Abstract
Background
Acute respiratory distress syndrome (ARDS) is a fatal outcome of severe sepsis. Machine learning models are helpful for accurately predicting ARDS in patients with sepsis at an early stage.
Objective
We aim to develop a machine-learning model for predicting ARDS in patients with sepsis in the intensive care unit (ICU).
Methods
The initial clinical data of patients with sepsis admitted to the hospital (including population characteristics, clinical diagnosis, complications, and laboratory tests) were used to predict ARDS, and screen out the crucial variables. After comparing eight different algorithms, namely, XG boost, logistic regression, light GBM, random forest, GaussianNB, complement NB, support vector machine (SVM), and K nearest neighbors (KNN), rebuilding a prediction model with the best one. When remodeling with the best algorithm, 10% was randomly selected to test, and the remaining was trained for cross-validation. Using the area under the curve (AUC), sensitivity, accuracy, specificity, positive and negative predictive value, F1 score, kappa value, and clinical decision curve to evaluate the model's performance. Eventually, the application in the model illustrated by the SHAP package.
Results
Ten critical features were screened utilizing the lasso method, namely, PaO2/PAO2, A-aDO2, PO2(T), CRP, gender, PO2, RDW, MCH, SG, and chlorine. The prior ranking of variables demonstrated that PaO2/PAO2 was the most significant variable. Among the eight algorithms, the performance of the Gaussian NB algorithm was significantly better than that of the others. After remodeling with the best algorithm, the AUC in the training and validation sets were 0.777 and 0.770, respectively, and the algorithm performed well in the test set (AUC = 0.781, accuracy = 78.6%, sensitivity = 82.4%, F1 score = 0.824). A comparison of the overlap factors with those of previous models revealed that the model we developed performs better.
Conclusion
Sepsis-associated ARDS can be accurately predicted early via a machine learning model based on existing clinical data. These findings are helpful for accurate identification and improvement of the prognosis in patients with sepsis-associated ARDS.
Keywords: Acute respiratory distress syndrome, ARDS, Machine learning, Algorithm, Sepsis, ICU
1. Introduction
Acute respiratory distress syndrome (ARDS) is an acute diffuse lung injury accompanied by further acute respiratory failure and is a clinical syndrome with a high incidence in critical illness. The outcome of ARDS depends on the severity of the lung injury at the early stage [1,2]. The clinical manifestations were respiratory distress and refractory hypoxemia with bilateral pulmonary infiltration, which was challenging to distinguish from cardiogenic pulmonary edema on imaging [[1], [2], [3], [4]]. The diagnosis of ARDS, therefore, relies solely on clinical criteria, as pathological measurements of lung injury are impractical in most patients [[4], [5], [6]]. The poor reliability of some criteria in the Berlin definition may lead to insufficient understanding by clinicians. Clinicians had a low recognition rate for mild and severe ARDS in the LUNG-SAFE study [5]. ARDS is common in critical illness but has not been fully recognized or treated. Sepsis is a life-threatening organ dysfunction induced by the host's dysregulated inflammatory response to various infections [[6], [7], [8], [9]] and is the most common risk factor for ARDS [[10], [11], [12], [13], [14]]. ARDS is associated with high morbidity, adverse outcomes, high mortality, and excessive medical costs in the ICU. A large-scale trial of moderate to severe ARDS involving 459 ICUs in 50 countries reported that the hospital mortality rate was 43% at 90 days [15]. The mortality rate of sepsis-associated ARDS is approximately 27%–37% [16]. Therefore, early dynamic prediction of sepsis-associated ARDS and corresponding treatment can effectively improve the clinical prognosis.
Sepsis-associated ARDS patients have high morbidity and mortality rates. Machine learning models are helpful for accurately predicting ARDS in patients with sepsis at an early stage. Currently, machine learning is the application of artificial intelligence in generating disease prediction models [[17], [18], [19]]. For instance, based on machine learning algorithms (gradient boosting, random forest, bootstrapping, minimum absolute shrinkage, and selection operators), classification variables are selected to identify ARDS phenotypes using existing clinical data [[20], [21], [22]]. Previously developed machine learning models were used to classify ARDS patients into hypoinflammatory and hyperinflammatory subphenotypes. Presently, regardless of the etiology or severity, ARDS patients are treated in a homogenous fashion [3]. The use of the novel P/FPE ratio for assessing ARDS severity after onset is significantly better than the use of the current PaO2/FiO2 criteria [23]. This approach can help manage patients with ARDS and provide more accurate, personalized treatment options for each severity of ARDS. However, some studies based on databases used to predict ARDS have deleted many laboratory parameters before model construction because more than 50% of the important data are missing (for example, oxygen partial pressure and carbon dioxide partial pressure). In addition, multiple database indices cannot be completely consistent, which inevitably leads to bias in the results and limits their application; therefore, a high-performance model that can predict ARDS in septic patients using only simple clinical indicators is needed.
Therefore, in the study, machine learning algorithms will be applied to develop an early prediction model for ARDS based on clinical data, and patient characteristics will be evaluated by interpreting the final model to early identify or exclude ARDS in patients with sepsis. High-risk patients with sepsis-associated ARDS may benefit from this model.
2. Methods
2.1. Research objects
This study included 279 patients, 18 years old or older, who met the criteria of ' Sepsis-3 ' [10] in the ICU ward of the Third Xiangya Hospital of Central South University from January 2013 to April 2022, and were diagnosed with sepsis and managed according to international guidelines. They are treated by the same group of doctors, the same group of first-line doctors have roughly the same level, and if they encounter difficult diseases, they are guided by an experienced superior doctor, so most patients receive treatment almost the same, which will not have a big difference in the results. Admission time was more than 24 h and was not diagnosed as ARDS within 24 h of admission. ARDS was defined according to the Berlin definition. Sepsis-associated ARDS was defined as ARDS occurring 24 h after admission in patients diagnosed with sepsis. Compared with sepsis, patients diagnosed with septic shock were excluded due to the different etiology, severity, and therapy, and they tend to be more severe. Patients who were diagnosed with ARDS before admission or within 24 h of admission, or had a history of chronic lung disease or pneumonectomy (such as bronchiolitis, pulmonary fibrosis, or pulmonary contusion), or had a high data loss rate or incomplete important clinical data were excluded. Only the data of the admission day were used as potential features. All included patients had sepsis as an admission diagnosis.
2.2. Data
The data were derived from medical records. Data include (1) demographic characteristics (2) clinical characteristics (previous disease history, admission/discharge diagnosis, course of disease, surgery/consultation, etc.) (3) complications (4) medication history (5) laboratory indicators (blood gas analysis, procalcitonin (PCT), C-reactive protein (CRP), bacterial (fungal) culture and identification, myocardial injury markers, liver and kidney function, electrolyte, blood routine, coagulation routine, urine sediment analysis) and other variables. When there was more than one data point available for a specific feature, we used only the first one. Crucial features were screened by the lasso method from 82 first recorded variables at admission. The primary outcome is to predict ARDS in patients with sepsis after admission.
2.3. Design
This study first utilizes multiple machine learning algorithms for data classification. These algorithms are as follows: XG Boost, logistic regression, light GBM, random forest, Gaussian NB, Complement NB, SVM, and KNN. Through the data set partitioning function, the randomization/allocation of the training and validation cohorts were performed according to the proportion of the sample, which is a commonly used function in cross-validation. In each training, 70% of the overall sample was selected for training, the remaining for validating, ensuring that the training samples selected for multiple model algorithms are consistent to better compare multiple models. The performance of the model is evaluated by the above indicators (AUC, etc.). The forest plot shows the ROC results of each algorithm to predict ' ARDS '.
The best algorithm is selected for remodeling through multiple algorithms comparison. It was chosen by comprehensively comparing the various indicators of the multiple models (AUC, accuracy, and Kappa index). During modeling with the best algorithm, 10% was randomly selected to test. The study was conducted strictly according to the TRIPOD checklist (supplemental materials).
2.4. Multiple machine learning algorithms
Each algorithm has its own specific situation [[24], [25], [26], [27]]. As an ensemble learning algorithm, XG Boost can efficiently process missing data to construct accurate prediction models [24]. Light GBM shows excellent performance in processing very large structured data sets with ultra-high training speed [25], but it is susceptible to the number of features and sample size. Random forest (RF) has high classification accuracy but requires a large amount of calculation [26,27]. Due to its stable classification efficiency and excellent performance on small-scale data, Gaussian NB is easy to implement and run quickly [28]. Using traditional logistic regression, more data were needed to obtain a classifier with similar performance to the Gaussian NB. KNN classifies samples by nearest neighbors. However, there are several limitations, such as sparse problems, imbalance problems, and noise problems [29]. SVM can be used for linear/nonlinear classification, solving machine learning problems with small samples, but is sensitive to parameter adjustment and function selection [30]. Complement NB is especially suitable for imbalanced data sets. Specifically, CNB uses the supplementary data of each category to calculate the weight of the model. Considering the overall indicators to find the most accurate model, comprehensively.
2.5. Model interpretation
The SHAP package regards all the features as ' contributors ' and generates a SHAP value. The SHAP value diagram and variable importance graph are used to show the contribution and importance ranking of each feature to the model, respectively.
2.6. Statistical analysis
The chi-square test and Mann-Whitney U test were used for the categorical variables and the quantitative variables, respectively. Analysis of the differences was conducted using the stats models 0.11.1 package (Python).
In this study, the critical features were screened by the lasso method, and the cross-validation method was used to eliminate features with a coefficient of 0. The KNN algorithm is used to fill the missing values. The lasso method obtains a simpler model by compressing partial regression coefficients to construct a penalty function. Therefore, it retains the advantages of subset shrinkage and is a biased estimation for processing multicollinearity data (The version of the package used by the algorithm is shown in Appendix 1). The predictive value and critical value of sepsis variables were determined by the receiver operating characteristic (ROC) curve.
2.7. Medical ethics approval
This was a retrospective study that received expedited approval and informed consent waiver from the Ethics Committee of the Third Xiangya Hospital, Central South University (protocol number 22310).
3. Results
3.1. Baseline characteristics
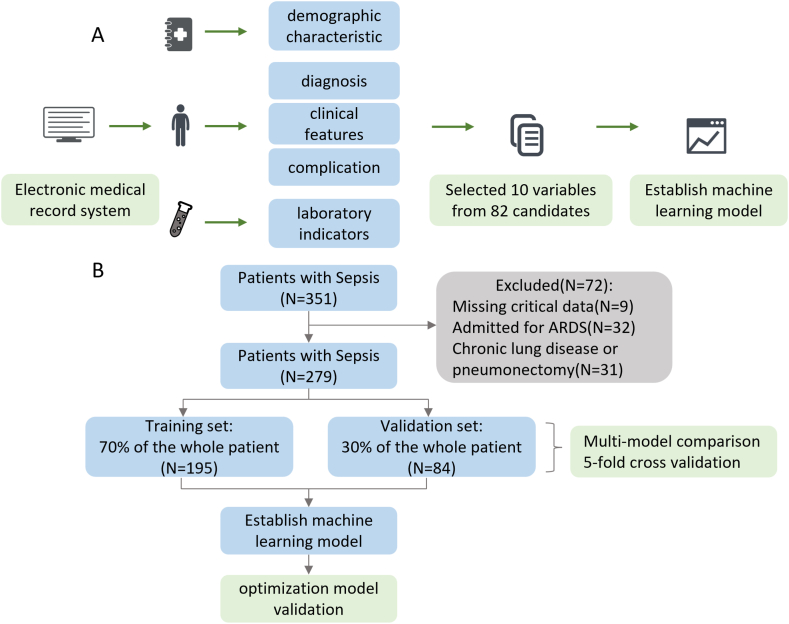
This study involved 279 patients. During the multiple model comparison, the training set and the validation set were 195 and 84 patients, respectively. The baseline characteristics of the population are summarized in Table 1. Consistent with previous studies, most of the patients were elderly, and the median age was 61 (range 48–69). Males accounted for 71.3% of the total population, which verified previous reports that male patients were more likely to suffer from sepsis due to smoking and other risk factors. Among the people with sepsis-related acute respiratory distress syndrome, 77.8% were male, indicating that they were more likely to develop ARDS. Statistically significant differences in gender (P < 0.05) were found in the dataset. It is not difficult to find that most patients received mechanical ventilation treatment, which is still a crucial treatment. In the population, there are 167 (59.86%) sepsis patients with ARDS and 112 (40.14%) sepsis patients without ARDS. These characteristic variables were statistically significant, including gender, prothrombin time activity (PTA), oxygen saturation, oxygen partial pressure (PO2), oxygen content, oxygen partial pressure in the temperature (PO2(T)), respiratory index, ratio of arterial to alveolar oxygen partial pressure (PaO2/PAO2), alveolar-arterial oxygen partial pressure difference (A-aDO2), FiO2, hemoglobin, mean corpuscular volume (MCV), red blood cell distribution width (RDW), lymphocyte count, C-reactive protein (CRP), lactate dehydrogenase (LDH), Creatine Kinase (CK), myoglobin, urine specific gravity (USG or SG), urine protein, potassium, and body temperature. In sepsis patients with ARDS, PaO2/PAO2 is the most important risk factor, followed by A-aDO2, PO2(T), CRP, gender, and PO2. Fig. 1A, B, is the schematic diagram of this study.
Table 1.
The baseline characteristics of the total population.
| Variables | deletion | Classification items | All (n = 279) | Non-ARDS (n = 112) |
ARDS (n = 167) |
statistic | P - value |
|---|---|---|---|---|---|---|---|
| Blood transfusion therapy (Yes = 1 No = 0), n (%) | 0 | 0 | 1 (0.358) | 1 (0.893) | 0 (0.000) | nan | nan |
| 1 | 278 (99.642) | 111 (99.107) | 167 (100.000) | ||||
| Gender male 1, female 0, n (%) | 0 | 0 | 80 (28.674) | 43 (38.393) | 37 (22.156) | 8.642 | 0.003 |
| 1 | 199 (71.326) | 69 (61.607) | 130 (77.844) | ||||
| FIB g/L, median [IQR] | 0 | nan | 3.320 [1.990,4.830] | 2.990 [1.830,4.470] | 3.530 [2.050,4.830] | −1.341 | 0.180 |
| APTT s, median [IQR] | 0 | nan | 43.100 [33.500,57.400] | 43.600 [32.800,55.900] | 42.700 [34.800,58.000] | −0.511 | 0.610 |
| INR, median [IQR] | 0 | nan | 1.290 [1.140,1.580] | 1.230 [1.100,1.510] | 1.310 [1.160,1.620] | −1.387 | 0.166 |
| PT s, median [IQR] | 0 | nan | 14.900 [13.200,18.100] | 14.200 [12.800,16.900] | 15.000 [13.300,18.500] | −1.485 | 0.138 |
| PTA %, mean (±SD) | 0 | nan | 62.489 ± 25.394 | 66.740 ± 26.214 | 59.638 ± 24.417 | 2.303 | 0.022 |
| D-dimer mg/L, median [IQR] | 0 | nan | 4.640 [2.210,10.290] | 4.797 [2.310,11.310] | 4.480 [2.100,10.250] | 0.702 | 0.483 |
| pH, median [IQR] | 0 | nan | 7.400 [7.330,7.454] | 7.401 [7.350,7.455] | 7.390 [7.310,7.450] | 1.401 | 0.161 |
| Mechanical Ventilation (No = 0 96 h = 1 ≥ 96 h = 2), median [IQR] | 0 | nan | 2.000 [1.000,2.000] | 2.000 [1.000,2.000] | 2.000 [1.000,2.000] | −0.354 | 0.664 |
| Age, median [IQR] | 0 | nan | 61.000 [48.000,69.000] | 62.000 [48.000,72.000] | 61.000 [48.000,68.000] | 0.590 | 0.555 |
| Cl mmol/L, median [IQR] | 0 | nan | 113.000 [108.000,117.739] | 113.000 [107.000,117.000] | 113.000 [108.487,118.000] | −0.759 | 0.448 |
| SO2c %, median [IQR] | 0 | nan | 96.757 [93.747,98.485] | 97.686 [95.900,99.000] | 95.800 [91.300,97.834] | 4.983 | <0.001 |
| Hb (BGA) g/L, median [IQR] | 0 | nan | 99.00 [83.05,116.08] | 98.00 [82.83,115.83] | 101.00 [85.18,119.00] | −1.001 | 0.317 |
| PO2 mmHg, median [IQR] | 0 | nan | 85.900 [72.900,117.000] | 102.509 [83.653,133.000] | 76.700 [65.400,98.100] | 5.577 | <0.001 |
| PCO2 mmHg, median [IQR] | 0 | nan | 31.900 [27.300,38.700] | 31.922 [27.200,37.111] | 31.900 [27.387,39.200] | −0.524 | 0.601 |
| oxygen content vol%, median [IQR] | 0 | nan | 14.900 [11.700,18.468] | 16.200 [12.200,18.468] | 14.200 [11.400,17.400] | 2.725 | 0.006 |
| PO2(T) mmHg, median [IQR] | 0 | nan | 100.000 [71.000,113.000] | 111.237 [95.600,134.000] | 80.300 [65.400,111.237] | 6.381 | <0.001 |
| PCO2(T)mmHg, median [IQR] | 0 | nan | 34.846 [28.100,37.300] | 34.846 [29.600,35.400] | 34.846 [27.500,38.700] | 0.028 | 0.978 |
| pH(T), median [IQR] | 0 | nan | 7.356 [7.330,7.420] | 7.356 [7.350,7.410] | 7.356 [7.320,7.421] | −0.058 | 0.955 |
| Lac mmol/L, median [IQR] | 0 | nan | 3.800 [1.700,5.298] | 4.400 [1.400,5.298] | 3.800 [2.100,5.298] | −0.480 | 0.630 |
| SB mmol/L, median [IQR] | 0 | nan | 17.610 [16.400,20.800] | 17.610 [17.500,20.600] | 17.610 [16.100,21.100] | 0.112 | 0.911 |
| AB mmol/L, median [IQR] | 0 | nan | 19.152 [17.600,21.600] | 19.152 [19.152,21.400] | 19.152 [17.100,21.600] | 0.911 | 0.360 |
| A-BE mmol/L, median [IQR] | 0 | nan | −6.498 [-8.200,-3.100] | −6.498 [-6.498,-3.200] | −6.498 [-8.400,-3.000] | 0.481 | 0.629 |
| S-BE mmol/L, median [IQR] | 0 | nan | −6.821 [-8.400,-3.200] | −6.821 [-6.821,-3.500] | −6.821 [-8.700,-3.200] | 0.671 | 0.500 |
| P50 mmHg, median [IQR] | 0 | nan | 27.452 [25.230,28.220] | 27.452 [25.430,27.770] | 27.400 [24.850,29.150] | 0.480 | 0.630 |
| AG mmol/L, median [IQR] | 0 | nan | 8.300 [0.925,13.300] | 5.600 [0.925,13.000] | 9.000 [0.925,13.300] | −1.202 | 0.227 |
| RI %, median [IQR] | 0 | nan | 129.000 [110.988,299.000] | 110.988 [66.000,129.000] | 201.000 [110.988,380.000] | −6.741 | <0.001 |
| PaO2/PAO2 mmHg, median [IQR] | 0 | nan | 44.200 [25.100,57.047] | 57.047 [43.700,60.200] | 34.600 [20.800,53.000] | 6.764 | <0.001 |
| A-aDO2 mmHg, median [IQR] | 0 | nan | 146.758 [124.800,225.400] | 146.758 [98.600,147.600] | 163.100 [139.300,299.500] | −4.920 | <0.001 |
| TCO2 vol%, median [IQR] | 0 | nan | 38.732 [34.400,42.800] | 38.732 [36.900,41.900] | 38.732 [33.800,43.100] | 0.427 | 0.669 |
| K mmol/L, median [IQR] | 0 | nan | 3.700 [3.252,4.300] | 3.500 [3.252,4.100] | 3.800 [3.252,4.300] | −2.288 | 0.021 |
| Ca mmol/L, median [IQR] | 0 | nan | 1.020 [1.010,1.120] | 1.020 [1.020,1.110] | 1.030 [0.990,1.120] | 0.076 | 0.940 |
| Hct (BGA) %, median [IQR] | 0 | nan | 34.800 [27.900,37.007] | 36.500 [27.900,37.007] | 34.200 [28.000,37.007] | 0.452 | 0.650 |
| FiO2, median [IQR] | 0 | nan | 42.256 [35.000,50.000] | 42.256 [34.000,42.256] | 42.256 [37.000,60.000] | −2.408 | 0.015 |
| TEMP °C, median [IQR] | 0 | nan | 37.000 [36.800,37.373] | 37.373 [37.000,37.373] | 37.000 [36.600,37.373] | 2.352 | 0.017 |
| Platelets 109/L, median [IQR] | 0 | nan | 107.000 [47.000,191.000] | 107.000 [45.000,179.000] | 107.000 [49.000,201.000] | −0.429 | 0.668 |
| Hct %, median [IQR] | 0 | nan | 31.200 [25.100,37.100] | 30.000 [23.800,37.300] | 31.400 [26.200,36.700] | −0.832 | 0.406 |
| Hemoglobin g/L, median [IQR] | 0 | nan | 98.000 [77.000,118.000] | 93.000 [69.000,116.000] | 102.000 [82.000,120.000] | −2.317 | 0.021 |
| WBC 109/L, median [IQR] | 0 | nan | 11.320 [6.350,17.880] | 11.840 [7.820,19.790] | 10.890 [6.050,16.660] | 1.586 | 0.113 |
| Na mmol/L, median [IQR] | 0 | nan | 138.000 [134.000,142.000] | 138.000 [134.000,141.077] | 138.000 [134.000,142.000] | −0.786 | 0.432 |
| MCHC g/L, median [IQR] | 0 | nan | 326.000 [316.000,337.000] | 326.000 [312.000,337.000] | 326.000 [318.000,335.000] | −0.351 | 0.726 |
| MCH pg, median [IQR] | 0 | nan | 30.700 [29.300,31.800] | 30.900 [29.300,33.000] | 30.500 [29.400,31.600] | 1.944 | 0.052 |
| MCV fL, median [IQR] | 0 | nan | 93.800 [88.200,98.000] | 94.364 [88.300,100.500] | 92.700 [88.100,96.400] | 2.030 | 0.042 |
| RBC 1012/L, median [IQR] | 0 | nan | 3.360 [2.780,4.040] | 3.210 [2.710,3.970] | 3.450 [2.890,4.040] | −1.158 | 0.247 |
| Monocyte count 109/L, median [IQR] | 0 | nan | 0.450 [0.200,0.810] | 0.490 [0.260,0.870] | 0.410 [0.190,0.710] | 1.896 | 0.058 |
| Basophils %, median [IQR] | 0 | nan | 0.100 [0.100,0.300] | 0.200 [0.100,0.400] | 0.100 [0.100,0.300] | 0.675 | 0.492 |
| Eosinophils %, median [IQR] | 0 | nan | 0.100 [0.000,0.504] | 0.200 [0.000,0.500] | 0.100 [0.000,0.504] | 1.010 | 0.299 |
| Monocyte %, median [IQR] | 0 | nan | 3.800 [2.000,6.600] | 4.000 [1.700,6.200] | 3.800 [2.300,6.800] | −0.899 | 0.369 |
| Lymphocyte %, median [IQR] | 0 | nan | 8.900 [4.300,16.000] | 10.672 [4.700,17.500] | 7.800 [4.200,15.200] | 1.810 | 0.070 |
| Neutrophils %, median [IQR] | 0 | nan | 85.100 [76.000,91.500] | 84.400 [73.900,90.700] | 86.100 [76.900,91.700] | −1.663 | 0.096 |
| RDW %, median [IQR] | 0 | nan | 14.100 [13.100,15.700] | 14.400 [13.100,16.100] | 14.000 [13.100,15.300] | 1.361 | 0.174 |
| Basophils count 109/L, median [IQR] | 0 | nan | 0.010 [0.000,0.030] | 0.020 [0.010,0.030] | 0.010 [0.000,0.030] | 0.997 | 0.310 |
| Eosinophils count 109/L, median [IQR] | 0 | nan | 0.010 [0.000,0.060] | 0.010 [0.000,0.060] | 0.010 [0.000,0.060] | 1.539 | 0.111 |
| Neutrophils count 109/L, median [IQR] | 0 | nan | 9.100 [5.420,14.690] | 9.910 [6.110,16.650] | 8.845 [5.170,14.380] | 1.325 | 0.185 |
| Lymphocyte count 109/L, median [IQR] | 0 | nan | 0.690 [0.400,1.220] | 0.880 [0.490,1.270] | 0.650 [0.370,1.090] | 2.713 | 0.007 |
| CRP mg/L, median [IQR] | 0 | nan | 127.115 [71.465,200.000] | 105.782 [61.869,167.803] | 145.480 [88.490,216.990] | −3.530 | <0.001 |
| platelet hyperplasia %, median [IQR] | 0 | nan | 0.137 [0.080,0.220] | 0.140 [0.090,0.220] | 0.130 [0.070,0.220] | 1.367 | 0.172 |
| MPV fL, mean (±SD) | 0 | nan | 10.693 ± 1.417 | 10.715 ± 1.230 | 10.678 ± 1.530 | 0.219 | 0.827 |
| PDW fL, median [IQR] | 0 | nan | 16.400 [15.900,16.900] | 16.300 [15.900,16.811] | 16.405 [15.900,16.900] | −0.813 | 0.416 |
| RDW (fL),median [IQR] | 0 | nan | 48.703 [44.436,54.300] | 50.900 [45.746,56.200] | 47.100 [44.148,51.700] | 2.641 | 0.008 |
| TP g/L, median [IQR] | 0 | nan | 50.500 [44.500,57.400] | 51.500 [45.100,58.800] | 50.000 [44.300,56.600] | 1.072 | 0.284 |
| DBil μmol/L, median [IQR] | 0 | nan | 8.100 [4.500,19.200] | 7.000 [3.500,18.400] | 9.000 [5.200,19.500] | −1.651 | 0.099 |
| TBil μmol/L, median [IQR] | 0 | nan | 15.900 [10.000,31.700] | 13.700 [9.000,28.900] | 17.200 [10.700,33.800] | −1.636 | 0.102 |
| AST, median [IQR] | 0 | nan | 62.000 [27.000,201.000] | 58.000 [29.000,180.000] | 66.000 [27.000,214.000] | −1.281 | 0.201 |
| ALT, median [IQR] | 0 | nan | 36.000 [16.000,111.000] | 29.000 [14.000,109.000] | 39.000 [18.000,111.000] | −1.133 | 0.257 |
| Urea mmol/L, median [IQR] | 0 | nan | 12.193 [7.410,18.670] | 11.660 [6.120,18.340] | 12.610 [8.260,18.800] | −1.186 | 0.236 |
| TBA μmol/L, median [IQR] | 0 | nan | 6.100 [3.000,16.800] | 5.900 [3.000,24.100] | 6.200 [3.100,14.900] | 0.298 | 0.766 |
| A/G, median [IQR] | 0 | nan | 1.300 [1.000,1.500] | 1.200 [0.900,1.500] | 1.300 [1.000,1.600] | −1.307 | 0.190 |
| Globulin g/L, median [IQR] | 0 | nan | 22.400 [18.200,27.200] | 23.500 [18.800,28.700] | 21.900 [18.000,25.825] | 1.610 | 0.108 |
| Albumin g/L, mean (±SD) | 0 | nan | 27.810 ± 6.598 | 28.285 ± 6.898 | 27.491 ± 6.368 | 0.983 | 0.326 |
| CK-MB, median [IQR] | 0 | nan | 35.000 [21.000,72.000] | 34.000 [18.154,58.000] | 37.000 [22.000,85.000] | −1.682 | 0.093 |
| LDH, median [IQR] | 0 | nan | 451.000 [287.000,863.000] | 363.446 [279.000,781.000] | 500.000 [305.000,953.000] | −2.132 | 0.033 |
| CK, median [IQR] | 0 | nan | 379.000 [108.000,1425.000] | 242.000 [86.000,837.000] | 521.000 [146.000,1730.000] | −2.633 | 0.008 |
| UA μmol/L, median [IQR] | 0 | nan | 347.000 [236.000,474.000] | 363.000 [245.000,472.000] | 346.000 [236.000,493.000] | 0.682 | 0.496 |
| Cre μmol/L, median [IQR] | 0 | nan | 135.000 [81.000,263.000] | 131.000 [80.000,264.000] | 140.000 [82.000,253.000] | −0.291 | 0.771 |
| Mb ng/ml, median [IQR] | 0 | nan | 564.000 [177.700,1199.000] | 370.800 [151.300,997.174] | 749.600 [263.600,1221.400] | −2.421 | 0.016 |
| PCT ng/ml, median [IQR] | 0 | nan | 7.579 [2.070,35.980] | 4.980 [1.230,32.910] | 10.350 [2.560,38.980] | −1.927 | 0.054 |
| TT s, median [IQR] | 0 | nan | 18.000 [16.300,21.100] | 18.200 [16.600,21.600] | 17.900 [16.100,20.800] | 1.381 | 0.168 |
| Glu, median [IQR] | 0 | nan | 0.000 [0.000,0.000] | 0.000 [0.000,0.000] | 0.000 [0.000,0.000] | −0.015 | 0.983 |
| SG, median [IQR] | 0 | nan | 1.018 [1.015,1.020] | 1.018 [1.015,1.020] | 1.020 [1.015,1.020] | −2.318 | 0.018 |
| Pro, median [IQR] | 0 | nan | 1.000 [0.000,1.000] | 1.000 [0.000,1.000] | 1.000 [0.000,1.000] | −2.040 | 0.016 |
APTT: activated partial thromboplastin time; TT: thrombin time; INR: international normalized ratio; PTA: prothrombin time activity; SO2c: oxygen saturation; Lac: lactic acid; SB: standard bicarbonate; AB: actual bicarbonate; S-BE: standard base excess; A-BE: actual base excess; AG: anion gap; RI: respiratory index; TBA: total bile acid; A/G: Albumin/Globulin; BGA: Blood Gas Analysis. nan, Not A number.
Fig. 1.
Workflow diagram of this study. (A) Data collection process. (B) Establishment of machine learning model and comparison of eight models.
3.2. Variable selection
With the lasso method, ten critical variables were selected: ' PaO2/PAO2 ', ' A-aDO2 ', ' PO2(T) ', ' CRP ', ' gender ', ' PO2 ', ' RDW ', ' mean red blood cell hemoglobin content (MCH) ', ' SG ', ' chlorine '.
3.3. Multi-algorithm models comparison
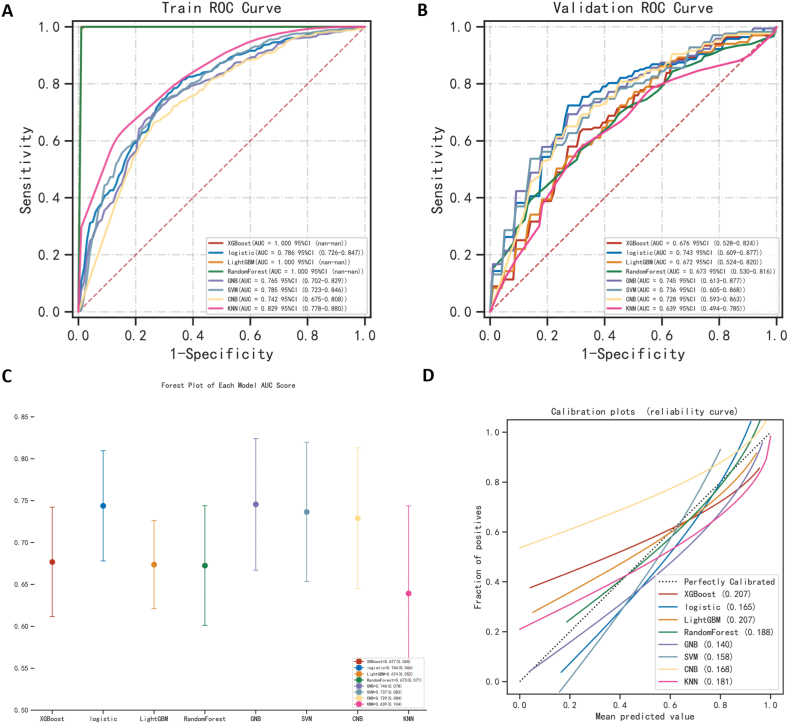
The classification of the data samples was attempted using eight machine-learning algorithms. When evaluating the kappa statistics on the training data, we found that the highest is the XG Boost algorithm (0.991). However, when comparing the kappa values, Gaussian NB had the highest consistency of kappa values on the two datasets, with only a 5.7% difference. Considering the overall indicators, Gaussian NB was found to be the most robust and accurate algorithm, with AUCs of 0.765 in the training set and 0.745 in the validation set, respectively (Fig. 2A and B). In addition, Gaussian NB is the best option when data is scarce [24]. This may be because it is a simple, fast, and highly scalable algorithm that performs well on small-scale data and is a suitable choice for binary classification problems. Furthermore, its cut-off value, sensitivity, accuracy, specificity, positive and negative predictive value, F1 score, and Kappa value were 0.728, 0.732, 0.734, 0.748, 0.812, 0.646, 0.769, and 0.460, respectively (Table 2). Alternatively, Table 2 and Supplemental Table 1 provide indexes for other machine learning algorithms. The forest plot (Fig. 2C) illustrates the ROC results of each model to predict ARDS. The clinical decision curve (Fig. 2D) shows the net benefit ability of each model.
Fig. 2.
Comparison of eight machine learning algorithms. (A, B) The ROC results of the models were established by eight machine learning algorithms in the training set and validation set. (C) A forest plot of each model AUC score built by eight machine learning algorithms. (D) Calibration plots of models built by eight machine learning algorithms.
Table 2.
Multi-model classification–training set results.
| Model | AUC(SD) | Cut off (SD) | Accuracy (SD) | Sensitivity (SD) | Specificity (SD) | Positive predictive value (SD) |
Negative predictive value (SD) |
F1 score (SD) | Kappa (SD) |
|---|---|---|---|---|---|---|---|---|---|
| XG Boost | 1.000 (0.000) | 0.873 (0.012) | 0.996 (0.000) | 1.000 (0.000) | 1.000 (0.000) | 1.000 (0.000) | 0.989 (0.000) | 1.000 (0.000) | 0.991 (0.000) |
| logistic | 0.789 (0.016) | 0.553 (0.036) | 0.744 (0.013) | 0.784 (0.038) | 0.694 (0.034) | 0.792 (0.012) | 0.679 (0.031) | 0.787 (0.016) | 0.469 (0.022) |
| Light GBM | 1.000 (0.000) | 0.567 (0.024) | 0.994 (0.002) | 0.997 (0.004) | 1.000 (0.000) | 1.000 (0.000) | 0.985 (0.005) | 0.998 (0.002) | 0.987 (0.005) |
| RandomForest | 1.000 (0.000) | 0.540 (0.037) | 0.987 (0.012) | 0.997 (0.004) | 0.998 (0.004) | 1.000 (0.000) | 0.970 (0.027) | 0.999 (0.002) | 0.974 (0.024) |
| GNB | 0.765 (0.017) | 0.728 (0.120) | 0.734 (0.012) | 0.732 (0.027) | 0.748 (0.036) | 0.812 (0.019) | 0.646 (0.018) | 0.769 (0.012) | 0.460 (0.023) |
| SVM | 0.787 (0.018) | 0.602 (0.031) | 0.733 (0.015) | 0.759 (0.052) | 0.705 (0.056) | 0.793 (0.021) | 0.659 (0.034) | 0.774 (0.020) | 0.451 (0.027) |
| KNN | 0.834 (0.020) | 0.680 (0.098) | 0.640 (0.075) | 0.743 (0.112) | 0.748 (0.127) | 0.916 (0.070) | 0.541 (0.059) | 0.810 (0.046) | 0.336 (0.104) |
| CNB | 0.742 (0.022) | 0.195 (0.388) | 0.700 (0.023) | 0.693 (0.062) | 0.721 (0.052) | 0.787 (0.020) | 0.609 (0.029) | 0.735 (0.034) | 0.395 (0.037) |
3.4. Best algorithm model
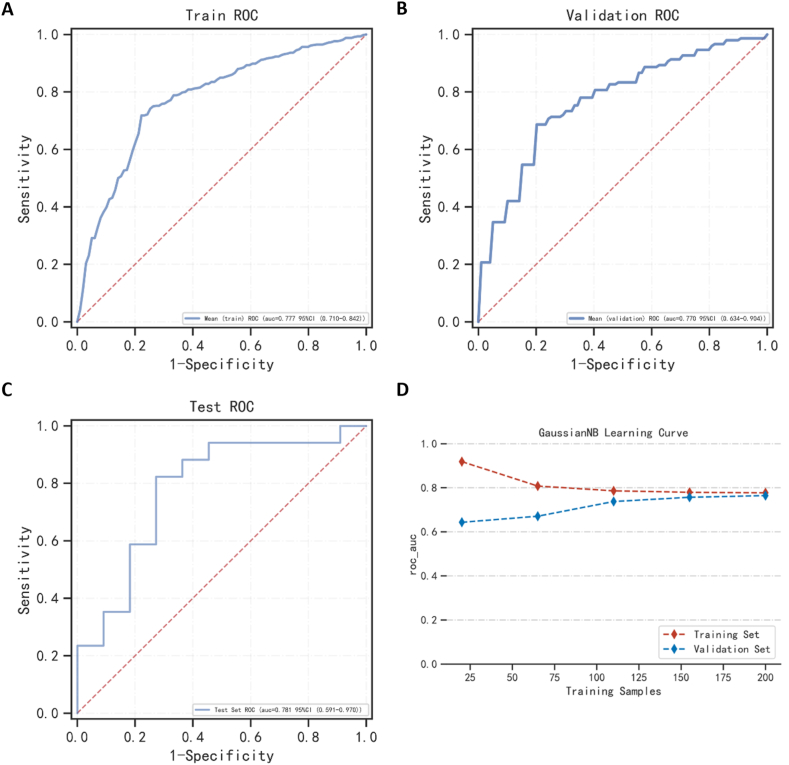
Through multi-model comparison, it was found that Gaussian NB performed best, and we used Gaussian NB to re-establish the prediction model for analysis. The AUCs of the training and validation sets were 0.777 and 0.770, respectively (Fig. 3A and B). The AUC of the final model in the test set was 0.781, and the accuracy was 78.6% (Fig. 3C–Table 3). What's more, once the sample size reached 175, the AUC reached a stable state (Fig. 3D). Training, validation, and test set evaluation indexes are shown in Supplemental Tables 2–3 and Table 3, respectively.
Fig. 3.
The performance of the model is built by the Gaussian NB algorithm. (A, B, C) The ROC result of the model was established by the Gaussian NB algorithm in the training set, validation set, and testing set. (D) The ROC result of the model was established by the Gaussian NB algorithm in the training set and the validation set according to the change in sample size.
Table 3.
Test set results of the best model.
| AUC | Cut off | Accuracy | Sensitivity | Specificity | Positive predictive value | Negative predictive value | F1 score |
|---|---|---|---|---|---|---|---|
| 0.781 | 0.562 | 0.786 | 0.824 | 0.727 | 0.824 | 0.727 | 0.824 |
3.5. Model interpretability
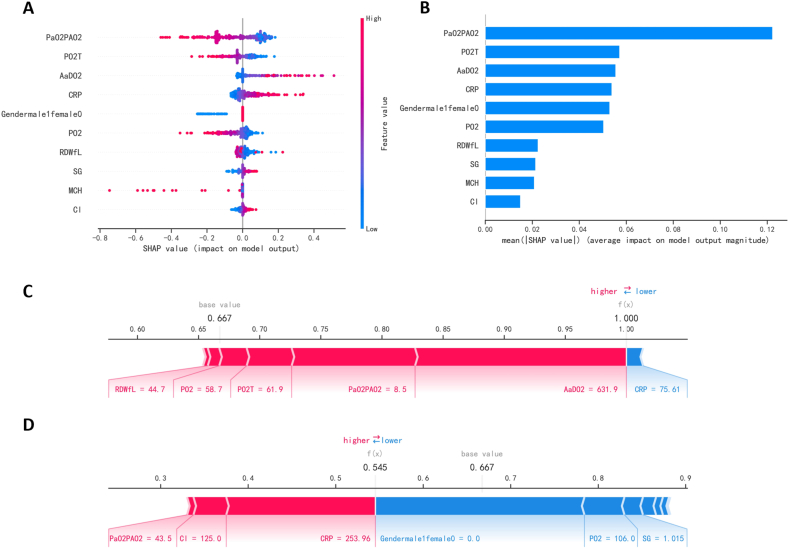
The SHAP diagram (Fig. 4A) depicts the role of each feature in the validation set in predicting ARDS. From blue to red, indicating that the abscissa's absolute value increases from small to large. When the abscissa is negative and the absolute value is larger, the possibility of negative prediction results is greater. In contrast, when the abscissa is positive and the absolute value is larger, the possibility of positive prediction results is greater. For example, the greater PaO2/PAO2, the less likely the patient is to develop ARDS, yet the patient is more likely to do so the larger of A-aDO2. The priority ranking of each variable (Fig. 4B) shows that PaO2/PAO2, A-aDO2, PO2(T), CRP, and gender are more relevant variables. In terms of features, PaO2/PAO2 is the most significant feature variable, followed by A-aDO2, PO2(T), CRP, and gender.
Fig. 4.
Interpretation of the model. (A) SHAP plot of 10 key variables. (B) Importance ranking chart of 10 key variables. (C, D) Show patients with positive (ARDS) and negative (NO-ARDS) predictions, respectively.
Two force diagrams exhibit how the features of the two cases affect the results (Fig. 4C and D). A patient who developed ARDS was predicted to be positive by the model (Fig. 4C). In this case, the longest red part is A-aDO2 (631.99 mmHg), which is the greatest contributor to ARDS in the patient. The second largest positive impact on the results is PaO2/PAO2 (8.5 mmHg), and the largest negative impact on the results is CRP (75.61 mg/L). Similarly, a patient who didn't develop ARDS was predicted to be negative (Fig. 4D). The three variables that possess the most positive effects are CRP (253.96 mg/L), Cl (125 mmol/L), and PaO2/PAO2 (43.5 mmHg). On the contrary, the most negative effects were gender (female), PO2 (106 mmHg), and urine specific gravity (1.015).
4. Discussion
This may be the first attempt to construct a clinical prediction model for ARDS in sepsis patients in the ICU with limited and easily available clinical data using machine learning. In this retrospective cohort study, we compared the baseline characteristics of sepsis patients and identified 10 clinical variables originating from readily available clinical data to establish a prediction model for ARDS. The results of this study and the established model could lead to early, accurate identification and personalized treatment of ARDS. Compared with several of the previous ARDS prediction models [31,32], our model performed better in terms of the overlap of several variables. The AUC of the overlapping variables for predicting ARDS incidence was only 0.626 in the training set (Supplemental Fig. 1), which was significantly lower than that of our model. On the other hand, the pathogenesis of COVID-19 and sepsis is not the same, so it is normal to predict ARDS with different variables. In addition, compared with other prediction models, ARDS can be predicted within 24 h before they reach the Berlin definition. Interestingly, the model requires fewer clinical indicators, which means that the patient's medical expenses can be saved to a large extent. Overall, the risk of ARDS in sepsis patients in the ICU can be predicted based on clinical variables alone, at least in selected populations with sepsis, and the model performed better than previous ARDS prediction models did, which is the novelty of our work.
Due to the SHAP values, our research becomes interpretable machine learning. Several features, such as PaO2/PAO2, A-aDO2, PO2, and gender, have been identified by previous risk score models [[33], [34], [35]]. Notably, several points in our study were not noted in previous models, namely, CRP, RDW (fL), MCH, and SG. These are significant characteristics neglected by traditional risk scores. Studies have shown that the negative predictive value of the CRP level remains reasonable when comparing patients with no sepsis vs. confirmed, possible, or uncertain sepsis [36]. Moreover, CRP levels in ARDS patients are generally high. ARDS is associated with the activation of inflammatory cells and the release of inflammatory factors. The RDW/albumin ratio is a predictive prognostic biomarker for ARDS patients [37]. In addition, sepsis can induce red blood cell dysfunction, as indicated by decreased mean corpuscular hemoglobin content and erythrocyte deformability. Mechanical ventilation is generally required for patients with sepsis-associated ARDS, and the PaO2/PAO2 ratio plays the most significant role in this model, as it reflects pulmonary ventilation function and helps to determine the severity of ARDS. Studies have shown that adjusting the mechanical ventilation settings according to the patient's condition is expected to improve lung function and clinical outcome [38]. Similarly, A-aDO2 is used to judge lung ventilation function and sensitively reflects lung oxygen uptake. The variable indicators involved in the model are not only easy to obtain but also highly representative.
Although this study developed and validated an early dynamic prediction model for sepsis-related ARDS, which provides some support for early clinical measures for high-risk patients, there are still some limitations, and additional work is needed. First, we opted to analyze patients admitted to ICUs in China. Due to differences in medical status, ICU conditions, and laboratory examination conditions among various countries, the results of this study may be more applicable to sepsis patients admitted to ICUs in China. Second, this was a single-center retrospective analysis. Inevitably, the results of the study will be biased due to differences in the diagnosis and treatment levels at each hospital. Our findings will be more reliable and can be extended to other regions through multiregional and multicenter cooperation in the future. In addition, the number of eligible patients was limited. In the future, we intend to include more patients in prospective studies to validate our findings. Finally, no imaging data were collected. Compared with comprehensive imaging and laboratory data, simple laboratory examination data are not detailed enough. Nevertheless, the cost of prediction and medical expenses can be saved for patients only by using the data. In the future, more measures will be integrated into the diagnostic system to achieve personalized treatment.
5. Conclusions
This study developed a machine learning model that can predict sepsis-associated ARDS early, exclusively utilizing clinically available data, and can guide clinicians to take appropriate preventive measures to improve the clinical prognosis of high-risk patients.
Data availability statement
Data will be made available on reasonable request to the corresponding authors.
Fundings
This work was supported by the National Natural Science Foundation of China (Nos. 82172832) and the Wisdom Accumulation and Talent Cultivation Project the Third Xiangya Hospital of Central South University (YX202108).
CRediT authorship contribution statement
Zhenzhen Jiang: Writing – review & editing, Writing – original draft, Data curation, Conceptualization. Leping Liu: Writing – review & editing, Validation, Methodology, Formal analysis. Lin Du: Formal analysis, Data curation. Shanshan Lv: Formal analysis, Data curation. Fang Liang: Methodology, Formal analysis, Data curation. Yanwei Luo: Writing – review & editing, Supervision, Funding acquisition, Conceptualization. Chunjiang Wang: Writing – review & editing, Supervision, Methodology, Conceptualization. Qin Shen: Writing – review & editing, Supervision, Methodology, Data curation.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
The authors acknowledge those patients for providing valuable clinical datathe Extreme Smart Analysis platform (https://www.xsmartanalysis.com/).
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.heliyon.2024.e28143.
Contributor Information
Chunjiang Wang, Email: wongcj@csu.edu.cn.
Qin Shen, Email: shenqin@csu.edu.cn.
Appendix A. Supplementary data
The following is/are the supplementary data to this article.
References
- 1.Meyer N.J., Gattinoni L., Calfee C.S. Acute respiratory distress syndrome. Lancet. 2021;398(10300):622–637. doi: 10.1016/S0140-6736(21)00439-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Villar J., et al. A clinical classification of the acute respiratory distress syndrome for predicting outcome and guiding medical therapy*. Crit. Care Med. 2015;43(2):346–353. doi: 10.1097/CCM.0000000000000703. [DOI] [PubMed] [Google Scholar]
- 3.Fan E., Brodie D., Slutsky A.S. Acute respiratory distress syndrome: advances in diagnosis and treatment. JAMA. 2018;319(7):698–710. doi: 10.1001/jama.2017.21907. [DOI] [PubMed] [Google Scholar]
- 4.Ranieri V.M., et al. Acute respiratory distress syndrome: the Berlin Definition. JAMA. 2012;307(23):2526–2533. doi: 10.1001/jama.2012.5669. [DOI] [PubMed] [Google Scholar]
- 5.Matthay M.A., et al. Acute respiratory distress syndrome. Nat. Rev. Dis. Prim. 2019;5(1):18. doi: 10.1038/s41572-019-0069-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Papazian L., et al. Diagnostic workup for ARDS patients. Intensive Care Med. 2016;42(5):674–685. doi: 10.1007/s00134-016-4324-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vincent J.L., et al. Sepsis definitions: time for change. Lancet. 2013;381(9868):774–775. doi: 10.1016/S0140-6736(12)61815-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kaukonen K.M., et al. Systemic inflammatory response syndrome criteria in defining severe sepsis. N. Engl. J. Med. 2015;372(17):1629–1638. doi: 10.1056/NEJMoa1415236. [DOI] [PubMed] [Google Scholar]
- 9.Dellinger R.P., et al. Surviving sepsis campaign: international guidelines for management of severe sepsis and septic shock. Crit. Care Med. 2012;41(2):580–637. doi: 10.1097/CCM.0b013e31827e83af. 2013. [DOI] [PubMed] [Google Scholar]
- 10.Singer M., et al. The Third international consensus definitions for sepsis and septic shock (Sepsis-3) JAMA. 2016;315(8):801–810. doi: 10.1001/jama.2016.0287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Guillen-Guio B., et al. Sepsis-associated acute respiratory distress syndrome in individuals of European ancestry: a genome-wide association study. Lancet Respir. Med. 2020;8(3):258–266. doi: 10.1016/S2213-2600(19)30368-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sheu C.C., et al. Clinical characteristics and outcomes of sepsis-related vs non-sepsis-related ARDS. Chest. 2010;138(3):559–567. doi: 10.1378/chest.09-2933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wheeler A.P., Bernard G.R. Acute lung injury and the acute respiratory distress syndrome: a clinical review. Lancet. 2007;369(9572):1553–1564. doi: 10.1016/S0140-6736(07)60604-7. [DOI] [PubMed] [Google Scholar]
- 14.Matthay M.A., et al. Future research directions in acute lung injury: summary of a National Heart, Lung, and Blood Institute working group. Am. J. Respir. Crit. Care Med. 2003;167(7):1027–1035. doi: 10.1164/rccm.200208-966WS. [DOI] [PubMed] [Google Scholar]
- 15.Bellani G., et al. Epidemiology, patterns of care, and mortality for patients with acute respiratory distress syndrome in intensive care units in 50 countries. JAMA. 2016;315(8):788–800. doi: 10.1001/jama.2016.0291. [DOI] [PubMed] [Google Scholar]
- 16.Auriemma C.L., et al. Acute respiratory distress syndrome-attributable mortality in critically ill patients with sepsis. Intensive Care Med. 2020;46(6):1222–1231. doi: 10.1007/s00134-020-06010-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Luo Y., et al. Machine learning based on routine laboratory indicators promoting the discrimination between active tuberculosis and latent tuberculosis infection. J. Infect. 2022;84(5):648–657. doi: 10.1016/j.jinf.2021.12.046. [DOI] [PubMed] [Google Scholar]
- 18.Fleuren L.M., et al. Machine learning for the prediction of sepsis: a systematic review and meta-analysis of diagnostic test accuracy. Intensive Care Med. 2020;46(3):383–400. doi: 10.1007/s00134-019-05872-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ming T., et al. Integrated analysis of gene Co-expression network and prediction model indicates immune-related roles of the identified biomarkers in sepsis and sepsis-induced acute respiratory distress syndrome. Front. Immunol. 2022;13 doi: 10.3389/fimmu.2022.897390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sinha P., et al. Development and validation of parsimonious algorithms to classify acute respiratory distress syndrome phenotypes: a secondary analysis of randomised controlled trials. Lancet Respir. Med. 2020;8(3):247–257. doi: 10.1016/S2213-2600(19)30369-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sinha P., Churpek M.M., Calfee C.S. Machine learning classifier models can identify acute respiratory distress syndrome phenotypes using readily available clinical data. Am. J. Respir. Crit. Care Med. 2020;202(7):996–1004. doi: 10.1164/rccm.202002-0347OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Maddali M.V., et al. Validation and utility of ARDS subphenotypes identified by machine-learning models using clinical data: an observational, multicohort, retrospective analysis. Lancet Respir. Med. 2022;10(4):367–377. doi: 10.1016/S2213-2600(21)00461-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sayed M., Riaño D., Villar J. Novel criteria to classify ARDS severity using a machine learning approach. Crit. Care. 2021;25(1):150. doi: 10.1186/s13054-021-03566-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hou N., et al. Predicting 30-days mortality for MIMIC-III patients with sepsis-3: a machine learning approach using XGboost. J. Transl. Med. 2020;18(1):462. doi: 10.1186/s12967-020-02620-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yan J., et al. LightGBM: accelerated genomically designed crop breeding through ensemble learning. Genome Biol. 2021;22(1):271. doi: 10.1186/s13059-021-02492-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sadozai H., et al. Distinct stromal and immune features collectively contribute to long-term survival in pancreatic cancer. Front. Immunol. 2021;12 doi: 10.3389/fimmu.2021.643529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gregory G.A., et al. Global incidence, prevalence, and mortality of type 1 diabetes in 2021 with projection to 2040: a modeling study. Lancet Diabetes Endocrinol. 2022;10(10):741–760. doi: 10.1016/S2213-8587(22)00218-2. [DOI] [PubMed] [Google Scholar]
- 28.Singh S.K., et al. Predicting sustainable arsenic mitigation using machine learning techniques. Ecotoxicol. Environ. Saf. 2022;232 doi: 10.1016/j.ecoenv.2022.113271. [DOI] [PubMed] [Google Scholar]
- 29.Zhang S., et al. Efficient kNN classification with different numbers of nearest neighbors. IEEE Transact. Neural Networks Learn. Syst. 2018;29(5):1774–1785. doi: 10.1109/TNNLS.2017.2673241. [DOI] [PubMed] [Google Scholar]
- 30.Zhou S. Sparse SVM for sufficient data reduction. IEEE Trans. Pattern Anal. Mach. Intell. 2022;44(9):5560–5571. doi: 10.1109/TPAMI.2021.3075339. [DOI] [PubMed] [Google Scholar]
- 31.Singhal L., et al. eARDS: a multi-center validation of an interpretable machine learning algorithm of early onset Acute Respiratory Distress Syndrome (ARDS) among critically ill adults with COVID-19. PLoS One. 2021;16(9) doi: 10.1371/journal.pone.0257056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Xu W., et al. Risk factors analysis of COVID-19 patients with ARDS and prediction based on machine learning. Sci. Rep. 2021;11(1):2933. doi: 10.1038/s41598-021-82492-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Brat R., et al. Lung ultrasonography score to evaluate oxygenation and surfactant need in neonates treated with continuous positive airway pressure. JAMA Pediatr. 2015;169(8) doi: 10.1001/jamapediatrics.2015.1797. [DOI] [PubMed] [Google Scholar]
- 34.Agarwal R., et al. Etiology and outcomes of pulmonary and extrapulmonary acute lung injury/ARDS in a respiratory ICU in North India. Chest. 2006;130(3):724–729. doi: 10.1378/chest.130.3.724. [DOI] [PubMed] [Google Scholar]
- 35.Luhr O.R., et al. The impact of respiratory variables on mortality in non-ARDS and ARDS patients requiring mechanical ventilation. Intensive Care Med. 2000;26(5):508–517. doi: 10.1007/s001340051197. [DOI] [PubMed] [Google Scholar]
- 36.Stocker M., et al. C-reactive protein, procalcitonin, and white blood count to rule out neonatal early-onset sepsis within 36 hours: a secondary analysis of the neonatal procalcitonin intervention study. Clin. Infect. Dis. 2021;73(2):e383–e390. doi: 10.1093/cid/ciaa876. [DOI] [PubMed] [Google Scholar]
- 37.Yang L., et al. Monocyte-to-lymphocyte ratio is associated with 28-day mortality in patients with acute respiratory distress syndrome: a retrospective study. J Intensive Care. 2021;9(1):49. doi: 10.1186/s40560-021-00564-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Barrot L., et al. Liberal or conservative oxygen therapy for acute respiratory distress syndrome. N. Engl. J. Med. 2020;382(11):999–1008. doi: 10.1056/NEJMoa1916431. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data will be made available on reasonable request to the corresponding authors.