 1 mM CTP, 1 mM GTP, 10 mM dithiothreitol, 100 μCi of [α-32P]UTP) was added to resuspended nuclei for a 30-min incubation at 30°C. RNase-free DNase (RQ1; Promega) was then added to the transcription reaction mix in 0.6 ml of HSB buffer (0.5 mM NaCl, 50 mM MgCl2, 2 mM CaCl2, 0.1 mM EDTA), which was then incubated for 5 min at 30°C. Proteinase K (200 μg at a concentration of 5μg/μl) was then added, and the mixture was incubated for 20 min at 42°C. The reaction product was phenol extracted and precipitated twice with isopropanol. The pellet was washed in 75% ethanol and resuspended in 100 μl of diethyl pyrocarbonate (DEPC)-treated water. Samples were counted with a scintillation counter, and the amounts used for hybridization are indicated in the figure legends. Samples were added to prehybridized filters (see below), and hybridization was for 24 to 48 h at 42°C.
Preparation of filters for nuclear run-on analysis.
Filters were prepared with 10 to 20 μg of either single-stranded M13 or double-stranded pBluescript DNA containing the appropriate cloned sequence. The DNAs, in a volume of 50 μl, were added to 50 μl of 20× SSC and transferred to a Genescreen Plus filter prewetted in 10× SSC by using a slot blot manifold. Filters were UV cross-linked and prehybridized (see “Northern blot analysis” above).
Probes for nuclear run-on analysis.
The probes used for the nuclear run-on assays were as follows: E1A, bp 1 to 1342 cloned into M13mp19 or pBluescript SK(+); anti-E1A, bp 1342 to 1 cloned into M13mp18; E1B, bp 2051 to 3331 cloned into pBluescript SK(+); E1B + pIX, bp 3328 to 3788 cloned into pSP6; MLP or E2B, bp 11555 to 13636 cloned into M13mp18 in either orientation; GAPDH (single strand), 1.3-kb PstI fragment of rat GAPDH cDNA cloned from pBS-KS(−) (17) into M13mp19; GAPDH (double strand), 1.4-kb EcoRI fragment of human GAPDH cDNA cloned into pGEM, courtesy of Kartik Krishnan (13).
RESULTS
MLP-deficient viruses overexpress the E2A protein late in infection.
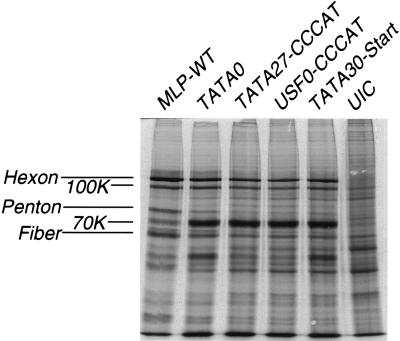
Previous studies have shown that viruses containing certain mutations in the promoter elements of the MLP exhibit a marked replication deficiency (34, 49, 50). In some mutants, this deficiency has been attributed to lowered RNA expression from the MLP, but direct measurements of late protein accumulation were not reported. To examine the protein profile, cells were infected with a set of mutant viruses containing different mutations in the MLP, all of which had significant transcriptional deficiencies. Cells were pulse labelled with [35S]methionine at 24 h p.i. and analyzed by SDS-PAGE. As shown in Fig. 2, all cells infected with viruses with MLP deficiencies demonstrate, as expected, a significant decrease in the synthesis of the major late proteins hexon, 100K, fiber, and penton base. In addition, a prominent band migrating at about 70 kDa is considerably elevated in all of the mutant-infected cell extracts. Thus, the protein phenotype of the deficient mutants is consistent regardless of the specific combination of MLP mutations. All mutant-infected cells synthesize both lowered levels of late proteins and elevated levels of the 70-kDa species.
FIG. 2.
Late protein synthesis in cells infected with a set of MLP mutant viruses and the MLP-WT virus. At 24 h p.i., cell proteins were labeled for 1 h and cell extracts were harvested, separated by SDS-PAGE, and prepared for autoradiography, as described in Materials in Methods. UIC, uninfected cell control.
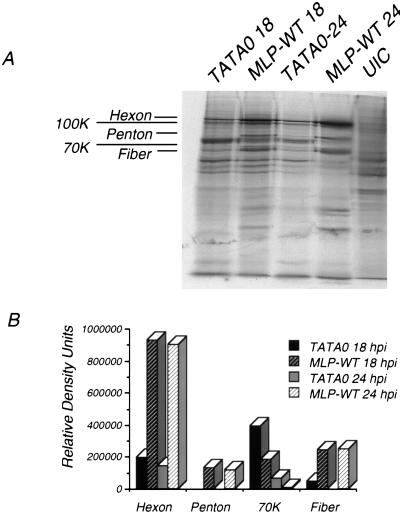
To examine the protein phenotype further, one of the mutants was chosen as a representative of the deficient set. Mutant TATA0 contains two mutations within the TATA box, is significantly deficient in replication, and reverts much less frequently to a wild-type phenotype than does TATA27::CCCAT (37a). As before, proteins in TATA0- and wild type-infected cells were analyzed by SDS-PAGE at 18 and 24 h p.i. (Fig. 3A). Quantitation of individual protein bands (Fig. 3B) revealed that the synthesis of hexon was reduced about fivefold at 18 and 24 h p.i. in the mutant-infected cells, compared with the wild type-infected cells. Synthesis of fiber was reduced by at least fourfold at 18 h p.i. and to nearly background levels at 24 h p.i., while measurements of penton base were indistinguishable from the background level. The 70-kDa protein was increased by 2.5-fold (18 h) and 8-fold (24 h) in TATA0-infected cells. (Fig. 2B; Table 1). Quantitation of other infections showed similar reductions in late proteins and increases in the 70-kDa protein (data not shown).
FIG. 3.
Late protein synthesis in TATA0- and MLP-WT-infected cells. At 18 or 24 h p.i., cells were labeled and cell extracts were harvested as described in Materials and Methods. (A) Autoradiogram of labeled proteins separated by SDS-PAGE. (B) Quantitation of relative band intensities, as determined with a PhosphorImager.
TABLE 1.
Summary of changes in gene expression in cells infected with TATA0, compared to MLP-WT
| Gene | Assay of levels of: | TATA0 infection
|
|
|---|---|---|---|
| Time p.i. (h) | Change in expression vs MLP-WT | ||
| E1A | mRNA (steady state) | 24 | 3× increase |
| Transcription | 22 | 3× increase | |
| E1B | mRNA (steady state) | 12 | Not detectable in MLP-WT |
| Transcription | 22 | 3× increase | |
| E2A | mRNA (steady state) | 12 | 4× increase |
| Protein (steady state) | 12 | 5× increase | |
| 18 | 5× increase | ||
| 24 | 15× increase | ||
| Protein synthesis | 18 | 2.5× increase | |
| 24 | 8× increase | ||
| E2B | Transcription | 22 | 3× increase |
| pIX | mRNA (steady state) | 12 | Not detectable in MLP-WT |
| Transcription | 22 | 3× increase | |
| E3 | mRNA (steady state) | 24 | 7–8× increase in MLP-WT |
| E4 | RNA splicing pattern | 12–24 | Severely altered |
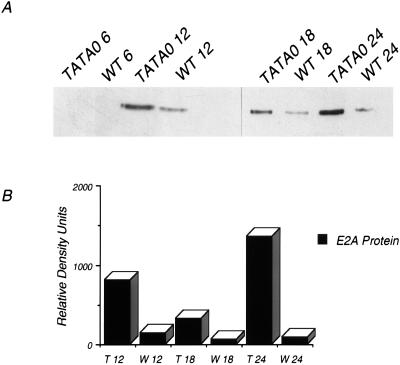
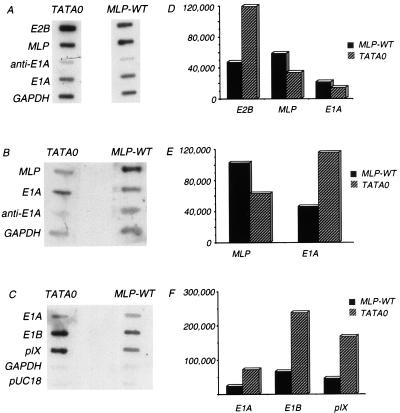
The identity of the 70-kDa protein was unknown, but several possible viral and cellular candidates were considered. Because it was expressed in both wild type- and mutant-infected cells, a viral protein was suspected, and its relative mobility suggested that it might be the single-stranded DNA binding protein DBP. Western blot analysis was performed to test this (Fig. 4A). At 12, 18, and 24 h p.i., levels of DBP in TATA0-infected cells were elevated between 5 and 15-fold compared with the wild type (Fig. 4B; Table 1). This corresponds to the increase in synthesis of the protein of similar relative mobility shown in Fig. 3A. Western blot analysis established that there is a significant increase in DBP expression at late times in mutant-infected cells and suggests that lowered MLP expression is accompanied by increased early gene 2 expression. Coupled with the previous observation that E1B RNA levels are elevated in MLP mutant-infected cells (50), this result suggests that expression of the MLP and of at least some early genes are inversely correlated.
FIG. 4.
E2A accumulation in TATA0- and MLP-WT-infected cells. At the indicated times p.i. (in hours), cell extracts were harvested and cell proteins were resolved via SDS-PAGE. (A) Western transfer and antibody detection of E2A. (B) Relative levels of E2A, as determined by densitometry.
Analysis of RNA levels in MLP mutant- and wild type-infected cells.
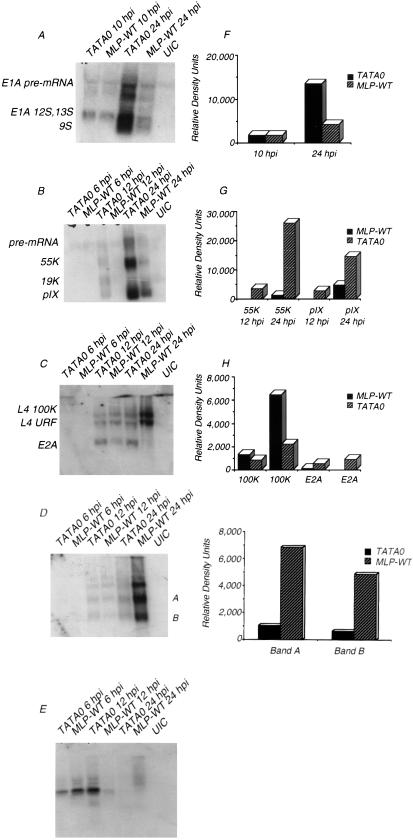
The results described above and previously suggest that decreases in MLP expression lead to corresponding increases in the expression of at least two early genes. It was of some interest to see if these effects could be extended to other early and late genes and to begin the analysis of the level at which these effects occur. Accordingly, a survey of quantitative changes to steady-state viral mRNAs was conducted by using Northern blot analysis. Probes capable of hybridizing to E1A, E1B, E2A, E3, and E4, as well as late gene pIX, were used in the analysis (Fig. 5; Table 1). At the indicated times both early and late in infection, total RNA was isolated from cells infected with MLP-WT or TATA0. Levels of RNA from several genes were overexpressed in TATA0-infected cells. E1A mRNA levels were equivalent at 10 h p.i. but at 24 h p.i. were overexpressed threefold by TATA0 (Fig. 5A and F; Table 1). At 12 h p.i., E1B and pIX mRNA was detected in cells infected with TATA0 but not with MLP-WT; at 24 h p.i., E1B 55K levels were 10 times higher and pIX levels were 4 times higher in TATA0-infected cells (Fig. 5B and G; Table 1). L4 mRNA accumulated to levels three times higher in MLP-WT-infected cells by 24 h p.i., and E2A levels were four times higher in TATA0-infected cells at 12 h p.i.; at 24 h p.i., E2A mRNA was detected in TATA0- but not in MLP-WT-infected cells (Fig. 5C and H; Table 1). A different effect was observed with E3 expression: at 12 h p.i., there was little difference in E3 region mRNA; at 24 h p.i., however, there was a greater increase in message levels in the MLP-WT-infected cells than in those infected with TATA0 (Fig. 5D and I; Table 1). Although the exact identification of the three prominent species of RNA is difficult since the E3 region of this virus is highly recombinant (7), we infer that the two lower bands (labeled A and B in Fig. 5D) are E3 species rather than MLP-specific RNAs, because on a longer exposure these bands are visible at 6 h p.i., before full MLP activation (data not shown). Nevertheless, the relative increase in expression of E3, compared with E1A, E1B, E2A, and pIX RNAs in wild-type as opposed to mutant infection, may reflect the fact that some E3 species are expressed from the MLP late in infection, a phenomenon previously demonstrated for the E3 11.6K gene (58). Thus, the increase in the wild-type infection may reflect the greater transcriptional abilities of the wild-type MLP. Northern analysis of the E4 region (Fig. 5E) showed yet a different change in gene expression due to reduced MLP activity: the use of a probe complementary to the 3′ end of all E4 messages revealed both qualitative and quantitative changes in the accumulation of E4 RNA species. This complex pattern of changes was not explored further.
FIG. 5.
Early and late message accumulation in TATA0- and MLP-WT-infected cells. At the indicated times p.i., total RNA was harvested and Northern blot analyses of early and late messages were performed as described in Materials and Methods. The blots were probed with the following random-primed double-stranded DNA sequences: bp 342 to 1342 (E1A) (A), bp 3328 to 3788 (E1B, pIX) (B), bp 23039 to 23912 (E2A, L4 100K, L4 URF) (C), bp 28879 to 29602 (E3) (D), and bp 31920 to 32423 (E4) (E). The corresponding relative levels of mRNA species (F to I) were determined by densitometry and corrected for total RNA levels by reprobing of stripped blots with either actin or GAPDH. The precise sequence identities of bands labeled A and B in panel D are unknown. The bands in panel E were not identified as to specific open reading frames and are thus left unlabeled.
With the results from the Northern analyses taken together, it is clear that changes to expression from the MLP are accompanied by profound changes in expression from many if not all of the other genes encoded in the adenovirus genome. One possibility arising from these results is that the MLP and/or the products of the MLTU play roles in the regulation of most other adenovirus genes at late times in infection. Methods to determine whether this role is active or merely contingent will be discussed later.
Comparison of relative early and late promoter strengths in TATA0- and MLP-WT-infected cells.
The results described above show that there are significant changes in the relative abundance of viral RNA species in cells infected with a virus having a severely deficient MLP. To determine the level of gene expression at which these changes are exerted, it is necessary to examine the first step, namely transcription initiation. This was examined by nuclear run-on experiments using nuclei harvested at 22 h p.i. Radiolabelled de novo transcripts were hybridized to DNA probes on membranes, and levels of hybridization were quantified. MLP promoter activity was reduced in TATA0-infected cells (Fig. 6A and B; Table 1) to some 60% of the wild-type level in the two experiments shown. In contrast, the relative rates of E1B, pIX, and E2B transcription were elevated about threefold (Fig. 6; Table 1). The relative rate of E1A transcription was also elevated some threefold in TATA0-infected cells in two of the three experiments represented by Fig. 6; the small decrease observed in one experiment (Fig. 6A and D) was not seen in two other experiments and is probably a loading artifact.
FIG. 6.
Analysis of relative promoter strengths in TATA0- and MLP-WT-infected cells. At 22 h p.i., nuclei were harvested and transcription reactions were performed in vitro in the presence of 100 μCi of [α-32P]UTP. De novo transcripts were isolated and hybridized to filters containing unlabeled DNA probes, as indicated to the left of panels A to C. Filters were washed and then exposed by autoradiography: 107 cpm were hybridized to single-stranded M13 DNA probes (A), 106 cpm were hybridized to single-stranded DNA probes (B), and 106 cpm were hybridized to double-stranded DNA probes (C). (D to F) Quantitation of the counts hybridized to each of the probes shown in panels A to C. The values were normalized as follows. First, background values (anti-E1A in panel A and pUC18 in panel C) were subtracted from each of the experimental values. Background values were not subtracted for the data in panel B because this had the effect of making the decrease in activity of the MLP and the increase in E1A activity from TATA0-infected nuclei anomolously high compared with all other experiments. After subtraction of background, the corrected values were normalized to the hybridization to GAPDH.
Despite the difficulties inherent in interpreting small changes in transcription initiation, we conclude from these results that the increased steady-state RNA levels observed in the Northern analysis (Fig. 5) are correlated with increased levels of initiation.
Complementation of the overexpression of early genes in MLP mutant-infected cells by wild-type coinfection.
The results presented above suggest that lowered expression from the MLP at late times in infection is correlated with an increased expression of other viral genes at the level of transcription initiation. These changes in expression could be exerted either in cis or in trans, and which of these two alternatives is correct has mechanistic implications. For example, a finding that changes in expression are mediated in cis would exclude mechanisms involving diffusible regulatory factors. On the other hand, a finding that the increases in early gene expression occur by a trans-acting mechanism would exclude such models as the occlusion of early promoters by active transcription from the MLP.
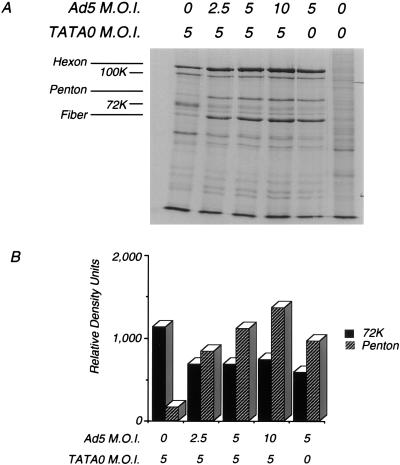
To address the issue of whether or not changes in gene expression are mediated in cis or in trans, cells were coinfected with wild-type adenovirus type 5 (Ad5-WT) and TATA0 at various input multiplicities, and measurements were made of representative viral proteins and steady-state RNA levels. In the first experiment, cells were infected with a constant multiplicity of TATA0 virus and with increasing amounts of Ad5-WT; at 24 h p.i., proteins were labeled for 1 h. Infected cell proteins were analyzed by SDS-PAGE. As shown in Fig. 7, the relative level of DBP synthesized in the TATA0-infected cells was reduced to that of the wild type upon coinfection with wild-type virus, and the relative level of a representative late protein, penton base, was increased to wild-type levels. This experiment suggests that the changes to early gene expression observed in TATA0-infected cells are mediated in trans. A similar experiment was performed with TATA0 and MLP-WT, and essentially identical results were obtained (data not shown).
FIG. 7.
E2A protein synthesis in TATA0-infected cells coinfected with Ad5-WT. Cells were infected with the indicated MOI of TATA0 and Ad5-WT; at 24 h p.i., cells were labeled with 100 μCi of [35S]methionine for 1 h and cell extracts were prepared. (A) Autoradiogram of the labeled proteins separated by SDS-PAGE. (B) Quantitation of relative intensities of the E2A and penton base proteins determined by densitometry.
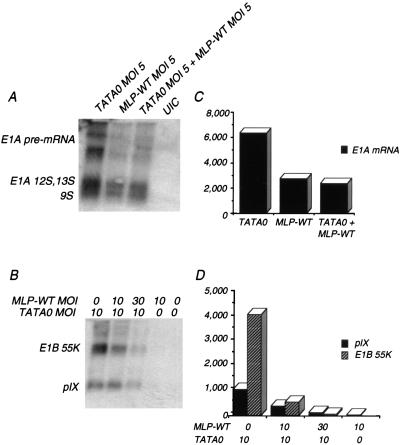
To extend these observations to other early genes and the pIX gene, we measured the steady-state levels of RNA in coinfected cells. Total cell RNA was harvested and Northern blot analysis was performed with probes complementary to E1A (Fig. 8A and C) or E1B and pIX (Fig. 8B and D). The results at the RNA level were similar to those observed at the protein level. Upon coinfection with an equivalent multiplicity of MLP-WT, E1A mRNA levels were reduced to wild-type levels. In TATA0-infected cells, E1B and pIX mRNA levels showed a dose-dependent reduction of mRNA to near wild-type levels upon coinfection with MLP-WT. These results demonstrate a global trans effect on changes in gene expression in MLP mutant-infected cells and will help us in further defining the mechanism(s) underlying these changes.
FIG. 8.
Early and late mRNA accumulation in infected cells coinfected with TATA0 and MLP-WT. Cells were infected with the indicated MOI of TATA0 and MLP-WT. At 20 h p.i. (A) or 24 h p.i. (B), total RNA was harvested from infected cells and analyzed by Northern blot hybridization. The random-primed double-stranded DNA probes were as follows: bp 906 to 1342 (E1A) (A); bp 3328 to 3788 (E1B, pIX) (B). (C and D) Quantitation of relative band intensities, as determined by PhosphorImager, corrected for loading of RNA by reprobing of stripped blots with a GAPDH probe.
DISCUSSION
Gene expression in adenovirus infection undergoes major quantitative and temporal changes during the viral replicative cycle. The gene regulation underlying these changes is comprised of an extensive network of gene products and cis-acting sequences, as summarized in Table 2. The most prominent temporal change in gene expression is the early-to-late switch in infection punctuated by the commencement of viral DNA replication. Although the phenomenon of the early-to-late switch, i.e., full activation of late genes with a corresponding decrease in early gene expression, has been described, the mechanistic forces involved in facilitating this change have yet to be demonstrated. It has been shown that full activation of the MLP is not only boosted by E1A and IVa2 but that there is also a requirement for the initiation, but not the maintenance, of DNA replication for full MLP activation (56). In addition, the activation and repression of early gene expression was shown to be mediated by a balance of labile protein activators and repressors (5, 11, 12, 27, 31, 40, 54).
TABLE 2.
Levels at which gene expression is regulated by adenovirus and factors involved in regulation
| Level of expression | Factor(s) involved |
|---|---|
| Transcriptiona | |
| Initiation | E1A, IVa2, E2A, E4 |
| Elongation | cis-acting sites |
| RNA processingb | |
| Splice site choice | cis-acting sites |
| mRNA transport | E1B, E4 |
| mRNA stability | E2A |
| Poly(A) site choice | cis-acting sites |
| Translationc | |
| Initiation | VA RNA |
| Efficiency | Tripartite leader, 100K |
Previous experiments in our laboratory have defined the cis-acting elements necessary for full activation of the MLP in the correct genomic context. As well as providing genetic evidence for the importance of specific promoter elements to the function of the MLP, viruses with mutant MLPs also can be used to explore the phenomenon of the reduction of early gene expression late in infection and to begin to answer questions as to the mechanism(s) underlying this poorly understood phenomenon. Using TATA0, an MLP mutant virus harboring a double mutation in the TATA box, we show that expression of the early genes E1A, E1B, and E2 and the late gene pIX is inversely correlated with expression from the MLP, as the former genes are overexpressed by the MLP mutant virus. The increases in expression of these genes ranged from some 3-fold to as much as 15-fold, depending on the specific gene and the level at which the measurement was made (Table 1). It should be emphasized that although the magnitude of increase varied with the level at which gene expression was measured (transcription initiation, steady-state level of RNA, protein synthesis, or protein accumulation), all measurements showed an increase. While subtle differences in the precise mechanisms underlying the increases for expression of specific genes cannot be ruled out, the uniformity of the trend suggests a single mechanism for the increases.
There were two exceptions to the general trend of increased expression of early genes and the late gene pIX in MLP-deficient viruses. Measurement of steady-state E3 RNA levels showed a marked increase in MLP-WT-infected cells compared with those infected with TATA0 (Fig. 5D and I). Although we have not identified the species expressed from the E3 region of this pair of viruses, the viral genomes are both derived from Ad2+ND1, an adenovirus type 2–simian virus 40 chimera (28). It is possible that some or all of the species are derived from spliced transcripts expressed from the MLP at late times in infection, as is the case for the 11.6-kDa protein, the adenovirus “death protein” ADP (57). If this interpretation is correct, then the increase in E3 species in the MLP-WT infection is precisely as predicted. The other exception is the anomalous behavior of RNAs expressed from E4 (Fig. 5E). In TATA0-infected cells, a transient increase in E4 species at 12 h p.i. is followed by virtual disappearance of species hybridizing to a 3′ probe. Similar results have been obtained in several other experiments (data not shown). It has been known for some time that the E4 transcription unit is subject to complex temporal regulation at the level of splicing (14). How lowered expression from the MLP could lead to the results shown in Fig. 5E is not known, although competition for limiting splicing machinery components might contribute to the observed phenotype.
The results discussed above show that lowered expression from the MLP at late times in infection leads to significant increases in expression from some early and late transcription units. These increases can be traced in part to increases in transcription initiation (Fig. 6) and raise the question as to the mechanisms(s) underlying these changes in gene expression. Given that four transcription units are affected, it seems likely that the mechanism is general rather than promoter specific. Hypotheses concerning mechanism would be clarified by knowledge of whether the mechanism operates in cis, in which case the increases in gene expression would not be affected by coinfection with the wild-type virus, or in trans, in which case the increases would be reversed by such coinfection. The results of such coinfections are presented in Fig. 7 and strongly support the idea that the mechanism operates in trans. Coinfection with the wild-type virus leads to a reduction in the levels of increase of 72K DBP and E1A, E1B, and pIX RNAs in mutant-infected cells. This result rules out simple cis-acting mechanisms, such as template-specific effects both upstream and downstream of the MLP.
The genetic results shown in Fig. 7 suggest some trans-mediated mechanism for the lowered levels of early gene expression in the wild type-infected cells. Furthermore, we can hypothesize that the lowered levels of such expression are important for the efficient completion of the viral replicative cycle. Two types of general models may account for these effects. “Passive” models include a promoter competition mechanism, in which the MLP competes with other promoters for rate-limiting components of the transcriptional machinery. For example, it is known that both the E2E and E2L promoters contain nonconsensus TATA box sequences (reviewed in reference 52). If the nonconsensus TACAAA sequence of the E2L promoter is altered to the consensus TATAAA, there is a large increase in in vitro promoter activity (23). Perhaps there is a biological advantage to maintaining a nonconsensus TATA box in the E2 promoters, because they may fail to compete with the MLP, which contains a consensus TATA box, at late times in infection. While a simple promoter competition model could account for the results with the E2 promoters, and perhaps the simple E1B and pIX promoters, it is hard to envisage such a mechanism operating at the E1A promoter, because it contains a strong enhancer element (3). Genetic experiments to test the promoter competition model at the E2L promoter are underway. The alternative “active” models propose that the MLTU encodes a diffusible factor that down-regulates other promoters. Possible candidates could include nonstructural proteins such as the L1 52,55K protein, the L3 23K protease, or the L4 100K or 33K protein. Extensive phenotypic characterization of temperature-sensitive mutants of the first three proteins has not included a detailed analysis of transcriptional effects, and this is currently under investigation. There are no reports of mutations in the 33K protein, a conserved phosphoprotein of unknown function (9) which accumulates in the nucleus at late times in infection (18). Recently, we created a virus in which the function of the 33K gene is mutated; although this virus has a pronounced growth defect, there are no detectable changes in early gene expression (16a). Thus, it is not a candidate for a putative MLTU-encoded repressor.
ACKNOWLEDGMENTS
This work was supported by grant R01 GM49070 from the NIGMS and by a core grant from the NCI to the Columbia Comprehensive Cancer Center (CA13696).
We thank Pat Munz for valuable technical assistance and the members of the virology group for discussions and advice.
REFERENCES
- 1.Babich A, Nevins J R. The stability of early adenovirus mRNA is controlled by the viral 72K DNA binding protein. Cell. 1981;26:371–379. doi: 10.1016/0092-8674(81)90206-3. [DOI] [PubMed] [Google Scholar]
- 2.Babiss L E, Ginsberg H S, Darnell J E. Adenovirus E1B proteins are required for accumulation of late viral mRNA and for effects on cellular mRNA translation and transport. Mol Cell Biol. 1985;5:2552–2558. doi: 10.1128/mcb.5.10.2552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Berk A J. Adenovirus promoters and E1A transactivation. Annu Rev Genet. 1986;20:45–79. doi: 10.1146/annurev.ge.20.120186.000401. [DOI] [PubMed] [Google Scholar]
- 4.Berk A J, Lee F, Harrison T, Williams J, Sharp P A. Pre-early adenovirus 5 gene product regulates synthesis of early viral messenger RNAs. Cell. 1979;17:935–944. doi: 10.1016/0092-8674(79)90333-7. [DOI] [PubMed] [Google Scholar]
- 5.Binger M H, Flint S J. Accumulation of early and intermediate mRNA species during subgroup C adenovirus productive infections. Virology. 1984;136:387–403. doi: 10.1016/0042-6822(84)90175-2. [DOI] [PubMed] [Google Scholar]
- 6.Bridge E, Hemström C, Pettersson U. Differential regulation of adenovirus late transcriptional units by the products of early region. Virology. 1991;183:260–266. doi: 10.1016/0042-6822(91)90138-2. [DOI] [PubMed] [Google Scholar]
- 7.Brunet L J, Babiss L E, Young C S H, Mills D R. Mutations in the adenovirus major late promoter: effects upon viability and transcription during infection. Mol Cell Biol. 1987;7:1091–1100. doi: 10.1128/mcb.7.3.1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Carter T H, Blanton R A. Possible role of the 72,000-dalton DNA binding protein in regulation of adenovirus type 5 early gene expression. J Virol. 1978;25:664–674. doi: 10.1128/jvi.25.2.664-674.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cauthen A N, Spindler K R. Sequence of the mouse adenovirus type-1 DNA encoding the 100-kDa, 33-kDa and DNA-binding proteins. Gene. 1996;168:183–187. doi: 10.1016/0378-1119(95)00715-6. [DOI] [PubMed] [Google Scholar]
- 10.Chang L-S, Shenk T. The adenovirus DNA-binding protein stimulates the rate of transcription directed by adenovirus and adeno-associated virus promoters. J Virol. 1990;64:2103–2109. doi: 10.1128/jvi.64.5.2103-2109.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Craig E A, Raskas H J. Effect of cycloheximide on RNA metabolism early in productive infection with adenovirus 2. J Virol. 1974;14:26–32. doi: 10.1128/jvi.14.1.26-32.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cross F R, Darnell J E., Jr Cycloheximide stimulates early adenovirus transcription if early gene expression is allowed before treatment. J Virol. 1983;45:683–692. doi: 10.1128/jvi.45.2.683-692.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Davis E, Krishnan K, Yan H, Newcomb E, Krolewski J J. A mutant form of p135tyk2, an interferon α inducible tyrosine kinase, can suppress tumorigenicity. Leukemia. 1996;10:543–551. [PubMed] [Google Scholar]
- 14.Dix I, Leppard K N. Regulated splicing of adenovirus type 5 E4 transcripts and regulated cytoplasmic accumulation of E4 mRNA. J Virol. 1993;67:3226–3231. doi: 10.1128/jvi.67.6.3226-3231.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dolph P J, Huang J, Schneider R J. Translation by the adenovirus tripartite leader: elements which determine independence from cap-binding complex. J Virol. 1991;64:2669–2677. doi: 10.1128/jvi.64.6.2669-2677.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ensinger M J, Ginsberg H S. Selection and preliminary characterization of temperature-sensitive mutants of type 5 adenovirus. J Virol. 1972;10:328–339. doi: 10.1128/jvi.10.3.328-339.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16a.Fessler, S. P., and C. S. H. Young. Unpublished results.
- 17.Fort P, Marty L, Piechaczyk M, El Sabrouty S, Dani C, Jennteur P, Blanchard J M. Various rat adult tissues express only one major mRNA species from the glyceraldehyde-3-phosphate-dehydrogenase multigene family. Nucleic Acids Res. 1985;13:1431–1442. doi: 10.1093/nar/13.5.1431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gambke C, Deppert W. Late nonstructural 100,000- and 33,000-dalton proteins of adenovirus type 2. I. Subcellular localization during the course of infection. J Virol. 1981;40:585–593. doi: 10.1128/jvi.40.2.585-593.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Giard D J, Aaronson S A, Todaro G J, Arnstein P, Kersey J H, Dosik H, Parks W P. In vitro cultivation of human tumors: establishment of cell lines derived from a series of solid tumors. J Natl Cancer Inst. 1973;51:1417–1423. doi: 10.1093/jnci/51.5.1417. [DOI] [PubMed] [Google Scholar]
- 20.Gilmartin G M, Hung S L, DeZazzo J D, Fleming E S, Imperiale M J. Sequences regulating poly(A) site selection within the adenovirus major late transcription unit influence the interaction of constitutive processing factors with the pre-mRNA. J Virol. 1996;70:1775–1783. doi: 10.1128/jvi.70.3.1775-1783.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hardy S, Engel D A, Shenk T. An adenovirus early region 4 gene product is required for induction of the infection-specific form of cellular E2F activity. Genes Dev. 1989;3:1062–1074. doi: 10.1101/gad.3.7.1062. [DOI] [PubMed] [Google Scholar]
- 22.Hayes B W, Telling G C, Myat M M, Williams J F, Flint S J. The adenovirus L4 100-kilodalton protein is necessary for efficient translation of viral late mRNA species. J Virol. 1990;64:2732–2742. doi: 10.1128/jvi.64.6.2732-2742.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Huang D-H, Horikoshi M, Roeder R G. Activation of the adenovirus EIIa late promoter by a single-point mutation which enhances binding of transcription factor IID. J Biol Chem. 1988;263:12596–12601. [PubMed] [Google Scholar]
- 24.Huang M-M, Hearing P. The adenovirus early region 4 open reading frame 6/7 protein regulates the DNA binding activity of the cellular transcription factor, E2F, through a direct complex. Genes Dev. 1989;3:1699–1710. doi: 10.1101/gad.3.11.1699. [DOI] [PubMed] [Google Scholar]
- 25.Huang M-M, Hearing P. Adenovirus early region 4 encodes two gene products with redundant effects in lytic infection. J Virol. 1989;63:2605–2615. doi: 10.1128/jvi.63.6.2605-2615.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jones N, Shenk T. An adenovirus type 5 early gene function regulates expression of other early viral genes. Proc Natl Acad Sci USA. 1979;76:3665–3669. doi: 10.1073/pnas.76.8.3665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Katze M G, Persson H, Philipson L. Control of adenovirus early gene expression: posttranscriptional control mediated by both viral and cellular gene products. Mol Cell Biol. 1981;1:807–813. doi: 10.1128/mcb.1.9.807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kelly T J, Jr, Lewis A M., Jr Use of nondefective adenovirus-simian virus 40 hybrids for mapping the simian virus 40 genome. J Virol. 1973;12:643–652. doi: 10.1128/jvi.12.3.643-652.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kreivi J-P, Zerivitz K, Akusjärvi G. Sequences involved in the control of adenovirus L1 alternative RNA splicing. Nucleic Acids Res. 1991;19:2379–2386. doi: 10.1093/nar/19.9.2379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Leong K, Lee W, Berk A J. High-level transcription from the adenovirus major late promoter requires downstream binding sites for late-phase-specific factors. J Virol. 1990;64:51–60. doi: 10.1128/jvi.64.1.51-60.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lewis J B, Mathews M B. Control of adenovirus early gene expression: a class of immediate early products. Cell. 1980;21:303–313. doi: 10.1016/0092-8674(80)90138-5. [DOI] [PubMed] [Google Scholar]
- 32.Li X-C, Huang W L, Flint S J. The downstream regulatory sequence of the adenovirus type 2 major late promoter is functionally redundant. J Virol. 1992;66:5685–5690. doi: 10.1128/jvi.66.9.5685-5690.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Logan J, Shenk T. In vivo identification of sequence elements required for normal function of the adenovirus major late transcriptional control region. Nucleic Acids Res. 1986;14:6327–6335. [PMC free article] [PubMed] [Google Scholar]
- 34.Lu H, Reach M D, Minaya E, Young C S H. The initiator element of the adenovirus major late promoter has an important role in transcription initiation in vivo. J Virol. 1997;71:102–109. doi: 10.1128/jvi.71.1.102-109.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lutz P, Kedinger C. Properties of the adenovirus IVa2 gene product, an effector of late-phase-dependent activation of the major late promoter. J Virol. 1996;70:1396–1405. doi: 10.1128/jvi.70.3.1396-1405.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lutz P, Rosa-Calatrava M, Kedinger C. The product of the adenovirus intermediate gene IX is a transcriptional activator. J Virol. 1997;71:5102–5109. doi: 10.1128/jvi.71.7.5102-5109.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mathews M B. Control of translation in adenovirus-infected cells. Enzyme. 1990;44:250–264. doi: 10.1159/000468763. [DOI] [PubMed] [Google Scholar]
- 37a.Minaya, E., S. P. Fessler, and C. S. H. Young. Unpublished results.
- 38.Nevins J R. Mechanism of activation of early viral transcription by the adenovirus E1a gene product. Cell. 1981;26:213–220. doi: 10.1016/0092-8674(81)90304-4. [DOI] [PubMed] [Google Scholar]
- 39.Nevins J R. Regulation of early adenovirus gene expression. Microbiol Rev. 1987;51:419–430. doi: 10.1128/mr.51.4.419-430.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nevins J R, Ginsberg H S, Blanchard J-M, Wilson M C, Darnell J E. Regulation of the primary expression of the early adenovirus transcription units. J Virol. 1979;32:727–733. doi: 10.1128/jvi.32.3.727-733.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nevins J R, Wilson M C. Regulation of adenovirus-2 gene expression at the level of transcription termination and RNA processing. Nature. 1981;290:113–118. doi: 10.1038/290113a0. [DOI] [PubMed] [Google Scholar]
- 42.Nordqvist K, Akusjärvi G. Adenovirus early region 4 stimulates mRNA accumulation via 5′ introns. Proc Natl Acad Sci USA. 1990;87:9543–9547. doi: 10.1073/pnas.87.24.9543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nordqvist K, Öhman K, Akusjärvi G. Human adenovirus encodes two proteins which have opposite effects on accumulation of alternatively spliced mRNAs. Mol Cell Biol. 1994;14:437–445. doi: 10.1128/mcb.14.1.437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.O’Connor R J, Hearing P. The C-terminal 70 amino acids of the adenovirus E4-ORF6/7 protein are essential and sufficient for E2F complex formation. Nucleic Acids Res. 1991;19:6579–6586. doi: 10.1093/nar/19.23.6579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pilder S, Moore M, Logan J, Shenk T. The adenovirus E1B-55K transforming polypeptide modulates transport or cytoplasmic stabilization of viral and host cell mRNAs. Mol Cell Biol. 1986;6:470–476. doi: 10.1128/mcb.6.2.470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Prescott J, Falck-Pedersen E. Sequence elements upstream of the 3′ cleavage site confer substrate strength to the adenovirus L1 and L3 polyadenylation sites. Mol Cell Biol. 1994;14:4682–4693. doi: 10.1128/mcb.14.7.4682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Prescott J C, Falck-Pedersen E. Varied poly(A) site efficiency in the adenovirus major late transcription unit. J Biol Chem. 1992;267:8175–8181. [PubMed] [Google Scholar]
- 48.Prescott J C, Liu L, Falck-Pedersen E. Sequence-mediated regulation of adenovirus gene expression by repression of mRNA accumulation. Mol Cell Biol. 1997;17:2207–2216. doi: 10.1128/mcb.17.4.2207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Reach M, Babiss L E, Young C S H. The upstream factor binding site is not essential for activation of transcription from the adenovirus major late promoter. J Virol. 1990;64:5851–5860. doi: 10.1128/jvi.64.12.5851-5860.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Reach M, Xu L-X, Young C S H. Transcription from the adenovirus major late promoter uses redundant activating elements. EMBO J. 1991;10:3439–3446. doi: 10.1002/j.1460-2075.1991.tb04908.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sandler A B, Ketner G. The metabolism of host RNAs in cells infected by an adenovirus E4 mutant. Virology. 1991;181:319–326. doi: 10.1016/0042-6822(91)90498-z. [DOI] [PubMed] [Google Scholar]
- 52.Sharp P A. Adenovirus transcription. In: Ginsberg H S, editor. The adenoviruses. New York, N.Y: Plenum Press; 1984. pp. 173–204. [Google Scholar]
- 53.Shaw A R, Ziff E B. Transcripts from the adenovirus-2 major late promoter yield a single early family of 3′ coterminal mRNAs and five late families. Cell. 1980;20:905–916. doi: 10.1016/0092-8674(80)90568-1. [DOI] [PubMed] [Google Scholar]
- 54.Shaw A R, Ziff E B. Selective inhibition of adenovirus type 2 early region II and III transcription by an anisomycin block of protein synthesis. Mol Cell Biol. 1982;2:789–799. doi: 10.1128/mcb.2.7.789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Shenk T, Flint S J. Transcriptional and transforming activities of the adenovirus E1A proteins. Adv Cancer Res. 1991;57:47–85. doi: 10.1016/s0065-230x(08)60995-1. [DOI] [PubMed] [Google Scholar]
- 56.Thomas G P, Mathews M B. DNA replication and the early to late transition in adenovirus infection. Cell. 1980;22:523–533. doi: 10.1016/0092-8674(80)90362-1. [DOI] [PubMed] [Google Scholar]
- 57.Tollefson A E, Scaria A, Hermiston T, Ryerse J S, Wold L J, Wold W S M. The adenovirus death protein (E3-11.6K) is required at very late stages of infection for efficient cell lysis and release of adenovirus from infected cells. J Virol. 1996;70:2296–2306. doi: 10.1128/jvi.70.4.2296-2306.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tollefson A E, Scaria A, Saha S K, Wold W S M. The 11,600-Mw protein encoded by region E3 of adenovirus is expressed early but is greatly amplified at late stages of infection. J Virol. 1992;66:3633–3642. doi: 10.1128/jvi.66.6.3633-3642.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tribouley C, Lutz P, Staub A, Kedinger C. The product of the adenovirus intermediate gene IVa2 is a transcriptional activator of the major late promoter. J Virol. 1994;68:4450–4457. doi: 10.1128/jvi.68.7.4450-4457.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Volkert F C, Young C S H. The genetic analysis of recombination using adenovirus overlapping terminal DNA fragments. Virology. 1983;125:175–193. doi: 10.1016/0042-6822(83)90072-7. [DOI] [PubMed] [Google Scholar]