Abstract
Primate foamy viruses (FVs) express, in addition to the 130-kDa envelope protein, a 170-kDa glycoprotein, which reacts with antisera specific for the envelope and Bel proteins. We determined the exact nature of this 170-kDa glycoprotein by using the molecularly cloned human FV (HFV). Radioimmunoprecipitation analysis of 293T cells transfected with appropriate expression constructs by using antisera specific for the HFV Env, Bel1, and Bel2 proteins, as well as reverse transcription-PCR analysis of HFV-infected cells, demonstrated that this protein is an Env-Bet fusion protein that is secreted into the supernatant. However, it is only loosely associated, or not associated, with viral particles. gp170 is generated by an alternatively spliced Env mRNA using a splice donor and splice acceptor pair localized within the env open reading frame (ORF), which is normally used to generate Bel1 and Bet transcripts derived from the internal promoter within the env ORF. gp170 is expressed at a level 30 to 50% of the Env precursor gp130. However, it alone does not confer infectivity to HFV particles, because capsids derived from proviruses expressing only the gp170 were not released into the supernatant. In contrast, viruses derived from proviral clones deficient in gp170 expression showed similar in vitro infectivity and replication kinetics to wild-type virus. Furthermore, both types of viruses were inactivated to a similar extent by neutralizing sera, indicating that shedding of gp170 probably does not affect the humoral immune response in the infected host.
Human foamy virus (HFV) is the prototype member of the family Spumavirinae, also referred to as foamy viruses (FVs). FVs are complex retroid viruses that exploit a unique replication strategy that has been discovered in recent studies (6, 25, 36, 37). In addition to the genes for the Gag, Pol, and Env proteins, FVs harbor open reading frames (ORFs) in the 3′ region of the genome that code for accessory proteins. The first ORF codes for a DNA-binding protein (Tas/Bel1) that is a potent trans activator of gene expression directed by promoters of the cognate virus (16, 17, 24, 31, 38).
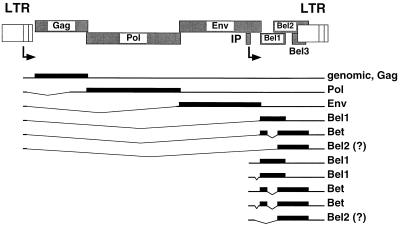
FV gene expression involves two promoters and several transcripts, some of which are multiply spliced (23, 27) (Fig. 1). The long terminal repeat (LTR) promoter directs the expression of the pregenomic RNA/Gag mRNA; single-spliced mRNAs for the Pol, Env, and Tas/Bel1 proteins; and a double-spliced mRNA for the Bet protein (27). Bet is a highly expressed accessory protein of unknown function made up of ORF-1- and ORF-2-encoded sequences (1, 14, 27). In the initial phase of replication, gene expression relies on the internal promoter (IP) located in the env gene upstream of the accessory ORFs (22). IP-directed transcripts can give rise to Tas/Bel1 and Bet proteins. It has been reported that these transcripts are often spliced in the untranslated leader sequence overlapping the env gene (22) (Fig. 1).
FIG. 1.
Known HFV mRNAs derived from the LTR or internal promoter coding for structural as well as accessory proteins. The mRNAs are indicated as lines with inserts for the deleted intron sequences, the coding regions of the individual mRNAs are shown as bars, and the resulting proteins are listed in the column to the right.
The morphogenesis of HFV appears to be unusual too. HFV capsids do not bud spontaneously across cellular membranes but require the presence of Env protein (7). The 130-kDa Env precursor protein is cleaved by a cellular protease into surface (SU) and transmembrane (TM) subunits during its transport to the cell membrane (10). However, due to the localization of a retention signal in the cytoplasmic domain, most of the 130-kDa HFV Env protein is retained in the endoplasmic reticulum in the absence of either the expression of other HFV structural genes or the inactivation of the endoplasmic reticulum retention signal (10, 11).
Beside the 130-kDa Env precursor, an even larger glycoprotein (170 kDa) has been detected in HFV-infected cells (29). This protein has been reported to cross-react with antibodies recognizing Env, Tas/Bel1, and Bet (9). However, neither mRNA nor any function of this protein has been described yet. In this study, we set about to characterize this enigmatic protein more closely.
MATERIALS AND METHODS
Eukaryotic expression constructs.
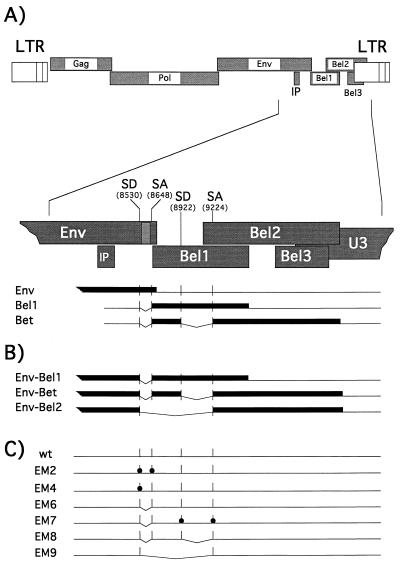
The expression construct pcHFVenv/bel1-3, containing the env, bel1, bel2, and bel3 ORFs, was generated by inserting a fragment of pHSRV2 (35) from the translation start of the env ORF (nucleotide [nt] 5719 relative to the genomic transcription start) to the SspI site (nt 10445) ∼35 bp downstream of the bel2 stop codon into pCDNA3.1+zeo (Invitrogen). Mutants of the parental construct were generated by recombinant PCR with primers harboring the desired mutations. All sequences derived by PCR were sequenced to confirm the introduction of the desired mutations and exclude additional offsite mutations. The following mutants of pcHFVenv/bel1-3 were generated: EM2 (SD/SA mutant), the splice donor (SD) (nt 8530) and splice acceptor (SA) (nt 8648) were inactivated by GT-to-GG and AG-to-AA exchanges, respectively; EM4 (SD mutant), the SD (nt 8530) consensus sequence was changed from GT to GG; and EM6 (Env-Bel1/Bet), deletion of the intron between SD (nt 8530) and SA (nt 8648). In pcHFVenv-bel1 (EM7), the sequences between SD (nt 8530) and SA (nt 8648) were deleted and the SD (nt 8922) and the SA (nt 9224) were inactivated by GT-to-GC and AG-to-TC mutations, respectively. pcHFVenv-bet (EM8) was created by deletion of the sequences between SD (nt 8530) and SA (nt 8648) and between SD (nt 8922) and SA (nt 9224), whereas in pcHFVenv-bel2 (EM9), all intervening sequences between SD (nt 8530) and SA (nt 9224) were deleted. The parental human cytomegalovirus immediate-early promoter-driven infectious proviral clone pcHSRV2 wt has been described recently (25). The individual mutant proviral clones were generated by replacing an 858-bp NheI-ClaI fragment of pcHSRV2 with the respective fragments from the pcHFVenv/bel1-3 constructs.
Generation of recombinant HFV supernatants and RIPA.
Viral supernatants containing the different recombinant viruses were generated by transfection of 293T cells (5) essentially as described previously (21). The viral supernatants were harvested 48 to 72 hours after transfection. Supernatants from independent transfections with the same plasmids were pooled and filtered (pore size, 0.45 μm). The supernatants were used immediately or stored at −80°C until use. For characterization of HFV protein expression by radioimmunoprecipitation analysis (RIPA), transiently transfected 293T cells were metabolically labeled with [35S]methionine for approximately 20 h. At 36 h after addition of the DNA, the cells were lysed in RIPA buffer (20 mM Tris [pH 7.4], 0.3 M NaCl, 1% [wt/vol] sodium deoxycholate, 1% [vol/vol] Triton X-100, 0.1% [wt/vol] sodium dodecyl sulfate) containing protease inhibitors. Viral proteins were precipitated as described previously (7) with HFV-positive chimpanzee sera or with rabbit antisera generated against recombinant HFV proteins and specific for SU (20), Bel1 (22), Bel1/Bet (1), and Bet/Bel2 (1). Secreted viral proteins were analyzed with filtered (pore size 0.45 μm) supernatant supplemented with 5× RIPA buffer to yield 1× RIPA buffer. Particle-associated proteins were detected after centrifugation through a 20% sucrose cushion as described previously (7).
RT-PCR analysis.
Human KMST-6 fibroblastoid cells (28) were infected with supernatants of known titer from transfected 293T cells, containing molecularly defined viruses, at a multiplicity of infection (MOI) of 0.5. Mock-infected cultures were incubated with the same amount of supernatant from 293T cells transfected with pCDNA3.1+zeo. At 7 days after infection, total RNA was isolated from the infected cultures with the RNeasy Kit (Qiagen) as specified by the manufacturer. A 1-μg portion of total RNA was used for each individual reverse transcription-PCR (RT-PCR) analysis with the Titan RT-PCR System (Boehringer) as specified by the manufacturer. The primers used were 391 (AAGAGCAGATTGAAAGAGCAAAAGC), hybridizing upstream of the transcription start of the internal promotor (nt 8419) within the env ORF, and 8 (CTGGACTCTTCTAGTAGCCCT), hybridizing downstream of the SA site (nt 9224) within the bel2 ORF. Aliquots of the amplification reaction products were separated by agarose gel electrophoresis.
Determination of virus titers and growth kinetics.
The growth kinetics of individual viruses were analyzed by infection of BHK-21 cells or of BHK-21 cells constitutively expressing the HFV transcriptional transactivator (BHK/Bel1 cells) (3) at a MOI of 0.05. Cell-free samples of the supernatant were collected over a period of 26 days and subjected to titer determination on BHK/LTRlacz cells as described previously (35).
Neutralization of HFV infection.
Neutralization of HFV infectivity by chimpanzee sera was analyzed as described previously with minor modifications (4). Briefly, 500 focus-forming units (FFU) of HSRV2 wt or HSRV2 EM4 virus stock generated by transient transfection of 293T cells was incubated with different antisera dilutions for 1 h at 37°C in a total volume of 100 μl prior to the addition to 2 × 103 BHK/LTRlacz indicator cells plated 24 h previously in 96-well plates. At 48 h later, the supernatants were aspirated, the cells were washed once with phosphate-buffered saline, and 100 μl of lysis buffer (Promega) was added. After a 15-min incubation at room temperature, 100 μl of 2× assay buffer (20 mM NaH2PO4, 80 mM Na2HPO4, 0.1 mM MnCl2, 2 mM MgSO4, 40 mM 2-mercaptoethanol, 4 mg of o-nitrophenyl-β-d-galactopyranoside [ONPG] per ml [pH 7.3]) was added and the plates were incubated at 37°C for an additional 6 to 12 h. The enzymatic reaction was terminated by the addition of 100 μl of 3 M Na2CO3, and the absorbance at 405 nm was determined in an enzyme-linked immunosorbent assay reader (Bio-Rad).
RESULTS
Origin of the Env-Bel fusion proteins.
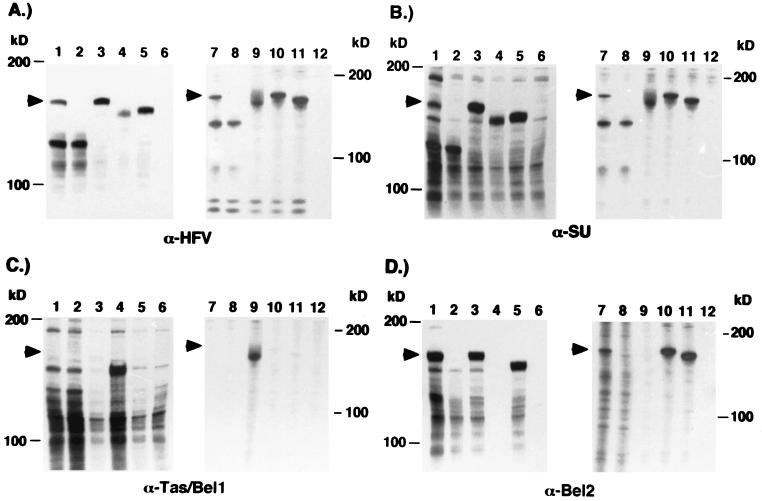
Computer-assisted analysis of the 3′ part of the HFV genome coding for the Env, Bel1, and Bet proteins (nt 5719 to 10445) indicated that alternatively spliced Env mRNA utilizing the SD (nt 8530) within the env ORF could potentially encode Env-Bel1, Env-Bet, and/or Env-Bel2 fusion proteins (Fig. 2A and B). Interestingly, all potential Env fusion proteins would lack the complete membrane-spanning domain (MSD) as well as the cytoplasmic domain of the TM subunit containing the ER retention signal (10), suggesting a targeting of these fusion proteins to the cellular secretory pathway. Furthermore, these SD and SA sites seem to be highly conserved in various FV isolates originating from different primates (data not shown). To examine whether such Env-Bel fusion proteins were indeed generated by the 3′ part of the HFV genome, a eukaryotic expression construct, pcHFVenv/bel1-3 wt, was generated, containing the complete HFV env ORF as well as the bel1-3 ORFs (Fig. 2C). In addition, expression constructs containing the potential cDNAs coding for Env-Bel1 (EM7), Env-Bet (EM8), and Env-Bel2 (EM9) were generated by joining the respective SA sites with the env SD site by recombinant PCR (Fig. 2B and C). 293T cells were transfected with these expression constructs, and subsequently lysates of metabolically labeled cells were examined by RIPA with antisera specific for all major HFV proteins (Fig. 3A), HFV Env (Fig. 3B), Bel1 (Fig. 3C), and Bel2 (Fig. 3D). Analysis with a chimpanzee serum recognizing all major HFV proteins showed the presence of the 130-kDa Env precursor and a protein of unknown origin of about 170 kDa (Fig. 3A, lane 1), which was expressed at 30 to 50% of the level of the Env precursor gp130. The 170-kDa protein could be precipitated by an antiserum specific for the HFV Env protein (Fig. 3B, lane 1), by an antiserum reactive with the C-terminal amino acids encoded by the bel2 ORF (Fig. 3D, lane 1), or by an antiserum recognizing sequences common to Bel1 and Bet (data not shown), but not by an antiserum specific for amino acids unique to the Bel1 protein (Fig. 3C, lane 1). These results showed that the coding capacity for the 170-kDa HFV protein resides within this part of the HFV genome. Furthermore, the lack of reactivity of the 170-kDa protein with a Bel1-specific antiserum indicated that it is not an Env-Bel1 fusion protein. The sizes of the Env-Bel fusion proteins generated by transfection of 293T cells with the potential cDNA expression constructs (Fig. 3, lanes 3 to 5) suggested that the nature of the 170-kDa Env reactive protein most probably is an Env-Bet rather than an Env-Bel2 fusion protein. This was further supported by the precipitation of gp170 with the Bel1/Bet-specific antiserum (data not shown).
FIG. 2.
Schematic illustration of alternatively spliced env mRNAs and constructs generated to characterize gp170. (A) HFV genome organization. The region spanning the 3′ part of the envelope ORF and the bel1-3 ORFs is enlarged. The env mRNA and the tas/bel1 and bet mRNA are indicated below. Coding regions are drawn as solid bars, noncoding regions are indicated by thin lines. The SD and SA sites normally used to generate the tas/bel1 and bet transcripts derived from the IP within the env ORF are indicated. (B) Illustration of the three potential Env-Bel fusion proteins generated by alternative splicing of the env mRNA by using the first SD site within the env ORF and the different SD and SA sites downstream. (C) Illustration of the different mutants. wt, wild-type HSRV2 sequences (35); EM2, inactivation of the SD (nt 8530) and SA (nt 8648) (indicated by dots); EM4, inactivation of the SD (nt 8530); EM6, Env-Bel1/Bet, intron deletion between SD (nt 8530) and SA (nt 8648); EM7, Env-Bel1, EM6 mutation combined with the inactivation of the SD (nt 8922) and SA (nt 9224); EM8, Env-Bet, EM6 mutation combined with the intron deletion between SD (nt 8922) and SA (nt 9224); EM9: Env-Bel2, deletion of all sequences between SD (nt 8530) and SA (nt 9224).
FIG. 3.
Identification of the 170-kDa Env-Bet fusion protein expressed from subgenomic or proviral constructs. 293T cells were transfected with the different expression constructs depicted in Fig. 2 and metabolically labeled, and cellular lysates were precipitated with antisera against HFV (α-HFV) (A), SU (α-SU) (B), Tas/Bel1 (α-Tas/Bel1) (C), and Bel2 (α-Bel2) (D). Lanes: 1, pcHFVenv/bel1-3 wt; 2, pcHFVenv/bel1-3 EM4; 3, pcHFVenv-bet (EM8); 4, pcHFVenv-bel1 (EM7); 5, pcHFVenv-bel2 (EM9); 6, mock, pCDNA3.1+zeo, 7, pcHSRV2 wt; 8, pcHSRV2 EM4; 9, pcHSRV2 EM7; 10, pcHSRV2 EM8; 11, pcHSRV2 EM9; 12, mock, pCDNA3.1+zeo. In lanes 1 and 2, fivefold more lysate was used than in lanes 3 to 6. The arrow indicates the 170-kDa Env-Bet fusion protein.
Next, we investigated whether the SD site (nt 8530) within the HFV env ORF is required to generate the 170-kDa protein. In HFV proviruses, this SD normally is used to generate spliced mRNAs coding for Bel1 and Bet proteins, derived from the HFV IP localized within the env ORF (22). SD sites contain a conserved GT dinucleotide which is essential for splicing to an SA site (reviewed in reference 12). Therefore, a mutant, pcHFVenv/bel1 EM4, was generated that has the SD (nt 8530) inactivated by changing the GT dinucleotide to GG, a modification that has been shown to abolish adenovirus splicing (26). When cell lysates of metabolically labeled 293T cells transfected with pcHFVenv/bel1-3 EM4 were examined by RIPA with different antisera, the 170-kDa protein was no longer detectable (Fig. 3, lanes 2). In contrast, the expression of the 130-kDa Env precursor was not affected by the inactivated SD site within the env ORF. The phenotype of the EM2 mutant, which has both the SD (nt 8530) and the SA (nt 8648) inactivated, was indistinguishable from that of the EM4 mutant (data not shown). These results showed that the SD site within the env ORF is used and is a prerequisite for the generation of the 170-kDa Env-Bet fusion protein. Furthermore, they indicated that an alternative splice mechanism is the origin for the env-bet mRNA.
Expression analysis of the Env-Bet fusion protein in the proviral context.
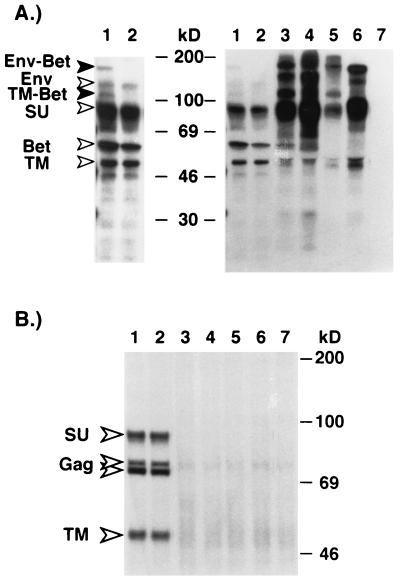
A CMV promoter-driven HFV proviral clone, pcHSRV2, has been described recently (25). This construct allowed the production of virus titers up to 105 FFU/ml by transient transfection of 293T cells. To analyze the generation of the HFV Env-Bet protein in the context of the expression of other HFV structural proteins and its influence on HFV replication in vitro, the SD mutation EM4 was introduced into the pcHSRV2 provirus, termed pcHSRV2 EM4. In addition, proviral constructs were generated containing various deletions spanning the SD and SA sites within the env and bel1/2 ORFs, resulting in the loss of expression of the 130-kDa Env protein and its cleavage products. pcHSRV2 EM6 is capable of expressing Env-Bel1 and Env-Bet proteins, whereas pcHSRV2 EM7, EM8, and EM9 can express only Env-Bel1, Env-Bet, and Env-Bel2 fusion proteins, respectively. Different recombinant viruses were generated by transfection of 293T cells and metabolically labeled, and HFV protein expression was examined by RIPA with different antisera in cell lysates (Fig. 3) and supernatants (Fig. 4A). Similar to the results obtained with the subgenomic expression constructs shown above, the analysis of cellular protein expression showed that the 170-kDa protein is precipitated by a chimpanzee serum reactive with all major HFV proteins (Fig. 3A, lane 7), by an Env-specific antiserum (Fig. 3B, lane 7), by a Bel2-specific antiserum (Fig. 3D, lane 7), or by an antiserum recognizing sequences common to Bel1 and Bet (data not shown) but not by a Bel1-specific rabbit serum (Fig. 3C, lane 7). In addition, they demonstrated that the inactivation of the SD site within the env ORF results in a loss of expression of the putative Env-Bet fusion protein (Fig. 3, lanes 8). Furthermore, the analysis of particle-associated HFV proteins indicated that the Env-Bet protein is not particle associated (Fig. 4B, lane 1), even though small amounts of the fusion protein and its cleavage products could be detected in the supernatant of pcHSRV2 wt-transfected cells (Fig. 4A, lane 1). However, it is possible that the 170-kDa Env-Bet fusion protein, which lacks the MSD of the HFV envelope, is loosely associated with HFV particles via interaction of its extracellular domains with the membrane-anchored 130-kDa Env protein and may be shed during purification of the HFV particles by ultracentrifugation through sucrose. Alternatively, the amounts of particle-associated Env-Bet fusion proteins might be to small to be detected by standard RIPA. This analysis also showed that in the absence of the 130-kDa wild-type envelope protein, no HFV particles were released into the supernatant, indicated by the absence of HFV Gag and Env proteins in the viral particle preparations of 293T cells transfected with pcHSRV2 EM6, EM7, EM8, and EM9 (Fig. 4B, lanes 3 to 6). Nevertheless, the different Env fusion proteins and their respective cleavage products were detectable in these supernatants (Fig. 4A, lanes 3 to 6), in even larger amounts than were found in the pcHSRV2 wt sample (Fig. 4A, lane 1). No apparent difference in the extra- and intracellular distribution of virus particles could be observed between wild-type virus and the EM4 mutant when examining Gag expression by RIPA (Fig. 3A, lanes 7 and 8; Fig. 4B, lanes 1 and 2) or the particle maturation by electron microscopy (data not shown).
FIG. 4.
Analysis of the particle association of the Env-Bet fusion protein. Following transfection with different proviral expression constructs, 293T cells were metabolically labeled, and the appearance of HFV Env proteins in the supernatant and associated with viral particles was analyzed. (A) RIPA of viral proteins secreted into the supernatant by using an anti-HFV chimpanzee serum. On the left is a longer exposure of lanes 1 and 2. (B) Particle-associated proteins after pelleting through 20% sucrose. Lanes: 1, pcHSRV2 wt; 2, pcHSRV2 EM4; 3, pcHSRV2 EM6; 4, pcHSRV2 EM7; 5, pcHSRV2 EM8; 6, pcHSRV2 EM9; 7, mock, pCDNA3.1+zeo.
Identification and isolation of Env-Bel mRNAs from HFV-infected human fibroblasts.
The results of the RIPA shown above suggested that the 170-kDa protein is an Env-Bet fusion protein. Furthermore, no Env-Bel1 or Env-Bel2 proteins were detected. To confirm the nature of the 170-kDa protein as an Env-Bet protein and to determine whether other possible Env-Bel fusion proteins not detected by the RIPA exist, we performed RT-PCR analysis on infected KMST-6 human fibroblast cultures. 293T cells were transfected with the proviral clones pcHSRV2, pcHSRV2 EM2, pcHSRV2 EM4, or pCDNA3.1+zeo as a control. Supernatants containing molecularly defined infectious viruses were harvested 48 h later and subjected to titer determination on BHK/LTRlacz cells. Subsequently, they were used to infect KMST-6 cells at a MOI of 0.5. Seven days after infection, total RNA was isolated from the infected cultures and used as template for an RT-PCR analysis. The primers used hybridize upstream of the transcription start of the internal promotor (nt 8419) within the env ORF (primer 391) and downstream of the SA site (nt 9224) within the bel2 ORF (primer 8) (Fig. 5A). Following amplification, aliquots of the reaction mixtures were separated by agarose gel electrophoresis (Fig. 5B). HFV-specific amplification products were detected only in HFV-infected cultures, and not in mock-infected cultures, whereas primers specific for the β-globin mRNA yielded amplification products in all samples (data not shown). All HFV-infected samples yielded fragments corresponding in size to the expected amplification products of 1,055 bp for the full-length pregenomic, gag, pol or env mRNA and of 757 bp for the Bel1-deleted ΔHFV (32) pregenomic RNA (Fig. 5B, lanes 2 to 4). Furthermore, in KMST-6 cells infected with wild-type HSRV2, an additional 637-bp fragment, corresponding in size to the putative env-bet mRNA, was detected whereas no fragments of 937 and 363 bp, corresponding to the putative env-bel1 and env-bel2 mRNA, respectively, could be detected (Fig. 5B, lane 2). Strikingly, the env-bet amplification product was present only in samples of KMST-6 cells infected by wild-type HSRV2 (Fig. 5B, lane 2) but was absent in samples of KMST-6 cells infected by the HSRV2 EM2 or HSRV2 EM4 mutant viruses harboring a defective SD site within the env ORF (Fig. 5B, lanes 3 and 4). Similar results were obtained with primers located further upstream of the transcription initiation site of the internal promoter within the env ORF and more 3′ of the SA (nt 9224) within the bel2 ORF (data not shown). All amplification products were cloned, and at least three individual clones of each insert size were sequenced. The sequencing data confirmed the nature of the amplification products as unspliced mRNA for the 1,055-bp fragment, ΔHFV mRNA for the 757-bp fragment and env-bet mRNA for the 637-bp fragment (data not shown). No fragments coding for potential Env-Bel1 or Env-Bel2 proteins could be isolated. Taken together, the results of the RT-PCR analysis of human fibroblasts infected with molecularly defined HSRV2 strains confirmed the findings of the protein analysis. The 170-kDa Env-reactive band present in cell lysates of wild-type-HFV-infected cells represents an Env-Bet fusion protein generated by alternative splicing of the env mRNA, using the SD-SA pairs located within the env ORF and the bel1 and bel2 ORFs.
FIG. 5.
RT-PCR analysis of RNA from fibroblastoid cells infected with HSRV2 wt or virus mutants. KMST-6 cells were infected with supernatants containing different molecularly defined HSRV2 mutants. Total RNA was prepared and analyzed by RT-PCR. (A) Schematic illustration of the RT-PCR analysis. The 5′ primer (primer 391) hybridizes upstream of the transcription initiation site of the IP (nt 8419), and the 3′ primer (primer 8) hybridizes downstream of the SA site (nt 9224) in the bel2 ORF. The expected sizes of the fragments amplified by the RT-PCR with the indicated primers are given in the left column. ΔHFV is the bel1 intron-deleted pregenomic mRNA (32). (B) Agarose gel electrophoresis of the amplicons obtained by RT-PCR of total RNA from cells infected with wild-type HSRV2 (lane 2), HSRV2 EM2 (lane 3), or HSRV2 EM4 (lane 4) or mock-infected cells (lane 5). Size standards are shown in lane 1. The faint additional PCR products probably represent heteroduplex molecules of the different amplification products.
Virus replication kinetics.
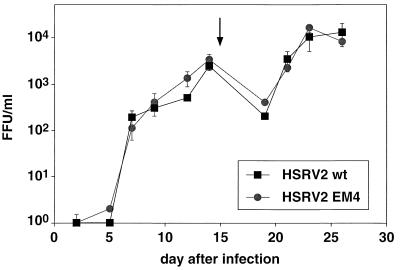
To determine whether the Env-Bet fusion protein influences the in vitro replication of HFV, we examined the replication kinetics with small amounts of input virus. BHK-21 or BHK/Bel1 cells (3) were infected with stocks of HSRV2 wt or HSRV2 EM4 of known titer at a MOI of 0.05, and secretion of virus into the supernatant was monitored for a period of 26 days by titer determination on indicator cells. The virus production was similar for both types of virus regardless of whether BHK-21 (Fig. 6) or BHK/Bel1 cells (data not shown) were used as targets. Sequencing of PCR-amplified proviral fragments spanning the SD (nt 8530) site on day 14 after infection revealed no signs of reversion of the SD mutation in HSRV2 EM4-infected cells (data not shown). Furthermore, complementation of replication-defective HFV-based vectors with either the 130-kDa Env protein alone or in combination with the 170-kDa Env-Bet fusion protein revealed no difference in the relative infectivity of the different viruses on various target cell types (data not shown). These results indicated that the expression of the Env-Bet protein by HFV does not result in enhanced virus release or higher relative infectivity in vitro.
FIG. 6.
Replication kinetics of different HSRV2 mutants on BHK-21 cells. Release of infectious virus by BHK-21 cells infected at an MOI of 0.05 with wild-type HSRV2 (HSRV2 wt) or virus lacking the 170-kDa Env-Bet fusion protein (HSRV2 EM4) is shown. The cultures were split 1:10 on day 15 (arrow).
Analysis of the influence of the Env-Bet fusion protein on virus neutralization.
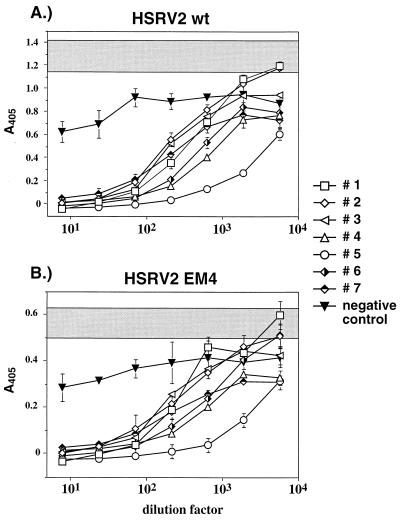
The shedding of envelope proteins or the secretion of envelope variants in vivo may affect the antiviral immune response of the host. To test whether such a mechanism applies to the 170-kDa Env-Bet protein, we examined the neutralization capacity of several chimpanzee sera on different recombinant HFVs in vitro. Supernatants containing wild-type or SD mutant virions were generated by transient transfection of 293T cells with pcHSRV2 wt and pcHSRV2 EM4, respectively. Subsequently, a specific amount of virus (500 FFU) was preincubated with different dilutions of chimpanzee sera shown to be reactive with all major HFV proteins (data not shown), before the addition to the indicator cells. At 48 h after the addition of the virus-serum mixture, the cells were lysed and the Tas/Bel1-dependent expression of β-galactosidase was determined in an enzymatic assay. Both viruses, HSRV2 wt and HSRV2 EM4, were neutralized by the different sera to a similar extent (Fig. 7). However, the overall amount of β-galactosidase produced by HSRV2 EM4 was reproducibly reduced about twofold, although the same amount of infectious virus, as determined by a focus formation assay on BHK/LTRlacz cells, was used as the inoculum. These results provide evidence that the generation of the 170-kDa Env-Bet protein may not be used to affect the host antiviral immune response, at least to the extent possible in this in vitro assay.
FIG. 7.
Inactivation of different HSRV2 mutants by neutralizing chimpanzee sera. The serum dilution is given on the x axis, and the absorbance at 405 nm (A405) is given on the y axis. The absorbance, including standard deviation, of samples in which the virus was incubated with growth medium is shaded. Seven different neutralizing chimpanzee sera (sera 1 to 7) and one control serum from a seronegative human donor (negative control) were used. (A) Neutralization of HSRV2 (HSRV2 wt). (B) Neutralization of virus lacking the 170-kDa Env-Bet protein (HSRV2 EM4).
DISCUSSION
HFV has been reported previously to express a protein of about 170 kDa that is recognized by Env- and Bel-specific antisera (9). In this work, we demonstrated by RIPA with various antisera that the 170-kDa reactive band is an Env-Bet fusion protein. Furthermore, using site-directed mutagenesis of splice site consensus sequences and RT-PCR analysis, we could show that it is generated by an alternatively spliced env mRNA that employs SD-SA pairs located within the env and bel1/2 ORFs, which are normally used to generate Bel1 and Bet transcripts derived from the internal promoter within the env ORF. This is by analogy to other retroviruses that express fusion proteins between env and accessory gene products. Human immunodeficiency virus (HIV), for example, generates a Tat-Env-Rev fusion protein termed Tev (2) and other Tat- and Rev-related Env fusion proteins (8, 13, 33) from alternatively spliced mRNAs, some of which retain the activities of the accessory gene products. What distinguishes the Env-Bet fusion protein from these is that the HFV gp170 contains the complete extracellular part of the Env protein whereas the former proteins contain only small segments of the env coding regions. Strikingly, the Env-Bet fusion protein lacks the MSD and cytoplasmic domain of the 130-kDa Env precursor protein, resulting in a targeting to the secretory pathway. Indeed, the Env-Bet fusion protein and its cleavage products, SU and TM-Bet, could be detected in the supernatant of transfected 293T cells (Fig. 4A). Interestingly, in the absence of expression of the 130-kDa Env protein, even larger amounts of the Env-Bet fusion protein and its cleavage product could be detected in the supernatant. The reason for this is unclear at present. However, we were not able to detect particle-associated Env-Bet protein. This implies that the Env-Bet protein plays no direct role in the virus entry process. This is further supported by the findings that FV vectors provided in trans with either gp130 alone or gp130 plus gp170 showed similar relative infectivity and that viruses lacking the Env-Bet protein displayed similar replication kinetics to wild-type virus in vitro. Taken together, the results presented in this report show that gp170 is dispensable for HFV replication in vitro. However, it remains to be seen how the different mutant viruses behave in vivo.
Interestingly, the SD-SA pair within the env ORF used to generate the Env-Bet fusion proteins is highly conserved within FV isolates from different primates, arguing for an important role of the Env-Bet fusion in the virus life cycle. The large amount of gp170 detected in infected cells relative to the gp130 Env precursor is a further argument in favor of the view that the Env-Bet fusion protein plays an important role in FV replication in vivo.
It has been suggested that Env protein shed by HIV particles may affect the host immune system (30). We therefore investigated whether wild-type HFV may be more resistant to neutralization by chimpanzee sera than is a mutant lacking gp170. However, we were unable to demonstrate any difference between HSRV2 wt and HSRV2 EM4 in a neutralization assay. The result implies that the Env-Bet protein is not used to counteract the host humoral immune response, at least as far as can be determined by this in vitro test.
gp170 may also exert its function through the Bet part of the fusion protein. Since no assay system for Bet is available yet and since the function of Bet is virtually unknown, this aspect could not be analyzed in the present study.
The real role of the Env-Bet protein in the FV life cycle will probably be revealed only by examination of different mutant viruses in vivo, as has been done for the HIV/SIV accessory protein Nef, which is also dispensable for in vitro growth (15, 19) but is necessary for high virus loads and establishment of productive infections in the host (18). It will be interesting to see whether there is a selection pressure to retain the expression of the Env-Bet protein in vivo. The use of a small-animal model for HFV infection (34) may be instrumental in shedding more light on the potential function of gp170 in the FV life cycle.
ACKNOWLEDGMENTS
We thank A. Saib and H. de The for communicating results prior to publication and R. M. Flügel for the Bel1 specific antiserum.
This work was supported by the EU (BMH4-CT97-2010), Bayerische Forschungsstiftung, and DFG (SFB 165). D.L. is supported by the virology fellowship program of the BMBF, Germany.
REFERENCES
- 1.Baunach G, Maurer B, Hahn H, Kranz M, Rethwilm A. Functional analysis of human foamy virus accessory reading frames. J Virol. 1993;67:5411–5418. doi: 10.1128/jvi.67.9.5411-5418.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Benko D M, Schwartz S, Pavlakis G N, Felber B K. A novel human immunodeficiency virus type 1 protein, Tev, shares sequences with Tat, Env, and Rev proteins. J Virol. 1990;64:2505–2518. doi: 10.1128/jvi.64.6.2505-2518.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bieniasz P D, Erlwein O, Aguzzi A, Rethwilm A, McClure M O. Gene transfer using replication-defective human foamy virus vectors. Virology. 1997;235:65–72. doi: 10.1006/viro.1997.8658. [DOI] [PubMed] [Google Scholar]
- 4.Bieniasz P D, Rethwilm A, Pitman R, Daniel M D, Chrystie I, McClure M O. A comparative study of higher primate foamy viruses, including a new virus from a gorilla. Virology. 1995;207:217–228. doi: 10.1006/viro.1995.1068. [DOI] [PubMed] [Google Scholar]
- 5.Du Bridge R B, Tang P, Hsia H C, Leong P M, Miller J H, Calos M P. Analysis of mutation in human cells by using an Epstein-Barr virus shuttle system. Mol Cell Biol. 1987;7:379–387. doi: 10.1128/mcb.7.1.379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Enssle J, Jordan I, Mauer B, Rethwilm A. Foamy virus reverse transcriptase is expressed independently from the gag protein. Proc Natl Acad Sci USA. 1996;93:4137–4141. doi: 10.1073/pnas.93.9.4137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fischer N, Heinkelein M, Lindemann D, Enssle J, Baum C, Werder E, Zentgraf H, Müller J G, Rethwilm A. Foamy virus particle formation. J Virol. 1998;72:1610–1615. doi: 10.1128/jvi.72.2.1610-1615.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Furtado M R, Balachandran R, Gupta P, Wolinsky S M. Analysis of alternatively spliced human immunodeficiency virus type-1 mRNA species, one of which encodes a novel tat-env fusion protein. Virology. 1991;185:258–270. doi: 10.1016/0042-6822(91)90773-5. [DOI] [PubMed] [Google Scholar]
- 9.Giron M L, Rozain F, Debons-Guillemin M C, Canivet M, Peries J, Emanoil-Ravier R. Human foamy virus polypeptides: identification of env and bel gene products. J Virol. 1993;67:3596–3600. doi: 10.1128/jvi.67.6.3596-3600.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Goepfert P A, Shaw K L, Ritter G D, Jr, Mulligan M J. A sorting motif localizes the foamy virus glycoprotein to the endoplasmic reticulum. J Virol. 1997;71:778–784. doi: 10.1128/jvi.71.1.778-784.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Goepfert P A, Wang G, Mulligan M J. Identification of an ER retrieval signal in a retroviral glycoprotein. Cell. 1995;82:543–544. doi: 10.1016/0092-8674(95)90026-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Green M R. Pre-mRNA splicing. Annu Rev Genet. 1986;20:671–708. doi: 10.1146/annurev.ge.20.120186.003323. [DOI] [PubMed] [Google Scholar]
- 13.Guatelli J C, Gingeras T R, Richman D D. Alternative splice acceptor utilization during human immunodeficiency virus type 1 infection of cultured cells. J Virol. 1990;64:4093–4098. doi: 10.1128/jvi.64.9.4093-4098.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hahn H, Baunach G, Bräutigam S, Mergia A, Neumann-Haefelin D, Daniel M D, McClure M O, Rethwilm A. Reactivity of primate sera to foamy virus Gag and Bet proteins. J Gen Virol. 1994;75:2635–2644. doi: 10.1099/0022-1317-75-10-2635. [DOI] [PubMed] [Google Scholar]
- 15.Hammes S R, Dixon E P, Malim M H, Cullen B R, Greene W C. Nef protein of human immunodeficiency virus type 1: evidence against its role as a transcriptional inhibitor. Proc Natl Acad Sci USA. 1989;86:9549–9553. doi: 10.1073/pnas.86.23.9549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.He F, Blair W S, Fukushima J, Cullen B R. The human foamy virus Bel-1 transcription factor is a sequence-specific DNA binding protein. J Virol. 1996;70:3902–3908. doi: 10.1128/jvi.70.6.3902-3908.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Keller A, Partin K M, Löchelt M, Bannert H, Flügel R M, Cullen B R. Characterization of the transcriptional trans activator of human foamy retrovirus. J Virol. 1991;65:2589–2594. doi: 10.1128/jvi.65.5.2589-2594.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kestler H W D, Ringler D J, Mori K, Panicali D L, Sehgal P K, Daniel M D, Desrosiers R C. Importance of the nef gene for maintenance of high virus loads and for development of AIDS. Cell. 1991;65:651–662. doi: 10.1016/0092-8674(91)90097-i. [DOI] [PubMed] [Google Scholar]
- 19.Kim S, Ikeuchi K, Byrn R, Groopman J, Baltimore D. Lack of a negative influence on viral growth by the nef gene of human immunodeficiency virus type 1. Proc Natl Acad Sci USA. 1989;86:9544–9548. doi: 10.1073/pnas.86.23.9544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lehrmann, H., D. Lindemann, S. Bräutigam, and A. Rethwilm. Expression of foamy virus envelope proteins by recombinant baculoviruses and generation of protein-specific antisera. Submitted for publication.
- 21.Lindemann D, Bock M, Schweizer M, Rethwilm A. Efficient pseudotyping of murine leukemia virus particles with chimeric human foamy virus envelope proteins. J Virol. 1997;71:4815–4820. doi: 10.1128/jvi.71.6.4815-4820.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Löchelt M, Flügel R M, Aboud M. The human foamy virus internal promoter directs the expression of the functional Bel 1 transactivator and Bet protein early after infection. J Virol. 1994;68:638–645. doi: 10.1128/jvi.68.2.638-645.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Löchelt M, Muranyi W, Flügel R M. Human foamy virus genome possesses an internal, Bel-1-dependent and functional promoter. Proc Natl Acad Sci USA. 1993;90:7317–7321. doi: 10.1073/pnas.90.15.7317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mergia A, Shaw K E, Pratt Lowe E, Barry P A, Luciw P A. Identification of the simian foamy virus transcriptional transactivator gene (taf) J Virol. 1991;65:2903–2909. doi: 10.1128/jvi.65.6.2903-2909.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Moebes A, Enssle J, Bieniasz P D, Heinkelein M, Lindemann D, Bock M, McClure M O, Rethwilm A. Human foamy virus reverse transcription that occurs late in the viral replication cycle. J Virol. 1997;71:7305–7311. doi: 10.1128/jvi.71.10.7305-7311.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Montell C, Fisher E F, Caruthers M H, Berk A J. Resolving the functions of overlapping viral genes by site-specific mutagenesis at a mRNA splice site. Nature. 1982;295:380–384. doi: 10.1038/295380a0. [DOI] [PubMed] [Google Scholar]
- 27.Muranyi W, Flügel R M. Analysis of splicing patterns of human spumaretrovirus by polymerase chain reaction reveals complex RNA structures. J Virol. 1991;65:727–735. doi: 10.1128/jvi.65.2.727-735.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Namba M, Nishitani K, Hyodoh F, Fukushima F, Kimoto T. Neoplastic transformation of human diploid fibroblasts (KMST-6) by treatment with 60Co gamma rays. Int J Cancer. 1985;35:275–280. doi: 10.1002/ijc.2910350221. [DOI] [PubMed] [Google Scholar]
- 29.Netzer K O, Rethwilm A, Maurer B, ter Meulen V. Identification of the major immunogenic structural proteins of human foamy virus. J Gen Virol. 1990;71:1237–1241. doi: 10.1099/0022-1317-71-5-1237. [DOI] [PubMed] [Google Scholar]
- 30.Parren W H, Burton D R, Sattentau Q J. HIV-1 antibody—debris or virion? Nat Med. 1997;3:366–367. doi: 10.1038/nm0497-366d. [DOI] [PubMed] [Google Scholar]
- 31.Rethwilm A, Erlwein O, Baunach G, Maurer B, ter Meulen V. The transcriptional transactivator of human foamy virus maps to the bel 1 genomic region. Proc Natl Acad Sci USA. 1991;88:941–945. doi: 10.1073/pnas.88.3.941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Saib A, Peries J, The H d. A defective human foamy provirus generated by pregenome splicing. EMBO J. 1993;12:4439–4444. doi: 10.1002/j.1460-2075.1993.tb06129.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Salfeld J, Göttlinger H G, Sia R A, Park R E, Sodroski J G, Haseltine W A. A tripartite HIV-1 tat-env-rev fusion protein. EMBO J. 1990;9:965–970. doi: 10.1002/j.1460-2075.1990.tb08195.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schmidt M, Niewiesk S, Heeney J, Aguzzi A, Rethwilm A. Mouse model to study the replication of primate foamy viruses. J Gen Virol. 1997;78:1929–1933. doi: 10.1099/0022-1317-78-8-1929. [DOI] [PubMed] [Google Scholar]
- 35.Schmidt M, Rethwilm A. Replicating foamy virus-based vectors directing high level expression of foreign genes. Virology. 1995;210:167–178. doi: 10.1006/viro.1995.1328. [DOI] [PubMed] [Google Scholar]
- 36.Weiss R A. Foamy viruses bubble on. Nature. 1996;380:201. doi: 10.1038/380201a0. [DOI] [PubMed] [Google Scholar]
- 37.Yu S F, Baldwin D N, Gwynn S R, Yendapalli S, Linial M L. Human foamy virus replication: a pathway distinct from that of retroviruses and hepadnaviruses. Science. 1996;271:1579–1582. doi: 10.1126/science.271.5255.1579. [DOI] [PubMed] [Google Scholar]
- 38.Zou J X, Luciw P A. The transcriptional transactivator of simian foamy virus 1 binds to a DNA target element in the viral internal promoter. Proc Natl Acad Sci USA. 1996;93:326–330. doi: 10.1073/pnas.93.1.326. [DOI] [PMC free article] [PubMed] [Google Scholar]