Summary
Pathological consequences of circadian misalignment, such as shift work, show considerable individual differences, but the lack of mechanistic understanding hinders precision prevention to prevent and mitigate disease symptoms. Here, we employed an integrative approach involving physiological, transcriptional, and histological phenotypes to examine inter-individual differences in pre-symptomatic pathological progression, preceding irreversible disease onset, in wild-type mice exposed to chronic jet-lag (CJL). We observed that CJL markedly increased the prevalence of hepatic steatosis with pronounced inter-individual differences. Stratification of individual mice based on CJL-induced hepatic transcriptomic signature, validated by histopathological analysis, pinpoints dysregulation of lipid metabolism. Moreover, the period and power of intrinsic behavioral rhythms were found to significantly correlate with CJL-induced gene signatures. Together, our results suggest circadian rhythm robustness of the animals contributes to inter-individual variations in pathogenesis of circadian misalignment-induced diseases and raise the possibility that these physiological indicators may be available for predictive hallmarks of circadian rhythm disorders.
Subject areas: Physiology, Molecular biology, Neuroscience, Omics, Transcriptomics
Graphical abstract
Highlights
-
•
CJL increases NAFLD prevalence in wild-type mice with distinct individual differences
-
•
Individual differences in hepatic RNA signatures mirror the pathological severity
-
•
Proneness to fatty liver is related to the robustness of circadian rhythms before CJL
Physiology; Molecular biology; Neuroscience; Omics; Transcriptomics
Introduction
Circadian rhythms are an essential physiological function for maintaining homeostasis in many organisms, including humans.1,2 The circadian clock, which controls the myriad of circadian rhythms, is a cell-autonomous timekeeping mechanism that exists in most cells throughout the body. The suprachiasmatic nucleus (SCN), the central clock, synchronizes various biological processes from the cellular to the organismal level with the environmental cycle associated with the Earth’s rotation.1,2 Circadian disruption often results in pathological alterations which can increase the risks of various diseases such as diabetes, metabolic syndrome, cardiovascular diseases, menstrual irregularities, infertility, mood disorders, and certain types of cancers.3 Several epidemiological studies have revealed that night shift work is associated with higher risks of these age- and lifestyle-related diseases including nonalcoholic fatty liver disease (NAFLD).4,5,6
With the global prevalence of the 24/7 lifestyle, shift work has become indispensable for the maintenance of public services and economic activity. It has been reported that approximately 10%–30% of the working population in countries around the world is engaged in shift work.7 Therefore, there is a pressing need to develop mitigating measures against circadian rhythm disorders. However, despite the accumulation of numerous epidemiological studies, valid preventive or interventional strategies targeting circadian rhythm disorders have not yet been established. An inherent challenge is that shift work schedules can elicit negative health consequences to different degrees among individuals, even those on the same schedule. Such inter-individual variations in tolerance to shift work are a major obstacle in developing effective measures for circadian rhythm disorders.7,8,9,10
In accordance, studies in laboratory animals further demonstrate adverse pathophysiological consequences due to genetic or environmental perturbations to the circadian clock.11,12,13,14 Recent studies revealed that wild-type C57BL/6J mice suffered a significantly abbreviated lifespan with nonalcoholic steatohepatitis (NASH)-like chronic inflammation and hepatocellular carcinoma in the liver when subjected to a long-term CJL paradigm.12,14 In contrast to the growing understanding of the adverse events occurring at the severe disease stage, the pre-symptomatic state induced by the circadian misalignment remains poorly characterized. Particularly, little is known regarding the pathophysiological mechanisms of inter-individual differences, which constitute a significant gap in knowledge required to develop preventive and precision approaches at the reversible stage.
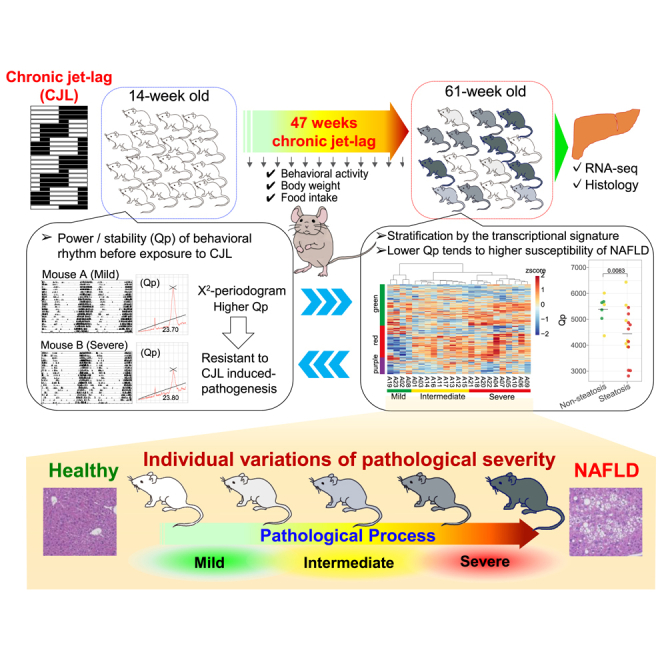
In the current study, we subjected wild-type male C57BL/6J mice to CJL characterized by 8-h advance every 4 days (referred to as ADV paradigm) for 47 weeks and investigated pathological alterations at the pre-symptomatic stage and underlying mechanisms for inter-individual differences.
We observed higher prevalence of hepatic steatosis in the ADV mice compared to the control mice under normal light-dark cycles (LD), and importantly highly diverged inter-individual variations in pathological severity. Comprehensive transcriptomic analysis of livers identified transcriptional signatures including lipid/fatty acid metabolism that are highly associated with the circadian light-dark cycles. The transcriptional signatures and histological analyses led to stratification of CJL mice to mild, intermediate, and severe groups. The free-running period length, known to be controlled by the SCN, of the mild mice was significantly shorter than that of the severe mice. Moreover, the power of the chi-square periodogram (Qp) of behavioral rhythms of the mild mice was significantly higher than that of the severe mice. Together, our integrative study illustrates that environmental adaptation involving SCN contributes to the inter-individual variations in circadian misalignment-induced pathophysiology.
Results
Mouse physiology altered by CJL
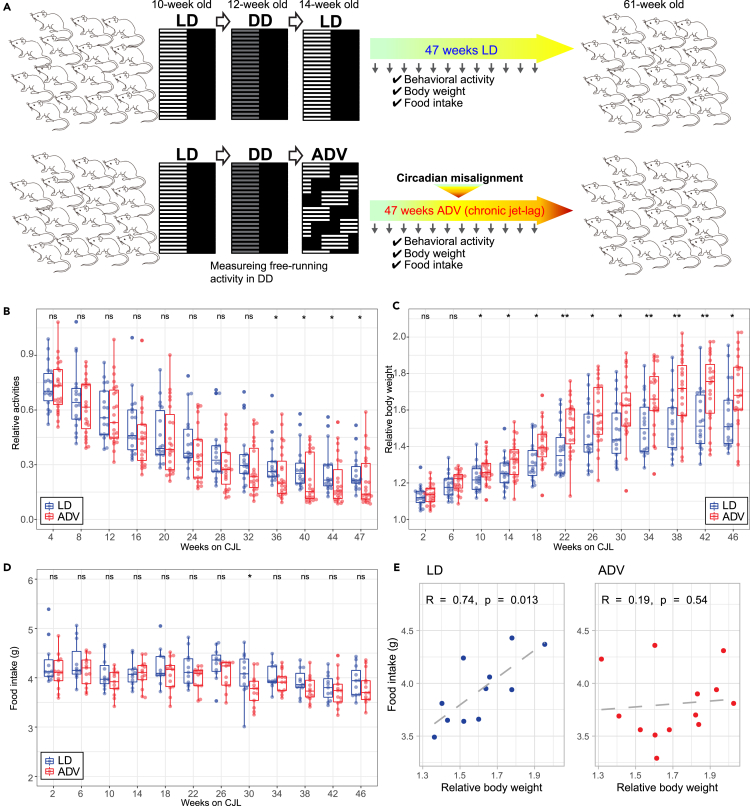
In this study, we exposed wild-type C57BL/6J male mice to the CJL paradigm as previously reported.14 Briefly, the experimental ADV mice underwent 8-h advance every 4 days over 47 weeks, whereas the control LD group was maintained under the normal 12 h:12 h light-dark condition with the light period starting at 8 a.m. local time (Figure 1A). Before initiating the CJL treatment, mice were entrained to the 12 h:12 h LD cycle for 2 weeks and then housed in constant darkness (DD) for 2 weeks to measure baseline free-running activities. No statistical difference was observed in circadian free-running period and the maximum power of the chi-square periodogram between the ADV and LD groups before the CJL (Figure S1A). The mean wheel-running activities in individual mice declined during aging (Figure S1B), consistent with previous reports.15 The ADV-conditioned mice showed significantly reduced activities compared with LD control mice after 36 weeks on CJL (Figure 1B), suggesting that the ADV condition accelerates aging-related reduction in behavioral activity. The ADV mice also gained considerably more weight than LD control mice after 10 weeks on CJL (Figure 1C), without increase in food intake compared to LD (Figure 1D). There was a significant association between food intake and body weight gain only in LD mice but not in ADV (Figure 1E). These results suggest that CJL markedly alters mouse physiology.
Figure 1.
Long-term circadian misalignment accelerates age-related phenotypes in mice
(A) Schematic illustration of the experimental design.
(B and C) Beeswarm and boxplots showing relative wheel-running activities (B) and relative body weight (C) in the LD (n = 18) and ADV mice (n = 23). The relative wheel-running activities were calculated as mean every 4 weeks during CJL and shown as the relative to mean value during the 3-week period before starting CJL. The body weights shown are relative values compared to 4 weeks before starting CJL.
(D) The mean daily food intake of mice in 2nd experiment.
(E) Scatterplots showing Spearman correlation between relative body weight and food intake in LD and ADV mice. For all boxplots in this paper: horizontal line, median; box, interquartile range (IQR); whiskers, most extreme value within ±1.5 × IQR. Two-sided Wilcoxon rank-sum test, ∗p < 0.05, ∗∗p < 0.01, ns: not significant.
Inter-individual variations in pathological consequences caused by CJL
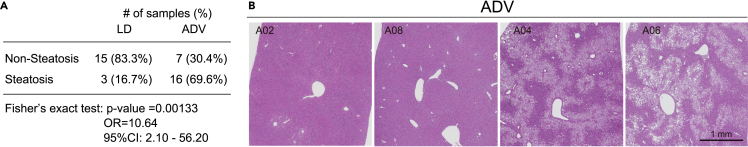
We next examined histopathological changes in the liver, a key target organ of circadian clock. H&E staining revealed that the 69.6% of ADV mice (16/23) were diagnosed with fatty liver without apparent fibrosis, compared with 16.7% of LD mice (3/18) (Figure 2A). Oil red O staining showed a severe accumulation of lipid droplets in the ADV mice (Figures S2A–S2C). The Fisher’s exact test revealed a significantly higher prevalence of steatosis in the ADV mice compared to LD (Figures 2A and S2D), consistent with previous results.12 Notably, there are pronounced individual differences in liver pathology among the ADV mice, where the livers in a subset of mice appeared relatively normal (Figure 2B). These results suggest that our CJL mouse cohort model exhibits not only pathophysiological alterations induced by circadian misalignment but also inter-individual variations as seen in human circadian rhythm disorders.
Figure 2.
Elevated likelihood of steatosis in the ADV mice
(A) The number of animals showed hepatic steatosis in LD and ADV. p value, odds ratio (OR), and 95% confidence interval (CI) for the OR are indicated from Fisher’s exact test.
(B) Representative photomicrographs of H&E-stained liver in ADV showing no and apparent fat depositions.
Transcriptional signatures associated with liver steatosis caused by CJL
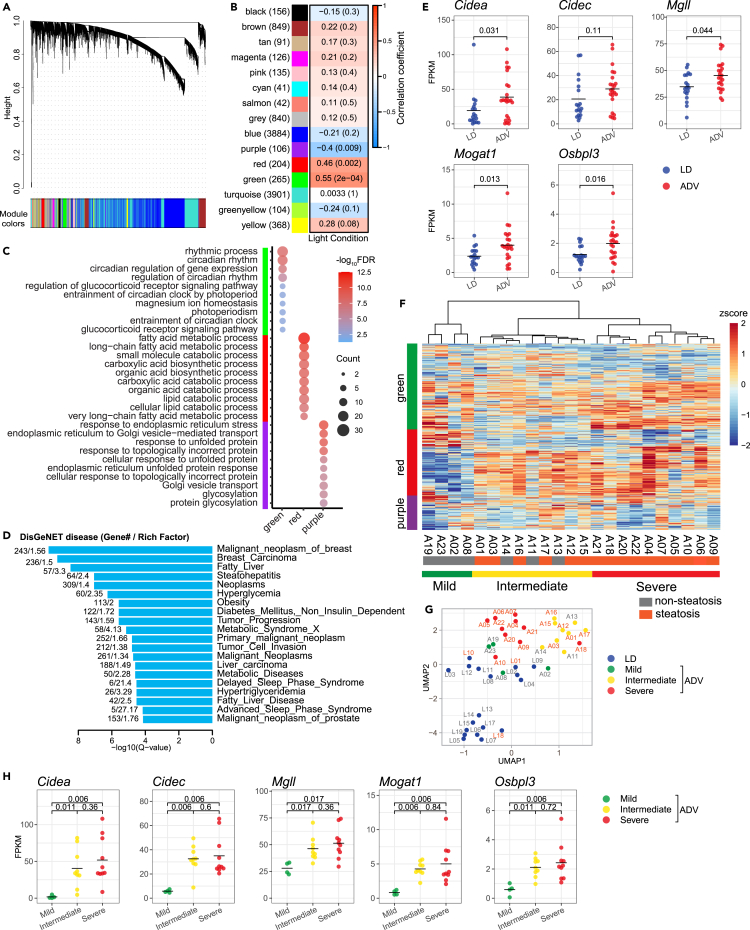
We next investigated pathophysiological mechanisms in the liver that may contribute to varying responses to circadian misalignment. To identify CJL-induced co-expression of functionally related gene modules, we conducted weighted gene co-expression network analysis (WGCNA).16 Specifically, we analyzed hepatic gene co-expression patterns by RNA sequencing (RNA-seq) analysis to determine gene network alterations associated with the light cycle conditions (ADV and LD). Out of 11,112 expressed genes analyzed, we generated 15 color-coded modules, three of which (purple, green, and red) were significantly correlated with light cycles (Figures 3A and 3B; Table S1). We performed gene ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis, which revealed specific pathways in these three modules, including circadian rhythm and rhythmic functions, lipid metabolism including fatty acids and carboxylic acid processes, and endoplasmic reticulum functions, respectively, for the green, red, and purple modules (Figure 3C; Table S2). We further examined human pathology associated with these gene networks using DisGeNET.17 These modules are closely correlated with fatty liver-related disease such as fatty liver, steatohepatitis, obesity, non-insulin dependent diabetes mellitus, metabolic syndrome, and metabolic disease in addition to circadian disorders including delayed sleep phase syndrome and advanced sleep phase syndrome (Figure 3D; Table S3). As expected, there was a considerable alteration in the expression of the fatty liver-associated genes (Figure 3E). These genes include Cidea and Cidec lipid droplet-associated genes important in various aspects of lipid metabolism which have been shown promoting hepatic lipid accumulation,18 Mgll encoding monoacylglycerol lipase,19 Mogat1 producing diacylglycerol from monoacylglycerol,20 and Osbpl3 encoding a lipid transfer protein.21 We previously reported that a longer-term (85 weeks) ADV paradigm promotes low-grade chronic inflammation with fibrosis in mouse liver similar to chronic steatohepatitis.14 Notably, the relevant immune pathways were not significantly activated in the liver of 47-week ADV mice compared with LD mice, suggesting that the circadian misalignment first perturbs metabolic homeostasis before the activation of inflammatory pathways (Figure S3).
Figure 3.
Identification of gene networks associated with fatty liver disease in CJL mice
(A) Clustering dendrogram of genes with dissimilarity based on the topological overlap together with the assigned module colors. Hierarchical cluster analysis was conducted to detect co-expression clusters with corresponding color assignments. Each color represents a module in the constructed gene co-expression network by WGCNA. X axis represents gene, and y axis represents the height of the gene tree.
(B) Heatmap of the correlation between module eigengenes and light condition of mice. Each cell contains the correlation coefficient and p value. The color scheme, from blue through white to red, indicates the levels of Pearson correlation, from low to high.
(C) Bubble plot of top 10 enriched GO terms for genes in the green, red, and purple modules. Color gradient ranging from red to blue corresponds to in order of increasing FDR, and the size of bubble indicates the gene count of each GO term.
(D) Top 20 enrichment terms of DisGeNET category in the green, red, and purple modules. Rich Factor indicates the ratio of number of observed genes to expected genes.
(E) Beeswarm plots of gene expression levels related to lipid metabolism. Black line denotes the mean values for each group. p values indicate those obtained by two-sided Wilcoxon rank-sum test.
(F) Heatmap showing the hierarchical clustering of ADV mice based on the gene expressions in the green, red, and purple modules. The clustering was performed using squared Euclidean distance and Ward linkage. The ADV mice are divided in three group (mild, intermediate, severe) as indicated. The mice with or without hepatic steatosis are indicated by gray and orange, respectively.
(G) UMAP plot based on the gene expressions in the green, red, and purple modules. The LD and ADV mice in the three groups are color-coded as indicated. The mice with or without hepatic steatosis are indicated by gray and orange, respectively, in the text labels.
(H) Beeswarm plots of gene expression levels related to lipid metabolism shown in (E). The ADV mice in the three groups are represented by colors as indicated. Black line denotes the mean values for each group. The adjusted p values indicated are based on the comparison of the expression level of mild, intermediate, and severe groups in ADV, by two-sided Wilcoxon rank-sum test corrected by Benjamini-Hochberg multiple testing.
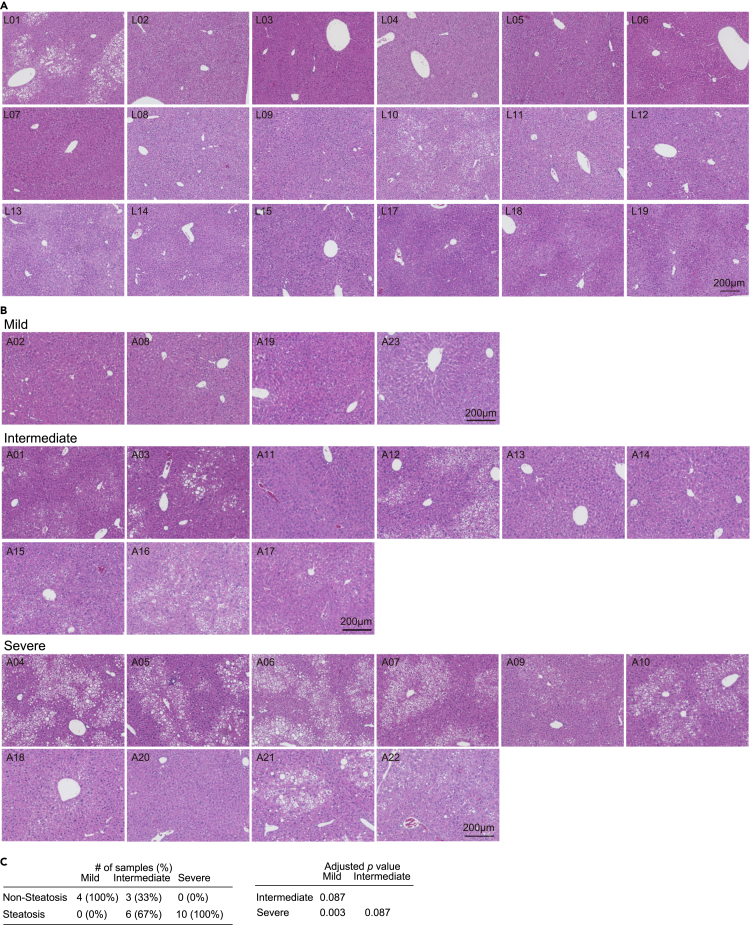
To delineate the pathological mechanisms underlying inter-individual differences, a non-biased clustering analysis using the gene expression patterns of the three modules was performed, and the ADV mice were classified into three predicted groups (mild, intermediate, and severe) based on the transcriptional signature (Figures 3F and 3G), where “severe” mice displayed more marked gene expression changes in the aforementioned fatty liver-associated genes, in contrast to subtle changes in the mild group (Figure 3H). These results underscore prominent roles of circadian rhythms and lipid metabolism during the systemic remodeling in response to CJL. Moreover, our histological analyses showed a significantly higher prevalence of fatty liver (10 out of 10) in the “severe” ADV mice than the mild group (0/4) (Figure 4). Together, these results demonstrated that the transcriptional signature-based stratification of CJL mice mirrors the inter-individual difference of the circadian misalignment-induced liver pathology.
Figure 4.
The transcriptional signature-based stratification of CJL mice mirrors the severity of liver steatosis
(A and B) Representative photomicrographs of H&E-stained livers in LD (A) and ADV (B) mice. The mild, intermediate, and severe groups in ADV are indicated.
(C) The number of ADV mice in the three groups showed hepatic steatosis. The adjusted p values are based on the pairwise comparisons using Fisher’s exact test corrected by Benjamini-Hochberg multiple testing.
Free-running circadian period and robustness related to the transcriptional signature-based stratification
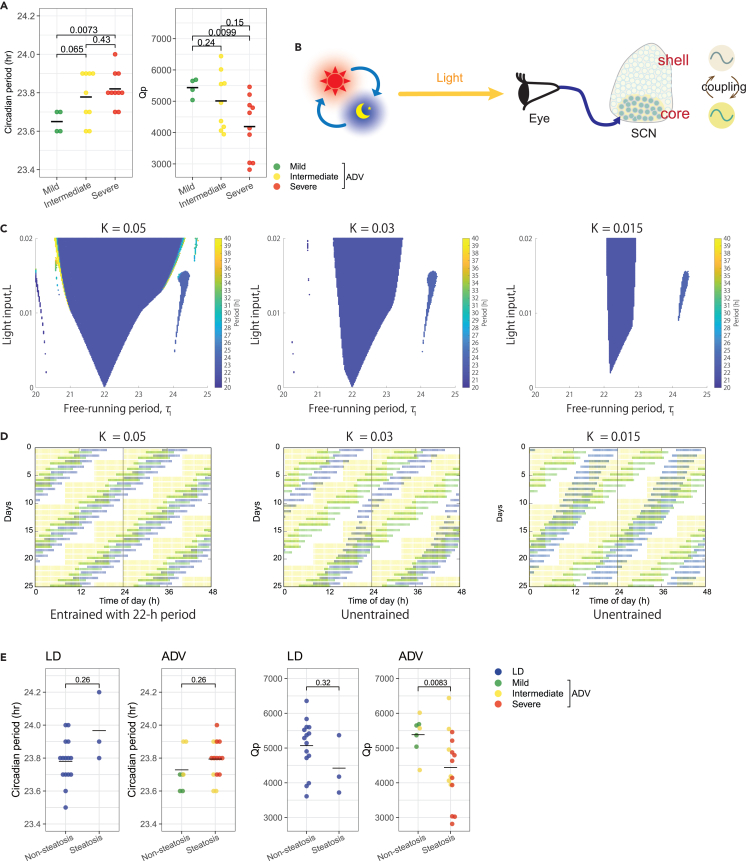
It has been reported that the chronotype of individuals may be a contributing factor for the shift work tolerance.3,7,8,9,10 In addition, a recent study suggests that the magnitudes of phase shifts are correlated with the circadian period in human, where shorter free-running circadian periods were associated with greater phase advance shifts,22 suggesting a connection between adaptation to circadian misalignment and the free-running period. Therefore, we compared the free-running period which was measured before initiating CJL in the stratified ADV mice (Figures 1A and S4) and found that the average free-running period of the mild mice was significantly shorter than that of the severe mice (Figure 5A). Moreover, the Qp, which has been widely used as a measure of the robustness of circadian rhythms,23 was also much higher in the mild mice compared to the severe group (Figure 5A), suggesting that mice having robust circadian rhythms are more resistant to pathophysiology as a result of chronic circadian misalignments.
Figure 5.
Free-running circadian period and Qp related to the severity of CJL-induced pathophysiology
(A) Beeswarm plots of the free-running period and Qp in the mild, intermediate, and severe ADV mice. Black line denotes the mean values for each group. The adjusted p values indicated are derived from two-sided Welch’s t test corrected by Benjamini-Hochberg multiple testing. The color represents the stratified groups (mild, intermediate, and severe).
(B) Mathematical model of the central circadian clock. The core and shell of the SCN are represented by coupled two oscillators. The light signal is input to the core oscillator.
(C) Entrainment property of the mathematical model to the ADV condition for three settings of the coupling strength (K = 0.05, 0.03, 0.015). The entrained period of the circadian system is drawn by varying the free-running period τ1 and the light input L.
(D) Actograms simulated for the three couplings. The free-running periods were set to (τ1, τ2)=(23.5 h, 23 h) for the core and shell oscillators, whereas the light input was fixed to L = 0.015. The yellow bar indicates the ADV schedule, while the green and blue bars represent activities of the core and the shell, respectively.
(E) Beeswarm plots of the free-running period and Qp in the non-steatosis and steatosis mice. Black line denotes the mean values for each group. The adjusted p values indicated are derived from two-sided Welch’s t test corrected by Benjamini-Hochberg multiple testing. The color represents the stratified groups (mild, intermediate, and severe) in addition to LD.
Next, a mathematical model was generated to simulate the effect of the free-running period and the Qp on the entrainment properties of the circadian system to the ADV condition. The central circadian system was modeled as two coupled oscillators, which represent the core and the shell of the SCN (Figure 5B). The system with high Qp was modeled by a strong coupling (K = 0.05), which tightly bonds the core and shell oscillators, whereas those with low Qp by a weak coupling (K = 0.03 or 0.015). Following ADV exposure, the core SCN oscillator received the light signal. By varying the free-running period and the light strength, the temporal windows of entrainment were identified. Strong coupling was associated with a large entrainment window centered at 22 h period (Figure 5C, left). This indicated that circadian rhythms with a 22 h period entrain favorably to ADV, whereas deviation from this free-running period reduces entrainability. In comparison, as the coupling is weakened, the entraining window is drastically narrowed especially for free-running period longer than 23.5 h (Figure 5C, middle) or 23 h (Figure 5C, right). The simulated actograms indicate that ADV elicits differential phase resetting in the core and shell oscillators at different free-running periods (Figure 5D). This implies that the circadian system characterized by a long free-running period and weak internal connection has a low entrainment capability to ADV. Such characteristic originated from the SCN determines the adaptivity of the circadian system to the ADV, which may influence the CJL-induced expression patterns of the liver-associated genes.
Since the free-running behavioral rhythm and its Qp are governed by the SCN,24,25,26 these behavioral signatures may be applied to predict the tolerance to CJL or shift work schedule. However, interestingly, the period and Qp alone are not fully sufficient to predict the development of hepatic steatosis caused by CJL (Figure 5E; Table S4), suggesting that additional factors may be involved in the inter-individual variations of pathophysiology induced by circadian misalignment.
Discussion
Tolerance to the shift work is known to display strong individual difference that is thought to result from varying adaptation to shift work.7,8,9,10 However, the physiological and biological bases of the shift work tolerance are not well characterized. In this study, utilizing an integrated pipeline of physiological, transcriptional, and histological phenotyping in individual wild-type C57BL/6J mice exposed to CJL, we illustrate the inter-individual variations in pathological progression during the pre-symptomatic state prior to transitioning to irreversible disease state such as NASH and hepatocellular carcinoma12,14 during chronic environmental misalignment.
Our histological examination demonstrated significant hepatic steatosis with no apparent fibrosis, suggesting nonalcoholic fatty liver (NAFL), the initial and reversible stage of liver disease, in the ADV mice at 47 weeks. Interestingly, even under the same ADV exposure, some mice showed severe fatty liver, but some appeared normal (see Figure 2), indicating individual differences. Hepatic transcriptomic analysis uncovered transcriptional signatures including genes involved in lipid/fatty acid metabolism associated with circadian misalignments. The stratification based on gene expression signatures revealed pronounced hepatic steatosis in the severely changed group of ADV mice but not in the mild group, supporting that the individual ADV mice respond differently in hepatic gene expression to CJL. Such inter-individual variability in phenotypic responses is a major source of within-group variation which can affect the power of animal experiments and the reproducibility of results.27 The exact reason for inter-individual variations is not well understood. However, it is generally accepted that inter-individual variation is the net result of a complex interaction of genetic and environmental factors.27,28 Our results suggest that, while CJL generally affects mice physiology, the magnitude of this effect varies between individuals. In humans, the naturally occurring characteristics in circadian phase entrainment are known as “chronotype.” Different people may synchronize differently to the same light-dark cycle, due to genetic variance as well as interaction with environmental factors.29
Our observation revealed that the SCN rhythmicity may contribute to the tolerance to CJL. Characteristics of behavioral activity rhythms, free-running period, and Qp of chi-square periodogram in mice before exposing to the CJL significantly associated with the transcriptional signature of the liver (see Figure 5A). Since the robustness of SCN rhythmicity reflects the behavioral activity rhythm in rodents,24,25,26 endogenous period length of the master oscillator and its robustness may correlate with the ability of adaptation to the CJL. Especially, it has been reported that the robustness of SCN rhythmicity is enhanced by the strength of intercellular coupling within the SCN.24 The mathematical model simulating the relationship between the strength of intra-SCN coupling and the ability to entrain to the shifted light condition mimicking ADV condition revealed that the stronger intra-SCN coupling predicted better adaptation to the ADV-like shifted light condition. These results support our hypothesis that the congenital nature of SCN may contribute to the ability of the adaptation to CJL condition. Decline of SCN rhythmicity in aged mice by the weakened intercellular coupling may be one of the reasons for the severe phenotype of aged rodents by exposure to the CJL condition.
The mathematical model of the SCN may also predict the impact of CJL on the liver. Supposing that the liver clocks are coupled to the SCN as peripherals, declined SCN rhythmicity due to aging may weaken the SCN output to the liver. Lowered capability of the SCN to adapt to CJL may further lead to dissociation of the liver clocks from the master clock and could potentially induce desynchronized oscillations among the liver cells. Such circadian desynchrony may induce the impairment of cellular homeostasis and enhance the serious consequences, e.g., hepatic steatosis.
In summary, our analysis of differential responses in individual wild-type mice to environmental perturbation reveals the impact of CJL and changes in circadian behavior as potential contributing factors on the progression of organ pathophysiology and disease. Our study unveils inter-individual variations in pathophysiological progression during the pre-symptomatic disease state which may be exploited in clinical settings for disease prognosis and prevention.
Limitations of the study
Although there are limitations in this study, such as only phase-advanced CJL was performed and we could not investigate the intra-SCN coupling, our findings provide speculation that the endogenous circadian rhythms (period length) and aging (intra-SCN coupling) may affect the tolerance to shift work, at least in part.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Chemicals, peptides, and recombinant proteins | ||
| TRIzol reagent | Thermo Fischer Scientific | Cat#15596026 |
| RNAlater | Thermo Fisher Scientific | Cat#AM7020 |
| Oil Red O solution | MUTO PURE CHEMICALS Co., Ltd., Tokyo, Japan | Cat#40491 |
| Critical commercial assays | ||
| RNeasy Mini Kit | QIAGEN | Cat#74106 |
| Deposited data | ||
| Raw and analyzed data | This paper | GEO: GSE245519 |
| Experimental models: Organisms/strains | ||
| Mouse: C57BL/6J | Japan SLC,Inc. | C57BL/6JJmsSlc |
| Software and algorithms | ||
| Clocklab Analysis 6 (v6.1.12) | ActiMetrics | RRID:SCR_014309 |
| R (programming language) 3.63, 4.2.0, 4.3.1 | R Project | https://www.r-project.org/about.html |
| Trimmomatic (0.39) | Bolger et al.30 | http://www.usadellab.org/cms/?page=trimmomatic |
| STAR aligner (2.7.2) | Dobin et al.31 | https://github.com/alexdobin/STAR |
| WGCNA R package | Langfelder and Horvath16 | https://cran.r-project.org/web/packages/WGCNA/index.html |
| Rethomics R package | Geissmann et al.32 | https://rethomics.github.io/ |
| clusterProfiler | Yu et al.33 | https://bioconductor.org/packages/release/bioc/html/clusterProfiler.html |
| SAMtools 1.7 | Li et al.34 | http://www.htslib.org/ |
| DESeq2 | Love et al.35 | https://bioconductor.org/packages/release/bioc/html/DESeq2.html |
| HOMER (v4.11) | Heinz et al.36 | http://homer.ucsd.edu/homer/ |
| AllEnricher | Zhang et al.37 | https://github.com/zd105/AllEnricher |
| biomaRt | Durinck et al.38 | https://bioconductor.org/packages/release/bioc/html/biomaRt.html |
| exact2x2 R package | Fay et al.39 | https://cran.r-project.org/web/packages/exact2x2/index.html |
| RVAideMemoire R package | Herve40 | https://cran.r-project.org/web/packages/RVAideMemoire/index.html |
| gage R package | Luo et al.41 | https://bioconductor.org/packages/release/bioc/html/gage.html |
| ImageJ 2.1.0/1.53c | NIH | https://imagej.nih.gov/ij/index.html |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Kazuhiro Yagita (kyagita@koto.kpu-m.ac.jp).
Materials availability
This study did not generate new unique reagents.
Data and code availability
-
•
All raw and processed RNA-seq datasets generated in this study have been deposited in the NCBI’s Gene Expression Omnibus (GEO) under accession number GSE245519 and are publicly available as of the date of publication. Microscopy data reported in this paper will be shared by the lead contact upon request.
-
•
This paper does not report any original code.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
Experimental model and subject details
Mice
Male C57BL/6J (10 weeks old) were purchased from SLC (Hamamatsu, Japan). Mice was housed separately in individual cages (170 × 350 × 145 mm) with a 120-mm diameter running wheel (SANKO, Osaka, Japan) in light-shield mouse housing boxes (1800 × 360 × 520 mm) at the room temperature of 25.0 ± 1.5°C with food and water available ad libitum, as described previously.14 The light intensity was set at approximately 200 lx within the cages. The mice were entrained to a 12:12-h light-dark cycle with an 8:00–20:00 light period for 2 weeks and then housed in the constant darkness (DD) for 2 weeks. After DD condition, mice were returned to normal light condition (8:00 to 20:00) and then divided into two groups of light-schedule conditions: LD-condition (light–dark condition with an 8:00–20:00 light period, n = 19) and ADV-condition (8-h phase advance once every 4 days, n = 23). Chronic jet lag (CJL) conditions were designed based on our previous studies.14,42 Body weight of each mouse was measured once every two weeks during these CJL experimental periods. The animal experiments were done twice consecutively with the same protocol starting from 2018 (ADV-condition; n = 10, LD-condition; n = 7) and 2020 (ADV-condition; n = 13, LD-condition; n = 12). One of the mice in the 2nd experiment, L16, was excluded from all analyses, since the wheel-running activities of L16 was drastically reduced in the middle of experiment with unknown reason and therefore we were not able to assess the locomotor activity of L16 accurately. Food intake measured in the 2nd experiment was to give mice a pre-weighted food pellet (CRF-1; Oriental Yeast Co., Ltd, Tokyo, Japan) in their home cage top hopper and weigh the remaining food once every two weeks. Tissue sampling was done from 9:30 to 18:00 in 47 weeks after chronic jet lag condition when ADV mice were in the same light-dark cycle as LD mice with an 8:00–20:00 light period. The blood was collected from mouse heart under deep terminal anesthesia by isoflurane. Then the mice were decapitated and the livers were collected. For RNA-seq analysis the tissues were incubated with RNAlater (Thermo Fisher Scientific) for several hours according to the manufacturer’s instructions and snap-frozen in liquid nitrogen. For histological analysis the tissues were fixed in 10% neutral buffered formalin. All experiments were approved by the Experimental Animals Committee, Kyoto Prefectural University of Medicine, and were performed in accordance with the institutional guidelines and Guidelines for Proper Conduct of Animal Experiments by the Science Council of Japan.
Method details
Behavioral analysis
The behavioral analysis of the mice exposed to the CJL conditions was performed as described in our previous reports.14,42 The wheel-running frequency was measured by counting the number of signals from a magnet sensor (59070-010, Littelfuse Inc., Chicago, IL, USA). Clocklab software (Actimetrics, Wilmette, IL, USA) was used to analyze the behavioral activity in wheel revolutions per 1-min bin. The mean activities every 4 weeks were calculated using the daily activities which calculated in R language with a cut off <0.1 CPM to remove extraordinarily low activity levels, according to visual inspection.43 Chi-square periodogram analysis was performed using Rethomics32 in R language based on an interval of 7 days beginning 7 days after the animals were housed in DD.
RNA-seq
Mouse livers were homogenized in TRIzol reagent (Thermo Fischer Scientific). Total RNA was extracted using RNeasy column (QIAGEN) according to the manufacturer’s instructions. Poly(A)-enriched stranded RNA sequencing was carried out by Macrogen Japan on Illumina NovaSeq 6000 with 101-bp paired-end reads. After adaptor sequences were trimmed using Trimmomatic,30 the sequence reads were mapped to the mouse genome (GRCm38/mm10) using STAR31 as described previously.14,44,45 Parameters used for Trimmomatic and STAR were ‘ILLUMINACLIP:TruSeq3-PE-2.fa:2:30:10 LEADING:20 TRAILING:20 SLIDINGWINDOW:4:15 MINLEN:36’ and ‘--outSAMmultNmax 1 --outSAMtype BAM SortedByCoordinate’, respectively. To obtain reliable alignments, the reads with a mapping quality of less than 10 were removed by SAM tools.34 The University of California, Santa Cruz (UCSC) known canonical gene set (55,421) were used for annotation, and the reads mapped to the exons were quantified using analyzeRNA.pl script in the Homer software36 with an option of –rpkm or -noadj for FPKM or raw read counts, respectively. Since the Homer treat each half of the read separately and count each as 0.5 reads for paired-end reads, the raw read counts were rounded to the nearest integer before transforming rlog using DESeq2.35 To report one isoform per locus (gene symbol), the highest expressed isoform was chosen. We assumed that a gene was expressed if there were more than 20 reads mapped on average in the exons of the gene. The expression level cutoff, average FPKM >0.5, was used for the downstream data analysis.
Weighted co-expression network analysis
The RNA-seq read counts were transformed to rlog using DESeq2 and subjected to the WGCNA46 using the Bioconductor R packages to build an unsigned network. We performed automatic network construction and module detection using the following major parameters: power = 7, TomType = ”unsigned”, minModulesize = 30, reassignThreshold = 0, mergeCutHeight = 0.25. Modules were arbitrarily labeled with different colors. Module eigengene represents the first principal component of a given module and the gene expression. The light conditions (LD and ADV) were converted to numerical data (0 and 1, respectively) and were used for trait data to construct module–trait relationships which were calculated by Pearson correlation between the module eigengene of each module and the light condition. Significance was determined by p < 0.01.
Functional enrichment analysis
Gene ontology (GO) and KEGG pathway enrichment analyses of genes in network modules were carried out using clusterprofiler33 in R language. AllEnricher37 was used for disease enrichment analysis over the diseases in DisGeNET.17 The input list of genes were converted to human genes using R/Bioconductor package biomaRt.38 For enrichment analysis based on KEGG brite functional hierarchy in Functree,47 p values for enrichment test were calculated using gage package41 and the FDR was calculated from the p value for multiple testing with Benjamini-Hochberg procedure.
Mathematical modeling
As a mathematical model for the SCN, two coupled oscillators, which represent the core and the shell, were simulated. Each oscillator is based on the amplitude-phase model48,49,50,51,52,53:
| (Equation 1) |
| (Equation 2) |
where r and φ represent radius and angle (or phase) of the polar coordinate. This single oscillator system has a limit cycle with amplitude A and angular frequency ω (or period τ = 2π/ω). The nearby trajectories converge to the limit cycle will a relaxation rate λ. By representing the single oscillator in the Cartesian (x, y) coordinate and coupling two of such systems, the model becomes
| (Equation 3) |
| (Equation 4) |
The indices i = 1 and i = 2 denote the core and shell oscillators, respectively, K represents strength of their coupling, and . The fourth term of Equation 3 applies only to the core oscillator (i = 1), which receives a light input:
| (Equation 5) |
according to the ADV condition. The angular frequency is determined as ωi = 2π/τi using the free-running period τi. Since the shell region is reported to run faster than the core region in the SCN,54 the period of the shell was set to be 0.5 h shorter than that of the core (i.e., τ2 = τ1-0.5). The other parameter values were set as A = 0.2, λ = 0.2 [1/h]. The coupling strength K was set in such a way that the core and the shell are synchronized with each other under free-running conditions. To describe the level of Qp (i.e., strength of the circadian rhythmicity), three values were set as K = 0.05, 0.03, 0.015. A large coupling simulates the case that the core and the shell are strongly synchronized with each other to give rise to a robust circadian oscillation (i.e., large Qp), whereas a small coupling produces a weak circadian oscillation (i.e., small Qp).
The entrainment property of the circadian system to the ADV condition was evaluated by a phase variability computed as follows. At every periodic point of the ADV schedule (the ADV schedule has a period of T = 264 h), the corresponding phase of the circadian oscillator was recorded as . The phase variability was computed as a standard deviation of the recorded circadian phases { If the circadian oscillator is entrained to the shift cycle, the phase variability should become zero (or close to zero). As another quantity to characterize the entrainment property, averaged oscillation period of the circadian system was computed as .
Histological analysis
Livers were fixed in 10% neutral buffered formalin at room temperature for 2 days. The haematoxylin and eosin (H&E) staining was performed by New Histo. Science Laboratory Co. (Tokyo, Japan) or Genostaff Co., Ltd. (Tokyo, Japan). The images were obtained with a BZ-X710 microscope (KEYENCE, Osaka, Japan). As for Oil Red O staining, livers were fixed in 10% neutral buffered formalin at room temperature overnight and equilibrated in 30% sucrose for overnight at 4°C. The livers embedded in OCT were sectioned at 7 μm with a cryostat (CM-1850, Leica, Germany) and dried. After being immersed in 60% isopropanol for 1 min, the sections were incubated in Oil Red O solution (MUTO PURE CHEMICALS Co., Ltd., Tokyo, Japan) for 15 min at 37°C and then immersed in 60% isopropanol for 2 min and rinsed with water. The stained samples were mounted using Aqua-Poly/Mount (Polysciences, Inc., PA, USA) and the images were acquired with a light microscope (BA210E, SHIMADZU RIKA Co., Tokyo, Japan). Lipid droplets with an area of more than 6 μm2 in each image (0.6 mm2) were counted and summed using ImageJ 2.1.0/1.53c. The case in which lipid droplets are found in 5% or more hepatocytes is defined as steatosis.55 The Fisher’s exact test using minimum likelihood was performed using exact2x2 package39 in R language. The pairwise comparisons using Fisher’s exact test were performed using RVAideMemoire40 package in R.
Quantification and statistical analysis
The statistical analyses were performed by R 3.6.3, 4.2.0 or 4.3.1. The significance was defined as p < 0.05 unless otherwise noted.
Acknowledgments
This work was supported in part by Grants-in-Aid for Scientific Research from the Japan Society for the Promotion of Science (22K18366, 21H02664, 19K22516) and the JST-Mirai (JPMJMI19D7) to K. Yagita, The Welch Foundation (AU-2127-20220331) and NIH (R35GM145232) to S.-H.Y., and NIH (R01AG065984) to Z.C.
Author contributions
Conceptualization, K. Yagita; methodology, N.K., Y.U., H.I., Y.T., Y.S., A.U., K. Yabumoto, T.S., A.S., and K. Yagita; investigation, N.K., Y.U., H.I., I.T., Y.T., Y.S., A.U., and N.M.; visualization, N.K. and Y.U.; funding acquisition, K. Yagita; project administration, N.K. and K. Yagita; supervision, S.-H.Y., C.Z., and K. Yagita; writing – original draft, N.K., Y.U., S.-H.Y., C.Z., and K. Yagita; writing – review and editing, all authors.
Declaration of interests
The authors declare no competing interests.
Published: February 5, 2024
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2024.108934.
Supplemental information
References
- 1.Bass J., Takahashi J.S. Circadian Integration of Metabolism and Energetics. Science. 2010;330:1349–1354. doi: 10.1126/science.1195027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pilorz V., Helfrich-Förster C., Oster H. The role of the circadian clock system in physiology. Pflugers Arch. 2018;470:227–239. doi: 10.1007/s00424-017-2103-y. [DOI] [PubMed] [Google Scholar]
- 3.Boivin D.B., Boudreau P., Kosmadopoulos A. Disturbance of the Circadian System in Shift Work and Its Health Impact. J. Biol. Rhythms. 2022;37:3–28. doi: 10.1177/07487304211064218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shanmugam V., Wafi A., Al-Taweel N., Büsselberg D. Disruption of circadian rhythm increases the risk of cancer, metabolic syndrome and cardiovascular disease. J. Local Global Health Sci. 2013;2013:3. [Google Scholar]
- 5.Huang H., Liu Z., Xie J., Xu C. Association between night shift work and NAFLD: a prospective analysis of 281,280 UK Biobank participants. BMC Publ. Health. 2023;23:1282. doi: 10.1186/s12889-023-16204-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kim K., Lee Y.-J., Kwon S.-C., Min Y.-S., Lee H.K., Baek G., Kim S.H., Jang E.-C. Correlation between shift work and non-alcoholic fatty liver disease among male workers in the steel manufacturing company of Korea: a cross-sectional study. Ann. Occup. Environ. Med. 2022;34:e33. doi: 10.35371/aoem.2022.34.e33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Alfonsi V., Scarpelli S., Gorgoni M., Pazzaglia M., Giannini A.M., De Gennaro L. Sleep-Related Problems in Night Shift Nurses: Towards an Individualized Interventional Practice. Front. Hum. Neurosci. 2021;15 doi: 10.3389/fnhum.2021.644570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Degenfellner J., Schernhammer E. Shift work tolerance. Occup. Med. 2021;71:404–413. doi: 10.1093/occmed/kqab138. [DOI] [PubMed] [Google Scholar]
- 9.Gentry N.W., Ashbrook L.H., Fu Y.-H., Ptáček L.J. Human circadian variations. J. Clin. Invest. 2021;131 doi: 10.1172/JCI148282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Saksvik I.B., Bjorvatn B., Hetland H., Sandal G.M., Pallesen S. Individual differences in tolerance to shift work – A systematic review. Sleep Med. Rev. 2011;15:221–235. doi: 10.1016/j.smrv.2010.07.002. [DOI] [PubMed] [Google Scholar]
- 11.Turek F.W., Joshu C., Kohsaka A., Lin E., Ivanova G., McDearmon E., Laposky A., Losee-Olson S., Easton A., Jensen D.R., et al. Obesity and metabolic syndrome in circadian Clock mutant mice. Science. 2005;308:1043–1045. doi: 10.1126/science.1108750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kettner N.M., Voicu H., Finegold M.J., Coarfa C., Sreekumar A., Putluri N., Katchy C.A., Lee C., Moore D.D., Fu L. Circadian Homeostasis of Liver Metabolism Suppresses Hepatocarcinogenesis. Cancer Cell. 2016;30:909–924. doi: 10.1016/j.ccell.2016.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hill A.M., Crislip G.R., Stowie A., Ellis I., Ramsey A., Castanon-Cervantes O., Gumz M.L., Davidson A.J. Environmental circadian disruption suppresses rhythms in kidney function and accelerates excretion of renal injury markers in urine of male hypertensive rats. Am. J. Physiol. Renal Physiol. 2021;320:F224–F233. doi: 10.1152/ajprenal.00421.2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Inokawa H., Umemura Y., Shimba A., Kawakami E., Koike N., Tsuchiya Y., Ohashi M., Minami Y., Cui G., Asahi T., et al. Chronic circadian misalignment accelerates immune senescence and abbreviates lifespan in mice. Sci. Rep. 2020;10:2569. doi: 10.1038/s41598-020-59541-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Valentinuzzi V.S., Scarbrough K., Takahashi J.S., Turek F.W. Effects of aging on the circadian rhythm of wheel-running activity in C57BL/6 mice. Am. J. Physiol. 1997;273:R1957–R1964. doi: 10.1152/ajpregu.1997.273.6.R1957. [DOI] [PubMed] [Google Scholar]
- 16.Langfelder P., Horvath S. WGCNA: an R package for weighted correlation network analysis. BMC Bioinf. 2008;9:559. doi: 10.1186/1471-2105-9-559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Piñero J., Ramírez-Anguita J.M., Saüch-Pitarch J., Ronzano F., Centeno E., Sanz F., Furlong L.I. The DisGeNET knowledge platform for disease genomics: 2019 update. Nucleic Acids Res. 2020;48:D845–D855. doi: 10.1093/nar/gkz1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Slayton M., Gupta A., Balakrishnan B., Puri V. CIDE Proteins in Human Health and Disease. Cells. 2019;8:238. doi: 10.3390/cells8030238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tardelli M., Bruschi F.V., Claudel T., Fuchs C.D., Auer N., Kunczer V., Stojakovic T., Scharnagl H., Habib A., Grabner G.F., et al. Lack of monoacylglycerol lipase prevents hepatic steatosis by favoring lipid storage in adipose tissue and intestinal malabsorption. J. Lipid Res. 2019;60:1284–1292. doi: 10.1194/jlr.M093369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shi Y., Cheng D. Beyond triglyceride synthesis: the dynamic functional roles of MGAT and DGAT enzymes in energy metabolism. Am. J. Physiol. Endocrinol. Metab. 2009;297:E10–E18. doi: 10.1152/ajpendo.90949.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Du X., Turner N., Yang H. The role of oxysterol-binding protein and its related proteins in cancer. Semin. Cell Dev. Biol. 2018;81:149–153. doi: 10.1016/j.semcdb.2017.07.017. [DOI] [PubMed] [Google Scholar]
- 22.Eastman C.I., Suh C., Tomaka V.A., Crowley S.J. Circadian rhythm phase shifts and endogenous free-running circadian period differ between African-Americans and European-Americans. Sci. Rep. 2015;5:8381. doi: 10.1038/srep08381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Brown L.A., Fisk A.S., Pothecary C.A., Peirson S.N. Telling the Time with a Broken Clock: Quantifying Circadian Disruption in Animal Models. Biology. 2019;8 doi: 10.3390/biology8010018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nakamura T.J., Nakamura W., Tokuda I.T., Ishikawa T., Kudo T., Colwell C.S., Block G.D. Age-Related Changes in the Circadian System Unmasked by Constant Conditions. eneuro. 2015;2 doi: 10.1523/ENEURO.0064-15.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nakamura T.J., Nakamura W., Yamazaki S., Kudo T., Cutler T., Colwell C.S., Block G.D. Age-Related Decline in Circadian Output. J. Neurosci. 2011;31:10201–10205. doi: 10.1523/JNEUROSCI.0451-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nakamura W., Yamazaki S., Nakamura T.J., Shirakawa T., Block G.D., Takumi T. In Vivo Monitoring of Circadian Timing in Freely Moving Mice. Curr. Biol. 2008;18:381–385. doi: 10.1016/j.cub.2008.02.024. [DOI] [PubMed] [Google Scholar]
- 27.Voelkl B., Altman N.S., Forsman A., Forstmeier W., Gurevitch J., Jaric I., Karp N.A., Kas M.J., Schielzeth H., Van de Casteele T., Würbel H. Reproducibility of animal research in light of biological variation. Nat. Rev. Neurosci. 2020;21:384–393. doi: 10.1038/s41583-020-0313-3. [DOI] [PubMed] [Google Scholar]
- 28.Lathe R. The individuality of mice. Genes Brain Behav. 2004;3:317–327. doi: 10.1111/j.1601-183X.2004.00083.x. [DOI] [PubMed] [Google Scholar]
- 29.Roenneberg T., Pilz L.K., Zerbini G., Winnebeck E.C. Chronotype and Social Jetlag: A (Self-) Critical Review. Biology. 2019;8 doi: 10.3390/biology8030054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bolger A.M., Lohse M., Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dobin A., Davis C.A., Schlesinger F., Drenkow J., Zaleski C., Jha S., Batut P., Chaisson M., Gingeras T.R. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Geissmann Q., Garcia Rodriguez L., Beckwith E.J., Gilestro G.F. Rethomics: An R framework to analyse high-throughput behavioural data. PLoS One. 2019;14 doi: 10.1371/journal.pone.0209331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yu G., Wang L.-G., Han Y., He Q.-Y. clusterProfiler: an R Package for Comparing Biological Themes Among Gene Clusters. OMICS A J. Integr. Biol. 2012;16:284–287. doi: 10.1089/omi.2011.0118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N., Marth G., Abecasis G., Durbin R., 1000 Genome Project Data Processing Subgroup The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Love M.I., Huber W., Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Heinz S., Benner C., Spann N., Bertolino E., Lin Y.C., Laslo P., Cheng J.X., Murre C., Singh H., Glass C.K. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol. Cell. 2010;38:576–589. doi: 10.1016/j.molcel.2010.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang D., Hu Q., Liu X., Zou K., Sarkodie E.K., Liu X., Gao F. AllEnricher: a comprehensive gene set function enrichment tool for both model and non-model species. BMC Bioinf. 2020;21:106. doi: 10.1186/s12859-020-3408-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Durinck S., Spellman P.T., Birney E., Huber W. Mapping identifiers for the integration of genomic datasets with the R/Bioconductor package biomaRt. Nat. Protoc. 2009;4:1184–1191. doi: 10.1038/nprot.2009.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fay M.P. Confidence intervals that match Fisher's exact or Blaker's exact tests. Biostatistics. 2010;11:373–374. doi: 10.1093/biostatistics/kxp050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Herve M. RVAideMemoire: Testing and Plotting Procedures for Biostatistics. 2023. https://CRAN.R-project.org/package=RVAideMemoire
- 41.Luo W., Friedman M.S., Shedden K., Hankenson K.D., Woolf P.J. GAGE: generally applicable gene set enrichment for pathway analysis. BMC Bioinformatics. 2009;10:161. doi: 10.1186/1471-2105-10-161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Minami Y., Ohashi M., Hotta E., Hisatomi M., Okada N., Konishi E., Teramukai S., Inokawa H., Yagita K. Chronic inflammation in mice exposed to the long-term un-entrainable light–dark cycles. Sleep Biol. Rhythms. 2018;16:63–68. [Google Scholar]
- 43.Siepka S.M., Takahashi J.S. Methods to record circadian rhythm wheel running activity in mice. Methods Enzymol. 2005;393:230–239. doi: 10.1016/S0076-6879(05)93008-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Umemura Y., Koike N., Ohashi M., Tsuchiya Y., Meng Q.J., Minami Y., Hara M., Hisatomi M., Yagita K. Involvement of posttranscriptional regulation of Clock in the emergence of circadian clock oscillation during mouse development. Proc. Natl. Acad. Sci. USA. 2017;114:E7479–E7488. doi: 10.1073/pnas.1703170114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Umemura Y., Koike N., Tsuchiya Y., Watanabe H., Kondoh G., Kageyama R., Yagita K. Circadian key component CLOCK/BMAL1 interferes with segmentation clock in mouse embryonic organoids. Proc. Natl. Acad. Sci. USA. 2022;119 doi: 10.1073/pnas.2114083119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhang B., Horvath S. A general framework for weighted gene co-expression network analysis. Stat. Appl. Genet. Mol. Biol. 2005;4 doi: 10.2202/1544-6115.1128. [DOI] [PubMed] [Google Scholar]
- 47.Uchiyama T., Irie M., Mori H., Kurokawa K., Yamada T. FuncTree: functional analysis and visualization for large-scale omics data. PLoS One. 2015;10 doi: 10.1371/journal.pone.0126967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Klotter K. Theoretical analysis of some biological models. Cold Spring Harb. Symp. Quant. Biol. 1960;25:189–196. doi: 10.1101/sqb.1960.025.01.017. [DOI] [PubMed] [Google Scholar]
- 49.Wever R. [On the mechanism of biological 24-hour periodicity] Kybernetik. 1962;1:139–154. doi: 10.1007/BF00289033. [DOI] [PubMed] [Google Scholar]
- 50.Pavlidis T. The Free-Run Period of Circadian Rhythms and Phase Response Curves. Am. Nat. 1973;107:524–530. [Google Scholar]
- 51.Daan S., Berde C. Two coupled oscillators: simulations of the circadian pacemaker in mammalian activity rhythms. J. Theor. Biol. 1978;70:297–313. doi: 10.1016/0022-5193(78)90378-8. [DOI] [PubMed] [Google Scholar]
- 52.Winfree A.T. Springer; 1980. The Geometry of Biological Time. [Google Scholar]
- 53.Kronauer R.E., Czeisler C.A., Pilato S.F., Moore-Ede M.C., Weitzman E.D. Mathematical model of the human circadian system with two interacting oscillators. Am. J. Physiol. 1982;242:R3–R17. doi: 10.1152/ajpregu.1982.242.1.R3. [DOI] [PubMed] [Google Scholar]
- 54.Noguchi T., Watanabe K., Ogura A., Yamaoka S. The clock in the dorsal suprachiasmatic nucleus runs faster than that in the ventral. Eur. J. Neurosci. 2004;20:3199–3202. doi: 10.1111/j.1460-9568.2004.03784.x. [DOI] [PubMed] [Google Scholar]
- 55.Chalasani N., Younossi Z., Lavine J.E., Charlton M., Cusi K., Rinella M., Harrison S.A., Brunt E.M., Sanyal A.J. The diagnosis and management of nonalcoholic fatty liver disease: Practice guidance from the American Association for the Study of Liver Diseases. Hepatology. 2018;67:328–357. doi: 10.1002/hep.29367. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
-
•
All raw and processed RNA-seq datasets generated in this study have been deposited in the NCBI’s Gene Expression Omnibus (GEO) under accession number GSE245519 and are publicly available as of the date of publication. Microscopy data reported in this paper will be shared by the lead contact upon request.
-
•
This paper does not report any original code.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.