Abstract
The nonstructural protein NS1 of autonomous parvoviruses is essential for viral DNA amplification and gene expression and is also the major cytopathic effector of these viruses. NS1 acts as nickase, helicase, and ATPase and upregulates P38-driven transcription of the capsid genes. We report here the identification of a novel cellular protein that interacts with NS1 from parvovirus H-1 and which we termed SGT, for small glutamine-rich tetratricopeptide repeat (TPR)-containing protein. The cDNA encoding full-length SGT was isolated through a two-hybrid screen with, as bait, the truncated NS1dlC69 polypeptide, which lacks the C-terminal transactivation domain of NS1. Full-length NS1 and SGT interacted in the two-hybrid system and in an in vitro interaction assay. Northern blot analysis revealed one major transcript of about 2 kb that was present in all rat tissues investigated. Rat sgt cDNA coded for 314 amino acids, and the protein migrated in sodium dodecyl sulfate-polyacrylamide gel electrophoresis with an apparent molecular mass of 34 kDa. SGT could be detected in both the nucleus and the cytoplasm of rat cells, as determined by indirect immunofluorescence analysis and Western blotting of fractionated cellular extracts with an affinity-purified antiserum raised against recombinant SGT (AC1.1). In H-1 virus-infected rat and human cells, compared to mock-infected controls, differences in the migration of SGT polypeptides were revealed after Western blot analysis of total cellular extracts. Moreover, the transient expression of NS proteins was sufficient to induce SGT modification. These results show that cellular SGT, which we have identified as an NS1-interacting protein, is modified by parvovirus infection as well as NS expression.
Autonomous parvoviruses have linear, single-stranded DNA genomes of approximately 5,000 nucleotides in size with palindromic hairpin ends. They have been found associated with tumor material and display oncosuppressive activity (37). The closely related parvoviruses H-1 and MVM express two nonstructural (NS) proteins during their life cycles. NS1 and NS2 have a common N terminus but show a difference in size and subcellular distribution. NS2 is present in different isoforms that have a molecular mass in the range of 25 kDa and are generated through alternative splicing events. NS2 polypeptides are mainly located in the cytoplasm; however, nonphosphorylated NS2 is also found in the nucleus of infected cells (9, 27). The 83-kDa NS1 protein accumulates in the nucleus and is involved in viral DNA replication and modulation of viral and cellular promoters (11, 44). NS1 displays various biochemical activities which are required for viral genome amplification, such as ATP binding and ATPase activity, covalent and noncovalent DNA binding, as well as helicase- and site-specific endonuclease activity (11). NS1 has been shown to form homo-oligomers, and homo-oligomerization is likely to be a prerequisite for NS1-dependent viral DNA replication (34).
NS1 transactivates the P38 promoter, which drives expression of the capsid genes (13, 35, 36). A transcriptional activation domain was mapped to the acidic C terminus of NS1 (26). In a recent study, it was shown that in the context of the whole viral genome, the SP1 and TATA boxes are required for NS1-mediated transactivation of P38. In minimal constructs, however, NS1-mediated transactivation requires the presence of NS1-binding sites within the transactivation response region (28). NS1 also interacts directly with the transcription factor SP1 (23). However, it has not yet been shown that an interaction between NS1 and SP1 is required for P38 transactivation, and thus the mechanism by which NS1 transactivates the P38 promoter is not yet fully understood (29).
NS1 seems to be the major effector of parvovirus-induced cytotoxicity, with NS2 enhancing cell killing in some but not all cell lines tested (4). It has been well documented that parvoviruses preferentially lyse transformed rather than nontransformed cells. Human keratinocytes, for example, are sensitized to H-1 virus infection after their malignant transformation (7). Moreover, cells that were transformed by oncogenic viruses or cellular oncogenes displayed an increased permissiveness for parvoviral replication (14, 38). The mechanisms utilized by NS1 and NS2 to exert their cytopathic effect are still elusive; however, various effects on the cell have been reported which could cause cell death. Cells infected with MVM virus were shown to be arrested in the S phase of the cell cycle, their DNA replication thus being blocked (31). NS1 is also known to modify the expression of cellular genes as well as the phosphorylation of some cellular proteins, one of which is β-tubulin. It may be speculated that these disturbances could also contribute to NS1-dependent cytotoxicity (1, 44).
Due to their restricted coding capacity, parvoviruses heavily depend on cellular helper functions for their life cycles (10). It is expected that some of these helper functions are mediated by direct interaction of cellular factors with the viral genome and/or with the regulatory proteins NS1 and NS2. NS1 interaction with cellular factors is also likely to be involved in NS1-induced cellular perturbances. Virus and host protein interactions are being extensively studied and have, for example, in the case of the adenovirus E1A protein, helped to unravel the mechanisms by which viral proteins influence the cell cycle (21, 46). The widespread use of the two-hybrid system, first described by Fields and Song in 1989 (16), has allowed the identification of interaction partners for many cellular as well as viral proteins. In this study, we aimed to identify cellular partners for NS1 and have applied the two-hybrid system by using NS1 from H-1 virus as a bait. NS-1 harbors an intrinsic transcription-activating region located at the C terminus of the protein (26). We have previously observed that full-length NS1, when expressed as a fusion protein with the DNA binding domain (BD) of GAL4 (GAL4-BD) was sufficient to activate reporter gene expression in the yeast two-hybrid system. A BD fusion protein with a truncated form of NS1, BD-NS1dlC69, in which the 69 C-terminal amino acids of the NS1 protein had been removed, no longer activated transcription in yeast. The BD-NS1dlC69 protein, however, retained the capacity to form oligomers with full-length NS1, indicating that it is stably expressed in yeast and that at least a fraction of it is properly folded (34). By using this truncated NS1 protein as bait, we were able to isolate a novel cellular protein. This interaction partner for NS1 was designated SGT, for small glutamine-rich tetratricopeptide repeat (TPR)-containing protein, according to the features of the protein. Furthermore, we show that this cellular protein is modified upon H-1 virus infection or transient expression of NS proteins.
MATERIALS AND METHODS
Bacterial and yeast strains.
The following bacterial strains were used. Escherichia coli sure (Stratagene) was used for the preparation of the cDNA library, HB101 was used for recovery of the activation domain (AD) plasmids, JM109 was used for subcloning steps, and BL21DE3 (Novagen) was used for expression of recombinant proteins.
The following yeast (Saccharomyces cerevisiae) strains were used: HF7c [mata ura3-52 his3-200 lys2-801 ade2-101 trp1-901 leu2-3,112 gal4-542 gal80-538 LYS2::GAL1-HIS3 URA3::(GAL4 17-mers)3-CYC1-lacZ] and SFY526 (mata ura3-52 his3-200 ade2-101 lys2-801 trp1-901 leu2-3,112 canr gal4-542 gal80-538 URA3::GAL1-lacZ).
Viruses and mammalian cell lines.
Parvovirus H-1 was prepared after infection of NBE (43) cells with a multiplicity of infection (MOI) of 5 PFU per cell and purified by cesium chloride gradient centrifugation, as previously described (6).
The cell lines FR3T3 (41) and FREJ4 (45) were propagated in Dulbecco’s modified Eagle’s medium supplemented with 1 mM sodium pyruvate and 10% donor calf serum. NBE cells were propagated in modified Eagle’s medium supplemented with 5% fetal calf serum; 293T cells (33) were propagated in Dulbecco’s modified Eagle’s medium supplemented with 10% fetal calf serum.
Transient transfection.
293T cells were transfected according to the standard calcium phosphate precipitation protocol (18) with 10 μg of plasmid DNA per 106 cells. Cells were washed after overnight incubation with the precipitate and grown for an additional 36 h prior to extraction.
Plasmids.
All plasmids were constructed and amplified according to standard procedures (39).
Vectors for the two-hybrid system. (i) pGBT9NS1 and pGADNS1.
pGBT9NS1 and pGADNS1 were produced in a two-step cloning. First, two PCR products for the H-1 virus NS1 coding sequence were generated with the following primer pairs: (i) 5′ gcggatcccgATGGCTGGAAACGCTTAC 3′ and 5′ AGTTTTGAATTCTGCCGCG 3′ and (ii) 5′ CGCGGCAGAATTCAAACT 3′ and 5′ gcgtcgacTTAGTCCAAGGTCAGCTC 3′. The initiation codon starting at nucleotide position 1 and the complement of the stop codon starting at position 2014 within the NS1 coding sequence are underlined. The EcoRI restriction site starting at position 823 within the NS1 coding region is given in italics. The sequences given in lowercase contain restriction sites, and the sequences given in uppercase correspond to parts of the NS1 coding region of H-1 virus. Both PCR products were sequentially inserted into a pBluescript SK+ vector (Stratagene) by utilizing the BamHI, EcoRI, and SalI sites. The resulting plasmid, pBlueNS1, was sequenced to ensure that no PCR-derived mutations were present. The NS1 coding region was excised with BamHI and SalI and inserted into the BamHI and SalI sites of the pGBT9 and the pGAD424 vectors (2).
(ii) pGBT9dlC69.
The construction of pGBT9dlC69 was described previously (34).
(iii) pGADNot.
pGADNot was generated for construction of the two-hybrid cDNA library as follows. An adaptor containing a NotI site, generated through annealing of the oligonucleotides 5′ GATCCGCGGCCGCG 3′ and 5′ TCGACGCGGCCGCG 3′, was introduced into the BamHI-SalI-restricted pGAD424 vector.
(iv) pGBT9SGT.
To construct pGBT9SGT, the SGT cDNA-containing insert was excised from the vector pGADNot-SGT through restriction with EcoRI and SalI and inserted into respective sites of the pGBT9 vector.
Vectors for SGT or NS1 protein expression in vitro in bacteria and mammalian cells. (i) pBlueSGT.
pBlueSGT was constructed for in vitro transcription and translation of rat SGT as follows. The SGT cDNA-containing insert was excised from the vector pGADNotSGT through restriction with EcoRI and NotI and religated into the pBluescript SK vector. Construction of pBlueNS1 was described above in the section on pGBT9NS1.
(ii) pET16NNSGT.
pET16NNSGT was constructed for expression of His-tagged rat SGT in bacteria as follows. First, a modified pBluescript SK vector, pBlueHNE, was generated by insertion of an adaptor, consisting of the annealed oligonucleotides 5′ AATTCGCCCATATGA 3′ and 5′ AGCTTCATATGGGCG 3′, into the EcoRI and HindIII sites of pBluescript SK. The EcoRI-NotI-restricted SGT cDNA insert was introduced into the pBlueHNE vector, and then the NdeI-NotI insert was isolated. This fragment was finally inserted into NdeI-NotI-restricted pET16NN, generated through the insertion of an adaptor, consisting of the annealed oligonucleotides 5′ TATGCCCGGGGCGGCCGCG 3′ and 5′ GATCCGCGGCCGCCCCGGGCA 3′, into the NdeI and the BamHI sites of pET16 (Novagene) to give rise to the vector pET16NNSGT.
(iii) pGSTSGT.
pGSTSGT was constructed for expression of a glutathione S-transferase (GST)-SGT fusion protein in bacteria as follows. The SGT cDNA insert was excised from pGADNot-SGT through restriction with XhoI and NotI. This fragment was inserted into the vector pGEX-5X-1 (Pharmacia) to allow the expression of an in-frame fusion between GST and SGT.
(iv) pXFlagNS1 and pXFlagdlC69.
pXFlagNS1 and pXFlagdlC69 were constructed for expression of NS1 and NS1dlC69 in mammalian cell lines as follows. An adaptor, consisting of the annealed oligonucleotides 5′ AATCCATGGCTGACTACAAGGACGACGATGACAAGAAGATCTCCG 3′ and 5′ TCGACGGAGATCTTCTTGTCATCGTCGTCCTTGTAGTCAGCCATG 3′ and coding for the M2 flag, was inserted into the EcoRI and SalI sites of the pX vector (32) to give rise to pXFlag. The NS1 and NS1dlC69 coding regions were inserted into the BamHI- and SalI-restricted pXFlag vector.
Generation of a FREJ4 cDNA plasmid library.
Total RNA was isolated from FREJ4 cells in a one-step protocol with acidic guanidinium thiocyanate, phenol, and chloroform (8) and used for poly(A)+ RNA purification with oligo(dT) coupled to magnetic beads according to the manufacturer’s instructions (Promega). Five micrograms of poly(A)+-RNA was used for cDNA synthesis (Timesaver kit and Directional Cloning Toolbox; Pharmacia). The cDNAs were cloned directionally into the vector pGADNot, which was digested with EcoRI and NotI. Approximately 5 × 106 independent colonies were amplified in E. coli sure (Stratagene). Bacteria were harvested and plasmid DNA was prepared by following a basic alkaline lysis protocol (39).
Two-hybrid screen.
The yeast strain HF7c, containing two reporter genes, HIS3 and lacZ, under the control of two independent promoters, was transformed according to a basic lithium acetate protocol (17) with the bait plasmid pGBT9-dlC69. The resulting strain expressing the bait was selected and grown in tryptophan-deficient (Trp−) synthetic complete medium (17). A 10-liter culture of the bait-expressing strain with an optical density at 600 nm of 0.6 was transformed with 2.5 mg of plasmid DNA from the FREJ4 cDNA library. Double transformants were grown on plates containing synthetic complete medium lacking Trp and Leu to select for the presence of the bait and library plasmids and lacking His to select for protein-protein interactions.
β-Galactosidase assay.
The β-galactosidase filter assays were performed according to a standard protocol (3). For the in vivo assays, fresh transformants were streaked onto selective dishes lacking Trp and Leu and supplemented with a final concentration of 100 mM sodium phosphate (pH 7) and 40 μg of X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside) per ml and grown for 48 h at 30°C.
Sequence analysis.
Sequencing of the SGT cDNA insert from the plasmid pGADSGT was performed on both strands by the dideoxy sequencing method (40) with T7 DNA polymerase, 35S-dATP, and synthetic oligonucleotides as primers. Analysis was done with the HUSAR (Heidelberg Unix Sequence Analysis Resource) package (42).
Purification of bacterially expressed His-tagged SGT, GST, and SGT fused to GST (GST-SGT).
Five hundred-milliliter cultures of the bacterial strain BL21DE3 containing the plasmid pET16NNSGT or of strain JM109 containing either pGEX-5X-1 or pGSTSGT were grown up to an optical density at 600 nm of 0.6. After induction for 5 h with 2 mM IPTG (isopropyl-β-d-thiogalactopyranoside), the cells were harvested and resuspended in NTN buffer (50 mM Tris [pH 7.5], 120 mM NaCl, 0.1% Nonidet P-40) and 0.2% lysozyme and incubated at room temperature until the cells were lysed. The lysates were cooled on ice, sonicated 60 times with short pulses at 30 W, and clear spun for 10 min at 10,000 rpm in an SS40 rotor (Sorvall) at 4°C. The His-tagged SGT-containing supernatant was incubated for 30 min at 4°C in a head-over-head shaker with 500 μl of a 1:2 suspension of Ni2+-agarose (Qiagen) in NTN buffer. The agarose was pelleted and washed four times with 50 ml of NTN buffer. The proteins bound to the Ni2+-agarose were packed in a column and eluted with 150 mM imidazole in NTN buffer. The purity of the His-tagged SGT protein was evaluated in Coomassie-stained polyacrylamide gels and was judged to be above 90%. Essentially the same procedure was applied to purify GST and GST-SGT proteins, except that a 1:1 suspension of glutathione-Sepharose (Pharmacia) was used for protein binding. Elution of the proteins was performed with NTN buffer containing 5 mM glutathione. The protein-containing fractions were then pooled and dialyzed in NTN buffer.
In vitro association.
To express NS1 protein in vitro, 1 μg of the pBlueNS1 plasmid was used in an in vitro transcription and translation system supplemented with [35S]methionine and T7 RNA polymerase, according to the instruction of the manufacturer (Promega). To express SGT, a similar reaction was performed with the plasmid pBlueSGT and T3 RNA polymerase. The 35S-labeled proteins were incubated for 20 min at 4°C with 50 μl of glutathione-Sepharose beads to which purified GST or GST-SGT had been adsorbed. The beads were pelleted in an Eppendorf centrifuge and washed five times with 1 ml of NTN. The supernatant was removed, and the proteins were resuspended in 2× loading dye (24). Samples were analyzed on sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) (10% polyacrylamide) gels, and associated proteins were visualized by autoradiography of the dried gels.
Generation and purification of a rabbit polyclonal antiserum, AC1.1, directed against bacterially expressed His-tagged SGT.
A rabbit was injected with a suspension containing 300 μg of bacterially expressed and purified His-tagged SGT polypeptide and 300 μl of complete Freund’s adjuvant. Five consecutive booster injections were given with 300 μg of His-tagged SGT and 300 μl of incomplete Freund’s adjuvant with a period of 28 days between each booster. Fourteen days after the last injection, the animal was bled.
Cyanogen bromide-activated Sepharose (Pharmacia) was pretreated according to the manufacturer’s instructions and equilibrated in buffer 1 (0.1 M Na-phosphate buffer, 0.5 M NaCl). Twenty milligrams of purified His-tagged SGT in buffer 1 was incubated with the activated Sepharose until 85% of the protein was coupled. The protein coupled to Sepharose was washed with 0.2 M glycine–NaOH (pH 8.0), followed by four washings alternating between buffer 2 (0.1 M Na-acetate [pH 4.0], 50 mM NaCl) and buffer 1. After being washed with 0.2 M glycine–NaOH (pH 2.1), the Sepharose was transferred to a column, and the column was equilibrated with buffer 3 (20 mM Tris [pH 7.5], 0.1 M NaCl, 0.5% Nonidet P-40). Five milliliters of anti-SGT polyclonal serum was applied by gravity flow to the column. After extensive washing with buffer 3, the antibodies were eluted with 0.2 M glycine–NaOH (pH 2.1) and immediately neutralized through the addition of Tris base. This particular antibody preparation is referred to as AC1.1.
Total cell extracts.
A total of 108 cells were harvested, washed with phosphate-buffered saline (PBS), and resuspended in hypotonic buffer (30 mM HEPES–NaOH [pH 7.5], 5 mM KCl, 7 mM MgCl2, 1 mM dithiothreitol) in a final volume of 1.5 ml. NaCl was added to a final concentration of 500 mM, and cells were extracted for 1 h on ice. After a clear spin in an Eppendorf centrifuge at full speed for 10 min, 2× loading dye (24) was added.
Cellular fractionation.
A total of 108 cells were harvested, washed with PBS, resuspended in a hypotonic buffer (20 mM HEPES–KOH [pH 7.5], 7.5 mM MgCl2, 0.1 mM dithiothreitol), run through Dounce homogenization 30 times, and incubated for 20 min on ice. After centrifugation in an Eppendorf centrifuge for 5 min at 10,000 rpm, the supernatant corresponding to the cytoplasmic fraction was removed. The pellet containing the nuclei was resuspended in hypotonic buffer and adjusted to a final NaCl concentration of 500 mM. After extraction for 30 min on ice, the nuclear extract was clear spun.
Immunofluorescence.
Cells were grown on coverslips, fixed in 1% formaldehyde for 10 min, and treated with methanol for 5 min, acetone for 2 min, and 1% saponin for 15 min. After being rinsed in PBS, cells were preincubated with 1% goat serum in PBS, followed by two washing steps, one with 350 mM NaCl, 0.2% Tween 20, and 0.2% Nonidet P-40 in PBS, the other with 2 mM MgCl2 in PBS. Primary and secondary antibodies were successively incubated for 1 h each, followed by three PBS washes. After incubation in DAPI (4′,6-diamidino-2-phenylindole) stain for 1 min, coverslips were mounted onto glass slides in Elvanol (polyvinyl alcohol; Mw, 77,000 to 79,000; ICN). The slides were examined with a ×63 oil immersion objective with a Leica microscope.
Northern blot analysis.
A commercially available blot of poly(A)+ RNAs from various rat tissues (Clontech) was probed with the 1.3-kb SGT cDNA insert or a 2-kb β-actin cDNA fragment, which had been labeled with 32P by random priming (15). After being washed according to the instructions given by the supplier, the filter was exposed to an X-ray film for the indicated time at −80°C.
Western blotting.
Protein extracts from an equivalent number of cells or aliquots of purified or in vitro-translated protein were fractionated by SDS-PAGE with the indicated percentages of polyacrylamide and transferred to a nitrocellulose membrane (Schleicher & Schuell). Membranes were blocked by an incubation for 30 min with 10% low-fat milk powder and 0.2% Tween 20 in PBS at room temperature. Incubation with the AC1.1 antibody preparation was performed with 1:1,000 dilutions. Washes were performed with 0.5% Tween 20 in PBS. The secondary antibody, peroxidase-conjugated goat anti-rabbit immunoglobulin heavy plus light chains (Ig H+L) (Dianova), was used at a 1:10,000 dilution. Detection with enhanced chemiluminescence was performed as recommended by the supplier (Amersham).
RESULTS
Isolation of a cDNA encoding an NS1dlC69-interacting protein.
In search of NS1-interacting cellular proteins, we performed a two-hybrid screen by using a truncated form of NS1, NS1dlC69, fused to the GAL4-BD as bait. We have chosen the H-1 virus-susceptible EJ-ras-transformed FR3T3 cell line, FREJ4 (45), as a source for poly(A)+ RNA, from which we generated a directionally cloned two-hybrid cDNA library in a pGAD424 vector derivative. This library was transformed into yeast, allowing the expression of rat cDNA-encoded proteins fused to the activation domain of GAL4 (GAL4-AD).
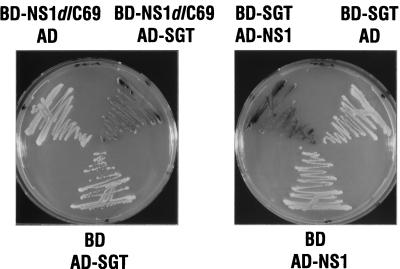
Independent double transformants (5 × 106) were plated on medium lacking leucine and tryptophan to select for the presence of both bait expression plasmids and cDNA library plasmids and were plated on medium lacking histidine to select for interactions between the bait, BD-NS1dlC69, and rat cDNA library-encoded proteins. From approximately 200 His-selectable clones that were visible after 4 days, 30 were also positive when assayed for β-galactosidase activity on filters. From these positive yeast clones, total cellular DNA was isolated with glass beads and phenol and transformed into E. coli HB101 by electroporation. The bacteria were grown on minimal medium lacking leucine to select for transformants containing the library plasmids. pGADNot plasmids containing cDNA inserts were purified and retransformed together with the BD-NS1dlC69-encoding plasmid or the empty pGBT9 vector into (i) the HF7c strain and reassayed for growth on histidine-deficient medium or for β-galactosidase activity and (ii) the SFY526 strain and assayed for β-galactosidase activity. Two independent cDNA clones were positive in both reporter strains in the presence of the bait and negative when only the BD was expressed. Activation of the β-galactosidase reporter in the SFY526 strain is shown for one of these clones in Fig. 1 (left panel). The coexpression of controls in which either BD-NS1dlC69 and AD or BD and AD-SGT were expressed was negative for interaction and therefore yielded white colonies. Taken together, our assays showed that both isolated cDNAs encode proteins interacting with the truncated NS1dlC69 protein, but not with the BD alone. Furthermore, by assaying for the two different reporter genes present in strain HF7c, we showed that activation occurred in a promoter-independent fashion and therefore is most likely mediated by protein-protein interaction.
FIG. 1.
In vivo β-galactosidase assay of SFY526 cells demonstrating the specific interaction between NS1dlC69 and SGT (left panel) and the interaction between full-length NS1 and SGT (right panel). SFY526 cells expressing the indicated protein pairs were streaked onto a selective dish (tryptophan and leucine negative) containing X-Gal and grown for 48 h at 30°C.
The cDNAs isolated encode a novel protein, SGT, that contains three TPRs and a glutamine-rich C terminus.
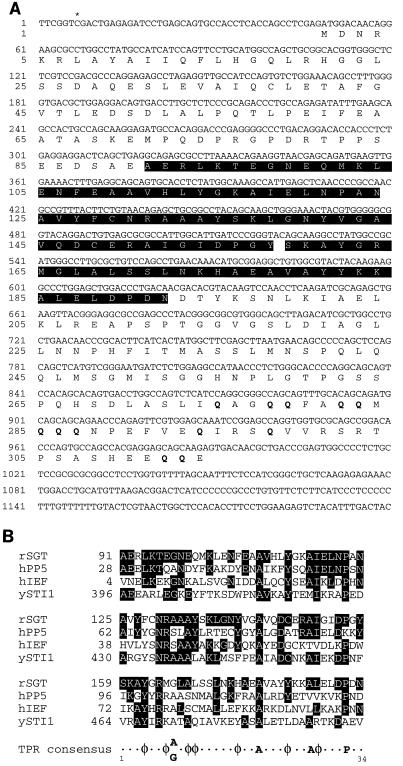
Sequence analysis revealed that the inserts of the two positive clones isolated had the same size (1.3 kb) and contained essentially the same sequence information. The inserts were, however, most probably generated through independent cDNA synthesis events, since one insert was 6 bp (i.e., 2 codons) shorter than the other at the 5′ end (Fig. 2A). We have named the cDNA-encoded protein SGT, for reasons outlined below. DNA sequence comparisons with the Blast N algorithm revealed that the SGT-encoding cDNA sequence was very similar to a previously identified rat EST (GenBank accession no. AA389221); however, no similarity to known genes was found.
FIG. 2.
(A) Nucleotide sequence of the cDNA encoding SGT protein. The deduced amino acid sequence of SGT is given below the nucleotide sequence. The sequence of the longer cDNA is shown, and the asterisk indicates the starting nucleotide of the sequence from the shorter cDNA insert. The three TPRs (aa 91 to 124, 125 to 158, and 159 to 193) are indicated by white letters. The glutamine residues that are clustered in the C terminus of the protein are given in boldface. This sequence has received accession no. AJ222724. (B) Alignment of SGT TPR motifs with equivalent TPR motifs of other proteins, the sequences of which were retrieved from databases: hPP5, human protein serine/threonine phosphatase PP5 (5); hIEF, the human transformation-sensitive protein IEF (20); ySTI1, the S. cerevisiae stress-inducible protein STI1 (30). Residues identical to the ones found in the TPRs of SGT are indicated by white letters. The position of the first amino acid residue of each TPR is given to the left. The TPR consensus given at the bottom was reported by Chen et al. (5). φ, amino acids with large hydrophobic side chains. rSGT, rat SGT.
When the cDNA sequence was translated into protein sequence, we found that it coded for 334 amino acids (Fig. 2A), which were fused in frame to the AD of GAL4. Searches in protein data libraries with the Blast P algorithm revealed that a part of SGT showed high similarity to serine/threonine phosphatases of the PP5/PPT1 family (5), the human transformation-sensitive protein IEF (20), and the S. cerevisiae stress-inducible STI1 protein (30), as well as many other proteins with unrelated functions. A detailed analysis indicated that in this homology region, SGT contains three TPR motifs in tandem array (amino acids [aa] 91 to 124, 125 to 158, and 159 to 193). An alignment of the three SGT TPR motifs with the TPR motifs of the other proteins giving the highest Blast P score is shown in Fig. 2B. Similarities are restricted to this region, which is located in the central part of SGT (Fig. 2A). Furthermore, the C-terminal part of SGT is glutamine rich, with 31% glutamine residues in the last 39 amino acids (aa 275 to 314, 12 glutamines out of 39 residues) (Fig. 2A). Hence, the cDNA product was designated SGT, for small, glutamine-rich, TPR-containing protein. The isoelectric point calculated is 4.9, reflecting the high percentage of negatively charged amino acid residues within this protein. Otherwise, besides putative myristylation, phosphorylation, and glycosylation sites, we have been unable to detect specific signatures in the SGT protein sequence that would have allowed a prediction as to the function of this novel protein.
SGT interacts with full-length NS1 in the two-hybrid system.
To determine whether, as with the NS1dlC69 mutant form, full-length NS1 also interacts with SGT, we exchanged bait and prey and thereby circumvented the intrinsic transactivation activity of full-length NS1 protein. We coexpressed BD-SGT together with AD-NS1 in SFY526 cells. This pair of proteins gave rise to blue colonies on X-Gal-containing plates, whereas the coexpression of BD-SGT and AD or of BD and AD-NS1 did not activate reporter gene expression and hence led to the formation of white colonies (Fig. 1, right panel). These results demonstrate that SGT also interacts with full-length NS1 in the two-hybrid system. They also show that SGT itself does not activate transcription, even when targeted to the promoter of the reporter gene through a fusion with the GAL4-BD.
The cDNA isolated through the two-hybrid screen encodes the full-length SGT protein.
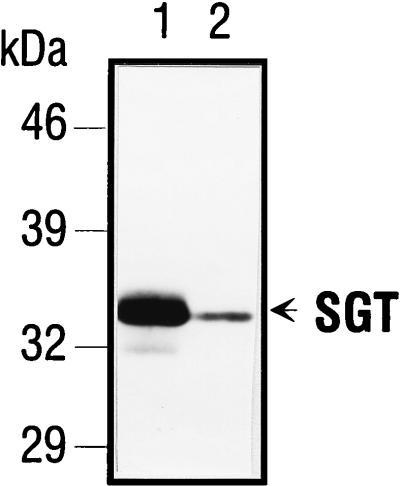
To determine whether or not the isolated cDNA clones encoded the full-length SGT protein, Western blot analyses were performed with an anti-SGT polyclonal serum (AC1.1) that was generated after immunization of a rabbit with purified, bacterially expressed His-tagged SGT. The serum was collected after the fifth booster immunization and subsequently purified via affinity chromatography. AC1.1 recognized both the SGT protein in extracts from FREJ4 cells (Fig. 3, lane 1) and protein generated through in vitro transcription and translation of the longer SGT cDNA clone (Fig. 3, lane 2). Under the separation conditions applied here, only one major SGT polypeptide was detected in FREJ4 cell extracts. This polypeptide comigrated with the in vitro-translated product, indicating that the methionine codon at nucleotide position 48 to 50 (Fig. 2A) after the GAL4-AD fusion site might actually serve as the initiation codon. This ATG is embedded in a Kozak sequence (A/GNNATGG, with ATG given in boldface) that is found at translation start sites of efficiently translated proteins (22). A UGA stop codon was found at position 990 to 992 in the SGT sequence. From these findings, we concluded that rat SGT contains 314 aa. The estimated molecular mass was 34 kDa, which agreed with the apparent molecular mass observed in SDS-PAGE (Fig. 3).
FIG. 3.
SGT cDNA encodes the full-length SGT protein. Total cellular extracts from FREJ4 cells (lane 1) and an SGT in vitro transcription and translation reaction (lane 2) were separated on an SDS–10% polyacrylamide gel and immunoblotted. The blot was incubated with a 1:1,000 dilution of the antirat SGT polyclonal serum AC1.1. SGT-specific polypeptides were detected after incubation with a peroxidase-conjugated goat anti-rabbit Ig H+L secondary antibody followed by an enhanced chemiluminescence reaction.
SGT-NS1 interaction is independent of yeast-specific factors.
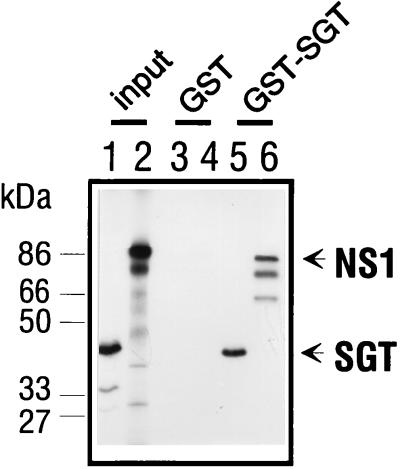
To confirm the specific interaction of NS1 and SGT, we performed an in vitro association experiment, a so-called GST pulldown assay, in which NS1 is specifically bound to a GST-SGT column. For this, a GST-SGT fusion protein was expressed in bacteria and bound to glutathione-Sepharose. Full-length in vitro-transcribed and -translated 35S-labeled NS1 was only retained by glutathione-Sepharose columns to which GST-SGT was bound (Fig. 4, lane 6). The lower-molecular-mass polypeptides likely represent internal NS1 in vitro translation products that also bound to GST-SGT, although we have no proof for this assumption. In reactions in which only the GST portion was present, no NS1 was retained by the column (Fig. 4, lane 4). These results indicate a direct physical interaction of NS1 and SGT not requiring the presence of any yeast-specific factors.
FIG. 4.
In vitro interaction assay. In vitro-transcribed and -translated 35S-labeled SGT (lanes 3 and 5) and NS1 (lanes 5 and 6) were allowed to interact with bacterially expressed GST (lanes 3 and 4) or GST-SGT (lanes 5 and 6) in NTN buffer (50 mM Tris [pH 7.5], 120 mM NaCl, 0.1% Nonidet P-40). The reaction mixtures were adsorbed to glutathione-Sepharose and washed with NTN buffer. The retained proteins were separated by SDS-PAGE in a 10% polyacrylamide gel and visualized through autoradiography. Lanes 1 and 2 contain a sample of the respective SGT and NS1 transcription and translation reaction mixtures. Molecular mass standards are given in kilodaltons.
SGT interacts with itself.
A model for the structure of TPRs has been suggested which predicts inter- and intramolecular interactions of proteins through this motif (19). To test if SGT was also able to form homo-oligomers in vitro, we have performed a GST pulldown assay with the bacterially expressed GST-SGT protein and in vitro-transcribed and -translated 35S-labeled SGT. When the GST-SGT fusion protein was bound to a glutathione-Sepharose column, it was able to retain SGT (Fig. 4, lane 5), whereas no SGT was retained on a glutathione-Sepharose column to which GST alone had been bound (Fig. 4, lane 3). This shows that SGT is indeed able to interact with itself in vitro and form homo-oligomers.
SGT mRNA is present in various rat tissues.
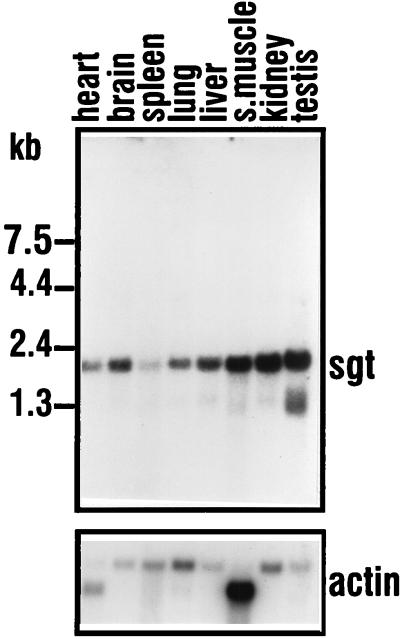
To determine the expression pattern of sgt mRNA, Northern blot analysis with poly(A)+ RNA from various rat tissues was performed. With the 1.3-kb sgt cDNA insert used as a probe, a major transcript of about 2 kb was detected in all tissues investigated (Fig. 5). Upon longer exposure, additional signals from lower-molecular-mass RNA species were detected (data not shown). The steady-state level of sgt mRNA showed only minor variations between the different tissues. When normalized to the β-actin signal, it appeared that testis and kidney displayed an approximately fivefold-increased steady-state level of sgt mRNA compared to lung or spleen. All other tissues showed intermediate mRNA levels. In summary, sgt transcripts are expressed ubiquitously, with no major differences in the amounts of transcript being found in the various rat tissues investigated.
FIG. 5.
Northern analysis of SGT in various rat tissues. A single blot was probed with the 1.3-kb SGT cDNA insert (upper panel) and with a 2-kb β-actin cDNA probe (lower panel). Exposure to an X-ray film was overnight for SGT and 5 h for actin at −80°C. s.muscle, skeletal muscle.
SGT is located in the cytoplasm and the nucleus of rat fibroblasts.
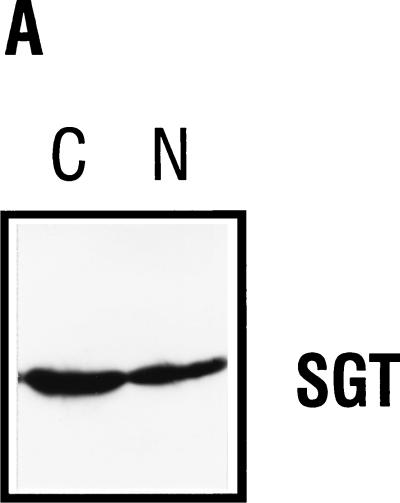
To determine the subcellular distribution of SGT, we have performed cellular fractionation experiments with FREJ4 cells and analyzed the nuclear and cytoplasmic fractions for the presence of SGT in Western blots by using the AC1.1 polyclonal serum. By this approach, SGT was detectable in both the nuclear and the cytoplasmic fractions (Fig. 6A).
FIG. 6.
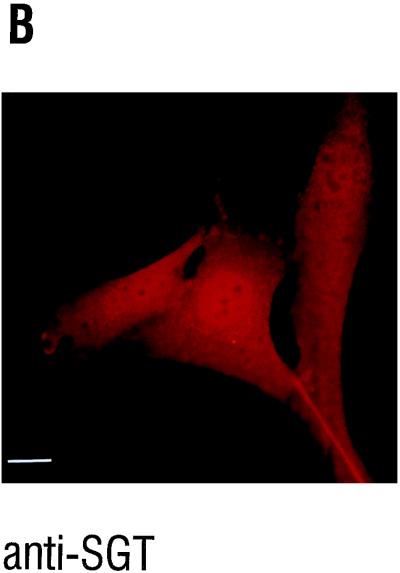
Subcellular localization of SGT. (A) Thirty micrograms of nuclear and cytoplasmic extracts from FREJ4 cells was separated in an SDS–10% polyacrylamide gel and subsequently immunoblotted. The blot was hybridized with a 1:1,000 dilution of the polyclonal serum AC1.1. SGT-specific peptides were detected after incubation with a peroxidase-conjugated goat antirabbit Ig H+L secondary antibody followed by an enhanced chemiluminescence reaction. (B and C) FR3T3 cells were grown on coverslips. Indirect immunofluorescence was conducted with the affinity-purified anti-SGT polyclonal antiserum AC1.1 (B) and DAPI staining of the same section (C). The white bar corresponds to 15 μm.
We also performed immunofluorescence microscopy experiments to determine the subcellular localization by an independent method. In FR3T3 cells, the parental cell line from which the FREJ4 line was established through transformation with an activated ras gene (45), both the nucleus and the cytoplasm were positive for SGT (Fig. 6B). For FREJ4 cells, the same distribution was observed (data not shown). No subcellular structure was preferentially stained. The only nonstained subcellular structures were the nucleoli. From these results, obtained by two independent methods, we conclude that SGT is located both in the cytoplasm and the nucleus of cultured rat cells.
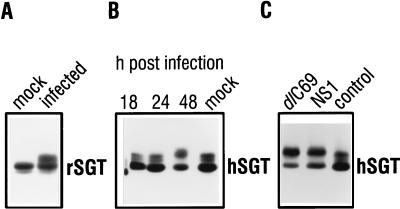
The pattern of SGT migration in SDS-PAGE is altered in H-1 virus-infected cells.
In order to understand the biological relevance of the SGT-NS1 interaction, it was of interest to study the effect parvovirus infection might have on the expression of SGT polypeptides. In Western blot analysis with the AC1.1 anti-rat SGT serum, the most striking difference was a change in the migration patterns of SGT between virus-infected and noninfected FREJ4 rat cells (Fig. 7A). Under the separation conditions applied, two differently migrating SGT polypeptides could be distinguished in extracts from mock-infected cells. In addition, only in extracts from infected cells could a third species with a reduced mobility be detected. Likewise, a shift in SGT migration was observed upon infection of human 293T cells (Fig. 7B). In the latter cells, an affinity-purified anti-SGT antibody raised against the recombinant human polypeptide (12) detected one major and two minor SGT polypeptides with apparent molecular masses of approximately 34 kDa in extracts from mock-infected cells. Forty-eight hours after infection of 293T cells with H-1 virus, the polypeptide which migrated at the middle position was no longer detected, and the amount of the polypeptide having the lowest mobility increased (Fig. 7B). At this time point, the first morphological changes indicating the onset of parvovirus-induced cell death were visible (data not shown). No change in migration patterns of human SGT was detectable 24 h postinfection.
FIG. 7.
SGT is modified after parvovirus infection. (A) Thirty micrograms of total extracts from FREJ4 cells that were either mock infected or infected with H-1 virus at an MOI of 5 PFU per cell and harvested 96 h postinfection were separated in an SDS–10% polyacrylamide gel and subsequently immunoblotted. The blot was incubated with a 1:1,000 dilution of the polyclonal serum AC1.1. SGT-specific peptides were detected after incubation with a peroxidase-conjugated goat antirabbit Ig H+L secondary antibody followed by an enhanced chemiluminescence reaction. (B and C) 293T cells were infected with H-1 virus at an MOI of 5 PFU per cell and harvested after 18, 24, and 48 h as indicated. Mock-infected cells were also harvested after 48 h (B). 293T cells were transfected by a standard calcium phosphate protocol with either the empty pXFlag expression vector (control) or vectors leading to the transient expression of NS1 or the mutant NS1dlC69 protein. Cell extracts were prepared through the addition of 2× concentrated loading dye (24), and an aliquot was separated in an SDS–10% polyacrylamide gel and immunoblotted. The blots were hybridized with a 1:1,000 dilution of the polyclonal EK1.1 serum and treated subsequently as indicated for panel A.
The 293T cell line can be transiently transfected with high efficiency. We have therefore transfected this cell line with the expression vectors pXFlagNS1 and pXFlagdlC69, which encode the full-size NS1 protein and its mutant version, NS1dlC69, respectively. Forty-eight hours posttransfection, the SGT polypeptides extracted from these cells also displayed the same altered migration pattern, as observed after infection (Fig. 7C). Control transfection with the empty expression vector gave rise to a pattern identical to the one observed after mock infection. Since NS2 expression was only detected after infection, but not after transfection of the NS1 and NS1dlC69 expression vectors (data not shown), we conclude that the presence of NS1 or NS1dlC69 is sufficient to bring about modification of SGT polypeptides, although we cannot exclude a contribution by NS2 proteins.
DISCUSSION
NS1-SGT interaction.
The NS1 proteins of autonomous parvoviruses are multifunctional proteins and the major cytopathic effectors of these viruses. The identification of cellular proteins that interact with NS1 is a first step toward improving our understanding of NS1-related cytotoxicity or other functions of this protein. We report here the identification of SGT, a cellular interaction partner for NS1. Interaction between full-length NS1 and SGT was demonstrated in the yeast two-hybrid system and further substantiated in an in vitro interaction assay. The latter observation argues for a direct interaction between both proteins. The subcellular localization of NS1 and SGT in mammalian cells is also consistent with an interaction between these proteins, since NS1 accumulates mostly in the nucleus of infected cells (36) and SGT was located in both the nucleus and the cytoplasm of the cell lines investigated. Even though the SGT-NS1 complex is stable enough to be detected in the two-hybrid system and in vitro, we were yet not able to demonstrate an interaction in vivo by a classical coimmunoprecipitation. There are certainly many possible reasons that could account for this, but since we observed a modification of SGT in the presence of NS1 in vivo, the possibility that this modification might affect SGT-NS1 complex stability also remains to be investigated.
SGT is modified after NS expression.
The effect of NS1 on the synthesis and phosphorylation of cellular proteins has been recently investigated in a cellular system in which NS1 expression was inducible (1). Under the conditions applied, NS1 from parvovirus MVMp was shown to specifically interfere with the synthesis of an as-yet-unidentified protein, p50, and of β-tubulin. NS1 further interfered with the phosphorylation of an also unidentified protein, p14. The effect on p14 took place early after induction of NS1 expression, and the mechanism by which p14 phosphorylation is inhibited is not yet understood. Interestingly, we have observed that SGT from rat and human cells was modified upon infection with H-1 virus. The observed SGT modification does not appear to be the consequence of a general cellular reaction triggered upon viral infection, since SGT was also modified when NS1 or the NS1dlC69 mutant was expressed after transfection of the respective expression vectors. This strongly suggests that the modification is dependent on the presence of NS proteins. A contribution by NS2 cannot be excluded, even though NS2 proteins were undetectable after transfection. We are currently analyzing the exact nature of the SGT modification. Treatment of cellular extracts with alkaline phosphatase prior to Western analysis indicated that the reduced mobility of the additional polypeptide species, found in H-1 virus-infected or NS1-expressing cells, could be due to phosphorylation (12). The protein sequence of SGT harbors several consensus sequences for Ser/Thr phosphorylation as well as one potential Tyr-phosphorylation site. However, additional posttranslational modifications of the SGT polypeptides remain possible. It is certainly of great interest to establish the order of events that leads from parvovirus infection and NS expression to SGT modification and cell death. It is at the moment unknown whether SGT modification is brought about through direct interaction with NS1. It may be speculated that the interaction between SGT and NS1 is altered after SGT modification. It can further be expected that the physical modification of SGT through the presence of NS proteins also results in a functional modification of SGT that might well contribute to the parvovirus life cycle and/or cytotoxicity.
SGT is a novel cellular protein.
The rather ubiquitous expression of sgt mRNA and SGT protein argues for a housekeeping function for SGT. Protein sequence analysis did not reveal any signature that would have allowed a prediction as to the potential function of SGT. SGT is a 34-kDa protein that is rather rich in glutamine residues at its C terminus. In addition, SGT contains three tandemly repeated TPR motifs. These motifs contain a stretch of 34 aa in which only a few positions are preferentially occupied by specific amino acids. They occur mostly clustered in tandem array, but in addition, isolated single or double motifs can be found in some TPR-containing proteins. TPRs have a high probability of forming amphipathic α-helices, and in the case of TPR1 and TPR3 of SGT, have a high probability of α-helix formation according to Chou-Fassman and Garnier-Robson prediction. TPRs are found in proteins from different organisms ranging from bacteria to humans (25). TPR-containing proteins are involved in cellular processes as diverse as cell cycle control, transcriptional repression, stress response, and protein transport, and no common biochemical activity could be attributed to these proteins. They are, however, commonly part of multiprotein complexes. A structural model was proposed based on circular dichroism and model fitting, known as the knob-and-hole model, that predicts inter- as well as intramolecular interactions (19). We have shown that SGT also forms homo-oligomers and are currently mapping the protein regions responsible for self-association and hetero-oligomerization with NS1. It can be expected that the TPRs are important for either or even both of these interactions. The identification of the SGT-NS1 interaction sites will allow the construction of mutants with impaired binding capacity to be used in order to evaluate the significance of SGT-NS1 interaction in the parvovirus life cycle.
ACKNOWLEDGMENTS
We thank the members of the laboratory for helpful discussions, especially N. Salomé and J. Cornelis, who also commented on the manuscript, as well as J. Nüesch and L. Deleu. The advice of P. Legrain (Institute Pasteur, Paris) on the use of the two-hybrid system is gratefully acknowledged. We thank R. Meyer (DKFZ) for assistance with the fluorescence microscope.
This work was supported by the German-Israeli Foundation for Scientific Research and Development.
REFERENCES
- 1.Anouja F, Wattiez R, Mousset S, Caillet-Fauquet P. The cytotoxicity of the parvovirus minute virus of mice nonstructural protein NS1 is related to changes in the synthesis and phosphorylation of cell proteins. J Virol. 1997;71:4671–4678. doi: 10.1128/jvi.71.6.4671-4678.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bartel P, Chien C T, Sternglanz R, Fields S. Elimination of false positives that arise in using the two-hybrid system. BioTechniques. 1993;14:920–924. [PubMed] [Google Scholar]
- 3.Breeden L, Nasmyth K. Regulation of the yeast HO gene. Cold Spring Harbor Symp Quant Biol. 1985;50:643–650. doi: 10.1101/sqb.1985.050.01.078. [DOI] [PubMed] [Google Scholar]
- 4.Caillet-Fauquet P, Perros M, Brandenburger A, Spegelaere P, Rommelaere J. Programmed killing of human cells by means of an inducible clone of parvoviral genes encoding non-structural proteins. EMBO J. 1990;9:2989–2995. doi: 10.1002/j.1460-2075.1990.tb07491.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen M X, McPartlin A E, Brown L, Chen Y H, Barker H M, Cohen P T. A novel human protein serine/threonine phosphatase, which possesses four tetratricopeptide repeat motifs and localizes to the nucleus. EMBO J. 1994;13:4278–4290. doi: 10.1002/j.1460-2075.1994.tb06748.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen Y Q, de Foresta F, Hertoghs J, Avalosse B L, Cornelis J J, Rommelaere J. Selective killing of simian virus 40-transformed human fibroblasts by parvovirus H-1. Cancer Res. 1986;46:3574–3579. [PubMed] [Google Scholar]
- 7.Chen Y Q, Tuynder M C, Cornelis J J, Boukamp P, Fusenig N E, Rommelaere J. Sensitization of human keratinocytes to killing by parvovirus H-1 takes place during their malignant transformation but does not require them to be tumorigenic. Carcinogenesis. 1989;10:163–167. doi: 10.1093/carcin/10.1.163. [DOI] [PubMed] [Google Scholar]
- 8.Chomczynski P, Sacchi N. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem. 1987;162:156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- 9.Cotmore S F, Tattersall P. Alternate splicing in a parvoviral nonstructural gene links a common amino-terminal sequence to downstream domains which confer radically different localization and turnover characteristics. Virology. 1990;177:477–487. doi: 10.1016/0042-6822(90)90512-p. [DOI] [PubMed] [Google Scholar]
- 10.Cotmore S F, Tattersall P. The autonomously replicating parvoviruses of vertebrates. Adv Virus Res. 1987;33:91–174. doi: 10.1016/s0065-3527(08)60317-6. [DOI] [PubMed] [Google Scholar]
- 11.Cotmore S F, Tattersall P. DNA replication in the autonomous parvoviruses. Semin Virol. 1995;6:271–281. [Google Scholar]
- 12.Cziepluch, C., et al. Unpublished data.
- 13.Doerig C, Hirt B, Beard P, Antonietti J P. Minute virus of mice nonstructural protein NS-1 is necessary and sufficient for trans-activation of the viral P39 promoter. J Gen Virol. 1988;69:2563–2573. doi: 10.1099/0022-1317-69-10-2563. [DOI] [PubMed] [Google Scholar]
- 14.Faisst S, Schlehofer J R, zur Hausen H. Transformation of human cells by oncogenic viruses supports permissiveness for parvovirus H-1 propagation. J Virol. 1989;63:2152–2158. doi: 10.1128/jvi.63.5.2152-2158.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Feinberg A P, Vogelstein B. A technique for radiolabeling DNA restriction endonuclease fragments to high specific activity. Anal Biochem. 1983;132:6–13. doi: 10.1016/0003-2697(83)90418-9. [DOI] [PubMed] [Google Scholar]
- 16.Fields S, Song O. A novel genetic system to detect protein-protein interactions. Nature. 1989;340:245–246. doi: 10.1038/340245a0. [DOI] [PubMed] [Google Scholar]
- 17.Gietz R D, Schiestl R H, Willems A R, Woods R A. Studies on the transformation of intact yeast cells by the LiAc/SS-DNA/PEG procedure. Yeast. 1995;11:355–360. doi: 10.1002/yea.320110408. [DOI] [PubMed] [Google Scholar]
- 18.Graham F L, van der Eb A J. Transformation of rat cells by DNA of human adenovirus 5. Virology. 1973;54:536–539. doi: 10.1016/0042-6822(73)90163-3. [DOI] [PubMed] [Google Scholar]
- 19.Hirano T, Kinoshita N, Morikawa K, Yanagida M. Snap helix with knob and hole: essential repeats in S. pombe nuclear protein nuc2+ Cell. 1990;60:319–328. doi: 10.1016/0092-8674(90)90746-2. [DOI] [PubMed] [Google Scholar]
- 20.Honore B, Leffers H, Madsen P, Rasmussen H H, Vandekerckhove J, Celis J E. Molecular cloning and expression of a transformation-sensitive human protein containing the TPR motif and sharing identity to the stress-inducible yeast protein STI1. J Biol Chem. 1992;267:8485–8491. [PubMed] [Google Scholar]
- 21.Kouzarides T. Transcriptional control by the retinoblastoma protein. Semin Cancer Biol. 1995;6:91–98. doi: 10.1006/scbi.1995.0012. [DOI] [PubMed] [Google Scholar]
- 22.Kozak M. An analysis of 5′-noncoding sequences from 699 vertebrate messenger RNAs. Nucleic Acids Res. 1987;15:8125–8148. doi: 10.1093/nar/15.20.8125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Krady J K, Ward D C. Transcriptional activation by the parvoviral nonstructural protein NS-1 is mediated via a direct interaction with Sp1. Mol Cell Biol. 1995;15:524–533. doi: 10.1128/mcb.15.1.524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 25.Lamb J R, Tugendreich S, Hieter P. Tetratrico peptide repeat interactions: to TPR or not to TPR? Trends Biochem Sci. 1995;20:257–259. doi: 10.1016/s0968-0004(00)89037-4. [DOI] [PubMed] [Google Scholar]
- 26.Legendre D, Rommelaere J. Terminal regions of the NS-1 protein of the parvovirus minute virus of mice are involved in cytotoxicity and promoter trans inhibition. J Virol. 1992;66:5705–5713. doi: 10.1128/jvi.66.10.5705-5713.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li X, Rhode S L., III Nonstructural protein NS2 of parvovirus H-1 is required for efficient viral protein synthesis and virus production in rat cells in vivo and in vitro. Virology. 1991;184:117–130. doi: 10.1016/0042-6822(91)90828-y. [DOI] [PubMed] [Google Scholar]
- 28.Lorson C, Burger L R, Mouw M, Pintel D J. Efficient transactivation of the minute virus of mice P38 promoter requires upstream binding of NS1. J Virol. 1996;70:834–842. doi: 10.1128/jvi.70.2.834-842.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lorson C, Pintel D J. Characterization of the minute virus of mice P38 core promoter elements. J Virol. 1997;71:6568–6575. doi: 10.1128/jvi.71.9.6568-6575.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nicolet C M, Craig E A. Isolation and characterization of STI1, a stress-inducible gene from Saccharomyces cerevisiae. Mol Cell Biol. 1989;9:3638–3646. doi: 10.1128/mcb.9.9.3638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Op De Beeck A, Caillet-Fauquet P. The NS1 protein of the autonomous parvovirus minute virus of mice blocks cellular DNA replication: a consequence of lesions to the chromatin? J Virol. 1997;71:5323–5329. doi: 10.1128/jvi.71.7.5323-5329.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pagano M, Pepperkok R, Verde F, Ansorge W, Draetta G. Cyclin A is required at two points in the human cell cycle. EMBO J. 1992;11:961–971. doi: 10.1002/j.1460-2075.1992.tb05135.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pear W S, Nolan G P, Scott M L, Baltimore D. Production of high-titer helper-free retroviruses by transient transfection. Proc Natl Acad Sci USA. 1993;90:8392–8396. doi: 10.1073/pnas.90.18.8392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pujol A, Deleu L, Nüesch J P F, Cziepluch C, Jauniaux J-C, Rommelaere J. Inhibition of parvovirus minute virus of mice replication by a peptide involved in the oligomerization of nonstructural protein NS1. J Virol. 1997;71:7393–7403. doi: 10.1128/jvi.71.10.7393-7403.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rhode S L., III trans-Activation of parvovirus P38 promoter by the 76K noncapsid protein. J Virol. 1985;55:886–889. doi: 10.1128/jvi.55.3.886-889.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rhode S L, III, Richard S M. Characterization of the trans-activation-responsive element of the parvovirus H-1 P38 promoter. J Virol. 1987;61:2807–2815. doi: 10.1128/jvi.61.9.2807-2815.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rommelaere J, Cornelis J J. Antineoplastic activity of parvoviruses. J Virol Methods. 1991;33:233–251. doi: 10.1016/0166-0934(91)90024-t. [DOI] [PubMed] [Google Scholar]
- 38.Salome N, van Hille B, Duponchel N, Meneguzzi G, Cuzin F, Rommelaere J, Cornelis J J. Sensitization of transformed rat cells to parvovirus MVMp is restricted to specific oncogenes. Oncogene. 1990;5:123–130. [PubMed] [Google Scholar]
- 39.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 40.Sanger F, Nicklen S, Coulson A R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Seif R, Cuzin F. Temperature-sensitive growth regulation in one type of transformed rat cells induced by the tsa mutant of polyoma virus. J Virol. 1977;24:721–728. doi: 10.1128/jvi.24.3.721-728.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Senger M, Glatting K H, Ritter O, Suhai S. X-HUSAR, an X-based graphical interface for the analysis of genomic sequences. Comput Methods Programs Biomed. 1995;46:131–141. doi: 10.1016/0169-2607(94)01610-r. [DOI] [PubMed] [Google Scholar]
- 43.Shein H M, Enders J F. Multiplication and cytopathogenicity of simian vacuolating virus 40 in cultures of human tissue. Proc Soc Exp Biol Med. 1962;109:495–503. doi: 10.3181/00379727-109-27246. [DOI] [PubMed] [Google Scholar]
- 44.Vanacker J M, Rommelaere J. Non-structural proteins of autonomous parvoviruses: from cellular effects to molecular mechanisms. Semin Virol. 1995;6:291–297. [Google Scholar]
- 45.Van Hille B, Duponchel N, Salome N, Spruyt N, Cotmore S F, Tattersall P, Cornelis J J, Rommelaere J. Limitations to the expression of parvoviral nonstructural proteins may determine the extent of sensitization of EJ-ras-transformed rat cells to minute virus of mice. Virology. 1989;171:89–97. doi: 10.1016/0042-6822(89)90514-x. [DOI] [PubMed] [Google Scholar]
- 46.Whyte P, Buchkovich K J, Horowitz J M, Friend S H, Raybuck M, Weinberg R A, Harlow E. Association between an oncogene and an anti-oncogene: the adenovirus E1A proteins bind to the retinoblastoma gene product. Nature. 1988;334:124–129. doi: 10.1038/334124a0. [DOI] [PubMed] [Google Scholar]