Abstract
The matrix protein of human immunodeficiency virus type 1 (HIV-1) has been reported to play a crucial role in the targeting of the Gag polyprotein precursor to the plasma membrane and in the incorporation of viral envelope glycoproteins into budding virions. In this report, we present evidence that mutation of a highly conserved Leu at matrix amino acid 20 blocks or markedly delays virus replication in a range of cell types, including T-cell lines, primary human peripheral blood mononuclear cells, and monocyte-derived macrophages. These mutations do not impair virus assembly and release, RNA encapsidation, or envelope glycoprotein incorporation into virions but rather cause significant defects in an early step in the virus life cycle, as measured by single-cycle infectivity assays and the analysis of viral DNA synthesis early postinfection. This infectivity defect is independent of the type of envelope glycoprotein carried on mutant virions; similar results are obtained in pseudotyping experiments using wild-type or truncated HIV-1 envelope glycoproteins, the amphotropic murine leukemia virus envelope, or the vesicular stomatitis G protein. Intriguingly, matrix residue 20 mutations also increase the apparent binding of Gag to membrane, accelerate the kinetics of Gag processing, and induce defects in endogenous reverse transcriptase activity without affecting virion density or morphology. These results help elucidate the function of matrix in HIV-1 replication.
The human immunodeficiency virus type 1 (HIV-1) matrix (MA) protein is initially synthesized as part of a polyprotein precursor, Pr55Gag, which is proteolytically cleaved by the viral protease (PR) to generate the mature Gag proteins: MA (p17), CA (p24), NC (p7), and p6. During translation, the MA domain of Pr55Gag is modified by the covalent attachment of a myristic acid moiety to the N-terminal Gly residue (60). In the virion, MA is located just inside the lipid bilayer of the viral envelope and is attached to the bilayer by a multipartite membrane binding domain (35; for reviews, see references 34, 42, and 71).
Two major functions for the HIV-1 MA protein have been clearly established. (i) MA is critical to the targeting of the Gag precursor to the plasma membrane. Mutation of the N-terminal Gly, which serves as the acceptor site for Gag myristylation, abolishes virus assembly in most systems (7, 30, 37, 56). Mutation of a highly basic domain near the N terminus of MA (residues 17 to 31) disrupts proper Gag targeting and virus assembly (24, 75, 78), and single amino acid changes between MA residues 84 and 88 redirect virus assembly to cytoplasmic compartments (30). A large deletion in MA also retargets assembly to the cytoplasm (20, 33). (ii) MA is required for efficient incorporation of the envelope (Env) glycoproteins into virions. Deletions and multiple-amino-acid substitutions throughout the majority of MA impair Env incorporation (17, 74), and single-amino-acid substitutions near the amino terminus of MA (e.g., at residues 10, 12, 30, and 34) abolish or significantly reduce Env incorporation (27, 28, 54).
It has also been suggested that MA plays a role in translocating the viral preintegration complex to the nucleus (9, 38, 66), although a significant amount of data is not consistent with this hypothesis (21, 23–25) (see Discussion). An additional role for retroviral MA proteins early in the virus life cycle has been proposed; however, the mechanism responsible for this function has not been elucidated. More than a decade ago, a set of deletions in the C terminus of murine leukemia virus (MuLV) MA was reported to dramatically reduce virus infectivity without affecting assembly and release, RNA encapsidation, or virion reverse transcriptase (RT) activity (15). More recently, mutations in an avian retrovirus (Rous sarcoma virus) which impair virus infectivity without affecting assembly and release were described (57). Several reports have also implicated HIV-1 MA in an early step in the virus life cycle prior to the completion of reverse transcription (11, 58, 73).
The events which immediately follow membrane fusion and the release of the viral nucleocapsid into the host cell cytoplasm, which are often referred to as uncoating, are poorly understood. In addition to the reports mentioned above implicating MA in these steps, mutation of other HIV-1 gene products has been observed to affect early, postfusion events. These gene products include Vif (62, 63, 67), Nef (2, 3, 12, 61), NC (4), and CA (16, 58, 68). Mutations in HIV-1 CA which prevent the incorporation of cyclophilin A into virions have also been reported to impair an early postentry step (6). In most of these studies, the synthesis of viral DNA at early time points postinfection was used as a marker for detecting early events.
During the course of our previous analysis of the MA basic domain (24), we noted that a nonbasic residue within this region, the Leu at MA amino acid 20, is remarkably well conserved among lentiviral MA proteins (53). We report here that mutation of this residue, in particular to Lys, causes marked replication defects in a range of cell types without impairing virus assembly and release, RNA encapsidation, or the incorporation of Env glycoproteins into virions. These mutants display significant infectivity defects in single-cycle experiments in CD4+ HeLa cells and T-cell lines, using the MAGI infectivity assay or molecular clones engineered to express luciferase postinfection. An early defect is also observed by PCR amplification of viral DNA at early time points after infection. The residue 20 mutants display several additional phenotypes, including accelerated Gag precursor processing, an apparent enhancement in Gag membrane binding, and reduced activity in endogenous RT (ERT) assays.
MATERIALS AND METHODS
Cells, viruses, and plasmids.
HeLa, CEM(12D-7), and MAGI cells were maintained as described previously (27). Jurkat, H9, MT-4, and A.301 cells were cultured in RPMI 1640 medium supplemented with 10% fetal bovine serum (FBS), 2 mM glutamine, and antibiotics. 293T cells were maintained in Dulbecco’s modified Eagle’s medium plus 10% FBS, 2 mM glutamine, and antibiotics. The isolation and culture of primary human monocyte-derived macrophages (MDM) have been described previously in detail (24). Human peripheral blood mononuclear cells (PBMC) were stimulated with 1 μg of phytohemagglutinin per ml for 3 days and maintained in RPMI 1640 medium plus 10% FBS, 2 mM glutamine, antibiotics, and 20 U of recombinant human interleukin-2 (Boehringer Mannheim) per ml. For infection of T-cell lines, PBMC, and MAGI cells, virus was derived from the T-cell line-tropic molecular clone pNL4-3 (1) and derivatives containing the amino acid 20 mutations. For macrophage infectivity analyses, we used the macrophage-tropic pNL4-3 derivative pNL(AD8) (24). As described previously, since replication of pNL4-3 is restricted in macrophages, this virus was included as a negative control (24). We used the following Env expression plasmids: for HIV-1 Env, pHenv (29); for amphotropic MuLV (ampho-MuLV) Env, pSVAMLVenv (45); for vesicular stomatitis virus G glycoprotein (VSV-G), pHCMV-G (72). pSVAMLVenv was obtained from N. Landau and D. Littman through the National Institutes of Health (NIH) AIDS Research and Reference Reagent Program; pHCMV-G was kindly provided by J. Burns. The luciferase-expressing clone pNL4-3.Luc.R−E− was obtained from N. Landau through the NIH AIDS Research and Reference Reagent Program.
Transfections and infections.
For most assays, virus stocks were prepared by HeLa transfection as described previously (26). In the case of the luciferase-expressing (pNLuc) clones, very low RT activity was obtained in HeLa cells relative to that obtained with pNL4-3. In contrast, the amounts of RT activity released into the media of pNLuc- and pNL4-3-transfected 293T cells were comparable. Virus stocks of the pNLuc and pNLuc/20LK clones were therefore prepared in 293T cells. Transfected cell supernatants were harvested, passed through a 0.45-μm-pore-size filter, normalized for RT activity, and used in infections as indicated.
Mutagenesis and cloning.
Our strategy for introducing MA mutations into pNL4-3 by site-directed mutagenesis has been described previously (30). The amino acid 20 mutations were introduced into the macrophage-tropic molecular clone pNL (AD8) (24) by cloning the env-containing SalI-BamHI fragment (pNL4-3 nucleotides [nt] 5785 to 8465) from pNL(AD8) into pNL4-3/20LE, pNL4-3/20LK, and pNL4-3/20LR. For use in pseudotyping experiments, an env− derivative of the 20LK mutant was constructed by cloning the KFS env frameshift mutation (22) into pNL4-3/20LK on the SalI-BamHI fragment. pNLuc and pNLuc/20LK were constructed by cloning the SalI-NcoI fragment (pNL4-3 nt 5785 to 10568) from pNL4-3.Luc.R−E− (13) into pNL4-3 and pNL4-3/20LK. Because the vpr mutation present in pNL4-3.Luc.R−E− is 5′ of the SalI site (13), pNLuc and pNLuc/20LK express full-length, functional vpr genes. The pHenvCTdel-144 HIV-1 Env expression vector expressing a 144-amino-acid-truncated form of the HIV-1 Env glycoprotein was constructed by introducing the SalI-BamHI fragment from pNLTr712 (69; kindly provided by V. Bosch) into pHenv (29). The protease-defective (PR−) version of pNL4-3 contains an Asp→Asn mutation at PR residue 25, which lies in the PR active site (47). To construct PR− versions of the 1GA and MA amino acid 20 mutants for use in membrane binding analyses, the SphI-EcoRI fragment (pNL4-3 nt 1443 to 5743) encompassing the PR coding region was introduced from pNL4-3/PR− into pNL4-3/1GA, pNL4-3/20LK, pNL4-3/20LR, and pNL4-3/20LE.
Pulse-chase analysis of Gag processing and virion release.
HeLa cells transfected with the indicated molecular clones were pulse-labeled for 30 min in [35S]Cys and [35S]Met. After the labeling period, the medium was replaced with 10% FBS–RPMI 1640, and the cells were cultured at 37°C for a chase time of 1, 3, or 6 h. At each time point, the cells were harvested and solubilized in lysis buffer. Labeled virions were recovered by spinning the supernatants in 1.5-ml Sarstedt tubes in a refrigerated Tomy microcentrifuge (26). The virion pellets and the supernatant from the microcentrifuge spin were mixed with lysis buffer. Recovery of the supernatant fraction permitted the evaluation of proteins that are released from the cell in a soluble, virion-free form. The three fractions (cell lysates, virion lysates, and supernatants) were then separately immunoprecipitated with AIDS patient serum and analyzed by sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (PAGE).
Radioimmunoprecipitation and Western blot analysis.
Methods used for metabolically labeling transfected HeLa cells, preparing cell and virion lysates, and immunoprecipitating viral proteins have been detailed previously (26, 70). For Western blotting, proteins were separated by SDS-PAGE (10% gel) and transferred to polyvinylidine difluoride membranes (Millipore). Membranes were incubated with a mixture of rabbit anti-gp120 antibody (a kind gift of K. Strebel) and AIDS patient sera (human HIV immunoglobulin [Ig]; obtained from NIH AIDS Research and Reference Reagent Program) as primary antibodies. Subsequently, membranes were incubated with a mixture of horseradish peroxidase-conjugated anti-rabbit Ig and anti-human Ig (Amersham). Antigens recognized by antibodies were detected with enhanced chemiluminescence reagents (Amersham).
RNA dot blot analysis.
Wild-type (wt) and mutant virions, produced by transient transfection of HeLa cells, were centrifuged for 45 min at 100,000 × g in an SW55 rotor (Beckman), and the pellets were resuspended in a buffer containing 10 mM Tris-HCl (pH 7.4), 100 mM NaCl, and 5 mM MgCl2. A 40-μl sample of each virus preparation, normalized for RT activity, was digested with 10 U of RNase-free DNase (Boehringer Mannheim) for 1 h at 37°C. Virions were disrupted by incubation with 1% Nonidet P-40 for 30 min at 37°C in the presence of 100 U of rRNasin (Promega), followed by the addition of 30 μl of 20× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) and 20 μl of formaldehyde on ice. The samples were heated to 60°C for 15 min, diluted with 2 volumes of cold 20× SSC, immobilized onto nylon membranes by using a dot blot manifold, and then hybridized overnight at 45°C to a 32P-labeled HIV-1-specific DNA probe. Membranes were washed twice with 2× SSC–0.1% SDS at 65°C for 20 min, dried, and exposed to X-ray film.
PCR assays.
Virus stocks were obtained by cotransfection of HeLa cells with the env− molecular clones pNL4-3KFS or pNL4-3KFS/20LK and an ampho-MuLV Env expression vector (pSVAMLVenv [45]) or an HIV-1 Env expression vector (pHenv [29]). Transfection supernatants, normalized for RT activity and adjusted to a 1-ml volume, were treated with 100 U of DNase (Boehringer Mannheim) for 1 h at 37°C in the presence of 10 mM MgCl2. Virus (5 × 105 RT cpm) was used to infect 106 CEM(12D-7) cells for 3 h at 37°C. Cells were washed three times in phosphate-buffered saline (PBS), resuspended in growth medium, and incubated at 37°C. At specified times postinfection, cells were washed in PBS, lysed, and digested with proteinase K as described previously (19). HIV-specific sequences were amplified in PCR mixtures containing approximately 10 μl of lysate, 0.2 mM deoxynucleoside triphosphates, 1 μM each oligonucleotide, 1.5 mM MgCl2, 10 mM Tris-HCl, 50 mM KCl, and 1.25 U of Taq polymerase (Boehringer Mannheim) in a 50-μl volume. Samples were amplified for 30 cycles, using an annealing temperature of 60°C. Oligonucleotide primer pairs used were as follows, with pNL4-3 positions indicated (53): long terminal repeat (LTR) (plus-sense primer, nt 9035 to 9055 [AGCTGTAGATCTTAGCCACTT]; minus-sense primer, nt 9541 to 9559 [AGGCTCAGATCTGGTCTAA]); Env (plus-sense primer, nt 6944 to 6966 [ACAGTACAATGTACACATGGAAT]; minus-sense primer, nt 7466 to 7498 [CTGCCACATGTTTATAAATTGTTTTATTCTGCA]); circles (plus-sense primer, nt 9035 to 9055 [AGCTGTAGATCTTAGCCACTT]; minus-sense primer, nt 635 to 653 [GTCCCTGTTCGGGCGCCAC]). PCR products were electrophoresed on 1% agarose gels in Tris-borate-EDTA (TBE) running buffer, denatured in 0.4 M NaOH–1.5 M NaCl, and transferred to Hybond-N+ (Amersham). PCR products were hybridized to 32P-labeled oligonucleotides and exposed to X-ray film. An oligonucleotide probe corresponding to pNL4-3 nt 421 to 524 was used to detect LTR and circle PCR products, and an oligonucleotide probe spanning nt 7150 to 7166 was used to detect envelope-specific products. To ensure that all reactions were performed within the linear range, PCR was done on a dilution series of cell lysates. PCR amplification of human β-globin sequence was performed as described above except that 2 μCi of [32P]TTP was included in each reaction. β-Globin primers (RS79/80) have been described previously (59). PCR products were electrophoresed as described above, and gels were dried and exposed to film.
ERT assays.
The ERT assay was performed by using a protocol modified from that described by Goncalves et al. (36). Virus, normalized for exogenous RT activity or p24 concentration, was pelleted at 14,000 × g for 90 min and permeabilized for 10 min at room temperature with the indicated detergent, followed by the addition of an ERT reaction mixture containing a final concentration of 50 mM Tris-HCl (pH 8.0), 2 mM magnesium acetate, 10 mM dithiothreitol, 0.1 mM dCTP, dGTP and dATP, and 10 μCi of [32P]TTP in a final volume of 100 μl. After 16 h of incubation at 37°C, 20 μg of RNase A was added, and samples were incubated for 1 h at 37°C and then digested with 20 μg of proteinase K for 3 h at 65°C. Reaction products were purified by using a Wizard PCR Prep DNA purification kit (Promega) and denatured in 0.1 M NaOH for 1 h at 37°C prior to electrophoresis in 1% agarose in TBE running buffer. 32P-labeled DNA molecular weight markers (Gibco BRL) were denatured and run in parallel. The gels were dried and exposed to film. As indicated by the size of the products obtained, these conditions allow reverse transcription to proceed beyond minus-strand strong-stop DNA. As a negative control in ERT assays, we used the RT active-site mutant RT/D186N, kindly provided by A. Engelman (19).
Luciferase assays.
Virus stocks were obtained by cotransfecting the luciferase-expressing env− molecular clone pNLuc or pNLuc/20LK with an ampho-MuLV or HIV-1 Env expression vector (see above). H9 or CEM(12D-7) cells (5 × 105) were incubated with 100 μl of luciferase-expressing virus in the presence of DEAE-dextran (20 μg/ml) for 3 h at 37°C; 1 ml of growth medium was added, and cells were incubated at 37°C for 48 h. Cells were washed twice with PBS and then lysed in 100 μl of reporter lysis buffer (Promega). Samples were subjected to one freeze-thaw cycle, and cell membranes were removed by centrifugation. Luciferase activity was measured following addition of 100 μl of substrate (Promega) to 10 μl of extract.
Membrane binding analysis.
HeLa cells were transfected with PR− versions of pNL4-3, MA amino acid 20 mutant derivatives, or the myristylation mutant pNL4-3/1GA (30). Two days posttransfection, cell fractionations were performed as follows. Cells were rinsed with ice-cold PBS, scraped, and centrifuged at 600 × g for 5 min. Cell pellets were washed once with 10 mM Tris-HCl (pH 7.4) containing 1 mM EGTA and once with 10 mM Tris-HCl (pH 7.4) containing 1 mM EDTA and then resuspended in 10 mM Tris-HCl (pH 7.4) containing 1 mM EDTA, 6% (wt/vol) sucrose, 0.2 mg of leupeptin per ml, and 0.2 mM phenylmethylsulfonyl fluoride. Cells were then disrupted by sonication in ice water. Lysis was monitored microscopically and continued until 80% of cells had broken. Lysates were centrifuged at 2,000 rpm in an Eppendorf Microfuge for 3 min to remove unbroken cells and nuclei. The resulting supernatants were not adjusted (no salt) or adjusted to 1 M NaCl (high salt) and centrifuged at 100,000 × g for 1 h in a Beckman SW55Ti rotor. Detergent treatments were performed by adjusting the mixture to 0.1% SDS prior to the 100,000 × g centrifugation. The supernatants, and the pellets resuspended in 10 mM Tris-HCl (pH 7.4) containing 1 mM EDTA, 6% (wt/vol) sucrose, and 1 M NaCl (for high-salt conditions), were subjected to Western blotting as described above. For sucrose density gradient analysis, the sonicated supernatants, adjusted to 1 M NaCl as described above, were loaded onto gradients composed of 20, 30, 40, 50, and 60% (wt/vol) sucrose in TE. The gradients were centrifuged for 16 h at 100,000 × g at 4°C in a Beckman SW55Ti rotor. Eleven 480-μl fractions were collected from the tops of the tubes for Western blot analysis.
RESULTS
Mutation of MA amino acid 20 causes replication defects in a variety of cell types.
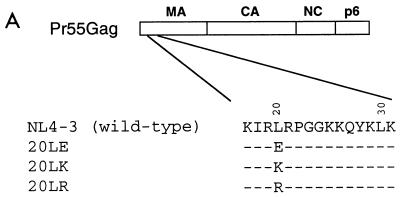
Sequence comparison of MA from primate lentiviruses (HIV-1, HIV-2, and simian immunodeficiency viruses from a range of species) reveals a remarkable degree of conservation of a Leu at MA amino acid 20 (53). Because of the high degree of conservation at this position, and its location in the MA highly basic domain (see the introduction), we sought to evaluate the role of this residue in HIV-1 replication. Three amino acid 20 mutations were introduced: Leu→Glu (20LE) (30), Arg (20LR), and Lys (20LK) (Fig. 1A). Following mutagenesis, these changes were introduced into the full-length, infectious molecular clone pNL4-3 (1) and the macrophage-tropic pNL4-3 derivative pNL(AD8) (24).
FIG. 1.
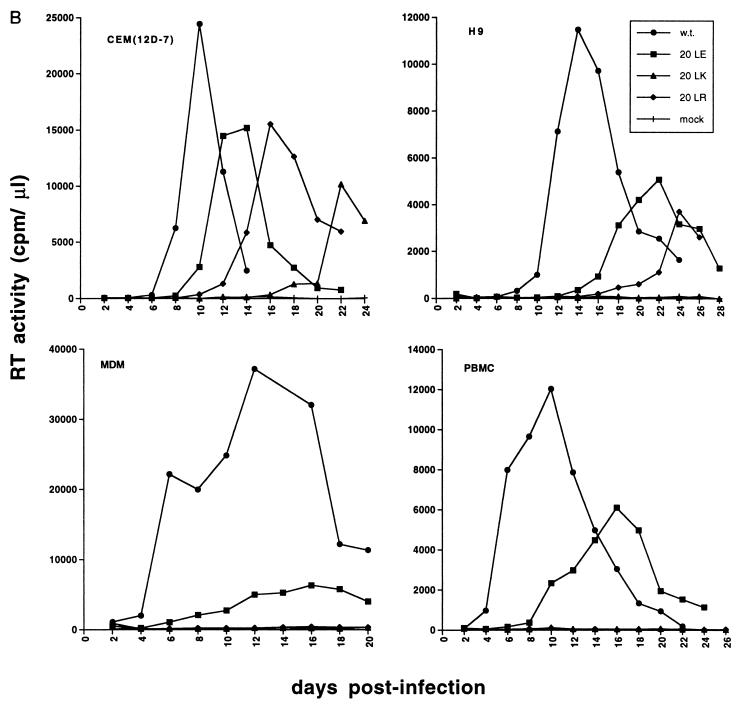
Mutagenesis and replication kinetics of MA amino acid 20 mutants. (A) The sequence of the highly basic domain of wt pNL4-3 is shown at the top. Beneath the wt sequence are indicated the residue 20 mutations. (B) Virus stocks obtained by transfection of HeLa cells with the indicated molecular clones were normalized for RT activity and used to infect the CEM(12D-7) or H9 T-cell line or primary MDM or PBMC. For infection of CEM(12D-7), H9, and PBMC, wt virus was pNL4-3; for MDM infection, wt virus was pNL(AD8). RT activity was monitored in the cell supernatant over time.
Virus stocks were prepared by transfection of HeLa cells and were used to infect a variety of T-cell lines, activated primary PBMC, and fully differentiated MDM. The results indicated that in T-cell lines [Jurkat, CEM(12D-7), H9, and MT-4], as well as in primary cell types (PBMC and MDM), the amino acid 20 mutations imposed significant replication defects (Fig. 1B and data not shown). This effect was particularly pronounced for the basic substitutions (i.e., 20LK and 20LR) and was most marked in the primary cells. In most PBMC and in all MDM donors tested, the 20LR and 20LK mutations completely blocked the establishment of a productive infection. In certain PBMC donors, a low level of virus replication was detected with all position 20 mutants (data not shown).
Amino acid 20 mutations do not impair virus production, Env incorporation, or RNA encapsidation.
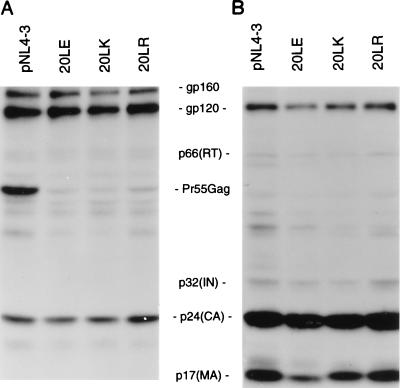
Previous results from our laboratory indicated that HIV-1 MA mutations can block virus replication by disrupting virus production or Env glycoprotein incorporation into virions (27, 28, 30, 54). To determine whether these aspects of virus assembly were affected by amino acid 20 mutations, we transfected HeLa cells in parallel with wt or mutant molecular clones, metabolically labeled with [35S]Cys, and immunoprecipitated cell- and virion-associated proteins with AIDS patient serum (see Materials and Methods). The results (Fig. 2) demonstrated that the amount of virion-associated protein produced from the transfected cells [p24(CA), p17(MA), gp120, p66(RT) and p32(IN)] was not significantly affected by the amino acid 20 mutations.
FIG. 2.
Radioimmunoprecipitation analysis of cell- and virion-associated proteins. HeLa cells were transfected with the indicated molecular clones and metabolically labeled with [35S]Cys. Cell (A) and virion (B) fractions were obtained and immunoprecipitated with AIDS patient serum (Materials and Methods). The positions of the Env glycoprotein precursor (gp160), the mature surface Env glycoprotein (gp120), p66(RT), the Gag precursor Pr55Gag, p32(IN), p24(CA), and p17(MA) are indicated.
The efficiency of virus assembly and release was also determined by measuring the amount of virion-associated RT activity present in the transfected cell supernatant. This analysis confirmed the lack of a significant effect on steady-state virus particle production and indicated that the amino acid 20 mutations did not affect exogenous RT activity present in mutant virions (data not shown). Together with the immunoprecipitation data (Fig. 2), the RT results also demonstrated that these substitutions did not impair the incorporation or subsequent processing of Pr160Gag-Pol in virions.
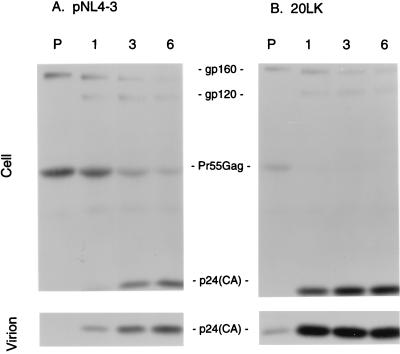
In contrast, the level of mutant Pr55Gag detected in the cell-associated fraction of transfected HeLa cells was significantly reduced relative to the wt level (Fig. 2A). This effect was also observed in CEM(12D-7) cells acutely infected with the 20LK mutant (data not shown). To investigate this issue further, we performed pulse-chase analysis of HeLa cells transfected in parallel with pNL4-3 (wt) or the 20LK mutant (Fig. 3). The results indicated that the residue 20 mutant Pr55Gag was processed to the mature Gag proteins more rapidly than wt and that the production of virion-associated p24(CA) was accelerated. No significant difference was observed in the profiles of soluble (non-virion-associated) proteins released from wt- and 20LK-transfected cells (data not shown). We also analyzed 20LE Gag processing kinetics in parallel with wt and 20LK. 20LE displayed an accelerated rate of Gag processing and virion release relative to wt, but the magnitude of the effect was less pronounced than observed with the 20LK mutant (data not shown). The results of these experiments (Fig. 2 and 3) indicate that whereas the steady-state level of virus production is not affected by the residue 20 mutations, these substitutions increase the rate of Pr55Gag processing and the kinetics of virion release.
FIG. 3.
Pulse-chase analysis of Gag processing and virion release. HeLa cells transfected with the wt or 20LK mutant molecular clone were metabolically labeled for 30 min with [35S]Cys and [35S]Met and then chased in cold medium for 1, 3, or 6 h. Cell- and virion-associated proteins were obtained and immunoprecipitated with AIDS patient serum (Materials and Methods). The positions of the viral proteins are indicated.
An effect of MA amino acid 20 mutations on RNA encapsidation appeared unlikely, since deletion of the entire MA domain did not block the amount of genomic viral RNA present in virions (46). However, to investigate this issue directly, we performed dot blot analysis on RNA derived from wt pNL4-3 or 20LK mutant particles. As controls, we used two mutants (in the 5′ untranslated region) previously demonstrated to reduce viral RNA encapsidation (51). As reported, the S1S3 and Δ42Δ21 mutants caused significant defects in the encapsidation of viral RNA. In contrast, the 20LK mutant virions contained wt levels of virion RNA (data not shown). These results suggest that the replication defects imposed by residue 20 mutations are not caused by impaired RNA encapsidation.
Single-cycle infectivity assays demonstrate a defect early in the virus life cycle.
To determine whether early steps in the virus life cycle might be affected by residue 20 mutations, we performed single-cycle infectivity analyses using the MAGI assay (44). The results indicated that the 20LE mutation caused a 2-fold reduction in virus infectivity (45% ± 10% of wt pNL4-3 activity) whereas both 20LK and 20LR mutations reduced infectivity approximately 10-fold (12% ± 1% and 12% ± 5%, respectively, of wt activity). These observations suggest that amino acid 20 mutations affect an early step in the virus life cycle.
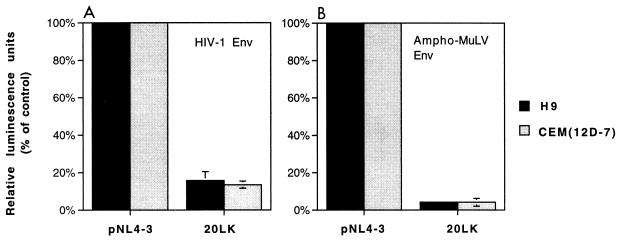
As an additional measure of virus infectivity, we made use of HIV-1 molecular clones modified to express luciferase following infection. We constructed derivatives of the env− clone pNL4-3.Luc.R−E− (13) in which the vpr defect was repaired (Materials and Methods) and cotransfected these molecular clones, expressing either wt or 20LK MA, with the HIV-1 Env expression vector pHenv (29); virus stocks were prepared and used to infect H9 and CEM(12D-7) cells. Forty-eight hours postinfection, cells were lysed and luciferase activity was measured. Consistent with the MAGI data (see above), the results of these experiments indicated that the 20LK mutation markedly reduced virus infectivity in a single round of replication (Fig. 4A).
FIG. 4.
Relative infectivities of 20LK in H9 and CEM(12D-7) T-cell lines. Virus stocks obtained by cotransfecting 293T cells with luciferase-expressing molecular clones and an HIV-1 or MuLV Env expression vector were used for infection of cells as indicated (Materials and Methods).
Infectivity defect imposed by amino acid 20 mutation is not reversed by gp41 truncation or pseudotyping with heterologous Env glycoproteins.
Previously, we and others demonstrated that the effect of MA mutations on Env incorporation could be reversed by truncating sequences within the long cytoplasmic domain of HIV-1 gp41 or by pseudotyping with heterologous retroviral Env glycoproteins containing short cytoplasmic tails (27, 28, 49). These and other results (14, 17, 74) suggested a direct interaction between MA and the long cytoplasmic tail of gp41. Although the residue 20 substitutions described here do not affect Env incorporation (Fig. 2), the possibility remained that these mutations might in some manner affect membrane fusion and virus entry by perturbing the putative interaction between MA and gp41. To evaluate this possibility, we tested the effect of gp41 truncation on the infectivity of 20LK mutant virus. The KFS env frameshift mutation (22) was introduced into pNL4-3 and pNL4-3/20LK as described in Materials and Methods. These env− molecular clones were then cotransfected in parallel with pHenv, which expresses the wt HIV-1 Env glycoprotein, or pHenvCTdel-144, which expresses a 144-amino-acid-truncated HIV-1 Env glycoprotein (27, 69). Virus stocks were harvested and tested in the MAGI infectivity assay. The results (Table 1) indicated that the length of the gp41 cytoplasmic tail did not influence the 20LK phenotype. A similar analysis was performed with the ampho-MuLV Env expression vector pSVAMLVenv (45). Again, the 20LK phenotype was independent of the Env glycoprotein used in the analysis.
TABLE 1.
Infectivities of pseudotyped 20LK virions in MAGI cells
| Virus | Pseudotyping Env | Relative infectious unitsa |
|---|---|---|
| pNL4-3KFS | wt HIV-1 | 100 |
| pNL4-3KFS/20LK | wt HIV-1 | 16 ± 6 |
| pNL4-3KFS | Truncated HIV-1 | 100 |
| pNL4-3KFS/20LK | Truncated HIV-1 | 22 ± 13 |
| pNL4-3KFS | Ampho-MuLV | 100 |
| pNL4-3KFS/20LK | Ampho-MuLV | 23 ± 11 |
| pNL4-3KFS | VSV-G | 100 |
| pNL4-3KFS/20LK | VSV-G | 19 ± 3 |
Quantitated by determining the number of blue cells after infection with wt or mutants. Pseudotyped virus stocks were obtained by cotransfection of HeLa cells with the indicated molecular clones and Env-expressing vectors (Materials and Methods) and were normalized for RT activity prior to infection. Data are averages from at least three independent assays ± standard deviation.
It was recently reported that the infectivity defect caused by mutation of the nef gene was suppressed by pseudotyping with VSV-G (2). This finding suggested that VSV-G, which directs entry via receptor-mediated endocytosis rather than by fusion at the plasma membrane (50), allowed HIV-1 to bypass the defect imposed by the nef mutation (2). To assess whether the 20LK entry defect might be similarly affected by pseudotyping with VSV-G, we cotransfected the env− clones pNL4-3KFS and pNL4-3KFS/20LK with pHCMV-G (72) and tested the infectivity of the pseudotyped virions in MAGI cells. We observed that the 20LK defect was not reversed by VSV-G (Table 1), suggesting that the 20LK block is independent of the route of HIV-1 entry.
To confirm that the 20LK infectivity defect was not suppressed by pseudotyping with heterologous Env glycoproteins, molecular clones pNLuc and pNLuc/20LK were cotransfected into HeLa cells with the ampho-MuLV Env expression vector. The resulting virus stocks were normalized for RT activity and used to infect H9 and CEM(12D-7) cells (Fig. 4B). These data again demonstrate that the 20LK phenotype is independent of the type of Env glycoprotein carried by the mutant virions.
The 20LK MA mutation impairs the synthesis of viral DNA postinfection.
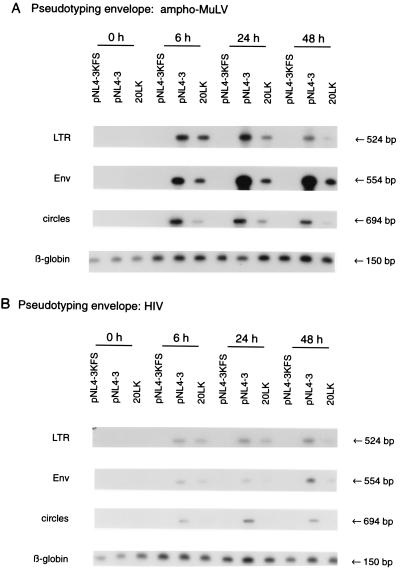
The MAGI and luciferase assays described above detect early events in the virus life cycle by measuring, directly or indirectly, the synthesis of virally encoded gene products after DNA integration. Thus, these assays cannot define the early step (reverse transcription, nuclear import, or integration) which is affected by these mutations. As a measure of early events prior to nuclear import, we assessed the amount of viral DNA synthesized at early time points postinfection. To ensure that only early events were measured (i.e., that second-round reinfections did not contribute to the signal measured), we performed the assays using pseudotyped virions. The env− pNL4-3KFS or pNL4-3KFS/20LK mutant clone was cotransfected with an ampho-MuLV (Fig. 5A) or HIV-1 (Fig. 5B) Env expression vector. The resulting virus was harvested, normalized for RT activity, and used to infect the CEM(12D-7) T-cell line. Nonpseudotyped pNL4-3KFS-derived virus was used as a negative control. At the indicated time points postinfection (Fig. 5), the cells were lysed and the viral DNA was PCR amplified by using primers specific for LTR or Env sequences or which amplify circular (nuclear) forms of viral DNA (Materials and Methods). To ensure that PCRs were performed within the linear template range, PCR was carried out on serially diluted infected cell lysates (data not shown). As an additional PCR control, we amplified cellular β-globin sequences. Amplified products were then electrophoresed on agarose gels and Southern blotted with HIV-1-specific probes. The results demonstrated that the 20LK mutant was significantly impaired in its ability to synthesize viral DNA postinfection. Interestingly, the magnitude of the effect appeared to be consistently greater at later time points (particularly 48 h) postinfection. These results are consistent with data presented above and support the hypothesis that MA residue 20 mutations cause a defect at an early step in the virus life cycle. In some assays (e.g., Fig. 5B), a greater difference was observed in the level of circular DNAs relative to Env- or LTR-specific products. However, since 20LK is deficient in synthesizing stable linear as well as circular DNAs, the data presented in Fig. 5 indicate that the 20LK-imposed defect is elicited prior to nuclear transport.
FIG. 5.
PCR amplification of viral DNA at early time points postinfection. Virus stocks obtained by cotransfection of env− molecular clones and ampho-MuLV (A) or HIV-1 (B) Env expression vectors were normalized for RT activity and used to infect the CEM(12D-7) T-cell line. At the indicated times postinfection, cells were lysed and viral DNA was amplified by PCR using primers whose positions and sequences are indicated in Materials and Methods. The amplified DNA was then electrophoresed on agarose gels and subjected to Southern blotting using HIV-1-specific probes. Nonpseudotyped virus served as a negative control (pNL4-3KFS). As a positive control for the PCRs, β-globin sequences were amplified from the same set of lysates. The size of each PCR product is shown on the right; on the left is indicated the type of product amplified with each pair of primers.
20LK virions are defective in ERT activity.
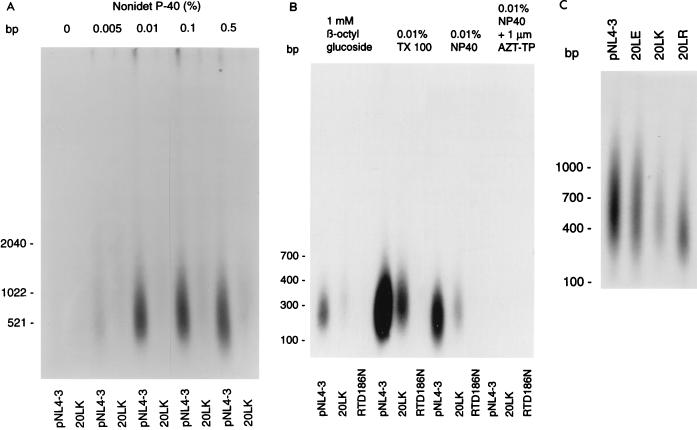
It was recently reported that mutation of Vif, which appears to cause a defect early in the virus life cycle when virus is produced in nonpermissive cells, also causes a defect in ERT activity (36). The ERT assay uses the viral genomic RNA as a template for reverse transcription, rather than an exogenous template, which is provided in standard (exogenous) RT assays. Because in ERT assays deoxynucleoside triphosphates must diffuse into the virion core following gentle disruption of the lipid bilayer with detergent, these assays not only measure reverse transcription per se but also provide a tool for probing virion structure and permeability. Under a range of concentrations of Nonidet P-40, a significant defect in 20LK ERT activity was observed (Fig. 6A). Similar defects were also observed when other detergents (Triton X-100 and β-octylglucoside) were used (Fig. 6B). As anticipated, an RT active-site mutant (RT/D186N [19]) showed no activity in these assays. As an additional negative control, zidovudine (AZT)-triphosphate (1 μM) was found to abolish all ERT activity.
FIG. 6.
ERT activities of amino acid 20 mutants. (A) Virions normalized for exogenous RT activity were permeabilized with different concentrations of Nonidet P-40. Similar results were obtained if virions were normalized for p24 content. (B) Virions were permeabilized with either 1 mM β-octylglucoside, 0.01% Triton X-100 (TX 100), or 0.01% Nonidet P-40 (NP40) as indicated. RT/D186N and the addition of 1 μM AZT-triphosphate (AZT-TP) were included as negative controls. (C) Comparison of 20LE, 20LK, and 20LR ERT activities. Virions were permeabilized with 0.01% Nonidet P-40.
The data presented in Fig. 1 indicate that MA residue 20 mutants display a hierarchy of phenotypes, with the 20LE mutant in general being the least, and 20LK being the most, affected. If the defect observed in ERT activity is biologically meaningful, one would predict that a parallel hierarchy would be observed in ERT assays. To test this prediction, we performed ERT assays using all three residue 20 mutants in parallel. The results (Fig. 6C) indicated that the severity of the biological defect observed in spreading viral infections was reflected in the ERT activity: the 20LE mutant showed small but reproducible reductions in ERT activity, whereas 20LK displayed a markedly more profound defect. The 20LR mutant was typically intermediate between 20LE and 20LK. This hierarchy of phenotypes was observed consistently in several assays.
20LK virions show no detectable structural defects.
As mentioned above, mutation of Vif has been reported to induce defects in both virus entry and ERT activity as well as morphological aberrations evident by electron microscopy (36, 40). In addition, mutations in HIV-1 MA and CA, which display apparent defects early in the virus life cycle, have been reported to show morphological differences relative to wt (58). To ascertain whether 20LK virions were morphologically distinct from wt virions, we performed electron microscopy on HeLa cells transfected with molecular clone pNL4-3 or pNL4-3/20LK. No morphological differences between wt and 20LK mutant virions were observed (data not shown).
We also measured the densities of wt and 20LK mutant virions by sucrose density gradient ultracentrifugation (41). The major peak of both wt and 20LK virion RT activity was observed at a density of approximately 1.16 g/ml, which is within the expected range for retroviral particles (5). This finding was confirmed in assays using metabolically labeled virions which were banded on sucrose gradients and analyzed by radioimmunoprecipitation (data not shown). These results indicate that the 20LK mutation did not affect virion morphology or density as measured by electron microscopy or sucrose gradient analysis.
MA residue 20 mutants display an apparent increase in Gag membrane binding.
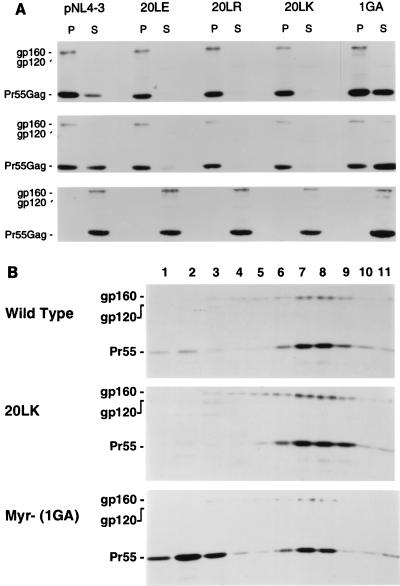
As mentioned previously, amino acid 20 is located within a cluster of basic amino acids which has been implicated in the binding of Gag to membrane. To evaluate a potential effect of MA amino acid 20 mutations on membrane binding, we performed cell fractionation and sucrose density analysis of cell-associated material derived from cells transfected with the residue 20 mutants. Because the very rapid rate of amino acid 20 mutant Gag processing (Fig. 3) would confound the assessment of Pr55Gag membrane binding, we analyzed wt and mutant Gag in the context of a PR active-site mutation (see Materials and Methods). The results of the cell fractionation experiments demonstrated that the amount of Pr55Gag present in the pelletable, membrane-containing fraction is significantly increased by all residue 20 mutations and that the amount detected in the supernatant fraction is reduced (Fig. 7A). As expected, the HIV-1 Env glycoproteins (the Env precursor gp160 and the mature surface glycoprotein gp120) localize primarily in the pellet (membrane) fraction. Both Gag and Env proteins were shifted from pellet to supernatant fractions by the addition of 0.1% SDS. These results were obtained reproducibly in several independent assays and are observed in the absence of NaCl and under high-salt (1 M NaCl) conditions. As a control, we also analyzed the 1GA myristylation mutant (30), which, as demonstrated previously (7), showed a reduced level of Pr55Gag in the pellet fraction and an increase in the supernatant fraction relative to wt.
FIG. 7.
Membrane binding analysis. HeLa cells transfected with a PR− version of pNL4-3, pNL4-3/20LK, or the myristylation mutant pNL4-3/1GA (30) were sonicated, fractionated, and subjected to centrifugation in 20 to 60% sucrose gradients (Materials and Methods). The positions of the Env glycoproteins (gp160 and gp120) and the Gag precursor Pr55Gag (Pr55) are indicated. (A) Cell fractionation analysis. Pellet fractions (P) and supernatant fractions (S) were subjected to SDS-PAGE and blotted with AIDS patient serum. Conditions used: no NaCl (top); 1 M NaCl (middle); no NaCl plus 0.1% SDS (bottom). (B) Sucrose gradient analysis. After centrifugation through 20 to 60% sucrose gradients, fractions were collected from the top, subjected to SDS-PAGE, and blotted with AIDS patient serum. Fraction 1, top of the gradient; fraction 11, bottom of gradient. The soluble proteins are found at the top of the gradient; membrane-bound proteins band primarily in fractions 6 to 9.
To investigate in more detail the effect of the 20LK mutation on membrane binding, we performed sucrose density gradient analysis of cell-associated material derived from pNL4-3/PR−, pNL4-3/PR−/20LK, and pNL4-3/PR−/1GA-transfected HeLa cells (Fig. 7B). Membrane-associated material banded in fractions 6 to 9 (as determined by peak levels of the Env glycoprotein precursor gp160); the soluble material remained near the top of the gradient (primarily in fractions 1 and 2). Western blot analysis of similar gradients with an anti-gp41 antibody indicated that, as expected, the gp41 peak cosediments with Gag in fractions 6 to 9 (data not shown). Consistent with the cell fractionation results (Fig. 7A), the 20LK mutation increased the amount of Pr55Gag in the membrane (6 to 9) relative to cytosolic (1 and 2) fractions. Again, as expected, the 1GA myristylation mutant showed increased soluble and decreased amounts of membrane-bound Gag relative to wt. Treatment of the lysates with detergent (0.5% Triton X-100 or 0.1% SDS) before gradient ultracentrifugation resulted in a nearly complete loss of Pr55Gag and gp41 from the putative membrane fractions (data not shown). These results argue that the amino acid 20 mutations cause an increase in the binding of Gag to membrane.
DISCUSSION
In this report, we describe a set of three single-amino-acid substitution mutations in MA residue 20 which cause significant defects in virus replication by impairing an early step in the life cycle. These mutations impose infectivity defects in single-cycle assays using the CD4+ HeLa (MAGI) cell line and in T-cell lines infected with viruses engineered to express luciferase following infection. The infectivity defects are independent of the type of Env glycoprotein present on mutant virions. Analysis of viral DNA synthesis by PCR at early time points postinfection indicates that the defect is manifested at an early, postentry step. In addition, the residue 20 mutations display impaired ERT activity, and cell fractionation and sucrose density gradient analyses suggest that they increase the binding of Gag to membrane. Since the position 20 mutants release wt levels of virus particles which appear normal in density, protein composition, and morphology yet display reduced infectivity in single cycle assays, we consider the defect imposed by MA residue 20 mutations to be at an early step in replication. However, as discussed below, the defect is probably influenced by events taking place during assembly.
The apparent involvement of MA amino acid 20 in membrane binding is of interest in the context of previous studies. Residue 20 lies in a highly basic domain of MA which is thought to face the lipid bilayer (39) and which has been implicated in the targeting of Gag to the plasma membrane (75, 78). It has been proposed (78) that the basic residues in this cluster interact with the negatively charged acidic phospholipids on the inner face of the plasma membrane, thereby promoting membrane binding. Perhaps the substitution of a charged residue for Leu at position 20 alters the conformation of the basic domain, thereby increasing its affinity for membrane. Interestingly, mutations in an analogous domain of the Rous sarcoma virus MA which affect virus replication without impairing assembly and release have recently been described (57). Thus, the mechanism by which the mutations described here interfere with infectivity may also be operative in other retroviral systems.
We emphasize that although the data presented in Fig. 7 strongly suggest that the residue 20 mutations increase Gag membrane binding, we cannot formally exclude other possible interpretations of these results. For example, similar data might be obtained if these mutations increased Gag multimerization into complexes which pelleted with membrane in the fractionation experiments and banded with membrane in the sucrose gradients. The treatment of samples with detergent prior to sucrose gradient analysis resulted in a shift of both Pr55Gag and the Env glycoproteins (both gp160 and gp41) from the putative membrane fractions, arguing that this material does in fact represent membrane-bound Gag. However, the detergent treatment could also potentially disrupt the putative Gag aggregates, resulting in their shift to the supernatant fraction or to the top of the gradients. It is also possible that the residue 20 mutations increase the association of Gag with the cytoskeleton and that cytoskeleton-bound Gag behaves like membrane-bound Gag in the assays used here. However, it has been reported that high-salt conditions similar to those used in our cell fractionation and gradient experiments disrupt the association of Gag with the cytoskeleton (18). The ill-defined nature of Gag multimerization, aggregation, membrane binding, and virus assembly makes direct evaluation of these issues difficult, in particular since Gag multimerization and membrane binding are likely to be closely linked processes. We also note that in cell fractionation assays (Fig. 7A) all three position 20 mutants showed similar effects on apparent membrane binding, despite their different biological phenotypes. This observation suggests that the membrane binding increase may not be critical to the phenotype of these mutants. Alternatively, differences may be too subtle to be detected in this analysis or may involve p17 rather than Pr55Gag membrane binding affinity. We are currently developing assays to distinguish between membrane-bound and multimeric Gag and to analyze p17 and Pr55Gag membrane binding and Gag multimerization in a kinetic fashion.
It has been proposed that after virus entry, HIV-1 MA plays a role in translocating the viral preintegration complex to the nucleus, thereby enabling HIV-1 to infect nondividing cells. Mutations in the highly basic domain near the N terminus of MA were reported to block or markedly reduce infectivity in nondividing cells, including primary MDM (9, 38, 66). Furthermore, it was reported that phosphorylation of a Tyr residue at the C terminus of MA was required for infectivity in macrophages (31). However, an extensive mutational analysis of the MA basic domain failed to confirm a role for these basic residues in infection of terminally differentiated MDM (24, 25). In addition, mutation of the C-terminal Tyr of MA had no effect on HIV-1 infectivity in MDM (23). Others also reported that a C-terminal Tyr mutant displayed wild-type replication kinetics in macrophages (8), and a recent publication by Fouchier et al. (21) confirmed our earlier finding that MA basic domain mutations do not specifically impair infectivity in MDM. This latter study also used a variety of biochemical techniques to demonstrate that neither intact MA nor the highly basic domain of MA possessed the ability to target heterologous proteins to the nucleus (21). These authors thus concluded that HIV-1 MA does not contain a nuclear localization sequence. The mutations reported here reduce infectivity in both dividing and nondividing cells, and defects are observed in viral DNA synthesis at an early step in reverse transcription prior to nuclear import.
Although what role, if any, HIV-1 MA plays in translocating the viral preintegration complex to the nucleus is unclear, other functions for MA early in the infection process can be envisioned. Whereas the majority of MA appears to associate tightly with the lipid bilayer of the viral envelope, several groups have reported the presence of some MA in both the viral core (32) and the preintegration complex (10, 52). The MA domain of Pr55Gag may therefore bind tightly to membrane during assembly and then, to some extent, dissociate from membrane after cleavage of Pr55Gag by the viral protease. We speculate that the affinity of MA for membrane must be precisely balanced: weakening membrane binding interferes with virus assembly in the producer cell (as is observed for mutations affecting MA myristylation) (7, 30, 37, 56); strengthening membrane binding (as apparently occurs with the residue 20 mutations) may disrupt or destabilize the core or preintegration complex, leading to defects in ERT activity in virions and virus entry in the target cell. It has been observed that Pr55Gag binds membrane more tightly than MA itself (55, 64, 79), suggesting that cleavage of Pr55Gag may trigger conformational changes in MA which reduce its affinity for membrane. It has also been proposed that phosphorylation of MA by kinase(s) present within the virion may induce a partial release of MA from membrane (8, 31, 32). In any case, MA may function to stabilize the viral core and direct appropriate uncoating events, thereby facilitating the synthesis of viral DNA postinfection. In the PCR experiments presented in Fig. 5, we observed that the difference in the amount of viral DNA present in wt- and 20LK-infected cells increased with time. This observation, which has also recently been made with both Vif (62) and NC (4) mutations, suggests that the residue 20 changes may destabilize the viral core or preintegration complex such that viral DNAs are degraded following reverse transcription. Instability of the preintegration complex would also be predicted to decrease the levels of circular (nuclear) DNAs if this complex is degraded before reaching the nucleus. This hypothesis is consistent with the observation that in some assays greater differences are observed in the synthesis of circular versus linear DNAs. A related model would propose that the increase in membrane binding affinity induced by residue 20 mutations causes the formation of a tightly packed shell of MA inside the lipid bilayer of the viral envelope; as a result, appropriate uncoating events do not take place following membrane fusion, and defects in ERT activity are evident in mutant virions.
Although a defect in ERT activity has been observed previously with other HIV-1 mutants (e.g., in Vif [36] and gp41 [76]), the biological ramifications of such a defect are unclear. A low level of reverse transcription has been detected in virions prior to infection (48, 65), and it has been suggested that the synthesis of DNA in virions increases virus infectivity (77). While a defect in virion reverse transcription may directly contribute to the reduced infectivity observed with MA amino acid 20 mutants, it is more likely that the results of these assays indicate the presence of a structural perturbation in the virion core which impairs uncoating steps after virus entry. Thus, regardless of the biological consequences of ERT activity, the assay provides information concerning the permeability and integrity of the viral core and as such represents a useful biochemical tool to probe virion structure. It is noteworthy that the extent of the ERT defect measured with the three residue 20 mutations parallels the defects observed in the ability of the mutants to establish a productive spreading infection; 20LE is the least and 20LK is the most affected.
Efforts are currently under way in our laboratory to further define the role of MA early in the HIV-1 life cycle. The recent identification of a viral revertant of 20LK (43) will assist in this effort.
ACKNOWLEDGMENTS
We thank M. A. Martin for continued enthusiastic support and critical review of the manuscript and R. Willey for comments on the manuscript and many helpful discussions. We acknowledge D. Gabuzda for helpful suggestions regarding the ERT assay, F. Maldarelli for assistance with membrane binding assays, and J. M. Orenstein for performing electron microscopy. We thank A. Engelman for the D186N RT mutant, M. S. McBride and A. Panganiban for the Δ42Δ21 and S1S3 RNA encapsidation mutants, and J. Burns for plasmid pHCMV-G. The following reagents were obtained through the NIH AIDS Research Reference and Reagent Program: pSVAMLVenv (from D. Littman and N. Landau), MAGI cells (from M. Emerman), pNL4-3.Luc.R−E− (from N. Landau), and HIV-1 patient Ig (from A. Prince).
R.E.K. was supported by an Australian Commonwealth AIDS Research Grant fellowship.
REFERENCES
- 1.Adachi A, Gendelman H E, Koenig S, Folks T, Willey R, Rabson A, Martin M A. Production of acquired immunodeficiency syndrome-associated retrovirus in human and non-human cells transfected with an infectious molecular clone. J Virol. 1986;59:284–291. doi: 10.1128/jvi.59.2.284-291.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aiken C. Pseudotyping human immunodeficiency virus type 1 (HIV-1) by the glycoprotein of vesicular stomatitis virus targets entry to an endocytic pathway and suppresses both the requirement for Nef and the sensitivity to cyclosporin A. J Virol. 1997;71:5871–5877. doi: 10.1128/jvi.71.8.5871-5877.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aiken C, Trono D. Nef stimulates human immunodeficiency virus type 1 proviral DNA synthesis. J Virol. 1995;69:5048–5056. doi: 10.1128/jvi.69.8.5048-5056.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Berthoux L, Pechoux C, Ottmann M, Morel G, Darlix J-L. Mutations in the N-terminal domain of human immunodeficiency virus type 1 nucleocapsid protein affect virion core structure and proviral DNA synthesis. J Virol. 1997;71:6973–6981. doi: 10.1128/jvi.71.9.6973-6981.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bolognesi D P, Luftig R, Sharper J H. Localization of RNA tumor virus polypeptides. I. Isolation of further virus substances. Virology. 1973;56:549–564. doi: 10.1016/0042-6822(73)90057-3. [DOI] [PubMed] [Google Scholar]
- 6.Braaten D, Franke E K, Luban J. Cyclophilin A is required for an early step in the life cycle of human immunodeficiency virus type 1 before initiation of reverse transcription. J Virol. 1996;70:3551–3560. doi: 10.1128/jvi.70.6.3551-3560.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bryant M, Ratner L. Myristoylation-dependent replication and assembly of human immunodeficiency virus type 1. Proc Natl Acad Sci USA. 1990;87:523–527. doi: 10.1073/pnas.87.2.523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bukrinskaya A G, Ghropade A, Heinzinger N K, Smithgall T E, Lewis R E, Stevenson M. Phosphorylation-dependent human immunodeficiency virus type 1 infection and nuclear targeting of viral DNA. Proc Natl Acad Sci USA. 1996;93:367–371. doi: 10.1073/pnas.93.1.367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bukrinsky M I, Haggerty S, Dempsey M P, Sharova N, Adzhubei A, Spitz L, Lewis P, Goldfarb D, Emerman M, Stevenson M. A nuclear localization signal within HIV-1 matrix protein that governs infection of non-dividing cells. Nature. 1993;365:666–669. doi: 10.1038/365666a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bukrinsky M I, Sharova N, McDonald T L, Pushkarskaya T, Tarpley W G, Stevenson M. Association of integrase, matrix, and reverse transcriptase antigens of human immunodeficiency virus type 1 with viral nucleic acids following acute infection. Proc Natl Acad Sci USA. 1993;90:6125–6129. doi: 10.1073/pnas.90.13.6125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Casella C R, Raffini L J, Panganiban A T. Pleiotropic mutations in the HIV-1 matrix protein that affect diverse steps in replication. Virology. 1997;228:294–306. doi: 10.1006/viro.1996.8355. [DOI] [PubMed] [Google Scholar]
- 12.Chowers M Y, Pandori M W, Spina C A, Richman D D, Guatelli J C. The growth advantage conferred by HIV-1 nef is determined at the level of viral DNA formation and is independent of CD4 downregulation. Virology. 1995;212:451–457. doi: 10.1006/viro.1995.1502. [DOI] [PubMed] [Google Scholar]
- 13.Connor R I, Chen B K, Choe S, Landau N R. Vpr is required for efficient replication of human immunodeficiency virus type 1 in mononuclear phagocytes. Virology. 1995;206:935–944. doi: 10.1006/viro.1995.1016. [DOI] [PubMed] [Google Scholar]
- 14.Cosson P. Direct interaction between the envelope and matrix proteins of HIV-1. EMBO J. 1996;15:5783–5788. [PMC free article] [PubMed] [Google Scholar]
- 15.Crawford S, Goff S P. Mutations in Gag proteins p12 and p15 of Moloney murine leukemia virus block early stages of infection. J Virol. 1984;49:909–917. doi: 10.1128/jvi.49.3.909-917.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dorfman T, Bukovsky A, Ohagen A, Hoglund S, Gottlinger H G. Functional domains of the capsid protein of human immunodeficiency virus type 1. J Virol. 1994;68:8180–8187. doi: 10.1128/jvi.68.12.8180-8187.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dorfman T, Mammano F, Haseltine W A, Gottlinger H G. Role of the matrix protein in the virion association of the human immunodeficiency virus type 1 envelope glycoprotein. J Virol. 1994;68:1689–1696. doi: 10.1128/jvi.68.3.1689-1696.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Edbauer C A, Naso R B. Cytoskeleton-associated Pr65gag and retrovirus assembly. Virology. 1983;130:415–426. doi: 10.1016/0042-6822(83)90096-x. [DOI] [PubMed] [Google Scholar]
- 19.Engelman A, Englund G, Orenstein J M, Martin M A, Craigie R. Multiple effects of mutations in human immunodeficiency virus type 1 integrase on viral replication. J Virol. 1995;69:2729–2736. doi: 10.1128/jvi.69.5.2729-2736.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Facke M, Janetzko A, Shoeman R L, Krausslich H-G. A large deletion in the matrix domain of the human immunodeficiency virus gag gene redirects virus particle assembly from the plasma membrane to the endoplasmic reticulum. J Virol. 1993;67:4972–4980. doi: 10.1128/jvi.67.8.4972-4980.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fouchier R A M, Meyer B E, Simon J H M, Fischer U, Malim M H. HIV-1 infection of non-dividing cells: evidence that the amino-terminal basic region of the viral matrix protein is important for Gag processing but not for post-entry nuclear import. EMBO J. 1997;16:4531–4539. doi: 10.1093/emboj/16.15.4531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Freed E O, Delwart E L, Buchschacher G L, Jr, Panganiban A T. A mutation in the human immunodeficiency virus type 1 transmembrane glycoprotein gp41 dominantly interferes with fusion and infectivity. Proc Natl Acad Sci USA. 1992;89:70–74. doi: 10.1073/pnas.89.1.70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Freed E O, Englund G, Maldarelli F, Martin M A. Phosphorylation of residue 131 of human immunodeficiency virus type 1 matrix is not required for macrophage infection. Cell. 1997;88:171–173. doi: 10.1016/s0092-8674(00)81836-x. [DOI] [PubMed] [Google Scholar]
- 24.Freed E O, Englund G, Martin M A. Role of the basic domain of human immunodeficiency virus type 1 matrix in macrophage infection. J Virol. 1995;69:3949–3954. doi: 10.1128/jvi.69.6.3949-3954.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Freed E O, Martin M A. HIV-1 infection of non-dividing cells. Nature. 1994;369:107–108. doi: 10.1038/369107b0. [DOI] [PubMed] [Google Scholar]
- 26.Freed E O, Martin M A. Evidence for a functional interaction between the V1/V2 and C4 domains of human immunodeficiency virus type 1 envelope glycoprotein gp120. J Virol. 1994;68:2503–2512. doi: 10.1128/jvi.68.4.2503-2512.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Freed E O, Martin M A. Virion incorporation of envelope glycoproteins with long but not short cytoplasmic tails is blocked by specific, single amino acid substitutions in the human immunodeficiency virus type 1 matrix. J Virol. 1995;69:1984–1989. doi: 10.1128/jvi.69.3.1984-1989.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Freed E O, Martin M A. Domains of the human immunodeficiency virus type 1 matrix and gp41 cytoplasmic tail required for envelope incorporation into virions. J Virol. 1996;70:341–351. doi: 10.1128/jvi.70.1.341-351.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Freed E O, Myers D J, Risser R. Mutational analysis of the cleavage sequence of the human immunodeficiency virus type 1 envelope glycoprotein precursor gp160. J Virol. 1989;63:4670–4675. doi: 10.1128/jvi.63.11.4670-4675.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Freed E O, Orenstein J M, Buckler-White A J, Martin M A. Single amino acid changes in the human immunodeficiency virus type 1 matrix protein block virus particle production. J Virol. 1994;68:5311–5320. doi: 10.1128/jvi.68.8.5311-5320.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gallay P, Swingler S, Aiken C, Trono D. HIV-1 infection of nondividing cells: C-terminal tyrosine phosphorylation of the viral matrix protein is a key regulator. Cell. 1995;80:379–388. doi: 10.1016/0092-8674(95)90488-3. [DOI] [PubMed] [Google Scholar]
- 32.Gallay P, Swingler S, Song J, Bushman F, Trono D. HIV nuclear import is governed by the phosphotyrosine-mediated binding of matrix to the core domain of integrase. Cell. 1995;83:569–576. doi: 10.1016/0092-8674(95)90097-7. [DOI] [PubMed] [Google Scholar]
- 33.Gallina A, Mantoan G, Rindi G, Milanesi G. Influence of MA internal sequences, but not of the myristylated N-terminus sequence, on the budding site of HIV-1 Gag protein. Biochem Biophys Res Commun. 1994;204:1031–1038. doi: 10.1006/bbrc.1994.2566. [DOI] [PubMed] [Google Scholar]
- 34.Gelderblom H R. Assembly and morphology of HIV: potential effect of structure on viral function. AIDS. 1991;5:617–638. [PubMed] [Google Scholar]
- 35.Gelderblom H R, Hausmann E H S, Ozel M, Pauli G, Koch M A. Fine structure of human immunodeficiency virus (HIV) and immunolocalization of structural proteins. Virology. 1987;156:171–176. doi: 10.1016/0042-6822(87)90449-1. [DOI] [PubMed] [Google Scholar]
- 36.Goncalves J, Korin Y, Zack J, Gabuzda D. Role of Vif in human immunodeficiency virus type 1 reverse transcription. J Virol. 1996;70:8701–8709. doi: 10.1128/jvi.70.12.8701-8709.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gottlinger H G, Dorfman T, Sodroski J G, Haseltine W A. Role of capsid precursor processing and myristylation in morphogenesis and infectivity of human immunodeficiency virus type 1. Proc Natl Acad Sci USA. 1989;86:5781–5785. doi: 10.1073/pnas.86.15.5781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Heinzinger N K, Bukrinsky M I, Haggerty S A, Ragland A M, Kewalramani V, Lee M-A, Gendelman H E, Ratner L, Stevenson M, Emerman M. The Vpr protein of human immunodeficiency virus type 1 influences nuclear localization of viral nucleic acids in nondividing host cells. Proc Natl Acad Sci USA. 1994;91:7311–7315. doi: 10.1073/pnas.91.15.7311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hill C P, Worthylake D, Bancroft D P, Christensen A M, Sundquist W I. Crystal structures of the trimeric human immunodeficiency virus type 1 matrix protein: implications for membrane association and assembly. Proc Natl Acad Sci USA. 1996;93:3099–3104. doi: 10.1073/pnas.93.7.3099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hoglund S, Ohagen A, Lawrence K, Gabuzda D. Role of vif during packing of the core of HIV-1. Virology. 1994;201:349–355. doi: 10.1006/viro.1994.1300. [DOI] [PubMed] [Google Scholar]
- 41.Huang M, Orenstein J M, Martin M A, Freed E O. p6Gag is required for particle production from full-length human immunodeficiency virus type 1 molecular clones expressing protease. J Virol. 1995;69:6810–6818. doi: 10.1128/jvi.69.11.6810-6818.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hunter E. Macromolecular interactions in the assembly of HIV and other retroviruses. Semin Virol. 1994;5:71–83. [Google Scholar]
- 43.Kiernan, R. E., and E. O. Freed. Unpublished results.
- 44.Kimpton J, Emerman M. Detection of replication-competent and pseudotyped human immunodeficiency virus with a sensitive cell line based on activation of an integrated β-galactosidase gene. J Virol. 1992;66:2232–2239. doi: 10.1128/jvi.66.4.2232-2239.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Landau N R, Page K A, Littman D R. Pseudotyping with human T-cell leukemia virus type 1 broadens the human immunodeficiency virus host range. J Virol. 1991;65:162–169. doi: 10.1128/jvi.65.1.162-169.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lee P P, Linial M L. Efficient particle formation can occur if the matrix domain of human immunodeficiency virus type 1 Gag is substituted by a myristylation signal. J Virol. 1994;68:6644–6654. doi: 10.1128/jvi.68.10.6644-6654.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Loeb D D, Swanstrom R, Everitt L, Manchester M, Stamper S E, Hutchinson C A., III Complete mutagenesis of the HIV-1 protease. Nature. 1989;340:397–400. doi: 10.1038/340397a0. [DOI] [PubMed] [Google Scholar]
- 48.Lori F, di Marzo Veronese F, de Vico A L, Lusso P, Reitz M S, Jr, Gallo R C. Viral DNA carried by human immunodeficiency virus type 1 virions. J Virol. 1992;66:5067–5074. doi: 10.1128/jvi.66.8.5067-5074.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mammano F, Kondo E, Sodroski J, Bukovsky A, Gottlinger H G. Rescue of human immunodeficiency virus type 1 matrix protein mutants by envelope glycoproteins with short cytoplasmic domains. J Virol. 1995;69:3824–3830. doi: 10.1128/jvi.69.6.3824-3830.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Matlin K S, Reggio H, Helenius A, Simons K. Pathway of vesicular stomatitis virus entry leading to infection. J Mol Biol. 1982;156:609–631. doi: 10.1016/0022-2836(82)90269-8. [DOI] [PubMed] [Google Scholar]
- 51.McBride M S, Panganiban A T. The human immunodeficiency virus type 1 encapsidation site is a multipartite RNA element composed of functional hairpin structures. J Virol. 1996;70:2963–2973. doi: 10.1128/jvi.70.5.2963-2973.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Miller M D, Farnet C M, Bushman F D. Human immunodeficiency virus type 1 preintegration complexes: studies of organization and composition. J Virol. 1997;71:5382–5390. doi: 10.1128/jvi.71.7.5382-5390.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Myers G, Hahn B H, Mellors J W, Henderson L E, Korber B, Jeang K-T, McCutchan F E, Pavlakis G N, editors. Human retroviruses and AIDS. A compilation and analysis of nucleic acid and amino acid sequences. Los Alamos, N.Mex: Los Alamos National Laboratory; 1995. [Google Scholar]
- 54.Ono A, Huang M, Freed E O. Characterization of human immunodeficiency virus type 1 matrix revertants: effects on virus assembly, Gag processing, and Env incorporation into virions. J Virol. 1997;71:4409–4418. doi: 10.1128/jvi.71.6.4409-4418.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ono, A., and E. O. Freed. Unpublished results.
- 56.Pal R, Reitz M S, Jr, Tschachler E, Gallo R C, Sarngadharan M G, Veronese F D M. Myristylation of gag proteins of HIV-1 plays an important role in virus assembly. AIDS Res Hum Retroviruses. 1990;6:721–730. doi: 10.1089/aid.1990.6.721. [DOI] [PubMed] [Google Scholar]
- 57.Parent L J, Wilson C B, Resh M D, Wills J W. Evidence for a second function of the MA sequence in the Rous sarcoma virus Gag protein. J Virol. 1996;70:1016–1026. doi: 10.1128/jvi.70.2.1016-1026.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Reicin A S, Ohagen A, Yin L, Hoglund S, Goff S P. The role of Gag in human immunodeficiency virus type 1 virion morphogenesis and early steps of the viral life cycle. J Virol. 1996;70:8645–8652. doi: 10.1128/jvi.70.12.8645-8652.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Saiki R K, Gelfand D H, Stoffel S, Scharf S J, Higuchi R, Horn G T, Mullis K B, Erlich H A. Primer-directed enzymatic amplification of DNA with a thermostable DNA polymerase. Science. 1988;239:487–491. doi: 10.1126/science.2448875. [DOI] [PubMed] [Google Scholar]
- 60.Schultz A M, Henderson L E, Oroszlan S. Fatty acylation of proteins. Annu Rev Cell Biol. 1988;4:611–647. doi: 10.1146/annurev.cb.04.110188.003143. [DOI] [PubMed] [Google Scholar]
- 61.Schwartz O, Marechal V, Danos O, Heard J-M. Human immunodeficiency virus type 1 Nef increases the efficiency of reverse transcription in the infected cell. J Virol. 1995;69:4053–4059. doi: 10.1128/jvi.69.7.4053-4059.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Simon J H M, Malim M H. The human immunodeficiency virus type 1 Vif protein modulates the postpenetration stability of viral nucleoprotein complexes. J Virol. 1996;70:5297–5305. doi: 10.1128/jvi.70.8.5297-5305.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sova P, Volsky D J. Efficiency of viral DNA synthesis during infection of permissive and nonpermissive cells with Vif-negative human immunodeficiency virus type 1. J Virol. 1993;67:6322–6326. doi: 10.1128/jvi.67.10.6322-6326.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Spearman P, Horton R, Ratner L, Kuli-Zade I. Membrane binding of human immunodeficiency virus type 1 matrix protein in vivo supports a conformational myristyl switch mechanism. J Virol. 1997;71:6582–6592. doi: 10.1128/jvi.71.9.6582-6592.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Trono D. Partial reverse transcripts in virions from human immunodeficiency and murine leukemia viruses. J Virol. 1992;66:4893–4900. doi: 10.1128/jvi.66.8.4893-4900.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.von Schwedler U, Kornbluth R S, Trono D. The nuclear localization signal of the matrix protein of human immunodeficiency virus type 1 allows the establishment of infection in macrophages and quiescent T lymphocytes. Proc Natl Acad Sci USA. 1994;91:6992–6996. doi: 10.1073/pnas.91.15.6992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.von Schwedler U, Song J, Aiken C, Trono D. Vif is crucial for human immunodeficiency virus type 1 proviral DNA synthesis in infected cells. J Virol. 1993;67:4945–4955. doi: 10.1128/jvi.67.8.4945-4955.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wang C-T, Barklis E. Assembly, processing, and infectivity of human immunodeficiency virus type 1 Gag mutants. J Virol. 1993;67:4264–4273. doi: 10.1128/jvi.67.7.4264-4273.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wilk T, Pfeiffer T, Bosch V. Retained in vitro infectivity and cytopathogenicity of HIV-1 despite truncation of the C-terminal tail of the env gene product. Virology. 1992;189:167–177. doi: 10.1016/0042-6822(92)90692-i. [DOI] [PubMed] [Google Scholar]
- 70.Willey R L, Bonifacino J S, Potts B J, Martin M A, Klausner R D. Biosynthesis, cleavage and degradation of the human immunodeficiency virus type 1 envelope glycoprotein gp160. Proc Natl Acad Sci USA. 1988;85:9580–9584. doi: 10.1073/pnas.85.24.9580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wills J W, Craven R C. Form, function, and use of retroviral Gag proteins. AIDS. 1991;5:639–654. doi: 10.1097/00002030-199106000-00002. [DOI] [PubMed] [Google Scholar]
- 72.Yee J-K, Miyanohara A, LaPorte P, Bouic K, Burns J C, Friedmann T. A general method for the generation of high-titer, pantropic retroviral vectors: highly efficient infection of primary hepatocytes. Proc Natl Acad Sci USA. 1994;91:9564–9568. doi: 10.1073/pnas.91.20.9564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Yu X, Yu Q-C, Lee T-H, Essex M. The C terminus of human immunodeficiency virus type 1 matrix protein is involved in early steps of the virus life cycle. J Virol. 1992;66:5667–5670. doi: 10.1128/jvi.66.9.5667-5670.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Yu X, Yuan X, Matsuda Z, Lee T-H, Essex M. The matrix protein of human immunodeficiency virus type 1 is required for incorporation of viral envelope protein into mature virions. J Virol. 1992;66:4966–4971. doi: 10.1128/jvi.66.8.4966-4971.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Yuan Y, Yu X, Lee T-H, Essex M. Mutations in the N-terminal region of human immunodeficiency virus type 1 matrix protein block intracellular transport of the Gag precursor. J Virol. 1993;67:6387–6394. doi: 10.1128/jvi.67.11.6387-6394.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Zhang H, Dornadula G, Alur P, Laughlin M A, Pomerantz R J. Amphipathic domains in the C terminus of the transmembrane protein (gp41) permeabilize HIV-1 virions: a molecular mechanism underlying natural endogenous reverse transcription. Proc Natl Acad Sci USA. 1996;93:12519–12524. doi: 10.1073/pnas.93.22.12519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Zhang H, Dornadula G, Pomerantz R J. Endogenous reverse transcription of human immunodeficiency virus type 1 in physiological microenvironments: an important stage for viral infection of nondividing cells. J Virol. 1996;70:2809–2824. doi: 10.1128/jvi.70.5.2809-2824.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Zhou W, Parent L J, Wills J W, Resh M D. Identification of a membrane-binding domain within the amino-terminal region of human immunodeficiency virus type 1 Gag protein which interacts with acidic phospholipids. J Virol. 1994;68:2556–2569. doi: 10.1128/jvi.68.4.2556-2569.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Zhou W, Resh M D. Differential membrane binding of the human immunodeficiency virus type 1 matrix protein. J Virol. 1996;70:8540–8548. doi: 10.1128/jvi.70.12.8540-8548.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]