Abstract
The aus (Oryza sativa L.) varietal group comprises of aus, boro, ashina and rayada seasonal and/or field ecotypes, and exhibits unique stress tolerance traits, making it valuable for rice breeding. Despite its importance, the agro-morphological diversity and genetic control of yield traits in aus rice remain poorly understood. To address this knowledge gap, we investigated the genetic structure of 181 aus accessions using 399,115 SNP markers and evaluated them for 11 morpho-agronomic traits. Through genome-wide association studies (GWAS), we aimed to identify key loci controlling yield and plant architectural traits.
Our population genetic analysis unveiled six subpopulations with strong geographical patterns. Subpopulation-specific differences were observed in most phenotypic traits. Principal component analysis (PCA) of agronomic traits showed that principal component 1 (PC1) was primarily associated with panicle traits, plant height, and heading date, while PC2 and PC3 were linked to primary grain yield traits. GWAS using PC1 identified OsSAC1 on Chromosome 7 as a significant gene influencing multiple agronomic traits. PC2-based GWAS highlighted the importance of OsGLT1 and OsPUP4/ Big Grain 3 in determining grain yield. Haplotype analysis of these genes in the 3,000 Rice Genome Panel revealed distinct genetic variations in aus rice.
In summary, this study offers valuable insights into the genetic structure and phenotypic diversity of aus rice accessions. We have identified significant loci associated with essential agronomic traits, with GLT1, PUP4, and SAC1 genes emerging as key players in yield determination.
Supplementary Information
The online version contains supplementary material available at 10.1186/s12284-024-00700-4.
Keywords: Rice, aus rice, GWAS, Yield, Agronomic Traits
Background
Asian cultivated rice (Oryza sativa L.) is a crucial staple food crop for more than one half of global population (Khush 2005). Being one of the earliest domesticated crops and a model organism, the phylogenetic and geographic origins of rice is well studied (Molina et al. 2011; Gross and Zhao 2014; Gutaker et al. 2020). Based on ecology, genetics and genomics, O. sativa is broadly classified into indica, aus, temperate japonica, tropical japonica and aromatic groups (Glaszmann 1987; Garris et al. 2005; Zhao et al. 2011; Wang et al. 2018). The aus group which was initially geographically assigned to South and West Asia (Glaszmann 1987), is now suggested to have originated from central India or Bangladesh based on comprehensive genomic data (Civáň et al. 2015). The aus group comprises of two seasonal ecotypes: aus and boro (Li et al. 2014; Alexandrov et al. 2015) distributed in both Bangladesh and India (Travis et al. 2015). The ‘boro’ indicates the cropping season spanning December to May, while ‘aus/ahu’ refers to April to August. Both these ecotypes have been traditionally selected to complete the life cycle in a short period and to have tolerance to abiotic stresses like drought, cold and heat. Furthermore, the aus group also includes deep-water cultivars (referred as ‘ashina’ in Glaszmann 1987) from Assam and Bangladesh, as well as the ‘rayada’ cultivars originating from a small geographical area along the Madhumati river in Bangladesh (Rubaiyath Bin Rahman and Zhang 2013). All these aus cultivar types have been categorized as circum-aus in Wang et al. (2018). In this paper, we have the term ‘aus’ in italic font to referrer to the genetic group, while ‘aus’ in non-italic font refers to seasonal ecotype.
Aus cultivars group has immense potential for utilization in breeding due to its tolerance many abiotic stress factors. While stress tolerance studies have traditionally focused on the Japonica variety Nipponbare, which benefits from available genetic resources and a reference genome, recent advancements in de novo reference genomes for aus cultivars like N22 and Kasalath offer valuable insights into aus-specific genes and pathways. Genetic analysis of the 3,000 Rice Genome Project (3K-RGP) accessions (Li et al. 2014) revealed a greater abundance of ‘private’ alleles in aus compared to other rice groups, particularly around major domestication genes like Sh4, sd1, Wx, and Rc that control traits such as grain shattering, semi-dwarf height, grain amylose content, and pericarp colour, respectively (Wang et al. 2018) Moreover, considerable population structural diversity within aus (Norton et al. 2018) can be exploited suitably for rice improvement.
A range of crucial stress tolerance genes such as OsSub1, SNORKELs, OsPSTOL1, and Dro1, were first reported in aus genotypes (Bin Rahman and Zhang 2018). Notably, these genes are absent from the Nipponbare reference sequence. This underlines the potential of aus germplasm for unveiling novel allelic variations associated with crucial agronomic traits to safeguard rice production from the progressive changes in global climate causing frequent extreme weather events like drought, flooding, and high temperature. With high-quality SNP data accessible for diverse rice germplasm panels like BAAP and 3 K-RGP (Rice SNP-seek database; https://snp-seek.irri.org), genome-wide association studies (GWAS) offers a compelling approach to uncover natural variations pertaining to agronomic, grain quality, and stress tolerance traits (Norton et al. 2018; Bhandari et al. 2020).
To date, the morphological diversity of aus rice has not been evaluated on a global scale. Therefore, it is interesting to explore the genetic basis of the phenotypic diversity which will help better utilization of aus germplasm in rice breeding. In this study we evaluated 181 aus rice from the 3 K-RGP panel for 42 agro-morphological traits with following objectives: (i) to understand how well the agro-morphological diversity correlates with the population genetic structure, and how it relates to the origin and distribution of aus cultivars, (ii) to determine the genetic factors associated with the agronomical features of aus rice using GWAS, and compare those with earlier reports on diverse rice germplasm.
Materials and Methods
Plant Materials and Growth Conditions
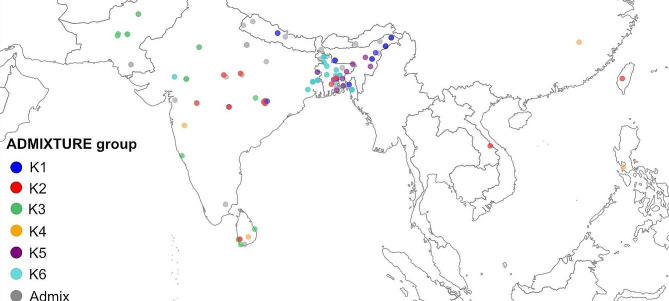
A total of 181 aus rice accessions from the 3000 Rice Genome Project (3 K-RGP) (Li et al. 2014) were included in the study (Dataset S1). Originally, there were 214 aus accessions in the 3 K-RGP, but we could obtain seeds of 181 accessions from the IRRI genebank. The passport data on the origin of accessions were obtained from Genesys (https://www.genesys-pgr.org/). The geographic distribution of the aus accessions is shown in Fig. 1.
Fig. 1.
Geographical distribution ofausrice accessions used in the study. A few accessions from other parts of the world have not been shown here. Each point indicates the geolocation of the cultivar as given in https://www.genesys-pgr.org. The colour of the points indicates to which aus sub-group the accession belongs (see Fig. 2) based on population structure analysis at K = 6
During the wet season (June-November) of 2020, 181 aus accessions were cultivated at Hazaribag, Jharkhand (23.9596 °N, 85.3739 °E, 600 m) under rainfed conditions. Each accession was grown under puddled transplanted conditions in a 2.5 m × 1.5 m plot with a spacing of 20 cm (row-to-row) and 15 cm (plant-to-plant) following an augmented block design. Three check varieties: Vandana, Sahbhagi Dhan and IR64 were included. Standard production practices were followed to manage the crop.
SNP Genotypic Data and Filtering
The genotypic data for the 181 accessions were obtained from the data repository of 3 K-RGP (https://snp-seek.irri.org/). The raw genotypic data we considered from 3 K-RP Base SNP dataset which contained 18,128,777 SNPs. The Base SNP dataset was originally created from ~ 29 million biallelic SNPs by removing SNPs with excess of heterozygous calls. The marker set was then filtered using nucleotide variation missing rate < 0.20 and a minor allele frequency (MAF) > 0.01 using PLINK (Purcell et al. 2007).
Population Structure
The MAF-filtered data was further thinned by applying a two-step LD pruning using PLINK (“indep-pairwise 10 kb 1 0.8” and “indep-pairwise 50 1 0.8”). The resulting set of 399, 115 SNP (referred to as 399 K set) was then used for population structure analysis.
The population structure of 181 aus genotypes was assessed using ADMIXTURE v. 1.3.0 (Alexander et al. 2009). Sub-populations from K = 1 to 8 were tested. Since the cross-validation error barely differed between K values beyond K = 6, we defined K = 6 as optimal clusters in aus germplasm (Fig. S1). An 80% threshold of cluster membership was used to assign cultivars into population sub-groups. Furthermore, population structure analysis of aus genotypes from India (71 accessions) and Bangladesh (76 accessions) was assessed independently using similar criteria. The Principal component analysis (PCA) for all accessions was done using TASSEL5 (Bradbury et al. 2007). A neighbour-joining tree was built by calculating the pairwise genetic distances between samples using the VCF2Dis software (https://github.com/BGI-shenzhen/VCF2Dis). From the genetic distance matrix, a neighbor-joining tree was built using the programme FastME (Lefort et al. 2015).
Phenotype Data Recording and Statistical Analysis
Forty-two phenotypic traits comprising of 11 agronomical and 30 qualitative morphological variables, were evaluated following standard procedure (IRRI 2013); for details see Dataset S2). Data on agronomical variables were recorded from 15 randomly chosen plants in each plot (excluding the border rows) and grain yield data was recorded from plot yield as yield per m2 after threshing and drying the seed to around 14% moisture content.
The phenotyping data for agronomic (quantitative) variables were analyzed using an augmented block design with the R package augmentedRCBD (R Core Team 2021). Adjusted means, range, skewness, kurtosis, coefficient of variation (CV), genetic coefficient of variation (GCV), phenotypic coefficient of variation (PCV), broad sense heritability (hBS) and frequency distribution were calculated.
The 181 aus accessions were classified into seven genetic clusters: K1 to K6, and ‘admix’, based on Admixture analysis results. A univariate analysis using a general linear model for quantitative traits was performed for seven genetic cluster in SPSS Statistics v.21 (IBM, Armonk, NY). The ANOVA was calculated to find the significance of difference among the groups means. In addition, the comparison of means within each group was carried out using Tukey’s test at P < 0.05.
To investigate the relationships among agronomical variables and the factors underlying the trait variation, a principal component analysis (PCA) was carried out for 11 traits using GraphPad Prism v. 9.0. The variables were standardized to have a mean of 0 and standard deviation (SD) of 1 before analysis, and the principal components (PCs) were selected based on parallel analysis which performs 1000 Monte Carlo simulations on “random data” of equal dimension to the input data, and finally selects PCs with eigen values greater than those from the simulations at 95% percentile level.
Genome-wide Association Analysis
The three principal components (PCs) obtained from PCA of 11 agronomical traits, along with the log10-transformed values of individual traits were included in GWAS (Dataset S3). We used a separate larger set of 458, 615 SNPs (referred as 458 K set) of 181 aus genotypes which was filtered from the base SNP set using the criteria: missingness (0.25) and MAF (0.05) and LD pruning (indep-pairwise 2 kb 1 0.8). The Fixed and Random Model Circulating Probability Unification (FarmCPU) model (Liu et al. 2016) in the R package of Genomic Association and Prediction Integrated Tool (GAPIT) (Wang and Zhang 2021) was used for GWAS analysis. The FarmCPU model is a multi-locus linear mixed model (MLMM) that improves statistical power and reduces both false positives and false negatives (Liu et al. 2016; Kaler et al. 2019). Population structure was accounted for by using a kinship matrix to reduce the occurrence of false positives and spurious associations. Quantile–quantile (Q-Q) plots of the estimated and observed P-values for marker–trait associations were generated to evaluate the model fit.
The critical P-value for explaining a significantly associated marker was the rather conservative Bonferroni correction, calculated by the–log10(p-value of 0.05/ΣSNPs), which corresponds to -log10(0.05/458,615) = 6.96. The percentage of total phenotypic variance (PVE) explained by significant MTAs was generated in GAPIT. The PVE of the markers is calculated in GAPIT as their corresponding variance divided by the total variance, which is the sum of residual variance and the variance of the associated markers, calculated using the R/lme4 package.
Linkage Disequilibrium and Prediction of Candidate Genes
Linkage disequilibrium (LD) decay was measured by correlation coefficients (r2) for all pairs of SNPs with a sliding window approach with the following parameters: -MaxDist 500-MAF 0.05-Het 0.88-Miss 0.999 using PopLDdecay v3.27 (Zhang et al. 2019). The LD decay distance was determined when the LD r2 fell to 0.1. Considering the LD decay distance, we defined the interval of significantly associated SNP(s) ± LD decay distance as QTL regions.
The identified QTL regions covered by significant SNPs were searched for candidate genes or QTLs using the Rice SNP seek database (https://snp-seek.irri.org/_jbrowse.) which integrates various databases like QTARO, Oryzabase and MSU databases. For trait-associated SNPs, contingency tables between SNP alleles and phenotype were made and visually inspected to examine the associations.
The LDBlockshow (Dong et al. 2020) was used to estimate the local LD blocks within the QTL/ gene. Gene haplotype analysis was performed using all SNPs within the coding sequence region ignoring the synonymous SNPs. Haplotype analyses were done for the aus germplasm as well as the 3,020 accessions in the 3 K-RG using the Rice SNP-Seek database. Significant phenotypic differences among the haplotypes were determined using Tukey’s multiple comparisons test in one-way ANOVA using GraphPad Prism v. 9.0.
Results
Genetic Structure and Subgroupings Within aus rice
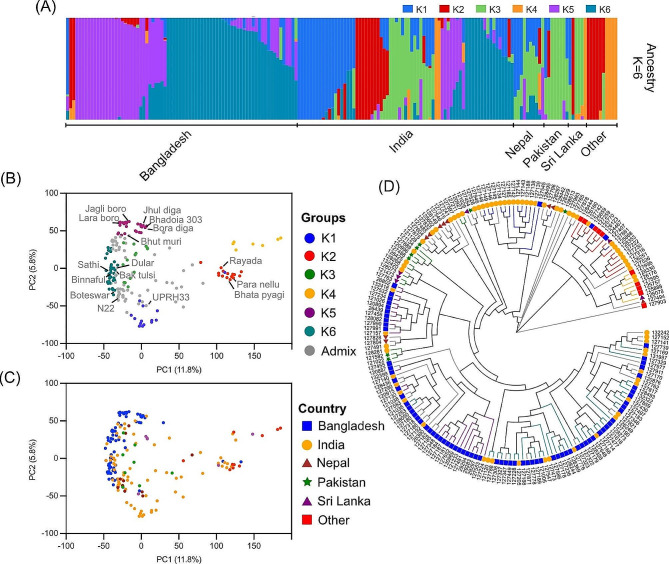
The patterns of the genetic structure of 181 aus accessions were analyzed using the 399 K SNP set. Using the cross-validation error values generated in ADMIXTURE by varying sub-groupings (K) from 1 to 8 (Fig. S1A), we found six subgroups (at ≥ 80% cut-off) designated as K1 to K6 (Fig. 2A). At K = 6, the aus germplasm from Bangladesh is mostly comprised of two clusters, whereas, the Indian germplasm exhibited a richer diversity with five clusters (Fig S1B-C; Dataset S1). The distinctness of six aus subgroups is also apparent in the PCA (Fig. 2B). To check the genetic differentiation of aus and boro ecotypes, we examined the clustering at K = 2. Interestingly, at this level, aus and boro ecotypes appeared to be genetically close as both were included in the same cluster (Dataset S1). However, at K = 6, the boro cultivars showed some degree of differentiation and all are grouped under K5 (Fig. 2B).
Fig. 2.
Population structure ofausrice.A The plot of ADMIXTURE subpopulation membership coefficients at K = 6. The cross-validation error values at different K indicates K = 6 as the most ideal sub-groups, B Biplot of first two PCA axes of 181 aus accessions colour coded according to the ADMIXTURE classification at K = 6, C Biplot of first two PCA axes of 181 aus accessions colour coded according to their geographical association. D NJ tree based on pairwise genetic distance. The accessions were colour coded according to the ADMIXTURE grouping (branch colour) as well as geographic origin as given in B and C
Genetic distance-based analysis revealed considerable geographical structuring (Figs. 1 and 2C and D). First, the genetic groups identified through ADMIXTURE analysis seem to be clustering geographically well. K1, K5 and K6 are close to each other on the map and these populations have low FST values (Table S1). K1 is mostly (87%) of Indian origin, while K5 and K6 are predominantly (86% and 75%, respectively) originating from Bangladesh. Overall, Among the subgroups, K3 and K6 are the closest (FST = 0.162), while K2 and K5 are the most distant (FST = 0.518).
Interestingly, although K3 has low overall FST with K1, K5 and K6, yet is geographically distant (located in North West India, Pakistan and Sri Lanka). As K3 included many early maturing drought-tolerant accessions (assessed in our separate study), it is possible that this groups was further extended geographically for their drought tolerance. The K2 represented by the rayada cultivar from Bangladesh along with the accessions from central India, Sri Lanka and the countries outside the Indian subcontinent had greater genetic distance than all other aus clusters. This is consistent with the previous reports that rayada cultivars are genetically distinct from most other aus cultivars (Wang et al. 2014). Overall, the geographical distribution and selection of accessions for specific ecologies seem to have played crucial roles in shaping the population structure of aus rice.
Overall Agro-morphological Variability of aus rice
The analysis of agronomic traits revealed high phenotypic variability within aus population (Table S2; Fig. S2). Deviations from the normality have been observed from most of the traits except 1000-grain weight and grain yield plot− 1. Overall, as compared to check varieties, the aus genotypes exhibited early heading, taller plant height, fewer tiller, longer flag leaves, lesser grain weight, as well as lower yield and harvest index. Accessions such as P335 (342), Vaikatharyan (305), ARC 10,100, Begum, AUS177, Han Nuo, Jashure aus, Malagkit, and ARC13276 recorded higher (> 200) grains panicle− 1. Although a single accession, AUS177, showed higher yield than the best check Sahbhagi dhan (1375.2 g), there were ten other accessions which showed a higher yield than the second best check i.e., IR64 (1200.59 g). Noteworthy accessions with higher yield and harvest index were R762, PR106, Rantnagiri 45 − 2, Jabor sail, I Kung Pao, N22, NCS 840, Bhut muri, Herath banda, and Narikel badi. The correlation among the traits in aus germplasm is shown in (Fig. S3). Largely, the late maturing genotypes seems to have greater values of plant height, panicle length, spikelet number, panicle weight and yield.
Phenotypic Diversity Among aus Genetic Subgroups
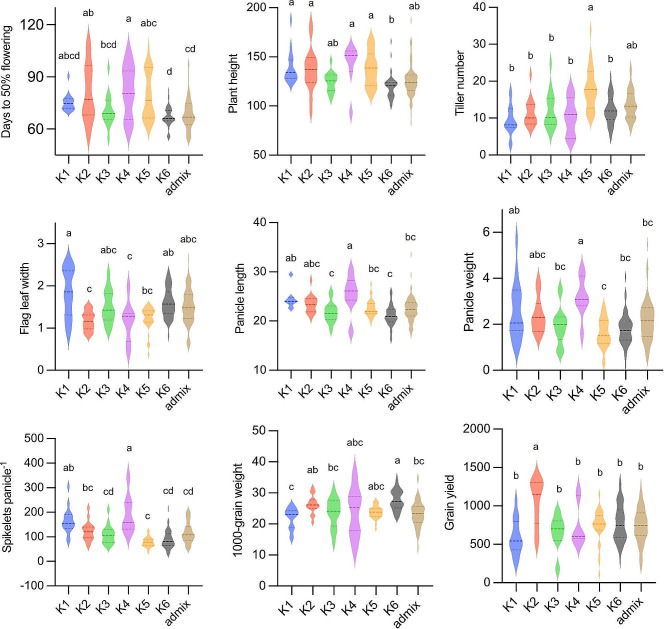
We have analyzed the agro-morphological differences between the aus subgroups identified through ADMIXTURE analysis. There are significant differences for most of the agronomical traits except flag leaf length and harvest index (Fig. 3; Dataset S4). However, for 30 qualitative traits, we have found considerable overlapping among the subgroups for many of the traits (Fig. S4-S5).
Fig. 3.
Differences for quantitative morphological traits among theaussubgroups. Multiple comparison of Trait means of subgroups was done using the Tukey’s HSD test. The trait means of subgroups with the same letters above the violin plots are not significantly different. The traits not significantly differing among the subgroups are not shown here
The K1, represented mostly by Assam rice accessions, is poor yielding due to lower tillering despite bearing higher number of spikelets per panicle. K2, the genetically most distinct subgroup, has the highest yield potential resulting from heavier panicles with higher spikelet numbers. K3 subgroup is early maturing with a moderate yield level. The accessions belonging to K4 have low yield potential largely due to delayed flowering and tall plants, despite having the most desirable panicle traits. The boro and deep-water cultivars grouped in K5 showed wide variation for days to heading, taller plants, high tillering ability, and inferior panicle traits resulting in a moderate level of grain yield. The K6 included drought tolerant cultivars having earliest to flowering, shortest plant height, and smaller panicles with lesser but heavier grains, resulting in a moderately high yield level (Fig. 3). Overall, the phenotypic variability of the genetic groups showed substantial linkage with their growing ecology or geographical distribution.
The frequency distribution of 30 qualitative morphological variables within different aus subgroups indicated considerable overlapping among the genetic groups for most of the traits. Purple colouration of basal leaf sheaths, internodes, apiculi, and auricles occurred in higher frequency in K6 and K5, and less in K4. The accessions of K5 and K6 subgroups showed spreading type plant architecture in higher frequency. The presence of awnned cultivars was noted in all subgroups, but occurred in higher frequency in K5, K3 and K4. Red seed coat colour is most frequent (90%) in K6. The glutinous endosperm was mostly frequent in the accessions of K1 and K2 which explained by their geographical distribution in Northeastern India and southeast Asian countries where glutinous rice is preferred.
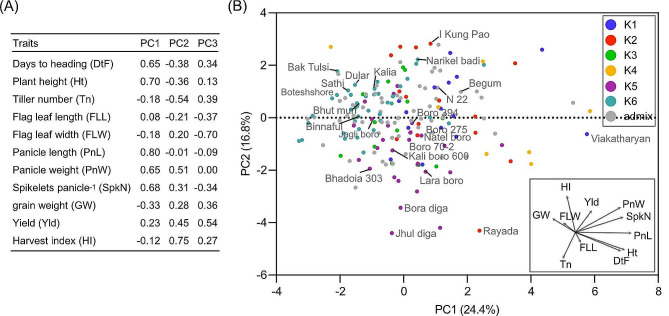
We performed a PCA using 11 quantitative agronomic traits to investigate the relationships among traits and the factors underlying the trait variation. Altogether, three principal components, PC1, PC2 and PC3, were selected which explained 24.4%, 16.8% and 13.9% of the trait variance, respectively (Fig. 4A). The PC1 explained variation in agro-morphological traits arising from plant architecture and flowering as it was positively loaded with days to flowering, plant height, panicle length, panicle weight and spikelets per panicle. This result suggested that accessions with high PC1 scores exhibited larger and heavier panicles, taller plants and longer days to heading. The grain weight and tiller number loaded negatively on PC1, indicating a trade-off relationship between grains per panicle and 1000-grain weight, as well as between tiller number and panicle size. Similarly, PC2 mostly explained variation in traits directly related to grain yield by showing positive loading by panicle weight, grain yield and harvest index. On PC2 days to heading and tiller number showed negative loading on PC2, suggesting a negative correlation between days to flowering and harvest index which is consistent with the observation that prolonged vegetative growth due to late heading leads to a reduced harvest index. The PC3 also explained variation for grain yield. The accessions with high PC3 scores mostly exhibited higher grain yield.
Fig. 4.
Principal component analysis for morpho-agronomic traits in 181auscultivars.A Summary of first three PCs for 11 traits. B PCA biplot showing the distribution of aus accessions based on the trait loadings (shown in the inset) on the first two PCs. The accessions were colour coded according to their classification into six genetic subgroups based on 399 K SNP dataset
The correspondence between the patterns of genetic and morphological diversity was checked by performing a biplot analysis of PC1 and PC2 (Fig. 4B). The boro and deep-water accessions (belonging to K6) were found to be morphologically distinct from the drought tolerant cultivars of K5. However, the genetic distance between these two groups is less (FST = 0.183). The rayada cultivar remained distinct from the rest of the aus both genetically and agronomically. The genetic distinctness of cultivars of K4 was also reflected in their morphological clustering. These results indicated that the selection of diverse stress tolerant and high yielding aus cultivars is feasible for breeding programmes.
GWAS for PC Scores
We conducted GWAS using the first three PCs (PC1 to PC3) as well as 11 traits using a 458 K SNP set to identify the key loci controlling agronomic characteristics of aus rice. The normality of the PCs and individual traits was checked using the Kolmogorov-Smirnov test. PC1 showed slight deviation from normality (P = 0.0117), while PC2 and PC3 showed normal distribution. Among the 11 morpho-agronomic traits, except for 1000-grain weight and yield plot− 1, we found significant deviations from the normal distribution. This result corroborates with earlier observations (Yano et al. 2019) that PCA can transform skewed data to a normal distribution, which is useful to improving the statistical power of GWAS.
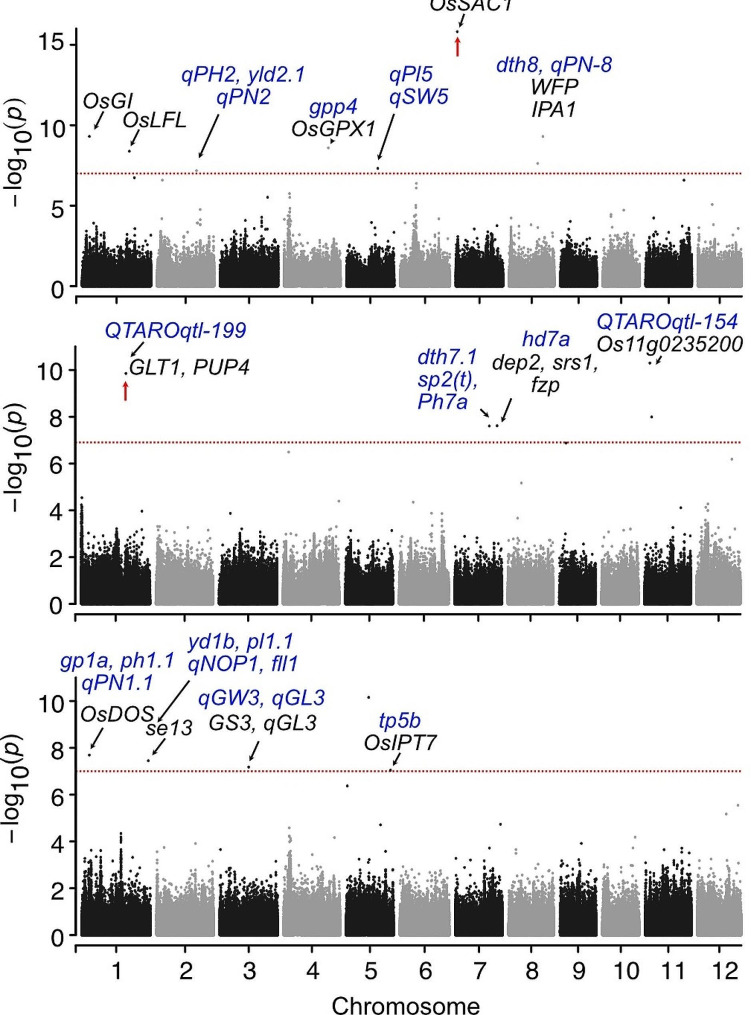
Except for PCs, grain weight and yield, we used log10-transformed values of the rest of the traits to conduct GWAS using the FarmCPU model with corrections for kinship bias (Fig. 5; Fig. S6-S7). GWAS from PCs identified 18 peaks (PC1 = 8, PC2 = 5 and PC3 = 5) with a -log10(P)- value that exceeded the Bonferroni cut-off. The significant associations detected for all three PCs are listed together along with their phenotypic variance (Table 1). Interestingly, some of the peaks detected for different PCs on Chr1 (Chromosome1), Chr5, Chr5, Chr8 and Chr11 were found to be coinciding, indicating that these peaks may represent a common flanking region (Table 1). We defined the QTL regions corresponding to the significant SNPs by expanding the upstream and downstream flanking regions according to the chromosome-wide LD decay distance analyzed in this study (Fig. S8).
Fig. 5.
GWAS for morpho-agronomic traits inausrice. Manhattan plots of PC1, PC2 and PC3 are shown. Horizontal red dotted lines represent the significant threshold for the study. The colocalization of previously identified QTLs (blue font) and genes (black font) are indicated by black arrows. The red arrows indicate the peaks we studied further
Table 1.
QTLs for morpho-agronomic traits identified in GWAS using principal components in 181 aus rice germplasm
| Trait | QTL | Chr | Flanking region (Mb) | Peak SNP position | Ref allele | -Log10(P) | MAF | Effect | PVE |
|---|---|---|---|---|---|---|---|---|---|
| PC1 (DtF, Ht, PnL, PnW, SpkN) | qPC1-1.1 | 1 | 4.36–4.68 | 4,521,776 | G | 9.30 | 0.10 | -0.69 | 5.06 |
| qPC1-1.2 | 1 | 29.49–29.81 | 29,651,634 | C | 8.38 | 0.21 | 0.53 | 1.47 | |
| qPC1-2.1 | 2 | 24.78–24.90 | 24,835,927 | G | 7.18 | 0.30 | -0.38 | 2.05 | |
| qPC1-4.1 | 4 | 27.76–27.85 | 27,801,414 | G | 8.59 | 0.20 | 0.37 | 2.05 | |
| qPC1-5.1 | 5 | 19.42–19.81 | 19,616,683 | G | 7.32 | 0.23 | 0.42 | 5.80 | |
| qPC1-7.1 | 7 | 0.73–1.05 | 888,054 | A | 15.81 | 0.16 | -1.04 | 7.97 | |
| qPC-8.1 | 8 | 17.81–18.19 | 18,001,305 | A | 7.63 | 0.16 | -0.56 | 5.13 | |
| qPC1-8.2 | 8 | 21.12–21.51 | 21,316,516 | T | 9.30 | 0.13 | 0.76 | 1.11 | |
| PC2 (PnW, Yld, HI) | qPC2-1.1 | 1 | 27.93–28.25 | 28,087,887 | T | 9.85 | 0.15 | 0.44 | 10.53 |
| qPC2-7.1 | 7 | 21.54–21.86 | 21,700,005 | T | 7.61 | 0.27 | 0.33 | 2.58 | |
| qPC2-7.2 | 7 | 26.45–26.77 | 26,611,742 | C | 7.62 | 0.12 | 0.50 | 1.23 | |
| qPC2-11.1 | 11 | 3.35–3.48 | 3,412,660 | G | 10.30 | 0.48 | -0.34 | 3.98 | |
| qPC2-11.2 | 11 | 4.49–4.62 | 4,552,359 | T | 7.99 | 0.15 | -0.54 | 1.60 | |
| PC3 (Tn, GW, Yld) | qPC3-1.1 | 1 | 4.62–4.94 | 4,789,451 | G | 7.69 | 0.07 | 0.46 | 6.20 |
| qPC3-1.2 | 1 | 41.66–41.98 | 41,821,709 | A | 7.45 | 0.33 | 0.39 | 6.51 | |
| qPC3-3.1 | 3 | 17.99–18.45 | 18,218,891 | C | 7.18 | 0.06 | -0.55 | 2.07 | |
| qPC3-5.1 | 5 | 13.96–14.35 | 14,156,143 | G | 10.16 | 0.17 | 0.48 | 5.86 | |
| qPC3-5.2 | 5 | 27.71–28.10 | 27,904,519 | C | 7.04 | 0.35 | 0.26 | 0.37 |
DtF, Days to 50% flowering; Ht, Plant height, PnL, Panicle length, PnW, Panicle weight, SpkN, Spikelets per panicle; Yld, Yield; HI, Harvest index; Tn, Tiller number, GW, 1000-grain weight; Chr, Chromosome; MAF, Minor allele frequency; PVE, Phenotypic variation explained %.
DtF, Days to 50% flowering; Ht, Plant height, PnL, Panicle length, PnW, Panicle weight, SpkN, Spikelets per panicle; Yld, Yield; HI, Harvest index; Tn, Tiller number, GW, 1000-grain weight; Chr, Chromosome; MAF, Minor allele frequency; PVE, Phenotypic variation explained %.
The PCA indicated that PC1 is representative of plant architecture and flowering, while both PC2 and PC3 are representative of grain yield. In this study, we focussed on the genetic loci responsible for plant architecture or grain yield as a trait rather than focusing on specific traits. Therefore, we focussed on the results of GWAS using PCs. The significant associations identified for each PC were colocalized with several previously reported QTLs for yield and agronomic traits when searched in databases (Fig. 5). Notably, the significant marker-trait associations (MTAs) identified in the current study were overlapping with many cloned genes such as, for PC1, OsGI (GIGANTEA) regulating days to heading, OsGPX1 (Plant Glutathione peroxidases 1) influencing plant height, spikelet number and root development, OsMADS15 influencing flowering time and plant architecture, and WFP/ IPA1 (WEALTHY FARMERS PANICLE/ IDEAL PLANT ARCHITECTURE 1) controlling yield and plant architecture. For PC2, important genes coincided with the index SNPs were OsGLT1 (NADH-glutamate synthase 1) controlling yield, dep2/SRS1 (DENSE AND ERECT PANICLE 2) regulating panicle size, fzp (frizzy panicle) influencing panicle and yield traits, and SP1 (Os11g0235200, SHORT PANICLE 1) for panicle traits.
Important genes identified for PC3 were: OsDOS (DELAY OF SENESCENCE) regulating crop maturity, SE13 (PHOTOSENSITIVITY 13) controlling heading date and yield, GS3 and qGL3 for grain traits, and OsIPT7 (Adenosine phosphate isopentenyltransferase 7) influencing yield traits. Furthermore, on Chr4 prominent peaks were detected at position ~ 3.540 Mb for both PC1 and PC3. This association corresponds to the QTL qSNP-4a and spp4-2, both reported for spikelets panicle− 1. On Chr6 another prominent peak was observed for PC1 at position ~ 10.116 Mb which has been identified as a QTL hotspot region harbouring QTLs like qSNP6 and gp6 (spikelet number), gw6 (1000-grain weight), qTN2-6-1 (tiller number at maturity), qPH2-6-1 and Ph6 (plant height at maturity), qGY6.1 (grain yield), and qHD6-1 (heading date). An important gene present in this region is Hd1 responsible for regulating photoperiodic flowering in rice. Furthermore, we observed that none of the GWAS signals identified for PCs were identified when performing GWAS using individual traits except for qPC1-2.1, which is a hit for spikelet number.
Phenotypic Effect of the Allelic Variations of MTAs
Among the eight QTLs identified for PC1, significant differences in PC scores were observed between the accession groups with the reference (‘ref’) and alternate allele for all QTLs except for qPC1-1.2 and qPC1-1.2, possibly due to their low MAF and smaller effect size, respectively (Fig. S9; Table 1). The loci identified for PC1 have shown significant allelic differences for days to heading and many of the plant architectural traits supporting our assumption for PC1 (Fig. S9). However, QTLs like qPC1-1.1, qPC1-5.1, qPC1-8.1 and qPC1-8.1 also explained the variation for grain yield. Except for qPC2-11.2, the rest of the QTLs identified for PC2 showed significant differences in PC2 scores (Fig. S10). All the PC2 QTL have explained variation for grain yield traits as expected from the trait loadings on PC2. Whereas, some of these QTL have also explained variation for flowering and architectural traits. We found significant differences in PC3 scores for two QTLs – qPC3-1.2, and qPC3-5.1. Both these QTL have explained variation for yield traits as we all as days to heading and spikelet number. Although the rest of the PC3 QTLs did not show significant variation in PC scores, but these QTLs have influenced 1000-grain weight and yield. Overall, these results indicated that the GWAS using PCs could identify QTLs for the traits which showed lesser loadings on a particular PC.
Haplotype Analysis of Potential QTLs
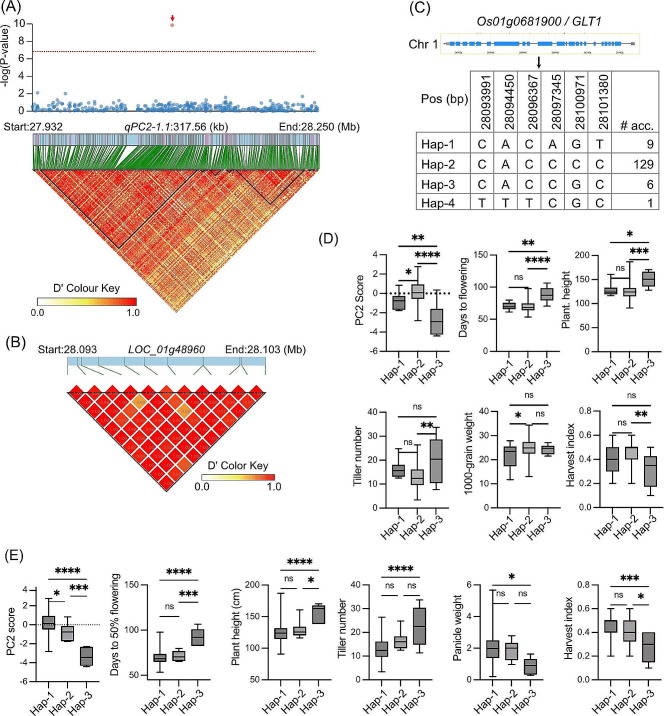
The qPC2-1.1 (peak SNP Chr1: 28,087,887) explained the highest variance for PC2 which represents grain yield, panicle weight and harvest index. We examined this QTL for genes that could influence grain yield and component traits. The QTL region was delineated to 27.93–28.25 Mb and contained 374 SNPs (Fig. 6A). In total, 58 genes including two retrotransposons are present in qPC2-1.1 (www.rapdb.dna.affrc.go.jp; Dataset S5). Among these genes, Os01g0681900, located 3,321 bp downstream of the peak SNP, and Os01g0680200, located 90.3 kb upstream of the peak SNP, were reported to be associated with grain yield and other yield-related traits.
Fig. 6.
Haplotype analysis withinqPC2-1.1. A Local Manhattan plot and LD heat map for qPC2-1.1 on Chr 1. The red arrow (Top panel) indicates the position of OsGLT1.B LD heat map of LOC_Os01g48960. C Structure and DNA polymorphism of OsGLT1. D Box plots of PC2 score and agro-morphological traits for three haplotypes Hap-1 (n = 9), Hap-2 (n = 126) and Hap-3 (n = 6) of OsGLT1. E Box plots of PC2 score and agro-morphological traits for three haplotypes Hap-1 (n = 116), Hap-2 (n = 11) and Hap-3 (n = 7) of OsPUP4. Box edges represent the 0.25 and 0.75 quantiles, with the median values shown within boxes. Whiskers extend to the most extreme point, which is no more than 1.5 times the interquartile range. Differences between the haplotypes were statistically tested using multiple comparisons with Tukey’s t test (ns, not significant, *, **, ***, and **** represent P value < 0.05, < 0.01, < 0.001, and < 0.0001, respectively)
Os01g0681900 (synonymous OsNADH-GOGAT1 or GLT1), annotated as glutamate synthase or NADH-DEPENDENT GULTAMATE SYNTHASE 1, influences grain yield by affecting panicle number, tiller number, tillering ability. It also regulates nitrogen-carbon metabolomes (Yang et al. 2016), and plays a key role in the transcriptional regulation of ammonium-responsive genes (Kojima et al. 2023). The LD plot based on 12 SNPs within GLT1 indicated strong linkage among the SNPs (Fig. 6B). We identified four haplotypes (named Hap-1 to Hap-4) using the six non-synonymous SNPs in the current 181 aus panel (Fig. 6C). Hap-2 was most frequent (present in 72% of the accessions), while Hap-4 was detected in a single accession. The average PC2 score for the accessions carrying Hap-2 was significantly higher than those with either Hap-1 or Hap-3 (Fig. 6D), indicating that GLT1 is associated with PC2. The haplotypes exhibited significant differences for most of the agro-morphological traits except for panicle weight and yield (P = 0.051) (Fig. 6D). The Hap-3 accessions showed significantly longer days to heading as well as taller plants than both Hap-1 and Hap-2 accessions (Fig. 6D). Although there were non-significant differences in grain yield among the haplotypes, Hap-2 showed considerably higher level of yield and recorded the highest 1000-grain weight and harvest index among the haplotypes. Interestingly, accessions with Hap-3 showed the highest tiller number but the lowest harvest index, indicating that those may had either low spikelet fertility or had produced many non-productive tillers. In the aus panel, Hap-1 was predominant in accessions from India, Bangladesh and Sri Lanka. While, Hap-3 accessions are mostly confined in Bangladesh and adjoining Assam. Grouping two ‘diga’ (known to be of deep-water ecology) cultivars in Hap-3 suggested that this haplotype is primarily unique to deep-water accessions. The average elongation ability under submergence stress of the six accessions belonging to Hap-3 was 149.3% (data from a separate study).
The other gene, Os01g0680200 / OsPUP4 (PURINE PERMEASE 4 syn. Big Grain 3), in qPC2-1.1 is reported to regulate grain size, along with several other traits like grain number, secondary branch number, tiller angle, days to heading, 1000-grain weight, and plant height. PUP4 was suggested to be involved in the long distance transport of cytokinin, by reinforcing cytokinin loading into vascular bundle cells (Xiao et al. 2019) (Dataset S5). We identified three haplotypes (Hap-1 to Hap-3) of this gene in the current aus genotypes based on three non-synonymous SNPs. Hap-1 was present in 90.7% while, Hap-3 was found in 3.5% of the aus panel. The haplotypes showed significant differences for PC2 scores along with days to heading, plant height, tiller number, panicle length and harvest index (Fig. 6E). Accessions possessing Hap-1 of PUP4 were early to flower, shorter in height, had lesser tillers with heavier panicles, and higher harvest index. Notably, the deep-water accessions having Hap-3 of PUP4 also carried the Hap-3 of GLT1. These accessions were considerably late maturing and taller with lightweight panicles.
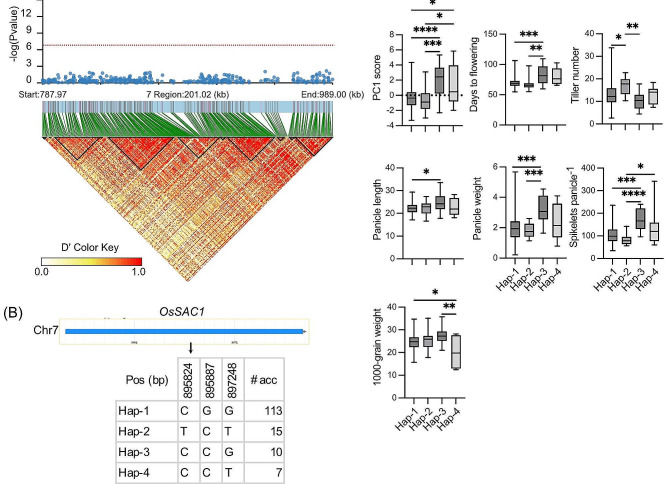
On Chr7 the qPC1-7.1 also explained high phenotypic variance and had a high effect size for PC1 which represents days to flowering and plant architectural traits. This QTL was delineated to 0.73–1.05 Mb and contained 48 genes (Dataset S6; https://rapdb.dna.affrc.go.jp/). Among the genes, Os07g0116300, annotated as OsSAC1 or SUGAR ACCUMULATION 1, is previously reported to influence grain yield, spikelet number, plant height, panicle length, 1000-grain weight, starch content, and photosynthetic rate (Zhu et al. 2017). The qPC1-7.1, identified in the present study, is a novel one as no QTLs for yield or plant architectural traits were earlier reported in this region (https://snp-seek.irri.org/), except qSS-7a for hybrid sterility (Li et al. 2008). LD block analysis of qPC1-7.1 based on 342 SNPs revealed 5 large and 6 small blocks (Fig. 7A). For SAC1, we identified six haplotypes using three non-synonymous SNPs within the gene. Out of these, each of the haplotypes 5 and 6 were detected in a single accession. Hence, we presented the results of haplotypes 1 to 4 (Hap-1 to Hap-4) (Fig. 7B). Hap-1 was the most frequent, detected in 77% of the accessions. While, Hap-4 was the least frequent and observed in 5% of the accessions. The accessions belonging to Hap-3 had the highest average PC1 score, and thereby, also had the highest values for days to flowering, panicle length, panicle weight, spikelets panicle− 1 (Fig. 7C). This haplotype also recorded the highest average 1000-grain weight. The accessions with OsSAC1 Hap4 showed wide variation for PC1 score and other traits like panicle length and 1000-grain weight. The accessions with Hap-1 and Hap-2 were early maturing but recorded lower values for panicle weight and spikelets panicle− 1. We found that OsSAC1 Hap-3 was prevalent in the accessions belonging to the genetic cluster K2 and K4. It has been shown that the K2 cluster, which also include the cultivar rayada, is characterized by heavier panicles with higher spikelet number. As well as both K2 and K4 mostly included longer duration accessions with longer and heavier panicles, and higher spikelets panicle− 1. The Hap-3 has been found to be geographically most widespread while Hap-2 is mostly confined in Bangladesh. Within the qPC1-7.1 another gene Os07g0119000 (annotated as OsMAPKKK11, MAPK Kinase Kinase 11) seems to have a role in rice growth and development (Duan et al. 2014; Guo et al. 2018, 2020), in addition to coordinating resistance to biotic and abiotic stress responses(Yamada et al. 2017; Chen et al. 2021) (Dataset S6).
Fig. 7.
Haplotype analysis withinqPC1-7.1. A Local Manhattan plot and LD heat map for qPC1-7.1 on Chr 7. The red arrow (Top panel) indicates the position of LOC_Os07g02520/ Os07g0116300.B Structure and polymorphism of OsSAC1. Four haplotypes were detected in 147 aus accessions. In the rest of the accessions either of the SNPs were missing or in heterozygous condition. (C) Box plots show significant variation among the haplotypes for PC1 score and agronomic traits. Box edges represent the 0.25 and 0.75 quantiles, with the median values shown within boxes. Whiskers extend to the most extreme point, which is no more than 1.5 times the interquartile range. Differences between the haplotypes were statistically tested using Tukey’s test (*, **, ***, and **** represent P value < 0.05, < 0.01, < 0.001, and < 0.0001, respectively)
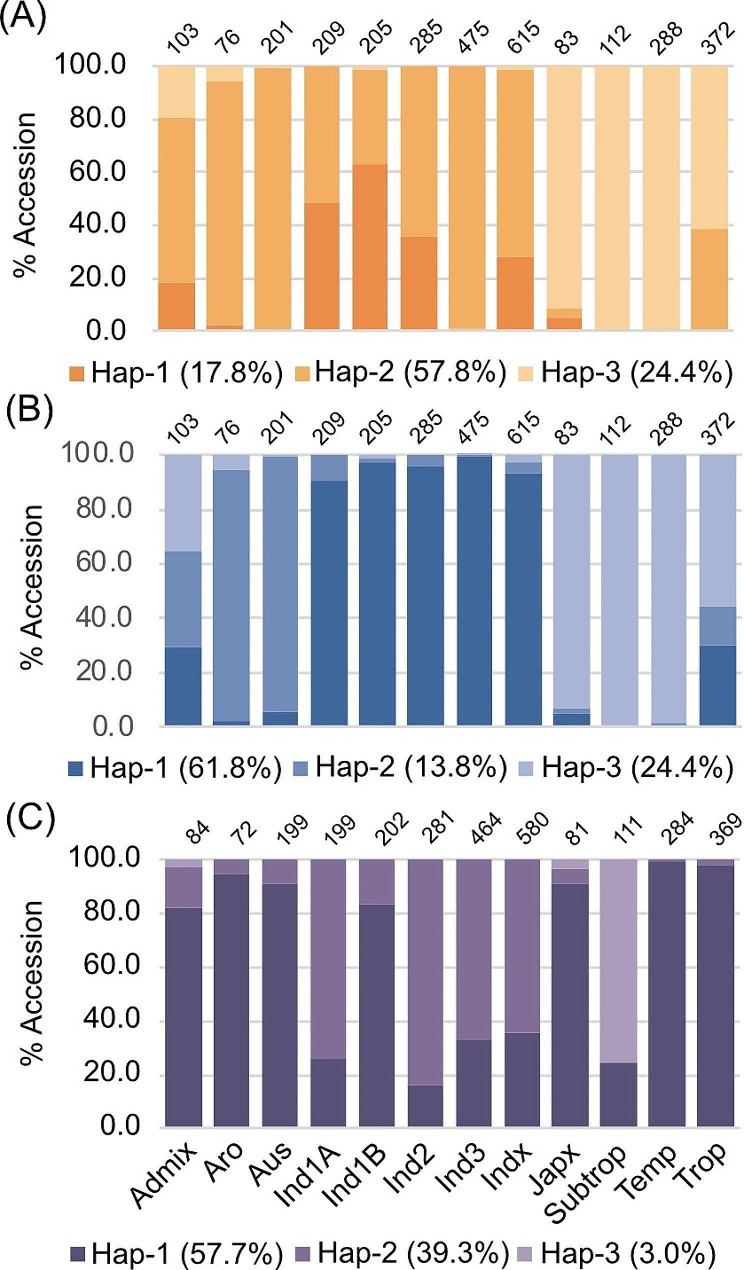
We further compared the haplotype frequency of GLT1, PUP4 and SAC1 among the 3,002 accessions from 3 K-RG panel using Rice SNP-seek web-based applications. Three haplotypes were detected for GLT1 based on 11 SNPs (out of which four appeared in our aus panel). The haplotypes showed differential distribution among the Oryza sativa subpopulations: Hap-1: indica; Hap-2: indica subgroups; aus + indica + aro + tropical japonica; and Hap-3: japonica subgroups (Fig. 8A). Similarly, three haplotypes were also detected for PUP4. The aus and aromatic subpopulations carry separate gene haplotypes than both indica and japonica subpopulations (Fig. 8B). For SAC1, out of three haplotypes, aus subpopulation mostly carries similar haplotypes found in aromatic, temperate japonica, tropical japonica, and indica 1B subpopulations (Fig. 8C).
Fig. 8.
Gene haplotype distribution in 3,000 rice genome (3 K-RG) panel. (A) frequency of three haplotypes of GLT1 in different rice subgroups. (B) frequency of three haplotypes of PUP4 in different rice subgroups. (C) frequency of three haplotypes of SAC1 on Chromosome 7 in different rice subgroups. The frequency of each haplotype in 3 K-RG panel is mentioned within the parentheses. Haplotype analysis was performed using the non-synonymous SNPs within the genes using the Rice SNP-Seek Database (https://snp-seek.irri.org/_snp.zul). Grouping of rice germplasm into 12 subgroups was done following Wang et al. (2018). Admix, admixture; Aro, aromatic; Aus, aus; Ind1A, indica 1 A; Ind 1B, indica 1B, Ind2, indica 2; Ind3, indica 3; Indx, indica admixture; Japx, japonica admixture; Subtrop, subtropical japonica; Temp, temperate japonica; Trop, tropical japonica. Number of accessions in each subpopulation is mentioned on the top of each bar
Discussion
Understanding the genetic structure and origins of morphological and developmental variations in O. sativa is vital for bolstering global food security. Extensive investigations on rice population structure have consistently identified two major varietal groups, denoted as sub-species: Indica and Japonica (Wang et al. 2018). This classification has ancient Chinese roots, known as Hisen/Sen and Keng/Geng, and was subsequently substantiated by morphological and serological distinctions, along with the presence of partial reproductive barriers (Kato 1928; Morishima and Oka 1981). The Japonica varietal group was further divided into tropical japonica and temperate japonica (Oka 1958). Additionally, the Javanica group emerged based on gross morphological distinctions and geographical distribution (Morishima and Oka 1960). Apart from these three primary groups, various minor varietal groups, such as aus, ashina, boro, rayada, basmati, and sadri, are cultivated across the Indian subcontinent. While these groups may lack significant morphological disparities compared to Indica and Japonica, recent studies employing isozyme loci and molecular marker systems have established some as distinct genetic groups within O. sativa (Glaszmann 1987; Garris et al. 2005; Wang et al. 2018).
Glaszmann’s (1987) comprehensive work categorized 1,688 O. sativa accessions into six varietal groups, encompassing two major (Indica and Japonica), two minor (aus and aromatic), and two satellite (ashina/deep-water and rayada). Subsequent studies, such as Garris et al. (2005), proposed a widely accepted classification consisting of indica, aus, aromatic, tropical japonica, and temperate japonica. However, these studies omitted many accessions from the satellite groups originating in Bangladesh and northeastern India (Group III and Group IV of Glaszmann 1987). Research into the population structure of Asian rice is often constrained by the limited representation of rice cultivars from specific ecological regions, which may obscure finer population structures. For instance, while the analysis of the 3 K-RG panel, comprising millions of SNPs, offered an enhanced resolution of within-species diversity, it failed to fully unravel the structure within the circum-aus group, encompassing aus, boro, ashina, and rayada types (Wang et al. 2018). However, another study incorporating various Chinese accessions successfully elucidated the structure within the circum-aus group (Wang et al. 2014).
In this study, we specifically focussed on comprehending the finer population structures within the circum-aus group by scrutinizing the aus/boro accessions from the 3 K-RGP dataset, an area where our understanding of population genomic diversity is quite limited. Our analysis unveiled the existence of six sub-groups, supporting the differentiation of previously identified aus subgroups, encompassing aus, ashina, and rayada types. Significantly, these sub-groups displayed distinct geographic patterns. In prior research by Travis et al. (2015), the genetic structuring of 345 aus cultivars, predominantly originating from Bangladesh, Assam, and eastern India, was examined using 384 SNPs. Travis et al. identified two distinct groups within the aus varietal category. Geographically, this group is distributed across south and west Asia, extending from Iran to Assam along the Himalayas (Glaszmann 1987), with its center of diversity situated in Bangladesh and the eastern to northeastern regions of India (Civáň et al. 2015). These rice varieties are cultivated under a wide range of hydric conditions, from irrigated regions in Pakistan to drought-prone uplands in Bangladesh and eastern India, as well as deep-water environments in Bangladesh and northeastern India. Consequently, they have developed numerous adaptive traits to thrive in these diverse environments, leading to genetic variations at the DNA level and intricate fine-scale population structures.
Our findings revealed that when assessing genetic differentiation between aus and boro ecotypes at K = 2, they appeared to be genetically close, sharing a cluster. However, at K = 6, boro cultivars exhibited some degree of differentiation, primarily forming a cluster under K5. This suggests that while there is genetic proximity between aus and boro ecotypes, finer genetic distinctions become apparent when examining a more detailed population structure. At both levels of structuring, rayada remained at a separate cluster, aligning with previous research findings (Wang et al. 2014; Travis et al. 2015). Interestingly, our study found that many deep-water cultivars, categorized as ashina, exhibited a stronger genetic affinity with boro cultivars, a phenomenon not previously reported. Furthermore, we observed that the majority of aus accessions originating from countries outside the Indian subcontinent tended to cluster separately, as illustrated in Fig. 1. Pairwise genetic distance calculations among the genetic clusters (K1-K6) revealed that drought-tolerant aus accessions (belonging to K3) may have dispersed further to regions like Sri Lanka and Pakistan, likely due to their stress tolerance. Similarly, rayada cultivars may have also extended beyond Bangladesh to central India. However we could not find a possible reason the spread of rayada types. Overall, there was a connection between the geographical distribution of aus sub-groups and agro-ecological diversity in Southern Asia. After comparing the distribution of the accessions along the five agro-ecological zones (FAO’s global agro-ecological zones modified by Gumma et al. 2022), we found that K1 is distributed in humid tropics (Zone 5). K2 and K3 are distributed along the Zones- 1, 2 and 3 representing arid tropics, semi-arid subtropics and semi-arid tropics, respectively. K6 is widespread in sub-humid tropics (Zone 4), while K5 is distributed along the transition zone of Zones- 4 and 5. Only the accessions of K4 are not confined to any agro-ecological zones.
When constructing phylogenetic trees from genome-wide data, it became evident that aus cultivars cluster within the Indica clade, indicating their greater genetic similarity with Indica rice. This genetic closeness aligns well with their morphological similarities. However, several studies have also suggested a distinct origin for indica and aus varietal groups (Schatz et al. 2014; Civáň et al. 2015). The aus cultivars form a distinct cluster from both indica and japonica when neighbor-joining trees are constructed from the ‘domestication sweep’ regions (Civáň et al. 2015), highlighting the potential for a separate origin of aus.
Based on this study and previous evidence, it appears that the circum-aus group represents an evolutionary development of aus rice. Further genome-wide surveys with increased sample sizes of aus, indica, and wild species from the Indian subcontinent could provide greater clarity on the origin of the aus group. Moreover, the wide agro-morphological variations observed within this group enhance the potential for utilizing aus accessions in rice breeding programs to create novel genetic variations for yield-related traits.
In our study, we employed high-density marker data to unravel the intricate genetics governing yield traits in aus rice. This was achieved through mixed-model GWAS analyses using principal component (PC) scores derived from 11 agro-morphological traits. PCA, a dependable approach for extracting the underlying variability from numerous correlated traits, was utilized to create PC scores, which can be considered as composite variables. The application of GWAS with PC scores has proven effective in reducing Type I error rates by circumventing multiple testing, a method employed in both human and plant systems (He et al. 2008). Additionally, PCA normalizes skewed individual trait data, improving the reliability of GWAS results (Goh and Yap 2009). Notably, in rice, GWAS using PC scores has demonstrated higher power in detecting loci that might be overlooked when using individual traits as dependents (Yano et al. 2019).
Our PCA analysis of 11 agro-morphological traits unveiled that PC1 explained 24% of the variation in flowering and plant architectural traits, while PC2 captured 17% of the variations in grain yield traits, such as panicle weight, yield, and harvest index. PC3 captured 14% of the variation for grain yield. Utilizing PC scores for GWAS proved to be more effective in identifying significant associations for yield-related traits compared to using individual traits for GWAS. Moreover, several peak SNPs coincided with previously reported QTLs and genes, strengthening the reliability of GWAS results. For instance, the genes associated with PC1 score were linked, either falling within or being in LD with OsGI, OsGPX1, OsMADS15, and IPA1, which regulate days to flowering, plant height, spikelet number, root development, panicle architecture, and grain yield. SNPs identified for PC2 were linked to genes such as OsGLT1, dep2, fzp, and SP1, while PC3 results revealed associations with several genes influencing grain size and yield, including OsDOS, SE13, GS3, GL3, and OsIPT7.
Further exploration of the identified QTLs (qPC2-1.1 and qPC1-7.1), explaining higher phenotypic variance with larger effect sizes, uncovered potential candidate genes for agro-morphological traits in aus rice germplasm. QTL qPC2-1.1, located on Chromosome 1, significantly explained variation for PC2, representing grain yield, panicle weight, and harvest index. Examination of gene models suggested that OsNADH-GOGAT1 (GLT1) and OsPUP4 are the likely candidate genes for this QTL. GLT1 influences various yield-related traits, and nitrogen-carbon metabolism by regulating ammonium-responsive genes (Funayama et al. 2013), and is expressed in roots, young leaves, and grains. Rice GLT1 mutants exhibit reduced tillering (Tamula et al., 2010). In aus rice, we detected four haplotypes of GLT1, with one particular haplotype (Hap-2) present in 72% of the accessions. This haplotype was associated with early maturation, higher 1000-grain weight, and increased harvest index.
Our analysis of the 3 K-RGP showed that aus rice carries a specific GLT1 haplotype, prevalent in indica (Ind3) accessions from southeast Asia, as well as in aromatic and tropical japonica rice. Haplotypes of GLT1 exhibit significant differentiation between rice varietal groups (Yang et al. 2016). Another potential gene within qPC2-1.1 is Big Grain 3/OsPUP4, which influences various agro-morphological traits. The PUP4 gene family includes 12 members involved in cytokinin transportation. Activating OsPUP4 results in increased grain size (Xiao et al. 2019). OsPUP7, a homolog of OsPUP4, regulates multiple phenotypic traits in rice, including plant height, grain size, and days to heading. Haplotypes of PUP4 in aus accessions differed for many traits, suggesting this gene is another candidate for qPC2-1.1. The distribution of PUP4 haplotypes also varies among indica, aus, and japonica.
The candidate gene for qPC1-7.1 appears to be OsSAC1, involved in regulating sugar partitioning in carbon-demanding juvenile leaves and leaf sheaths. Although the function of OsSAC1 is not fully characterized, this gene likely contributes to building the carbon skeleton in rice plants. Previously, no QTLs for flowering or plant architectural traits were reported in the qPC1-7.1 region, highlighting the effectiveness of GWAS using PC scores in identifying QTLs not detectable through individual traits. Haplotype frequencies of OsSAC1 vary between indica and japonica, with the dominant gene haplotype in aus rice being the most frequent in temperate- and tropical-japonica, along with modern indica varieties. Overall, from the distribution of haplotype frequencies of OsGLT1, OsPUP4, and OsSAC1, it is evident that aus rice possesses different gene haplotypes compared to most indica rice. These findings support the recent hypothesis of O. sativa evolution, suggesting separate origins of indica, aus, and japonica in different geographic regions, with aromatic rice likely originating from hybridization between aus and japonica (Civáň et al. 2015; Civáň et al. 2019). It would be interesting to study whether the haplotypes we have identified in aus also has the same effects in other varietal groups, or whether they are modified by these differing genetic backgrounds; examining this issue may help in providing new breeding material for a wider range of rice populations.
Conclusions
Our investigation focused specifically on unravelling the finer population structures within the circum-aus varietal group. We unveiled the existence of six aus sub-groups, and highlighted their distinct geographic patterns. Our findings emphasized the genetic proximity between aus and boro ecotypes, while revealing the genetic distinctness of rayada cultivars. Notably, many deep-water cultivars (categorized as ashina), displayed a strong genetic affinity with boro cultivars. Furthermore, our research revealed that aus accessions from countries outside the Indian subcontinent tend to cluster separately, indicating genetic distinctions.
GWAS analyses with PC scores derived from 11 agro-morphological traits proved more effective in identifying significant associations for yield-related traits compared to using individual traits in GWAS. Our investigations unveiled the potential candidate genes for QTLs, such as OsNADH-GOGAT1 (GLT1), OsPUP4, and OsSAC1, offering valuable insights into the genetic basis of grain yield and other agronomical traits.
In conclusion, this research contributes to a deeper understanding of the genetic intricacies of aus rice and offers insights into its evolutionary history. These findings are important for future rice breeding programs aimed at improving yield-related traits and enhancing global food security.
Electronic Supplementary Material
Below is the link to the electronic supplementary material.
Acknowledgements
We thank Dr. Arunava Pattanayak, Former Director, ICAR-Indian Institute of Agricultural Biotechnology for critically reading the manuscript and suggesting improvements. We express our gratitude to the Director, ICAR-National Rice Research Institute, Cuttack, Odisha, and the Director, ICAR Research Complex for NEH Region, Umiam, Meghalya for providing necessary support for the study.
Abbreviations
- GWAS
Genome-wide association study
- PCA
Principal component analysis
- PC
Principal component
- 3 K-RGP
3,000 Rice Genome Project
- BAAP
Bengal Assam Aus Panel
- LD
Linkage disequilibrium
- Chr
Chromosome
- MAF
Minor allele frequency
- DtF
Days to 50% flowering
- Ht
Plant height
- PnL
Panicle length
- PnW
Panicle weight
- SpkN
Spikelets per panicle
- Yld
Yield
- HI
Harvest index
- Tn
Tiller number
- GW
1000-grain weigh
- PVE
Phenotypic variation explained
- MTA
Marker-trait association
- QTL
Quantitative trait locus
Author Contributions
SR, KC and UN designed the project; PS, MB, JK, BCV, PSH and DB performed phenotyping; SR, SG, AB, SS, and PS analysed the data; SR and SG wrote the paper; MDP, PCK and NPM provided directions for the study and performed corrections of the manuscript. All authors read and approved the final manuscript.
Funding
This study was funded by National Agricultural Science Fund, Indian Council of Agricultural Research, India (Project No. NASF/GTR-8032/2020-21).
Data Availability
All data generated or analysed during this study are included in this published article and its supplementary information files.
Declarations
Ethics approval and consent to participate
Not applicable.
Competing Interests
The authors declare no competing interests.
Consent for publication
Not applicable.
Footnotes
Correspondence:
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- Alexander DH, Novembre J, Lange K. Fast model-based estimation of ancestry in unrelated individuals. Genome Res. 2009;19:1655–1664. doi: 10.1101/gr.094052.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexandrov N, Tai S, Wang W, et al. SNP-Seek database of SNPs derived from 3000 rice genomes. Nucleic Acids Res. 2015;43:D1023–D1027. doi: 10.1093/nar/gku1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhandari A, Sandhu N, Bartholome J et al (2020) Genome-Wide Association Study for Yield and Yield Related Traits under Reproductive Stage Drought in a diverse indica-aus Rice Panel. 10.1186/s12284-020-00406-3. Rice 13: [DOI] [PMC free article] [PubMed]
- Bin Rahman ANMR, Zhang J (2018) Preferential Geographic distribution pattern of abiotic stress tolerant rice. 10.1186/s12284-018-0202-9. Rice 11: [DOI] [PMC free article] [PubMed]
- Bradbury PJ, Zhang Z, Kroon DE, et al. TASSEL: software for association mapping of complex traits in diverse samples. Bioinformatics. 2007;23:2633–2635. doi: 10.1093/bioinformatics/btm308. [DOI] [PubMed] [Google Scholar]
- Chen J, Wang L, Yuan M (2021) Update on the roles of Rice MAPK cascades. Int J Mol Sci 22. 10.3390/ijms22041679 [DOI] [PMC free article] [PubMed]
- Civáň P, Craig H, Cox CJ, Brown TA. Three geographically separate domestications of Asian rice. Nat Plants. 2015;1:15164. doi: 10.1038/nplants.2015.164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Civáň P, Ali S, Batista-Navarro R et al (2019) Origin of the aromatic group of cultivated rice (Oryza sativa L.) traced to the Indian subcontinent. Genome Biol Evol 11:832–843 [DOI] [PMC free article] [PubMed]
- R Core Team (2021) R: A language and environment for statistical computing
- Dong S-S, He W-M, Ji J-J, et al. LDBlockShow: a fast and convenient tool for visualizing linkage disequilibrium and haplotype blocks based on variant call format files. Brief Bioinform. 2020;22:bbaa227. doi: 10.1093/bib/bbaa227. [DOI] [PubMed] [Google Scholar]
- Duan P, Rao Y, Zeng D, et al. SMALL GRAIN 1, which encodes a mitogen-activated protein kinase kinase 4, influences grain size in rice. Plant J. 2014;77:547–557. doi: 10.1111/tpj.12405. [DOI] [PubMed] [Google Scholar]
- Funayama K, Kojima S, Tabuchi-Kobayashi M, et al. Cytosolic glutamine Synthetase1;2 is responsible for the primary assimilation of ammonium in Rice roots. Plant Cell Physiol. 2013;54:934–943. doi: 10.1093/pcp/pct046. [DOI] [PubMed] [Google Scholar]
- Garris AJ, Tai TH, Coburn J, et al. Genetic structure and diversity in Oryza sativa L. Genetics. 2005;169:1631–1638. doi: 10.1534/genetics.104.035642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glaszmann J-C. Isozymes and classification of Asian rice varieties. Theor Appl Genet. 1987;74:21–30. doi: 10.1007/BF00290078. [DOI] [PubMed] [Google Scholar]
- Goh L, Yap VB. Effects of normalization on quantitative traits in association test. BMC Bioinformatics. 2009;10:1–8. doi: 10.1186/1471-2105-10-415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gross BL, Zhao Z (2014) Archaeological and genetic insights into the origins of domesticated rice. Proceedings of the National Academy of Sciences 111:6190–6197 [DOI] [PMC free article] [PubMed]
- Gumma MK, Thenkabail PS, Teluguntla P, et al. Multiple agricultural cropland products of South Asia developed using Landsat-8 30 m and MODIS 250 m data using machine learning on the Google Earth Engine (GEE) cloud and spectral matching techniques (SMTs) in support of food and water security. GIsci Remote Sens. 2022;59:1048–1077. doi: 10.1080/15481603.2022.2088651. [DOI] [Google Scholar]
- Guo T, Chen K, Dong N-Q, et al. GRAIN SIZE AND NUMBER1 negatively regulates the OsMKKK10-OsMKK4-OsMPK6 cascade to coordinate the trade-off between grain number per panicle and grain size in rice. Plant Cell. 2018;30:871–888. doi: 10.1105/tpc.17.00959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo T, Lu Z-Q, Shan J-X, et al. ERECTA1 acts upstream of the OsMKKK10-OsMKK4-OsMPK6 cascade to control spikelet number by regulating cytokinin metabolism in rice. Plant Cell. 2020;32:2763–2779. doi: 10.1105/tpc.20.00351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutaker RM, Groen SC, Bellis ES, et al. Genomic history and ecology of the geographic spread of rice. Nat Plants. 2020;6:492–502. doi: 10.1038/s41477-020-0659-6. [DOI] [PubMed] [Google Scholar]
- He L-N, Liu Y-J, Xiao P, et al. Genomewide linkage scan for combined obesity phenotypes using principal component analysis. Ann Hum Genet. 2008;72:319–326. doi: 10.1111/j.1469-1809.2007.00423.x. [DOI] [PubMed] [Google Scholar]
- IRRI . SES: standard evaluation system for Rice. 5. Manila, Philippines: International Rice Research Institute; 2013. [Google Scholar]
- Kaler AS, Gillman JD, Beissinger T, Purcell LC. Comparing different statistical models and Multiple Testing Corrections for Association Mapping in Soybean and maize. Front Plant Sci. 2019;10:1794. doi: 10.3389/fpls.2019.01794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato A. On the affinity of rice varieties as shown by the fertility of rice plants. Centr Agric Inst Kyushu Imp Univ. 1928;2:241–276. [Google Scholar]
- Khush GS. What it will take to feed 5.0 billion rice consumers in 2030. Plant Mol Biol. 2005;59:1–6. doi: 10.1007/s11103-005-2159-5. [DOI] [PubMed] [Google Scholar]
- Kojima S, Minagawa H, Yoshida C et al (2023) Coregulation of glutamine synthetase1;2 (GLN1;2) and NADH-dependent glutamate synthase (GLT1) gene expression in Arabidopsis roots in response to ammonium supply. Front Plant Sci 14. 10.3389/fpls.2023.1127006 [DOI] [PMC free article] [PubMed]
- Lefort V, Desper R, Gascuel O. FastME 2.0: a Comprehensive, Accurate, and fast Distance-based phylogeny inference program. Mol Biol Evol. 2015;32:2798–2800. doi: 10.1093/molbev/msv150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Xu P, Deng X, et al. Identification of four genes for stable hybrid sterility and an epistatic QTL from a cross between Oryza sativa and Oryza glaberrima. Euphytica. 2008;164:699–708. doi: 10.1007/s10681-008-9684-7. [DOI] [Google Scholar]
- Li J-Y, Wang J, Zeigler RS. The 3,000 rice genomes project: new opportunities and challenges for future rice research. Gigascience. 2014;3:8. doi: 10.1186/2047-217X-3-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Huang M, Fan B, et al. Iterative usage of fixed and Random Effect Models for Powerful and efficient genome-wide Association studies. PLoS Genet. 2016;12:e1005767. doi: 10.1371/journal.pgen.1005767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molina J, Sikora M, Garud N, et al. Molecular evidence for a single evolutionary origin of domesticated rice. Proc Natl Acad Sci. 2011;108:8351–8356. doi: 10.1073/pnas.1104686108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morishima H, Oka H-I (1960) The pattern of interspecific variation in the genus Oryza: its quantitative representation by statistical methods. Evol (N Y) 153–165
- Morishima H, Oka H-I. Phylogenetic differentiation of cultivated rice, XXII. Numerical evaluation of the indica-japonica differentiation. Japanese J Breed. 1981;31:402–413. doi: 10.1270/jsbbs1951.31.402. [DOI] [Google Scholar]
- Norton GJ, Travis AJ, Douglas A, et al. Genome wide association mapping of grain and straw biomass traits in the rice bengal and assam aus panel (baap) grown under alternate wetting and drying and permanently flooded irrigation. Front Plant Sci. 2018;9:1–18. doi: 10.3389/fpls.2018.01223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oka HI. Intervarietal variation and classification of cultivated rice. Indian J Genet Plant Breed. 1958;18:79–89. [Google Scholar]
- Purcell S, Neale B, Todd-Brown K, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubaiyath Bin Rahman ANM, Zhang J. Rayada specialty: the forgotten resource of elite features of rice. Rice. 2013;6:1–10. doi: 10.1186/1939-8433-6-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schatz MC, Maron LG, Stein JC, et al. Whole genome de novo assemblies of three divergent strains of rice, Oryza sativa, document novel gene space of aus and indica. Genome Biol. 2014;15:1–16. doi: 10.1186/s13059-014-0506-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Travis AJ, Norton GJ, Datta S, et al. Assessing the genetic diversity of rice originating from Bangladesh, Assam and West Bengal. Rice. 2015;8:35. doi: 10.1186/s12284-015-0068-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Zhang Z. GAPIT Version 3: Boosting Power and Accuracy for Genomic Association and Prediction. Genomics Proteom Bioinf. 2021;19:629–640. doi: 10.1016/j.gpb.2021.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C-H, Zheng X-M, Xu Q, et al. Genetic diversity and classification of Oryza sativa with emphasis on Chinese rice germplasm. Heredity (Edinb) 2014;112:489–496. doi: 10.1038/hdy.2013.130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W, Mauleon R, Hu Z, et al. Genomic variation in 3,010 diverse accessions of Asian cultivated rice. Nature. 2018;557:43–49. doi: 10.1038/s41586-018-0063-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao Y, Liu D, Zhang G, et al. Big Grain3, encoding a purine permease, regulates grain size via modulating cytokinin transport in rice. J Integr Plant Biol. 2019;61:581–597. doi: 10.1111/jipb.12727. [DOI] [PubMed] [Google Scholar]
- Yamada K, Yamaguchi K, Yoshimura S, et al. Conservation of chitin-induced MAPK signaling pathways in rice and Arabidopsis. Plant Cell Physiol. 2017;58:993–1002. doi: 10.1093/pcp/pcx042. [DOI] [PubMed] [Google Scholar]
- Yang X, Nian J, Xie Q, et al. Rice Ferredoxin-Dependent Glutamate Synthase regulates Nitrogen-Carbon metabolomes and is genetically differentiated between japonica and indica subspecies. Mol Plant. 2016;9:1520–1534. doi: 10.1016/j.molp.2016.09.004. [DOI] [PubMed] [Google Scholar]
- Yano K, Morinaka Y, Wang F, et al. GWAS with principal component analysis identifies a gene comprehensively controlling rice architecture. Proc Natl Acad Sci. 2019;116:21262–21267. doi: 10.1073/pnas.1904964116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C, Dong S-S, Xu J-Y, et al. PopLDdecay: a fast and effective tool for linkage disequilibrium decay analysis based on variant call format files. Bioinformatics. 2019;35:1786–1788. doi: 10.1093/bioinformatics/bty875. [DOI] [PubMed] [Google Scholar]
- Zhao K, Tung C-W, Eizenga GC, et al. Genome-wide association mapping reveals a rich genetic architecture of complex traits in Oryza sativa. Nat Commun. 2011;2:467. doi: 10.1038/ncomms1467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu X, Shen W, Huang J, et al. Mutation of the OsSAC1 gene, which encodes an endoplasmic reticulum protein with an unknown function, causes Sugar Accumulation in Rice leaves. Plant Cell Physiol. 2017;59:487–499. doi: 10.1093/pcp/pcx203. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data generated or analysed during this study are included in this published article and its supplementary information files.